Abstract
The mutual relationship between the intestinal microbiota and its mammalian host is influenced by diet. Consumption of various nutrients affects the structure of the microbial community and provides substrates for microbial metabolism. The microbiota can produce small molecules that are absorbed by the host and affect many important physiological processes. Age-dependent and societal differences in the intestinal microbiota could result from differences in diet. Examples include differences in the intestinal microbiota of breast- vs formula-fed infants, or differences in microbial richness in individuals consuming an agrarian plant-based vs a Western diet, which is high in meat and fat. We review how diet affects the structure and metabolome of the human intestinal microbiome, and may contribute to health or pathogenesis of disorders such as coronary vascular disease and inflammatory bowel diseases.
Keywords: Intestine, Microbiota, Diet, Inflammation
Diet and the Early Intestinal Microbiota
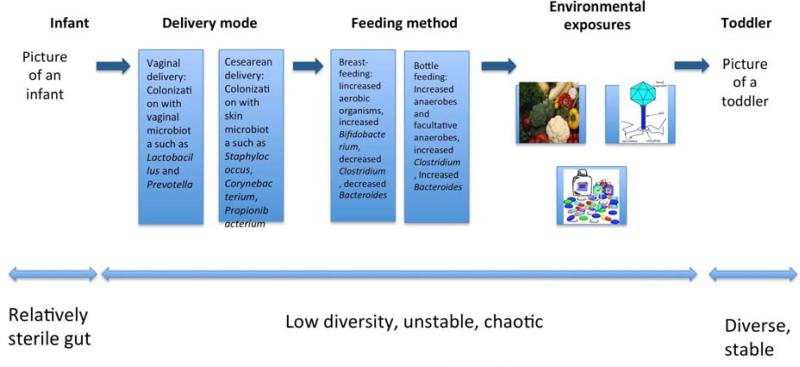
A healthy human fetus develops in an environment that is thought to be largely sterile1. Significant colonization of the gut begins immediately after delivery although, remarkably, bacteria can be found in an infant's first meconium2. The initial intestinal colonization pattern depends upon mode of delivery1. Infants born vaginally are initially colonized by bacterial taxa found in the vagina, such as Lactobacillus and Prevotella, whereas infants who are born by Cesarean section are initially colonized by bacteria found in the skin microbiota2. After this primary inoculation, infants are regularly exposed to microbes, and diversity increases rapidly3. The initial colonization pattern is thought to be chaotic, and a growing body of literature has shown that environmental exposures early in life, including diet, are responsible for these fluctuations (Figure 1). Characterization of the intestinal microbiota in a single infant, over a period of 2.5 years, showed how the bacterial taxa changed with life events such as illnesses, dietary changes, and antibiotic treatment. Interestingly, the greatest change in the composition of the infant's intestinal microbiota occurred with the introduction of solid foods. There was also a shift towards a more stable, adult-like microbiota with weaning3. Ultimately, the intestinal microbiota of the young resemble that of the adult by approximately the age of 34 (Figure 1).
Figure 1. Development of the human intestinal microbiota and the effects of environmental exposures.
Colonization of the early intestinal microbiota depends on multiple environmental exposures. The taxa that colonize the relatively sterile newborn intestine are largely determined by the mode of delivery. The intestinal microbiota also develops differently determined by differences in infant feeding (breast feeding vs formula feeding). Throughout infancy and early childhood, the microbiota changes with dietary alterations, infections, and exposure to antibiotics. The intestinal microbiota of the infant is characterized by instability and low levels of diversity. However, by the toddler years, the intestinal microbiota is similar in diversity and stability to that of adults.
Within the first year of life, there are significant inter-individual differences in the composition of the intestinal microbiota, yet some similarities exist. Similarities among individual infants can be attributed to the major taxonomic groups associated with the infant diet. Multiple studies have established differences in the composition of the intestinal microbiota based on whether infants are breast- or formula-fed5-7. Indeed, this introduces the concept of a potential association between Infant diets, the composition of the intestinal microbiota, and health (Figure 1).
Breast milk is the ideal food for infants, as it meets all their nutritional and physiologic demands. Over the years, there have been improvements in our understanding of the composition of human milk and subsequently, complex formulas have been developed that have a similar nutrient value. For mothers who are unable to breast-feed, infant formulas are now considered to be acceptable alternatives to breast milk. However, formula is not a perfect substitute for human milk. For example, there are important bioactive compounds in human milk that contribute to physiologic functions such as absorption and digestion of nutrients, immune protection, and defense against potentially pathogenic gut microbes8, 9. The effects of these bioactive compounds are difficult to replicate.
Human milk oligosaccharides (MOS) are bioactive compounds that are the third largest component of human milk10. MOS are indigestible glycans that remain whole as they travel through the intestinal tract to the colon, where they nourish specific colonic bacteria. It is believed that MOS benefit the infant by functioning as prebiotics and selectively promoting growth of members of the genus Bifidobacterium. Some of the differences in the composition of the intestinal microbiota in breast-fed vs formula-fed infants are thought to be secondary to the effects of MOS on these bacteria—many studies have shown an increased proportion of Bifidobacteria in breast-fed infants 6, 7, 11 compared to formula-fed infants. An increased proportion of Bifidobacteria in the microbiota of breast-fed infants may be associated with health. For example, these bacteria have been linked to fortification of the gut mucosal protection through activities against pathogens12, 13. Bifidobacteria are also thought to modulate the intestinal immune system and have been shown to increase the production of immunoglobulin A14. Many aspects of the mechanisms by which Bifidobacteria produce these effects are yet to be determined and more research is needed in this area. Recent studies have shown that select strains of Bifidobacteria possess specialized enzymes capable of metabolizing MOS glycans15-17. Genome sequencing of numerous strains, particularly Bifidobacterium longum subsp. Infantis, has confirmed the presence of genes that allow these organisms to metabolize mammalian carbohydrates 18.
