Abstract
Pseudomonas aeruginosa has a wide ecological distribution that includes natural habitats and clinical settings. To analyze the population structure and distribution of P. aeruginosa, a collection of 111 isolates of diverse habitats and geographical origin, most of which contained a genome with a different SpeI macrorestriction profile, was typed by restriction fragment length polymorphism based on 14 single nucleotide polymorphisms (SNPs) located at seven conserved loci of the core genome (oriC, oprL, fliC, alkB2, citS, oprI, and ampC). The combination of these SNPs plus the type of fliC present (a or b) allowed the assignment of a genetic fingerprint to each strain, thus providing a simple tool for the discrimination of P. aeruginosa strains. Thirteen of the 91 identified SNP genotypes were found in two or more strains. In several cases, strains sharing their SNP genotype had different SpeI macrorestriction profiles. The highly virulent CHA strain shared its SNP genotype with other strains that had different SpeI genotypes and which had been isolated from nonclinical habitats. The reference strain PAO1 also shared its SNP genotype with other strains that had different SpeI genotypes. The P. aeruginosa chromosome contains a conserved core genome and variable amounts of accessory DNA segments (genomic islands and islets) that can be horizontally transferred among strains. The fact that some SNP genotypes were overrepresented in the P. aeruginosa population studied and that several strains sharing an SNP genotype had different SpeI macrorestriction profiles supports the idea that changes occur at a higher rate in the accessory DNA segments than in the conserved core genome.
Pseudomonas aeruginosa is a gram-negative bacterium present in soil and aquatic environments (42). In addition, this bacterial species is also an important opportunistic pathogen for humans, animals, and plants. It can produce severe infections in immunocompromised hosts (29) and is the major factor for morbidity and mortality in cystic fibrosis patients (13). Its ecological diversity is probably related to its great metabolic versatility. Its pathogenic ability derives from the presence of several cell-associated and secreted virulence factors, such as elastase, exotoxin A, phospholipase, and alkaline protease, among others (8, 26, 47). P. aeruginosa uses a type III secretion system to directly deliver several effector proteins into the cytoplasm of the host cell, a key step in the cytotoxic and invasion processes (12, 15, 34, 49). Remarkably, the genes encoding all of these virulence factors are not clustered in pathogenicity islands but are rather dispersed in the bacterial chromosome (45).
An important question regarding P. aeruginosa is whether the strains isolated from infected patients correspond to specialized strains adapted to clinical habitats or whether the virulence of this bacterial species results from a set of traits that are present in most or all strains from any environment. Several data support the view that the strains isolated from nonclinical environments (referred to hereafter as environmental strains) are indistinguishable from clinical isolates in terms of several genotypic, taxonomic, or metabolic properties (2, 6, 10, 11, 17, 32, 33). Furthermore, a recent whole-genome analysis of the presence or absence of strain-specific genes within a set of 18 strains isolated from clinical and nonclinical habitats revealed no correlation between genome content and infection type as well as a remarkable conservation of genes, including those encoding most known virulence factors (48).
Current knowledge indicates that the P. aeruginosa genome is made up of a mosaic of a conserved core and variable accessory segments (10, 14, 17, 31, 41). The core genome is characterized by a conserved synteny of genes and a low average nucleotide substitution rate (about 0.5%). Only 2.5% of the coding sequences exhibit significantly higher sequence diversity. Clone- or strain-specific genome islands define the variable part of the chromosome (4, 10, 16, 19, 21, 32, 36) and lead to fluctuations in the genome size, which can range from 5.2 to 7 Mbp (36).
Single nucleotide polymorphism (SNP) genotyping of six genes of the core genome in a collection of 19 environmental and clinical P. aeruginosa strains and a restriction fragment length polymorphism (RFLP) analysis of their chromosomes revealed a high sequence conservation in four of the genes and a high frequency of recombination within the chromosome, leading to a random association of alleles (17). This indicates that the P. aeruginosa gene pool is in linkage equilibrium. Due to the frequent exchange of genomic islands and accessory DNA segments, P. aeruginosa populations would consist of a series of equivalent genotypes formed by related strains (termed clones), which form a net-like population structure (17). A different collection of 73 environmental and clinical isolates collected from diverse sources was studied by combining the analysis of some phenotypic traits, the DNA sequences of three genes, and amplified fragment length polymorphism pattern analysis (28). The results showed that the isolates could be grouped into clusters but that the clusters obtained by the different experimental approaches were not always congruent. Amplified fragment length polymorphism analyses suggested the existence of groups of strains (clones) having a related chromosome structure, although the relatedness and organization of this clonal population were obscured by DNA rearrangements and insertion of large DNA segments at conserved regions of the otherwise highly conserved chromosomal backbone. The detection of clonal complexes suggested a transient epidemic-like spread of certain genotypes. This epidemic distribution of P. aeruginosa populations is more clearly detected when the strains analyzed are collected in a small region or in a specific environment (3, 22, 27).
A clear view on the structure and dynamics of P. aeruginosa populations is relevant both to understand the biology of this ubiquitous bacterial species in different habitats and to perform epidemiological studies to trace nosocomial infections (27). Current analyses on this topic have been performed focusing either on a few markers on many strains or on many markers on only a few strains (3, 6, 10, 17, 22, 27, 28, 48). However, a consistent picture of P. aeruginosa populations would benefit from the analysis of large numbers of strains by using many, rather than a few, genetic markers and by using highly discriminative tests. To this end, 111 strains were selected, mainly from our own collections of more than 5,000 isolates, to gather a group containing strains from a broad range of sources and habitats (both clinical and nonclinical environments) and in which most of the strains represented a unique clone according to the SpeI macrorestriction fragment fingerprint, which is the current “gold standard” for genotyping P. aeruginosa (25). We have looked for SNPs in seven conserved loci of the core genome in these 111 strains. This has led to find a combination of 14 SNPs which, together with the type of fliC gene variant present (a or b), allows the assignment of a highly discriminative signature to individual strains. Assuming linkage equilibrium, we could expect free recombination of loci and one unique SNP marker genotype per clone. However, in some cases, an SNP genotype was shared by two or more strains. The possible meaning of these findings is discussed.
MATERIALS AND METHODS
Bacterial strains and culture media.
The P. aeruginosa strain collection used consisted of 111 isolates obtained from patients with different pathologies and from various countries (87 strains), from different nonclinical environments (termed environmental strains, 20 isolates), or from other unknown sources. A detailed description is provided in Table 1. The strains were kindly provided by F. Baquero (Ramón y Cajal Hospital, Madrid, Spain) and I. Attrée (CHU—Grenoble, Grenoble, France) or belonged to our own collections. Some strains were purchased from the American Type Culture Collection or Colección Española de Cultiros Tipo. Bacterial strains were cultivated in Luria-Bertani medium (35) unless otherwise stated.
TABLE 1.
