Abstract
Mutants of Escherichia coli lacking all of the known saturable K+ transport systems, “triple mutants,” require elevated K+ concentrations for growth. K+ transport activity in such mutants, called TrkF activity, has low substrate specificity and a low rate that increases with increasing external pH. Attempts to isolate mutants requiring even higher concentrations of K+ failed, implying that either TrkF is essential or is composed of multiple minor K+ transport activities. Instead, we sought mutations that allowed triple mutants to grow at lower K+ concentrations. Mutations so identified include ones altering MscL, the large mechanosensitive channel, or Opp, the oligopeptide permease. However, a possible contribution of wild-type Opp and MscL to TrkF activity was not proven. In contrast, expression of wild-type ProP, TrkG, and TrkH proteins increased uptake when encoded on multicopy plasmids. In all of these situations, the driving force for K+ appeared to be the transmembrane electric potential, and in most cases substrate specificity was low; these are characteristics of TrkF activity. These results support the view that TrkF is composed of multiple, “aberrant” K+ transport activities, i.e., paths that, regardless of their physiological function, allow K+ to cross the cell membrane by a uniport process.
Wild-type strains of Escherichia coli accumulate K+ by three independent saturable systems, Trk (formerly TrkA, or TrkG and TrkH), Kup (formerly TrkD), and Kdp. Any one of these three systems is sufficient for growth in medium containing concentrations of K+ in the low millimolar range (4, 35). Strains which carry null mutations in all three systems, here referred to as triple mutants, require high concentrations of K+ for growth. At pH 7 in minimal medium, 25 to 30 mM K+ allows only half the maximal growth rate that is attained with 100 mM K+ or more. The rate of K+ uptake in triple mutants is linearly proportional to the external K+ concentration (26). Rb+ and Cs+, congeners of K+, are taken up as readily as K+ in the triple mutants, whereas each of the saturable systems discriminates against Rb+ and Cs+ (5, 6, 27).
K+ uptake in triple mutants was attributed to a system called TrkF. Attempts to obtain mutants that required even more K+ for growth were not successful. This could indicate that TrkF, at least in the triple mutant background, is essential for growth. Or, TrkF could represent the sum of multiple, minor K+ transport activities. Genetic inactivation of any one of these redundant activities would result in a reduction of K+ uptake too small to yield a discernible change in the K+ requirement for growth. Therefore, a complementary approach was taken by searching for genetic changes that would allow triple mutants to grow in medium containing 5 mM K+. Our analysis suggests that changes that increase the rate of K+ uptake can result from transport energized by the transmembrane electrical potential through paths for which K+ is not the physiological substrate. By analogy, we suggest that uptake via TrkF represents the same sort of aberrant transport through a variety of systems for which K+ is not the physiological substrate.
MATERIALS AND METHODS
Plasmids. (i) pJD101.
All plasmids constructed in this work are derivatives of pJD101, an EcoRV-PvuII deletion derivative of pBR322 in which the EcoRI-HindIII region has been replaced with the multiple cloning site of M13mp21. The EcoRV-PvuII deletion removes the tet gene, whose product mediates a low rate of K+ uptake (11).
(ii) pEB49.
This pJD101 derivative contains a chromosomal BamHI-EcoRV fragment obtained from Kohara clone λ252 (28) encoding cls (formerly nov [25]) cloned between the BamHI and EcoRI sites in the multiple cloning site. The BglII-HindIII fragment encoding cls (25) is replaced by a HindIII-AvaI fragment containing the kanamycin resistance gene from pEG5005 (14).
(iii) pEB54.
A fragment of approximately 4 kb from Kohara clone λ251 (28) that extends from an EcoRV site in oppA to a BamHI site in oppF was cloned in SmaI-BamHI-digested pJD101 to create pEB53. pEB54 was made from pEB53 by replacing the 2-kb PvuII fragment encoding oppB and parts of oppA and oppC with a HindIII-AvaI fragment containing the kanamycin resistance gene from pEG5005 oriented with its direction of transcription the same as that of the opp operon.
(iv) Overexpression of native genes: pEBGC11, pEBGC13, pEBGC30, pJD301, and pDC1.
A clone carrying the proP gene resulted from an attempt to clone a UV-induced stk mutation (“suppressor of transport of K+”) in strain TK2420(μcts pEG5005). Using mini-Mu in vivo cloning (14), lysates were used to transfect the Mu lysogenic strain TK2313(μ+), after which the transfectants were selected for growth at 5 mM K+. A plasmid was isolated from one transfectant, digested with HindIII, ligated into HindIII-digested pJD101, and transformed into TK2420 with selection for kanamycin resistance to create the plasmid pEBGC10. The removal of a 3.3-kb KpnI fragment from pEBGC10 resulted in pEBGC11. A 4.7-kb BamHI fragment, encoding ProP (8), was subcloned into pJD101 to create pEBGC13. Further subcloning experiments and DNA sequencing revealed that the growth at 5 mM K+ was conferred by the wild-type proP; no mutation was found. This result was confirmed using pDC1, a different proP clone (8).
A similar attempt to clone an independent stk mutation resulted in the cloning of a HindIII fragment, to create the plasmid pEBGC1. The insert was mapped to trkG using the E. coli gene-mapping membrane (Takara Biochemical) described by Lee et al. (20). This result was confirmed using pEBGC30, a derivative of ptrkG/pGH27 (16) from which the BamHI fragment was deleted. The role of the TrkH protein, a functional homolog of the TrkG protein (31), was studied using pJD301, a derivative of JD101 carrying the entire trkH gene along with its promoter as an EcoR1 fragment of pWE101 (9).
Strains.
