Abstract
Mycoplasma mobile glides on surfaces at up to 7 μm/s by an unknown mechanism. We studied the energetics that power gliding by using a novel, growth medium-free system. We found that cells could glide in defined media if the glass substrate is preconditioned by exposure to horse serum. The active component that potentiates gliding is sensitive to proteinase K treatment. We used the defined medium system to test the effect of various inhibitors, ionophores, and poisons on motility of M. mobile. Valinomycin, carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP), N,N′-dicyclohexylcarbodiimide, phenamil, amiloride, rifampin, and puromycin had no short-term effects on gliding. We also confirmed that we were able to modulate the membrane potential with valinomycin and FCCP by using a potential-sensitive dye. Shifting the pH likewise had no effect on motility. These results rule out the use of conventional ion motive forces to power gliding. Arsenate had a dramatic inhibitory effect on gliding, and both the speed and the fraction of cells moving tracked ATP levels. Sodium orthovanadate had a slight but significant inhibitory effect on gliding. Taken together, these results suggest that the motor system of M. mobile is likely an ATPase or is directly coupled to an ATPase.
While studies of locomotion in swimming bacteria are well advanced, investigations of gliding motility remain comparatively limited. Yet gliding motility, defined as a smooth translocation over a solid surface, is represented frequently throughout the eubacterial phylogenetic tree and in some instances has been associated with pathogenicity (25). Even several species of mycoplasmas, some of the simplest bacteria known in terms of size and genomic content, are known to perform gliding motility (14, 32).
The mycoplasmas are wall-less bacteria characterized by small physical dimensions and genome sizes (32). Among the mycoplasmas, the fish pathogen Mycoplasma mobile demonstrates extremely robust gliding motility (16, 34). M. mobile is one of the flask-shaped mycoplasmas (approximately 1.0 × 0.3 μm) and has a genome of approximately 780 kbp (4). It has always been observed to glide in the direction of the “head” (corresponding to the tapered end of the cell) without reversals or pauses at speeds of up to 7 μm/s (34). It can tow an erythrocyte, roughly 10 times its size, without significant loss in speed and has been measured to exert up to 27 pN of force (28, 33). Some recent progress at uncovering the molecular mechanism of gliding in M. mobile has been made, including localization of the gliding apparatus to the head region of its flask-like cell body and isolation of mutants with altered gliding phenotypes (29, 30, 41). However, little is known about the prerequisites or energy source for gliding in M. mobile.
Flagellated bacteria use ion motive forces to power their flagellar motors. For instance, Escherichia coli uses a proton motive force (ΔpH), while various Vibrio species use a sodium motive force (ΔpNa) to drive their polar (but not lateral) flagella (3, 18, 19, 24). The energy source for motility has been investigated in some genera of gliding bacteria as well. In the case of the Flavobacteria, gliding motility appears to be powered by ΔpH, while the social motility system of the Myxobacteria relies on type IV pili and, therefore, ATP hydrolysis (25).
The mycoplasmas seem to lack any form of respiration and generate ATP through fermentation of sugars and substrate-level phosphorylation (32). It is known that mycoplasmas can generate a transmembrane potential (ΔΨ) ranging from −28 to −48 mV (negative inside the cell) and a ΔpH ranging from −52 to −72 mV (37). Therefore, it would seem that mycoplasmas could use either an ion motive force or direct utilization of ATP to power motility. We set out to develop a medium-free system in which we could study gliding of M. mobile on glass and determine its energy source.
MATERIALS AND METHODS
Reagents.
Heart infusion broth and yeast extract were from Becton Dickinson (Sparks, Md.). 3,3′-dipropylthiadicarbocyanine iodide (DiSC3[5]) was from Molecular Probes (Eugene, Oreg.). The ENLITEN ATP measurement system was from Promega (Madison, Wis). All other reagents were from Sigma-Aldrich (St. Louis, Mo.). Water was 18 MΩ deionized (dH2O).
Strains.
M. mobile strain 163K (ATCC 43663) was grown to an optical density at 600 nm (OD600) of 0.07 to 0.10 in plastic tissue culture flasks at 22°C in Aluotto medium consisting of 10% inactivated horse serum, 2.1% beef heart infusion broth, and 0.56% yeast extract adjusted to pH 7.8 and supplemented with 50 mg of ampicillin/liter and 250 mg of thallium acetate/liter (1).
Preparation of coverslips.
Circular glass coverslips were subjected to the following sequence of treatments (all at room temperature with gentle agitation): 10 min in saturated ethanolic KOH, four 5-min changes in dH2O, 15 min in inactivated horse serum, and three 5-min changes in dH2O. The coverslips were then left to dry in a laminar flow hood and stored at room temperature until use, resulting in a preparation that was stable for at least 4 weeks. Note that fetal bovine serum can also be used with equal effectiveness.
Protease treatment.
Prepared coverslips were digested overnight with 20 mg of proteinase K/ml (or dH2O as a control) at 42°C in a humid environment and washed with four 5-min changes in dH2O.
Buffers.
The following buffers were used: phosphate-buffered saline (PBS; 150 mM NaCl, 50 mM sodium phosphate [pH 8.0]), PBS/G (PBS [pH 8.0] plus 10 mM glucose), PBS-K/G (140 mM NaCl, 10 mM KCl, 50 mM sodium phosphate, pH 8.0 [or other pH as specified], 10 mM glucose), ArBS-K/G (140 mM NaCl, 7.5 mM KCl, 47.5 mM sodium arsenate, 2.5 mM potassium arsenate [pH 8.0], 10 mM glucose), and valinomycin buffer (100 mM NaCl, 50 mM KCl, 50 mM sodium phosphate [pH 8.0], 10 mM glucose).
Motility assay.