SNP- genotypes of P. aeruginosa strains from different origins
| Strain | Origin (reference)a | Sample type (source)b | SNP at genec:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| oriC (AlwNI, 267, T→C) | oprL (BstUI, 396, T→C) |
alkB2
|
citS
|
oprl |
ampC
|
fliC type | |||||||||||
| MspA1I (471, G→A) | Fnu4HI (1047, A→G) | AciI (533, A→G) | MniI (896, G→C) | AciI (195, T→C) | MspI (789, G→A) | MnI (883, C→T) | BstUI (1060, C→T) | HinfI (1150, G→A) | AlwNI (1166, G→C) | AgeI (1585, T→C) | BsrI (1639, C→A) | ||||||
| PAO1 | AU (44) | Clin (1) | T | T | G | A | A | G | T | G | C | C | G | G | T | C | b |
| RR1 | ES (50) | Env (9) | — | C | A | G | — | C | C | — | — | — | — | — | — | A | a |
| CECT 119 | ES | Env (10) | C | C | — | G | — | — | C | — | — | T | A | — | — | A | b |
| 1.1A1 | CH (7) | Clin (2) | C | C | A | G | — | C | — | A | — | — | — | — | — | — | a |
| 4.1A1 | CH (7) | Clin (2) | — | — | — | — | — | C | — | — | — | — | — | — | — | A | b |
| 22D10 | CH (7) | Clin (2) | C | C | — | — | — | C | — | — | — | — | — | — | — | A | a |
| 7B5 | CH (7) | Clin (2) | C | C | A | — | G | — | C | — | — | — | — | — | — | b | |
| 18F7 | CH (7) | Clin (2) | — | — | — | — | — | — | — | — | — | — | — | — | C | — | b |
| 19G12 | CH (7) | Clin (2) | C | C | — | G | G | — | C | — | — | T | — | C | ND | ND | a |
| 36D5 | CH (7) | Clin (2) | C | C | — | G | G | — | C | — | — | — | — | — | ND | ND | a |
| 60B5 | CH (7) | Clin (2) | — | — | — | — | — | C | C | — | T | — | — | — | C | — | a |
| 036SIC95 | CH | Clin (2) | — | — | A | G | — | C | — | A | T | — | — | — | — | A | b |
| 017CHI96 | CH | Clin (2) | — | C | A | G | — | — | — | A | T | T | A | — | — | A | a |
| 050SIC98-1-1 | CH | Clin (2) | C | — | — | G | — | C | C | A | T | — | — | — | C | — | a |
| 039SIC93 | CH | Clin (2) | — | ND | A | G | G | C | C | A | ND | — | — | — | ND | ND | a |
| 005BS92 | CH | Clin (2) | — | C | — | — | — | C | — | A | ND | T | A | — | — | A | b |
| 030SIC94-1-2 | CH | Clin (2) | — | — | A | — | — | C | C | A | ND | T | A | — | — | A | a |
| 030CHI91-1-1 | CH | Clin (2) | C | C | — | G | G | — | C | A | ND | — | — | C | ND | ND | a |
| 038SIC92 | CH | Clin (2) | C | C | A | G | G | — | C | A | — | — | — | C | ND | ND | a |
| 057SIC97-3-1 | CH | Clin (2) | C | C | A | G | — | C | C | A | ND | — | — | — | ND | ND | b |
| 020SIC93-6-1 | CH | Clin (2) | — | C | — | G | — | C | C | A | T | — | — | — | — | A | b |
| 892 | DE (17) | Clin (3,4) | — | — | A | G | G | C | C | — | — | — | — | C | — | A | b |
| K9 | DE (17) | Clin (3,4) | — | — | A | — | G | C | — | A | — | T | A | — | C | — | a |
| G7 | DE (17) | Clin (3,4) | — | C | A | G | — | C | C | — | — | T | A | — | — | A | b |
| DSM1128 | US (17) | Clin (11) | — | C | — | G | G | C | C | ND | ND | ND | ND | ND | — | — | a |
| DM | DE (17) | Clin (3,4) | C | C | A | G | — | C | — | — | — | — | — | C | — | — | a |
| HJ2 | DE (17) | Clin (3,4) | — | — | — | — | — | C | C | — | T | — | — | — | C | — | a |
| 63741 | DE (17) | Clin (1) | — | — | A | G | G | C | C | — | — | — | — | C | — | A | b |
| ATCC 10145 | CR (17) | Unkn | — | C | A | — | — | — | C | A | — | — | A | — | C | A | a |
| ATCC 15691 | A (17) | Clin (5) | — | C | — | G | G | C | C | — | — | T | A | — | — | A | a |
| ATCC 33356 | DE (17) | Clin (6) | C | C | — | — | — | C | C | — | — | T | A | — | — | A | a |
| ATCC 33818 | PC (17) | Env (9) | — | C | — | G | — | C | C | — | — | — | A | — | C | — | b |
| ATCC 21776 | JP (17) | Env (8) | — | ND | A | G | G | — | C | — | T | — | — | — | — | A | b |
| H2 | DE (17) | Env (7) | — | — | A | G | — | — | C | — | — | — | — | C | — | A | a |
| PAK | JP | Clin | — | C | A | — | G | — | C | — | — | T | A | — | ND | ND | a |
| ZW 30 | A | Clin (3) | C | — | — | G | G | — | C | — | — | — | A | C | ND | ND | b |
| ZW 31 | A | Clin (3) | C | — | A | G | — | C | C | — | — | — | — | — | ND | ND | a |
| ZW 41 | IT | Clin (3) | — | — | — | — | — | — | — | — | — | T | A | — | — | A | b |
| ZW 43 | IT | Clin (3) | C | C | — | G | G | — | C | A | — | — | — | C | ND | ND | b |
| ZW 49 | IT | Clin (3) | C | — | A | G | — | — | C | — | — | ND | A | — | C | — | a |
| ZW 64 | SE | Clin (3) | — | — | A | G | — | — | C | A | — | T | A | — | — | A | a |
| ZW 77 | UK | Clin (3) | — | C | ND | G | — | — | — | — | — | — | — | — | — | A | b |
| ZW 79 | IR | Clin (3) | — | — | — | — | — | C | C | — | T | — | — | — | C | — | a |
| ZW 81 | UK | Clin (3) | C | C | A | — | G | — | C | — | — | — | — | — | ND | ND | a |
| ZW 83 | UK | Clin (3) | — | C | — | ND | — | C | C | A | — | — | — | — | — | A | b |
| ZW 85 | UK | Clin (3) | C | C | — | — | G | — | C | — | — | — | — | — | — | A | b |
| ZW 88 | UK | Clin (3) | — | — | A | G | — | C | C | — | — | — | — | — | — | A | a |
| ZW 92 | FR | Clin (3) | C | C | A | G | — | C | C | — | T | — | — | — | C | — | b |
| ZW 98 | NL | Clin (3) | C | ND | — | G | — | — | C | — | — | — | — | — | — | A | a |
| ZW 117 | A | Clin (3) | — | — | — | — | — | C | — | — | — | — | — | — | — | A | b |
| ZW 119 | PL | Clin (3) | C | C | A | G | G | — | C | — | — | — | — | C | ND | ND | a |
| BST1 | DE | Clin (3) | C | C | — | G | — | — | C | ND | — | — | — | C | ND | ND | a |
| KB1 | DE | Clin (3) | — | — | — | — | — | — | C | — | — | T | A | — | — | A | a |
| MF6 | DE | Clin (3) | C | — | A | — | — | C | C | — | — | — | A | — | — | A | a |
| PD1 | DE | Clin (3) | C | C | A | G | — | — | C | — | — | T | A | — | — | A | a |
| RP1 | DE | Clin (3) | — | — | — | — | — | C | C | — | T | — | — | — | C | — | a |
| SS1 | DE | Clin (3) | — | C | A | G | — | C | C | — | — | T | A | — | — | A | b |
| A 5670 | DE | Clin (5) | — | C | — | G | G | C | C | — | — | T | A | — | — | A | a |
| A 5803 | DE | Clin (2) | C | C | A | G | G | — | C | — | — | — | — | — | ND | ND | a |
| AL 5846 | DE | Clin (5) | C | C | A | — | G | — | C | — | — | — | — | — | — | A | a |
| 2733/92 | DK | Clin (3) | — | — | A | G | G | C | C | — | T | — | — | — | C | — | a |
| VA27260 | DE | Clin (3) | — | — | A | G | — | — | — | A | — | T | A | — | — | A | a |
| VA26232 | DE | Clin (3) | C | C | — | G | — | — | C | — | — | — | — | — | ND | — | a |
| GR2248 | GR | Clin | — | C | — | G | — | C | C | — | T | T | A | — | — | A | b |
| 2813A/92 | DK | Clin (3) | — | C | — | — | — | — | — | — | — | — | — | — | — | A | b |
| PT36 | DE | Env (10) | C | C | A | — | G | — | C | — | — | — | — | — | — | A | a |
| 641/HD 11/ml | DE | Env (10) | — | — | A | G | — | — | C | — | — | — | A | — | — | A | a |
| PT20 | DE | Env (10) | — | — | A | G | — | C | C | — | — | — | — | — | — | A | a |
