Abstract
Many group I introns encode endonucleases that promote intron homing by initiating a double-stranded break-mediated homologous recombination event. In this work we describe intron homing in Bacillus subtilis phages SPO1 and SP82. The introns encode the DNA endonucleases I-HmuI and I-HmuII, respectively, which belong to the H-N-H endonuclease family and possess nicking activity in vitro. Coinfections of B. subtilis with intron-minus and intron-plus phages indicate that I-HmuI and I-HmuII are required for homing of the SPO1 and SP82 introns, respectively. The homing process is a gene conversion event that does not require the major B. subtilis recombination pathways, suggesting that the necessary functions are provided by phage-encoded factors. Our results provide the first examples of H-N-H endonuclease-mediated intron homing and the first demonstration of intron homing initiated by a nicking endonuclease.
Group I introns constitute a phylogenetically widespread class of self-splicing intervening RNA sequences that share a common secondary structure and splicing pathway. In many cases these introns encode site-specific DNA endonucleases. Rather than being involved in the splicing process, these endonucleases promote the transfer of the intron from an intron-plus gene to an intron-minus gene, in a process known as homing (9). Therefore, the intron-encoded endonucleases that mediate this process are referred to as homing endonucleases.
Group I intron homing was first described for the ω intron (LSU-1) of the mitochondrial large ribosomal rRNA gene of Saccharomyces cerevisiae (reviewed in reference 9). Genetic analyses showed that LSU-1 intron propagation is by unidirectional gene conversion that depends on expression of the intronic open reading frame (19), whose protein product was subsequently shown to be a site-specific DNA endonuclease (7). Genetic similarities between homing of the LSU-1 intron and other gene conversion events in yeast suggested that group I intron homing occurs via the double-stranded break (DSB) repair mechanism (33, 35). Homing has been experimentally demonstrated in other biological systems, such as those of bacteriophages, protist nuclei, and algal chloroplasts (4, 26, 28).
Typically, a DSB is generated by the intron-encoded endonuclease at a specific site in the intronless gene, close to the site of intron insertion. The DSB in the recipient allele is resected by host-encoded exonucleases, leaving single-stranded 3′ ends that invade the homologous region of the intron-containing donor duplex. After D-loop formation the 3′ ends function as primers in a repair process that uses the complementary intron-containing DNA strands as template. Distance-dependent coconversion of flanking exon sequences observed with intron homing is due to exonucleolytic degradation of the cleaved recipient DNA and possibly to branch migration during recombination. The homing process is completed by cleavage and ligation of the two Holliday junctions resulting in resolution of the two intron-containing alleles (2).
Homing endonucleases have been classified into four major families based on conserved amino acid sequence motifs, denoted LAGLIDADG, GIY-YIG, His-Cys box, and H-N-H, respectively (3). The H-N-H endonucleases I-HmuI and I-HmuII, encoded by group I introns in the DNA polymerase genes of Bacillus subtilis phages SPO1 and SP82, respectively, are distinct from typical intron-encoded endonucleases in that they cut only one strand of their DNA substrate (14). Furthermore, both enzymes cleave intron-containing substrates in addition to intronless versions of their cognate genes, and each enzyme prefers the DNA of the heterologous phage as a substrate. I-HmuII is required to exclude the SPO1 intron and flanking genetic markers from the progeny of mixed infections, a process that was interpreted as an intron replacement event but that proceeded by the same mechanism as intron homing (14). However, it has not been directly demonstrated that intron replacement or homing can be initiated by a nicking endonuclease. Furthermore, rather than resulting from a gene conversion event, the observed exclusion of the SPO1 intron and flanking markers during coinfection with SP82 could have been due to general phage exclusion mediated by I-HmuII.
In this work we demonstrate homing of the group I introns in phages SPO1 and SP82 into intronless target sites. As is the case with events that are initiated at DSBs in phage T4 (4), homing is shown to be a unidirectional, nonreciprocal gene conversion event. Intron homing is dependent on the function of the intronic H-N-H DNA endonucleases I-HmuI and I-HmuII and does not require the major B. subtilis recombination pathways.
MATERIALS AND METHODS
Bacterial and bacteriophage strains and growth conditions.
B. subtilis CB312 was used for propagation of phages SPO1 and SP82 in Luria-Bertani (LB) medium. SPO1sus14 mutants were propagated on CB313 (sup3), a lysine-inserting ochre suppressor strain (14). B. subtilis YB886 and recombination-defective derivatives recA (YB1015), recF15 addA5 (BG143), recF15 recH342 (BG137), recO1 recH342 (BG443), recU1 recH342 (BG437), and recS1 recH342 (BG435) were obtained from Juan C. Alonso, Universidad Autonoma de Madrid, Spain, and are described in reference 11.
SPO1I-HmuIoc and SP82I-HmuIIoc contain ochre (UAA) in place of lysine (AAG) at the fourth codon of the SPO1 and SP82 intron endonuclease genes, respectively (14). To construct the intronless phage SPO1ΔI, a lysate of SPO1I-HmuIoc was serially diluted and spotted onto a top agar lawn of B. subtilis CB312 containing plasmid pWO1ΔI. The area corresponding to the highest dilution that exhibited continuous lysis was excised, crushed, and washed with buffered saline, and the supernatant was collected. Phages with an intron deletion were identified by plaque hybridization using oligonucleotide SPpolΔI, spanning the intron insertion site, as a probe. Phages were plated out overnight and transferred to Hybond-N nylon membrane (Amersham). Membranes were placed on a series of solution-saturated (solution was either 0.5 M NaOH, 1.5 M NaCl; 1 M Tris-HCl [pH 7.5], 1.5 M NaCl; or 2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]) Whatman 3MM filter papers for 5 min each. Dried and UV-irradiated membranes were prehybridized in a solution containing 3× Denhardt's, 3× SCC, 0.1% sodium dodecyl sulfate (SDS), and 50 μg of salmon sperm DNA/ml at 65°C for 1 h and were hybridized overnight at 50°C in a solution of 3× Denhardt's, 6× SCC, 0.1% SDS using 32P-end-labeled SPpolΔI (5 × 106 cpm). Membranes were washed twice in 6× SSC at 60°C and once in 1× SSC at 50°C and were exposed to BIOMAX film (Kodak). Phages from positive plaques were isolated, diluted, and checked for purity by plaque hybridization.
