Abstract
Nontypeable Haemophilus influenzae (NTHi) is a common cause of localized respiratory tract disease and initiates infection by colonizing the nasopharynx. Approximately 75 to 80% of NTHi clinical isolates produce proteins that belong to the HMW family of adhesins, which are believed to facilitate colonization. The prototype HMW adhesins are designated HMW1 and HMW2 and were identified in NTHi strain 12. HMW1 and HMW2 are 71% identical and 80% similar overall, yet display differing cellular binding specificities. In the present study we set out to define more clearly the relationships between HMW1 and HMW2 and other members of the HMW family of adhesins. PCR analysis of 49 epidemiologically distinct isolates revealed that all strains possessing hmw genes as determined by Southern analysis contain two hmw loci in conserved, unlinked physical locations on the chromosome. Functional analysis of the HMW adhesins produced by three unrelated strains demonstrated that each isolate possesses one protein with HMW1-like adherence properties and another with HMW2-like adherence properties. These findings suggest that the hmw1 and hmw2 loci may have arisen via a gene duplication event in an ancestral strain. In addition, they support the hypothesis that the distinct binding specificities of HMW1 and HMW2 emerged early and have persisted over time, suggesting an ongoing selective advantage.
Nontypeable Haemophilus influenzae (NTHi) strains are commensal organisms in the nasopharynx and are also a frequent cause of localized respiratory tract disease, including otitis media, conjunctivitis, sinusitis, pneumonia, and exacerbations of chronic bronchitis (13, 29, 38). The pathogenesis of NTHi disease begins with colonization of the nasopharynx, followed by contiguous spread within the respiratory tract. Successful colonization requires that the organism overcome the mucociliary escalator, a task accomplished in part by adherence to respiratory epithelium (29, 38). NTHi adherence is mediated by both pilus and nonpilus adhesins. In experiments with cultured human epithelial cells, the major nonpilus adhesins were found to be HMW1/HMW2 and Hia (20, 22, 33, 35). Based on examination of several collections of epidemiologically distinct NTHi strains, approximately 75 to 80% of isolates produce HMW1/HMW2-like proteins, while most of the remaining isolates produce Hia (7, 20, 36). Of note, isolates produce either HMW1/HMW2-like proteins or Hia, but not both (7, 20, 36).
The HMW adhesins were first identified as major targets of the human serum antibody response during acute otitis media (4). The prototype proteins are designated HMW1 and HMW2 and are produced by NTHi strain 12, the strain from which they were originally cloned and sequenced (5). HMW1 and HMW2 are encoded by separate chromosomal loci, with each locus consisting of three genes, designated hmwA, hmwB, and hmwC. The hmwA genes encode the surface-exposed adhesins (HMW1 and HMW2), and the hmwB and hmwC genes encode accessory proteins required for processing and secretion of the adhesins (5, 6, 15, 16, 32, 35). Based on the predicted amino acid sequences, HMW1 and HMW2 exhibit 71% identity and 80% similarity, while HMW1B/HMW2B and HMW1C/HMW2C are 99 and 97% identical, respectively (5, 6). Despite the high degree of amino acid sequence homology, HMW1 and HMW2 differ in their binding specificities in assays assessing adherence to a variety of cultured epithelial cell types. In assays with isogenic strain 12 derivatives lacking one or both of the HMW proteins and with Escherichia coli transformants expressing HMW1 or HMW2, HMW1 mediates high levels of adherence to most human epithelial cell lines examined while HMW2 mediates appreciable levels of adherence to only a subset of epithelial cell lines (11, 33, 35).
Investigation to date has focused primarily on HMW1 and HMW2 from strain 12, and thus relatively little is known about the HMW adhesins produced by other NTHi isolates. In a recent study, we examined 59 epidemiologically distinct NTHi isolates by Southern analysis and found that 47 (80%) had sequence that hybridized with a 5′ fragment of hmw1A. Of these 47, 45 were capable of adherence to Chang epithelial cells and expressed at least one protein that reacted with an HMW-specific antiserum (36). In a similar study, van Schilfgaarde et al. examined 58 NTHi isolates obtained from patients with otitis media or chronic obstructive pulmonary disease and from healthy volunteers for the ability to adhere to two epithelial cell types (40). Thirty-two of the isolates were capable of adherence to two different epithelial cell lines, and 23 of the 32 were HMW protein or hmw gene positive. Additional experimentation revealed five different adherence patterns based on the inhibiting effect of dextran sulfate (40). However, neither of these studies addressed whether diverse NTHi isolates typically contain two distinct HMW adhesins and whether these proteins have binding properties similar to those described for the prototype HMW1 and HMW2 adhesins of strain 12.
In the present study we set out to define more clearly the evolutionary and functional relationships between the NTHi strain 12 HMW1 and HMW2 proteins and other members of the HMW family of adhesins. Using a PCR approach, we found that all strains possessing sequence homologous to hmw1A contain two hmw loci. These loci are present at conserved, but unlinked, locations on the chromosome. Further analysis revealed that heterogeneous strains express one protein with HMW1-like adherence properties and another with HMW2-like adherence properties. These findings support the hypothesis that the distinct cellular binding specificities of HMW1 and HMW2 emerged early in evolution and have persisted over time, suggesting ongoing selective pressure.
MATERIALS AND METHODS
Culture and storage conditions.
H. influenzae strains were grown on chocolate agar supplemented with 1% IsoVitaleX or in brain heart infusion broth supplemented with hemin and NAD and were stored at −80°C in brain heart infusion broth with 20% glycerol. E. coli strains were grown on Luria-Bertani (LB) agar or in LB broth and were stored at −80°C in LB broth plus 50% glycerol. For E. coli, antibiotic concentrations used to select for plasmids included ampicillin at 100 μg/ml, kanamycin at 50 μg/ml, and chloramphenicol at 30 μg/ml. For H. influenzae, kanamycin was used at 25 μg/ml to select for transformants.
Bacterial strains.