| PT12 | DE | Env (10) | C | C | A | G | G | — | C | — | — | — | — | — | — | A | a |
| PT22 | DE | Env (10) | C | C | — | G | — | C | C | — | T | — | — | — | C | — | a |
| PT6 | DE | Env (10) | — | — | A | ND | — | C | — | — | — | T | A | — | — | A | b |
| PT2 | DE | Env (10) | C | C | — | — | G | — | C | — | — | — | — | — | ND | ND | b |
| ATCC 33348 | PC | Clin | — | — | A | G | — | C | C | — | — | — | — | — | — | A | a |
| ATCC 33364 | PC | Clin | C | C | ND | G | — | — | C | — | — | — | — | — | ND | ND | a |
| ATCC 14886 | PC | Env (8) | C | C | — | G | — | C | C | — | T | — | — | — | C | — | a |
| ATCC 21472 | PC | Env (8) | — | — | A | G | G | — | C | — | T | — | — | — | — | A | b |
| ATCC 33988 | US | Env | — | C | — | G | — | C | C | — | T | — | — | — | C | — | b |
| DSM 939 | PC | Env (10) | — | — | — | — | — | — | C | — | — | — | A | — | — | A | a |
| DSM 288 | PC | Unkn | — | — | — | — | — | C | — | A | — | T | A | — | — | A | b |
| DSM 1253 | PC | Unkn | — | — | — | — | — | C | — | A | — | T | A | — | — | A | b |
| VA 27081 | DK | Clin (3) | — | — | — | — | — | C | — | — | — | — | — | — | — | A | a |
| NAG 96 | ES | Clin (3) | C | C | A | G | — | C | C | — | — | ND | — | C | — | A | a |
| JPR 135 | ES | Clin (3) | — | — | A | G | — | C | C | — | — | T | — | — | ND | ND | b |
| JPR 6 | ES | Clin (3) | — | C | A | G | — | — | — | — | — | — | — | C | — | A | b |
| CDR 67 | ES | Clin (3) | C | C | — | G | G | C | — | — | — | — | — | C | ND | ND | a |
| MOG 111 | ES | Clin (3) | C | C | — | ND | G | ND | C | — | — | — | — | — | ND | ND | a |
| MOG5 | ES | Clin (3) | — | — | A | G | — | C | C | — | — | — | — | — | — | A | a |
| JMSMA 7 | ES | Clin (3) | C | C | A | G | G | — | C | — | — | — | — | C | ND | ND | a |
| JMSMA125 | ES | Clin (3) | C | ND | ND | — | G | — | C | — | — | — | — | C | ND | ND | a |
| RRG139 | ES | Clin (3) | C | C | — | G | G | — | C | — | — | — | — | C | ND | ND | a |
| JMAM76 | ES | Clin (3) | C | C | — | G | ND | — | C | — | — | — | — | — | — | A | a |
| AAR114 | ES | Clin (3) | — | — | A | G | — | C | C | A | — | T | A | — | — | — | b |
| NCC 81 | ES | Clin (3) | — | — | — | — | — | C | C | — | T | — | — | — | C | — | a |
| NHP 11 | ES | Clin (3) | — | C | — | G | — | C | C | A | — | — | — | — | — | A | b |
| DGA 138 | ES | Clin (3) | C | C | A | G | — | C | C | — | T | — | — | — | C | — | b |
| ASL 27 | ES | Clin (3) | — | — | A | G | — | C | C | — | — | — | — | — | — | — | b |
| VSF 17 | ES | Clin (3) | — | — | A | — | G | C | — | A | — | T | A | — | C | — | a |
| SFQ 47 | ES | Clin (3) | — | — | — | — | — | — | — | — | — | — | — | — | — | — | b |
| BTGE 45 | ES | Clin (3) | C | — | A | G | — | C | C | A | — | T | A | — | — | A | a |
| FM 915 | ES | Clin (3) | — | — | A | G | — | C | C | — | — | — | — | — | — | A | a |
| MW 28 | ES | Clin (3) | C | C | — | — | G | — | C | — | — | — | — | — | ND | ND | b |
| CHA | FR (46) | Clin (3) | C | C | — | G | — | C | C | — | T | — | — | — | C | — | a |
| ATCC 15522 | ESSO | Env (8) | — | C | A | — | — | C | — | — | — | — | — | — | — | A | a |
| ATCC 15524 | ESSO | Env (8) | — | C | A | — | — | C | — | — | — | — | — | — | — | A | a |
| ATCC 15528 | ESSO | Env (8) | — | C | A | — | — | C | — | — | — | — | — | — | — | A | a |
| 3D10 | CH (7) | Clin | — | — | — | — | — | — | — | — | — | — | — | — | — | — | b |
| 18F8 | CH | Clin | — | — | — | — | — | — | — | — | — | — | — | — | — | — | b |
| SG1 | DE (17) | Clin (3) | C | C | — | — | — | — | C | — | T | — | — | — | — | — | a |
| SG31 | DE (17) | Env (10) | C | C | — | — | — | — | C | — | T | — | — | — | — | — | a |
| GR2052 | GR | Clin | — | — | A | G | — | C | C | — | — | — | A | — | — | A | b |
| GR2057 | GR | Clin | — | — | A | G | — | C | C | — | — | — | A | — | — | A | b |
Origin abbreviations: AU, Australia; A, Austria; CH, Switzerland; CR, Czech Republic; DE, Germany; DK, Denmark; ES, Spain; FR, France; GR, Greece; IR, Ireland; IT, Italy; JP, Japan; NL, The Netherlands; PL, Poland; SE, Sweden; US, United States of America; PC, public collection; ESSO, Esso Oil Co.
Abbreviations: Env, environmental; Clin, Clinical; Unkn, unknown origin. Sources: 1, burn patient; 2, tracheal aspirate from an intubated patient; 3, cystic fibrosis patient; 4, sputum; 5, wound; 6, human feces; 7, catheter; 8, soil; 9, mushroom; 10, water; 11, ear infection.
The location of the SNPs at each gene is indicated both by its position and by the restriction enzyme used in the RFLP analysis. Numbering of the SNPs at oriC, citS, oprl, and ampC is according to the method described in reference 17. Numbering of the SNPs at alkB2 and oprL is given relative to the translation start site. Nucleotides that differ from that of PAO1 in each SNP are shown. The SNPs detected in strain PAO1 match to those present in the strain whose complete genome has been sequenced (45). ND, not determined; —, same nucleotide as in strain PAO1.
RAPD genotyping.
Genotyping by the random amplified polymorphic DNA (RAPD) method was performed as described previously (7). One isolate per RAPD type was further typed by analyzing the SpeI macrorestriction pattern of its genome by pulsed-field gel electrophoresis (PFGE) as described below.
PFGE.
Samples were analyzed as described in reference 33, with minor modifications. P. aeruginosa strains were grown overnight at 37°C in Luria-Bertani medium, centrifuged for 10 min at 1,500 × g, and suspended in 75 mM NaCl and 25 mM EDTA (pH 7.4) to a concentration of 5 × 109 cells/ml. The cell suspension was mixed 1:1 with 2% (wt/vol) low-melting-point agarose to prepare the agarose plugs. The embedded cells were digested for 48 h at 56°C with proteinase K (1 mg/ml in 0.5 M EDTA [pH 9.5], 1% [vol/vol] N-lauryl-sarkosine). Agarose plugs were equilibrated and stored in 10 mM EDTA and 10 mM Tris-HCl (pH 7.4) at 4°C until used. One-third to one-half of a plug was equilibrated in SpeI buffer (50 mM NaCl, 6 mM Tris-HCl [pH 7.5], 10 mM MgCl2). Digestion was performed overnight at 37°C in 90 μl of enzyme buffer with 4 U of SpeI, 0.1 mg of bovine serum albumin/ml, and 5 mM dithiothreitol. Separation of DNA fragments was performed by PFGE in a CHEF-DR III apparatus (Bio-Rad), with a 1.5% (wt/vol) agarose gel, 0.5× Tris-borate-EDTA buffer, and a linear ramping from 8 to 50 s for 24 h, 12 to 25 s for 22 h, and 1 to 14 s for 14 h. DNA fragments were visualized by ethidium bromide staining. Genotypes were defined from the evaluation of SpeI fragment patterns as described in reference 30.
