Abstract
Numerous bacteria and mammalian cells harbor two enzymes, phosphopentomutase (PPM) and 2-deoxyribose 5-phosphate aldolase (DERA), involved in the interconversion between nucleosides and central carbon metabolism. In this study, we have examined the presence of this metabolic link in the hyperthermophilic archaeon, Thermococcus kodakaraensis KOD1. A search of the genome sequence of this strain revealed the presence of a closely related orthologue (TK2104) of bacterial DERA genes while no orthologue related to previously characterized PPM genes could be detected. Expression, purification, and characterization of the TK2104 protein product revealed that this gene actually encoded a DERA, catalyzing the reaction through a class I aldolase mechanism. As PPM activity was detected in T. kodakaraensis cells, we partially purified the protein to examine its N-terminal amino acid sequence. The sequence corresponded to a gene (TK1777) similar to phosphomannomutases within COG1109 but not COG1015, which includes all previously identified PPMs. Heterologous gene expression of TK1777 and characterization of the purified recombinant protein clearly revealed that the gene indeed encoded a PPM. Both enzyme activities could be observed in T. kodakaraensis cells under glycolytic and gluconeogenic growth conditions, whereas the addition of ribose, 2-deoxyribose, and 2′-deoxynucleosides in the medium did not lead to a significant induction of these activities. Our results clearly indicate the presence of a metabolic link between pentoses and central carbon metabolism in T. kodakaraensis, providing an alternative route for pentose biosynthesis through the functions of DERA and a structurally novel PPM.
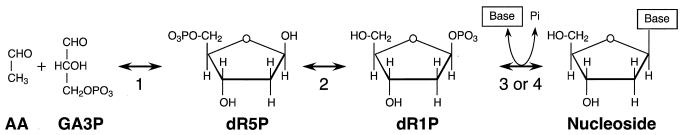
In a number of bacteria and in mammalian cells, there exists a direct metabolic link between the ribose moiety of (deoxy)nucleosides and central carbon metabolism, independent of the pentose phosphate cycle (Fig. 1) (3, 9, 26). Two enzymes provide this link: the phosphopentomutases (PPMs) and 2-deoxyribose 5-phosphate aldolases (DERAs). PPM isomerizes (deoxy)ribose 1-phosphate to (deoxy)ribose 5-phosphate. Subsequently, DERA cleaves the pentose phosphate to acetaldehyde and glyceraldehyde 3-phosphate, allowing further metabolism to obtain carbon and energy. The (deoxy)ribose 1-phosphate is supplied by various nucleoside phosphorylases, such as thymidine phosphorylase and purine nucleoside phosphorylase, which release the pentose moiety from (deoxy)ribonucleosides, producing (deoxy)ribose 1-phosphate. These phosphorylases are known to have a major role in the interconversion of nucleosides as well as the salvage of purine and pyrimidine bases (4, 14, 17). Because the phosphorylases, PPM, and DERA are all reversible, the pathway in total enables both the catabolism and biosynthesis of nucleosides.
FIG. 1.
Schematic diagram of the metabolic link described in this study. The enzymes involved in the biosynthesis and catabolism of nucleosides are shown (1 to 4). 1, DERA; 2, PPM; 3, thymidine phosphorylase; 4, purine nucleoside phosphorylase; AA, acetaldehyde; GA3P, glyceraldehyde 3-phosphate.
As various bacteria can grow on a variety of (deoxy)nucleosides or (deoxy)nucleotides and their derivatives as carbon and energy sources, a catabolic role of this pathway has been indicated (20, 21, 24, 26, 30). It has been shown that an addition of these substrates to the growth medium causes an induction of these enzymes while the presence of glucose represses their expression (5, 21, 26, 30). Moreover, examination of mutant strains of Escherichia coli lacking these proteins revealed that they failed to grow on these substrates, strongly indicating the catabolic nature of this pathway (15, 19). On the other hand, this pathway in mammalian cells seems to play a biosynthetic role. The inhibition of DERA in these cells led to a specific inhibition of labeled purine and pyrimidine incorporation into DNA, whereas RNA and protein synthesis were not affected (9).
While the presence of these enzymes has been identified in various bacteria and mammalian cells, very little is known about this metabolic link in the Archaea. There has been only one indication of its presence, that from a report on the DERA from Aeropyrum pernix (22), an aerobic hyperthermophilic archaeon. However, very few archaeal genomes harbor the DERA gene. Moreover, there has been no evidence that suggests the presence of a PPM in the Archaea. In fact, a PPM orthologue cannot even be found on the genome of A. pernix.
Thermococcus kodakaraensis KOD1 is a hyperthermophilic archaeon isolated from a solfatara on Kodakara Island, Kagoshima, Japan (2, 18). The strain is a strict anaerobe and grows heterotrophically on a variety of organic substrates including starch, pyruvate, amino acids, and peptides (2, 13). Along with Pyrococcus furiosus, Pyrococcus abyssi, and Pyrococcus horikoshii, T. kodakaraensis is a member of the order Thermococcales. We have recently determined the entire genome sequence of this strain (unpublished data) and are examining the presence or absence of various metabolic pathways in this strain. In the search for candidate genes that encode the enzymes mentioned above, we observed candidate genes on the T. kodakaraensis genome that were presumed to encode thymidine phosphorylase and purine nucleoside phosphorylase. We also found a candidate gene for DERA, which could not be found on the Pyrococcus genomes. In contrast, as in the case of A. pernix, we could not identify a PPM orthologue on the genome classified in COG1015 (cluster of orthologous genes [COG]), in which all previously known PPMs are included.
In this study, we have examined the presence of this pathway in T. kodakaraensis KOD1, focusing on the identification and characterization of PPM and DERA. By examining PPM activity in T. kodakaraensis and partially purifying the enzyme, a structurally novel PPM was identified in this strain. Biochemical characterization of the protein, along with the DERA of this strain, clearly indicate the presence of a metabolic link between central carbon metabolism and pentose biosynthesis and catabolism in T. kodakaraensis.