Another difference in the composition of the intestinal microbiota in breast-fed vs formula-fed infants is that aerobic organisms seem to be more prevalent in the feces of breast-fed infants, whereas anaerobic and facultatively anaerobic organisms, which preferentially use anerobic glycolysis, are more commonly found in the feces of formula-fed infants6. In terms of taxonomy, multiple studies have shown a decrease in Clostridia colonization of breast-fed infants19, 20 including colonization with Clostridium difficile 21. Bacteroides are also present in a lower concentration in breast-fed infants7, 20. This is particularly interesting because certain Bacteroides strains are able to digest MOS,22 suggesting a potential competitive relationship between Bifidobacterium and Bacteroides in breast-fed infants.
Given the health benefits associated with breast feeding, there are ongoing attempts to develop infant formulas that are more similar in composition and function to breast milk. Oligosaccharide-enriched formulas have been developed, and infants fed these specialized formulas have been shown to harbor greater numbers of bifidobacteria in the feces 5,23. Breast milk also differs from formula in the type and quantity of protein and other nutrients. Interestingly, in a recent examination of the intestinal microbiota of healthy infants, a formula that was more similar to breast milk, in terms of protein composition and amount of phosphate, was found to increase proportions of Bifidobacterium similar to a formula that was supplemented with these bacteria24. Ultimately, alterations in infant formulas that lead to a more “breast-fed–like” composition of the intestinal microbiota may be beneficial, although additional evidence is needed25. Recent technologies to assess the structure and function of the infant intestinal microbiome will allow a deeper understanding of the relationship between diet, early intestinal colonization patterns, and health. This may be particularly important given the belief that early environmental exposures affect predisposition to immune-mediated diseases later in life26, 27.
Diet Helps Shape the Composition of the Intestinal Microbiota
The co-evolution of humans and our intestinal microbiota has led to our inter-dependent, mutualistic relationship. Food sources have guided the evolution of Homo sapiens. A comparison of the intestinal microbiota between different primates and mammals found that humans clustered more closely with other primates than non-primates. Interestingly, diet was the most important determinant where human microbiota samples were most similar to samples from omnivorous primate species28. The variety of foods in an omnivorous diet, as well as the free-living nature of our species, could affect how our diet determines the intestinal microbiota. Alterations in the taxonomy of the intestinal microbiota in humans may be relatively modest compared to the more-pronounced effects observed in rodents, in which there are significant phylum-level alterations in response to changes in fiber, fat, and simple carbohydrate consumption29, 30. Short-term dietary interventions in healthy humans lead to statistically significant and rapid alterations in the composition of the intestinal microbiota, but the magnitude of the effect is modest relative to inter-subject variability in the intestinal microbiota, and changes in taxonomy are not consistent among individuals30. However, extreme short-term diets, such as those devoid of any carbohydrates, have been shown to have a more-pronounced effect on the human microbiota31.
Nevertheless, significant and meaningful alterations in the intestinal microbiota have been associated with alterations in diet, primarily influenced by the consumption of dietary fiber from fruits, vegetables, and other plants. In controlled dietary experiments in humans, variations in intake of resistant starch or non-starch polysaccharide altered levels of specific bacterial taxa such as Ruminococuccus bromii and Eubacterium rectale32. These taxa were shown to selectively metabolize specific insoluble carbohydrate substrates, based on in vitro analyses of human fecal samples33. Model systems have demonstrated that an important function of the intestinal microbiome is its ability to metabolize complex carbohydrates and polysaccharides, collectively termed glycans, which leads to production of short-chain fatty acids through fermentation34. Sources of glycans for intestinal microbiome metabolism are derived not only from diet but also from mucus produced by the host. Remarkably, intestinal bacteria have the capacity to alter substrate utilization depending on the source of substrate abundance. For example, the abundant intestinal bacterium Bacteroides thetaiotaomicron, undergoes changes in gene expression that allow it to predominantly metabolize host-derived glycans when dietary sources of these molecules are unavailable35.
Globally, different diets driven among different populations help shape the taxonomy of their intestinal microbiomes. In a landmark study, De Fillipo et al. demonstrated that the composition of the intestinal microbiota differs significantly between children living in a rural African village in Burkina Faso and those living in Europe36. The microbiota of children in Burkina Faso had greater amounts of Prevotella, lower amounts of Bacteroides, overall greater microbial richness, and produced higher levels of short-chain fatty acids than the microbiota of European children. It would be reasonable to speculate that the agrarian diet of Burkina Faso (rich in carbohydrate content, fiber, and non-animal protein) compared with the Western diet (high in animal protein, sugar, starch, and fat and low in fiber) has a predominant role in these observed differences. The increased production of short-chain fatty acids is a functional consequence of the agrarian diet and supports this concept.