SNP analysis by DNA sequencing.
To search for SNPs in alkB2, this gene was sequenced in 20 strains (listed below). To this end, genomic DNA was PCR amplified by using either AlkB2-1 and AlkB2-2 or AlkB2-3 and RR1-20 as primers (Table 2). The DNA fragments obtained were sequenced with the same primers as above.
TABLE 2.
Primers used for PCRs and endonucleases used to analyze SNPs at the indicated loci by RFLP
| Locus | Primer | Primer sequencea | PCR fragment size (bp) | Endonuclease used in RFLP analysisb | SNP detected (position)c | Presence of the restriction site in strain PAO1 |
|---|---|---|---|---|---|---|
| oriC | oriC dir | AGAACCTGACGGCCGTTCTT | 632 | AlwNI | T→C (267) | No |
| oriC inv | AAGTTCCACGGACACGGATAT | |||||
| oprL | oprL dir | GCGCTACCTGGTGCTGCAGCG | 114 | BstUI | T→C (396) | No |
| oprL inv | GCGACGGTTCTGAGCCCAGGA | |||||
| alkB2 | RR1-20 | CGATCTGCTCTGCCAGGCTG | 820 | MspA1I | G→A (471) | No |
| AlkB2-3 | CTTGTAGGCGTGCGGGAGGA | |||||
| alkB2 | AlkB2-2 | GTGCGCGGCCACCATGTACA | 779 | Fnu4HI | A→G (1047) | No |
| AlkB2-1 | TTTCCGCACAGGGCTCCACG | |||||
| citS | Cit6 | CCATCGCGGCTACCCCATCG | 872 | AciI | A→G (533) | No |
| Cit3 | GCTTCGTTCGCGCCCCCATG | MnlI | G→C (896) | No | ||
| oprI | oprI dir | ATTTCCCCGGCTGGGAGATTGC | 341 | AciI | T→C (195) | No |
| oprl inv | CGAGGGACCGGTTTTCAACAGG | |||||
| ampC | 1162 | GGTCGAACCAATCTCTGCTCC | 762 | MspI | G→A (789) | Yes |
| 1161 | CATGAGCCGTTCGAACGGCTG | MnlI | C→T (883) | Yes | ||
| ampC | 1160 | CCTGACCCAGGACAAGATGC | 489 | BstUI | C→T (1060) | Yes |
| 1428 | AGGTTGGCATCGACGAAGCGC | HinfI | G→A (1150) | No | ||
| AlwNI | G→C (1166) | No | ||||
| ampC | 1427 | CAGTACGCCCAGGGCTATGGC | 519 | AgeI | T→C (1585) | Yes |
| 1163 | GGCCTTCAGCGGCACCTTGC | BsrI | C→A (1639) | No | ||
| fliC | Fla1 | GCCTGCAGAACGCCAACC | 997 or 1018 (type a) | |||
| Fla2 | GGCAGCTGGTTGGCCTG | 1300 (type b) |
Primers for oriC, citS, and ampC are based on those described in reference 40.
Some of the PCR fragments served to analyze more than one SNP by RFLP. In those cases, more than one endonuclease is indicated for each fragment and the absence or presence of the corresponding target in strain PAO1 is indicated in the adjacent column.
The position of each SNP at the corresponding locus is indicated as specified in Table 1.
SNP analysis by RFLP.
Boiled colonies of each of the 111 strains were subjected to PCR with adequate primers to amplify DNA fragments containing the SNP to be analyzed (Table 2). PCR was performed by using the Ready-to-go PCR beads kit (Amersham) as specified by the supplier. Amplified DNA was digested with the restriction enzyme discriminating each SNP, and the fragments generated were analyzed by agarose gel electrophoresis and ethidium bromide staining. To detect the SNP in the oprL gene, an artificial restriction site was created in the direct primer and mismatch PCR (5) was performed. In brief, the primer was designed so that its 3′ end is 1 bp upstream from the SNP, and it contains a mismatch close to the 3′ end, which generates a restriction site for BstUI if the sequence contains a cytosine at the site of the SNP but not if the base is a thymidine. Therefore, the presence or absence of the BstUI site allows for the analysis of the SNP.
Analysis of fliC.
P. aeruginosa strains contain one of two variants of the fliC gene (a type and b type), which encode flagellins of different molecular weights (1). Therefore, a PCR approach was undertaken to identify which variant was present in each strain. The oligonucleotides used for PCR, indicated in Table 2, rendered DNA fragments of 997 or 1,018 bp (a type fliC) or of 1,300 bp (b type fliC).
Nucleotide sequence accession number.
The EMBL GenBank accession numbers for alkB2 are as follows: strain RR1, AJ33602; strain 892, AJ633605; strain K9, AJ633603; strain G7, AJ633606; strain SG1, AJ633604; strain DSM 1128, AJ633607; strain DM, AJ633608; strain HJ2, AJ633609; strain 63741, AJ633610; strain SG31, AJ633611; strain ATCC 10145, AJ633612; strain ATCC 15691, AJ633613; strain ATCC 33356, AJ633614; strain ATCC 33818, AJ633615; strain PAK, AJ633616; strain 19G12, AJ633617; strain CECT 119, AJ633618; strain ATCC 15524, AJ633619; strain CHA, AJ633620.
RESULTS
Strain panel analyzed.
The investigated strain panel (Table 1) consists of 20 strains available from public culture collections and 91 isolates from the local collections of the collaborating laboratories and their partners. The Geneva collection, which mainly consists of contemporary sequential isolates from tracheal aspirates of intubated patients treated at intensive care units (7), was first screened by RAPD genotyping. One strain per RAPD type was then analyzed in its SpeI macrorestriction fragment pattern to ensure that only unique clones were incorporated into the common panel. Two isolates with the same RAPD and SpeI profile were also included as internal controls (strains 3D10 and 18F8). Strains from the Hannover collection were selected to include all abundant clones, such as C, J, K, M, and TB (33), and all clones found in more than one habitat, considering a source and geographic origin as diverse as possible, as well as the whole collection period from 1982 onwards. About 1,600 of the more than 3,000 P. aeruginosa isolates of the Hannover collection have been typed by macrorestriction fragment pattern analysis. For the purpose of this study, unique SpeI genotypes were each represented by just one strain. In other words, even the abundant clones were represented by one isolate. Pairs of strains which were previously known to have the same SNP combination at oriC, citS, ampC, and oprL and which contained related SpeI genotypes (17) were included as internal controls only in a few cases. The Madrid collection (strains NAG96 to MVV28 in Table 1) also included isolates with unique RFLP macrorestriction profiles. The panel was completed with other strains from diverse origins.
Determination of SNPs in selected genes.
Seven loci were analyzed for SNPs suitable for discrimination of P. aeruginosa strains. The loci were chosen according to several criteria: they should be highly conserved, they should belong to different categories (regulatory, structural, or metabolic genes), and they should be evenly distributed through the P. aeruginosa genome. Several SNPs had already been described at oriC (the origin of replication), citS (citrate synthase), ampC (chromosomal beta-lactamase), oprI (outer membrane protein OprI), fliC (flagellin) (17), and oprL (28). A subset of these reported SNPs in which the less frequent sequence variant was present in more than 15% of the analyzed strains were selected for our analyses. Sequencing of alkB2 in P. aeruginosa strain RR1 had shown the presence of several SNPs relative to strain PAO1 (24). To determine whether any of these SNPs could be useful to our study, alkB2 was sequenced in 20 P. aeruginosa strains (strains and EMBL GenBank accession numbers are indicated in Materials and Methods). Forty-two SNPs were found. Thirty-nine of them were present in less than 15% of the strains analyzed and were therefore discarded. One of them (at position 1017, with numbering relative to the translation start site) was present in all but the reference strain PAO1 and was discarded as well. Two SNPs were detected that changed in more than 15% of the 20 strains, namely G→A at position 471 and A→G at position 1047. These two SNPs were chosen for further analyses. Therefore, the complete set of SNPs selected to analyze the P. aeruginosa population structure included one at oriC, two at citS, one at oprI, seven at ampC, one at oprL, and two at alkB2 (Table 1). P. aeruginosa strains are known to encode either a- or b-type flagellins that differ by 35% in primary structure (1, 39). Since these two fliC variants (a type and b type) can be used as a discrimination test (39, 40), the presence of each variant was also evaluated as a discrimination tool.