MATERIALS AND METHODS
Strains, plasmids, and media.
T. kodakaraensis KOD1 was isolated from a solfataric hot spring on the shore of Kodakara Island, Kagoshima, Japan (2, 18). Cells were grown in either a nutrient-rich MA-YT medium (0.48 and 2.64% of Marine Art SF agents A and B, respectively [Senju Pharmaceuticals, Osaka, Japan], 0.5% yeast extract, and 0.5% tryptone) or a minimal ASW-AA medium (23). E. coli strain DH5α was used for gene cloning. E. coli strain BL21(DE3) CodonPlus-RIL (Stratagene, La Jolla, Calif.) and the vector pET-21a (Novagen, Madison, Wis.) were used for gene expression.
DNA manipulation and sequence analyses.
Restriction enzymes and DNA polymerase were purchased from Toyobo (Osaka, Japan) and Takara Shuzo (Kyoto, Japan). Genomic and plasmid DNAs were isolated by using DNA isolation kits from QIAGEN (Hilden, Germany). DNA ligation reactions were performed with a DNA ligation kit (Toyobo). DNA sequencing was performed with the ABI PRISM BigDye terminator cycle sequencing ready reaction kit (Applied Biosystems, Foster City, Calif.). Sequence analyses were performed by using DNASIS software (Hitachi Software, Yokohama, Japan). Database homology searches were performed by using the Basic Local Alignment Search Tool program (1). Multiple-sequence alignment and phylogenetic analysis were performed with the ClustalW program (28) provided by the DNA Data Bank of Japan.
Partial purification of PPM activity from T. kodakaraensis.
All purification steps were performed at room temperature with columns purchased from Amersham Biosciences (Little Chalfont, Buckinghamshire, United Kingdom) unless mentioned otherwise. T. kodakaraensis cells were cultivated in MA-YT medium with the addition of 0.5% soluble starch (Nacalai Tesque, Kyoto, Japan). Cells were harvested and lysed by sonication on ice water. After ultracentrifugation of the cell lysate at 110,000 × g for 70 min at 4°C, the supernatant, containing the PPM activity, was loaded onto an anion-exchange column (Resource Q) which was equilibrated with buffer A (50 mM sodium phosphate buffer [pH 7.0]). Proteins were eluted with a linear gradient of 0 to 1.0 M sodium chloride. Fractions with PPM activity were dialyzed against buffer A and loaded onto a Mono Q HR 5/5 column equilibrated with buffer A. Proteins were eluted with a linear gradient of 0 to 1.0 M sodium chloride. Fractions exhibiting PPM activity were dialyzed against 2 M ammonium sulfate and applied to a hydrophobic column (Resource ISO) equilibrated with 2 M ammonium sulfate at pH 7.0. The bound proteins were eluted with a linear gradient of 2.0 to 0 M ammonium sulfate at pH 7.0. Fractions carrying PPM activity were dialyzed against buffer A and applied to a hydroxyapatite column (Bio-Scale CHT-I; Bio-Rad, Hercules, Calif.). Fractions exhibiting PPM activity were concentrated by using Centricon YM-30 (Millipore Corporation, Bedford, Mass.) and further purified with a gel filtration column (Superdex 200 HR 10/30) equilibrated with buffer A containing 150 mM sodium chloride. Protein concentration was determined with a bicinchoninic acid protein assay kit (Pierce, Rockford, Ill.). N-terminal amino acid sequencing was performed with a protein sequencer (model 491 cLC; Applied Biosystems).
Expression of TK2104 and TK1777 genes in E. coli.
TK2104 and TK1777 genes were amplified by PCR and inserted into pUC118. An NdeI site was introduced in the 5′-flanking region of the genes. After confirming the sequence, the NdeI-EcoRI restriction fragment was inserted into the pET-21a expression vector at the corresponding sites. The resulting plasmids, pET-TK2104 and pET-TK1777, were introduced to E. coli strain BL21(DE3) CodonPlus-RIL. Cells were grown at 37°C in Luria-Bertani medium containing ampicillin (50 μg/ml) until the optical density at 660 nm reached 0.5. Gene expression was induced by the addition of 0.2 mM (final concentration) isopropyl-β-d-thiogalactopyranoside (IPTG), and incubation at 37°C was carried out for another 4 h.
Purification of recombinant TK2104 and TK1777.
Cells were harvested by centrifugation at 6,000 × g for 10 min at 4°C and washed with 50 mM bicine buffer (pH 8.0). The cell pellet was resuspended in the same buffer, and the cells were then disrupted by sonication in ice water. Soluble and insoluble fractions were separated by centrifugation (15,000 × g for 30 min at 4°C). The soluble fraction containing the recombinant TK2104 or TK1777 was incubated at 85°C for 20 min and centrifuged at 15,000 × g for 30 min at 4°C to remove heat-labile proteins from the host E. coli. All purification steps were performed at room temperature unless mentioned otherwise. The soluble fractions exhibiting DERA or PPM activity were partially purified with Resource Q as described above with 50 mM bicine buffer (pH 8.0) instead of buffer A. Fractions with activity were dialyzed against 1.8 M ammonium sulfate and loaded onto Resource ISO equilibrated with 1.8 M ammonium sulfate. The bound proteins were eluted with a linear gradient of 1.8 to 0 M ammonium sulfate (pH 8.0). Fractions exhibiting activity were dialyzed against 50 mM Tris-HCl containing 150 mM sodium chloride (pH 8.0), concentrated, and further purified with Superdex 200 HR 10/30 as described above. The apparent homogeneity of the proteins was examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Apparent molecular masses of the purified proteins were estimated by gel filtration (Superdex 200 HR 10/30) with HMW and LMW gel filtration calibration kits (Amersham Biosciences).
Enzyme activity measurements.