SPO1(sus14,I-HmuIoc) was obtained from a cross of SPO1I-HmuIoc with SPO1sus14. SPO1(sus14,ΔI) was obtained by crossing SPO1(sus14,I-HmuIoc) with SPO1ΔI.
Plasmids.
pWO1ΔI was constructed to have a longer target for homologous recombination than the intronless sequence in plasmid pHGO1ΔI (14). The 284-bp fragment of the intronless DNA polymerase gene in pHGO1ΔI was amplified by PCR with primers S2819 and S2835, and the PCR product was digested with EcoRI and SnaBI. The intron-containing EcoRI/SnaBI fragment of plasmid pHαEP1 (13) was replaced by the intronless EcoRI/SnaBI fragment of pHGO1ΔI to create pHαEP1ΔI. A 1.0-kb EcoRI/HindIII fragment was excised from pHαEP1ΔI and ligated into the corresponding sites of the Escherichia coli-B. subtilis shuttle vector pMS1 (29) to generate pWO1ΔI.
Phage crosses.
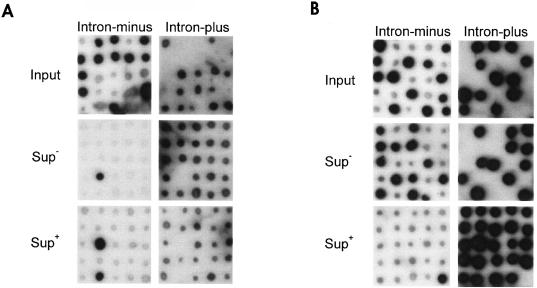
B. subtilis CB312 or CB313 cells were grown at 37°C to an optical density at 540 nm of 0.4 in LB broth and were infected with a multiplicity of infection (MOI) of 4 for each phage. After 15 min, SPO1 antiserum (which is also effective against SP82) was added to inactivate unabsorbed phage. Twenty minutes postinfection the culture was diluted 10−4 into prewarmed LB and was incubated for 3 h at 37°C. For infection with sus14 mutants, phage progeny were plated on CB313 to allow growth of all phages. Plaques were picked onto lawns of either CB312 or CB313 to determine the frequency of wild-type phages. sus14 mutants are able to form plaques on CB313 but not CB312. Intron-minus and intron-plus phages were distinguished by plating phages on a CB313 lawn and subsequent plaque hybridization with oligonucleotide SPpolΔI (complementary to the intron insertion site) and K4O (complementary to the ochre mutation in the endonuclease). Because oligonucleotide K4O has a 2-nucleotide mismatch with the wild-type sequence, it also permits discrimination between wild-type and ochre alleles of the intron-encoded endonuclease genes (Fig. 1).
FIG. 1.
Exclusion of intron-minus allele in progeny of SPO1 and SPO1ΔI coinfections. Plaques from progeny of mixed infections of SPO1ΔI and either wild-type SPO1 (A) or SPO1I-HmuIoc (B) on B. subtilis CB312 (Sup−) or CB313 (Sup+) were replicated, lifted onto nylon membranes, and probed with end-labeled oligonucleotides specific for the intron-minus (SPpolIΔI) and intron-plus (K4O) DNA polymerase gene. Note that the oligonucleotide, although having two mismatches with the wild-type sequence, still exhibits specific hybridization to wild-type introns under these conditions.
Plaque hybridization.
Phages from mixed infections were plated out overnight, and plaques were transferred to Hybond-N nylon membranes (Amersham). Membranes were placed on a series of solution-saturated (with one of the following solutions: 0.5 M NaOH, 1.5 M NaCl; 1 M Tris-HCl [pH 7.5], 1.5 M NaCl; or 2× SSC) 3MM filter papers for 5 min each. DNA on air-dried membranes was fixed by UV cross-linking. Membranes were prehybridized for an hour at 60°C in a solution containing 3× SSC, 1× Denhardt's reagent, 0.1% SDS, and 50 μg of denatured salmon sperm DNA/ml. Hybridization was carried out for 12 h at 45°C in a solution of 3× SSC, 1× Denhardt's reagent, 0.1% SDS, and end-labeled oligonucleotide. Membranes were washed three times in 3× SSC and 0.1% SDS at 45°C and once in 1× SSC and 0.1% SDS at 36°C, and then they were exposed to X-ray film.
Oligonucleotides.
The following oligonucleotides were used: 82c1, 5′-TCTCTTTCAGTATAATCACGAG; K4O, 5′-GAATATGGAATGGTAAGACATTAAAGG; S2819, 5′-CAGACAAGTATACGTAACTCTAACTG; S2835, 5′-CATGGAATTCCGTAAGGCAAACC; SPpolΔI, 5′-AGTAGTAATAGAGCCTAACGCTCA(A/G)CAATTC.
RESULTS
To study homing of introns encoding DNA endonucleases that cut only one DNA strand, an intronless gene 31 (DNA polymerase) of bacteriophage SPO1 was generated by plasmid-to-phage transfer. Plaque morphology of intronless phage SPO1ΔI was indistinguishable from the wild type, and the burst sizes of both phages were comparable (data not shown).