An important aspect to take into account when using SNPs for strain typing is whether the loci and the SNPs considered show genetic linkage or are rather randomly associated (this is, they are in linkage equilibrium). Previous work had shown that oriC, citS, ampC, oprI, and fliC are not genetically linked (17). Furthermore, the seven SNPs selected at ampC have a random association as well (40). As will be shown below, the complete set of SNPs analyzed in this work are also in linkage equilibrium. Therefore, the selected SNPs can be used as a way to discriminate between strains.
Analysis of the selected SNPs in the complete P. aeruginosa strain collection (composed of 111 strains) was performed by RFLP of PCR-amplified DNA, since in most cases, one of the variants of the SNP corresponds to a target for a restriction enzyme which is missing in the alternative variant. In the case of oprL, where such a target was not available, an artificial restriction site was created at one of the oligonucleotides used in the PCR. The results of the RFLP analyses are shown in Table 1. The profile of SNPs for each strain is referred to as the SNP genotype. Strain PAO1 was taken as the reference strain, and its sequence for each SNP is indicated in Table 1. For the other strains, the sequence is shown only when it differs from PAO1. The genes citS, ampC, oprI, and fliC, as well as the oriC locus, had been previously fully sequenced in some of the strains tested, namely strains 892, K9, G7, DM, 63741, ATCC 10145, DSM 1128, HJ2, ATCC 15691, ATCC 33356, ATCC 33818, ATCC 21776, H2, SG1, and SG31 (17). The results obtained by RFLP were fully consistent with the reported sequence data, indicating that the method is reliable. Furthermore, in the case of the 20 strains in which the alkB2 gene had been sequenced, the RFLP method also gave consistent results.
Linkage analysis and frequency of SNPs.
To analyze the possible linkage among all of the SNPs characterized, we determined the index of association (IA), which is a measure of linkage disequilibrium (38). IA is defined as VO/(VE−1), where VO is the observed variance and VE the expected variance of the mean number of SNPs at which two P. aeruginosa strains differ. Calculations were performed by using the software available at http://www.mlst.net. The value of IA was estimated by generation of 1,000 randomized data sets under the assumption of random association of loci. Using each genotype as a unit, as described before (17), the value of IA was 0.161, which indicates no evidence of association among the different SNPs analyzed, with a significance level of P < 0.001. The SNP set can therefore be used for strain typing.
An additional important requisite for an SNP to be useful in strain typing is that the less-frequent variant should be present in a significant fraction of the clones, with 50% being the ideal value. Table 3 shows the frequency of each SNP in the P. aeruginosa collection screened. The less-frequent variants of the SNPs selected in this study were present in 15 to 49% of the strains, values that are high enough to consider these SNPs suitable for strain typing. Since no linkage was detected among the SNPs characterized, the theoretical discrimination efficiency of this set of SNPs can be estimated by multiplying the accumulated frequencies of all of the SNPs considered. If this hypothesis is correct, the discrimination ability of this set of SNPs ranges from 2.7 × 10−3 in the worst case (a strain with all of the most frequent SNPs) to 1.5 × 10−8 in the best case (a strain with all of the less-frequent SNPs).
TABLE 3.
Frequencies of SNPs
| Gene | SNP | Position no. | Enzyme | % of strains the same as PAO1a | % of strains not the same as PAO1b |
|---|---|---|---|---|---|
| oriC | T→C | 267 | AlwNI | 59 | 41 |
| oprL | T→C | 396 | BstUI | 44 | 56 |
| alkB2 | G→A | 471 | MspA1I | 49 | 51 |
| alkB2 | A→G | 1047 | Fnu4HI | 38 | 62 |
| citS | A→G | 533 | AciI | 69 | 31 |
| citS | G→C | 896 | MnlI | 45 | 55 |
| oprI | T→C | 195 | AciI | 25 | 75 |
| ampC | G→A | 789 | MspI | 79 | 21 |
| ampC | C→T | 883 | MnlI | 80 | 20 |
| ampC | C→T | 1060 | BstUI | 77 | 23 |
| ampC | G→A | 1150 | HinfI | 71 | 29 |
| ampC | G→C | 1166 | AlwNI | 85 | 15 |
| ampC | T→C | 1585 | AgeI | 78 | 22 |
| ampC | C→A | 1639 | BsrI | 35 | 65 |
fliC type b (the same as in PAO1) appeared in 39% of the strains.
fliC type a (not the same as in PAO1) appeared in 61% of the strains.
Shared genotypes.
Comparison of the SNP profiles of the 111 isolates analyzed showed that several strains shared the SNP and fli genotypes (Fig. 1). In some cases, this finding was to be expected. For example, the strain pairs 892 and 63741, or SG1 and SG31, had been introduced as internal controls because they were known in advance to share 100% nucleotide sequence identity in the loci oriC, citS, oprI, ampC, fliC, and pilA and to share highly related or identical SpeI fragment fingerprints (17). Similarly, strains 3D10 and 18F8 were known to share the same RAPD profile and were isolated at the same hospital. In other cases, the shared SNP genotypes appear in strains that have been isolated from a single site (strains ATCC 15522, ATCC 15524 and ATCC 15528) (Table 1). For all other strains, however, shared SNPs profiles suggest unanticipated relatedness, since the SNP set used had a minimum estimated discrimination ability of 2.7 × 10−3. In most cases, the strains sharing the SNP genotype had very different origins and had been isolated in different countries, in different years, and even from different habitats (environmental and clinical). This is the case with strains PAO1 (reference strain, isolated in the 1950s in Australia), the clonal variants 3D10 and 18F8 (both from Switzerland), and strain SFQ47 (from Spain). Similarly, strains PT22 (a freshwater isolate from Germany), CHA (a highly virulent clinic isolate from a cystic fibrosis patient in France), and ATCC 14886 (of environmental origin) also share the same SNP profile. The last example is one of the most interesting, as it shows that environmental strains may cluster with highly virulent clinical isolates, reinforcing the idea that P. aeruginosa strains may not only survive in different habitats but keep their metabolic and infective properties independent of the habitat where they are found.
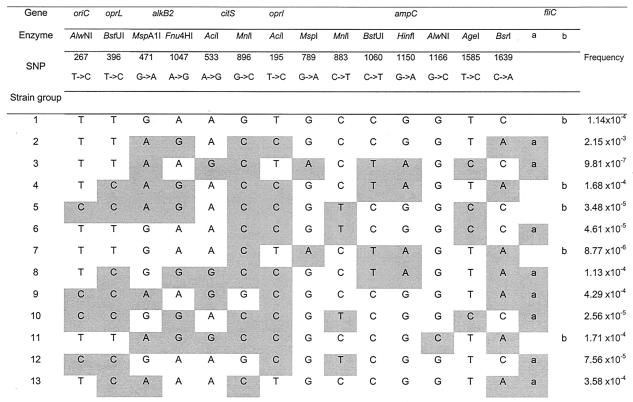
FIG. 1.