The 2-deoxyribose 5-phosphate (dR5P) cleavage activity was routinely measured by conversion of acetaldehyde (one of the products formed by cleavage of dR5P) to ethanol by oxidation of NADH (Oriental Yeast, Tokyo, Japan) in the presence of excess alcohol dehydrogenase (Oriental Yeast). To determine the enzyme activity at a higher temperature, a discontinuous enzyme assay was adopted. The assay mixture (200 μl) containing 20 mM sodium citrate buffer (pH 4.0) (Nacalai), 1 mM EDTA (Nacalai), and 10 mM dR5P (Sigma, St. Louis, Mo.) was preincubated at the respective temperature for 1 min, and the reaction was initiated by the addition of enzyme. After incubation for 1 min, the reaction was stopped in ice water, and the pH was adjusted to 8.0 with 100 μl of 1 M Tris-HCl. Following this, 0.4 mM NADH and 2 U of alcohol dehydrogenase were added, and the final volume was adjusted to 1 ml. After incubation at 25°C for 3 min, the decrease in absorbance at 340 nm was measured with an Ultrospec 3000 spectrophotometer (Amersham Biosciences). Product formation was proportional to the incubation time under these conditions. When the effect of pH on the enzyme activity was examined, all buffers were prepared so that their pHs would reflect accurate values at 95°C. To measure aldolase activity for other substrates, the first reaction was the same as described above. For the coupling reaction, 0.4 mM NADH, 2 U of triosephosphate isomerase (Sigma), and 2 U of glycerol 3-phosphate dehydrogenase (Sigma) were added and the mixture was incubated at 25°C for 3 min, followed by measurement of the decrease in absorbance at 340 nm.
For PPM activity, 2-deoxyribose 1-phosphate (dR1P) (Sigma) was used as a substrate. The initial assay mixture contained 25 mM Tris-HCl, 10 mM MgCl2, 50 μM glucose 1,6-bisphosphate (Sigma), 5 mM dR1P, and 2 μg of enzyme in a final volume of 200 μl. After incubation, enzymes were removed by ultrafiltration with Microcon YM-10 (Millipore), and a second reaction was performed by adding 20 μl of 1 M sodium citrate (Nacalai) and 15 μg of purified DERA of this strain (DERATk) (see below). After incubation at 75°C for 5 min, the reaction was stopped by cooling on ice. Acetaldehyde was quantified with alcohol dehydrogenase as described above. The amount of dR5P was calculated from the amount of acetaldehyde by using the equilibrium constant of DERATk obtained from control experiments with various concentrations of dR5P.
For phosphoglucomutase activity, formation of glucose 6-phosphate from glucose 1-phosphate was measured in a discontinuous assay. The initial reaction mixture (200 μl) consisted of 100 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 50 μM glucose 1,6-bisphosphate (Sigma), 5 mM glucose 1-phosphate (Sigma), and 1 μg of purified enzyme. The coupling reaction was performed by adding 800 μl of water, 0.5 mM NADP, and 2 U of glucose 6-phosphate dehydrogenase, and the absorbance was measured at 340 nm after incubation at 25°C for 3 min. Phosphofructose mutase activity was measured in a similar manner with the presence of an additional 2 U of phosphoglucose isomerase (Sigma) in the coupling reaction mixture. Phosphomannomutase activity was examined with the addition of 2 U of phosphomannose isomerase (Sigma) and 2 U of phosphoglucose isomerase in the coupling reaction mixture. To examine phosphoglucosamine mutase activity, glucosamine 1-phosphate (Sigma) was used as a substrate. The product glucosamine 6-phosphate was quantitatively converted to 6-phosphogluconate via fructose 6-phosphate and glucose 6-phosphate in the presence of glucosamine 6-phosphate deaminase (from this strain) (unpublished data), phosphoglucose isomerase, and glucose 6-phosphate dehydrogenase. The assay mixture contained 100 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 50 μM glucose 1,6-bisphosphate, 5 mM glucosamine 1-phosphate (Sigma), 2 U of glucosamine 6-phosphate deaminase, and 1 μg of purified enzyme in a final volume of 200 μl. The coupling reaction was initiated by the addition of 0.5 mM NADP, 2 U of phosphoglucose isomerase, and 2 U of glucose 6-phosphate dehydrogenase. After incubation at 25°C for 3 min, the formation of NADPH was measured at 340 nm. Phosphoacetylglucosamine mutase activity was measured as described elsewhere (11). Concerning phosphoglycerate mutase activity, the first reaction mixture contained 100 mM Tris-HCl, 10 mM MgCl2, 50 μM 2,3-diphosphoglycerate, 5 mM 3-phosphoglycerate, 1 mM ADP, and 2 μg of purified enzyme in a final volume of 200 μl. The coupling reaction was initiated by the addition 0.5 mM NADH and 2 U each of enolase (Sigma), pyruvate kinase (Sigma), and lactate dehydrogenase (Sigma). After 3 min of incubation at 25°C, the decrease in NADH was measured at 340 nm.
Gene nomenclature.
The locus numbers used in this study are based on the present draft of annotation for the genome of T. kodakaraensis.
Nucleotide sequence accession number.
The nucleotide sequence data for the TK2104 and TK1777 genes reported in this paper will appear in the DDBJ, EMBL, and GenBank DNA databases under the accession numbers AB092961 and AB126239, respectively.
RESULTS
Identification of candidate genes on the T. kodakaraensis genome encoding PPM and DERA.
First, we searched the T. kodakaraensis genome for candidate genes encoding PPM and DERA. The two activities would provide a metabolic link between the ribose moieties of nucleosides and central carbon metabolism (Fig. 1). One open reading frame (TK2104) was found whose translated product was similar to known DERA proteins classified in COG0274. TK2104 encoded a protein of 224 amino acids with a molecular mass of 24,504 Da, and the deduced sequence displayed 57, 42, 33, and 32% identity to the enzymes from Bacillus subtilis (P39121), A. pernix (D72474), Salmonella enterica serovar Typhimurium (AAL23382), and E. coli (P00882), respectively. The residues essential for catalytic activity in DERA from E. coli K167, K201, and D102 (numbering according to the E. coli sequence) (10) were completely conserved in the TK2104 protein. In contrast, we could not find any orthologues related to the known bacterial PPMs (COG1015). On the other hand, we observed the presence of four open reading frames encoding proteins homologous to known phosphomannomutases and phosphoglucomutases classified in COG1109.