Nontypeable H. influenzae strains 12, 5, and 15 are clinical isolates recovered from patients with acute otitis media. Strain 12 is the strain from which the hmw loci were first cloned and sequenced. Derivatives of strain 12 and strain 5 that lack expression of HMW1, HMW2, or both HMW1 and HMW2 have been described previously (35). Derivatives of strain 15 that lack expression of the HMW1-like, the HMW2-like, or both the HMW1-like and HMW2-like adhesins were constructed by transformation with pHMW1-16 (35) linearized with XbaI and then selecting for kanamycin-resistant colonies. The presence of single or double kanamycin inserts was confirmed by PCR, by Southern analysis using a labeled kanamycin cassette as a probe, and by Western analysis of whole-cell sonicates using anti-HMW antiserum. As a source of additional NTHi strains, we used a collection of 47 epidemiologically and genetically diverse clinical isolates that have been characterized by multilocus enzyme electrophoresis (25) and are known to contain genomic sequence that hybridizes with an intragenic fragment of hmw1A (36). Within this collection, 64% of strains were cultured from middle ear effusions, 32% of strains were recovered from blood, and 4% were recovered from cerebrospinal fluid (25). NTHi strain 11 is the strain from which hia was first cloned and sequenced and lacks hmw genes (7). H. influenzae strain Rd is a nonencapsulated former serotype d laboratory strain that has been sequenced in its entirety and lacks the hmw genes (31). DH5α is a laboratory strain of E. coli that is nonadherent to Chang, HaCaT, HEp-2, and NCI-H292 epithelial cells (Life Technologies).
Recombinant DNA methods.
Chromosomal DNA extractions, DNA ligations, restriction endonuclease digestions, and gel electrophoresis were performed according to standard techniques (30). Plasmids were introduced into E. coli by electroporation (12). H. influenzae was made competent for transformation by the MIV method of Herriott et al. (17).
PCR analysis of hmw loci.
Nucleotide sequences of the regions upstream of the hmw1 and hmw2 gene clusters were determined by sequencing lambda phage clones from a lambda library of NTHi strain 12 (5). In NTHi strain 12, open reading frame (ORF) HI1679 is located upstream of the hmw1 locus, and ORF HI1598 is located upstream of the hmw2 locus (ORFs HI1679 and HI1598 were originally identified in H. influenzae strain Rd). In order to determine the presence and physical locations of the hmw loci in other strains, PCR assays were performed using chromosomal DNA as the template and a 5′ primer which anneals to the 3′ end of ORF HI1679 (corresponding to nucleotides 361 to 385 of the strain Rd HI1679 coding sequence) or the 3′ end of ORF HI1598 (corresponding to nucleotides 451 to 475 of the strain Rd HI1598 coding sequence) and a 3′ primer which anneals to conserved sequence in the 5′ region of both hmw1A and hmw2A (corresponding to nucleotides 490 to 514 of the strain 12 hmw1 and hmw2 loci) (Fig. 1). PCR conditions included initial denaturation at 92°C for 2 min, followed by 30 cycles of denaturation at 92°C for 1 min, annealing at 60°C for 1 min, and extension at 72°C for 1 min and 30 s, with a final extension at 72°C for 7 min. Products were resolved by electrophoresis on 1% agarose gels. NTHi strain 12 served as a positive control, and strains 11 and Rd served as negative controls. Initially, PCR analysis of strain 3184A yielded a product with the HI1679-hmwA primer set but not with the HI1598-hmwA primer set, and PCR analysis of strain 1276 was negative with both primer sets. To determine whether sequence variation at the primer binding sites was the reason for the lack of product, additional primers were used, namely primer HI1679#2 (corresponding to nucleotides 195 to 219 of the strain Rd HI1679 coding sequence), primer HI1598#2 (corresponding to nucleotides 652 to 673 of the strain Rd HI1598 coding sequence), and primer hmwA#2 (corresponding to nucleotides 384 to 409 of the strain 12 hmw1 and hmw2 loci) (Fig. 1).
FIG. 1.
Schematic representation of PCR primer location used for analysis of numbers and chromosomal locations of hmw loci from 49 epidemiologically and genetically distinct NTHi clinical isolates. Diagram is based on locations and organization of the hmw1 and hmw2 loci in NTHi strain 12. Small arrows represent primers used in PCR analysis. Arrowheads designate direction of ORF transcription. HI1679 and HI1598 are designations based on the published sequence of H. influenzae strain Rd.
DNA and protein sequence analysis.
DNA sequencing was performed using the BigDye terminator cycle sequencing kit following the manufacturer's instructions (Applied Biosystems, Foster City, Calif.).
Comparisons of the amino acid sequences of the HMW adhesins from NTHi strains 12, 5, and 15 were performed using the BLAST 2 sequence (bl2seq) interface on the National Center for Biotechnology Information (NCBI) website. The following parameters were used for amino acid comparisons: Alignments, BlastP; Matrix, Blosum62; Open gap, 11; extension gap penalties, 1; gapx_dropoff, 50; filter, off.
For generation of phylogenetic trees, sequences were first aligned by the using T-Coffee method (26). Phylogenetic trees were constructed with MEGA version 2.1 (21), using neighbor joining with Poisson-corrected distances. Trees were unrooted or midpoint rooted, and bootstrapping was performed using 1,000 replicates.
Construction of plasmids containing hmwA genes from NTHi isolates.
The hmwA genes from NTHi strains 12, 5, and 15 were amplified by using primer HI1679-BglII (primer HI1679 with a BglII site engineered on the 5′ end) or primer HI1598-BglII (primer HI1598 with a BglII site engineered on the 5′ end) as a 5′ primer and primer hmwB-BglII or primer hmwB-SalI (corresponding to nucleotides 5154 to 5175 of the strain 12 hmw1 locus and nucleotides 4973 to 4995 of the strain 12 hmw2 locus) as a 3′ primer. The PCR products were digested with the appropriate restriction enzyme(s) and ligated into BamHI- or BamHI/SalI- digested pACYC184 (9). The presence of the insert was confirmed by PCR and restriction analysis. Plasmids containing hmwA genes downstream of either HI1598 or HI1679 were transformed into DH5α containing pHMW1BC, which encodes the hmw1B and hmw1C accessory genes, as described previously (32).
Adherence assays.