Shared SNP profiles. The locations of the SNPs at each gene are indicated both by position and by the restriction enzyme used in the RFLP analysis, identified in Table 1. Strain groups are as follows: 1, PAO1, 3D10, SFQ47, and 18F8; 2, ZW88 and PT20; 3, K9 and VSF17; 4, G7 and SS1; 5, ZW92 and DGA138; 6, RP1, HJ2, ZW79, NCC81, and 60B5; 7, DSM 288 and DSM 1253; 8, ATCC 15691 and A5670; 9, AL5846 and PT36; 10, PT22, CHA, and ATCC 14886; 11, 892 and 63741; 12, SG1 and SG31; 13, ATCC 15522, ATCC 15524, and ATCC 15528. Changes relative to strain PAO1 are shaded. The expected frequency of each SNP combination is indicated in the rightmost column, obtained by multiplying the frequencies at which each of the 14 individual SNPs appears in the strain collection analyzed, plus the type of fliC, as deduced from Table 3.
To further validate the significance of the shared SNP profiles, oriC, oprL, alkB2, citS, oprI, ampC, and fliC were fully sequenced in strains CHA and ATCC 14886. Interestingly, the sequences of all these loci were identical in the two strains. This corroborates again the accuracy of the RFLP method used to identify SNPs. Even more, the fact that these two strains share not only the SNPs analyzed but also the full sequence of the analyzed genes verifies that the two strains are highly related.
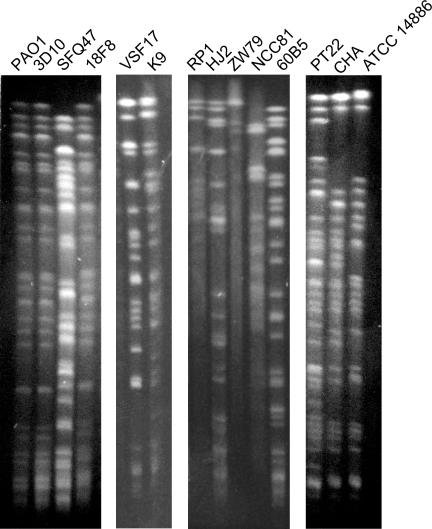
PFGE analysis.
To gain additional information on the genome organization of the strains sharing the SNP genotype, the SpeI fragment patterns of strains having the same SNP genotype were compared on adjacent lanes by PFGE (Fig. 2). Strains PAO1, 3D10, and 18F8, which share the same SNP profile, also show the same macrorestriction pattern. It was expected that strains 3D10 and 18F8 share the same SpeI macrorestriction pattern, since they are clonal variants intentionally introduced as an internal control in the typing assays. However, it was not expected that they would share the pattern with strain PAO1. Strain SFQ47, which shares the SNP genotype with PAO1, 3D10, and 18F8, has a different PFGE profile. Similarly, strains RP1 (clinical, Germany; clone J), HJ2 (clinical, Germany; clone M), ZW79 (clinical, United Kingdom), NCC81 (clinical, Spain), and 60B5 (clinical, Switzerland) share the same SNP genotype but have different SpeI macrorestriction patterns. Strains PT22 (environmental, Germany), CHA (clinical, France), and ATCC 14886 (environmental) show PFGE profiles that, although related in their fragment pattern, differ in several bands in spite of having the same SNP genotype. Finally, strains VSF17 (isolated in Spain) and K9 (Germany), which also share the SNP profile, have different macrorestriction patterns. Similar PFGE assays (data not shown) indicated that strain pairs G7 (clinical, Germany; clone G) and SS1 (clinical, Germany; clone K), DSM 288 and DSM 1253, ATCC 15691 (type strain) and A5670 (clinical, Germany), and PT20 (environmental strain, Germany) and ZW88 (clinical, United Kingdom) have different macrorestriction profiles, although they share identical SNP genotypes.
FIG. 2.
SpeI macrorestriction profiles of strains sharing the same SNP genotypes, performed by PFGE. The strains analyzed corresponded to groups 1 (PAO1, 3D10, SFQ47, and 18F8), 2 (VSF17 and K9), 6 (RP1, HJ2, ZW79, NCC81, and 60B5) and 10 (PT22, CHA, and ATCC 14886), as defined in the legend to Fig. 1.
DISCUSSION
Analysis of the evolution of bacterial populations is an important task for understanding their epidemiology and ecological behavior. To that goal, powerful molecular tools capable of discriminating between different isolates are required. In the case of P. aeruginosa, PFGE is the current gold standard. However, this method is time-consuming and difficult to apply to the study of large numbers of isolates. Furthermore, results from different laboratories are not always easy to compare (25). Multilocus sequence typing (and derived techniques like SNP typing) has been shown to be a powerful technique with several bacterial species (9, 23, 43). An SNP typing method to discriminate P. aeruginosa isolates was performed on the basis of the sequence of 7 conserved loci in 20 strains (17, 28; this work). The 14 SNPs analyzed mapped at oriC (1 SNP), oprL (1 SNP), alkB2 (2 SNPs), citS (2 SNPs), oprI (1 SNP), and ampC (7 SNPs). In addition, the variant of fliC, a or b, present in each strain was scored. The selected SNPs, many of which had already been described (17, 28), are not genetically linked (17, 40; this work), and in all cases, the less-frequent SNP variant was present in more than 15% of the sequenced strains. A simple RFLP method was designed to analyze the SNPs in a collection of 111 strains obtained from different origins and habitats. This allowed the definition of an SNP-based genetic fingerprint for each strain. For each SNP, the less-frequent variant was observed to be present in 15% (ampC) to 49% (alkB2) of the analyzed strains. Therefore, and considering the lack of linkage among them, the selected SNPs are useful for strain genotyping. A hypothetical phenotype containing the most frequent variant of each SNP should appear with a frequency of 2.7 × 10−3 (a value obtained by multiplying the frequencies at which the most frequent variant of each SNP appears, as deduced from the data in Table 3). This means that that RFLP method used should allow discrimination between P. aeruginosa strains with an accuracy of more than 99.97%.
A detailed analysis of the results obtained showed that several strains shared the same SNP genotype. Some of the strains sharing the same SNP profile had different SpeI macrorestriction profiles. In other cases, the SpeI macrorestriction profiles were clearly related, although they differed in several bands. An interesting case is that of isolates PT22, ATCC 14886, and CHA, which shared their SNP genotype and had different, although related, SpeI macrorestriction profiles. Strains CHA and ATCC 14886 shared not only the SNP genotypes but the full sequence of all the loci analyzed in this work, which highlights the relatedness of these two strains. Strain CHA is a highly virulent clinical isolate, whereas strains PT22 and ATCC 14886 were isolated from nonclinical (environmental) habitats. This observation supports the view that P. aeruginosa isolates that thrive in nonclinical habitats have probably all needed traits to infect mammals (2). Another interesting example is that of strains 3D10, 18F8, SFQ47, and PAO1, which are clinical isolates from different countries. On the whole, 13 groups of strains sharing SNP genotypes were found, 8 of which (groups 1, 2, 3, 4, 6, 7, 8, and 10) (Fig. 1) include strains that have different SpeI macrorestriction profiles. Strains K9 and VSF17 shared an SNP genotype that should appear with a very low theoretical frequency, 9.8 × 10−7, a value that suggests that this finding is not casual. Previous analyses have demonstrated that some PFGE-defined clones have widespread distribution (33). Indeed, two of the five strains from group 6 (Fig. 1), which have different SpeI genotypes but the same SNP genotype, belong to the most abundant clones in our strain collection (strains RP1 and HJ2, which belong to clones J and M, respectively). This collection includes isolates from numerous sources of diverse geographical origin (33). It is thus conceivable that these overrepresented genotypes define strains that are widely distributed. Notably, the PAO1 genotype is also overrepresented. Strain PAO1 was originally isolated form a burned patient in Australia 50 years ago (45). This suggests that the most prevalent strains are maintained in the environment for at least several decades, which may be the consequence of a better ecological adaptation. It should be noted that the bias deliberately introduced in our strain collection to avoid clonal variants has to be taken into account when using our results to evaluate the P. aeruginosa population structure. It is clear that our collection underscores the possible epidemic structure of natural populations, but we still detected the presence of some prevalent strain types that had a highly related core sequence (same SNP genotype), although they differed to a substantial extent in the SpeI macrorestriction profile.