Heterologous gene expression of TK2104 and characterization of the recombinant protein.
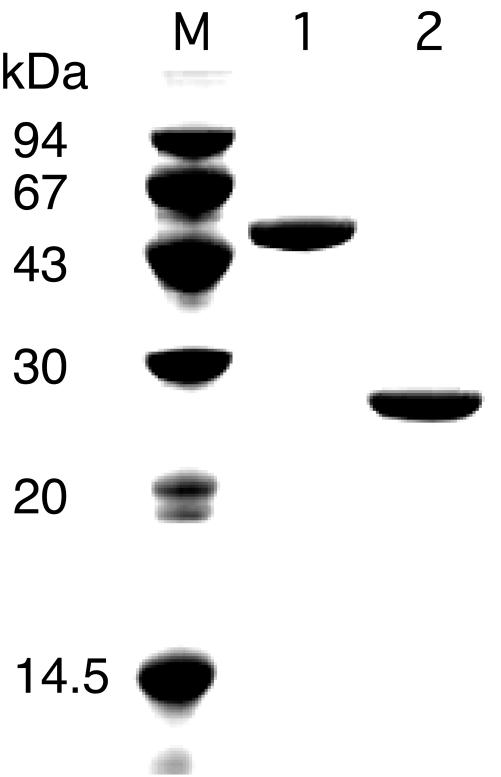
To characterize the protein product of TK2104 and to determine whether the enzyme was a true DERA, we expressed the gene in E. coli. The recombinant protein was purified to apparent homogeneity by heat treatment at 85°C for 20 min followed by anion-exchange and gel filtration chromatographies (Fig. 2, lane 2). The molecular mass of recombinant TK2104 estimated by SDS-PAGE agreed with that calculated from the deduced amino acid sequence. Furthermore, the N-terminal 10 amino acid residues of the purified protein were identical to the deduced amino acid sequence of the gene, confirming that we had obtained the recombinant TK2104 protein. The molecular mass of the TK2104 protein was determined to be 53 kDa by gel filtration chromatography. Taking into account the molecular mass of the subunit (24.5 kDa), this result indicated that the TK2104 protein exists in a dimeric form.
FIG. 2.
SDS-PAGE analysis of the purified protein products of TK1777 and TK2104. Lane M, molecular mass markers; lane 1, purified recombinant TK1777; lane 2, purified recombinant TK2104.
Examination of DERA activity.
The purified protein was dialyzed against 50 mM bicine buffer (pH 8.0) containing 10 mM EDTA and used for further analysis. The dR5P cleavage activity was assayed by measuring the production of acetaldehyde with alcohol dehydrogenase or by measuring the production of glyceraldehyde 3-phosphate with triosephosphate isomerase and glycerophosphate dehydrogenase. The purified protein proved to exhibit significant DERA activity (see below) in both systems and was therefore designated DERATk.
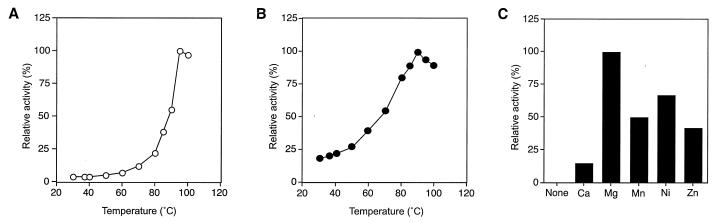
We examined the effect of temperature and pH on DERA activity. The enzyme displayed maximal activity at pH 4.0, and the optimal temperature was 95°C (Fig. 3A). As in the case of other aldolases (12, 29), the presence of citrate was found to enhance the activity of DERATk. Activities were much higher in citrate buffer than those observed in acetate buffers at equivalent pH, with a 2.8-fold activation at the optimal pH of 4.0 (data not shown). The specific activity of DERATk under optimal conditions was 285 ± 5 μmol min−1 mg−1. The thermostability of the recombinant protein was examined at pH 4.0 and 7.0. The enzyme retained over 80% of its activity after 120 min at 90°C. The half-life of the enzyme was 40 min in boiling water at pH 7.0 while it was 20 min at pH 4.0. Kinetic analysis was also carried out, and the enzyme catalyzed the cleavage reaction following Michaelis-Menten kinetics with a Km value of 0.81 ± 0.07 mM toward dR5P, and a kcat value of 116 ± 4 s−1 subunit−1 at 95°C (Table 1).
FIG. 3.
(A) Temperature profile of DERATk activity. Activity was examined at the optimal pH of 4.0. (B) Temperature profile of PPMTk activity. Activity was examined at the optimal pH of 7.5. (C) Effect of metal cations on PPMTk enzyme activity. A chloride salt of each metal cation was added at a final concentration of 1 mM, and PPM activity was examined at 90°C.
TABLE 1.
Kinetic parameters of DERATk and PPMTk reactionsa
| Protein | Substrate | Vmax (μmol min−1 mg−1) | Km (mM) | kcat (s−1 subunit−1) | kcat/Km (mM−1 s−1 subunit−1) |
|---|---|---|---|---|---|
| DERATk | dR 5P | 285 ± 5 | 0.81 ± 0.07 | 116 ± 4 | 143 |
| PPMTk | dR 1P | 210 ± 10 | 2.5 ± 0.2 | 173 ± 9 | 69 |
Activity measurements for DERATk and PPMTk were performed at 95 and 90°C, respectively.
Effects of NaBH4, EDTA, and metal cations on DERATk.
