Abstract
The genome of the Neodiprion sertifer nucleopolyhedrovirus (NeseNPV), which infects the European pine sawfly, N. sertifer (Hymenoptera: Diprionidae), was sequenced and analyzed. The genome was 86,462 bp in size. The C+G content of 34% was lower than that of the majority of baculoviruses. A total of 90 methionine-initiated open reading frames (ORFs) with more than 50 amino acids and minimal overlapping were found. From those, 43 ORFs were homologous to other baculovirus ORFs, and 29 of these were from the 30 conserved core genes among all baculoviruses. A NeseNPV homolog to the ld130 gene, which is present in all other baculovirus genomes sequenced to date, could not be identified. Six NeseNPV ORFs were similar to non-baculovirus-related genes, one of which was a trypsin-like gene. Only one iap gene, containing a single BIR motif and a RING finger, was found in NeseNPV. Two NeseNPV ORFs (nese18 and nese19) were duplicates transcribed in opposite orientations from each other. NeseNPV did not have an AcMNPV ORF 2 homolog characterized as the baculovirus repeat ORF (bro). Six homologous regions (hrs) were located within the NeseNPV genome, each containing small palindromes embedded within direct repeats. A phylogenetic analysis was done to root the tree based upon the sequences of DNA polymerase genes of NeseNPV, 23 other baculoviruses, and other phyla. Baculovirus phylogeny was then constructed with 29 conserved genes from 24 baculovirus genomes. Culex nigripalpus nucleopolyhedrovirus (CuniNPV) was the most distantly related baculovirus, branching to the hymenopteran NeseNPV and the lepidopteran nucleopolyhedroviruses and granuloviruses.
Viruses in the family Baculoviridae are large, rod-shaped, and enveloped and have circular double-stranded DNA genomes. They are the largest and most widely studied group of insect viruses. This family is subdivided into two genera, the granuloviruses (GVs), with small occlusion bodies that generally contain a single virion, and the nucleopolyhedroviruses (NPVs), with occlusion bodies that include multiple virions. The most widely studied group of baculoviruses is the lepidopteran NPV, which has been divided into groups I and II since early phylogenetic studies using the polyhedrin gene (81). The subdivision of NPVs I and II has been maintained when all the conserved genes in the baculoviruses have been used in phylogenetic analysis (32, 60). The Neodiprion sertifer nucleopolyhedrovirus (NeseNPV) infects the European pine sawfly from the order Hymenoptera. It has single nucleocapsids within an envelope, like the other NPVs that infect nonlepidopteran insects (22).
To date, a total of 23 baculovirus genomes are available in GenBank, some of which are close variants. From these, 22 infect insects from the order Lepidoptera and 1 infects mosquitoes from the order Diptera. The comparative study of these baculovirus genomes reflects their diversity, with a size range from 99,657 bp in Adoxophyes orana GV (AdorGV) (78) to 178,733 bp in Xestia c-nigrum GV (XcGV) (30); number of open reading frames (ORFs) from 109 in Culex nigripalpus NPV (CuniNPV) (1) to 181 in XcGV; number of homologous regions (hrs) from 17 in Spodoptera litura NPV (SpltMNPV) (65) to none in Cydia pomenella GV (CpGV) and Adoxophyes orana GV (AdorGV) (53, 78). The number of conserved genes reported among lepidopteran baculoviruses varies, as more baculovirus genomes became available (31). However, when CuniNPV is included in the analysis, the number is reduced to 30 genes (32).
NeseNPV has been used to control the European pine sawfly, N. sertifer, one of the most harmful insects to the coniferous forests of the northern hemisphere (41). Sawfly larvae live gregariously and resemble caterpillars. Originally from Europe, N. sertifer was accidentally brought to North America around 1925. Before the concern of viral registration, NeseNPV was successfully used by foresters, Christmas tree growers, and private landowners. Later, the virus was registered as Neochek-S and became available in the United States for use against the European pine sawfly (34). Nucleopolyhedroviruses of sawflies have been reported in 25 species (34). These viruses are highly host specific and, in contrast to most lepidopteran NPVs, replicate only in the epithelial cells of the larval midgut (22). Infection by the virus results in a reduction of larval feeding and, eventually, complete suppression of appetite long before death.
Few genes of NeseNPV had been previously sequenced (Liu and Maruniak, unpublished data; GenBank accession no. AF121349). Rohrmann et al. (72) published a description of the N-terminal sequence of the NeseNPV polyhedrin protein. The complete polyhedrin amino acid sequence was later obtained (Harris and Possee, unpublished data) and used in phylogenetic studies that placed NeseNPV as the outgroup baculovirus (11, 81).
Insight into the origin and subsequent evolution of the family Baculoviridae can be obtained by comparing the similarities and differences in the genomic contents and gene distribution of distantly related viruses. The comparison may also elucidate some properties of the baculovirus morphogenesis, structure, or genomic information that are essential for effective propagation and infectivity. In this work, we describe the genome sequence and the genetic organization of the hymenopteran baculovirus Neodiprion sertifer NPV.
MATERIALS AND METHODS
Virus, DNA purification, cloning, and sequencing.
Neodiprion sertifer NPV was obtained as a crude preparation of Neochek-S stocks from James Slavicek and John Podgewaite. The composition of this preparation was approximately 0.05% polyhedral inclusion bodies of NeseNPV and 99.95% milled N. sertifer insect parts. NeseNPV DNA was purified from this preparation as described by Maruniak (55). The dry preparation was homogenized in a solution containing 10 mM Tris, 1 mM EDTA, 2% sodium dodecyl sulfate (SDS), and 1% ascorbic acid. After centrifugation at 10,000 × g for 10 min, the pellet was resuspended in 0.5 M NaCl and then rinsed with water by another cycle of homogenization and centrifugation. The sample, resuspended in water, was layered on a 40 to 63% continuous sucrose gradient. The polyhedra that banded at approximately 55% was disrupted with a dilute alkaline solution (0.1 M sodium carbonate, 0.17 M NaCl, 0.01 M EDTA) and layered on a 40 to 56% continuous sucrose gradient. The band, formed at the top quarter of the tube, containing the single nucleocapsid virions, was treated with 1% SDS and 1 mg/ml proteinase K. After two phenol and three ether extractions, the viral DNA was dialyzed against TE (0.01 M Tris, 0.001 M EDTA). Approximately 4 μg of DNA was obtained per gram of Neochek-S preparation. The viral DNA was digested with HindIII and EcoRI separately and cloned in pGEM 7Z plasmid (Promega). This resulted in the construction of NeseNPV HindIII and a partial EcoRI libraries. Initially, the sequencing of each NeseNPV clone was done using the T7 and SP6 primers from the vector. Then, EZ::TN transposons (Epicentre) were inserted in the clones to randomly distribute primer binding sites (forward and reverse) present in the transposon. Colonies carrying the antibiotic resistance trait from the transposons were screened using several restriction endonucleases (RENs) to ensure that only one transposon was inserted per clone and to map its location within the viral fragment. This approach facilitated the assembly of problematic regions, such as highly repetitive sequences or those suspected of having secondary structures. Specific primers were synthesized, and additional sequences were obtained by primer walking to fill in gaps. Other strategies to sequence the genome comprised subcloning and PCR amplification of some regions. Sequencing reactions were done in a ThermoCycler machine (MJ Research) with the ABI PRISM BigDye Terminator sequencing ready reaction kit (Applied Biosystems) and sent to the University of Florida Interdisciplinary Center for Biotechnology Research to be run in an automated DNA sequencer.
Multiplex PCR.
Because no clonal isolate of NeseNPV was available, a multiplex PCR procedure was used to arrange the cloned fragments, to determine if the whole genome was cloned, and to detect genome variants. The physical map of the NeseNPV DNA was assembled with a modification of the optimized multiplex PCR method (74). The genomic viral DNA was used as a template for PCRs that would allow the correct order of the clones in the genome to be determined after sequencing the PCR products. Using the initial sequence data from all the HindIII clones, unique primers were synthesized within the first 300 bp that annealed with the DNA fragment toward the end of the fragment (end-out primers). The primers were first tested by sequencing the corresponding clone. The primers were then combined in groups of three per pool in such a way that primers from opposite ends of the same fragment were together in the same pool. All possible pairs of pool combination were used in PCRs using the genomic NeseNPV DNA as the template under the following conditions: 1.5 mM MgCl2, 0.5 mM deoxynucleoside triphosphates, 1 ng of genomic NeseNPV DNA, 10 pmol of each primer, 1 U of Elongase (Invitrogen), for a final 25-μl reaction volume. Cycling conditions were those described by Moraes and Maruniak (59). Two microliters of each PCR product were electrophoresed on a 0.7% agarose gel. When no PCR products were obtained, it meant that none of the six primers from the two pools tested amplified the DNA under those conditions and, hence, their corresponding fragments were in the wrong orientation or too distantly separated in the genome to amplify the DNA. When one DNA band was obtained, the PCR product was sequenced using the pooled primers previously used for the PCR amplification. When one DNA band was seen in all the lanes containing a particular pool of primers, the common pool of primers was used to sequence that PCR product. If more than one DNA band was obtained in a single lane, the pools involved were further divided in groups of two primers each and the PCR was repeated. The sequences obtained were compared to previous data from the HindIII library. Overlapping sequences permitted the location of the correct order of the HindIII fragments on the NeseNPV physical map. The order of the HindIII fragments was further confirmed by repeating the PCR amplification using the genomic DNA as template and only the primer pair from adjacent fragments identified by the multiplex method.
DNA sequence analysis.