The strains used are listed in Table 1. TK2420 cls::kan and TK2420 opp::kan contain chromosomal replacements of cls and oppA to -C, respectively, with a kanamycin resistance cassette. This was accomplished using pEB49 and pEB54, respectively, which were linearized with BamHI and transformed into JC7623 with selection for kanamycin resistance. The correct integration was verified by a loss of ampicillin resistance, cotransduction with galU and trpC and, in the case of TK2420 opp::kan, the acquisition of resistance to tri-l-ornithine (3). The polA(Ts) mutation was introduced by cotransduction with the rha mutation in the TK strains. The markers in the region of opp were introduced by cotransduction with the trpB83::Tn10 mutation or with a pyrF mutation, which itself was introduced by cotransduction with trpB83::Tn10.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Genotypea | Sourceb |
|---|---|---|
| Strains | ||
| CAE169 | Hfr Hayes thi thr Δ(kdpFAB)5 trkD1 trkA405 | |
| CAG12099 | zee3129::Tn10 | Madisonc |
| CAG12016 | zci506::Tn10d | Madison |
| CHE52 | Hfr KL16 thi proC zba::Tn10e | |
| JC411-6 | his | D. Fraenkel |
| JC7623 | recB21 recC22 sbcB15 | CGSC |
| KL708 | F-141 | CGSCf |
| KLF23/KL181 | F-123 | CGSC |
| LHB2001 | Δ(trkA) | E. Bakker |
| NK5151 | trpB83::Tn10 | CGSC |
| SY634 | polA34(Ts) | CGSC |
| TK2205 | F−thi rha lacZ nagA Δ(kdpFAB)5 trkA405 trkD1 | |
| TK2247 | F−thi rha lacZ nagA nadA Δ(trkA-mscL′) trkD1 | |
| TK2313 | F−thi rha lacZ nagA Δ(kdpFABCDE)81 trkA405 trkD1 endA | |
| TK2383 | TK2205 stkA1 (mscL, Asn15Asp) | |
| TK2386 | TK2420 Δ(oppA-C)::kan | |
| TK2387 | TK2420 polA34(Ts) | |
| TK2403 | F−thi rha lacZ nagA Δ(kdpFAB)5 trkD1 trkA409 galU | |
| TK2420 | F−thi rha lacZ nagA Δ(kdpFAB)5 Δ(trkA-mscL′) trkD1 | |
| TK2444 | TK2420 trkG::kan | |
| TK2453 | TK2420 cls::kan | |
| TK2459 | TK2420 stkB1 (oppB, Arg191Pro) | |
| TK2460 | TK2420 stkB2 (oppB, Arg191Gly) | |
| TK2461 | TK2420 stkB3 (oppC, Arg201Ser) | |
| TK2463 | TK2420 stkB4 (oppC, Arg201Cys) | |
| Plasmids | ||
| pEG5005 | Mu d5005 kan | M. Casadaban |
| pB10b | mscL | C. Kung |
| pDC1 | proP | J. Wood |
Only the genotype of the markers used is given for most strains.
Source or reference. No entry means that the strain is from the laboratory collection or was isolated in this work.
From the Singer et al. collection of mapping strains (32).
This insertion was formerly zch506.
zab::Tn10, an insertion isolated in this lab, is 95% cotransduced with proC.
Coli Genetics Stock Center at Yale University.
Media and growth conditions.
Minimal phosphate-buffered medium of pH 7.0 and approximately 0.2 osM and complex media of high or moderate K+ concentration, KML and ML, respectively, have been described previously (13). Solid medium contained Bacto-agar (Difco) at 15 g · liter−1. Glucose, at 0.2% in minimal liquid medium and 1% in minimal agar, was the carbon source unless otherwise specified. The media used to measure growth at different pH values were of the same osmolarity with the appropriate changes in the ratio of dibasic to monobasic phosphate salts. The antibiotics carbenicillin and kanamycin were used at 50 and 30 μg · ml−1, respectively.
Genetic methods.
Mapping by conjugal Hfr × F− crosses was performed at 30°C in KML or ML medium, depending on the K+ tolerance of the strains involved, using a donor-to-recipient ratio of 1:2 and a concentration of donor of about 108 ml−1. In the conjugal mapping of stk mutations, the selection was either for the inheritance of a Tn10 insertion brought in by an auxotrophic Hfr strain or for the prototrophic derivative of an auxotrophic strain mutation that created a suitable Tn10 insertion brought in by transduction. Time-of-entry measurements, as well as the linkage with different markers, were used to establish the approximate locations of the stk mutations. The transductional linkage to nearby markers was determined with P1 phage as described elsewhere (12), except that the lysates were made in liquid cultures of 6 to 8 ml of KML or ML medium at 37°C.
Cloning of stkB mutations.
The stkB mutations were transduced into TK2420 galU trpB::Tn10 polA(Ts), selecting for tryptophan prototrophy and scoring for the ability to grow at 5 mM K+ and to use galactose as the carbon source. The resulting TK2420 stkB polA(Ts) strains were transformed with pEB54. A single transformant was grown overnight at 30°C in 5 ml of minimal medium containing 120 mM K+, 50 μg of carbenicillin/ml, and 0.02% glucose, after which 0.2% glucose was added and the incubation was continued at 37°C for 6 h. A 100-μl aliquot of this culture was spread on a 115 mM K+ minimal medium plate containing 50 μg of carbenicillin/ml and a 6-mm disk with 200 μg of tri-l-ornithine and incubated overnight at 37°C. A single colony growing in the halo around the disk was purified on an identical plate. In our hands, the polA(Ts) mutation was leaky in minimal medium even at 42°C, so that the plasmids which cannot be replicated in a polA mutant could still be recovered from cultures grown at this temperature. Therefore, the galU-trp region was transduced into TK2420 galU trpB::Tn10 polA(Ts) (carrying no plasmid), selecting for growth on galactose and scoring for tryptophan prototrophy and carbenicillin resistance. Plasmids that excised from the genome upon cis-recombination were isolated from a single transductant and transformed into TK2420. The transformants were selected for carbenicillin resistance and scored for kanamycin sensitivity, which indicated that the kanamycin cassette had been exchanged for the genomic PvuII fragment encoding oppB, oppC, and part of oppA. Since these plasmids did not allow growth in 5 mM K+, the presence of stkB was verified by plating 100 μl of an overnight culture of a transformant on minimal medium plates containing 5 mM K+. The presence of stkB in the plasmid increased the frequency of the appearance of stk mutants from <10−6 to >10−5.