Class I aldolases form a Schiff base intermediate between an active-site lysine residue and the carbonyl group of the donor substrate (6), while class II enzymes utilize divalent metal cations (16). The former group of aldolases is known to be inhibited by the addition of NaBH4, whereas the latter group is inhibited by EDTA (27). The primary sequence of DERATk indicated that the enzyme was a class I aldolase. Purified DERATk was treated with NaBH4 (100 mM) in the presence and absence of dR5P (30 mM). As expected, 98 and 52% of cleavage activity was abolished after treatment with NaBH4 (20 h at 4°C) in the presence and absence of dR5P, respectively. In contrast, no inhibitory effects were observed with the addition of EDTA at 10 mM, confirming that DERATk was a class I aldolase.
We further examined the effects of divalent metal cations on the enzyme activity. No notable effects were observed when Mg2+, Ca2+, and Mn2+ were present in the reaction mixture at a concentration of 1 mM. The presence of 1 mM Zn2+ led to a slight decrease (35%) in enzyme activity (data not shown). The results indicate that the activity of DERATk is independent of divalent metal cations.
Substrate specificity of DERATk.
We examined whether DERATk was specific toward the substrate dR5P. Enzyme activities toward the following substrates were investigated: 2-deoxyribose, ribose 5-phosphate, arabinose 5-phosphate, fructose 1,6-bisphosphate, and fructose 6-phosphate. We observed aldolase activity only with the substrate dR5P, indicating that DERATk was a true archaeal DERA with strict substrate specificity.
Partial purification of PPM of T. kodakaraensis.
As T. kodakaraensis was found to harbor a DERA, we further examined the presence of PPM. As mentioned above, the genome sequence did not indicate the presence of a PPM gene. We therefore analyzed the enzyme activity in the cell extracts of T. kodakaraensis. With a coupled assay using DERATk, we were able to detect significant levels of PPM activity in the cell extract of starch-grown cells. The PPM was purified 14-fold by anion-exchange, hydrophobic, hydroxyapatite, and gel filtration column chromatographies. A 50-kDa protein was found to correspond well with the results of activity measurements through each purification step. When we analyzed the N-terminal sequence of the protein band, we obtained the sequence X-Arg-Leu-Phe-Gly-X-Ala, which corresponds to the sequence found in TK1777 (Met-Arg-Leu-Phe-Gly-Thr-Ala), one of the four orthologues related to phosphomannomutase and phosphoglucomutase.
Gene expression of TK1777 and purification of recombinant protein.
To elucidate the enzymatic activity of the TK1777 protein, we expressed the gene in E. coli and purified the recombinant protein. High levels of recombinant protein were obtained in a soluble form, and purification was performed by heat treatment, anion-exchange, hydrophobic, and gel filtration chromatographies (Fig. 2, lane 1). The N-terminal amino acid sequence of the purified recombinant protein was determined and was found to correspond to that of the deduced sequence from the TK1777 gene. Gel filtration chromatography indicated that the molecular mass of the purified protein was approximately 210 kDa, indicating a tetrameric assembly.
Substrate specificity of recombinant TK1777 protein.
Using the purified recombinant protein, we measured the PPM activity of the TK1777 protein by using the substrate dR1P. We also measured mutase activities with various phosphorylated substrates. The substrates were glucose 1-phosphate, mannose 1-phosphate, fructose 1-phosphate, N-acetylglucosamine 1-phosphate, glucosamine 1-phosphate, and 3-phosphoglycerate. Mutase activity towards these substrates was examined at a concentration of 5 mM. The TK1777 protein exhibited a significant level of mutase activity towards dR1P (210 μmol min−1 mg−1). Among the other substrates, we found trace levels of activity with glucose 1-phosphate and mannose 1-phosphate. Even when the concentration of glucose 1-phosphate was increased to 30 mM, we could only detect activity levels of approximately 3 μmol min−1 mg−1, much lower than that towards dR1P. The enzyme also displayed trace activity towards glucosamine 1-phosphate (0.2 μmol min−1 mg−1). No mutase activity was detected with fructose 1-phosphate, N-acetylglucosamine 1-phosphate, and 3-phosphoglycerate. Taking into account the substrate specificity of the protein, we designated the TK1777 protein product PPMTk.
Effect of metal cations, pH, and temperature on enzyme activity.
Purified PPMTk was dialyzed against 25 mM Tris-HCl buffer (pH 8.0) containing 5 mM EDTA and used for further analysis. Activity measurements were performed in a linked assay coupled with DERA and alcohol dehydrogenase. In the absence of metal ions, PPMTk did not display PPM activity, indicating that the activity was dependent on metal ions. At a concentration of 1 mM, the presence of Mg2+ led to the highest levels of enzyme activity (Fig. 3C). Besides Mg2+, we also found that Ni2+ (70%), Mn2+ (50%), and Zn2+ (45%) could support PPM activity, although to a lower extent.
We examined the effect of pH and temperature on PPM activity. At a fixed temperature of 90°C, PPMTk displayed maximal activity at pH 7.5. Under our assay methods, PPMTk displayed maximum activity at 90°C (Fig. 3B), with a specific activity of 210 ± 10 μmol min−1 mg−1. The half-life of the recombinant protein at 100°C was 90 min. We also carried out kinetic analysis of the PPM activity and found that the reaction followed Michaelis-Menten kinetics with the kinetic parameters shown in Table 1.
DERA and PPM activities in T. kodakaraensis KOD1.
Cell growth of T. kodakaraensis in the nutrient-rich MA-YT medium strictly requires the presence of elemental sulfur (S0) (2). However, in the presence of pyruvate or starch, T. kodakaraensis actively utilizes these substrates as carbon and energy sources, and the requirement for S0 is relieved (2). We inoculated T. kodakaraensis cells in MA-YT medium (without S0) supplemented with sodium pyruvate (0.2%), starch (0.2%), ribose (0.2%), deoxyribose (0.2%), or a mixture of 2′-deoxyguanosine and 2′-deoxycytidine (0.1% each). While rapid cell growth and high cell yields were observed in the presence of pyruvate or starch, no cell growth was observed with the other substrates.