The complete NeseNPV genome was assembled with the Sequencher 4.1 program (Gene Codes) using the sequence data from every individual clone and the multiplex PCR experiments. Every base of each DNA strand was sequenced a minimum of three times; however, due to the fact that the location of every sequence had been previously determined, either by transposon mapping or by primer walking, the assembly process was not random. The ORFs encoding 50 amino acids or more and showing minimum overlapping, as has been the criteria for baculoviruses (5), were found using the ORF Finder program (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) (77). BLASTP 2.2.6 (http://www.ncbi.nlm.nih.gov/BLAST) (3), conserved domain database (CDD) (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) search, run by default in parallel with BLASTP and PSI-BLAST (4) programs and Smith-Waterman similarity search (http://coe02.ucalgary.ca/algo-sw/sw_aa.shtml), were used to compare all of the possible ORFs to the amino acid sequence database. The following criteria were used to accept NeseNPV ORFs as baculovirus homologs: (i) During BLASTP comparison, an E value of 0.1 or less was accepted; (ii) when the BLAST list showed similarity with related ORFs from several baculoviruses, it was considered a NeseNPV homolog even if the E value of some members of the list were higher than 0.1; (iii) if an hr overlapped with an ORF, that ORF was not considered for the analysis. For NeseNPV ORFs that showed random matches with other baculovirus ORFs (represented by an E value over 0.1), forced alignments were performed to reveal if there was significant homology not evident by BLAST or Smith-Waterman similarity search (http://coe02.ucalgary.ca/). This was also done with NeseNPV ORFs that had signal peptides and transmembrane domains in order to search for an ld130 homolog candidate. CLUSTALW (http://www.ebi.ac.uk/clustalw/) was used for this purpose and for several other amino acid alignments. Some ORF overlapping was permitted, especially when both ORFs were homologous to other baculoviruses. The putative coding regions were numbered as NeseNPV ORFs. The sequences 160 nucleotides upstream of each ORF start codon were analyzed for early and late transcriptional motifs. Late motifs contained the sequence (A/G/T)TAAG (71). Early motifs were defined as either TATA alone (designated “e” in Table 1) or TATA with the CA(T/G)T sequence, 20 to 40 nucleotides downstream (“E” in Table 1). To find possible transmembrane domains in the NeseNPV putative proteins, the TMHMM server 2.0 was used (http://www.cbs.dtu.dk/services/TMHMM-2.0). To examine the NeseNPV ORFs for N-terminal signal peptides, SignalP 2.0 prediction server (61) was used (http://www.cbs.dtu.dk/services/SignalP). Tandem Repeats Finder (http://c3.biomath.mssm.edu/trf.html) was used to locate the hr's (6). The alignment parameters used were 2, 7, and 7 for match, mismatch, and indels, respectively, a minimum alignment score of 50 to report repeats, and a maximum period size (repeat size) of 500. Several programs from the Genetics Computer Group (Wisconsin Package Version 10.2) (18) were used to locate the palindrome sequences of NeseNPV hr's and to determine the G+C ratio of each hr and of the entire NeseNPV genome
TABLE 1.
NeseNPV ORFsa
| ORF | NeseSNPV
|
AcMNPV
|
LdMNPV
|
SpltMNPV
|
MacoMNPV-A
|
HzSNPV
|
CuniSNPV
|
CpGV
|
Best match with NsSNPV, % ID (score) total no. of aa | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Position | aa | Prom | Gene | ORF | Identity | ORF | Identity | ORF | Identity | ORF | Identity | ORF | Identity | ORF | Identity | ORF | Identity | ||
| 1 | 1 > 741 | 246 | L | polyhedrin | 8 | 49 (236) 245 | 1 | 48 (235) 245 | 1 | 53 (235) 249 | 1 | 52 (233) 246 | 1 | 50 (236) 246 | 1 | 38 (235) 248 | NeabNPV polh, 92 (239) 246 | ||
| 2 | 896 > 1483 | 195 | E | ||||||||||||||||
| 3 | 1512 < 2678 | 388 | |||||||||||||||||
| 4 | 2798 > 2968 | 56 | e | ||||||||||||||||
| 5 | 3089 > 4402 | 437 | e, L | mtase1 | 69 | 33 (178) 262 | 65 | 32 (189) 313 | 111 | 32 (186) 275 | 65 | 36 (195) 274 | HzSNPV orf 65, 36 (195) 274 | ||||||
| 6 | 4512 > 4853 | 113 | |||||||||||||||||
| 7 | 5338 > 6114 | 258 | E, L | Trypsin-like protein | |||||||||||||||
| 8 | 6222 > 6830 | 202 | e, L | ||||||||||||||||
| 9 | 7068 < 8318 | 416 | |||||||||||||||||
| 10 | 8390 < 8560 | 56 | |||||||||||||||||
| 8604 < 9147 | hr1 | ||||||||||||||||||
| 11 | 9277 > 10662 | 461 | E | ||||||||||||||||
| 11066 11859 | hr2 | ||||||||||||||||||
| 12 | 12144 < 12536 | 130 | L | ||||||||||||||||
| 13 | 12729 < 13169 | 146 | e | ||||||||||||||||
| 14 | 13364 < 13540 | 58 | E, L | ||||||||||||||||
| 15 | 13557 < 13721 | 54 | e | ||||||||||||||||
| 16 | 14281 > 16194 | 637 | e | [dr1]@ | |||||||||||||||
| 17 | 16442 < 16987 | 181 | L | iap | 27 | 30 (179) 286 | 139 | 26 (165) 155 | 64 | 139 | 36 (175) 276 | 106 | 34 (173) 268 | 17 | 38 (179) 275 | CpGV iap-3, 38 (179) 275 | |||
| 17045 17803 | hr3 | ||||||||||||||||||
| 18 | 18125 < 19030 | 301 | |||||||||||||||||
| 19265 19352 | dr2 | ||||||||||||||||||
| 19424 20131 | hr4 | ||||||||||||||||||
| 19 | 20513 > 21436 | 307 | |||||||||||||||||
| 21698 22093 | hr5 | ||||||||||||||||||
| 20 | 22307 > 23119 | 270 | e, L | ||||||||||||||||
| 21 | 23274 < 23543 | 89 | e, L | ||||||||||||||||
| 22 | 23886 < 24641 | 251 | e | dbp | 25 | 22 (194) 316 | 47 | 19 (101) 257 | 30 | 155 | 22 (201) 328 | 24 | 81 | 23 (121) 290 | AdhoSNPV dbp, 21 (252) 322 | ||||
| 23 | 24631 < 24933 | 100 | e | lef-11 | 37 | 45 | 34 | 24 (93) 144 | 149 | 32 (93) 120 | 32 | 30 (99) 127 | 58 | MacoMNPV-B lef-11, 33 (97) 124 | |||||
| 24 | 24940 < 25698 | 252 | e, L | p33 | 92 | 29 (187) 259 | 94 | 23 (218) 251 | 83 | 26 (232) 255 | 96 | 31 (218) 252 | 83 | 28 (251) 254 | 14 | 31 (136) 370 | 93 | 31 (178) 251 | MacoNPV-B orf 95, 31 (218) 252 |
| 25 | 25970 > 26482 | 170 | E, L | 93 | 95 | 84 | 29 (82) 182 | 95 | 22 (83) 161 | 84 | 26 (82) 162 | 92 | 25 (108) 161 | SpltMNPV orf 84, 29 (82) 182 | |||||
| 26 | 26562 > 27218 | 218 | L | ||||||||||||||||
| 27 | 27470 > 27643 | 57 | |||||||||||||||||
| 28 | 28065 < 30806 | 913 | E | dnapol | 65 | 28 (894) 984 | 83 | 26 (872) 1014 | 69 | 27 (892) 1022 | 115 | 27 (911) 998 | 69 | 28 (948) 1020 | 91 | 22 (719) 1138 | 111 | 28 (894) 1051 | MacoNPV-B dna-pol, 27 (913) 1000 |
| 29 | 30805 > 33183 | 792 | E | ||||||||||||||||
| 30 | 33256 > 33492 | 78 | E | ||||||||||||||||
| 31 | 33701 < 34900 | 399 | e | alk-exo | 133 | 27 (431) 419 | 157 | 26 (306) 420 | 109 | 28 (372) 408 | 54 | 27 (397) 395 | 117 | 25 (416) 428 | 54 | 28 (232) 367 | 125 | 26 (387) 398 | PxGV alk-exo, 29 (401) 378 |
| 32 | 34907 > 35614 | 235 | E | 106 | 140 | 24 (154) 246 | 101 | 23 (154) 236 | 71 | 21 (218) 216 | 104 | 52 | LdMNPV orf 140, 24 (154) 246 | ||||||
| 33 | 35632 > 36840 | 402 | e, L | p45 | 103 | 21 (386) 387 | 104 | 19 (400) 389 | 93 | 20 (306) 373 | 83 | 19 (399) 377 | 94 | 18 (397) 377 | 83 | 18 (387) 439 | AdorGV orf 69, 20 (410) 396 | ||
| 34 | 36824 > 37177 | 117 | L | ||||||||||||||||
| 35 | 37205 > 38311 | 368 | e, L | p40 | 101 | 102 | 91 | 85 | 92 | 85 | XcGV orf 93, 20 (383) 372 | ||||||||
| 36 | 38369 > 38629 | 86 | e, L | p6.9 | 100 | 101 | 90 | 86 | 91 | 23 | 86 | AdorGV p6.9, 56 (55) 55 | |||||||
| 37 | 38732 < 39133 | 133 | E | ||||||||||||||||
| 38 | 39165 > 40184 | 339 | E | odv-e56 | 148 | 37 (346) 376 | 14 | 39 (346) 356 | 17 | 36 (350) 371 | 6 | 35 (347) 373 | 15 | 38 (339) 354 | 102 | 34 (266) 361 | 18 | 37 (351) 355 | LdMNPV odv-e56, 39 (346) 356 |
| 39 | 40232 > 40810 | 192 | E, L | ||||||||||||||||
| 40 | 40815 < 42338 | 507 | e, L | lef-9 | 62 | 35 (484) 516 | 64 | 35 (487) 496 | 59 | 38 (481) 498 | 124 | 34 (490) 505 | 56 | 37 (484) 519 | 59 | 20 (407) 590 | 117 | 34 (491) 499 | HzNPV lef-9, 37 (484) 519 |
| 41 | 42280 < 42723 | 147 | e, L | 68 | 29 (104) 192 | 80 | 66 | 33 (109) 132 | 112 | 32 (121) 121 | 66 | 33 (103) 133 | 58 | 27 (103) 137 | 114 | 28 (114) 126 | AdorGV orf 97, 36 (80) 125 | ||
| 42 | 42884 > 44218 | 444 | e | ||||||||||||||||
| 43 | 44220 < 44597 | 125 | E, L | ||||||||||||||||