Comparisons were made of gliding speeds of cells in a given buffer and cells in the same buffer containing the compound to be tested, referred to as control buffer and test buffer, respectively. Cells (diluted to an OD600 of 0.025 in fresh medium [1 ml]) were centrifuged at room temperature for 10 min at 10,000 × g, washed in 1 ml of control buffer or test buffer, and centrifuged again in the same manner. The final pellet was suspended in 50 μl of the control buffer or test buffer. Six microliters of this suspension was pipetted onto a prepared coverslip and incubated at room temperature for 15 min in a humid environment. During that time, a flow chamber (5) was assembled with a spare coverslip ringed with Apiezon-L grease (GEC-Alsthom Ltd., Manchester, United Kingdom) and equilibrated by continuous flow with control or test buffer for at least 10 min. Coverslips with adhered cells were then ringed with grease and inverted onto the flow chamber. All observations were made at room temperature (∼22°C). Control buffer or test buffer was drawn into the flow chamber at the rate of 100 μl/min for 10 min to remove nonadherent cells, and then the motion of the remaining cells was recorded (with the flow left on). Cells were visualized with a Nikon Optiphot microscope equipped with a 60× oil immersion phase-contrast objective, a Zeiss optovar set to 1.25×, and a 10× projection lens. Recordings were made with a charge-coupled device camera connected to a Mini-DV recorder (Sony GV-D1000, New York, N.Y.). In the standard assay, 10-s recordings were made every 10 min over the course of 30 min. Time zero was defined to occur at the time of the first recording. Video was captured to a computer with Adobe (San Jose, Calif.) Premier software at a rate of 10 frames/s. The speed of gliding mycoplasmas was computed for frames 40 to 59 of each 10-s (100-frame) time point by using a particle tracking program written by Darnton et al. (8). Slight modifications were made to Darnton's program to output the proper statistics (the MATLAB scripts are available at http://www.rowland.org/labs/bacteria/index.html). Speeds of 30 to 50 cells were measured at each time point in each experiment. The compound being tested was considered to have a significant effect on motility only if a comparison of the test and control populations by a two-tailed Student's t test had a P value of <0.05 at all time points studied.
pH shift.
Cells were prepared in PBS-K/G (pH 8.0) and then shifted to PBS-K/G at the desired pH after the first recording was taken. The second recording was taken at t = 5 min.
Arsenate and ATP.
Measurements of motility parameters and ATP levels were made in parallel by slightly modifying the standard motility assay. Eleven 1-ml aliquots of cells at an OD600 of 0.025 were washed in PBS-K/G as described above. Cells in one tube were resuspended in 50 μl of PBS-K/G and used for the flow chamber experiments where they were treated as described above, while the remaining 10 tubes were washed an additional time in PBS-K/G to simulate the wash experienced by the cells in the flow chamber. At time zero of the experiment, the buffer feeding the flow chamber was either switched to ArBS-K/G or allowed to remain in PBS-K/G in the case of the control. Concurrently, each of the remaining 10 tubes was resuspended in 100 μl of ArBS-K/G or PBS-K/G, depending on the experiment. Ten-second recordings were made every 10 min as was done before, but the time course was extended to 90 min because there was no preincubation with test buffer. At each time point, one tube of cells was extracted for ATP by adding 2 μl of trichloroacetic acid to the tube and vortexing and then adding 12 μl of 1.5 M Tris base to neutralize the acid (12). The sample was then diluted with 342 μl of 5 mM Tris buffer (pH 7.75) and frozen at −20°C until the ATP assay was performed. In some cases, further dilution of the sample with 5 mM Tris (pH 7.75) was necessary to get the sample within the range of the assay standard curve. For the ATP assay, 10 μl of each sample was added to a glass vial, and then 100 μl of ENLITEN rL/L reagent was added to the tube and mixed by repeated pipetting. Measurements were taken exactly 15 s after the addition of the rL/L reagent in a home-built luminometer consisting of a photomultiplier tube connected to a current-to-voltage converter and a chart recorder (27). Two measurements were made for each sample and were compared to a standard curve of ATP constructed from duplicated measurements of samples of ATP ranging from 31.3 to 500 fmol.
Membrane potential.
Cells were grown as described above, and a sufficient amount of culture was centrifuged at 10,000 × g such that resuspension in 2 ml of test buffer resulted in an OD600 of 0.175 to 0.200. This suspension was placed in a cuvette, and DiS-C3-[5] was added from an EtOH stock to a final concentration of 150 nM (final EtOH, 0.05%). Fluorescence was monitored at excitation and emission wavelengths of 625 and 667 nm, respectively, in a Fluoromax fluorimeter (Spex, Edison, N.J.) equipped with a magnetic stirring cell. The dye was allowed to partition into the cells until a stable signal was obtained (4 to 5 min), and then the test substance (valinomycin, carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone [FCCP], or N,N′-dicyclohexylcarbodiimide [DCCD]) was added. Changes in the fluorescence signal were observed within a few seconds of the addition of the test substance if changes were observed at all.
Isolation of proteins that bind to glass from horse serum.
Approximately 100 μl of glass beads (425 to 600 μm in diameter) were prepared in exactly the same way as the coverslips described above. After drying, the beads were boiled in Laemmli sodium dodecyl sulfate-polyacrylamide gel electrophoresis buffer (36). The resulting sample was loaded to a sodium dodecyl sulfate-4 to 12% polyacrylamide gel electrophoresis gradient gel (Invitrogen, Carlsbad, Calif.) and subjected to electrophoresis. Major bands were excised and subjected to in-gel tryptic digestion and identification by ion trap mass spectrometry (13).
RESULTS
Conditioning of glass.