We next grew T. kodakaraensis cells in a synthetic, minimal ASW-AA medium based on amino acids (23). In the presence of S0, this medium meets the minimal demands for cell growth. To the ASW-AA medium with S0, we added sodium pyruvate, starch, ribose, deoxyribose, or a mixture of 2′-deoxyguanosine and 2′-deoxycytidine at concentrations described above. Compared to the ASW-AA medium without supplementation of any substrate, we detected an increase in DERA and PPM activities when pyruvate (a gluconeogenic substrate) or starch (a glycolytic substrate) was present in the medium, with slightly higher levels observed with pyruvate (Table 2). Although bacterial DERA and PPM activities have been reported to be highly induced in the presence of deoxyribose, ribonucleosides, or exogenous DNA (5, 21, 26, 30), we could only detect slight increases of DERA and PPM activities in T. kodakaraensis, which were lower than those observed in cells grown with pyruvate or starch (Table 2).
TABLE 2.
Enzyme levels of DERA and PPM in T. kodakaraensis KOD1
| Carbon source(s) | Enzyme activity (μmol min−1 mg−1) of:
|
|
|---|---|---|
| DERA | PPM | |
| Amino acids | 0.25 ± 0.05 | 0.22 ± 0.02 |
| Amino acids + pyruvate (0.2%) | 0.52 ± 0.02 | 0.71 ± 0.04 |
| Amino acids + starch (0.2%) | 0.43 ± 0.03 | 0.63 ± 0.05 |
| Amino acids + ribose (0.2%) | 0.31 ± 0.04 | 0.22 ± 0.01 |
| Amino acids + 2-deoxyribose (0.2%) | 0.31 ± 0.04 | 0.38 ± 0.06 |
| Amino acids + dG (0.1%) + dC (0.1%)a | 0.38 ± 0.03 | 0.17 ± 0.01 |
dG, 2-deoxyguanosine; dC, 2-deoxycytidine.
DISCUSSION
In this study, we have detected PPM and DERA activities in the cells of the hyperthermophilic archaeon T. kodakaraensis and have identified the genes responsible for these activities. In particular, the PPM of T. kodakaraensis was a structurally novel enzyme, previously annotated as a phosphomannomutase in COG1109. The results clearly indicate a metabolic link between the ribose moiety of nucleosides and the glycolytic pathway in T. kodakaraensis. As we have also detected orthologues of various nucleoside phosphorylases on the T. kodakaraensis genome (unpublished data), the chances are high that nucleoside metabolism is also linked to central carbon metabolism via this pathway in this strain.
At present, we cannot conclude the physiological role of this pathway in T. kodakaraensis. However, as we could detect both PPM and DERA activities in cells grown under various conditions, the pathway seems to be of physiological importance. Growth experiments indicated that T. kodakaraensis could not utilize ribose, 2-deoxyribose, or nucleosides as carbon and energy sources. Further, the addition of ribose, 2-deoxyribose, or nucleosides to a minimal medium did not lead to a high induction of PPMTk and DERATk. These results suggest that T. kodakaraensis cannot assimilate these pentose compounds, possibly due to the incapability to uptake these compounds into the cells. Therefore, a biosynthetic role of these enzymes in generating pentoses can be postulated.
The pathways involved in pentose biosynthesis and degradation are not well known in hyperthermophilic archaea (8, 31). The usual pathway found in bacteria and eukaryotes is the pentose phosphate pathway, consisting of an oxidative branch and a nonoxidative branch. The oxidative branch, with enzymes such as glucose 6-phosphate dehydrogenase and gluconate 6-phosphate dehydrogenase, generates pentoses from hexoses. However, genes encoding these key enzymes have not been identified on the genomes of hyperthermophilic archaea (31), including T. kodakaraensis. Concerning the nonoxidative branch, genes presumed to encode several enzymes of this branch have been found in various hyperthermophilic archaea, including transketolase, ribose 5-phosphate isomerase, and ribulose 5-phosphate 3-epimerase (31), implying that this branch may be responsible for pentose biosynthesis in these organisms. DNA microarray analysis in P. furiosus supports this presumption, as two genes corresponding to subunits of putative transketolases, along with genes involved in histidine and aromatic amino acid synthesis, are up-regulated on maltose-based medium compared to peptide-based medium (25). Ribose 5-phosphate is a precursor for the synthesis of these amino acids, and a coinciding induction of these genes suggests that the transketolases are involved in providing the pentose precursor. As these orthologues are commonly observed on the T. kodakaraensis genome, the same mechanism may also be utilized in this strain. However, the results of this study have indicated that, at least in this organism, there exists an additional pathway capable of producing pentoses from C2 and C3 carbon compounds.
The PPM of T. kodakaraensis was a structurally novel enzyme and belonged to COG1109, and not COG1015, in which most bacterial PPMs are included. This implies that some of the proteins within COG1109 that have previously been annotated as phosphomannomutases or related phosphohexomutases in various genome sequences may indeed be PPMs. Sequence alignment of various phosphohexomutases and the structure of phosphoglucomutase from rabbits (7) reveal three conserved sequence motifs that are necessary for enzyme activity (32). The sequence TXSHNP contains the active-site serine residue, the DXDXDR motif is involved in metal binding, and the sequence GEXS participates in sugar binding. Interestingly, although the first two motifs are found in PPMTk, the third motif involved in substrate binding is absent. This may reflect the similar enzyme (mutase) activities and the distinct substrate specificities of the phosphohexomutases and PPMs.