| 44 | 44599 < 44847 | 82 | e, L | ||||||||||||||||
| 45 | 44850 < 45911 | 353 | e | vlf-1 | 77 | 31 (333) 379 | 86 | 29 (337) 378 | 74 | 34 (344) 384 | 106 | 31 (335) 380 | 73 | 34 (336) 414 | 18 | 23 (313) 358 | 106 | 28 (362) 378 | SpltMNPV vlf-1, 34 (344) 384 |
| 46 | 45889 < 46146 | 85 | e, L | 78 | 31 (100) 109 | 87 | 75 | 105 | 74 | 27 (91) 110 | 105 | AdhoGV orf 62, 34 (88) 107 | |||||||
| 47 | 46169 < 47107 | 312 | e | gp 41 | 80 | 27 (231) 409 | 88 | 23 (288) 323 | 76 | 23 (287) 330 | 104 | 26 (288) 333 | 75 | 27 (285) 322 | 33 | 104 | 24 (226) 289 | HzSNPV gp41, 27 (285) 322 | |
| 48 | 47006 > 47536 | 176 | L | 81 | 34 (168) 233 | 89 | 34 (181) 219 | 77 | 36 (183) 232 | 103 | 37 (101) 173 | 76 | 39 (174) 241 | 106 | 28 (123) 186 | 103 | 35 (177) 191 | HaSNPV orf 71, 39 (174) 241 | |
| 49 | 47747 < 48907 | 386 | E | p47 | 40 | 25 (392) 401 | 48 | 27 (413) 390 | 36 | 25 (415) 422 | 145 | 26 (398) 397 | 35 | 26 (400) 412 | 73 | 68 | 30 (280) 460 | PxGV p47, 28 (390) 386 | |
| 48982 49468 | hr6 | ||||||||||||||||||
| 50 | 49560 < 51464 | 634 | e, L | p74 | 138 | 39 (647) 645 | 27 | 40 (572) 672 | 21 | 39 (576) 657 | 160 | 37 (639) 657 | 19 | 37 (643) 688 | 74 | 35 (645) 681 | 60 | 37 (597) 688 | RouMNPV p74, 39 (647) 645 |
| 51 | 51464 < 51892 | 142 | L | ||||||||||||||||
| 52 | 51944 > 52606 | 220 | E | Zinc finger protein | |||||||||||||||
| 53 | 52648 > 53808 | 386 | E | 119 | 22 (172) 181 | 58 | 22 (177) 195 | HaSNPV orf 57, 24 (182) 195 | |||||||||||
| 54 | 53817 > 54371 | 184 | |||||||||||||||||
| 55 | 54374 > 55528 | 384 | E, L | pif-2 | 22 | 46 (375) 382 | 119 | 44 (374) 407 | 135 | 45 (374) 425 | 48 | 43 (373) 419 | 136 | 45 (378) 383 | 38 | 44 (381) 403 | 48 | 43 (375) 372 | AcMNPV orf 22, 46 (375) 382 |
| 56 | 55553 > 55900 | 115 | e, L | ||||||||||||||||
| 57 | 55903 > 56505 | 200 | e | lef-2 | 6 | 26 (179) 210 | 137 | 31 (99) 216 | 114 | 25 (171) 210 | 14 | 25 (161) 211 | 121 | 29 (189) 238 | 25 | 28 (83) 225 | 41 | 28 (50) 171 | HzSNPV lef-2, 29 (189) 238 |
| 58 | 56535 < 57227 | 230 | e | lef-5 | 99 | 29 (230) 265 | 100 | 26 (337) 278 | 89 | 32 (170) 302 | 87 | 31 (237) 273 | 90 | 33 (155) 315 | 88 | 20 (208) 266 | 87 | 29 (222) 242 | PxGV lef-5, 31 (224) 247 |
| 59 | 57218 > 58132 | 304 | e | 38K | 98 | 29 (319) 320 | 99 | 30 (311) 322 | 88 | 30 (315) 304 | 88 | 37 (236) 301 | 89 | 33 (317) 321 | 87 | 29 (303) 303 | 88 | 32 (296) 343 | HaSNPV orf 86, 33 (317) 321 |
| 60 | 58122 < 58634 | 170 | E, L | 96 | 24 (172) 173 | 98 | 27 (166) 173 | 87 | 30 (172) 170 | 92 | 31 (167) 172 | 88 | 30 (172) 173 | 90 | 27 (169) 202 | 89 | 24 (160) 161 | SpltMNPV orf 87, 30 (172) 170 | |
| 61 | 58621 > 62052 | 1143 | E | helicase | 95 | 22 (1094) 1221 | 97 | 25 (868) 1218 | 86 | 23 (1135) 1235 | 93 | 29 (382) 1212 | 87 | 25 (1024) 1253 | 89 | 21 (817) 1332 | 90 | 21 (1076) 1131 | SeMNPV helicase, 22 (1157) 1222 |
| 62 | 62060 < 63493 | 477 | E | lef-4 | 90 | 27 (492) 464 | 93 | 23 (507) 485 | 82 | 27 (503) 475 | 98 | 27 (485) 454 | 82 | 28 (496) 461 | 96 | 22 (403) 497 | 95 | 26 (507) 480 | XcGV lef-4, 28 (474) 447 |
| 63 | 63477 > 64802 | 441 | E, L | 142 | 23 (443) 477 | 20 | 24 (433483) | 11 | 26 (460) 469 | 167 | 24 (456) 461 | 9 | 24 (454) 468 | 30 | 18 (296) 474 | 15 | 22 (374) 457 | SpltMNPV orf 11, 26 (460) 469 | |
| 64 | 64813 > 65049 | 78 | e, L | ||||||||||||||||
| 65 | 65077 > 65328 | 83 | e, L | odv-e18 | 143 | 19 | 12 | 166 | 28 (74) 83 | 10 | 14 | XcGV odv-e18, 34 (58) 83 | |||||||
| 66 | 65354 > 66136 | 260 | e, L | odv-ec27 | 144 | 23 (278) 290 | 18 | 21 (261) 283 | 13 | 24 (287) 283 | 165 | 23 (252) 278 | 11 | 22 (271) 284 | 32 | 97 | BmNPV odv-ec27, 23 (278) 290 | ||
| 67 | 66171 > 66497 | 108 | e, L | 145 | 30 (75) 77 | 17 | 30 (90) 92 | 14 | 29 (88) 93 | 164 | 30 (84) 92 | 12 | 23 (71) 92 | 9 | 26 (93) 101 | EppoNPV orf 127, 32 (82) 95 | |||
| 150 | 25 (87) 99 | ||||||||||||||||||
| 68 | 66492 < 67064 | 190 | E | lef-1 | 14 | 31 (189) 266 | 123 | 27 (183) 234 | 129 | 31 (181) 231 | 35 | 29 (161) 215 | 128 | 31 (186) 245 | 45 | 28 (194) 235 | 74 | 32 (191) 235 | PxGV lef-1, 34 (189) 251 |
| 69 | 67200 > 67772 | 190 | e, L | 115 | 27 (196) 204 | 143 | 29 (148) 203 | 107 | 29 (149) 200 | 68 | 28 (194) 203 | 101 | 31 (194) 199 | 46 | 28 (152) 203 | 35 | 30 (186) 199 | AdhoNPV orf 108, 32 (195) 198 | |
| 70 | 67944 > 69017 | 357 | E | 109 | 22 (396) 390 | 107 | 24 (322) 366 | 96 | 26 (349) 361 | 80 | 27 (282) 356 | 97 | 24 (337) 361 | 69 | 55 | 28 (223) 326 | SpltMNPV orf 96, 26 (349) 361 | ||
| 71 | 69017 > 69232 | 71 | E, L | ||||||||||||||||
| 72 | 69225 > 69581 | 118 | E | Regulat. of chrom. condensation | |||||||||||||||
| 73 | 69702 > 70070 | 122 | e, L | Regulat. of chrom. condensation | |||||||||||||||
| 74 | 70351 > 70518 | 55 | e | ||||||||||||||||
| 75 | 70605 > 71426 | 273 | e | ||||||||||||||||
| 76 | 71462 > 71644 | 60 | e, L | ||||||||||||||||
| 77 | 71645 > 72499 | 284 | e | ||||||||||||||||
| 78 | 73026 > 73457 | 143 | e | ||||||||||||||||
| 79 | 73525 < 75084 | 519 | L | pif | 119 | 33 (541) 530 | 155 | 39 (342) 530 | 124 | 38 (360) 526 | 49 | 33 (536) 529 | 114 | 31 (543) 528 | 29 | 32 (536) 523 | 75 | 30 (547) 538 | AcMNPV orf 119, 33 (541) 530 |
| 80 | 75093 < 75563 | 156 | e, L | 53 | 54 | 45 | 23 (81) 137 | 137 | 44 | 24 (125) 136 | 134 | AdhoNPV orf 47, 24 (118) 138 | |||||||
| 81 | 75562 > 78102 | 846 | E | lef-8 | 50 | 30 (882) 876 | 51 | 31 (855) 874 | 38 | 30 (903) 918 | 141 | 32 (885) 878 | 38 | 30 (885) 901 | 26 | 23 (891) 922 | 131 | 32 (900) 873 | PhopGV orf 121, 32 (860) 827 |
| 82 | 78104 > 78733 | 209 | E | ||||||||||||||||
| 83 | 78726 > 79163 | 145 | e | Densovirus capsid protein | |||||||||||||||
| 84 | 79180 > 81654 | 824 | e, L | vp91 capsid | 83 | 31 (658) 847 | 91 | 27 (751) 864 | 79 | 27 (779) 861 | 101 | 27 (627) 809 | 78 | 30 (642) 816 | 35 | 25 (702) 741 | 101 | 26 (464) 665 | AdhoNPV vp91.capsid, 32 (521) 821 |
| 85 | 81661 < 82593 | 310 | e, L | vp1054 | 54 | 23 (293) 365 | 57 | 28 (316) 332 | 49 | 21 (281) 352 | 133 | 24 (289) 336 | 48 | 27 (214) 351 | 8 | 22 (190) 329 | 138 | 23 (291) 332 | LdMNPV vp 1054, 28 (316) 332 |
| 86 | 82716 > 82937 | 73 | e, L | ||||||||||||||||
| 87 | 82945 > 83880 | 311 | e, L | ||||||||||||||||
| 84022 84047 | dr3 | ||||||||||||||||||
| 88 | 84084 > 84758 | 224 | e, L | [dr4]@ | |||||||||||||||
| 89 | 84855 < 85793 | 312 | L | vp39 | 89 | 28 (210) 347 | 92 | 25 (235) 352 | 81 | 28 (257) 302 | 99 | 23 (266) 325 | 81 | 21 (244) 293 | 24 | 96 | 23 (259) 285 | SpltMNPV vp39, 28 (257) 302 | |
| 90 | 85792 > 86370 | 192 | E, L | Putative phosphotransferase | |||||||||||||||
Identity = % amino acid identity obtained by BLAST (number of amino acids considered in the comparison) total number of amino acids of the compared ORF. @, dr within ORF. aa, no. of amino acids; Prom, promoter type (see Materials and Methods); Regulat., regulator; chrom., chromosome; > or <, transcriptional direction.
Phylogenetic analysis.
In order to root a tree of the 24 baculoviruses sequenced to date, a DNA polymerase phylogeny was inferred using the dna-pol genes from those baculoviruses and from 23 other organisms representing different Phyla which had a high score and the lowest e-values in tblastx searches against baculovirus DNA polymerases. The unclassified double-stranded DNA Heliothis zea virus 1 (Hz-1) (GenBank accession no. NC_004156) and several other DNA virus DNA polymerases were included for comparison. A maximum-likelihood tree from a data set with 2,397 amino acid sites including 47 taxa was calculated with the parallel version 5.2 of the TREE-PUZZLE program (http://www.tree-puzzle.de/) using default settings but assuming a variable rate of evolution among sites following a gamma distribution approximated by 8 categories, with the shape parameter (alpha) estimated from the data. Likewise, to have the maximum evidence for a baculovirus phylogeny, a single data set with 16,803 amino acid sites (proteon) comprising 29 conserved genes found in 24 baculovirus genomes was also analyzed with TREE-PUZZLE. The DNA polymerase tree adjacency pattern was then used to orient in time (by rooting) the protein tree.