Several other reports confirm the effects of diet on human intestinal microbiota taxonomy. The inverse relationship between Prevotella and Bacteroides has been reproduced in studies comparing the intestinal microbiota of residents of agrarian societies in South America and Bangladesh with residents of industrialized societies4, 37. The MetaHit consortium has proposed that individuals can be classified as having intestinal microbiota predominantly composed of Prevotella or Bacteroides; a third group has higher proportions of Ruminococcus, compared with the others. These categories have been named “enterotypes”38. There has been considerable discussion about the discreteness of enterotypes—some datasets support the existence of these categories whereas others do not.39 Detection of enterotypes depends upon the computational approach used to analyze datasets40. A better term might be enterogradient, based on whether Bacteroides or Prevotella dominate the intestinal microbial community—it appears that these 2 genera generally do not exist in equal proportions in the human intestine 41.
An analysis of the NIH-sponsored Human Microbiome Project dataset revealed that organisms that are phylogenetically related and functionally similar tend to co-exist within the same environment, consistent with niche-driven community structures. However, Bacteroides and Prevotella are taxonomically and functionally similar genera, yet do not co-exist in the intestine; this is an exception to the model, possibly resulting from competition within the same niche41. Nonetheless, a greater proportion of Prevotella in the human intestinal microbiota is a marker of residence in an agrarian culture, whereas a greater proportion of Bacteroides is associated with residence in more-industrialized regions38. Associations between diet and bacterial taxonomy, based on answers to dietary questionnaires collected over long periods of time, indicate that diet affects the proportions of Prevotella vs Bacteroides in U.S. populations30. However, additional long-term dietary interventional studies are needed to determine the magnitude of the effects of diet on these taxonomic associations, compared with other environmental factors (Figure 2). Having associated the presence of stable gut microbial communities with long-term dietary patterns, it will be important to determine how these relate to diseases that differ in incidence between industrialized and agrarian regions.
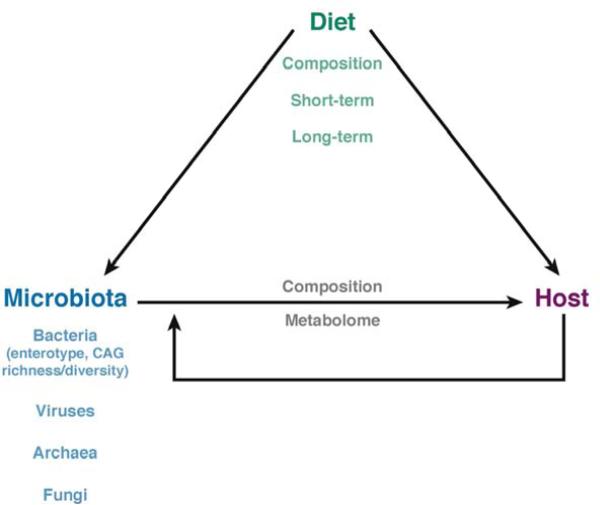
Figure 2. Interactions among diet, the intestinal microbiota, and the host.
Diet can directly affect the host or it can have an indirect effect through, for example, the intestinal microbiota. The composition and duration of diet can affect not only on bacteria in the intestine, but also viruses, Archaea, and fungi. Existing bacteria also affect these other microoganisms in a number of ways (co-abundance groups, enterotypes, richness/diversity). In addition to the composition of the intestinal microbiota, diet affects its production of metabolites, which can influence host physiology. Finally, the direct effects of diet on the host can produce changes that alter the intestinal microbiota.
A number of studies have associated increased microbial richness, at either the taxonomic or gene level, with diets higher in fruits, vegetables, and fiber42-44. This association has been reported in elderly individuals; differences in the taxonomy of the intestinal microbiota were associated with residence in different environments. The most extreme differences were observed in community residence vs long-term residential care, attributed to differences in diet—community residents typically consume diets higher in fiber and lower in fat42. This study also associated specific taxa with diet. Diet-associated bacterial clades, called “co-abundance groups”, included Bacteroides and Prevotella (Figure 2). Co-abundance groups were associated with the health status of elderly individuals—diets higher in fruits, vegetable, and fiber (associated with community residence) were linked to lower levels of frailty. Interestingly, only long-term alterations in environment and diet were associated with the composition of the microbiota, supporting observations previously made in studies performed with dietary questionnaires30. Consistent with these results, increased consumption of fruits and vegetables in higher-fiber diets and energy-restricted diets were both associated with increased bacterial gene richness (Figure 2)43, 44. In these studies, increased bacterial gene richness was also associated with human health, whereas lower bacterial richness was associated with obesity, insulin resistance, dylipidemia, and inflammatory disorders.
Diet has also been associated with other types of microbes in the gut, such as archaea, fungi, and bacteriophage (Figure 2)45-47. Although further studies are needed to determine how diet might affect these microbes, it is tempting to speculate that their interactions might affect human physiology. For example, carbohydrate consumption is associated with the proportion of the archaeon, Methanobrevibacter,46 in the intestinal microbiota. Methanobrevibacter can increase the production of short-chain fatty acids by metabolizing hydrogen, a byproduct of bacterial carbohydrate fermentation 48. Similarly, diet might also affect intestinal fungal communities, which have been associated with pathogenesis of inflammatory bowel diseases (IBD)49. Fungi are recognized by immune receptors such as Dectin1. A recent study49 demonstrated that mice lacking Dectin1 had increased susceptibility to chemically induced colitis, due to their altered responses to indigenous fungi. Interestingly, a polymorphism in the gene encoding Dectin1 (CLEC7A) was associated with a severe form of ulcerative colitis in humans49.