We also examined the presence of PPM and DERA in other archaeal strains. A database search of sequences from hyperthermophilic archaea reveals the presence of more than one gene that is annotated as a phosphomannomutase on each genome. For example, P. furiosus, P. abyssi, and P. horikoshii have three orthologues of COG1109, whereas two are found on the Methanococcus jannaschii and A. pernix genomes. Similarly, the T. kodakaraensis genome harbors four open reading frames, including TK1777. Among the four orthologues in T. kodakaraensis, three were very closely related to those of the Pyrococcus strains, enabling us to distinguish the corresponding genes in each organism. Interestingly, TK1777 proved to be the unique orthologue present only in T. kodakaraensis and not in the Pyrococcus species. In terms of DERA, we found that very few genomes of hyperthermophilic archaea harbor a DERA orthologue and were found only in A. pernix and Pyrobaculum aerophilum (Table 3). Orthologues were found in some moderately thermophilic archaea, including Methanobacterium thermoautotrophicum and Thermoplasma strains. None was found in the closely related Pyrococcus genomes, consistent with the absence of the PPM gene in these strains. This observation and the relatively rare presence of DERA genes suggest that the pathway found in T. kodakaraensis may not be widely distributed among (hyperthermophilic) archaea. At this stage, we cannot determine whether the genes noted as phosphomannomutases in P. aerophilum and A. pernix correspond to the PPM identified in this study. However, we would like to note that among the two orthologues in A. pernix, APE2433 displays significantly higher similarity to PPMTk than the other orthologue, APE0317. Experimental analyses of PPM and DERA in other hyperthermophilic organisms will help to determine the distribution of this pathway as well as to better understand pentose biosynthesis in the Archaea. Another important point that needs to be clarified is the specific function(s) of the multiple phosphomannomutase orthologues found in the various genomes of hyperthermophilic archaea.
TABLE 3.
Presence of orthologue genes involved in pentose catabolism and biosynthesis in various thermophilic and hyperthermophilic archaeaa
| Organism | Locus for:
|
|||
|---|---|---|---|---|
| DERA | PPM | Thymidine phosphorylase | Purine nucleoside phosphorylases | |
| Crenarchaeota | ||||
| Aeropyrum pernix | APE2437 | − | − | APE0993, APE2105 |
| Pyrobaculum aerophilum | PAE1231 | − | − | PAE1476, PAE3111 |
| Sulfolobus solfataricus | − | − | − | SSO1519, SSO2706 |
| Sulfolobus tokodaii | − | − | − | ST0975, ST2449 |
| Euryarchaeota | ||||
| Archaeoglobus fulgidus | − | − | AF1341, AF1342 | − |
| Halobacterium sp. strain NRC-1 | VNG1859G | − | − | VNG0893G, VNG1850G |
| Methanobacterium thermoautotrophicum | MTH818 | − | − | − |
| Methanococcus jannaschii | − | − | MJ0667 | − |
| Methanosarcina acetivorans | − | − | MA3242 | − |
| Methanosarcina mazei | − | − | MM0087 | − |
| Methanopyrus kandleri | − | − | − | − |
| Pyrococcus abyssi | − | − | PAB1982 | − |
| Pyrococcus furiosus | − | − | PF1607 | − |
| Pyrococcus horikoshii | − | − | PH1598 | − |
| Thermococcus kodakaraensis | +b | −c | + | + |
| Thermoplasma acidophilum | Ta0684 | − | − | − |
| Thermoplasma volcanium | TVN0175 | − | − | − |
Sequences with high similarity to the DERA, PPM, thymidine phosphorylase, and purine nucleoside phosphorylase of E. coli were examined. Locus names are indicated when orthologue genes were found in the genome sequences. The absence of orthologues is indicated by −. The presence of orthologues in T. kodakaraensis is indicated by +.
Enzyme activity was confirmed in this study.
A novel PPM was identified in this study.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Atomi, H., T. Fukui, T. Kanai, M. Morikawa, and T. Imanaka. Description of Thermococcus kodakaraensis sp. nov., a well studied hyperthermophilic archaeon previously reported as Pyrococcus sp. KOD1. Archaea, in press. [DOI] [PMC free article] [PubMed]
- 3.Barsky, D. L., and P. A. Hoffee. 1983. Purification and characterization of phosphopentomutase from rat liver. Biochim. Biophys. Acta 743:162-171. [DOI] [PubMed] [Google Scholar]
- 4.Barsotti, C., M. G. Tozzi, and P. L. Ipata. 2002. Purine and pyrimidine salvage in whole rat brain. Utilization of ATP-derived ribose-1-phosphate and 5-phosphoribosyl-1-pyrophosphate generated in experiments with dialyzed cell-free extracts. J. Biol. Chem. 277:9865-9869. [DOI] [PubMed] [Google Scholar]
- 5.Barth, P. T., I. R. Beacham, S. I. Ahmad, and R. H. Pritchard. 1968. The inducer of the deoxynucleoside phosphorylases and deoxyriboaldolase in Escherichia coli. Biochim. Biophys. Acta 161:554-557. [DOI] [PubMed] [Google Scholar]
- 6.Blom, N., and J. Sygusch. 1997. Product binding and role of the C-terminal region in class I d-fructose 1,6-bisphosphate aldolase. Nat. Struct. 4:36-39. [DOI] [PubMed] [Google Scholar]
- 7.Dai, J. B., Y. Liu, W. J. Ray, Jr., and M. Konno. 1992. The crystal structure of muscle phosphoglucomutase refined at 2.7-angstrom resolution. J. Biol. Chem. 267:6322-6337. [PubMed] [Google Scholar]
- 8.Galperin, M. Y., L. Aravind, and E. V. Koonin. 2000. Aldolases of the DhnA family: a possible solution to the problem of pentose and hexose biosynthesis in archaea. FEMS Microbiol. Lett. 183:259-264. [DOI] [PubMed] [Google Scholar]