Adherence assays were performed as described previously (35). Briefly, Chang (human conjunctiva; ATCC CCL20.2 [Wong-Kilbourne derivative, clone 1-5c-4]), HaCaT (derived from human keratinocytes) (8), HEp-2 (human laryngeal epidermoid carcinoma; ATCC CCL 23), and NCI-H292 (human lung mucoepidermoid carcinoma; ATCC CRL1848) cells were seeded into wells of 24-well tissue culture plates and grown to confluency. Bacteria were inoculated into broth and allowed to grow to a density of approximately 2 × 109 CFU per ml. Approximately 2 × 107 CFU was inoculated onto viable epithelial cell monolayers, and plates were gently centrifuged at 165 x g for 5 min to facilitate contact between the bacteria and the epithelial cells. After incubation for 30 min at 37°C in 5% CO2, monolayers were rinsed four times with phosphate-buffered saline to remove nonadherent organisms. Trypsin-EDTA (0.05% trypsin, 0.5% EDTA) was added to the wells to release epithelial cells and adherent bacteria. Dilutions of adherent organisms were plated on solid medium to determine the number of adherent bacteria per monolayer. Accordingly, 100% adherence corresponds to ∼2 × 107 CFU per monolayer. Percent adherence was calculated by dividing the number of adherent CFU per well by the number of inoculated CFU. Each strain was examined in triplicate in a given assay, and assays were performed a minimum of three times.
Inhibition of adherence with MAA.
Inhibition of HMW1 and HMW1-like mediated adherence with Maackia amurensis agglutinin (MAA) was carried out essentially as described previously (37). Briefly, epithelial cells were seeded into wells of a 24-well tissue culture plate. Confluent monolayers were fixed with 2% glutaraldehyde in 0.1 M sodium phosphate buffer (pH 7.4) at 4°C for 2 h and then rinsed three times with Tris-buffered saline. Fixed monolayers were incubated with or without MAA (Sigma) at a concentration of 5 μg/ml in PBS-1% bovine serum albumin-0.25% Triton X-100 at 4°C overnight. The MAA solution was removed and replaced with serum-free minimal essential medium, and adherence assays were carried out as described above.
Western blots.
Whole-cell sonicates of DH5α transformants expressing HMW proteins and of NTHi derivatives were prepared by resuspending bacterial pellets in 10 mM HEPES, pH 7.4, and sonicating to clarity. Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using 7.5 or 10% polyacrylamide gels. Western blotting was performed with guinea pig polyclonal antiserum GP75, which was raised against purified HMW1 and recognizes both HMW1 and HMW2 from NTHi strain 12.
Whole-cell (dot) immunoblots.
In order to examine HMW1 and HMW2 associated with the bacterial surface in a qualitative manner, whole-cell immunoblotting was performed using guinea pig polyclonal antiserum GP75, as previously described (16).
Nucleotide sequence accession numbers.
Nucleotide sequences of the hmw1A and hmw2A genes from NTHi strains 5 and 15 have been submitted to GenBank and assigned the accession numbers AY497551, AY497552, AY497554, AY497553, respectively.
RESULTS
Examination of the number and physical location of hmw loci in genetically distinct NTHi strains.
In previous work, Southern hybridization studies performed on a collection of genetically diverse NTHi isolates described by Musser et al. revealed that approximately 80% of the isolates contain sequence that hybridizes with an intragenic fragment of hmw1A (25, 36). In the current study, we were interested in determining whether all of these strains contain two hmw loci and whether the physical locations of the hmw loci are conserved. In addition, we elected to examine NTHi strains 5 and 15. NTHi strain 12 lambda library clones containing the hmw1 and hmw2 loci were used as templates to determine sequences upstream of the gene clusters (5). Comparison of these upstream sequences to the genome of H. influenzae strain Rd (the laboratory strain that has been sequenced in its entirety) revealed that the hmw1 locus in strain 12 is situated downstream of an ORF corresponding to HI1679 of Rd and that the hmw2 locus of strain 12 is located downstream of an ORF corresponding to HI1598 of Rd. Based on this information, we designed primers specific for HI1598 and HI1679 and for a region conserved in hmw1A and hmw2A (Fig. 1) and performed PCR. Initially, 47 of 49 strains (96%) examined were positive for hmw loci downstream of HI1679 and HI1598 using primer sets HI1679-hmwA and HI1598-hmwA. Strains Rd and 11 are known to lack sequence homologous to the hmw1A locus of strain 12 and served as negative controls. PCR analysis of strain 3184A yielded a product with the HI1679-hmwA primer set but not with the HI1598-hmwA primer set, and strain 1276 yielded no product with either primer set. To determine whether sequence heterogeneity at the primer binding sites might be the reason for the inability to amplify the intergenic regions in these two strains, additional primers were designed based on sequence upstream of the original primers (Fig. 1), and reactions were repeated. With the new primer sets (HI1679#2-hmwA#2 and HI1598#2-hmwA#2), PCR products were generated for strains 3184A and 1276, indicating the presence of two hmw loci in these strains and suggesting that sequence variation and a lack of primer binding is a plausible explanation for the preliminary negative results. These data indicate that genetically diverse NTHi strains with sequence homologous to hmw1A by Southern hybridization uniformly contain two hmw loci (Table 1). Furthermore, the two hmw loci are present at conserved, but physically distinct, locations on the chromosome.
TABLE 1.
PCR analysis of hmw loci from diverse NTHi isolates
| PCR primer set | No. of strains (%) |
|---|---|
| HI1679-hmwA | 49 (100%) |
| HI1598-hmwA | 49 (100%) |
| ∼0.8-kb product with HI1679-hmwA | 35 (71%) |
| ∼1.7-kb product with HI1679-hmwA | 14 (29%) |
| ∼1.0-kb product with HI1598-hmwA | 49 (100%) |
Interestingly, the PCR product corresponding to the region between HI1679 and the hmw locus was ∼1.7 kb in 29% (14 of 49) of the strains and ∼0.8 kb in the remaining 71% (35 of 49) of the strains (Table 1). Nucleotide sequencing was carried out on the ∼1.7-kb PCR product from eight strains and on the ∼0.8-kb PCR product from five strains, and BlastX analysis was performed using the NCBI interface. All 1.7-kb sequences had evidence of transposase gene remnants. As an example, the HI1679-hmwA intergenic region in strain 3219C is predicted to encode sequence that shares 57% identity and 70% similarity with a 95-amino-acid region (E value, 5e-22) of IS200 transposase A of Helicobacter pylori (data not shown). In contrast, the ∼0.8-kb products shared no homology with transposases. PCR products generated with the HI1598-hmwA primer set were all ∼1.0 kb. Products from five strains were sequenced, and none had sequence homologous to transposase or insertion sequence element genes.