GeneParity analysis.
The gene order of the NeseNPV genome was compared to those of six other baculovirus genomes: AcMNPV (representing group I lepidopteran NPVs), LdMNPV, MacoMNPV-A and HzSNPV (group II lepidopteran NPVs), CpGV (lepidopteran GV), and CuniNPV (dipteran NPV). For this, a modified version of the GeneParity plot method (33) was used. Each complete genome was compared individually to that of NeseNPV. Minor modifications were done to two of the genomes, relative to the original publication. For AcMNPV (5), the comparison was made starting with the polyhedrin gene. For CuniNPV (1), the order of lef-5, 38K, ac96, and helicase in the genome was used, not the ORF number. A gene cluster was defined as any group of genes (two or more) that appeared consecutively in NeseNPV and in the comparative genome. However, some viruses do not have all of the genes in a cluster because they may be present in another location of the genome or just absent. Therefore, relaxed clusters were also determined as those in which at least two genes were consecutive in both genomes compared, but a certain number of genes (a maximum of five in one case for CuniNPV) may have been inserted between genes that appear consecutively in NeseNPV.
Nucleotide sequence accession number.
The NeseNPV genome sequence has been deposited in GenBank under accession no. AY430810.
RESULTS AND DISCUSSION
Multiplex PCR.
Due to the lack of an insect colony or insect cell culture susceptible to NeseNPV, the DNA obtained for this project was not from a homogeneous isolate. The approach to circumvent this problem was to sequence the ends of all the NeseNPV HindIII and EcoRI clones obtained. After confirming that the terminal sequences of the submolar bands were nearly the same as those of the major bands, we continued sequencing the major bands. Some single nucleotide polymorphisms were detected while sequencing several clones of the same fragment and also when comparing the sequences obtained from the multiplex PCR adjacent to the HindIII recognition sites. From a total of 10,900 bases (13% of the total genome) analyzed for nucleotide differences from the consensus sequence, only 68 bases (0.6%) had nucleotide polymorphisms (data not shown). The colinear position and orientation of the HindIII clones in the NeseNPV physical map were assembled using the Multiplex PCR data (Fig. 1). A 330-bp fragment (HindIII-M) that was not originally cloned was sequenced and mapped during the multiplex PCR procedure. In addition, a 4.5-kb region found to be unclonable, located inside HindIII-D fragment, was PCR amplified and then sequenced. Multiplex PCR has been originally used to rapidly close gaps in whole-genome shotgun sequencing projects (74). It proved to be a very convenient and reliable method to ensure the correct location and position of the NeseNPV cloned fragments. It also allowed us to detect genetic variation, which is present in NeseNPV.
FIG. 1.

Linear map of the HindIII sites of the NeseNPV genome. The number and transcriptional direction of each ORF are labeled as a black arrow. Known baculovirus homologs are labeled in bold by the NeseNPV ORF number. Nonbaculovirus homologs are indicated in parentheses. The homologous regions (hr) are shown as triangles, and the direct repeats (dr) are shown as circles (•).
Nucleotide sequence analysis.
The genome of NeseNPV was 86,462 bp in size. Since baculovirus genomes range in size from 99,657 bp in AdorGV (78) to 178,733 bp for XcGV (30), NeseNPV, and other Neodiprion baculoviruses (B. M. Arif, personal communication), have the smallest genomes of baculoviruses sequenced thus far. Nucleotide number 1 of the genome was considered to be the A of the polyhedrin gene (polh) ATG start codon as has been conventionally adopted (30, 36, 75), and the successive nucleotides were numbered in the direction of the polh gene (Table 1). A total of 90 methionine-initiated ORFs with 50 or more amino acids and minimal overlapping of adjacent ORFs were found in NeseNPV. This is the smallest number of ORFs found in baculoviruses to date (Table 2). A total of 14 ORFs overlapped adjacent ORFs, and the maximum overlapping found was between NeseNPV ORF 40 (nese40 = lef-9) and nese41 (ac68) with 58 nucleotides shared. The distribution of the orientations for the predicted genes was uneven, with 54 ORFs (60.7%) oriented clockwise and 35 ORFs (39.3%) in the opposite direction. This is different from the other baculoviruses that present a more even distribution. The complete nucleotide sequence of NeseNPV was determined to have a G+C content of 34%. Currently, AdorGV presents the lowest G+C content of the sequenced baculoviruses with 34.5% (78). Therefore, the NeseNPV genome has the lowest G+C content of baculoviruses sequenced to this date (Table 2).
TABLE 2.
Characteristics of baculovirus genomesa
| Baculovirus | Classification | Genome size (bp) | Total ORFs | C+G (%) | No. of hr's | No. of bro | ORFs homolog to NeseNPV | Reference | GenBank accession no. |
|---|---|---|---|---|---|---|---|---|---|
| AcMNPV | I-NPV Lepidopt. | 133,894 | 154 | 41 | 8 | 1 | 42 | 5 | NC_001623 |
| OpMNPV | I-NPV Lepidopt. | 131,990 | 152 | 55 | 5 | 3 | 41 | 2 | NC_001875 |
| BmNPV | I-NPV Lepidopt. | 128,413 | 136 | 40 | 7 | 5 | 42 | 25 | NC_001962 |
| EppoNPV | I-NPV Lepidopt. | 118,584 | 136 | 41 | 5 | 1 | 51 | 35 | NC_003083 |
| CfMNPV | I-NPV Lepidopt. | 129,609 | 145 | NC_004778 | |||||
| CfDEFNPV | I-NPV Lepidopt. | 131,158 | 149 | NC_005137 | |||||
| SeMNPV | II-NPV Lepidopt. | 135,611 | 139 | 44 | 6 | 0 | 42 | 36 | NC_002169 |
| LdMNPV | II-NPV Lepidopt. | 161,046 | 163 | 58 | 13 | 16 | 41 | 42 | NC_001973 |
| SpltMNPV | II-NPV Lepidopt. | 139,342 | 141 | 42 | 17 | 2 | 42 | 65 | NC_003102 |
| HaSNPV | II-NPV Lepidopt. | 131,403 | 135 | 39 | 5 | 3 | 43 | 13 | NC_002654 |
| MacoNPV-A | II-NPV Lepidopt. | 155,060 | 169 | 42 | 4 | 8 | 43 | 46 | NC_003529 |
| MacoNPV-B | II-NPV Lepidopt. | 158,482 | 168 | 40 | 4 | 7 | 43 | 43 | NC_004117 |
| HzSNPV | II-NPV Lepidopt. | 130,869 | 139 | 39 | 5 | 3 | 43 | 14 | NC_003349 |
| RoMNPV | II-NPV Lepidopt. | 131,526 | 146 | 39 | 9 | 0 | 42 | 28 | NC_004323 |
| AdhoNPV | II-NPV Lepidopt. | 113,220 | 125 | 36 | 4 | 4 | 41 | 60 | NC_004690 |
| HaSNPV-C1 | II-NPV Lepidopt. | 130,760 | 134 | NC_003094 | |||||
| XcGV | GV Lepidoptera | 178,733 | 181 | 41 | 9 | 7 | 42 | 30 | NC_002331 |
| PxGV | GV Lepidoptera | 100,999 | 120 | 41 | 4 | 0 | 41 | 29 | NC_002593 |
| CpGV | GV Lepidoptera | 123,500 | 143 | 45 | 0 | 1 | 41 | 53 | NC_002816 |
| AdorGV | GV Lepidoptera | 99,657 | 119 | 35 | 0 | 0 | 41 | 78 | NC_005038 |
| PhopGV | GV Lepidoptera | 119,217 | 130 | NC_004062 | |||||
| CrleGV | GV Lepidoptera | 110,907 | 129 | NC_005068 | |||||
| CuniNPV | NPV Diptera | 108,252 | 109 | 51 | 4 | 6 | 29 | 1 | NC_003084 |
| NeseNPV | NPV Hymenoptera | 86,462 | 90 | 34 | 6 | 0 | This paper | AY430810 |
AcMNPV, Autographa californica MNPV; OpMNPV, Orgyia pseudotsugata MNPV; BmNPV, Bombyx mori NPV; EppoNPV, Epiphyas postvittana MNPV; CfMNPV, Choristoneura fumiferana MNPV; CfDEFNPV, defective CfNPV; SeMNPV, Spodoptera exigua MNPV; LdMNPV, Lymantria dispar MNPV; SpltMNPV, Spodoptera litura MNPV; HaSNPV, Helicoverpa armigera SNPV; MacoNPV-A, Mamestra configurata NPV-90/2; MacoNPV-B, Mamestra configurata NPV-96B; HzSNPV, Helicoverapa zea SNPV; RoMNPV, Rachiplusia ou MNPV; AdhoNPV, Adoxophyes honmai SNPV; HaSNPV-C1, Helicoverpa armigera SNPV-clone C1; XcGV, Xestia c-nigrum GV; PxGV, Plutella xylostella GV; CpGV, Cydia pomonella GV; AdorGV, Adoxophyes orana GV; PhopGV, Phthorimeae operculella GV; CrleGV, Cryptophlebia leucotreta GV; CuniNPV, Culex nigripalpus NPV; NeseNPV, Neodiprion sertifer NPV. homolog., homologous; Lepidopt., Lepidoptera.
From the 90 NeseNPV ORFs, 43 were homologous to ORFs from other baculoviruses (see “best match” column in Tables 1 and 3), and 29 of them belonged to the core of 30 conserved genes among all baculoviruses (32). The only gene not identified in NeseNPV from the 30 conserved genes was the ld130 gene, which codes for the fusion or F protein. The homology between NeseNPV ORFs and other baculoviruses ORFs was low, ranging from 18 to 56%, and the average amino acid identity between the NeseNPV ORFs and their best match homologs was 32.7% (Table 1). Even the polyhedrin gene, which is a highly conserved gene among baculoviruses, had an amino acid identity of 38 to 53%. The highest homology was found with the polyhedrin gene of another Neodiprion baculovirus, the Neodiprion abietis nucleopolyhedrovirus (NeabNPV) (GenBank accession no. AAM95580) with a 92% amino acid identity. Six NeseNPV ORFs were highly similar to non-baculovirus-related genes, and the other 41 ORFs were not considered homologs to other genes because of low amino acid sequence similarity and therefore are considered unique to NeseNPV. A similar range of low amino acid conservation was observed by Afonso et al. (1) when comparing the CuniNPV genome with the those of Lepidoptera baculoviruses, although a higher number of conserved baculovirus ORFs was present in NeseNPV. In these viruses, which are not closely related, similar functional genes may be present that are simply not identified by the amino acid identity. Additional research will be necessary to reveal their function. To facilitate the cross-reference of conserved genes, Table 1 presents the amino acid identity of the baculovirus genes with homology to NeseNPV. Also shown are the corresponding ORFs of other baculoviruses that were indirectly homologous to NeseNPV by homology between other baculoviruses; however, no amino acid identity information is shown under those circumstances.