In composite, studies associating diet with the intestinal microbiome have consistently associated agrarian-based diets (high in fruits, vegetables, and fiber) with distinct bacterial taxa, an increase in bacterial richness at the taxonomic and gene level, and better health, compared with Western diets. Together with the well-described health benefits of plant-based diets typical of vegetarians and vegans50, 51, it is likely that the intestinal microbiome associated with residence in industrialized areas results from the Western diet. The features of the intestinal microbiome of people on Western diets are associated with the increasing incidence of diseases such as obesity, coronary vascular disease, metabolic syndrome, and certain malignancies. At a minimum, the intestinal microbiome might be a useful biomarker of long-term consumption of healthy or unhealthy diets. More intriguing is the possibility that diet-induced alterations in the intestinal microbiome contribute to disease development. In support of this concept, intestinal microbiome-dependent effects of diet have been associated with risk of coronary vascular disease and intestinal inflammation.
Effects of Diet on the Microbial Metabolome
Although diet affects the composition and/or richness of the intestinal microbiota, perhaps more important are its effects on the microbial metabolome. Diet can alter the functional metabolism of the intestinal microbiome at a genomic level. For example, studies in Japanese populations have shown that following consumption of seaweeds, genes that encode enzymes that metabolize marine red algae are transferred from marine-associated bacteria to specific bacterial taxa in the intestinal microbiome52. Furthermore, many molecules in foods are substrates for the intestinal microbiota, which then produce small molecules that, after metabolism in the liver, affect host physiology (Figure 2)53. For example, indigestible carbohydrates in the diet are fermented by the intestinal microbiota to produce short-chain fatty acids, which regulate immune function54, 55, intestinal hormone production, and lipogenesis56.
The intestinal microbiota may also contribute to development of atherosclerosis by producing metabolites of the dietary lipid phosphatidylcholine that are associated with the risk for coronary vascular disease. Using a targeted approach to identify plasma metabolites associated with coronary vascular disease in patients, Wang et al. identified a novel pathway linking dietary lipid intake, the intestinal microbiota, and atherosclerosis (Figure 3)57, 58. Foods rich in phosphatidylcholine are a major source of choline. Catabolism of choline by the intestinal microbiota results in the formation of the gas, trimethylamine (TMA), which is metabolized by the liver into trimethylamine oxide (TMAO)—a small molecule that is strongly associated with the increased risk for coronary vascular disease in humans. TMAO was also found to promote the development of atherosclerosis in animal models, providing the first link between dietary lipid intake, the intestinal microbiota, and the risk for the development of atherosclerosis57. A similar pathway has been identified for conversion of dietary carnitine, which is high in red meat, into TMAO59. Importantly, omnivorous subjects produced more TMAO than vegans or vegetarians following ingestion of L-carnitine, via an intestinal microbiota-dependent mechanism59.
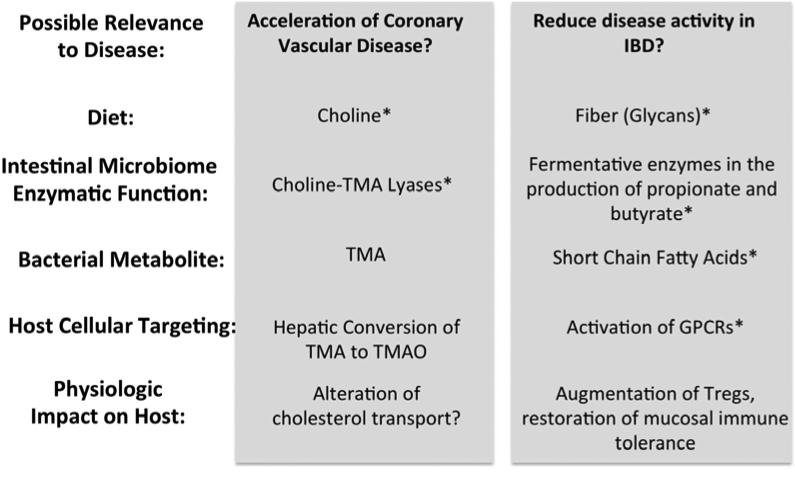
Figure 3. Pathways by which diet could affect health, through the production of metabolites by the intestinal microbiome.
Specific components of diet can serve as substrates for the intestinal microbiome, allowing it to produce specific metabolites that interact with mammalian cells, some in a receptor-mediated fashion. These processes affect host physiology and the development of diseases such as atherosclerosis and the inflammatory bowel diseases. *Possible therapeutic targets.
Researchers have identified the bacterial gene family responsible for the conversion of choline into TMA; these are called choline TMA lyases60. Most of the nonpathogenic bacterial taxa that express genes encoding these lyases are found in 3 of the 4 major phyla of the human intestinal microbiome. Using this information, it might be possible to develop technologies to quantify patients’ risk for heart disease due to consumption of choline, based on proportions of bacteria in the gut that have a choline TMA lyase gene (Figure 3). It might also be possible to reduce or remove bacteria that express TMA lyase from the intestine. Drugs might be developed to inhibit TMA lyase activity in bacteria, or medical foods might be created that reduce the production of TMA by bacteria from the diet.