Homing of the SPO1 intron was examined by coinfecting B. subtilis with SPO1ΔI and wild-type SPO1 or with SPO1I-HmuIoc (a phage with a suppressible nonsense mutation in the intron-encoded endonuclease I-HmuI), each at an MOI of 4. Progeny phages and the input phage mixtures used for these coinfections were plated and analyzed by plaque hybridization for the presence or absence of the intron (Fig. 1), as summarized in Table 1. In the progeny of crosses between SPO1 and SPO1ΔI the intron-plus allele was observed with a frequency of about 95%, regardless of the bacterial host used. In contrast, coinfection of B. subtilis CB312 (Sup−) with SPO1I-HmuIoc and SPO1ΔI resulted in a transmission of intron-plus and intron-minus markers near the input ratio. However, when the nonsense mutation in the gene for I-HmuI was suppressed (and the endonuclease activity was restored), the frequency of the intron-plus gene in the progeny was comparable to that obtained for coinfections with wild-type SPO1. These results demonstrated that the change in frequency of intron-plus versus intron-minus phages in the progeny was dependent on I-HmuI.
TABLE 1.
Dependence of intron inheritance on I-HmuI
| Phage cross | Hosta | No. | Distribution of genetic markers in progeny
|
|
|---|---|---|---|---|
| I+ | IΔ | |||
| SPO1 × SPO1ΔI | None | 1 | 46 | 54 |
| 2 | 56 | 44 | ||
| Sup− | 1 | 95 | 5 | |
| 2 | 96 | 4 | ||
| Sup+ | 1 | 92 | 4 | |
| 2 | 94 | 6 | ||
| SPO1I-HmuIoc × SPO1ΔI | None | 1 | 44 | 56 |
| 2 | 53 | 47 | ||
| Sup− | 1 | 48 | 52 | |
| 2 | 46 | 54 | ||
| Sup+ | 1 | 96 | 2 | |
| 2 | 93 | 7 | ||
CB312 is Sup−; CB313 is Sup+; none indicates phage mixture before addition of cells.
Intron homing versus phage exclusion.
The similarities in burst sizes and plaque morphologies of SPO1 and SPO1ΔI argued against a selective growth advantage for the intron-containing phage. However, the high frequency of intron-plus alleles in the progeny of mixed infections might be the result of exclusion of the SPO1ΔI phage caused by I-HmuI rather than intron homing. This issue was addressed by using phages with a suppressible nonsense mutation (sus14-1) in gene 14 (27), which provided a traceable genetic marker in mixed infections (Table 2). Gene 14 is about 40 kb away from gene 31 (30) and is not likely to be influenced by marker exclusion events originating at the intron insertion site in gene 31 (14, 31).
TABLE 2.
SPO1 intron homing is a gene conversion event
| Phage cross | Host | No. | Distribution of genetic markers in progenya
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| sus14,I+ | wt14,ΔI | wt14,I+ | sus14,ΔI | I+ | ΔI | sus14 | wt14 | |||
| SPO1sus14 × SPO1ΔI | None | 1 | 54 | 46 | 0 | 0 | 54 | 46 | 54 | 46 |
| 2 | 41 | 59 | 0 | 0 | 41 | 59 | 41 | 59 | ||
| 3 | 34 | 66 | 0 | 0 | 34 | 66 | 34 | 66 | ||
| Sup− | 1 | 52 | 15 | 32 | 1 | 84 | 16 | 53 | 47 | |
| 2 | 49 | 10 | 40 | 1 | 89 | 11 | 50 | 50 | ||
| 3 | 44 | 5 | 51 | 0 | 95 | 5 | 56 | 44 | ||
| Sup+ | 1 | 58 | 3 | 33 | 0 | 91 | 8 | 58 | 41 | |
| 2 | 52 | 3 | 45 | 0 | 97 | 3 | 52 | 48 | ||
| 3 | 43 | 4 | 53 | 0 | 96 | 4 | 43 | 57 | ||
| SPO1(sus14,I-HmuIoc) × SPO1ΔI | None | 1 | 47 | 53 | 0 | 0 | 47 | 53 | 47 | 53 |
| 2 | 35 | 65 | 0 | 0 | 35 | 65 | 35 | 65 | ||
| Sup− | 1 | 28 | 53 | 11 | 6 | 37 | 59 | 36 | 64 | |
| 2 | 41 | 47 | 5 | 7 | 46 | 54 | 48 | 52 | ||
| Sup+ | 1 | 53 | 7 | 40 | 0 | 93 | 7 | 53 | 47 | |
| 2 | 47 | 6 | 46 | 1 | 93 | 7 | 48 | 52 | ||
| SPO1 × SPO1(sus14,ΔI) | None | 1 | 0 | 0 | 65 | 35 | 65 | 35 | 35 | 65 |
| 2 | 0 | 0 | 48 | 52 | 48 | 52 | 52 | 48 | ||
| Sup− | 1 | 31 | 0 | 69 | 0 | 100 | 0 | 31 | 69 | |
| 2 | 33 | 0 | 65 | 2 | 98 | 2 | 35 | 65 | ||
| Sup+ | 1 | 30 | 2 | 65 | 3 | 95 | 5 | 33 | 67 | |
| 2 | 32 | 0 | 65 | 3 | 97 | 3 | 35 | 65 | ||
| SPO1I-HmuIoc × SPO1(sus14,ΔI) | None | 1 | 0 | 0 | 69 | 31 | 69 | 31 | 69 | 31 |
| 2 | 0 | 0 | 55 | 45 | 55 | 45 | 55 | 45 | ||
| Sup− | 1 | 8 | 6 | 50 | 36 | 58 | 42 | 56 | 44 | |
| 2 | 6 | 9 | 43 | 41 | 49 | 50 | 47 | 42 | ||
| Sup+ | 1 | 34 | 0 | 62 | 4 | 96 | 4 | 62 | 38 | |
| 2 | 29 | 1 | 67 | 3 | 96 | 4 | 68 | 32 | ||
wt14 indicates wild-type gene 14. I+ and ΔI indicate the presence or absence of introns, respectively.