TABLE 3.
Baculovirus genes present in NeseNPV genome
| Gene group | Genes conserved in all baculoviruses | Genes conserved in all lepidopteran | Genes not conserved |
|---|---|---|---|
| Replication genes | dnapol (ac65, nese28) | dbp (ac25, nese22) | |
| lef-1 (ac14, nese68) | |||
| lef-2 (ac6, nese57) | |||
| helicase (ac95, nese61) | |||
| Transcription genes | lef-4 (ac90, nese62) | lef-11 (ac37, nese23) | mtase1 (ac69, nese5) |
| lef-5 (ac99, nese58) | |||
| lef-8 (ac50, nese81) | |||
| lef-9 (ac62, nese40) | |||
| p47 (ac40, nese49) | |||
| vlf-1 (ac77, nese45) | |||
| Structural protein genes | pif (ac119, nese79) | polh (ac8, nese1) | |
| pif-2 (ac22, nese55) | odv-e18 (ac143, nese65) | ||
| gp41 (ac80, nese47) | |||
| odv-e56 (ac148, nese38) | |||
| odv-ec27 (ac144, nese66) | |||
| p6.9 (ac100, nese36) | |||
| p74 (ac138, nese50) | |||
| vp91-capsid (ac83, nese84) | |||
| vp39 (ac89, nese89) | |||
| vp1054 (ac54, nese85) | |||
| Auxiliary genes | alk-exo (ac133, nese31) | iap (ac27, nese17) | |
| Unknown functional genes | 38K (ac98, nese59) | P40 (ac101, nese35) | |
| p33 (ac92, nese24) | P45 (ac103, nese33) | nese53 (hz58) | |
| ac68 (nese41) | ac53 (nese80) | ||
| ac81 (nese48) | ac78 (nese46) | ||
| ac96 (nese60) | ac93 (nese25) | ||
| ac109 (nese70) | ac106 (nese32) | ||
| ac115 (nese69) | ac145/ac150 (nese67) | ||
| ac142 (nese63) |
DNA replication genes.
NeseNPV had the following genes considered essential for baculovirus DNA replication by Lu et al. (49): late expression factors lef-1 (nese68), lef-2 (nese57), helicase (nese61), and dnapol (nese28); but it lacked the immediate-early gene ie-1 and the late expression factor lef-3, also absent in CuniNPV (1). The ie-1 gene, however, is poorly conserved even among lepidopteran baculoviruses (36). Those four DNA replication genes present in NeseNPV are found in all baculoviruses sequenced (32), indicating that they are very important for DNA replication. Other genes identified in NeseNPV have been characterized as being involved in the processing of mature DNA (45, 56, 80) are vlf-1 (nese45) and alk-exo (nese31); and in DNA binding (57, 76), dbp (nese22) and p6.9 (nese36).
Regulatory genes.
Gene expression in baculoviruses follows a temporal cascade. Four genes capable of transregulating early viral gene expression that occurs prior to viral DNA replication have been identified: ie-0, ie-1, ie-2, and pe38 (23). NeseNPV does not have significant homology to any of these early transregulating genes of baculoviruses. On the other hand, it had homology with various late transcription activator genes (50, 51), including the late expression factors lef-4 (nese62), lef-5 (nese58), lef-8 (nese81), lef-9 (nese40), lef-11 (nese23), p47 (nese49), and the very late factor vlf-1 (nese45). From these, the four genes encoding the viral RNA polymerase subunits (lef-4, lef-8, lef-9, and p47), forming the simplest eukaryotic DNA-directed RNA polymerase (27), are present in all the sequenced baculoviruses, including CuniNPV (1, 32). Another gene involved in regulation of late expression was nese5, homologous to ac69 recently named mtase1 for its methyltransferase activity (79). Although mtase1 has no demonstrable effect on viral replication, it has been shown to stimulate transient late gene expression (44). NeseNPV lacks lef-6, lef-10, lef-12, and 39K that are late regulatory genes (51). However, Lin and Blissard (47), using a lef-6 null AcMNPV virus, have shown that lef-6 is not essential for viral DNA replication nor for late gene transcription but probably is important in accelerating the infection cycle of AcMNPV. In support of this finding, NeseNPV does not have lef-6, indicating that it is not needed for replication of this and perhaps other viruses. The two other lef genes absent from NeseNPV, lef-10 and lef-12, are not conserved in all baculoviruses (32). The protein kinase (pk-1) gene homolog, involved in very late gene expression (20), was not found in NeseNPV.
Structural genes.
From the 15 conserved structural genes of lepidopteran baculoviruses (32), only 12 were found to have homology with NeseNPV (Table 3). The structural polyhedrin or granulin gene homologs are present in all baculoviruses with the exception of CuniNPV. NeseNPV polyhedrin (nese1) had the highest amino acid homology to the Neodiprion baculovirus NeabNPV polyhedrin with a 92% amino acid identity, but when compared to the lepidopteran polyhedrins/granulins, the homology ranged from 53% with SpltMNPV to 38% with CpGV (Table 1). The NeseNPV basic DNA binding protein p6.9 (nese36) had a 56% amino acid identity with AdorGV p6.9 but was not associated by BLAST with any other baculovirus p6.9 gene, even though it had a high arginine (29%) and serine (27%) concentration and the RRPGRPR conserved motif identified in other p6.9 proteins (76). The size of the predicted NeseNPV p6.9 protein was 86 amino acids (aa), bigger that all other p6.9 proteins except for those of HaSNPV (76) and HzSNPV (14). Nese36, however, had a high amino acid identity with splicing factor proteins from organisms such as insects, mouse, and human. The occlusion-derived viral envelope odv-e18 (nese65), odv-e56 (nese38), and odv-ec27 (nese66) homologs in NeseNPV had amino acid identities of 34% (with XcGV), 39% (with LdMNPV) and 23% (with BmNPV), respectively (Table 1). The capsid-associated protein genes vp39 (nese89) and vp91 capsid (nese84) were also present. Although the identity of the NeseNPV vp39 homolog averaged only 25% with other vp39 of baculoviruses, it contained a total of six cysteines of the eight conserved in AcMNPV and OpMNPV (24). From those, four were in conserved locations that could generate disulfide bonds important for the protein structure. The vp1054 (nese85), a viral structural protein present in both occlusion-derived virus (ODV) and budded virus (BV) required for nucleocapsid assembly (63), and the glycoprotein gp41 (nese47), found only in ODV but required for the egress of nucleocapsids from the nucleus during the BV synthesis (64), were found in NeseNPV. The average identity with other vp1054 and gp41 proteins was 24% and 25%, respectively. The final NeseNPV structural gene found in all lepidopteran baculoviruses was p74 (nese50) with an average amino acid identity of 38%. This gene product, found associated with ODVs, is essential for the primary infection of midgut cells of insect larvae during oral infection (21). Another gene, associated with the ODV envelope of Spodoptera littoralis MNPV (SpliNPV), having a similar function as p74, is the ac119 homolog called the per os infectivity factor or pif gene (39). Nese79 had a 33% amino acid identity with ac119 and 34% with SpliNPV pif. A second pif gene (pif-2 = SeMNPV ORF 35), has recently been described for SeMNPV (70). Nese55 had a 35% amino acid identity with Se35 and 46% with its AcMNPV homolog, ac22. Both pif and pif-2 genes have been conserved in all the baculoviruses sequenced (32). Genes not identified in NeseNPV were p10, pk-1, odv-e66, odv-e25, and neither ld130 nor gp64. The p10 gene, responsible for the formation of fibrillar structures and associated with the occlusion body, is also absent in CpGV (53) and CuniNPV (1). CuniNPV also lacks odv-e66 and pk-1 homologs. A major difference between NeseNPV and other baculoviruses is the lack of a definitive gp64 or ld130 homolog that code for envelope fusion proteins. These functionally analogous proteins (37) present in BV envelopes are capable of inducing cell fusion and are involved in viral attachment (52, 68). All the baculoviruses sequenced to date have the ld130 homolog (32), which codes for the F protein, and the members of the group I NPVs also have gp64 (8, 9, 58). NeseNPV does not have either of these gene homologs. NeseNPV infections of sawfly larvae are restricted to the insect midgut epithelial cells, as happens with baculoviruses that infect other Hymenoptera, Diptera, Coleoptera, Thysanura and Trichoptera hosts (22). In NeseNPV, the function of viral transmission between the epithelial cells may lie in another gene. To find probable candidates for such role, all the NeseNPV ORFs were tested for transmembrane domains and N-terminal signal peptides. The following NeseNPV ORFs had both a signal peptide and a transmembrane domain: Nese26, nese55 (pif-2), nese60 (ac96 homolog), nese65 (odv-e18), nese67 (ac145 homolog), nese69 (ac115 homolog), nese71, nese79 (pif) and nese84 (vp91 capsid). These and other NeseNPV ORFs that had a transmembrane domain, an N-terminal signal peptide or a similar size to baculovirus ld130 homologs were aligned individually with the ld130 homologs from several baculoviruses. However in all searches, the amino acid identity was never higher than 15% and considered random amino acid matches.
Inhibitors of apoptosis.
Apoptosis or programmed cell death is known to happen in vertebrates and invertebrate organisms as a response to a series of inductions, including pathogen attacks (15). Baculoviruses have genes capable of suppressing cellular apoptosis to maintain viral replication. The family of genes called inhibitor of apoptosis, iap, which was first identified in CpGV (17), is found in most baculoviruses sequenced to date with the exception of CuniNPV. NeseNPV contains an iap homolog (nese17) with an amino acid identity of 38% to CpGV iap-3. Several baculoviruses have more than one iap gene in their genomes, while NeseNPV has only one. Homologs to iap usually have two baculovirus iap repeats (BIR) and a C-terminal zinc finger (7). Nese17 has only one BIR motif and the zinc finger motif, similar to what is found in SpltMNPV (65) and LdMNPV (42), and also in some iap-2s such as MacoMNPV and RoMNPV (28, 46). NeseNPV iap had a higher homology with the iap-3 gene family proposed by Luque et al. (53) based on their iap phylogenetic analysis. Until biological activity is determined for nese17, it will simply be called iap. Other apoptosis inhibitor genes include p35, found originally in AcMNPV (16), and Slp49 of SpliNPV, which is a p35 homolog that blocks apoptosis induced by infection of Sf9 cells with an AcMNPV p35-deficient mutant (19, 82, 69). NeseNPV did not have any homology with either p35 or Slp49 genes, indicating that either its iap is the only gene responsible for any apoptotic suppressor activity or a very low amino acid homology has made it difficult to locate a p35 or a Slp49 homolog. However, since it has been shown that not all iap's are capable of preventing cell death (54), the biological activity of the NeseNPV iap homolog will have to be experimentally confirmed.