Mapping and sequencing of stkB mutations.
Marker rescue experiments, in which fragments of plasmids containing stkB mutations were screened for those allowing the recovery of the Stk phenotype, were positive with either a 201-bp BstXI-SmaI fragment (oppB) or a 650-bp NruI-DraIII fragment (oppC). These fragments were sequenced in their entirety on both strands using the Sequenase 2.0 kit (U.S. Biochemical Corp.) and oligonucleotides flanking these fragments (DNA Synthesis Facility, Howard Hughes Medical Institute at the University of Chicago). In all cases, a single mutation was found.
Transport measurements.
Transport data were typically reproducible within 10% of the measured value (see Table 3), and the error between duplicate experiments never exceeded 20%. The transport of K+ was measured using flame photometry in cells depleted of K+ by treatment with 10 mM 2,4-dinitrophenol as described previously (26). The routine buffer for the transport measurements was 70 mM Na-phosphate buffer, pH 7.0, and the routine carbon and energy source was glucose at 2 g · liter−1. For the measurements at other pH values, cells after dinitrophenol treatment were washed twice with 0.1 M NaCl and then transferred to isosmotic buffers at the desired pH, routinely Na-phosphate buffer but in some cases as noted Na-HEPES or Na-piperazine-N,N′-bis(2-ethanesulfonic acid) buffer. The uptake of K+ or its congeners was initiated by adding a suitable mixture of 0.1 M KCl or the Cl− salts of other cations. The concentration of K+ or its congeners was varied by replacement with Na+, so that the total monovalent cation concentration remained constant. K+ efflux experiments were performed by filtering log-phase cells grown at 30°C in minimal medium containing 115 mM K+, or 5 mM K+ in the case of the stkA mutant, and washing the cells with K+-free 70 mM Na-phosphate buffer (pH 7), followed by suspension in the same buffer containing glucose (0.1%), followed by a suitable dilution with 0.1 M NaCl or chloride salts of other monovalent cations and incubation at 30°C. The samples for the measurements of cell K+ were collected and analyzed in the same way as were those for the uptake experiments.
TABLE 3.
Effect of multicopy genes on K+ uptake in strain TK2420
| Strain | Gene on plasmid | K+ uptakea at external K+ concn (mM) of:
|
|
|---|---|---|---|
| 5 | 40 | ||
| TK2420 | 2.5 ± 0.3b (n = 7) | ||
| TK2420(pJD101) | Control | 2.6 | |
| TK2420(pEBGC11) | proP | 0.7 | 6.4 |
| TK2420(pEBGC13) | proP | 0.9 | 6.8 |
| TK2420(pEBGC30) | trkG | 6.3 | |
| TK2420(pJD301) | trkH | 4.2 | |
Initial rate of K+ uptake with glucose as growth and transport substrate, in micromoles per gram (dry weight) per minute. No entry means that measurement was not done.
For the parental strain, TK2420, data are the mean ± standard deviation.
The transport of Rb+ when K+ was not present was measured by flame photometry, since the K+ filter of the photometer allows the specific emission line from Rb+ to pass with a sensitivity for Rb+ of about 7% of that for K+.
RESULTS
Mutations that allowed the triple K+ transport mutant E. coli strain TK2205 to grow on medium containing 5 mM K+ arose spontaneously at a frequency in the range of 10−6 to 10−7, a rate that significantly increased after UV mutagenesis. Such stk mutants could be divided into two categories on the basis of their growth phenotypes. A minority, from 2 to 10% in several batches, failed to grow on plates containing 115 mM K+. The majority grew well at low as well as high K+ concentrations. Preliminary mapping by Hfr crosses indicated that mutations that allowed growth at high K+ occurred in at least three loci that were widely separated on the chromosome of E. coli, while those that did not permit growth at high K+ could arise in at least three other loci that were not closely linked. Two mutations, stkA and stkB, the first ones of each type to be isolated, are described here. The other stk mutations were neither mapped in detail nor otherwise analyzed.
The stkA mutant TK2383.
The site of one type of mutation responsible for sensitivity to high K+, a locus initially referred to as stkA, was between the rpsL and aroE loci and was 90% cotransduced by P1 with the latter. The location of the stkA mutation in mscL was confirmed by complementation of the stkA mutation for growth at 115 mM K+ by both multicopy plasmid pB10b carrying mscL (24) and single-copy plasmid F-141, carrying the trkA405 mutation but wild type for mscL. The mscL gene was amplified from the chromosome of TK2383 by PCR and sequenced. The obtained sequence was identical to that published for mcsL (33), except for a transition mutation of AAC to GAC, replacing asparagine 15 with aspartic acid. This mutation has been described as one of a series of mscL mutations deleterious to growth (24). This result explained why stkA mutants were readily obtained from TK2205, which has the slightly leaky missense mutation in trkA, but never from trkA deletion strain TK2420. This deletion also removed the upstream region of the adjacent gene mscL, including its promoter and the coding region for the seven amino-terminal residues (15, 30), thus abolishing the activity of the MscL protein (33).
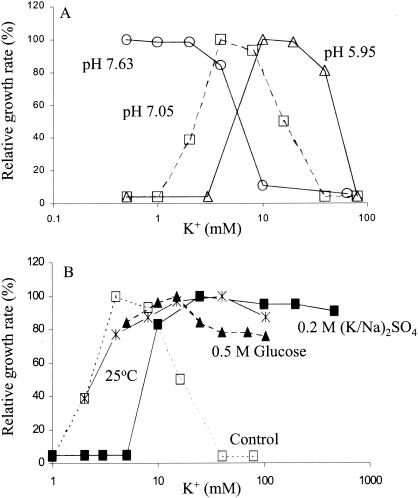
The K+ dependence of growth of the stkA mutant varied with pH, temperature, and osmolarity. The K+ requirement for growth was shifted to higher concentrations as the culture pH value was reduced (Fig. 1A). Failure to grow at elevated K+ concentrations was largely reversed during growth at lower temperature or in medium of elevated osmolarity achieved by either salt or sugar (Fig. 1B). However, this suppressive effect was only partial, as the mutant still grew at 5 mM K+ at 26°C, in glucose high-osmolarity medium and at 10 mM K+ in NaCl high-osmolarity medium, conditions under which the parental strain TK2205 did not grow (26).