In pilot experiments for this project, we noticed that M. mobile cells would glide on glass surfaces if they were pelleted by centrifugation and then simply resuspended in a pure buffer test medium (e.g., PBS/G). However, when an intervening wash step was included, cells would adhere to glass in aggregates but would not glide. Evidently, in the former case, an essential component from the medium that potentiates gliding was carried over from the pellet, while in the latter case, the extra wash step eliminated the component. We tested the major components of the growth medium (horse serum, beef heart infusion, and yeast extract) by adding each one back to a washed culture and found that only horse serum restored motility. We subsequently found that a simple pretreatment of glass with fresh horse serum was enough to potentiate the motility of washed cells. Therefore, the factor that potentiates gliding on glass surfaces is native to horse serum and is not a factor produced by mycoplasmas in culture. Subsequent to this discovery, we adopted the coverslip pretreatment method described in Materials and Methods.
Identity of conditioning factor.
We subjected pretreated coverslips to overnight incubation with either proteinase K or water. The coverslips were then washed with water to eliminate the protease so that gliding could be tested. We found that coverslips treated overnight with proteinase K no longer facilitated gliding motility of a washed culture, whereas the control coverslips treated with water alone still potentiated gliding of mycoplasmas. However, the coverslips treated with proteinase K still allowed adherence of cells to the glass with approximately the same number of cells stuck as is found in the control. We therefore conclude that a protein or combination of proteins either directly or indirectly mediates the ability of M. mobile cells to glide on glass surfaces.
As albumin is the most abundant protein known in serum, we pretreated coverslips as described above using a solution of bovine serum albumin (1 mg/ml) in place of the horse serum; note that the pretreatment of coverslips with fetal bovine serum also potentiated gliding. This type of pretreatment did not potentiate gliding of washed cells, so a protein or proteins other than albumin are probably involved in conditioning the glass surface. We attempted to isolate this component(s) by identifying the major species that bound to glass from horse serum. Mass spectrometry was able to identify albumin (as expected), immunoglobulin, and casein as the top three proteins bound to glass beads. However, attempts to coat glass coverslips with solutions of these proteins (1 mg/ml each) did not potentiate motility. Rather, immunoglobulin and casein abolished cytadherence of the cells, while albumin caused clumps of nonmotile cells to adhere to the glass surface. The actions of these proteins in combination have not yet been tested.
Gliding of M. mobile in artificial media.
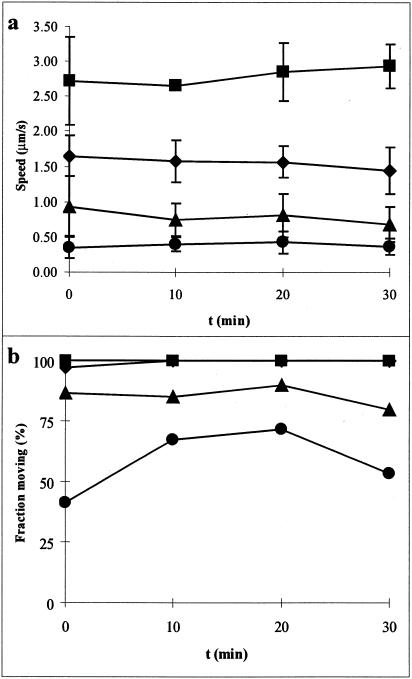
The development of the pretreatment procedure for glass coverslips allowed us to study motility in artificial media (e.g., simple buffer systems). Movies of M. mobile gliding can be seen at http://www.rowland.org/labs/bacteria/showmovie.php?mov=mycoplasma_mobile. A comparison of the basic buffers used in subsequent experiments is shown in Fig. 1. While none of the artificial media used resulted in gliding speeds as rapid as those in growth medium (Fig. 1a), most media supported motility of >80% of the cells (Fig. 1b). The omission of glucose from the medium caused a substantial decrease in both speed and percentage of cells that were moving. The sole source of ATP generation in most mycoplasmas is through substrate-level phosphorylation during glycolysis, although some mycoplasmas can use arginine as an energy source (32). This indicates that ATP synthesis may play a key role in the energetics of mycoplasma motility. We found that sucrose could also be used in place of glucose as an energy source in motility experiments (data not shown), indicating the ability of M. mobile to metabolize sucrose.
FIG. 1.
Gliding in growth and artificial media. (a) Gliding speed of motile cells. Only cells moving at speeds greater than 0.25 μm/s are considered. Error bars represent one standard deviation. (b) Percentage of cells moving at speeds greater than 0.25 μm/s. Symbols: ▪, growth medium; ♦, PBS-K/G; ▴, PBS/G; •, PBS.
Gliding speeds of mycoplasma in medium containing Na+ and glucose (PBS/G) were consistently around 1 μm/s, but the cells appeared thin and elongated. The inclusion of a small amount of K+ (10 mM KCl) in the medium caused a 33 to 50% increase in gliding speed, and the cells looked more like those in growth medium. However, the addition of too much K+ (>100 mM) caused cells to swell and become coccus shaped, which inhibited motility but not glass binding (data not shown). Mycoplasmas are known to selectively accumulate K+ to over 200 mM against a gradient (37). This fact, combined with these observations on gliding in artificial media of different ionic compositions, suggests that K+ plays an important role in osmoregulation and shape determination in M. mobile and, in turn, that osmoregulation and shape determination are important factors in gliding motility.
We also tested the ability of cells to glide in a medium in which almost all cations were completely impermeant to the membrane (200 mM choline chloride, 200 μM HEPES [pH 7.9], 10 mM glucose, and final Na+ of about 160 μM due to titration with NaOH). We found that >80% of cells glided in this medium, achieving speeds of up to 2.6 ± 0.46 μm/s. However, gliding cells tended to fall off of the glass surface after a short period of observation (10 min) for reasons that are not yet understood.