Progeny were plated on B. subtilis CB313 (Sup+), which can support the growth of all phages, and were analyzed by plaque hybridization for the presence or absence of the intron. The proportion of sus14 mutants in the burst was determined by the ability of individual plaques to replicate on a Sup− lawn. For each experiment the mixture of phages used for infection was analyzed to determine the ratio of input phages.
When B. subtilis CB312 or CB313 was coinfected with approximately equal multiplicities of SPO1sus14 and SPO1ΔI, intron-containing phages dominated the burst. While the frequency of the intron-plus allele increased from about 50% in the input phages to 90 to 95% in the progeny, the frequency of the sus14 mutant allele in the progeny remained largely unchanged compared to that of the input phage mixture (Table 2, columns 5 to 8). The increase in the intron frequency was mostly due to the appearance of intron-containing phages with the wild-type gene 14 (column 3), while the frequency of intronless SPO1 phages decreased compared to that of the input mixture (column 2).
Mixed infections of the Sup− host with SPO1(sus14,I-HmuIoc) and SPO1ΔI resulted in essentially equal transmission frequencies for the intron and sus14 alleles (Table 2, columns 5 to 8). When endonuclease activity was restored on the Sup+ host, the frequency of the intron approached 95% (columns 5 and 6) with most of the increase coming, once again, from intron-containing phages with wild-type gene 14 (columns 2 and 3).
Similar results were obtained in reciprocal crosses in which the intronless phage carried the sus14 mutation (Table 2). The predominance of the intron in the burst was dependent on a functional intron-encoded endonuclease, and the frequency of the sus14 allele in the burst was comparable to that of the input phages. The inheritance of the intron and the sus14 mutation are not linked in these crosses.
Homing of the SP82 intron into the intronless SPO1 DNA polymerase gene.
The intron endonuclease-dependent exclusion of SPO1 markers in the progeny coinfections with SP82 (14) suggested that the SP82 intron is likely to be capable of homing into the intronless SPO1 homologue. Homing of the SP82 intron was examined by mixed infection of B. subtilis CB312 and CB313 with the intronless phage SPO1(sus14,ΔI) and SP82 wild-type or SP82I-HmuIIoc, a phage with a suppressor-sensitive nonsense mutation in the intronic endonuclease I-HmuII (14).
As in crosses with wild-type SPO1 (Table 2), the progeny of mixed infections of SPO1(sus14,ΔI) and SP82 showed a predominance of intron-containing phages (Table 3, columns 5 and 6). The frequency of the sus14 mutant allele in the burst was largely unchanged compared to that of the input (columns 7 and 8). With the exception of the SP82I-HmuIoc × SPO1(sus14,ΔI) cross on the Sup+ host, the high transmission frequency of the intron was due to the appearance of intron-containing SPO1sus14 (column 3), while the frequency of SPO1(sus14,ΔI) declined compared to that in the input phages (column 2). The high-frequency transmission of the intron in mixed infections of the Sup+ host with SP82I-HmuIIoc and SPO1sus14 compared to that on the Sup− host showed that the predominance of the intron-containing phages in the burst is dependent on the activity of the endonuclease (columns 2, 3, 5, and 6). The predominance of phages with wild-type gene 14 in infections of the Sup− host was likely due to lack of progeny from cells that were singly infected with sus14 phages, which require complementation by the wild-type allele.
TABLE 3.
SP82 intron homing is a gene conversion event
| Phage cross | Host | No. | Distribution of genetic markers in progenya
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| wt14,I+ | sus14,ΔI | sus14,I+ | wt14,ΔI | I+ | ΔI | wt14 | sus14 | |||
| SP82 × SPO1(sus14,ΔI) | None | 1 | 56 | 44 | 0 | 0 | 56 | 44 | 56 | 44 |
| 2 | 58 | 42 | 0 | 0 | 58 | 42 | 58 | 42 | ||
| Sup− | 1 | 68 | 3 | 28 | 1 | 96 | 4 | 69 | 31 | |
| 2 | 54 | 3 | 42 | 0 | 96 | 3 | 54 | 45 | ||
| Sup+ | 1 | 53 | 7 | 39 | 0 | 92 | 7 | 53 | 46 | |
| 2 | 30 | 23 | 44 | 1 | 74 | 24 | 31 | 67 | ||
| SP82I-HmuIoc × SPO1(sus14,ΔI) | None | 1 | 54 | 45 | 0 | 0 | 54 | 45 | 54 | 45 |
| 2 | 45 | 54 | 0 | 0 | 45 | 54 | 45 | 54 | ||
| Sup− | 1 | 60 | 30 | 3 | 7 | 63 | 37 | 67 | 33 | |
| 2 | 59 | 26 | 5 | 10 | 64 | 36 | 69 | 31 | ||
| Sup+ | 1 | 74 | 0 | 25 | 1 | 99 | 1 | 75 | 25 | |
| 2 | 83 | 0 | 16 | 1 | 99 | 1 | 84 | 16 | ||
See Table 2, footnote a.