FIG. 1.
The K+ dependence of growth of the stkA mutant as a function of medium pH, osmolarity, and temperature. (A) Effect of pH of the growth medium. Growth in phosphate-buffered media of pH 5.95 (▵), 7.05 (□), and 7.63 (○) was measured at 37°C as described in Materials and Methods. Growth rate is plotted relative to the highest rate observed at each pH value, which was 0.85 h−1 at pH 5.95, 0.71 h−1 at pH 7.05, and 0.54 h−1 at pH 7.63. (B) Effect of osmolarity and temperature on growth. The mutant was grown under standard conditions, at pH 7.05 and 37°C (□), at a temperature of 25°C (*), at 37°C in medium of elevated osmolarity by addition of 0.5 M glucose (▴), or at 37°C in medium of high osmolarity by addition of a 0.2 M concentration of a mixture of the sulfate salts of Na+ or K+ to achieve the desired final K+ concentration (▪). Data were plotted as described for panel A. The maximum growth rates were 0.71 h−1 under standard conditions, 0.31 h−1 at 25°C, 0.50 h−1 in the high-osmolarity glucose medium, and 0.58 h−1 in the high-osmolarity sulfate medium.
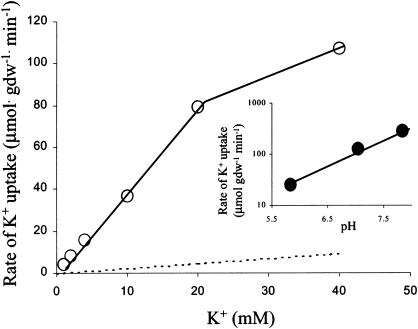
The initial rate of K+ uptake in K+-depleted cells of the stkA mutant was increased approximately 15-fold compared to that of the parental strain, TK2205, and was proportional to the external K+ concentration up to 20 mM K+ (Fig. 2). The rate of K+ uptake increased 3.9-fold per unit increase in pH (Fig. 2, inset). This dependence was analogous to that demonstrated for the parental strain (22). The effect of pH on K+ uptake readily explained the reason more K+ was needed and was tolerated at low rather than at high pH (Fig. 1A).
FIG. 2.
Kinetics of K+ uptake by the stkA mutant. The initial rate of K+ uptake (○) was measured at the stated K+ concentrations in K+-depleted cells at pH 7.05, as described in Materials and Methods. For comparison, the linear kinetics of the uptake by parental strain TK2205 are indicated by the dotted line; since transport rates in the wild-type strain via Trk are so much larger (Km of 1.5 mM; Vmax of 300 to 500 μmol · min−1 · g (dry weight)−1 [28]), they are not shown. The inset shows the effect of pH on uptake at 20 mM K+. Uptakes at pH 5.83 and 7.05 were performed in phosphate buffer; that at pH 7.84 was done in HEPES buffer.
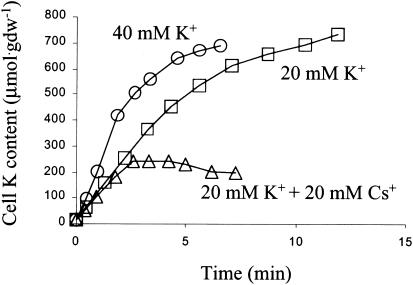
The stkA mutant also took up Rb+ and Cs+ rapidly. Rb+ uptake was 67 and 107 μmol · g (dry weight)−1 · min−1 at 15 and 30 mM Rb+, respectively. Indirect methods were used to assess uptake of other ions for which we did not have access to tracers (e.g., NH4+). When K+ and another monovalent cation were both present at 20 mM, the initial rate of K+ uptake was about the same as when only K+ was present, but net uptake ceased earlier, similar to when K+ alone was present at 40 mM (Fig. 3). While the level of K+ approached a steady state when only K+ was present, it began to fall when both K+ and Cs+ were present. Furthermore, this plateau both seemed to be reached earlier and was clearly less than half of the cell K+ content when only K+ was present. This implies that Cs+ efflux was slower or that Cs+ influx was faster than that of K+.
FIG. 3.
Effect of Cs+ on extent of K+ accumulation by the stkA mutant. The strain was depleted of K+, and uptake of K+ was measured at 20 mM K+ (□), at 40 mM K+ (○), and when 20 mM K+ and Cs+ (▵) were added at time zero.
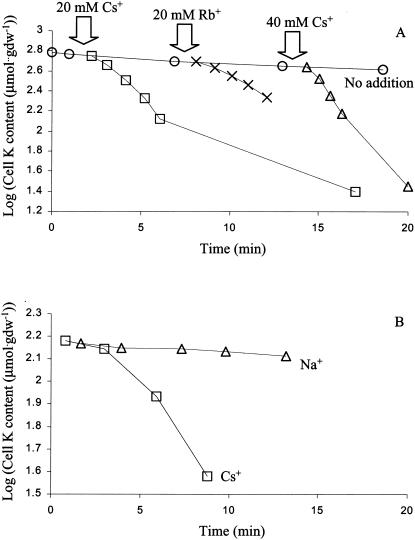
The effects of other ions on the rate of K+ efflux were measured in K+-replete cells in K+-free buffer, conditions where reentry of K+ was negligible. The addition of Cs+ or Rb+ provoked a rapid efflux of K+, whose initial rate was approximately proportional to the concentration of the added ion (Fig. 4A), but no monovalent cations other than Cs+ and Rb+ provoked rapid K+ efflux. Experiments in which Tris, Na+, or NH4+ were added did not produce convincing increases in K+ efflux (data not shown). Concomitant with the loss of K+, the cell contained presumably more of the ion that provoked K+ efflux. When Cs+ was added to cells which contained less than half of their normal pools of K+, rapid efflux started only after a lag of several minutes (Fig. 4B). These results suggest that K+ efflux was not directly coupled to uptake of Cs+ or Rb+, but rather was an indirect effect of the uptake of Cs+ or Rb+.