These are important concepts for scientists trying to use knowledge gained from our understanding of the intestinal microbiome to prevent and/or treat diseases. A similar paradigm could be developed for treatment of IBD. For example, production of short-chain fatty acids by the intestinal microbiome increases mucosal immune tolerance through the activation of G-protein coupled receptors (GPCRs) and the subsequent activation of T regulatory cells61 (Figure 3). Therefore, therapeutic strategies could include alterations in diet and/or the composition of the intestinal microbiota to increase bacterial fermentation to increase the production of short-chain fatty acids. Alternatively, it might be possible to develop small molecule activators of GPCRs.
Diet has a role in development of many human gastrointestinal diseases (colon cancer, non-alcoholic fatty liver disease and steatosis, irritable bowel syndrome, celiac sprue) and several of these are also associated with alterations in the gut microbiota. However, a direct link between diet, the intestinal microbiome, and disease pathogenesis has not been demonstrated for most of these illnesses, even in animal models. For example, diets rich in fiber have been shown to increase the production of short-chain fatty acids by the intestinal microbiota, and butyrate has been shown to have anti-tumorigenic properties. Conversely, diets rich in red meat that have been associated with an increased risk for the development of colon cancer in epidemiology studies, and might promote the growth of sulfate-reducing bacteria that produce hydrogen sulfide, a genotoxic agent. High-fiber diets lead to the increased production of hydrogen by bacterial fermentation, which contributes to the symptoms of gas and bloating in patients with irritable bowel syndrome. These associations are discussed in greater depth in other reviews in this special issue.
The best examples for how diet can alter the gut microbiota to contribute to the pathogenesis of a digestive disease come from studies of IBD. Crohn's disease (CD) and ulcerative colitis (UC) are characterized by chronic, relapsing inflammation of the gastrointestinal tract. The pathogenesis of IBD involves an abnormal immune response, likely to environmental factors, in genetically susceptible individuals62. The composition of the intestinal microbiota is thought to be an important factor in pathogenesis, because patients with IBD and healthy individuals have been reported to have differences in intestinal microbiota63. Because of the effects that diet can have on the structure and function of the intestinal microbiome, it is reasonable to suspect that diet may be involved in the pathogenesis of IBD, or even a potential therapeutic target.
Several studies have examined the associations between dietary patterns and the incidence of IBD64, 65. A systematic review concluded that diets with high levels of total fats, polyunsaturated fatty acids, omega-6 fatty acids, and meat were associated with an increased risk of CD and UC; high fiber and high fruit intake were associated with a decreased risk of CD, whereas high vegetable intake was associated with a decreased for UC65. In a prospective study, Jowett et al. found that patients who reported consuming higher amounts of meat, eggs, protein, and alcohol were more likely to experience a relapse of UC66. The association was much stronger for red and processed meats than for other meats. The results of these studies are broadly consistent with those from epidemiology studies associating IBD with industrialized nations67, 68 and the consumption of a Western diet.
Consumption of milk fat has been shown to alter bile acid composition, promoting expansion of the sulphite-reducing pathobiont Bilophila wadsworthia and exacerbating colitis in IL10 knockout mice69. In this model system, the exacerbation of colitis may involve a direct effect of Bilophila wadsworthia, which promote a T helper 1 cell-mediated immune response and/or the production of hydrogen sulfide, a toxic small molecule that disrupts intestinal epithelial barrier function70. These findings support the effects of diet on the structure and metabolome of the gut microbiome and its importance in disease pathogenesis. Together with recent studies characterizing the impact of the diet on the human intestinal microbiota30, 31, 43, it is tempting to speculate that altering the gut microbiota community structure and/or its metabolome via consumption of an agrarian plant-based vs a Western diet might increase or decrease, respectively the risk for IBD. As for non-intestinal diseases, it was recently shown that mice fed high fiber diets are protected from allergic inflammation in the lung through a mechanism involving production of propionate and the activation of GPR4171. Although IBD does not develop through an allergic disease process, this study demonstrates that diet can alter immune responsiveness by modifying the metabolome of the intestinal microbiota.
Because dietary antigens can stimulate the mucosal immune system, bowel rest with total parenteral nutrition (TPN) has been used to treat certain patients with IBD72. In the 1980's, TPN emerged as an important treatment for moderate to severe CD. In a prospective study of 30 patients with CD treated with bowel rest and TPN, 25 (83%) achieved initial remission, but relapse was common73. A subsequent randomized controlled trial evaluating various nutritional interventions for patients with CD showed that bowel rest was not a major factor in achieving remission74. Despite the conflicting evidence, bowel rest with TPN may improve symptoms, at least in the short-term, in patients presenting with a severe exacerbation of CD. It is possible that bowel rest alters the gut microbiota in a way that reduces symptoms of IBD, because fasting has been shown to have an effect on the gut microbiota in mice75.