In summary, the data suggest that the SP82 intron was transferred into the SPO1 intronless DNA polymerase gene by a gene conversion event analogous to homing of the SPO1 intron. Furthermore, the unidirectional and nonreciprocal transfer of the intron is dependent on the intronic endonuclease.
Gene conversion at the cleavage site.
The sequence differences of the DNA polymerase genes of SPO1 and SP82 around the cleavage site of I-HmuII permitted determination of the directionality of conversion in SPO1. Intron-containing phages with the sus14 mutation from the burst of a coinfection with SP82 and SPO1(sus14,ΔI) (Table 3, Sup−, cross 2) were probed with an end-labeled oligonucleotide complementary to the SP82 sequence spanning the I-HmuII cleavage site (Fig. 2). The hybridization result showed that 41 out of 42 phages carried the SP82-specific sequence (data not shown). With the exception of recombinant phages that were the result of homologous recombination [estimated to be about 5, based on the recombination frequency of about 12.5% in crosses of SP82I-HmuIIoc and SPO1(sus14,ΔI) in Sup− cells (Table 3), where there is no functional intron endonuclease], the remaining recombinants likely arose from intron homing events. The presence of the SP82 sequence in these phages suggests that in almost all cases of intron acquisition by SPO1(sus14,ΔI) the SPO1 sequence, immediately upstream and downstream of the I-HmuII cleavage site, had been converted.
FIG. 2.
Nucleotide sequence alignment around the I-HmuII cleavage site. The sequence of the top (coding) strand is shown, with the cleavage site of I-HmuII on the bottom strand indicated by an arrow. Differences from the SP82 sequence are highlighted. Oligonucleotide SP82c1 has eight mismatches to the SPO1 sequence, four on each side of the cleavage site, requiring bidirectional conversion to hybridize with SPO1 DNA.
Homing is independent of major host recombination pathways.
The mobile introns of E. coli bacteriophage T4 provided a useful system to thoroughly analyze the functional cis and trans requirements for intron homing (6). Genetic and biochemical analyses allowed for elucidation of a precise definition of the recombination events that result in intron inheritance and the determination of host and phage functions required for intron mobility (18, 25). In comparison to phage T4, genes of SPO1 involved in replication and recombination have been poorly or not at all studied. In B. subtilis, several recombination and repair genes, belonging to five epistatic groups, have been genetically characterized (11). The following single and double mutants were used to study homing requirements based on their capacity to impair homologous recombination and recombinational repair in B. subtilis: recA, recF15 addA5, recF15 recH342, recO1 recH342, recU1 recH342, and recS1 recH342.
The six B. subtilis recombination mutants and their parental strain, YB886, were coinfected with SPO1(sus14,ΔI) and SPO1 or SPO1I-HmuIoc (Table 4). Intron-containing phages dominated the progeny of mixed infections with SPO1, whereas the high transmission frequency of the intron allele was not observed in crosses with SPO1I-HmuIoc (columns 5 and 6). In comparison, the frequency of sus14 in the progeny remained largely unchanged compared to that of the input phages (columns 7 and 8). The slight increase of wild-type gene 14 in SPO1I-HmuIoc crosses with wild-type SPO1 was likely due to a relatively high proportion of singly infected cells in this experiment. Because the six B. subtilis recombination mutants and their parental strain are Sup−, sus14 phage would produce no progeny in the absence of complementation by the wild-type gene.
TABLE 4.
Intron homing into SPO1(sus14,ΔI) in recombination-deficient B. subtilis
| Intron donor | Host | No. | Distribution of genetic markers in progenya
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| wt14,I+ | sus14,IΔ | sus14,I+ | wt14,IΔ | I+ | IΔ | wt14 | sus14 | |||
| SPO1 | none | 1 | 70 | 30 | 0 | 0 | 70 | 30 | 70 | 30 |
| 2 | 54 | 46 | 0 | 0 | 54 | 46 | 54 | 46 | ||
| Wild type | 1 | 73 | 4 | 23 | 0 | 96 | 4 | 73 | 27 | |
| 2 | 83 | 1 | 15 | 1 | 98 | 2 | 84 | 16 | ||
| recA | 1 | 78 | 2 | 19 | 1 | 97 | 3 | 79 | 21 | |
| 2 | 68 | 4 | 27 | 1 | 95 | 5 | 69 | 31 | ||
| recF addA | 1 | 80 | 2 | 16 | 2 | 96 | 4 | 82 | 18 | |
| 2 | 59 | 3 | 34 | 2 | 93 | 5 | 61 | 37 | ||
| recF recH | 1 | 79 | 2 | 19 | 0 | 98 | 2 | 79 | 21 | |
| 2 | 63 | 3 | 34 | 0 | 97 | 3 | 63 | 37 | ||
| recO recH | 1 | 76 | 3 | 21 | 0 | 97 | 3 | 76 | 24 | |
| 2 | 67 | 2 | 31 | 0 | 98 | 2 | 67 | 33 | ||
| recU recH | 1 | 79 | 0 | 20 | 0 | 99 | 0 | 79 | 20 | |
| 2 | 70 | 2 | 28 | 0 | 98 | 2 | 70 | 30 | ||
| recS recH | 1 | 61 | 2 | 37 | 0 | 98 | 2 | 61 | 39 | |
| 2 | 88 | 0 | 11 | 1 | 99 | 1 | 89 | 11 | ||
| SPO1I-HmuIoc | none | 1 | 61 | 39 | 0 | 0 | 61 | 39 | 61 | 39 |
| Wild type | 2 | 47 | 53 | 0 | 0 | 47 | 53 | 47 | 53 | |
| 1 | 73 | 14 | 3 | 9 | 76 | 23 | 82 | 17 | ||
| 2 | 81 | 8 | 3 | 6 | 84 | 14 | 87 | 11 | ||
| recA | 1 | 62 | 20 | 3 | 15 | 65 | 35 | 77 | 23 | |
| 54 | 35 | 3 | 7 | 57 | 42 | 61 | 38 | |||
| recF addA | 1 | 53 | 27 | 5 | 15 | 58 | 42 | 68 | 32 | |
| 2 | 48 | 33 | 10 | 8 | 58 | 41 | 56 | 43 | ||
| recF recH | 1 | 56 | 34 | 2 | 8 | 58 | 42 | 64 | 36 | |
| 2 | 47 | 37 | 9 | 6 | 56 | 43 | 53 | 46 | ||
| recO recH | 1 | 56 | 30 | 10 | 3 | 66 | 33 | 59 | 40 | |
| 2 | 63 | 27 | 7 | 2 | 70 | 29 | 65 | 34 | ||
| recU recH | 1 | 66 | 19 | 1 | 12 | 67 | 31 | 78 | 20 | |
| 2 | 53 | 29 | 12 | 5 | 65 | 34 | 58 | 41 | ||
| recS recH | 1 | 59 | 26 | 4 | 9 | 63 | 35 | 68 | 30 | |
| 2 | 81 | 11 | 2 | 6 | 83 | 17 | 87 | 13 | ||
See Table 2, footnote a.