- 9.Groth, D. P., and N. Jiang. 1966. The role of deoxyribose 5-phosphate aldolase in the synthesis of deoxyribonucleotide in mammalian cells. Biochem. Biophys. Res. Commun. 22:62-68. [DOI] [PubMed] [Google Scholar]
- 10.Heine, A., G. DeSantis, J. G. Luz, M. Mitchell, C. H. Wong, and I. A. Wilson. 2001. Observation of covalent intermediates in an enzyme mechanism at atomic resolution. Science 294:369-374. [DOI] [PubMed] [Google Scholar]
- 11.Hofmann, M., E. Boles, and F. K. Zimmermann. 1994. Characterization of the essential yeast gene encoding N-acetylglucosamine-phosphate mutase. Eur. J. Biochem. 221:741-747. [DOI] [PubMed] [Google Scholar]
- 12.Imanaka, H., T. Fukui, H. Atomi, and T. Imanaka. 2002. Gene cloning and characterization of fructose-1,6-bisphosphate aldolase from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Biosci. Bioeng. 94:237-243. [DOI] [PubMed] [Google Scholar]
- 13.Imanaka, T., and H. Atomi. 2002. Catalyzing “hot” reactions: enzymes from hyperthermophilic archaea. Chem. Rec. 2:149-163. [DOI] [PubMed] [Google Scholar]
- 14.Karbownik, M., H. Modrzejewska, R. J. Reiter, K. Zasada, J. Greger, and A. Lewinski. 2000. Pyrimidine and purine salvage deoxyribonucleoside metabolism in hepatic and renal homogenates from rats pretreated with propylthiouracil or l-thyroxine. Neuroendocrinol. Lett. 21:51-55. [PubMed] [Google Scholar]
- 15.Karlström, O. 1968. Mutants of Escherichia coli defective in ribonucleoside and deoxyribonucleoside catabolism. J. Bacteriol. 95:1069-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marsh, J. J., and H. G. Lebherz. 1992. Fructose-bisphosphate aldolases: an evolutionary history. Trends Biochem. Sci. 17:110-113. [DOI] [PubMed] [Google Scholar]
- 17.Mascia, L., M. Cappiello, S. Cherri, and P. L. Ipata. 2000. In vitro recycling of α-d-ribose 1-phosphate for the salvage of purine bases. Biochim. Biophys. Acta 1474:70-74. [DOI] [PubMed] [Google Scholar]
- 18.Morikawa, M., Y. Izawa, N. Rashid, T. Hoaki, and T. Imanaka. 1994. Purification and characterization of a thermostable thiol protease from a newly isolated hyperthermophilic Pyrococcus sp. Appl. Environ. Microbiol. 60:4559-4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Munch-Petersen, A. 1970. Deoxyribonucleoside catabolism and thymine incorporation in mutants of Escherichia coli lacking deoxyriboaldolase. Eur. J. Biochem. 15:191-202. [DOI] [PubMed] [Google Scholar]
- 20.Mura, U., F. Sgarrella, and P. L. Ipata. 1978. Utilization of exogenous purine compounds in Bacillus cereus. Translocation of the ribose moiety of inosine. J. Biol. Chem. 253:7905-7909. [PubMed] [Google Scholar]
- 21.Robertson, B. C., P. Jargiello, J. Blank, and P. A. Hoffee. 1970. Genetic regulation of ribonucleoside and deoxyribonucleoside catabolism in Salmonella typhimurium. J. Bacteriol. 102:628-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakuraba, H., H. Tsuge, I. Shimoya, R. Kawakami, S. Goda, Y. Kawarabayasi, N. Katunuma, H. Ago, M. Miyano, and T. Ohshima. 2003. The first crystal structure of archaeal aldolase. Unique tetrameric structure of 2-deoxy-d-ribose-5-phosphate aldolase from the hyperthermophilic archaea Aeropyrum pernix. J. Biol. Chem. 278:10799-10806. [DOI] [PubMed] [Google Scholar]
- 23.Sato, T., T. Fukui, H. Atomi, and T. Imanaka. 2003. Targeted gene disruption by homologous recombination in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Bacteriol. 185:210-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schuch, R., A. Garibian, H. H. Saxild, P. J. Piggot, and P. Nygaard. 1999. Nucleosides as a carbon source in Bacillus subtilis: characterization of the drm-pupG operon. Microbiology 145:2957-2966. [DOI] [PubMed] [Google Scholar]
- 25.Schut, G. J., S. D. Brehm, S. Datta, and M. W. W. Adams. 2003. Whole-genome DNA microarray analysis of a hyperthermophile and an archaeon: Pyrococcus furiosus grown on carbohydrates or peptides. J. Bacteriol. 185:3935-3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sgarrella, F., F. P. A. Poddie, M. A. Meloni, L. Sciola, P. Pippia, and M. G. Tozzi. 1997. Channelling of deoxyribose moiety of exogenous DNA into carbohydrate metabolism: role of deoxyriboaldolase. Comp. Biochem. Physiol. B 117:253-257. [DOI] [PubMed] [Google Scholar]
- 27.Stribling, D., and R. N. Perham. 1973. Purification and characterization of two fructose diphosphate aldolases from Escherichia coli (Crookes' strain). Biochem. J. 131:833-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomson, G. J., G. J. Howlett, A. E. Ashcroft, and A. Berry. 1998. The dhnA gene of Escherichia coli encodes a class I fructose bisphosphate aldolase. Biochem. J. 331:437-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tozzi, M. G., F. Sgarrella, D. Barsacchi, and P. L. Ipata. 1984. Induction of deoxyribose-5-phosphate aldolase of Bacillus cereus by deoxyribonucleosides. Biochem. Int. 9:319-325. [PubMed] [Google Scholar]
- 31.Verhees, C. H., S. W. M. Kengen, J. E. Tuininga, G. J. Schut, M. W. W. Adams, W. M. de Vos, and J. van der Oost. 2003. The unique features of glycolytic pathways in Archaea. Biochem. J. 375:231-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Videira, P. A., L. L. Cortes, A. M. Fialho, and I. Sá-Correia. 2000. Identification of the pgmG gene, encoding a bifunctional protein with phosphoglucomutase and phosphomannomutase activities, in the gellan gum-producing strain Sphingomonas paucimobilis ATCC 31461. Appl. Environ. Microbiol. 66:2252-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]