Other conserved genes.
Other NeseNPV ORFs had homology with baculovirus genes, the biological functions of which have not yet been determined. These are listed in Table 3. Nese67 was homologous to ac145 (30%) and ac150 (25%).
Duplicated genes.
Two NeseNPV ORFs (nese18 and nese19) were duplicates, positioned in opposite orientation with a homologous region (hr4) and a small direct repeat (dr2) separating them (Fig. 1). NeseNPV does not have a copy of the AcMNPV ORF 2 homolog, characterized as the baculovirus repeat ORF (bro) (38). Other baculoviruses usually have one or more copies of bro genes with the exception of SeMNPV (36), PxGV (29), and AdorGV (78), which, like NeseNPV, have no bro homologs.
Nonbaculovirus homologs.
Six NeseNPV ORFs had high homology with known proteins not related to baculoviruses (Table 1). Amino acid identity as high as 40% was obtained between nese7 and the family of trypsin-like proteases from insects belonging to the orders Lepidoptera, Diptera, Coleoptera, and Siphonaptera. Homology was also obtained with the trypsin of other arthropods and mammals. Figure 2, shows the alignment of nese7 with trypsins of other insects and a mammal (cow). Conserved amino acid sequences are shown in boxes. The trypsin catalytic triad (histidine, aspartic acid, and serine) and the conserved cysteines from the trypsin-like proteins (73) were present in nese7 and are indicated in Fig. 2. The size of the predicted protein (258 aa) was comparable to that of other trypsins. The maximum-likelihood quartets method suggests that the trypsin-like gene of the NeseNPV may be of invertebrate origin and in 35% of the time its sister taxa was the trypsin of a Coleoptera, Diaprepes abbreviata (data not shown). This putative gene has not been reported in any DNA virus including baculoviruses. Neodiprion baculoviruses, however, seem to have it present, since Neodiprion lecontei (Basil Arif, personal communication) also has this gene. It will be interesting to determine if this gene was derived from the insect host, since no hymenopteran trypsin was found in the database search to make a comparison. Nese52 had four C2H2 zinc finger domains and significant homology with C2H2 zinc finger proteins from insects and mammals. Although on average, only the first half of this ORF presented the homology, this type of protein has not been previously described in baculoviruses. Two NeseNPV ORFs, nese72 and nese73, had homology with the regulator of chromosome condensation (RCC1) proteins from a wide group of organisms such as microsporidia, nematodes, fruit flies, frogs, rodents and humans. Nese83 had up to 39% amino acid identity with the capsid protein of several insect densoviruses. Finally, nese90 was homologous to a large number of phosphotransferases.
FIG. 2.
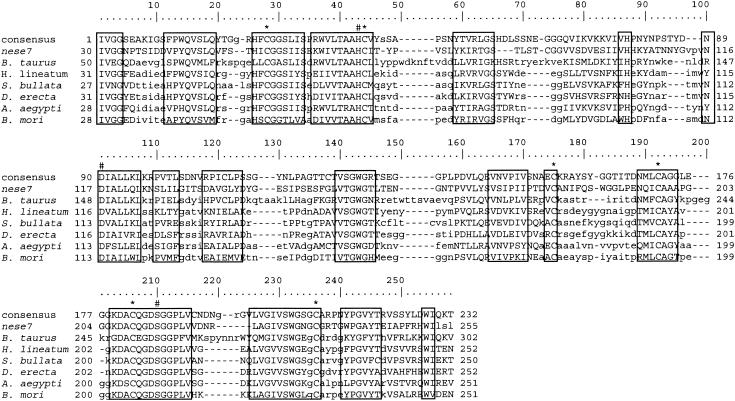
Alignment of NeseNPV ORF 7 (nese7) and trypsins from other organisms. Bos taurus, cow; Hypoderma lineatum, early cattle grub; Sarcophaga bullata, grey fleshfly; Drosophila erecta, a fruit fly; Aedes aegypti, yellow fever mosquito; Bombyx mori, silkworm. Regions highlighted as conserved amino acids by the conserved domain database (CDD) are boxed with a consensus sequence indicated on the top line. # represents the trypsin catalytic triad, histidine, aspartic acid, and serine. * indicates the conserved cysteines.
Absent genes.
NeseNPV did not present any of the known baculovirus genes involved in nucleotide metabolism or auxiliary genes such as the ribonucleotide reductases (rr1, rr2), dUTPase (dutpase), proliferating cell nuclear antigen (pcna), protein tyrosine phosphatase (ptp), ubiquitin, superoxidase dismutase (sod), conotoxin-like peptide (ctl), cathepsin, chitinase, ecdysteroid UDP-glucosyltransferase (egt), fibroblast growth factor (fgf), actin rearrangement-inducing factor-1 (arif-1), and enhancin. However, these genes are not conserved among baculoviruses. The exception is the ld130 gene, which has been shown to be present in all baculoviruses (32) and was not found in NeseNPV. Seven NeseNPV ORFs had a very low homology to baculovirus genes even after forcing alignment with the suspected baculovirus homolog, and so for the present analysis, they were considered random similarities between the viruses. However, since some of these NeseNPV ORFs had predicted amino acid sizes very close to those for the baculovirus gene products, and since some of these ORFs may have real functional similarities that the amino acid sequence identity did not support, a listing of such ORFs was considered useful. Under that group were ie-2 (ac151 = 408 aa) (nese9 = 416 aa), cg30 (ac88 = 264 aa) (nese12 = 130 aa), odv-e25 (ac94 = 228 aa) (nese26 = 218 aa), ie-1 (ac147 = 582 aa) (nese82 = 209 aa) and lef-3 (ac67 = 385 aa) (nese87 = 311 aa). Two AcMNPV homologs with unknown functions were also under this group: ac66 (808 aa) (nese29 = 792 aa), and ac75 (133 aa) (nese43 = 125 aa).
hrs.
Six hrs were located within the NeseNPV genome (Fig. 3), each containing small (5- or 6-bp) perfect palindromes, which can be extended up to 19-bp imperfect palindromes, embedded within a series of direct repeats. The nucleotide sequence has been aligned to show the size of the repeat units, and when needed, gaps were introduced to optimize the alignment (Fig. 3A). The number of hrs in other baculoviruses ranges from four in CuniNPV, PxGV and MacoNPV (1, 29, 46) to 17 in SpltNPV (65). Baculoviruses reported without typical hrs are CpGV (53) and AdorGV (78). NeseNPV hrs varied in length from 396 (hr5) to 794 (hr2) nucleotides (Table 4), and the combined size of the six hrs was 3,669 bp or 4.3% of the genome. This percentage was significantly higher than those for the hrs from CuniNPV (0.8%) and AdhoNPV (1.1%) (1, 60) but lower than those for several other baculovirus hrs, including SpltMNPV (65), HzSNPV (14), HaSNPV (13), and PxGV (29), which comprise approximately 6% of their genomes. NeseNPV hrs consist of repeats of 45 bp (hr5 and hr6), 65 bp (hr2), or alternating 65 bp and 45 bp units (hr1, hr3, and hr4) (Fig. 3B). The number of repeat units in each hr ranged from 9 to 14 and the length and number of times they occur are presented in Table 4. The average G+C content of the hrs was 49.8%, significantly higher than the 34% for the complete NeseNPV genome.
FIG. 3.
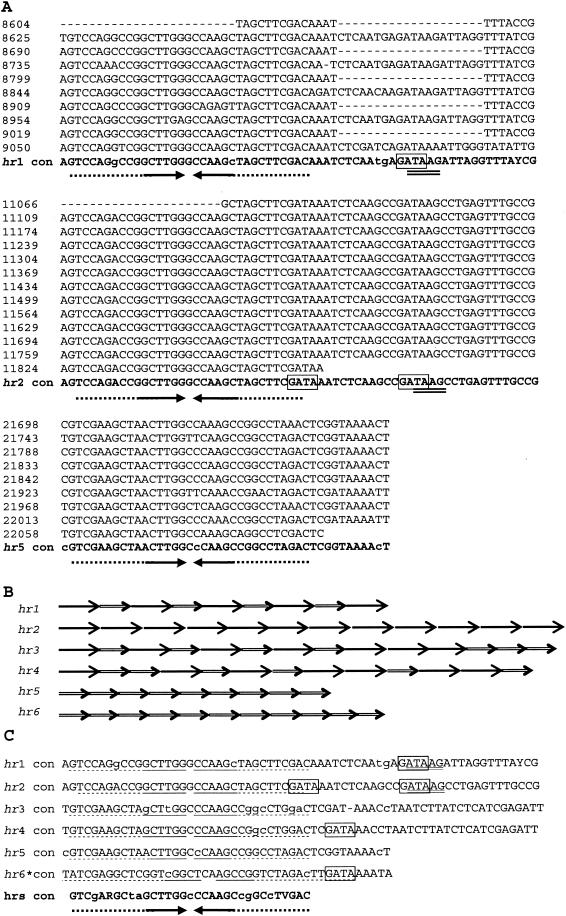
(A) hrs of NeseNPV. The nucleotide sequence of three NeseNPV hrs are aligned to exemplify the direct repeats found in units of 65 bp (hr2), 45 bp (hr5), or alternating 65 and 45 bp (hr1). The consensus sequence of the hrs is shown in bold, with uppercase letters representing 75% or more conserved nucleotides (60). Gaps (−) were inserted to optimize the alignment of the repeat units. The numbers on the left hand side of each hr indicate the nucleotide position in the genome. For each hr, the nucleotides forming a perfect palindrome (10 to 12 nucleotides) are marked with a solid arrow. Imperfect palindromes, formed by 70% or more complementary bases, are indicated by dots that extend 31 to 40 bases. The late promoter motif ATAAG and the potential GATA factor-binding site are double underlined and boxed, respectively. (B) Schematic representation of the repeats units of either 65 bp (solid arrows) or 45 bp (double-line arrows) found in each hr. (C) Comparison of the consensus sequences of the six NeseNPV hrs with the perfect palindrome region underlined and the extended imperfect palindrome dotted. An asterisk in hr6 indicates that the repeat sequence is different from the other hrs.