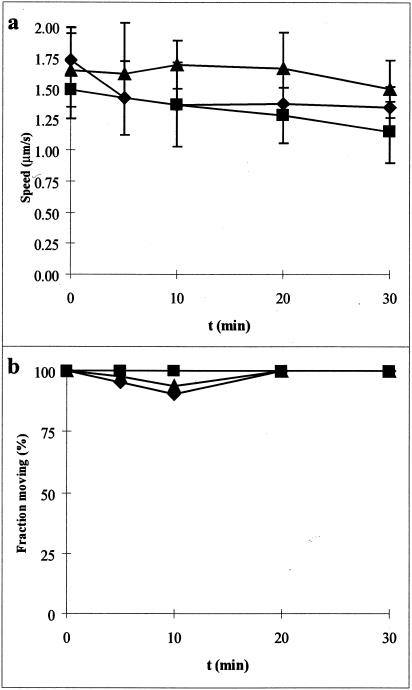
Effects of external pH.
If a proton motive force is involved in gliding, changing the ΔpH might have an effect on motility. We prepared all cells in pH 8.0 buffer and then subsequently shifted them to a lower pH in the flow chamber. Shifting the external pH of the medium did not have a significant effect on speed or percentage of motile cells (Fig. 2).
FIG. 2.
Effect of shifting pH on gliding motility. All samples were prepared in PBS-K/G (pH 8.0) and then shifted to the indicated pH at t = 1 min. (a) Gliding velocity; (b) fraction of cells moving. Symbols: ▪, pH 8; ♦, pH 7; ▴, pH 6.
Effects of ionophores, inhibitors, and poisons.
We examined the effects of several compounds known to interfere with various cellular processes that have been shown to be involved in the motility of other organisms. In all cases, motility and the percentage of motile cells were observed over a 30-min period and compared to a carefully matched buffer control that did not contain the compound in question. Buffer formulations are listed in Table 1.
TABLE 1.
Buffer formulations used in this studya
| Test substance (concn in buffer) | Mode of action | Base buffer | Solvent (%) | Matched control buffer |
|---|---|---|---|---|
| Valinomycin (10 μM) | K+ ionophore | Valinomycin buffer | MeOH (0.1) | Valinomycin buffer + 0.1% MeOH |
| FCCP (10 μM) | Protonophore | PBS/G | MeOH (0.1) | PBS/G + 0.1% MeOH |
| Arsenate (50 μM) | ATP poison | ArBS-K/G | Water | PBS-K/G |
| DCCD (10 μM) | F0F1 ATPase inhibitor | PBS/G | EtOH (0.025) | PBS/G + 0.025% EtOH |
| Sodium orthovanadate (1 mM) | P-type ATPase inhibitor | PBS-K/G | Water | PBS-K/G |
| Phenamil (10 μM) | Sodium channel blocker | PBS/G | MeOH (0.1) | PBS/G + 0.1% MeOH |
| Amiloride (10 μM) | Sodium channel blocker | PBS/G | Water | PBS/G |
| Rifampin (5 μg/ml) | RNA synthesis inhibitor | PBS/G | MeOH (0.1) | PBS/G + 0.1% MeOH |
| Puromycin (20 μM) | Protein translation inhibitor | PBS/G | Water | PBS/G |
Stock solutions were made of the test substance (for non-water-soluble compounds) in the solvent listed. The stock solution was then added to the base buffer, resulting in the final solvent concentration shown, where applicable. Abbreviations: MeOH, methanol; EtOH, ethanol.
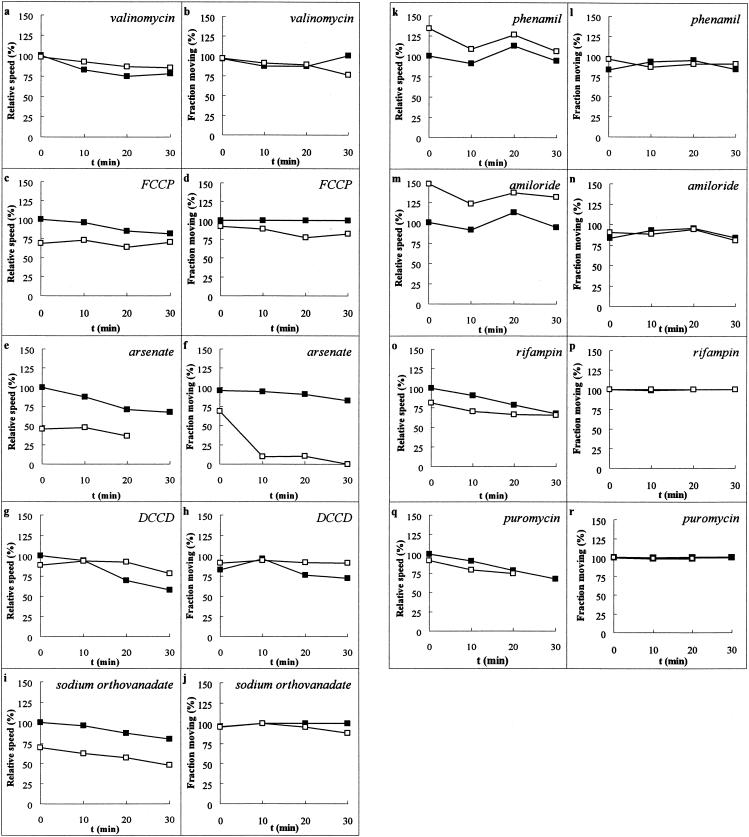
The addition of the potassium ionophore valinomycin to cells in the presence of high external K+ would be expected to clamp ΔΨ at a lower-than-normal value (37, 38). Valinomycin had no significant effect on motility compared to the control (Fig. 3a and b), even in the presence of 50 mM external potassium. This result was interesting, considering that high amounts of potassium caused cells to become coccoid and stop gliding (see above). However, at the end of the 30-min observation period, nearly all cells were still moving at a rate comparable to that of the matched control. Similarly, the protonophore FCCP did not cause a significant decrease in either gliding speed or percentage of cells that were gliding (Fig. 3c and d). FCCP is reported to abolish ΔpH in various mycoplasmas (37), and the inclusion of FCCP in the medium would be expected to short circuit any proton-driven motor by providing an alternate conduction path for protons and driving ΔpH toward 0.