Crosses with wild-type SPO1 resulted in a larger number of intron-containing recombinant phages than did crosses with SPO1I-HmuIoc (Table 4, column 3), suggesting that, analogous to T4 intron mobility, intron homing in SPO1 is independent of the major host recombination pathways. Appearance of recombinant phages in the progeny of crosses with SPO1I-HmuIoc (columns 3 and 4) further suggests that none of the Rec− strains impaired general recombination during mixed infections. This implies that, as is also the case for homing in T4, phage-encoded factors are mainly involved in SPO1 intron homing.
DISCUSSION
This work showed that in coinfections of B. subtilis with intronless SPO1 and intron-containing phage SP82 or SPO1, both introns exhibit homing into the intronless DNA polymerase gene. Crosses with phages carrying suppressor-sensitive mutations in gene 14 further indicated that intron homing is a gene conversion event likely involving a DNA repair process. The endonuclease-dependent predominance of the intron in the progeny, with the sus14 allele remaining at about the same frequency as in the parent phages, argues for homing of the intron by unidirectional conversion of the intron-minus DNA polymerase gene. The high proportion of recombinant intron-containing phages containing the gene 14 marker of the recipient further supports this duplicative nonreciprocal process.
I-HmuII has previously been shown to be responsible for exclusion of the SPO1 intron and flanking genetic markers from the progeny of mixed infections of B. subtilis with SPO1 and SP82, a process that was suggested to involve recombinational repair (14). Because marker exclusion and intron homing are both initiated by single-stranded cleavage by I-HmuII, these events probably share the same mechanism. In both cases cleavage of the DNA of one phage by the intron-encoded endonuclease of the other likely results in replacement of sequences surrounding the cut site by sequences of the intact phage genome. The directionality of the transfer of sequences would be dependent on the cleavage specificity of the intron-encoded endonuclease. Although capable of cleaving intron-containing substrates in vitro, I-HmuI has a preference for the SP82 intronless gene, whereas I-HmuII prefers the SPO1 sequence, regardless of the presence or absence of an intron (14). The ability of I-HmuII to promote marker exclusion (intron replacement) and intron homing suggests that the endonuclease is a highly versatile mobile element.
Our experimental data indicate that in homing and marker exclusion the conversion process radiates in both directions from the cut site. The bidirectional conversion of the I-HmuII cleavage site in crosses of SPO1ΔI and SP82 is consistent with results obtained by Stewart and Franck (31). In their study of the predominance of SP82 over SPO1 in mixed infections, the exclusion was clustered around gene 31 (DNA polymerase gene), reaching from genes 29 to 32. It remains to be seen whether there is directional bias in the coconversion over this interval.
Intron homing in these phages is a puzzling result. Although in this work the nature of the break in vivo was not established, I-HmuI and I-HmuII have a nicking activity in vitro (14). Unlike DNase I and protein gpII of bacteriophage f1, which are both nicking endonucleases in reactions with Mg2+ but which generate DSBs in the presence of Mn2+ (5, 16, 23), I-HmuII retains nicking activity even in the presence of divalent transition metal ions (M. Landthaler and D. A. Shub, unpublished data).
Conversely, homing initiated by group I intron-encoded endonucleases has been shown to involve a DSB of the intron-minus allele close to the intron insertion site (9). This is thought to promote DSB repair resulting in the transfer of the intron, which is accompanied by coconversion of flanking genetic markers (2). In the case of I-HmuI and I-HmuII, either a nick is sufficient to initiate the mobility of these introns in vivo or second-strand cleavage is achieved by some intron endonuclease-independent component. For group II introns, in which homing is mechanistically different from that of group I introns, the excised intron RNA cleaves the coding strand by reverse splicing into the DNA and the endonuclease subsequently nicks the template strand (34). The similarity of group I and group II intron endonucleases, sharing the H-N-H nuclease motif (15), suggested that second-strand cleavage by group I intron RNA might also be involved in the mobility of these phage introns. However, the nicking H-N-H endonuclease I-TwoI, in the nrdE-I2 intron of staphylococcal phage Twort, cleaves the coding strand, arguing against involvement of intron RNA in second-strand cleavage (22).
Single-strand breaks have been suggested to initiate recombination processes, and Strathern and coworkers (32) observed a stimulation of recombination when they expressed the gene II product of bacteriophage f1, which has a strand-specific nicking activity, in yeast. However, the authors could not rule out the possibility that a small fraction of DSBs actually promoted the increase in recombination. It has been shown that single-strand breaks generated by restriction endonucleases are repaired by a RecA- and RecB-dependent pathway in E. coli (17).