TABLE 4.
Structure of NeseNPV homologous regions (hr's) and direct repeats (dr's)
| Name | Length of repeat units/no. of times repeated | Position in genome | Repetitive region (total bp) | G+C (%) |
|---|---|---|---|---|
| hr1 | 65 bp/5; 45 bp/4 | 8604-9128 | 525 | 49.6 |
| hr2 | 65 bp/12 | 11066-11859 | 794 | 53.7 |
| hr3 | 65 bp/7; 45 bp/7 | 17045-17803 | 759 | 47.6 |
| hr4 | 65 bp/7; 45 bp/6 | 19424-20131 | 708 | 49.7 |
| hr5 | 45 bp/9 | 21698-22093 | 396 | 51.0 |
| hr6 | 45 bp/11 | 48982-49468 | 487 | 46.9 |
| dr1 | 120 bp/2 | 14748-14987 | 240 | 40.2 |
| dr2 | 29 bp/3 | 19265-19352 | 87 | 42.0 |
| dr3 | 13 bp/2 | 84022-84047 | 26 | 29.6 |
| dr4 | 48 bp/2 | 84617-84715 | 98 | 34.3 |
In addition to the hrs of NeseNPV, four small repetitive regions not recognized as hrs were identified. These regions were called direct repeats (drs), with dr1 consisting of two direct repeats of 120 bp, dr2 of three repeats of 29 bp, dr3 of two repeats of 13 bp, and dr4 of two repeats of 48 bp (Table 4). When compared with the hr nucleotide composition, the dr's were found to be A-T rich, especially dr3 (70.4%) and dr4 (65.7%). Two of the dr's (dr1, dr4) were located inside NeseNPV ORFs, nese16 and nese88, respectively. The same has been described for AdorGV (78), wherein the repeats may cause these putative ORFs to not be functional.
Baculovirus hrs have been characterized to be enhancers of RNA II-mediated transcription of early genes (26, 48, 62). hrs have also been characterized as origins of DNA replication (66, 67) and are typically dispersed around the genome. This dispersion is suggested to speed the rate at which replication occurs by initiation at several sites of the genome (67). However, in NeseNPV the hrs were located predominantly in the first half of the circular genome, with hr1 at 8.6 to 9.1 kb (9.95 to 10.58 map units) and hr6 at 49.0 to 49.5 kb (56.7 to 57.2 map units). Furthermore, the distance between hr5 and hr6 was 26.9 kb, which reveals a further clustering of hrs in the first quarter of the genome (Fig. 1; Table 4). Clustering of the hrs has also been observed in CuniNPV, with its hrs concentrated in 35% of the genome.
A unique feature observed in NeseNPV hrs was the presence of multiple copies of the sequences ATAAG and GATA (Fig. 3A and 3C). Baculovirus late promoter motifs are (A/G/T)TAAG; however, TAAG is the essential element (51). Although the late promoter element TAAG is found in baculoviruses, it is generally not located inside of an hr. Interestingly, a GATA element that binds host transcription factors (40) is located close to, and in hr1 and hr2 overlapping, each of the TAAG late promoter motifs within the hrs. Multiple GATA elements have been found in the Hz-1 PAT1 persistence-associated transcript, which does not encode a protein (12). The sequence of PAT1 also displays abundant direct and inverted repeats. No ORFs are predicted for the NeseNPV hrs either, indicating that they could have a yet-uncharacterized regulatory function similar to that of the GATA elements of Hz-1 PAT1. hrs are a distinctive feature of the majority of baculovirus genomes and have been shown to be essential for regulatory function (26, 62, 66, 67). The function of the NeseNPV hrs as origins of replication or enhancers of transcription has yet to be determined experimentally.
GeneParity.
The comparison of the gene order of NeseNPV and six other genomes showed that from the 43 NeseNPV baculovirus homolog genes, 42 were in AcMNPV, 41 in LdMNPV, 43 in MacoNPV-A, 43 in HzSNPV, 41 in CpGV, and 29 in CuniNPV. Eight clusters of two or more genes were found; however, not all of the genomes had all these clusters (Fig. 4). Clusters 1, 2, 3, and 5 were absent in CuniNPV, while clusters 6, 7, and 8 were present only in CuniNPV. Relaxed clusters were determined to try to recognize regions where genes may have been added or deleted between genes that were consecutive in a more ancestral lineage of baculovirus. For example, cluster 2 (p45, p40, and p6.9) was considered relaxed because in NeseNPV as well as in the other genomes there is an ORF between p45 and p40. Cluster 3 (vlf-1, ac78, gp41, and ac81) was considered relaxed only in AcMNPV because there is an extra ORF between ac78 and gp41 not found in any of the other baculovirus genomes analyzed. Cluster 4 (lef-5, 38K, ac96, helicase, and lef-4) was the most conserved cluster. It was considered relaxed to accommodate the addition of lef-4 to a cluster that has already been shown to be conserved (32). Five genes exist between helicase and lef-4 in CuniNPV, while the other baculoviruses have either three or four genes. However, not only was the order of the genes in cluster 4 conserved, but also their relative transcriptional direction. Cluster 5 (ac142, odv-e18, odv-ec27, and ac148) was relaxed because in NeseNPV there is an extra ORF between ac142 and odv-e18, which does not occur in the five other genomes.
FIG. 4.
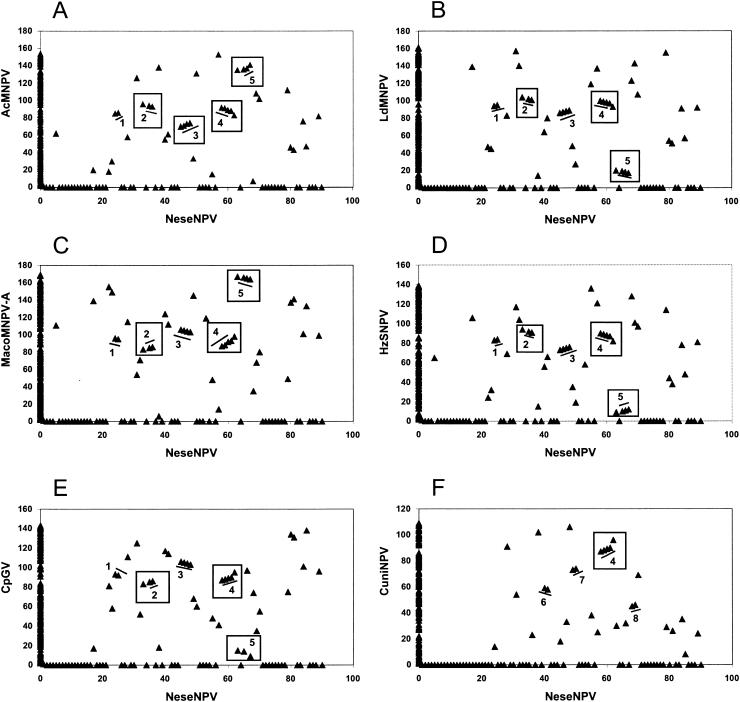
GeneParity plots comparing the NeseNPV gene order to six other baculoviruses: AcMNPV (A), LdMNPV (B), MacoMNPV-A (C), HzSNPV (D), CpGV (E), and CuniNPV (F). Eight clusters are underlined and/or boxed in the graphs, which include the following genes or AcMNPV homologs: 1, ac92, ac93; 2, p45, p40, p6.9; 3, vlf-1, ac78, gp41, ac81; 4, lef-5, 38K, ac96, helicase and lef-4; 5, ac142, odv-e18, odv-ec27*, ac148; 6, lef-9, ac68; 7, p47, p74; and 8, lef-1, ac115. Asterisks represent genes that are not part of a specific cluster in certain genomes. Straight lines mark clusters of two or more genes (underlined above) that are sequential in both NeseNPV and the genome being compared. Relaxed clusters that include genes that are not sequential in the compared genome are indicated with a box.
GeneParity plots have been used to compare conservation of genomic organization. The GV genomes maintain a strong colinearity between them (29, 53, 78), but they are not strongly colinear with NPVs such as AcMNPV. The comparison between lepidopteran NPVs show some genome arrangements, such as inversions. However, the order of a significant number of baculovirus genes is conserved (33, 36, 46, 65). In contrast, NeseNPV had few common clusters with other baculoviruses even after accepting relaxed clusters.
The presence of conserved gene clusters can result from a physical constraint preventing their separation, as suggested by Herniou et al. (32) about lef-5, 38K, ac96, and helicase (cluster 4 in the present analysis). Another explanation for the conservation of clusters could be gene overlapping. Cluster 3 had genes with overlapping coding regions. In genes transcribed on opposite strands and away from each other, such as the genes from clusters 1 and 4, the promoter region or start codon of one gene could be included in the continuous gene and so the physical separation of these two genes may cause their inactivation. Therefore, the continuity of the gene cluster would be maintained in the evolution of the baculovirus lineages.
Baculovirus phylogeny.
The DNA polymerase was used previously to investigate the relationship among distantly related organisms and DNA viruses (10). Therefore, the DNA polymerase of the baculovirus was used in order to help establish the position of the root for a baculovirus tree by rooting it with 23 DNA polymerases from widely diverse phylogenetic origins. It was necessary to include these diverse taxa to determine whether there were different DNA polymerase lineages for baculoviruses and the Hz-1 virus. The results (Fig. 5a) indicated that 75% of the time the DNA polymerases did split into viral and nonviral. Moreover, the NeseNPV DNA polymerase was more related to those from the lepidopteran NPV and GV than the DNA polymerase of the CuniNPV from Diptera. The Hz-1 virus DNA polymerase possibly does not belong to a baculovirus monophyletic group including CuniNPV, NeseNPV, and the lepidopteran NPVs and GVs. These results suggested that possibly CuniNPV was the appropriate rooting outgroup for the monophyletic baculovirus group, which was also supported by the overall level of genomic similarity to NeseNPV and to the other NPVs and GVs (e.g., CuniNPV does not have a polyhedrin/granulin ortholog). The same adjacency pattern, indicating that the CuniNPV is the most ancient lineage, was obtained by midpoint rooting of the maximum-likelihood tree or by UPGMA clustering after excluding all nonbaculovirus DNA polymerases from the analysis (data not shown). Once the cladogenetic events were oriented in time by using the DNA polymerase gene, indicating the split of the CuniNPV lineage from the ancestral stem leading to the NeseNPV and the other lepidopteran NPVs and GVs, a maximum-likelihood tree was estimated for the proteons including 29 conserved genes for 24 baculoviruses and used the CuniNPV as outgroup. Figure 5b shows a tree for the proteons which agrees with the DNA polymerase placement of CuniNPV and NeseNPV. As for the DNA polymerase gene phylogeny, the same adjacency pattern was also obtained without an explicit choice of outgroup by midpoint rooting and UPGMA clustering. Additionally, the group I NPV, group II NPV, and GV clades are delineated using both proteons and the DNA polymerase in the phylogenetic analysis.