Our results show that the use of RFLP for SNP genotyping can be a very powerful technique to discriminate between strains, but it is still unable to discriminate among highly related strains (for example, those having the same SNP profile but different macrorestriction patterns). This can have practical consequences when attempting to use this method to perform epidemiological studies to monitor an outbreak of a particular strain. To this end, the discriminatory ability should be improved even more by the addition of other markers or traits, for example, the presence or absence of genomic islands or of genes known to be present in some but not all P. aeruginosa strains.
The accumulation of molecular data in recent years and the growing evidence of the occurrence of horizontal gene transfer among bacteria in nature (reviewed in reference 37), have led to consideration that bacterial populations are not invariably clonal but range from the highly sexual Neisseria gonorrhoeae to the almost strictly clonal Salmonella (38). In this work, we have used both PFGE and SNP genotyping to analyze the P. aeruginosa population structure in a large and heterogeneous collection of strains. The sequence analysis of the loci studied here indicate that the core genome of P. aeruginosa is highly stable during evolution, since very few changes (even at the third position of the amino acid codons) were detected in strains isolated from different places and within a 50-year time lapse (17; this work). A similar result has been recently obtained for Escherichia coli with an in vitro model of bacterial evolution (20). On the other hand, the results obtained with the PFGE analysis support the view that, during evolution, large fragments of DNA (genome islands) can be excised from the genome or integrated into it, which then results in different macrorestriction patterns when analyzed by PFGE (16, 32) and confers genome diversity to this bacterial species (19). The population structure of P. aeruginosa is still under discussion, although recent data point to the idea that this species can display an epidemic population structure (28). The conservation of the SNP genotypes and the divergence of SpeI macrorestriction patterns in strains sharing the same SNP profile agree with the idea that the core genome of P. aeruginosa is highly conserved and that its evolution and structure rely more on acquisition, loss, and rearrangements of genome islands and genome islets than on point mutations. Therefore, and as noted earlier (17, 28), RFLP analysis of the chromosome and SNP analysis of individual genes measure different evolutionary forces. Horizontal gene transfer has an important role in bacterial adaptation to different habitats. In particular, P. aeruginosa populations can exchange large DNA blocks that integrate at specific sites (4, 17-19, 21, 32, 36). P. aeruginosa is characterized by its great biochemical and ecological versatility, and the genome size can vary as much as 30% in different isolates. In other words, horizontal gene transfer may have a more important role than point mutations on the adaptation of P. aeruginosa to different habitats.
Acknowledgments
We are indebted to F. Baquero and I. Attrée for providing P. aeruginosa strains. We are grateful to S. Jansen, U. Laabs, and L. Yuste for excellent technical assistance.
This work was supported by grant QLK2-CT-2001-01339 from the Vth framework program of the EU and by grants BIO2000-0939 and BIO2001-1081 from the Spanish Ministry of Science and Technology. C.v.D was supported by grant FN 3231-51940.97 from the Swiss National Foundation and grant OFES 01.0122 from the Swiss Federal Office for Education and Science. P.G. was supported by a grant from the Mukoviszidose e.V.
REFERENCES
- 1.Allison, J. S., M. Dawson, D. Drake, and T. C. Montie. 1985. Electrophoretic separation and molecular weight characterization of Pseudomonas aeruginosa H-antigen flagellins. Infect. Immun. 49:770-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alonso, A., F. Rojo, and J. L. Martínez. 1999. Environmental and clinical isolates of Pseudomonas aeruginosa show pathogenic and biodegradative properties irrespective of their origin. Environ. Microbiol. 1:421-430. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong, D., S. Bell, M. Robinson, P. Bye, B. Rose, C. Harbour, C. Lee, H. Service, M. Nissen, M. Syrmis, and C. Wainwright. 2003. Evidence for spread of a clonal strain of Pseudomonas aeruginosa among cystic fibrosis clinics. J. Clin. Microbiol. 41:2266-2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arora, S. K., M. Bangera, S. Lory, and R. Ramphal. 2001. A genomic island in Pseudomonas aeruginosa carries the determinants of flagellin glycosylation. Proc. Natl. Acad. Sci. USA 98:9342-9347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Athma, P., N. Fidahusein, and M. Swift. 1995. Single base polymorphism linked to the ataxia-telangiectasia locus is detected by mismatch PCR. Biochem. Biophys. Res. Commun. 210:982-986. [DOI] [PubMed] [Google Scholar]
- 6.Cabrol, S., A. Olliver, G. B. Pier, A. Andremont, and R. Ruimy. 2003. Transcription of quorum-sensing system genes in clinical and environmental isolates of Pseudomonas aeruginosa. J. Bacteriol. 185:7222-7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dénervaud, V., P. TuQuoc, D. Blanc, S. Favre-Bonté, V. Krishnapillai, C. Reimmann, D. Haas, and C. van Delden. 2004. Colonization of intubated patients by Pseudomonas aeruginosa: characterization of cell-to-cell signaling deficient strains. J. Clin. Microbiol. 42:554-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Döring, G., M. Maier, E. Muller, Z. Bibi, B. Tümmler, and A. Kharazmi. 1987. Virulence factors of Pseudomonas aeruginosa. Antibiot. Chemother. 39:136-148. [DOI] [PubMed] [Google Scholar]
- 9.Enright, M. C., and B. G. Spratt. 1999. Multilocus sequence typing. Trends Microbiol. 7:482-487. [DOI] [PubMed] [Google Scholar]
- 10.Ernst, R. K., D. A. D'Argenio, J. K. Ichikawa, M. G. Bangera, S. Selgrade, J. L. Burns, P. Hiatt, K. McCoy, M. Brittnacher, A. Kas, D. H. Spencer, M. V. Olson, B. W. Ramsey, S. Lory, and S. I. Miller. 2003. Genome mosaicism is conserved but not unique in Pseudomonas aeruginosa isolates from the airways of young children with cystic fibrosis. Environ. Microbiol. 5:1341-1349. [DOI] [PubMed] [Google Scholar]
- 11.Foght, J. M., D. W. Westlake, W. M. Johnson, and H. F. Ridgway. 1996. Environmental gasoline-utilizing isolates and clinical isolates of Pseudomonas aeruginosa are taxonomically indistinguishable by chemotaxonomic and molecular techniques. Microbiology 142:2333-2340. [DOI] [PubMed] [Google Scholar]
- 12.Frank, D. W. 1997. The exoenzyme S regulon of Pseudomonas aeruginosa. Mol. Microbiol. 26:621-629. [DOI] [PubMed] [Google Scholar]
- 13.Govan, J. R., and J. W. Nelson. 1992. Microbiology of lung infection in cystic fibrosis. Br. Med. Bull. 48:912-930. [DOI] [PubMed] [Google Scholar]
- 14.Heuer, T., C. Bürger, G. Maass, and B. Tümmler. 1998. Cloning of prokaryotic genomes in yeast artificial chromosomes: application to the population genetics of Pseudomonas aeruginosa. Electrophoresis 19:486-494. [DOI] [PubMed] [Google Scholar]
- 15.Holder, I. A., A. N. Neely, and D. W. Frank. 2001. Type III secretion/intoxication system important in virulence of Pseudomonas aeruginosa infections in burns. Burns 27:129-130. [DOI] [PubMed] [Google Scholar]