However, a nick in DNA can result in a DSB even in the absence of another nucleolytic activity. Single-strand interruptions could lead to a collapse of the replication fork during DNA replication, creating a DSB (20, 21). In Saccharomyces pombe a recombination event is induced during mating-type switching when the replication fork encounters a chromosomal imprint in one of the sister chromatids at the mat1 locus. Biochemical characterization suggests that the imprint is a single-strand break (1) or an alkali-labile modification (8) in the DNA. Interestingly, Glassberg and coworkers (12) mapped an origin of replication in gene 32, the gene downstream of the SPO1 DNA polymerase gene. The incision by I-HmuI and I-HmuII on the coding strand would interrupt the template for lagging-strand synthesis.
Regardless of the detailed mechanism of the initiation event, intron homing occurred in host backgrounds with deficiencies in the major recombination genes. In addition to genes involved in general DNA metabolism, 14 B. subtilis recombination genes have been characterized and classified into five epistatic groups, with recA being the central player. Except in recA-deficient strains, homologous recombination does not change more than fourfold relative to the wild type when cells have single recombination functions impaired (11). Mutations in recA and double mutations in recF addA (addAB is the E. coli recBC homolog), recF recH, recO recH, recU recH, and recS recH were shown to have drastic effects on homologous recombination. Using these strains as hosts for coinfections of wild-type SPO1 and SPO1(sus14,ΔI) impaired neither intron homing nor general recombination between phage genes. Though crosses in these strains indicate that the respective Bacillus rec genes are not required for intron homing, redundancy in the recombination machinery and the potential presence of previously uncharacterized rec genes leave open possible participation of Bacillus gene products in intron homing.
On the other hand, the data favor the view that the genes involved in intron homing and general recombination may be encoded by the phages SPO1 and SP82. In the case of phage T4, intron homing has been shown to occur in the context of phage recombination-dependent DNA replication, which requires a number of phage replication and recombination functions. These include the strand transferase (UvsX), single-strand DNA binding protein (gp32), DNA polymerase (gp43), helicase (gp41), DNA ligase (gp30), and a putative exonuclease complex (gp46/gp47) (6, 18, 25). According to the model, the DSB is processed by exonucleases producing a double-strand gap with single-stranded 3′ tails for strand invasion of the homologous intron-plus allele. Repair synthesis using an intron-plus strand as a template results in intron inheritance in which two processes have been implicated, the classic DSB repair pathway and the synthesis-dependent strand annealing pathway (24).
Intron homing in SPO1 and SP82 presents an experimental system to study the genes and processes involved in recombination of these phages and would allow an interesting comparison between intron homing in phages of gram-positive and that of gram-negative bacteria. Perhaps such a comparison would offer insights into why group I introns are seemingly more successful in phages infecting gram-positive hosts, as suggested by their relative abundance in these genomes (10). Furthermore, DNA endonucleases with nicking activity, like I-HmuI and I-HmuII, could provide a useful tool to study the role of DNA single-strand breaks as recombinogenic hot spots in bacterial and eukaryotic genomes.
Acknowledgments
We thank J. C. Alonso for providing bacterial strains and David Edgell for critical reading of the manuscript.
This work was supported by a grant from the National Institutes of Health (GM-37746).
REFERENCES
- 1.Arcangioli, B. 1998. A site− and strand-specific DNA break confers asymmetric switching potential in fission yeast. EMBO J. 17:4503-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belfort, M., and P. S. Perlman. 1995. Mechanisms of intron mobility. J. Biol. Chem. 270:30237-30240. [DOI] [PubMed] [Google Scholar]
- 3.Belfort, M., and R. J. Roberts. 1997. Homing endonucleases: keeping the house in order. Nucleic Acids Res. 25:3379-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell-Pedersen, D., S. M. Quirk, M. Aubrey, and M. Belfort. 1989. A site-specific endonuclease and co-conversion of flanking exons associated with the mobile td intron of phage T4. Gene 82:119-126. [DOI] [PubMed] [Google Scholar]
- 5.Campbell, V. W., and D. A. Jackson. 1980. The effect of divalent cations on the mode of action of DNase I. The initial reaction products produced from covalently closed circular DNA. J. Biol. Chem. 255:3726-3735. [PubMed] [Google Scholar]
- 6.Clyman, J., and M. Belfort. 1992. Trans and cis requirements for intron mobility in a prokaryotic system. Genes Dev. 6:1269-1279. [DOI] [PubMed] [Google Scholar]
- 7.Colleaux, L., L. d'Auriol, M. Betermier, G. Cottarel, A. Jacquier, F. Galibert, and B. Dujon. 1986. Universal code equivalent of a yeast mitochondrial intron reading frame is expressed into E. coli as a specific double strand endonuclease. Cell 44:521-533. [DOI] [PubMed] [Google Scholar]
- 8.Dalgaard, J. Z., and A. J. Klar. 1999. Orientation of DNA replication establishes mating-type switching pattern in S. pombe. Nature 400:181-184. [DOI] [PubMed] [Google Scholar]