FIG. 4.
Effects of Cs+ and Rb+ on net K+ efflux from K+-loaded cells of the stkA mutant. (A) Cells in the logarithmic phase of growth were filtered and suspended in pH 7 Na+-phosphate buffer. At the arrows, Cs+ at 20 mM (□), Cs+ at 40 mM (▵), or Rb+ at 20 mM (X) was added and total cell K+ was measured as described in Materials and Methods. (B) Efflux of K+ is delayed in partially K+-depleted cells. Cells depleted of K+ as described in Materials and Methods were allowed to take up K+ at 20 mM for 3 min, after which they were filtered and, at time zero, suspended in K+-free buffer containing only Na+ (▵) or to which 20 mM Cs+ had been added (□).
In general, the stkA mutation was deleterious to growth (data not shown). The cells made smaller colonies on complex medium. Growth yields in 5 mM K+ minimal medium at pH 7 and 37°C on limiting amounts of glucose, glycerol, or dl-lactate were only 52, 65, and 63%, respectively, of those of parent strain TK2205. In pH 5.9 phosphate-buffered medium containing 16 mM K+, growth yields on glucose and glycerol were higher, 85 and 96%, respectively, of those for the parent strain.
The diameter of the open McsL channel has been estimated to be 30 to 40 Å in diameter, a size large enough to allow molecules of the size of amino acids as well as much larger ones to pass through (7). The mutation did not seem to alter the size of the channel, based on its conductance in patch-clamp experiments (24). We could not detect leakage of several amino acids or of pyrimidines or pyrimidine precursors as tested by cross-feeding of mutants requiring arginine, proline, methionine, leucine, or uracil (data not shown). The mutant did cross-feed a gltA mutant, which requires a Krebs cycle intermediate, glutamate or α-ketoglutarate, but this effect was similar to cross-feeding of the gltA mutant by wild-type strains. Thus, leakage of amino acids and similar metabolic intermediates did not appear to explain the reduced growth yield of the mutant. It seemed more likely that inappropriate leakage of protons was responsible, resulting in a partially uncoupled phenotype.
The stkB mutants.
About 10% of stk mutants, referred to as stkB mutations, contained a mutation linked to the zci506::Tn10 insertion, which is in oppC. The zci506::Tn10 insertion made strains resistant to tri-l-ornithine (23), as did the stkB1 mutation, indicating that this mutation was also in the opp operon. The stkB mutations were dominant in diploids. Each of the 13 independently isolated stkB mutations was cloned and its DNA sequence determined. As shown in Table 2, these strains represented only four different mutations, and they altered only two residues: replacing arginine 191 in OppB with either proline or glycine or the homologous arginine 201 in OppC with either cysteine or serine. These genes encoded two very similar membrane-spanning components of Opp, being almost identical in length and sharing 25% sequence identity and 40% similarity.
TABLE 2.
Growth of auxotrophic stkB mutants on peptides and inhibition by tri-l-ornithinea
| Compound | Zone of inhibition or growth (mm)
|
|||||
|---|---|---|---|---|---|---|
| TK2420 | stkB1 (oppB R191P) | stkB2 (oppB R191G) | stkB3 (oppC R202S) | stkB4 (oppC R202C) | Δ(oppA-C)::kan | |
| Orn-Orn-Orn | 9 | 0 | 9 | 4 | 8 | 0 |
| Gly-His-Gly | 17 | 3 | 18 | 10 | 17 | 1 |
| Gly-Pro-Ala | 13 | 4.5 | 12 | 12 | 14 | 0 |
| Gly-Trp-Gly | 9 | 0 | 4 | 5 | 6 | 0 |
Strains were streaked on 115 mM K+ minimal glucose agar medium radially from a 6-mm disc containing 200 μg of tri-l-ornithine or 50 μg of the other peptides. The zone of inhibition by tri-l-ornithine or zone of growth produced by the other peptides is shown. Derivatives of the six strains, each auxotrophic for the italicized amino acid in the peptide, were used to test the ability of the other peptides to enter the cell. Auxotrophy for His was introduced by cotransduction with zee3129::Tn10, for Pro by cotransduction with zda::Tn10, and for Trp by introducing trpB83::Tn10 while retaining the stk or opp mutation. The auxotrophic derivatives of the strains all had very similar zones of growth (±1 mm) extending from discs containing 50 μg of the free amino acid.
The four stkB mutations had different effects on the peptide transport properties of Opp (Table 2). Only one was completely resistant to inhibition by tri-l-ornithine, and all transported Gly-Trp-Gly less efficiently than the wild type. However, two were not impaired in transport of tri-l-ornithine, and three transported the Gly-Pro-Ala peptide as well as the wild type. These results indicated that these two arginine residues had an important role in the substrate specificity of the Opp system. The opp deletion control confirmed that all three peptides required the Opp system for entry.
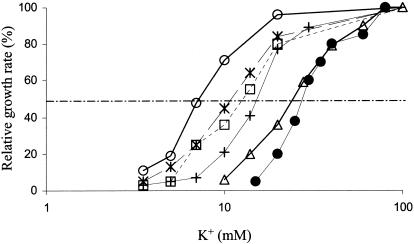
The four stkB mutants were screened by determining the effect of medium K+ concentration on their rate of growth (Fig. 5). Most subsequent studies were done only with the stkB2 mutant, since it had the largest effect on growth and transport. The growth rate increased approximately linearly with the logarithm of the K+ concentration over the range where growth rate varied rapidly with K+ concentration (Fig. 5), as has been reported for other strains (see Fig. 8 in reference 26). Each curve could be described by the concentration at which the growth rate was half of that at high K+. By this criterion, each of the stkB mutants had a slightly different effect on the K+ dependence of growth. The effect of pH, tested only in the stkB2 mutant, showed the same requirement for higher K+ concentrations at lower pH already noted in the stkA mutant above and in the parental TK2420 strain (22, 26).