FIG. 3.
Effect of ionophores, inhibitors, and poisons on gliding. Various agents were added (Table 1), and gliding was compared to that of a matched control. The graphs on the left-hand side of each pair show the speed of the cells that were moving at speeds greater than 0.25 μm/s relative to the initial speed of the cells in the matched control. The graphs on the right-hand side of each pair show the fraction of cells moving. (a and b) 10 μM valinomycin; (c and d) 10 μM FCCP; (e and f) 10 mM arsenate (note that no 30′ test time point is shown [e] because no cells were moving at speeds greater than 0.25 μm/s); (g and h) 10 μM DCCD; (i and j) 1 mM sodium orthovanadate; (k and l) 10 μM phenamil; (m and n) 10 μM amiloride; (o and p) 5 μg of rifampin/ml; (q and r) 20 μM puromycin (note that the 30-min test time point was not taken due to technical difficulties). Filled squares are matched control preparations. Open squares are test preparations. Error bars are omitted for clarity, but standard deviations in speeds were generally ±25%.
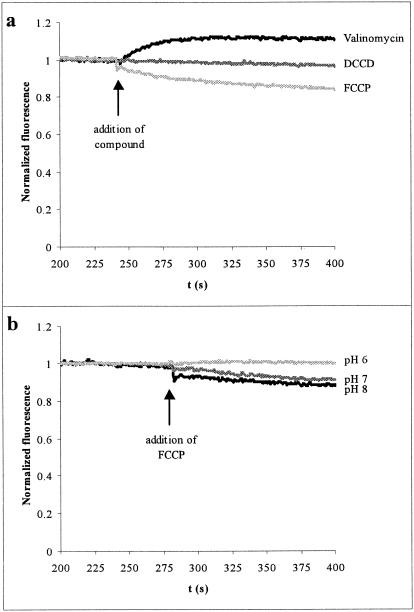
We studied the effect of valinomycin and FCCP on the membrane potential (ΔΨ) of M. mobile by using the positively charged fluorescent indicator DiSC3[5]. This compound partitions between the cell and the external medium at higher intracellular concentrations when the inside of the cell is more negative. At higher concentrations, the dye dimerizes or aggregates, quenching the fluorescence (44, 45). Thus, a decrease in fluorescence indicates membrane hyperpolarization. Under the conditions of the gliding experiment, the addition of valinomycin to the cells caused a depolarization of the membrane, while the addition of FCCP caused a slight hyperpolarization of the membrane (Fig. 4a). These shifts occurred within seconds after addition of either compound, so their effects must have been manifested during the gliding experiments.
FIG. 4.
Effect of selected substances on membrane potential. Cell suspensions were incubated with DiSC3[5] for 4 to 5 min, and then the indicated compound was added. Data are shown from 200 to 400 s of the experiment and are normalized to the average of data from the first 50 s of observations in the time period shown. Downward deflections indicate hyperpolarization. Upward deflections indicate depolarization. (a) Effect of 3 μM valinomycin, 5 μM FCCP, or 20 μM DCCD in PBS-K/G (pH 8.0). The valinomycin preparation contained 50 mM KCl to match the gliding experiments. (b) Effect of FCCP at various pHs of PBS-K/G.
Mycoplasmas are expected to have an internal K+ of about 200 mM (37). The addition of valinomycin in the presence of 50 mM external K+ (the concentration used in the gliding experiment) would be expected to clamp ΔΨ at RT/F ln (50 mM K+ext/200 mM K+int) = −35 mV, where R is the gas constant, T is the absolute temperature, and F is the Faraday. We believe that the normal ΔΨ of M. mobile is greater than −76 mV because an analogous experiment that we performed with 10 mM external K+ still induced depolarization upon the addition of valinomycin. Thus, in the gliding experiment, ΔΨ was reduced by more than a factor of two.
The experiment also shows that FCCP affects ΔΨ as well as ΔpH (37). The sign and magnitude of the shift in ΔΨ depends on the external pH. At a higher pH (pH 7.0 or 8.0), FCCP caused a hyperpolarization, while at a lower pH (pH 6.0), FCCP caused a depolarization (Fig. 4b). Gliding motility was not affected by external pH in this range (Fig. 2). Therefore, by employing valinomycin and FCCP, we were able to collapse both ΔΨ and ΔpH (and therefore modulate ΔpH) without significant effect on gliding motility. Thus, the proton motive force does not power gliding in M. mobile.