Examination of the binding specificities of HMW adhesins from diverse NTHi clinical isolates.
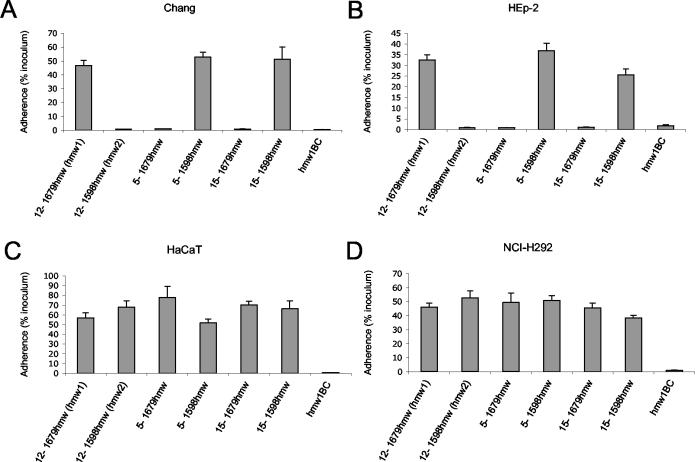
In an effort to understand the functional relationships between HMW1 and HMW2 from strain 12 and other members of the HMW family of adhesins, we examined the binding properties of the HMW adhesins from two additional clinical isolates, using HMW1 and HMW2 from strain 12 as controls. The hmwA genes located downstream of HI1679 and HI1598 were PCR amplified from NTHi strains 12, 5, and 15 and cloned into pACYC184. These plasmid constructs were coexpressed with pHMW1BC in DH5α. Expression of the adhesin was verified by Western blotting (Fig. 2A), and localization of the adhesin on the bacterial surface was verified by whole-cell dot immunoblot analysis using a polyclonal antiserum that recognizes HMW1 and HMW2 from strain 12 (Fig. 2B). Expression levels of the HMW adhesins from strains 12, 5, and 15 were similar. The binding specificity of each HMW adhesin was then examined in adherence assays with a panel of epithelial cells. Consistent with previous results, strain 12 HMW1 (12-1679hmw) mediated high-level adherence to Chang, HEp-2, HaCaT, and NCI-H292 epithelial cells, while strain 12 HMW2 (12-1598hmw) mediated high-level adherence to HaCaT and NCI-H292 cells but negligible adherence to Chang and HEp-2 cells (Fig. 3) (11, 33, 35). The adhesins cloned from strains 5 and 15 exhibited a similar pattern, with one adhesin mediating high-level adherence to all four cell lines (5-1598hmw and 15-1598hmw) and the second adhesin (5-1679hmw and 15-1679hmw) mediating high-level adherence to HaCaT and NCI-H292 cells and negligible adherence to Chang and HEp-2 cells (Fig. 3).
FIG. 2.
Expression and surface localization of HMW adhesins expressed in DH5α. (A) Western blot of whole-cell sonicates of DH5α expressing 12-1679hmw, 12-1598hmw, 5-1679hmw, 5-1598hmw, 15-1679hmw, or 15-1598hmw coexpressed with hmw1BC were separated via SDS-PAGE, transferred to nitrocellulose, and probed with GP75 (polyclonal antiserum raised against HMW1 from strain 12). The blot shows the fully processed, mature adhesins. (B) Surface localization of the adhesins is shown via whole-cell dot immunoblotting of the strains depicted in panel A, probing with GP75.
FIG. 3.
Adherence properties of HMW adhesins from NTHi strains 12, 5, and 15 expressed in DH5α. Graphs represent adherence, as a percentage of the inoculum, to Chang cells (A), HEp-2 cells (B), HaCaT cells (C), and NCI-H292 cells (D). Values represent the averages ± standard errors of three experiments, each performed in triplicate.
In previous work, our group found that HMW1 interacts with glycoprotein receptors containing α2,3-linked sialic acid on Chang and HaCaT epithelial cells (37). As a consequence, preincubation of epithelial cells with the lectin MAA results in a marked decrease in HMW1-mediated adherence, reflecting the fact that MAA binds to α2,3-linked sialic acid (11, 37). In contrast, MAA has minimal effect on HMW2-mediated adherence (11). In order to obtain additional evidence that the HMW adhesins from strains 5 and 15 exhibit either HMW1-like or HMW2-like binding specificity, we used MAA to specifically inhibit HMW1-mediated adherence. As shown in Fig. 4A, preincubation of Chang cells with MAA resulted in an ∼80% reduction in strain 12 HMW1-mediated adherence. Similarly, adherence of E. coli derivatives expressing 5-1598hmw and 15-1598hmw to Chang cells was reduced by ∼80% when compared with untreated controls (Fig. 4A). In experiments with HaCaT cells, MAA again inhibited adherence by strain 12 HMW1 (12-1679hmw), 5-1598hmw, and 15-1598hmw by ∼80% (Fig. 4B). DH5α expressing strain 12 HMW2 (12-1598hmw), 5-1679hmw, or 15-1679hmw adhered at high levels to HaCaT cells in the absence of MAA and exhibited only a small reduction in adherence in the presence of MAA (∼25% reduction compared to the untreated control) (Fig. 4B). This partial reduction in adherence may reflect nonspecific steric effects of MAA on accessibility of the HMW2 receptor. These studies indicate that adherence by one of the HMW adhesins expressed by a given strain is significantly inhibited in the presence of MAA, while adherence by the other adhesin is only minimally affected, effectively distinguishing the two adhesins from each other.
FIG. 4.
Inhibition of adherence with MAA. Monolayers of Chang cells (A) and HaCaT cells (B) were untreated (hatched bars) or preincubated with 5 μg of MAA/ml (shaded bars). Adherence of DH5α expressing HMW adhesins from NTHi strains 12, 5, and 15 was assessed in a 30-min adherence assay. Adherence is expressed as a percentage of the inoculum. Values represent the averages ± standard errors from three experiments, each performed in triplicate.
Taken together, the patterns of adherence to Chang, HEp-2, HaCaT, and NCI-H292 cells and the MAA inhibition phenotypes indicate that strains 5 and 15 express one HMW adhesin with HMW1-like binding properties and a second HMW adhesin with HMW2-like binding properties. Interestingly, in both strains 5 and 15, the hmw1-like locus lies downstream of HI1598 and the hmw2-like locus resides downstream of HI1679, opposite to the arrangement in strain 12.