FIG. 5.
Relationship of growth rate and medium K+ concentration for the four stkB mutants compared to that of control strain TK2420. Growth at pH 7.05 is shown for TK2420 (•), stkB1 (*), stkB2 (○), stkB3 (□), and stkB4 (+); growth of stkB2 at pH 6.55 is also shown (▵). Growth rate was plotted as the percentage of that at K+ concentrations of 100 mM or higher; maximum growth rates at pH 7.05 were 0.87, 0.91, 0.89, and 0.82 h−1 for the stkB1, stkB2, stkB3, and stkB4 mutants, respectively, and 0.76 h−1 for the stkB2 mutant at pH 6.55. Half-maximal rates of growth (dashed line) were achieved at 11, 7, 13, and 16 mM K+ for the stkB1, stkB2, stkB3, and stkB4 mutants at pH 7, respectively, and at 23 mM K+ for stkB2 at pH 6.55. Growth rates of wild-type strains of E. coli expressing Trk have been reported to remain at their maximum rate of growth until the medium K+ concentration is well below 1 mM (28).
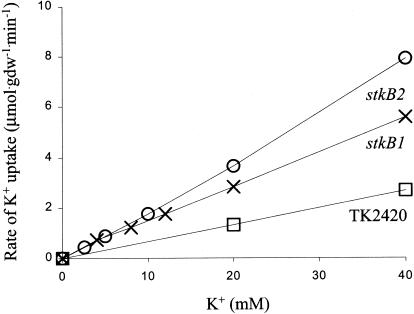
Initial rates of K+ uptake in K+-depleted stkB mutants were linear with external K+ concentration over the range tested, up to 40 mM (Fig. 6). The rate of uptake in the stkB2 mutant was somewhat greater than that in the stkB1 mutant, consistent with the finding that the stkB2 mutant required less K+ to achieve rapid growth than did the stkB1 mutant (Fig. 5). We did screening assays with the other two stkB mutants, and in each case the rate at 40 mM was 8 times that at 5 mM K+, within experimental error. The effect of pH was again characteristic: the initial rate of K+ uptake at 40 mM in the stkB1 mutant was 1.4, 4.3, and 8.2 μmol · g (dry weight)−1 · min−1 at pH 6.06, 6.95, and 7.48, respectively; the rate increased 3.5-fold per unit increase in pH.
FIG. 6.
Dependence of the initial rate of K+ uptake on external K+ concentration in K+-depleted cells of the stkB1 (X) and stkB2 (○) mutants. For comparison, data for strain TK2420 (□), the parental strain, are also shown; transport rates in wild-type strains of E. coli via Trk were 2 orders of magnitude larger (Km of 1.5 mM; Vmax of 300 to 500 μmol · min−1 · g (dry weight)−1 [28]).
The linear kinetics of transport in the stkB mutants suggested a very low apparent affinity and, hence, low selectivity. We tested this by examining the ability of Cs+ to stimulate K+ efflux. In experiments exactly like those shown in Fig. 4, addition of 40 mM Cs+ to the stkB2 mutant resulted in K+ efflux with a half-time of 8.2 min, compared to the rate of 53 min in the control containing only Na+. Efflux was slower than was the case for the stkA mutant, in keeping with the much lower rate of uptake in the stkB mutant. A similar stimulation of K+ efflux by Cs+ occurred in the parental strain, TK2420, with a half-time of 19 min after addition of 40 mM Cs+, compared to a half-time of 63 min in the control.
Suppression by overexpression of wild-type genes.
In the course of attempts to clone different stk mutants, we obtained two clones in multicopy plasmids that allowed strain TK2420 to grow on minimal medium containing 5 mM K+. Since the stk mutants were in a triple mutant background, already established K+ transporter genes were not cloned. The DNA fragments, initially isolated by in vivo mini-Mu cloning as described in Materials and Methods, were recloned in pJD101 for detailed analysis. Hybridization to the Kohara (19) clones as described in Materials and Methods and restriction enzyme analysis identified the genes carried. One of the clones carried the proP gene encoding a proton motive force-driven proline and glycine betaine transporter, while the other carried the trkG gene. The effect of the proP clone was due to the wild-type gene and not a mutation, since only the entire gene had the effect and overlapping fragments did not give rise to recombinants that grew on 5 mM K+ medium. The same was true for the trkG clone, since the same effect was observed with a derivative of the original trkG plasmid (16).
The ProP- and TrkG-expressing plasmids resulted in a modest increase in K+ uptake (Table 3). Linear dependence of K+ uptake on external K+ was examined only in the strain that overexpressed ProP. Rate of uptake at 40 mM K+ was, on average, 8.4-fold that at 5 mM K+, within experimental error of the expected 8-fold increase in rate, indicating linearity with external K+. In view of the effect of the trkG clone, we also tested a plasmid carrying trkH, a trkG homolog (31). That clone also increased K+ uptake modestly.
DISCUSSION
The goal of this study was to clarify the nature of the K+ transport system of E. coli called TrkF by isolating suppressor mutations that increased the rate of K+ uptake in mutants defective in saturable systems for K+ uptake. Past unsuccessful attempts to obtain mutants requiring even more K+ for growth led to the hypothesis that TrkF could be the sum of a multitude of minor K+ transport activities. In this work we identified mutations and genes that, when present in multicopy form, increased K+ uptake. The number of systems found in this way, many of which were unrelated to K+ transport, and the low K+ transport activity of each of them supported this hypothesis. Inevitably though, the low K+ transport activities prevented a direct contribution of each gene in single copy to TrkF to be estimated; attempts to do so revealed no significant difference between TK2420 and strains in which trkG, trkH, or oppA-C had been deleted (data not shown). However, all systems studied here showed the same lack of specificity, a marked dependence on external pH, and general lack of saturability that were characteristic of the TrkF system, thus providing further evidence, albeit indirect, of their role as part of TrkF.