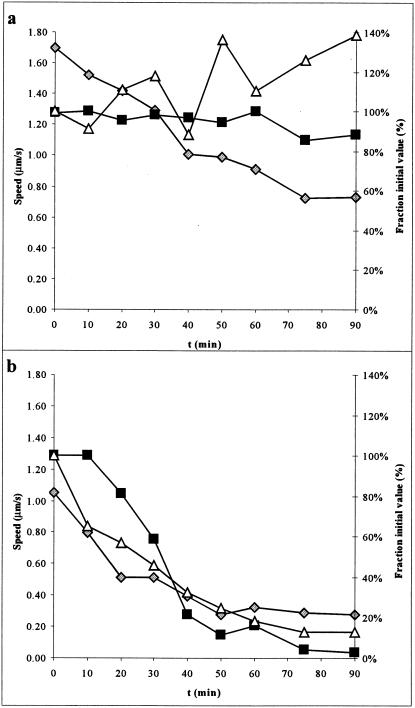
Arsenate ions compete with phosphate ions in substrate-level phosphorylation of ADP, and the resulting ADP-As molecule rapidly hydrolyzes (43). Therefore, we replaced the phosphate in our buffer with arsenate to study its effect. Arsenate caused a rapid and statistically significant drop in both the speed of gliding and the percentage of cells moving in the experiment (Fig. 3e and f). The drop in speed and fraction of cells moving is likely due to a specific effect of arsenate rather than simply the removal of phosphate since we were able to observe motility for extended periods of time in Tris- or TES [N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid]-based buffers (data not shown). Presumably, the cells have a phosphate or ATP reserve that is rapidly abolished in the presence of arsenate. To further examine the effect of arsenate, we measured both ATP levels and gliding parameters from the onset of arsenate addition over an extended period of time. We found that both gliding speed and fraction of cells moving tracked the levels of ATP present in the cell (correlation coefficient, r2 = 0.94 and 0.88, respectively) (Fig. 5). Thus, ATP must be an important energy source for gliding motility in M. mobile.
FIG. 5.
Effect of arsenate on gliding motility and ATP levels. Parallel preparations were assayed for motility and ATP. (a) PBS-K/G (pH 8.0); (b) ArBS-K/G (50 mM arsenate). Filled squares indicate the fraction of cells moving (right axis), gray diamonds indicate gliding speed (left axis), and open triangles indicate the fraction of ATP remaining compared to the value at t = 0 (right axis). Note that error bars have been omitted for clarity. Standard deviations in speeds were generally ±20%, while standard deviations for ATP values were <5%.
Given the demonstrated importance of ATP for motility, we tested several ATPase inhibitors to see whether they might be involved in gliding motility. DCCD is an inhibitor of the F0F1-type proton translocating ATPase and is also reported to collapse ΔpH in some mycoplasmas (21, 22, 37). This compound had no effect on gliding motility (Fig. 3g and h) and lends further evidence that the proton motive force does not drive motility. The P-type Na+ translocating ATPase inhibitor sodium orthovanadate caused a small but statistically significant decrease in gliding speed but did not reduce the percentage of cells gliding in a meaningful way (Fig. 3i and j) (39). To further examine the role of Na+ gradients in gliding motility, we tested the sodium channel blockers phenamil and amiloride. Both of these compounds can abolish swimming motility in Vibrio species that utilize a sodium motive force to power flagellar rotation (2, 18). Neither of these compounds affected gliding motility in M. mobile (Fig. 3k to n), and it is therefore also unlikely that a sodium motive force (ΔpNa) drives motility in M. mobile in the same manner that Vibrio species utilize ΔpNa.
Finally, we tested to see whether short-term biosynthesis of protein was required for motility. We thought that this might be a reasonable possibility given that some microorganisms are reported to move by continuous extrusion of biomolecules or by continuous shedding of a protein trail during gliding motility (11, 40). We attempted to shut off transcription or translation in separate experiments. The transcriptional inhibitor rifampin prevents RNA synthesis, while the protein synthesis inhibitor puromycin mimics aminoacyl-tRNA and aborts translation (23). These compounds had no effect on motility over the 30-min span of the experiment (Fig. 3o to r).
DISCUSSION
While there have been many remarkable observations of gliding motility of M. mobile, the source of energy for this phenomenon has not been convincingly demonstrated (15, 16, 28-30, 33-35). One study did examine the effects of various substances on M. mobile gliding (including some which overlap those discussed here), but that effort was directed at longer-term effects and did not specifically address the issue of energetics (31). We studied the energy source by employing techniques and substances that were useful for investigating motility in many swimming bacteria. We hoped that elucidation of the energy source could guide further experimentation as to the mechanism of gliding motility.
We surmised that the energy for gliding motility in M. mobile might by supplied by an ion motive force (such as ΔpH or ΔpNa) or that it could come directly from the hydrolysis of ATP. Many swimming bacteria (such as E. coli) utilize ΔpH as the source of power for motility, although some (such as Vibrio alginolyticus) use ΔpNa (24, 26). While mycoplasmas are certainly capable of and in fact do generate a proton motive force, it does not seem to supply the energy for gliding motility in this case. Rather, our observations suggest that motility is directly powered by the hydrolysis of ATP. Of the various compounds that we studied, arsenate was the only chemical that we found that had a dramatically significant effect on motility. We were able to successfully manipulate ΔΨ and ΔpH of the cell but without effect on motility. However, we did observe a strong real-time correlation between ATP levels, gliding speed, and percentage of cells moving. We note that changes in ATP levels should eventually affect ΔΨ and ΔpH; however, this effect would occur on a time scale much longer than that for the changes induced by the addition of valinomycin or FCCP (compare Fig. 5b and 4a). We propose that the motor apparatus in M. mobile, which has yet to be identified, is most likely an ATPase or is directly coupled to an ATPase. This idea is strengthened by our observation that sodium orthovanadate caused a small but significant decrease in gliding speed. Vanadyl (VO43−) ions are well-known inhibitors of P-type Na+/K+ translocating ATPases from a variety of organisms and are usually effective at concentrations 10- to 100-fold lower than those used here (6, 7). The ATPase motor system of M. mobile might be marginally sensitive to vanadate or might be derived from a P-type ATPase. This proposal may be helpful in targeting protein classes in M. mobile for further study with respect to gliding. Alternatively, it has been demonstrated that some sugar transport ABC-type ATPases are sensitive to vanadate, and the effect of vanadate in this experiment might be simply to limit the supply of fermentable sugar inside the cell (20).