- 9.Dujon, B. 1989. Group I introns as mobile genetic elements: facts and mechanistic speculations. Gene 82:91-113. [DOI] [PubMed] [Google Scholar]
- 10.Edgell, D. R., M. Belfort, and D. A. Shub. 2000. Barriers to intron promiscuity in bacteria. J. Bacteriol. 182:5281-5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernández, S., S. Ayora, and J. C. Alonso. 2000. Bacillus subtilis homologous recombination: genes and products. Res. Microbiol. 151:481-486. [DOI] [PubMed] [Google Scholar]
- 12.Glassberg, J., M. Franck, and C. R. Stewart. 1977. Multiple origins of replication for Bacillus subtilis phage SPO1. Virology 78:433-441. [DOI] [PubMed] [Google Scholar]
- 13.Goodrich-Blair, H., V. Scarlato, J. M. Gott, M. Q. Xu, and D. A. Shub. 1990. A self-splicing group I intron in the DNA polymerase gene of Bacillus subtilis bacteriophage SPO1. Cell 63:417-424. [DOI] [PubMed] [Google Scholar]
- 14.Goodrich-Blair, H., and D. A. Shub. 1996. Beyond homing: competition between intron endonucleases confers a selective advantage on flanking genetic markers. Cell 84:211-221. [DOI] [PubMed] [Google Scholar]
- 15.Gorbalenya, A. E. 1994. Self-splicing group I and group II introns encode homologous (putative) DNA endonucleases of a new family. Protein Sci. 3:1117-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenstein, D., and K. Horiuchi. 1989. Double-strand cleavage and strand joining by the replication initiator protein of filamentous phage f1. J. Biol. Chem. 264:12627-12632. [PubMed] [Google Scholar]
- 17.Heitman, J., T. Ivanenko, and A. Kiss. 1999. DNA nicks inflicted by restriction endonucleases are repaired by a RecA- and RecB-dependent pathway in Escherichia coli. Mol. Microbiol. 33:1141-1151. [DOI] [PubMed] [Google Scholar]
- 18.Huang, Y. J., M. M. Parker, and M. Belfort. 1999. Role of exonucleolytic degradation in group I intron homing in phage T4. Genetics 153:1501-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacquier, A., and B. Dujon. 1985. An intron-encoded protein is active in a gene conversion process that spreads an intron into a mitochondrial gene. Cell 41:383-394. [DOI] [PubMed] [Google Scholar]
- 20.Kuzminov, A. 1995. Collapse and repair of replication forks in Escherichia coli. Mol. Microbiol. 16:373-384. [DOI] [PubMed] [Google Scholar]
- 21.Kuzminov, A. 2001. Single-strand interruptions in replicating chromosomes cause double-strand breaks. Proc. Natl. Acad. Sci. USA 98:8241-8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Landthaler, M., U. Begley, N. C. Lau, and D. A. Shub. 2002. Two self-splicing group I introns in the ribonucleotide reductase large subunit gene of Staphylococcus aureus phage Twort. Nucleic Acids Res. 30:1935-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Melgar, E., and D. A. Goldthwait. 1968. Deoxyribonucleic acid nucleases. II. The effects of metals on the mechanism of action of deoxyribonuclease I. J. Biol. Chem. 243:4409-4416. [PubMed] [Google Scholar]
- 24.Mueller, J. E., J. Clyman, Y.-J. Huang, M. M. Parker, and M. Belfort. 1996. Intron mobility in phage T4 occurs in the context of recombination-dependent DNA replication by way of multiple pathways. Genes Dev. 10:351-364. [DOI] [PubMed] [Google Scholar]
- 25.Mueller, J. E., D. Smith, and M. Belfort. 1996. Exon coconversion biases accompanying intron homing: battle of the nucleases. Genes Dev. 10:2158-2166. [DOI] [PubMed] [Google Scholar]
- 26.Muscarella, D. E., and V. M. Vogt. 1989. A mobile group I intron in the nuclear rDNA of Physarum polycephalum. Cell 56:443-454. [DOI] [PubMed] [Google Scholar]
- 27.Okubo, S., T. Yanagida, D. J. Fujita, and B. M. Olsson-Wilhelm. 1972. The genetics of bacteriophage SPO1. Biken J. 15:81-97. [PubMed] [Google Scholar]
- 28.Remacle, C., and R. F. Matagne. 1993. Transmission, recombination and conversion of mitochondrial markers in relation to the mobility of a group I intron in Chlamydomonas. Curr. Genet. 23:518-525. [DOI] [PubMed] [Google Scholar]
- 29.Sayre, M. H., and E. P. Geiduschek. 1988. TF1, the bacteriophage SPO1-encoded type II DNA-binding protein, is essential for viral multiplication. J. Virol. 62:3455-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stewart, C. R. 1988. Bacteriophage SPO1, p. 477-515. In R. Calendar (ed.), The bacteriophages, vol. I. Plenum Press, New York, N.Y. [Google Scholar]
- 31.Stewart, C. R., and M. Franck. 1981. Predominance of bacteriophage SP82 over bacteriophage SPO1 in mixed infections of Bacillus subtilis. J. Virol. 38:1081-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strathern, J. N., K. G. Weinstock, D. R. Higgins, and C. B. McGill. 1991. A novel recombinator in yeast based on gene II protein from bacteriophage f1. Genetics 127:61-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szostak, J. W., T. L. Orr-Weaver, R. J. Rothstein, and F. W. Stahl. 1983. The double-strand-break repair model for recombination. Cell 33:25-35. [DOI] [PubMed] [Google Scholar]
- 34.Yang, J., S. Zimmerly, P. S. Perlman, and A. M. Lambowitz. 1996. Efficient integration of an intron RNA into double-stranded DNA by reverse splicing. Nature 381:332-335. [DOI] [PubMed] [Google Scholar]
- 35.Zinn, A. R., and R. A. Butow. 1985. Nonreciprocal exchange between alleles of the yeast mitochondrial 21S rRNA gene: kinetics and the involvement of a double-strand break. Cell 40:887-895. [DOI] [PubMed] [Google Scholar]