Examination of the binding properties of the HMW1-like and HMW2-like adhesins expressed in H. influenzae.
To address whether the HMW1-like and HMW2-like adhesins behave the same in H. influenzae as they do in E. coli, we compared NTHi strains 12, 5, and 15 and isogenic mutants lacking the HMW1-like protein, the HMW2-like protein, or both. As shown in Fig. 5, Western analysis using a polyclonal antiserum reactive with both HMW1 and HMW2 confirmed the phenotypes of all of these strains. Examination of the wild-type strains in assays with Chang, HEp-2, HaCaT, and NCI-H292 cells revealed strain-to-strain variation in the levels of adherence (ranging from ∼30 to ∼75% of the inoculum) (Fig. 6). Despite this variation, comparison of hmw2 and hmw1 hmw2 mutants allowed assessment of the contribution of the HMW1-like adhesins to adherence, and comparison of hmw1 and hmw1 hmw2 mutants allowed assessment of the contribution of the HMW2-like adhesins to adherence. Consistent with our results with E. coli transformants, the HMW1-like adhesins from all three strains mediated high-level adherence to Chang, HEp-2, HaCaT, and NCI-H292 cells, and the HMW2-like adhesins from all three strains mediated high-level adherence to HaCaT and NCI-H292 cells (Fig. 6). Interestingly, while E. coli transformants expressing the HMW2-like adhesins were unable to adhere to Chang and HEp-2 cells above background levels (Fig. 3A and B), in H. influenzae the HMW2-like proteins mediated appreciable levels of adherence to these two cell types (ranging from ∼10 to ∼50% of the inoculum) (Figs. 6A and 6B). Derivatives lacking both the HMW1-like protein and the HMW2-like protein adhered at negligible levels to all four cell lines, emphasizing the important role of these proteins in adherence.
FIG. 5.
Western blot of NTHi isogenic derivatives. Whole-cell sonicates of the parent strain (wt) and 1598hmw, 1679hmw, and 1598hmw 1679hmw derivatives of NTHi strains 12, 5, and 15 were separated via SDS-7.5% PAGE, transferred to nitrocellulose, and probed with GP75 (polyclonal antiserum raised against HMW1 from strain 12). The blot shows the fully processed, mature adhesins.
FIG. 6.
Adherence properties of isogenic hmw derivatives in NTHi strains 12, 5, and 15. Graphs represent adherence of isogenic derivatives of strain 12 (open bars), strain 5 (shaded bars) and strain 15 (hatched bars), as a percent of the inoculum, to Chang cells (A), HEp-2 cells (B), HaCaT cells (C), and NCI-H292 cells (D). Values represent the averages ± standard errors of three experiments each performed in triplicate. hmw2, hmw1, and hmw1 hmw2 designations are based on phenotypes of the adhesins as established for Fig. 3 and 4.
Examination of the relationship between HMW protein sequence and binding specificity.
As it appears that hmw-containing strains express one adhesin with HMW1-like binding specificity and a second adhesin with HMW2-like binding specificity, we reasoned that protein sequence might be predictive of binding specificity. The mature HMW adhesins of strains 12, 5, and 15 (corresponding to fully processed adhesins lacking the signal sequence and cleaved at the predicted propeptide cleavage site) (5) were found to vary considerably in length both within individual strains and between different strains. The strain 15 HMW mature adhesins range in size from 992 amino acids (15-1679hmw) to 1,180 amino acids (15-1598hmw), a >15% size difference, while the strain 5 adhesins are 1,158 amino acids (5-1679hmw) and 1,161 amino acids (5-1598hmw) in length, a 0.26% difference.
Using the BLAST interface of the NCBI website, pairwise comparisons of each predicted amino acid sequence were made, allowing comparison of each sequence to all others. Designations of HMW1 or HMW2 were made based on the analysis of adherence properties presented in Fig. 3, 4, and 6. When amino acid sequences corresponding to the predicted mature, fully processed adhesin were compared, amino acid similarity and identity between HMW1-like adhesins (∼72 and ∼62%, respectively) and between HMW2-like adhesins (∼69 and ∼59%, respectively) were slightly greater than the similarity and identity observed between sequences of HMW1-like and HMW2-like adhesins (∼63 and ∼52%, respectively) (Table 2). In a previous study, Dawid et al. established that binding activity of strain 12 HMW1 and HMW2 resides in an ∼360-amino-acid region, corresponding to the region of maximal dissimilarity between these proteins (11). The binding domains of the strain 5 and strain 15 HMW adhesins were defined as the amino acid sequences corresponding to the known HMW1 and HMW2 binding domains obtained by using ClustalW alignment. The HMW1-like binding domains were found to share ∼74% similarity and ∼63% identity, and the HMW2-like binding domains were found to share ∼69% similarity and ∼54% identity. In contrast, comparison of the HMW1-like and the HMW2-like binding domains revealed ∼50% amino acid similarity and ∼35% identity. These observations suggest conservation of sequence critical for a particular binding specificity.
TABLE 2.
Pairwise comparisons of predicted amino acid sequences of HMW adhesins from NTHi strains 12, 5, and 15
| Comparisona | % Similarity ± SD | % Identity ± SD | % Gaps ± SD |
|---|---|---|---|
| HMW1 vs. HMW1 matureb | 72 ± 4 | 62 ± 6 | 8 ± 1 |
| HMW2 vs. HMW2 mature | 69 ± 5 | 59 ± 5 | 11 ± 6 |
| HMW1 vs. HMW2 mature | 63 ± 7 | 52 ± 7 | 12 ± 5 |
| HMW1 vs. HMW1 binding domainc | 74 ± 4 | 63 ± 5 | 2 ± 1 |
| HMW2 vs. HMW2 binding domain | 69 ± 3 | 54 ± 3 | 2 ± 1 |
| HMW1 vs. HMW2 binding domain | 50 ± 2 | 35 ± 2 | 7 ± 1 |
Comparisons were done using the BLAST interface on the NCBI website.
Mature refers to the protein sequence after cleavage at the known and predicted cleavage sites (corresponding to the peptide bond between amino acids 441 and 442 of strain 12 HMW1 and HMW2).