All of the conditions described can be considered aberrant, since they either did not reflect the physiological functions of the systems involved or allowed entry through a system whose normal substrate is different from K+. Both the TrkG and TrkH proteins normally mediate K+ uptake, but only in the presence of other components such as the TrkA peripheral membrane protein (10, 13). When present in high gene dosage in the absence of TrkA, they allowed slow entry of K+.
The stkA mutant in which the MscL channel was altered was another example of movement of K+ through a system that accepts it, but where the system acted in an unphysiological way. The wild-type channel normally opens only when turgor pressure is excessively high, allowing internal osmotic solutes to leave rapidly and thus reduce turgor to acceptable levels (21). The N15D mutation has been characterized as leading to opening of the channel at a pressure some 20% lower than that needed to open the wild-type channel (24). The result was intermittent opening when turgor pressure was normal, allowing small molecules and ions to move down their electrochemical gradients. Since there was a strong driving force for cations to enter, rapid uptake of K+ occurred. In addition, the mutant had reduced carbon source growth yields, consistent with the idea that it was partially uncoupled due to a high rate of proton leakage into the cell through the channel. This is the only case here reported where K+ uptake was not linearly dependent on the external concentration of K+. We do not believe this necessarily represented saturability; when K+ uptake became very rapid and there was a large influx of protons as well, the cells could not export protons at a sufficient rate to maintain the membrane potential. We suggest it is the reduced membrane potential that caused a lower rate of K+ uptake.
All of the other situations that led to increased K+ uptake involve systems whose substrates do not resemble K+. Proline and peptides resemble K+ only to the extent that at physiological pH imino or amino groups are cationic and similar in size and can be considered examples of illicit transport. This term was initially coined to describe uptake by a peptide transport system of histidinol-P when the latter was coupled to a peptide (2). This example is better described as uptake of a substrate analog. A subsequent study found that mutants defective in cyclic AMP regulation were more resistant to a number of antibiotics (1). Since many sugar transport systems are under such control, it suggested that sugar transport systems were mediating illicit transport of some antibiotics. However, some of the enzymes of oxidative metabolism, including ones that pump protons and hence create the proton motive force, are also under cyclic AMP regulation (17, 34). An alternative explanation implicates a reduced membrane potential as a major mechanism in the increased resistance of mutants lacking cyclic AMP regulation to some antibiotics.
Some other examples of illicit transport of K+ have been reported. The tetracycline resistance gene of plasmid pBR322 mediated a low rate of K+ uptake (11). A number of clones from an alkalophilic bacillus complemented a triple K+ transport mutant to growth at moderate K+ concentrations (18). Since none of the genes identified appeared to be components of K+ transport systems, they presumably mediated illicit transport of K+. An N-terminal 135-amino-acid fragment of the KdpA gene has been reported to modestly reduce the K+ requirement for growth of a triple mutant (29). This fragment included only 2 of the 10 predicted membrane spans and only one of four regions implicated in specificity for K+ (6), so it was unlikely to retain any specificity and hence is most likely to be another example of illicit transport of K+.
Acknowledgments
E.T.B. was sponsored by The Netherlands Organization for Scientific Research. Part of this work was supported by grant DCB-8704059 from the National Science Foundation and by grant GM22323 from the National Institute for General Medical Sciences of the National Institutes of Health.
We thank George Canas for technical assistance, Steven Dorus for sequencing the stkA mutation, Janet Wood for the proP plasmid, pDC1, Ching Kung for the mscL plasmid, pB10b, Steven Short for providing the sequence of the E. coli opp operon prior to publication, Malcolm Casadaban for the mini-Mu cloning strains, Evert Bakker for strain LHB2001, the E. coli Genetics Stock Center at Yale for various strains, and Lucia Rothman-Denes and members of her lab for their hospitality to allow completion of this study.
REFERENCES
- 1.Alper, M. D., and B. N. Ames. 1978. Transport of antibiotics and metabolite analogs by systems under cyclic AMP control: positive selection of Salmonella typhimurium cya and crp mutants. J. Bacteriol. 133:149-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ames, B. N., G. F.-L. Ames, J. D. Young, D. Tsuchiya, and J. Lecocq. 1973. Illicit transport: the oligopeptide permease. Proc. Natl. Acad. Sci. USA 70:456-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrews, J. C., and S. A. Short. 1985. Genetic analysis of Escherichia coli oligopeptide transport mutants. J. Bacteriol. 161:484-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bakker, E. P. 1993. Low-affinity K+ uptake systems, p. 253-276. In E. P. Bakker (ed.), Alkali cation transport systems in procaryotes. CRC Press, Boca Raton, Fla.
- 5.Bossemeyer, D., A. Schlösser, and E. P. Bakker. 1989. Specific cesium transport via the Escherichia coli Kup (TrkD) K+ uptake system. J. Bacteriol. 171:2219-2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buurman, E. T., K. T. Kim, and W. Epstein. 1995. Genetic evidence for two sequentially occupied K+ binding sites in the Kdp transport ATPase. J. Biol. Chem. 270:6678-6685. [DOI] [PubMed] [Google Scholar]
- 7.Cruickshank, C. C., R. F. Minchin, A. C. Le Dain, and B. Martinac. 1997. Estimation of the pore size of the large-conductance mechanosensitive ion channel of Escherichia coli. Biophys. J. 73:1925-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Culham, D. E., B. Lasby, A. G. Marangoni, J. L. Milner, B. A. Steer, R. W. van Nues, and J. M. Wood. 1993. Isolation and sequencing of Escherichia coli gene proP reveals unusual structural features of the osmoregulatory proline/betaine transporter, ProP. J. Mol. Biol. 229:268-276. [DOI] [PubMed] [Google Scholar]
- 9.Dosch, D. C. 1984. A study of the Trk transport system of Escherichia coli. Ph.D. thesis. The University of Chicago, Chicago, Ill.