M. mobile is also remarkable for its robust gliding motility compared to that in other mycoplasmas (14). Here, we show that motility in M. mobile is facilitated in part by protein components from the growth medium that condition glass surfaces to enable gliding. While our attempts to specifically identify the protein component by mass spectrometry were unsuccessful, we suspect that the essential protein(s) might act as a surfactant, allowing transient cytadherence that would be necessary for gliding motility. Lipids might also be involved.
The mechanism of gliding motility in bacteria remains a mystery, and it is likely that several different schemes are employed in different genera. Variations on the basic mechanism might exist even within the closely grouped mycoplasmas. For instance, several large proteins implicated in surface adherence and motility in M. mobile have been identified (41). Mycoplasma pneumoniae, another gliding mycoplasma with a similar flask-shaped morphology, lacks any orthologs of these proteins. It is possible that there are different effectors of gliding motility even in these two closely related organisms. Unfortunately, genetic manipulations have not yet been possible in M. mobile despite much success with mutagenesis techniques in other mycoplasmas (9, 10, 17, 42). Perhaps the upcoming report of the genome sequence of M. mobile combined with the power of comparative genomics will help to shed light on the mechanism(s) of gliding in the mycoplasmas (J. Jaffe et al., submitted for publication). Ultimately, it will be important to understand this new class of motor and its mechanism at the molecular level.
Acknowledgments
We express our gratitude to Woody Hastings for the use of his luminometer and fluorimeter.
This work was supported by NIH grant number AI16478 to H.C.B.
There are no known competing financial interests involved in this work.
REFERENCES
- 1.Aluotto, B. B., R. G. Wittler, C. O. Williams, and J. E. Faber. 1970. Standardized bacteriologic techniques for the characterization of Mycoplasma species. Int. J. Syst. Bacteriol. 20:35-58. [Google Scholar]
- 2.Atsumi, T., Y. Maekawa, H. Tokuda, and Y. Imae. 1992. Amiloride at pH 7.0 inhibits the Na(+)-driven flagellar motors of Vibrio alginolyticus but allows cell growth. FEBS Lett. 314:114-116. [DOI] [PubMed] [Google Scholar]
- 3.Atsumi, T., L. McCarter, and Y. Imae. 1992. Polar and lateral flagellar motors of marine Vibrio are driven by different ion-motive forces. Nature 355:182-184. [DOI] [PubMed] [Google Scholar]
- 4.Bautsch, W. 1988. Rapid physical mapping of the Mycoplasma mobile genome by two-dimensional field inversion gel electrophoresis techniques. Nucleic Acids Res. 16:11461-11467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berg, H. C., and S. M. Block. 1984. A miniature flow cell designed for rapid exchange of media under high-power microscope objectives. J. Gen. Microbiol. 130:2915-2920. [DOI] [PubMed] [Google Scholar]
- 6.Cantley, L. C., Jr., L. Josephson, R. Warner, M. Yanagisawa, C. Lechene, and G. Guidotti. 1977. Vanadate is a potent (Na,K)-ATPase inhibitor found in ATP derived from muscle. J. Biol. Chem. 252:7421-7423. [PubMed] [Google Scholar]
- 7.Cantley, L. C., Jr., M. D. Resh, and G. Guidotti. 1978. Vanadate inhibits the red cell (Na+, K+) ATPase from the cytoplasmic side. Nature 272:552-554. [DOI] [PubMed] [Google Scholar]
- 8.Darnton, N., L. Turner, K. Breuer, and H. C. Berg. 2004. Moving fluid with bacterial carpets. Biophys. J. 86:1863-1870. [DOI] [PMC free article] [PubMed]
- 9.Dhandayuthapani, S., W. G. Rasmussen, and J. B. Baseman. 1999. Disruption of gene mg218 of Mycoplasma genitalium through homologous recombination leads to an adherence-deficient phenotype. Proc. Natl. Acad. Sci. USA 96:5227-5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hedreyda, C. T., K. K. Lee, and D. C. Krause. 1993. Transformation of Mycoplasma pneumoniae with Tn4001 by electroporation. Plasmid 30:170-175. [DOI] [PubMed] [Google Scholar]
- 11.Hoiczyk, E., and W. Baumeister. 1998. The junctional pore complex, a prokaryotic secretion organelle, is the molecular motor underlying gliding motility in cyanobacteria. Curr. Biol. 8:1161-1168. [DOI] [PubMed] [Google Scholar]
- 12.Karl, D. M. 1980. Cellular nucleotide measurements and applications in microbial ecology. Microbiol. Rev. 44:739-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kinter, M., and N. Sherman. 2000. Protein sequencing and identification using tandem mass spectrometry. Wiley Interscience, New York, N.Y.
- 14.Kirchhoff, H. 1992. Motility, p. 289-306. In J. Maniloff (ed.), Mycoplasmas: molecular biology and pathogenesis. American Society for Microbiology, Washington, D.C.