Binding domain refers to the protein sequence corresponding to the binding domains of strain 12 HMW1 and HMW2 as defined by Dawid et al. (11).
To extend this analysis, we generated phylogenetic trees. Sequences were aligned by the using T-Coffee method (26), and phylogenetic trees of the mature HMW adhesins and the HMW binding domains were made by using neighbor joining with Poisson-corrected distances implemented in MEGA version 2.1 (21). Initially, unrooted trees were generated with 1,000 bootstrap replicates, revealing separation of the HMW1-like mature proteins from the HMW2-like mature proteins and the HMW1-like binding domains from the HMW2-like binding domains (data not shown). With this information in mind, for the sake of clarity, midpoint-rooted trees were generated. As shown in Fig. 7A, the mature HMW1 protein sequences clustered together with a bootstrap confidence level of 100, and the mature HMW2 protein sequences clustered together with bootstrap confidence levels of 100 and 99. Similarly, the sequences of the HMW1-like binding domains clustered together with bootstrap confidence levels of 100 and 86, and the sequences of the HMW2-like binding domains clustered together with a bootstrap confidence level of 98 (Fig. 7B).
FIG. 7.
Phylogenetic analysis of HMW mature and binding domain amino acid sequences from NTHi strains 12, 5, and 15. Midpoint rooted phylogenetic trees were constructed with MEGA version 2.1 (21) using the neighbor joining method with Poisson-corrected distances. Bootstrap confidence values are shown at the branches. Scale bars represent the number of amino acid substitutions per site. (A) Phylogenetic tree based on the mature HMW amino acid sequences as defined in Table 2. (B) Phylogenetic tree based on the binding domain of the HMW adhesins from strains 12, 5, and 15 as defined in Table 2.
DISCUSSION
In this study, we analyzed a collection of 49 epidemiologically and genetically distinct NTHi isolates containing sequences homologous to an intragenic fragment of hmw1A from strain 12. In all isolates, we found evidence of two hmw loci, with one locus downstream of ORF HI1598 and the other locus downstream of ORF HI1679. In addition, we examined the adherence properties of the HMW adhesins from three different strains and in all three identified one adhesin with HMW1-like binding specificity and a second adhesin with HMW2-like binding specificity.
The presence of two closely related hmw loci in individual isolates of NTHi suggests that a gene duplication event occurred. Furthermore, the finding that the hmw loci are present at conserved physical locations on the chromosome in all hmw-containing isolates examined to date suggests that movement of the duplicated locus may have occurred early in the evolution of NTHi. In this context, it is interesting to note the similarity between our observations with the HMW adhesins and studies of the Helicobacter pylori BabA and BabB adhesins, which share striking homology with each other and appear to have arisen by an analogous ancestral gene duplication event (1, 28).
It is notable that transposon remnants are present between HI1679 and the downstream hmw locus in nearly 30% of strains, indicating a possible mechanism for acquisition of the original hmw locus or movement of the duplicated hmw locus. Using the genomic sequence of H. influenzae strain Rd for orientation, the strain 12 hmw1 locus is flanked upstream by HI1679 and downstream by HI1680. In contrast, the strain 12 hmw2 locus is flanked upstream by HI1598 and downstream by unrecognizable sequence, at least over the first ∼225 nucleotides. However, closer examination of the hmw2 downstream sequence reveals a 31-nucleotide stretch that corresponds to the end of HI1680. This finding suggests that the HI1679-HI1680 hmw junction may represent the location of the progenitor hmw locus, with duplication including the end of HI1680.
Based on examination of strains 12, 5, and 15, it appears that chromosomal location of a given hmw locus is not predictive of the binding specificity of the associated adhesin. In particular, in strain 12 the hmw1-like locus is downstream of HI1679 and the hmw2-like locus is downstream of HI1598, while in NTHi strains 5 and 15 the opposite is true. Given that the HMW adhesins share a high degree of amino acid identity at their N-terminal and C-terminal ends, it is possible that the strain-to-strain variability in physical locations of the hmw1-like and the hmw2-like loci is due to swapping of coding sequence for the binding domains, potentially via an intrastrain recombination event, as has also been proposed to explain the opposite locations of the H. pylori babA and babB genes in strains J99 and 26695 (1, 2, 18).
Recognizing that the strain 12 HMW1 and HMW2 adhesins have distinct cellular binding specificities, we wondered whether all adhesins in the HMW family can be classified as HMW1-like or HMW2-like based on binding properties. Our analysis of E. coli transformants expressing the hmw loci from NTHi strains 5 and 15 indicates that both of these isolates possess one protein with HMW1-like adherence properties and another with HMW2-like adherence properties, suggesting that the HMW adhesins indeed fall into two distinct subfamilies. Consistent with these results, phylogenetic analysis of the binding domains of seven additional HMW adhesins reveals clustering into an HMW1-like group and an HMW2-like group (See Fig. S1 in the supplemental material). Nevertheless, given the relatively limited scope of our studies, we cannot exclude the possibility that hybrid HMW proteins exist, analogous to the Moraxella catarrhalis UspA2H protein, which is present in approximately 20% of M. catarrhalis isolates and possesses the properties of both the UspA1 adhesin and the UspA2 serum resistance factor (23).
In adherence assays examining E. coli transformants, HMW2 mediated negligible adherence to Chang and HEp-2 cells. In contrast, in assays examining H. influenzae derivatives, HMW2 was capable of promoting appreciable adherence to Chang and HEp-2 cells. In considering this discrepancy, it is possible that levels of HMW2 expression are higher in H. influenzae than in E. coli. However, comparison of strains by Western analysis failed to support this idea. In a recent report, our group demonstrated that HMW1 and HMW2 are glycosylated (15). It is possible that the HMW adhesins are differentially glycosylated when expressed in E. coli and that these differences influence the ability of the adhesin to interact with some, but not all, cell types. Alternatively, H. influenzae may possess a coadhesin or some other factor that augments HMW2-mediated adherence to certain cell types, either by interacting cooperatively with the HMW2 receptor or by triggering up-regulation of the HMW2 receptor. Along these lines, in Bordetella pertussis the RGD motif in filamentous hemagglutinin interacts with the leukocyte response integrin/integrin-associated protein receptors on monocytes and up-regulates expression of CR3, allowing enhanced binding between a separate domain on filamentous hemagglutinin and the CR3 receptor (19). Similarly, in H. pylori, chronic infection of the gastric mucosa stimulates upregulation of sialyl-Lewis x antigen, which is recognized by the SabA adhesin, resulting in intimate adherence (24).