- 10.Dosch, D. C., G. L. Helmer, S. H. Sutton, F. F. Salvacion, and W. Epstein. 1991. Genetic analysis of potassium transport loci in Escherichia coli: evidence for three constitutive systems mediating uptake of potassium. J. Bacteriol. 173:687-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dosch, D. C., F. F. Salvacion, and W. Epstein. 1984. Tetracycline resistance element of pBR322 mediates potassium transport. J. Bacteriol. 160:1188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Epstein, W., and M. Davies. 1970. Potassium-dependent mutants of Escherichia coli K-12. J. Bacteriol. 101:836-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Epstein, W., and B. S. Kim. 1971. Potassium transport loci in Escherichia coli K-12. J. Bacteriol. 108:639-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Groisman, E. A., and M. J. Casadaban. 1986. Mini-Mu bacteriophage with plasmid replicons for in vivo cloning and lac gene fusing. J. Bacteriol. 168:357-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamann, A. 1991. Kaliumtransport bei Escherichia coli: molekularbiologische Untersuchungen der Komponenten der niedrigaffinen Kaliumaufnamesysteme. Ph.D. thesis. Osnabrück University, Osnabrück, Germany.
- 16.Helmer, G. A. 1982. A genetic and kinetic analysis of the Trk transport system of Escherichia coli K-12. Ph.D. thesis. The University of Chicago, Chicago, Ill.
- 17.Hempfling, W. P., and D. K. Beeman. 1971. Release of glucose repression of oxidative phosphorylation in Escherichia coli B by cyclic adenosine-3′-5′-monophosphate. Biochem. Biophys. Res. Commun. 45:924-930. [DOI] [PubMed] [Google Scholar]
- 18.Ito, K., B. Cooperberg, and T. A. Krulwich. 1997. Diverse genes of alkalophilic Bacillus firmus OF4 complement K+-uptake-deficient Escherichia coli including a ftsH homolog. Extremophiles 1:22-28. [DOI] [PubMed] [Google Scholar]
- 19.Kohara, Y., K. Akiyama, and K. Isono. 1987. The physical map of the whole E. coli chromosome: application of a new strategy for the rapid analysis and sorting of a large genomic library. Cell 50:495-508. [DOI] [PubMed] [Google Scholar]
- 20.Lee, C. H., Y. Kohara, K. Akiyama, C. L. Smith, W. J. Craigen, and C. T. Caskey. 1988. Rapid and precise mapping of the Escherichia coli release factor genes by two physical approaches. J. Bacteriol. 170:4537-4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levina, N., S. Tötemeyer, N. R. Stokes, P. Lewis, M. A. Jones, and I. R. Booth. 1999. Protection of Escherichia coli cells against extreme turgor by activation of the MscS and MscL mechanosensitive channels: identification of the genes required for MscS activity. EMBO J. 18:1730-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malli, R., and W. Epstein. 1998. Expression of the Kdp ATPase is consistent with regulation by turgor pressure. J. Bacteriol. 180:5102-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nichols, B. P., O. Shafiq, and V. Meiners. 1998. Sequence analysis of Tn10 insertion sites in a collection of Escherichia coli strains used for genetic mapping and strain construction. J. Bacteriol. 180:6408-6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ou, X., P. Blount, R. J. Hoffman, and C. Kung. 1998. One face of a transmembrane helix is crucial in mechanosensitive channel gating. Proc. Natl. Acad. Sci. USA 95:11471-11475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rakonjac, J., M. Milic, D. Ajdic-Predic, D. Santos, R. Ivanisevic, and D. J. Savic. 1992. nov: a new genetic locus that affects the response of Escherichia coli K-12 to novobiocin. Mol. Microbiol. 6:1547-1553. [DOI] [PubMed] [Google Scholar]
- 26.Rhoads, D. B., F. B. Waters, and W. Epstein. 1976. Cation transport in Escherichia coli. VIII. Potassium transport mutants. J. Gen. Physiol. 67:325-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rhoads, D. B., A. Woo, and W. Epstein. 1977. Discrimination between Rb+ and K+ by Escherichia coli. Biochim. Biophys. Acta 469:45-51. [DOI] [PubMed] [Google Scholar]
- 28.Rudd, K. E. 1998. Linkage map of Escherichia coli K-12, edition 10: the physical map. Microbiol. Mol. Biol. Rev. 62:985-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sardesai, A. A., and J. Gowrishankar. 2001. Improvement in K+-limited growth rate associated with expression of the N-terminal fragment of one subunit (KdpA) of the multisubunit Kdp transport system in Escherichia coli. J. Bacteriol. 183:3515-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schlösser, A., A. Hamann, D. Bossemeyer, E. Schneider, and E. P. Bakker. 1993. NAD+ binding to the Escherichia coli K+-uptake protein TrkA and sequence similarity between TrkA and domains of a family of dehydrogenases suggest a role for NAD+ in bacterial transport. Mol. Microbiol. 9:533-543. [DOI] [PubMed] [Google Scholar]
- 31.Schlösser, A., M. Meldorf, S. Stumpe, E. P. Bakker, and W. Epstein. 1995. TrkH and its homolog, TrkG, determine the specificity and kinetics of cation transport by the Trk system of Escherichia coli. J. Bacteriol. 177:1908-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singer, M., T. A. Baker, G. Schnitzler, S. M. Deischel, M. Goel, W. Dove, K. J. Jaacks, A. D. Grossman, J. W. Erickson, and C. A. Gross. 1989. A collection of strains containing genetically linked alternating antibiotics resistance elements for genetic mapping of Escherichia coli. Microbiol. Rev. 53:1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sukharev, S. I., P. Blount, B. Martinac, F. R. Blattner, and C. Kung. 1994. A large-conductance mechanosensitive channel in Escherichia coli encoded by mscL alone. Nature 368:265-268. [DOI] [PubMed] [Google Scholar]
- 34.Takahashi, Y. 1975. Effect of glucose and cyclic adenosine 3′,5′-monophosphate in the synthesis of succinate dehydrogenase and isocitrate lyase in Escherichia coli. J. Biochem. 78:1097-1100. [DOI] [PubMed] [Google Scholar]
- 35.Walderhaug, M. O., D. C. Dosch, and W. Epstein. 1987. Potassium transport in bacteria, p. 84-130. In B. P. Rosen and S. Silver (ed.), Ion transport in procaryotes. Academic Press, New York, N.Y.