- 15.Kirchhoff, H., U. Boldt, R. Rosengarten, and A. Klein-Struckmeier. 1987. Chemotactic response of a gliding mycoplasma. Curr. Microbiol. 15:57-60. [Google Scholar]
- 16.Kirchhoff, H., and R. Rosengarten. 1984. Isolation of a motile mycoplasma from fish. J. Gen. Microbiol. 130:2439-2445. [DOI] [PubMed] [Google Scholar]
- 17.Knudtson, K. L., and F. C. Minion. 1993. Construction of Tn4001lac derivatives to be used as promoter probe vectors in mycoplasmas. Gene 137:217-222. [DOI] [PubMed] [Google Scholar]
- 18.Kojima, S., T. Atsumi, K. Muramoto, S. Kudo, I. Kawagishi, and M. Homma. 1997. Vibrio alginolyticus mutants resistant to phenamil, a specific inhibitor of the sodium-driven flagellar motor. J. Mol. Biol. 265:310-318. [DOI] [PubMed] [Google Scholar]
- 19.Kojima, S., K. Yamamoto, I. Kawagishi, and M. Homma. 1999. The polar flagellar motor of Vibrio cholerae is driven by an Na+ motive force. J. Bacteriol. 181:1927-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landmesser, H., A. Stein, B. Bluschke, M. Brinkmann, S. Hunke, and E. Schneider. 2002. Large-scale purification, dissociation and functional reassembly of the maltose ATP-binding cassette transporter (MalFGK2) of Salmonella typhimurium. Biochim. Biophys. Acta 1565:64-72. [DOI] [PubMed] [Google Scholar]
- 21.Linker, C., and T. H. Wilson. 1985. Cell volume regulation in Mycoplasma gallisepticum. J. Bacteriol. 163:1243-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linker, C., and T. H. Wilson. 1985. Sodium and proton transport in Mycoplasma gallisepticum. J. Bacteriol. 163:1250-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madigan, M., J. Martinko, and J. Parker. 2003. Brock biology of microorganisms, 10th ed. Prentice Hall, Upper Saddle River, N.J.
- 24.Manson, M. D., P. Tedesco, H. C. Berg, F. M. Harold, and C. Van der Drift. 1977. A protonmotive force drives bacterial flagella. Proc. Natl. Acad. Sci. USA 74:3060-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McBride, M. J. 2001. Bacterial gliding motility: multiple mechanisms for cell movement over surfaces. Annu. Rev. Microbiol. 55:49-75. [DOI] [PubMed] [Google Scholar]
- 26.McCarter, L. L. 2001. Polar flagellar motility of the Vibrionaceae. Microbiol. Mol. Biol. Rev. 65:445-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitchell, G. W., and J. W. Hastings. 1971. A stable, inexpensive, solid-state photomultiplier photometer. Anal. Biochem. 39:243-250. [DOI] [PubMed] [Google Scholar]
- 28.Miyata, M., W. S. Ryu, and H. C. Berg. 2002. Force and velocity of Mycoplasma mobile gliding. J. Bacteriol. 184:1827-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyata, M., and A. Uenoyama. 2002. Movement on the cell surface of the gliding bacterium, Mycoplasma mobile, is limited to its head-like structure. FEMS Microbiol. Lett. 215:285-289. [DOI] [PubMed] [Google Scholar]
- 30.Miyata, M., H. Yamamoto, T. Shimizu, A. Uenoyama, C. Citti, and R. Rosengarten. 2000. Gliding mutants of Mycoplasma mobile: relationships between motility and cell morphology, cell adhesion and microcolony formation. Microbiology 146:1311-1320. [DOI] [PubMed] [Google Scholar]
- 31.Piper, B., R. Rosengarten, and H. Kirchhoff. 1987. The influence of various substances on the gliding motility of Mycoplasma mobile 163K. J. Gen. Microbiol. 133:3193-3198. [DOI] [PubMed] [Google Scholar]
- 32.Razin, S., D. Yogev, and Y. Naot. 1998. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 62:1094-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosengarten, R., M. Fischer, H. Kirchhoff, G. Kerlen, and K.-H. Seack. 1988. Transport of erythrocytes by gliding cells of Mycoplasma mobile 163K. Curr. Microbiol. 16:253-257. [Google Scholar]
- 34.Rosengarten, R., and H. Kirchhoff. 1987. Gliding motility of Mycoplasma sp. nov. strain 163K. J. Bacteriol. 169:1891-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosengarten, R., A. Klein-Struckmeier, and H. Kirchhoff. 1988. Rheotactic behavior of a gliding mycoplasma. J. Bacteriol. 170:989-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Schiefer, H. G., and U. Schummer. 1982. The electrochemical potential across mycoplasmal membranes. Rev. Infect. Dis. 4(Suppl.):S65-S70. [DOI] [PubMed] [Google Scholar]
- 38.Schummer, U., and H. G. Schiefer. 1983. Electrophysiology of mycoplasma membranes. Yale J. Biol. Med. 56:413-418. [PMC free article] [PubMed] [Google Scholar]
- 39.Shirvan, M. H., and S. Rottem. 1993. Ion pumps and volume regulation in mycoplasmas, p. 261-292. In S. Rottem and I. Kahane (ed.), Subcellular biochemistry: mycoplasma cell membranes. Plenum Press, New York, N.Y. [DOI] [PubMed]
- 40.Stewart, M. J., and J. P. Vanderberg. 1988. Malaria sporozoites leave behind trails of circumsporozoite protein during gliding motility. J. Protozool. 35:389-393. [DOI] [PubMed] [Google Scholar]
- 41.Uenoyama, A., A. Kusumoto, and M. Miyata. 2004. Identification of a 349-kilodalton protein (Gli349) responsible for cytadherence and glass binding during Mycoplasma mobile gliding. J. Bacteriol. 186:1537-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Voelker, L. L., and K. Dybvig. 1996. Gene transfer in Mycoplasma arthritidis: transformation, conjugal transfer of Tn916, and evidence for a restriction system recognizing AGCT. J. Bacteriol. 178:6078-6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Voet, D., and J. G. Voet. 1990. Biochemistry, 2nd ed. John Wiley & Sons, New York, N.Y.
- 44.Waggoner, A. S. 1979. Dye indicators of membrane potential. Annu. Rev. Biophys. Bioeng. 8:47-68. [DOI] [PubMed] [Google Scholar]
- 45.Waggoner, A. S. 1979. The use of cyanine dyes for the determination of membrane potentials in cells, organelles, and vesicles. Methods Enzymol. 55:689-695. [DOI] [PubMed] [Google Scholar]