There appears to be some strain-to-strain variation in the levels of adherence among NTHi derivatives expressing the HMW adhesins, variation that is less apparent when the same adhesins are expressed in E. coli. The variation is most marked in assays with HEp-2 cells examining NTHi derivatives expressing only HMW2, with levels ranging from ∼10% (strain 5) to ∼50% (strain 15). The differences in adherence were not due to variation in the quantity of protein, as examination of the promoter region of the hmwA genes in strains 12, 5, and 15 revealed a similar number of 7-bp repeats, which are known to influence the quantity of HMW protein produced (data not shown) (10). Differential glycosylation of the HMW adhesins by NTHi may provide a partial explanation for the variation in adherence patterns between strains and is currently under investigation. Alternatively, the observed differences in adherence of the NTHi derivatives may reflect variation in other surface molecules typically capable of promoting low-level adherence, such as lipopolysaccharide, OapA, and Hap (27, 34, 39). Beyond promoting adherence, these surface molecules may interfere sterically with HMW-mediated adherence.
Based on sequence data and phenotypic analysis, it appears that the HMW proteins are under two forms of selective pressure, with different consequences. On the one hand, the HMW adhesins stimulate an antibody response that selects for amino acid sequence divergence (4). At the same time, all strains appear to have one HMW adhesin with HMW1-like adhesive properties and a second HMW adhesin with HMW2-like adhesive properties, suggesting selective pressure to maintain both adhesive specificities. In this context, it is noteworthy that efforts to develop a vaccine effective against NTHi disease have been impeded by extensive genetic diversity among strains and among potential vaccine antigens (13, 14). However, Barenkamp has demonstrated that purified HMW1 and HMW2 are partially protective against challenge by strain 12 in the chinchilla otitis media model and may be useful as components of a multicomponent NTHi vaccine (3). One possibility would be to include the binding domains of both HMW1 and HMW2, aiming to stimulate antibody that blocks adherence.
In summary, all NTHi clinical isolates examined to date with hmw sequences by Southern analysis harbor two hmw loci. These two loci encode adhesins with distinct cellular binding specificities, similar to those described for NTHi strain 12. We speculate that the hmw1 and hmw2 loci were generated by a gene duplication event that occurred early in the evolution of NTHi. This duplication and subsequent divergence of the hmw sequences resulted in HMW1-like and HMW2-like binding properties, providing a broader adhesive potential for the organism. Expression of both HMW1-like and HMW2-like adhesins has persisted over time, suggesting that broader adhesive potential is associated with an ongoing selective advantage.
Supplementary Material
Acknowledgments
We thank Sheena Loosmore for providing the nucleotide sequences of the NTHi strain 15 hmw1A and hmw2A genes and Awdhesh Kalia for assistance with phylogenetic analysis and helpful discussions.
This work was supported by NIH grant RO1-DC-02873 to J.W.S., by NIH grant RO1-AI-48066 to S.J.B., and by NIH training grant T32HL007317 to A.Z.B.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org.
Dedicated to the memory of Katie Burmeister.
REFERENCES
- 1.Alm, R. A., J. Bina, B. M. Andrews, P. Doig, R. E. Hancock, and T. J. Trust. 2000. Comparative genomics of Helicobacter pylori: analysis of the outer membrane protein families. Infect. Immun. 68:4155-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 3.Barenkamp, S. J. 1996. Immunization with high-molecular-weight adhesion proteins of nontypeable Haemophilus influenzae modifies experimental otitis media in chinchillas. Infect. Immun. 64:1246-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barenkamp, S. J., and F. F. Bodor. 1990. Development of serum bactericidal activity following nontypeable Haemophilus influenzae acute otitis media. Pediatr. Infect. Dis. J. 9:333-339. [DOI] [PubMed] [Google Scholar]
- 5.Barenkamp, S. J., and E. Leininger. 1992. Cloning, expression, and DNA sequence analysis of genes encoding nontypeable Haemophilus influenzae high-molecular-weight proteins related to filamentous hemagglutinin of Bordatella pertussis. Infect. Immun. 60:1302-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barenkamp, S. J., and J. W. St. Geme III. 1994. Genes encoding high-molecular-weight adhesion proteins of nontypeable Haemophilus influenzae are part of gene clusters. Infect. Immun. 62:3320-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barenkamp, S. J., and J. W. St. Geme III. 1996. Identification of a second family of high-molecular-weight adhesion proteins expressed by non-typable Haemophilus influenzae. Mol. Microbiol. 19:1215-1223. [DOI] [PubMed] [Google Scholar]
- 8.Boukamp, P., R. T. Petrussevska, D. Breitkreutz, J. Hornung, A. Markham, and N. E. Fusenig. 1988. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Biol. 106:761-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dawid, S., S. J. Barenkamp, and J. W. St. Geme III. 1999. Variation in expression of the Haemophilus influenzae HMW adhesins: a prokaryotic system reminiscent of eukaryotes. Proc. Natl. Acad. Sci. USA 96:1077-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dawid, S., S. Grass, and J. W. St. Geme III. 2001. Mapping of binding domains of nontypeable Haemophilus influenzae HMW1 and HMW2 adhesins. Infect. Immun. 69:307-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foxwell, A. R., J. M. Kyd, and A. W. Cripps. 1998. Nontypeable Haemophilus influenzae: pathogenesis and prevention. Microbiol. Mol. Biol. Rev. 62:294-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilsdorf, J. R. 1998. Antigenic diversity and gene polymorphisms in Haemophilus influenzae. Infect. Immun. 66:5053-5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grass, S., A. Z. Buscher, W. E. Swords, M. A. Apicella, S. J. Barenkamp, N. Ozchlewski, and J. W. St Geme III. 2003. The Haemophilus influenzae HMW1 adhesin is glycosylated in a process that requires HMW1C and phosphoglucomutase, an enzyme involved in lipooligosaccharide biosynthesis. Mol. Microbiol. 48:737-751. [DOI] [PubMed] [Google Scholar]