Exclusive enteral nutrition (EEN) with elemental, semi-elemental, and defined formula diets have been widely studied for their ability to induce remission in patients with CD and are considered first-line therapy in certain parts of the world76, 77. These diets are also effective in maintaining remission78. EEN is an alternative to potent pharmacological agents and has no serious associated side effects. However, nutritional therapy has been shown to be effective for patients with CD, although its mechanism of action has not been characterized. Interestingly, there does not appear to be major differences in efficacy of EEN based on the composition of the formula. A Cochrane meta-analysis found similar efficacy of formulas with variable degrees of protein hydrolysis in treating CD79. Modulation of gut microbiota composition has been proposed although supporting data are sparse80. Literature on this subject suggests that there is a profound change in the fecal microbiota following EEN therapy80, 81. Clearly, additional studies are needed to better characterize the impact of EEN on the gut microbiota and its association with CD activity in longitudinal cohorts. Such studies may not only lead to the discovery of biomarkers to predict therapeutic efficacy, but may also provide new insights into bacterial–host interactions involved in disease pathogenesis. Ultimately, EEN may be a therapeutic tool that can help us define a diet that can be used to treat and/or maintain remission in patients with IBD.
Future Directions
A large amount of data indicate the importance of the diet in establishing the composition and metabolome of the human intestinal microbiome. Functional studies in animal models, together with descriptive association studies in humans, provide evidence for the role of diet in disease pathogenesis, through its effects on intestinal microbes. The dietary factors that predominate are age-dependent; breast milk vs infant formula are determinants in early life, whereas agrarian vs a Western diets determine the composition of the microbiota at later stages of development. The challenge moving forward will be to provide evidence for dietary influences on the intestinal microbiome that have meaningful effects on human physiology. As an easily modifiable environmental factor, dietary interventions that have substantial effects on human health, through modification of the intestinal microbiome, may ultimately provide powerful approaches to disease prevention and therapy.
Acknowledgements
Supported by NIH grants UH2/3 DK083981, RO1 DK089472, and R01 GM103591 (G.D.W.) and a NASPGHAN Foundation Fellow to Faculty Transition Award (L.G.A.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have no conflicts of interest to declare.
References
- 1.Dominguez-Bello MG, Blaser MJ, Ley RE, et al. Development of the human gastrointestinal microbiota and insights from high-throughput sequencing. Gastroenterology. 2011;140:1713–9. doi: 10.1053/j.gastro.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107:11971–5. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koenig JE, Spor A, Scalfone N, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4578–85. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–7. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Penders J, Thijs C, Vink C, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118:511–21. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 6.Stark PL, Lee A. The microbial ecology of the large bowel of breast-fed and formula-fed infants during the first year of life. J Med Microbiol. 1982;15:189–203. doi: 10.1099/00222615-15-2-189. [DOI] [PubMed] [Google Scholar]
- 7.Yoshioka H, Iseki K, Fujita K. Development and differences of intestinal flora in the neonatal period in breast-fed and bottle-fed infants. Pediatrics. 1983;72:317–21. [PubMed] [Google Scholar]
- 8.Le Huerou-Luron I, Blat S, Boudry G. Breast- v. formula-feeding: impacts on the digestive tract and immediate and long-term health effects. Nutr Res Rev. 2010;23:23–36. doi: 10.1017/S0954422410000065. [DOI] [PubMed] [Google Scholar]
- 9.Hernell O. Human milk vs. cow's milk and the evolution of infant formulas. Nestle Nutr Workshop Ser Pediatr Program. 2011;67:17–28. doi: 10.1159/000325572. [DOI] [PubMed] [Google Scholar]
- 10.LoCascio RG, Desai P, Sela DA, et al. Broad conservation of milk utilization genes in Bifidobacterium longum subsp. infantis as revealed by comparative genomic hybridization. Appl Environ Microbiol. 2010;76:7373–81. doi: 10.1128/AEM.00675-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balmer SE, Wharton BA. Diet and faecal flora in the newborn: breast milk and infant formula. Arch Dis Child. 1989;64:1672–7. doi: 10.1136/adc.64.12.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lievin V, Peiffer I, Hudault S, et al. Bifidobacterium strains from resident infant human gastrointestinal microflora exert antimicrobial activity. Gut. 2000;47:646–52. doi: 10.1136/gut.47.5.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukuda S, Toh H, Hase K, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–7. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 14.Ouwehand A, Isolauri E, Salminen S. The role of the intestinal microflora for the development of the immune system in early childhood. Eur J Nutr. 2002;41(Suppl 1):I32–7. doi: 10.1007/s00394-002-1105-4. [DOI] [PubMed] [Google Scholar]
- 15.LoCascio RG, Ninonuevo MR, Freeman SL, et al. Glycoprofiling of bifidobacterial consumption of human milk oligosaccharides demonstrates strain specific, preferential consumption of small chain glycans secreted in early human lactation. J Agric Food Chem. 2007;55:8914–9. doi: 10.1021/jf0710480. [DOI] [PubMed] [Google Scholar]