- 16.Kiewitz, C., K. Larbig, J. Klockgether, C. Weinel, and B. Tümmler. 2000. Monitoring genome evolution ex vivo: reversible chromosomal integration of a 106 kb plasmid at two tRNALys gene loci in sequential Pseudomonas aeruginosa airway isolates. Microbiology 146:2365-2373. [DOI] [PubMed] [Google Scholar]
- 17.Kiewitz, C., and B. Tümmler. 2000. Sequence diversity of Pseudomonas aeruginosa: impact on population structure and genome evolution. J. Bacteriol. 182:3125-3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kresse, A. U., S. D. Dinesh, K. Larbig, and U. Römling. 2003. Impact of large chromosomal inversions on the adaptation and evolution of Pseudomonas aeruginosa chronically colonizing cystic fibrosis lungs. Mol. Microbiol. 47:145-158. [DOI] [PubMed] [Google Scholar]
- 19.Larbig, K. D., A. Christmann, A. Johann, J. Klockgether, T. Hartsch, R. Merkl, L. Wiehlmann, H. J. Fritz, and B. Tümmler. 2002. Gene islands integrated into tRNAGly genes confer genome diversity on a Pseudomonas aeruginosa clone. J. Bacteriol. 184:6665-6680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lenski, R. E., C. L. Winkworth, and M. A. Riley. 2003. Rates of DNA sequence evolution in experimental populations of Escherichia coli during 20,000 generations. J. Mol. Evol. 56:498-508. [DOI] [PubMed] [Google Scholar]
- 21.Liang, X., X. Q. Pham, M. V. Olson, and S. Lory. 2001. Identification of a genomic island present in the majority of pathogenic isolates of Pseudomonas aeruginosa. J. Bacteriol. 183:843-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lomholt, J. A., K. Poulsen, and M. Kilian. 2001. Epidemic population structure of Pseudomonas aeruginosa: evidence for a clone that is pathogenic to the eye and that has a distinct combination of virulence factors. Infect. Immun. 69:6284-6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marín, M. M., L. Yuste, and F. Rojo. 2003. Differential expression of the components of the two alkane hydroxylases from Pseudomonas aeruginosa. J. Bacteriol. 185:3232-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moore, J. E., C. E. Goldsmith, J. S. Elborn, P. G. Murphy, P. H. Gilligan, S. Fanning, and G. Hogg. 2003. Towards “molecular Esperanto” or the Tower of Babel? (the need for harmonization of techniques for genotyping clinical isolates of Pseudomonas aeruginosa isolated from patients with cystic fibrosis). J. Clin. Microbiol. 41:5347-5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nicas, T. I., and B. H. Iglewski. 1985. The contribution of exoproducts to virulence of Pseudomonas aeruginosa. Can. J. Microbiol. 31:387-392. [DOI] [PubMed] [Google Scholar]
- 27.Pirnay, J. P., D. De Vos, C. Cochez, F. Bilocq, J. Pirson, M. Struelens, L. Duinslaeger, P. Cornelis, M. Zizi, and A. Vanderkelen. 2003. Molecular epidemiology of Pseudomonas aeruginosa colonization in a burn unit: persistence of a multidrug-resistant clone and a silver sulfadiazine-resistant clone. J. Clin. Microbiol. 41:1192-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pirnay, J. P., D. De Vos, C. Cochez, F. Bilocq, A. Vanderkelen, M. Zizi, B. Ghysels, and P. Cornelis. 2002. Pseudomonas aeruginosa displays an epidemic population structure. Environ. Microbiol. 4:898-911. [DOI] [PubMed] [Google Scholar]
- 29.Quinn, J. P. 1998. Clinical problems posed by multiresistant nonfermenting gram-negative pathogens. Clin. Infect. Dis. 27(Suppl. 1):S117-S124. [DOI] [PubMed] [Google Scholar]
- 30.Römling, U., B. Fiedler, J. Bosshammer, D. Grothues, J. Greipel, H. von der Hardt, and B. Tümmler. 1994. Epidemiology of chronic Pseudomonas aeruginosa infections in cystic fibrosis. J. Infect. Dis. 170:1616-1621. [DOI] [PubMed] [Google Scholar]
- 31.Römling, U., J. Greipel, and B. Tümmler. 1995. Gradient of genomic diversity in the Pseudomonas aeruginosa chromosome. Mol. Microbiol. 17:323-332. [DOI] [PubMed] [Google Scholar]
- 32.Römling, U., K. D. Schmidt, and B. Tümmler. 1997. Large genome rearrangements discovered by the detailed analysis of 21 Pseudomonas aeruginosa clone C isolates found in environment and disease habitats. J. Mol. Biol. 271:386-404. [DOI] [PubMed] [Google Scholar]
- 33.Römling, U., J. Wingender, H. Müller, and B. Tümmler. 1994. A major Pseudomonas aeruginosa clone common to patients and aquatic habitats. Appl. Environ. Microbiol. 60:1734-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roy-Burman, A., R. H. Savel, S. Racine, B. L. Swanson, N. S. Revadigar, J. Fujimoto, T. Sawa, D. W. Frank, and J. P. Wiener-Kronish. 2001. Type III protein secretion is associated with death in lower respiratory and systemic Pseudomonas aeruginosa infections. J. Infect. Dis. 183:1767-1774. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Schmidt, K. D., B. Tümmler, and U. Römling. 1996. Comparative genome mapping of Pseudomonas aeruginosa PAO with P. aeruginosa C, which belongs to a major clone in cystic fibrosis patients and aquatic habitats. J. Bacteriol. 178:85-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith, J. M., E. J. Feil, and N. H. Smith. 2000. Population structure and evolutionary dynamics of pathogenic bacteria. Bioessays 22:1115-1122. [DOI] [PubMed] [Google Scholar]
- 38.Smith, J. M., N. H. Smith, M. O'Rourke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 90:4384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spangenberg, C., T. Heuer, C. Bürger, and B. Tümmler. 1996. Genetic diversity of flagellins of Pseudomonas aeruginosa. FEBS Lett. 396:213-217. [DOI] [PubMed]
- 40.Spangenberg, C., T. C. Montie, and B. Tümmler. 1998. Structural and functional implications of sequence diversity of Pseudomonas aeruginosa genes oriC, ampC and fliC. Electrophoresis 19:545-550. [DOI] [PubMed] [Google Scholar]
- 41.Spencer, D. H., A. Kas, E. E. Smith, C. K. Raymond, E. H. Sims, M. Hastings, J. L. Burns, R. Kaul, and M. V. Olson. 2003. Whole-genome sequence variation among multiple isolates of Pseudomonas aeruginosa. J. Bacteriol. 185:1316-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spiers, A. J., A. Buckling, and P. B. Rainey. 2000. The causes of Pseudomonas diversity. Microbiology 146:2345-2350. [DOI] [PubMed] [Google Scholar]
- 43.Spratt, B. G. 1999. Multilocus sequence typing: molecular typing of bacterial pathogens in an era of rapid DNA sequencing and the internet. Curr. Opin. Microbiol. 2:312-316. [DOI] [PubMed] [Google Scholar]
- 44.Stanisich, V. A., and B. W. Holloway. 1972. A mutant sex factor of Pseudomonas aeruginosa. Genet. Res. 19:91-108. [DOI] [PubMed] [Google Scholar]
- 45.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 46.Toussaint, B., I. Delic-Attree, and P. M. Vignais. 1993. Pseudomonas aeruginosa contains an IHF-like protein that binds to the algD promoter. Biochem. Biophys. Res. Commun. 196:416-421. [DOI] [PubMed] [Google Scholar]
- 47.Van Delden, C., and B. H. Iglewski. 1998. Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg. Infect. Dis. 4:551-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolfgang, M. C., B. R. Kulasekara, X. Liang, D. Boyd, K. Wu, Q. Yang, C. G. Miyada, and S. Lory. 2003. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 100:8484-8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yahr, T. L., L. M. Mende-Mueller, M. B. Friese, and D. W. Frank. 1997. Identification of type III secreted products of the Pseudomonas aeruginosa exoenzyme S regulon. J. Bacteriol. 179:7165-7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yuste, L., M. E. Corbella, M. J. Turiégano, U. Karlson, A. Puyet, and F. Rojo. 2000. Characterization of bacterial strains able to grow on high molecular mass residues from crude oil processing. FEMS Microbiol. Ecol. 32:69-75. [DOI] [PubMed] [Google Scholar]