- 16.Grass, S., and J. W. St. Geme III. 2000. Maturation and secretion of the non-typable Haemophilus influenzae HMW1 adhesin: roles of the N-terminal and C-terminal domains. Mol. Microbiol. 36:55-67. [DOI] [PubMed] [Google Scholar]
- 17.Herriott, R. M., E. M. Meyer, and M. Vogt. 1970. Defined nongrowth media for stage II development of competence in Haemophilus influenzae. J. Bacteriol. 101:517-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ilver, D., A. Arnqvist, J. Ogren, I. M. Frick, D. Kersulyte, E. T. Incecik, D. E. Berg, A. Covacci, L. Engstrand, and T. Boren. 1998. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science 279:373-377. [DOI] [PubMed] [Google Scholar]
- 19.Ishibashi, Y., S. Claus, and D. A. Relman. 1994. Bordetella pertussis filamentous hemagglutinin interacts with a leukocyte signal transduction complex and stimulates bacterial adherence to monocyte CR3 (CD11b/CD18). J. Exp. Med. 180:1225-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krasan, G. P., D. Cutter, S. L. Block, and J. W. St. Geme III. 1999. Adhesin expression in matched nasopharyngeal and middle ear isolates of nontypeable Haemophilus influenzae from children with acute otitis media. Infect. Immun. 67:449-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 22.Laarmann, S., D. Cutter, T. Juehne, S. J. Barenkamp, and J. W. St Geme III. 2002. The Haemophilus influenzae Hia autotransporter harbours two adhesive pockets that reside in the passenger domain and recognize the same host cell receptor. Mol. Microbiol. 46:731-743. [DOI] [PubMed] [Google Scholar]
- 23.Lafontaine, E. R., L. D. Cope, C. Aebi, J. L. Latimer, G. H. McCracken, and E. J. Hansen. 2000. The UspA1 protein and a second type of UspA2 protein mediate adherence of Moraxella catarrhalis to human epithelial cells in vitro. J. Bacteriol. 182:1364-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahdavi, J., B. Sonden, M. Hurtig, F. O. Olfat, L. Forsberg, N. Roche, J. Angstrom, T. Larsson, S. Teneberg, K. A. Karlsson, S. Altraja, T. Wadstrom, D. Kersulyte, D. E. Berg, A. Dubois, C. Petersson, K. E. Magnusson, T. Norberg, F. Lindh, B. B. Lundskog, A. Arnqvist, L. Hammarstrom, and T. Boren. 2002. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. Science 297:573-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Musser, J. M., S. J. Barenkamp, D. M. Granoff, and R. K. Selander. 1986. Genetic relationships of serologically nontypable and serotype b strains of Haemophilus influenzae. Infect. Immun. 52:183-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Notredame, C., D. G. Higgins, and J. Heringa. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302:205-217. [DOI] [PubMed] [Google Scholar]
- 27.Prasadarao, N. V., E. Lysenko, C. A. Wass, K. S. Kim, and J. N. Weiser. 1999. Opacity-associated protein A contributes to the binding of Haemophilus influenzae to Chang epithelial cells. Infect. Immun. 67:4153-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pride, D. T., R. J. Meinersmann, and M. J. Blaser. 2001. Allelic variation within Helicobacter pylori babA and babB. Infect. Immun. 69:1160-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rao, V. K., G. P. Krasan, D. R. Hendrixson, S. Dawid, and J. W. St Geme III. 1999. Molecular determinants of the pathogenesis of disease due to non-typable Haemophilus influenzae. FEMS Microbiol. Rev. 23:99-129. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., and D. W. Russel. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Setlow, J. K., D. C. Brown, M. E. Boling, A. Mattingly, and M. P. Gordon. 1968. Repair of deoxyribonucleic acid in Haemophilus influenzae. I. X-ray sensitivity of ultraviolet-sensitive mutants and their behavior as hosts to ultraviolet-irradiated bacteriophage and transforming deoxyribonucleic acid. J. Bacteriol. 95:546-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.St. Geme, J. W., III, and S. Grass. 1998. Secretion of the Haemophilus influenzae HMW1 and HMW2 adhesins involves a periplasmic intermediate and requires the HMWB and HMWC proteins. Mol. Microbiol. 27:617-630. [DOI] [PubMed] [Google Scholar]
- 33.St. Geme, J. W., III. 1997. Insights into the mechanism of respiratory tract colonization by nontypable Haemophilus influenzae. Pediatr. Infect. Dis. J. 16:931-935. [DOI] [PubMed] [Google Scholar]
- 34.St. Geme, J. W., III, M. L. de la Morena, and S. Falkow. 1994. A Haemophilus influenzae IgA protease-like protein promotes intimate interaction with human epithelial cells. Mol. Microbiol. 14:217-233. [DOI] [PubMed] [Google Scholar]
- 35.St. Geme, J. W., III, S. Falkow, and S. J. Barenkamp. 1993. High-molecular-weight proteins of nontypeable Haemophilus influenzae mediate attachment to human epithelial cells. Proc. Natl. Acad. Sci. USA 90:2875-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.St. Geme, J. W., III, V. V. Kumar, D. Cutter, and S. J. Barenkamp. 1998. Prevalence and distribution of the hmw and hia genes and the HMW and Hia adhesins among genetically diverse strains of nontypeable Haemophilus influenzae. Infect. Immun. 66:364-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.St. Geme, J. W., III. 1994. The HMW1 adhesin of nontypeable Haemophilus influenzae recognizes sialylated glycoprotein receptors on cultured human epithelial cells. Infect. Immun. 62:3881-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.St. Geme, J. W., III. 2001. The pathogenesis of nontypable Haemophilus influenzae otitis media. Vaccine 19:S41-S50. [DOI] [PubMed] [Google Scholar]
- 39.Swords, W. E., B. A. Buscher, K. Ver Steeg Ii, A. Preston, W. A. Nichols, J. N. Weiser, B. W. Gibson, and M. A. Apicella. 2000. Non-typeable Haemophilus influenzae adhere to and invade human bronchial epithelial cells via an interaction of lipooligosaccharide with the PAF receptor. Mol. Microbiol. 37:13-27. [DOI] [PubMed] [Google Scholar]
- 40.van Schilfgaarde, M., P. van Ulsen, P. Eijk, M. Brand, M. Stam, J. Kouame, L. van Alphen, and J. Dankert. 2000. Characterization of adherence of nontypeable Haemophilus influenzae to human epithelial cells. Infect. Immun. 68:4658-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.