- 16.Marcobal A, Barboza M, Froehlich JW, et al. Consumption of human milk oligosaccharides by gut-related microbes. J Agric Food Chem. 2010;58:5334–40. doi: 10.1021/jf9044205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sela DA, Li Y, Lerno L, et al. An infant-associated bacterial commensal utilizes breast milk sialyloligosaccharides. J Biol Chem. 2011;286:11909–18. doi: 10.1074/jbc.M110.193359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zivkovic AM, German JB, Lebrilla CB, et al. Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4653–8. doi: 10.1073/pnas.1000083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stark PL, Lee A. Clostridia isolated from the feces of infants during the first year of life. J Pediatr. 1982;100:362–5. doi: 10.1016/s0022-3476(82)80430-7. [DOI] [PubMed] [Google Scholar]
- 20.Harmsen HJ, Wildeboer-Veloo AC, Raangs GC, et al. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J Pediatr Gastroenterol Nutr. 2000;30:61–7. doi: 10.1097/00005176-200001000-00019. [DOI] [PubMed] [Google Scholar]
- 21.Stark PL, Lee A, Parsonage BD. Colonization of the large bowel by Clostridium difficile in healthy infants: quantitative study. Infect Immun. 1982;35:895–9. doi: 10.1128/iai.35.3.895-899.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marcobal A, Barboza M, Sonnenburg ED, et al. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe. 2011;10:507–14. doi: 10.1016/j.chom.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Veereman-Wauters G, Staelens S, Van de Broek H, et al. Physiological and bifidogenic effects of prebiotic supplements in infant formulae. J Pediatr Gastroenterol Nutr. 2011;52:763–71. doi: 10.1097/MPG.0b013e3182139f39. [DOI] [PubMed] [Google Scholar]
- 24.Hascoet JM, Hubert C, Rochat F, et al. Effect of formula composition on the development of infant gut microbiota. J Pediatr Gastroenterol Nutr. 2011;52:756–62. doi: 10.1097/MPG.0b013e3182105850. [DOI] [PubMed] [Google Scholar]
- 25.Braegger C, Chmielewska A, Decsi T, et al. Supplementation of infant formula with probiotics and/or prebiotics: a systematic review and comment by the ESPGHAN committee on nutrition. J Pediatr Gastroenterol Nutr. 2011;52:238–50. doi: 10.1097/MPG.0b013e3181fb9e80. [DOI] [PubMed] [Google Scholar]
- 26.Olszak T, An D, Zeissig S, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–93. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Virgin HW, Todd JA. Metagenomics and personalized medicine. Cell. 2011;147:44–56. doi: 10.1016/j.cell.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ley RE, Hamady M, Lozupone C, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–51. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turnbaugh PJ, Ley RE, Mahowald MA, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–31. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 30.Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–8. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2013 doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walker AW, Ince J, Duncan SH, et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2011;5:220–30. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leitch EC, Walker AW, Duncan SH, et al. Selective colonization of insoluble substrates by human faecal bacteria. Environ Microbiol. 2007;9:667–79. doi: 10.1111/j.1462-2920.2006.01186.x. [DOI] [PubMed] [Google Scholar]
- 34.Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol. 2012;10:323–35. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sonnenburg JL, Xu J, Leip DD, et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–9. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 36.De Filippo C, Cavalieri D, Di Paola M, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107:14691–6. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin A, Bik EM, Costello EK, et al. Distinct distal gut microbiome diversity and composition in healthy children from Bangladesh and the United States. PLoS One. 2013;8:e53838. doi: 10.1371/journal.pone.0053838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–80. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lozupone CA, Stombaugh JI, Gordon JI, et al. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–30. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koren O, Knights D, Gonzalez A, et al. A guide to enterotypes across the human body: meta-analysis of microbial community structures in human microbiome datasets. PLoS Comput Biol. 2013;9:e1002863. doi: 10.1371/journal.pcbi.1002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Faust K, Sathirapongsasuti JF, Izard J, et al. Microbial co-occurrence relationships in the human microbiome. PLoS Comput Biol. 2012;8:e1002606. doi: 10.1371/journal.pcbi.1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Claesson MJ, Jeffery IB, Conde S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–84. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- 43.Cotillard A, Kennedy SP, Kong LC, et al. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500:585–8. doi: 10.1038/nature12480. [DOI] [PubMed] [Google Scholar]
- 44.Le Chatelier E, Nielsen T, Qin J, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–6. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 45.Dollive S, Peterfreund GL, Sherrill-Mix S, et al. A tool kit for quantifying eukaryotic rRNA gene sequences from human microbiome samples. Genome Biol. 2012;13:R60. doi: 10.1186/gb-2012-13-7-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoffmann C, Dollive S, Grunberg S, et al. Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLoS One. 2013;8:e66019. doi: 10.1371/journal.pone.0066019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Minot S, Sinha R, Chen J, et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 2011;21:1616–25. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Samuel BS, Gordon JI. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proc Natl Acad Sci U S A. 2006;103:10011–6. doi: 10.1073/pnas.0602187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iliev ID, Funari VA, Taylor KD, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012;336:1314–7. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Craig WJ. Nutrition concerns and health effects of vegetarian diets. Nutr Clin Pract. 2010;25:613–20. doi: 10.1177/0884533610385707. [DOI] [PubMed] [Google Scholar]
- 51.Craig WJ. Health effects of vegan diets. Am J Clin Nutr. 2009;89:1627S–1633S. doi: 10.3945/ajcn.2009.26736N. [DOI] [PubMed] [Google Scholar]
- 52.Hehemann JH, Correc G, Barbeyron T, et al. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010;464:908–12. doi: 10.1038/nature08937. [DOI] [PubMed] [Google Scholar]
- 53.Holmes E, Li JV, Marchesi JR, et al. Gut microbiota composition and activity in relation to host metabolic phenotype and disease risk. Cell Metab. 2012;16:559–64. doi: 10.1016/j.cmet.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 54.Maslowski KM, Vieira AT, Ng A, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–6. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith PM, Howitt MR, Panikov N, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–73. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Samuel BS, Shaito A, Motoike T, et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc Natl Acad Sci U S A. 2008;105:16767–72. doi: 10.1073/pnas.0808567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Z, Klipfell E, Bennett BJ, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tang WH, Wang Z, Levison BS, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med. 2013;368:1575–84. doi: 10.1056/NEJMoa1109400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koeth RA, Wang Z, Levison BS, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19:576–85. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Craciun S, Balskus EP. Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc Natl Acad Sci U S A. 2012;109:21307–12. doi: 10.1073/pnas.1215689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tremaroli V, Backhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–9. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 62.Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med. 2009;361:2066–78. doi: 10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peterson DA, Frank DN, Pace NR, et al. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host Microbe. 2008;3:417–27. doi: 10.1016/j.chom.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chapman-Kiddell CA, Davies PS, Gillen L, et al. Role of diet in the development of inflammatory bowel disease. Inflamm Bowel Dis. 2010;16:137–51. doi: 10.1002/ibd.20968. [DOI] [PubMed] [Google Scholar]
- 65.Hou JK, Abraham B, El-Serag H. Dietary intake and risk of developing inflammatory bowel disease: a systematic review of the literature. Am J Gastroenterol. 2011;106:563–73. doi: 10.1038/ajg.2011.44. [DOI] [PubMed] [Google Scholar]
- 66.Jowett SL SC, Pearce MS, et al. nfluence of dietary factors on the clinical course of ulcerative colitis: a prospective cohort study. Gut. 2004;53:1479–84. doi: 10.1136/gut.2003.024828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Molodecky NA, Soon IS, Rabi DM, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142:46–54. e42. doi: 10.1053/j.gastro.2011.10.001. quiz e30. [DOI] [PubMed] [Google Scholar]
- 68.Lashner BA. Epidemiology of inflammatory bowel disease. Gastroenterol Clin North Am. 1995;24:467–74. [PubMed] [Google Scholar]
- 69.Devkota S, Wang Y, Musch MW, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature. 2012;487:104–8. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sartor RB. Gut microbiota: Diet promotes dysbiosis and colitis in susceptible hosts. Nat Rev Gastroenterol Hepatol. 2012;9:561–2. doi: 10.1038/nrgastro.2012.157. [DOI] [PubMed] [Google Scholar]
- 71.Trompette A, Gollwitzer ES, Yadava K, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014 doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- 72.Graham TO, Kandil HM. Nutritional factors in inflammatory bowel disease. Gastroenterol Clin North Am. 2002;31:203–18. doi: 10.1016/s0889-8553(01)00022-x. [DOI] [PubMed] [Google Scholar]
- 73.Muller JM, Keller HW, Erasmi H, et al. Total parenteral nutrition as the sole therapy in Crohn's disease--a prospective study. Br J Surg. 1983;70:40–3. doi: 10.1002/bjs.1800700116. [DOI] [PubMed] [Google Scholar]
- 74.Greenberg GR, Fleming CR, Jeejeebhoy KN, et al. Controlled trial of bowel rest and nutritional support in the management of Crohn's disease. Gut. 1988;29:1309–15. doi: 10.1136/gut.29.10.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Crawford PA, Crowley JR, Sambandam N, et al. Regulation of myocardial ketone body metabolism by the gut microbiota during nutrient deprivation. Proc Natl Acad Sci U S A. 2009;106:11276–81. doi: 10.1073/pnas.0902366106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sandhu BK, Fell JM, Beattie RM, et al. Guidelines for the Management of Inflammatory Bowel Disease in Children in the United Kingdom. J Pediatr Gastroenterol Nutr. 2010 doi: 10.1097/MPG.0b013e3181c92c53. [DOI] [PubMed] [Google Scholar]
- 77.Caprilli R, Gassull MA, Escher JC, et al. European evidence based consensus on the diagnosis and management of Crohn's disease: special situations. Gut. 2006;55(Suppl 1):i36–58. doi: 10.1136/gut.2005.081950c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Takagi S, Utsunomiya K, Kuriyama S, et al. Effectiveness of an 'half elemental diet' as maintenance therapy for Crohn's disease: A randomized-controlled trial. Aliment Pharmacol Ther. 2006;24:1333–40. doi: 10.1111/j.1365-2036.2006.03120.x. [DOI] [PubMed] [Google Scholar]
- 79.Zachos M, Tondeur M, Griffiths AM. Enteral nutritional therapy for inducing remission of Crohn's disease. Cochrane Database Syst Rev. 2001:CD000542. doi: 10.1002/14651858.CD000542. [DOI] [PubMed] [Google Scholar]
- 80.Leach ST, Mitchell HM, Eng WR, et al. Sustained modulation of intestinal bacteria by exclusive enteral nutrition used to treat children with Crohn's disease. Aliment Pharmacol Ther. 2008;28:724–33. doi: 10.1111/j.1365-2036.2008.03796.x. [DOI] [PubMed] [Google Scholar]
- 81.Lionetti P, Callegari ML, Ferrari S, et al. Enteral nutrition and microflora in pediatric Crohn's disease. JPEN J Parenter Enteral Nutr. 2005;29:S173–5. doi: 10.1177/01486071050290S4S173. discussion S175-8, S184-8. [DOI] [PubMed] [Google Scholar]