FIG. 5.
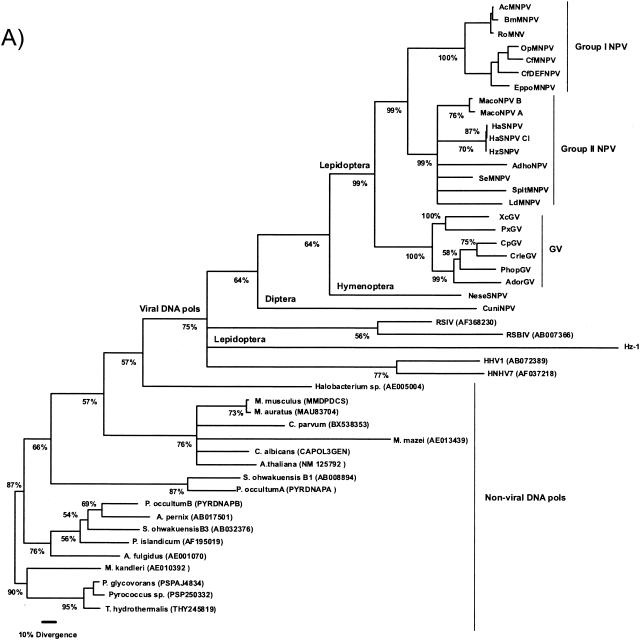
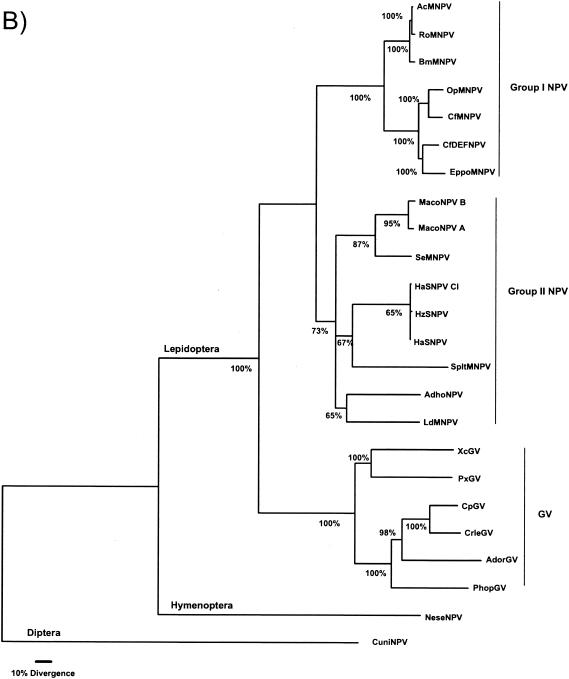
DNA polymerase phylogeny (A) indicates distinct lineages when all sequenced baculoviruses and the Hz-1 DNA polymerases are included. The root position of the DNA polymerase gene suggested that the lepidopteran NPV and GV share a common ancestral lineage with the NeseNPV after its split from the one leading to the CuniNPV. (B) A global maximum-likelihood phylogeny for 29 conserved genes from 24 baculoviruses supports the DNA polymerase tree in its separation of NeseNPV and CuniNPV. The numbers near nodes indicate the percentage of time a partition was found by the quartet method and is considered the level of support for the tree. Only nodes appearing more than 50% of the time were resolved.
NeseNPV is different from other baculoviruses in several notable areas: genome size, smaller number of ORFs, low GC content, hr location, and a general low homology with other baculoviruses. An indirect way of finding important genes is by their conservation among the baculoviruses genomes. In that regard, genes and ORFs also present in distantly related members of the Baculoviridae family, such as those infecting insects from the orders Diptera and Hymenoptera, can be used to predict genes essential for the virus replication cycle. However, there can be a series of other genes used by those viruses that differ from the more studied group of lepidopteran baculoviruses, since the virus-host relationship is very important in the evolutionary history of several organisms, especially for viruses that use the host machinery for their propagation. It will be very informative to compare NeseNPV, which was isolated from Old World N. sertifer insects, to NeleNPV, which infects New World N. lecontei insects, and also other sawfly baculoviruses. This will give a better understanding of the evolutionary history of host-derived genes, such as that for trypsin, and others found in the genomes of hymenopteran baculoviruses. We may also obtain information on the mutation rates of baculoviruses that have diverged over an extensive amount of time. The value of the genome information from hymenopteran baculoviruses will provide a foundation for the evolutionary history of the lepidopteran and dipteran baculoviruses as well. Ultimately this new information and comparison could result in the establishment of a new genus within the family Baculoviridae.
Acknowledgments
This work was supported by United States Department of Agriculture grant 2001-35302-11006.
We thank Ronit Keisari for technical help, Basil Arif and Hilary Lauzon for sharing information about the Neodiprion lecontei NPV, and D. Boucias, Q. Li, and Pamela Howell for comments and corrections on the manuscript.
Footnotes
Florida Agricultural Experiment Station journal series no. R-09762.
REFERENCES
- 1.Afonso, C. L., E. R. Tulman, Z. Lu, C. A. Balinsky, B. A. Moser, J. J. Becnel, D. L. Rock, and G. F. Kutish. 2001. Genome sequence of a baculovirus pathogenic for Culex nigripalpus. J. Virol. 75:11157-11165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahrens, C. H., R. L. Russell, C. J. Funk, J. T. Evans, S. H. Harwood, and G. F. Rohrmann. 1997. The sequence of the Orgyia pseudotsugata multinucleocapsid nuclear polyhedrosis virus genome. Virology 229:381-399. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ayres, M. C., S. C. Howard, J. Kuzio, M. Lopez-Ferber, and R. D. Possee. 1994. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology 202:586-605. [DOI] [PubMed] [Google Scholar]
- 6.Benson, G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Birnbaum, M. J., R. J. Clem, and L. K. Miller. 1994. An apoptosis-inhibiting gene from a nuclear polyhedrosis virus encoding a polypeptide with Cys/His sequence motifs. J. Virol. 68:2521-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blissard, G. W., and G. F. Rohrmann. 1989. Location, sequence, transcriptional mapping and temporal expression of the gp64 envelope glycoprotein gene of the Orgyia pseudotsugata multicapsid nuclear polyhedrosis virus. Virology 170:537-555. [DOI] [PubMed] [Google Scholar]
- 9.Blissard, G. W., and J. R. Wenz. 1992. Baculovirus gp64 envelope glycoprotein is sufficient to mediate pH-dependent membrane fusion. J. Virol. 66:6829-6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braithwaite, D. K., and J. Ito. 1993. Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res. 21:787-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bulach, D. M., C. A. Kumar, A. Zaia, B. Liang, and D. E. Tribe. 1999. Group II nucleopolyhedrovirus subgroups revealed by phylogenetic analysis of polyhedrin and DNA polymerase gene sequences. J. Invertebr. Pathol. 73:59-73. [DOI] [PubMed] [Google Scholar]
- 12.Chen, H.-H., F.-Y. Tsai, and C.-T. Chen. 2001. Negative regulatory regions of the PAT1 promoter of Hz-1 virus contain GATA elements which associate with cellular factors and regulate promoter activity. J. Gen. Virol. 82:313-320. [DOI] [PubMed] [Google Scholar]
- 13.Chen, X., W. F. J. Ijkel, R. Tarchini, X. Sun, H. Sandbrink, H. Wang, S. Peters, D. Zuidema, R. K. Lankhorst, J. M. Vlak, and Z. Hu. 2001. The sequence of the Helicoverpa armigera single nucleocapsid nucleopolyhedrovirus genome. J. Gen. Virol. 82:241-257. [DOI] [PubMed] [Google Scholar]
- 14.Chen, X., W.-J. Zhang, J. Wong, G. Chun, A. Lu, B. F. McCutchen, J. K. Presnail, R. Herrmann, M. Dolan, S. Tingey, Z. H. Hu, and J. M. Vlak. 2002. Comparative analysis of the complete genome sequences of Helicoverpa zea and Helicoverpa armigera single-nucleocapsid nucleopolyhedroviruses. J. Gen. Virol. 83:673-684. [DOI] [PubMed] [Google Scholar]
- 15.Clem, R. J. 1997. Regulation of programmed cell death by baculoviruses, p. 237-266. In L. K. Miller (ed.), The baculoviruses. Plenum Press, New York, N.Y.
- 16.Clem, R. J., M. Fechheimer, and L. K. Miller. 1991. Prevention of apoptosis by a baculovirus gene during infection of insect cells. Science 254:1388-1390. [DOI] [PubMed] [Google Scholar]
- 17.Crook, N. E., R. J. Clem, and L. K. Miller. 1993. An apoptosis-inhibiting baculovirus gene with a zinc-finger-like motif. J. Virol. 67:2168-2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du, Q., D. Lehavi, O. Faktor, Y. Qi, and N. Chejanovsky. 1999. Isolation of an apoptosis suppressor gene of the Spodoptera littoralis nucleopolyhedrovirus. J. Virol. 73:1278-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan, X., K. Thirunavuarasu, and R. F. Weaver. 1996. Temperature-sensitive mutations in the protein kinase (pk-1) gene of Autographa californica nuclear polyhedrosis virus that blocks very late gene expression. Virology 224:1-9. [DOI] [PubMed] [Google Scholar]
- 21.Faulkner, P., J. Kuzio, G. V. Williams, and J. A. Wilson. 1997. Analysis of p74, a PDV envelope protein of Autographa californica nucleopolyhedrovirus required for occlusion body infectivity in vivo. J. Gen. Virol. 78:3091-3100. [DOI] [PubMed] [Google Scholar]
- 22.Federici, B. A. 1997. Baculovirus pathogenesis, p. 33-56. In L. K. Miller (ed.), The baculoviruses. Plenum Press, New York, N.Y.
- 23.Friesen, P. D. 1997. Regulation of baculovirus early gene expression, p. 141-166. In L. K. Miller (ed.), The baculoviruses. Plenum Press, New York, N.Y.
- 24.Funk, C. J., S. C. Braunagel, and G. F. Rohrmann. 1997. Baculovirus structure, p. 7-27. In L. K. Miller (ed.), The baculoviruses. Plenum Press, New York, N.Y.
- 25.Gomi, S., K. Majima, and S. Maeda. 1999. Sequence analysis of the genome of Bombyx mori nucleopolyhedrovirus. J. Gen. Virol. 80:1323-1337. [DOI] [PubMed] [Google Scholar]
- 26.Guarino, L. A., and M. D. Summers. 1986. Interspersed homologous DNA of Autographa californica nuclear polyhedrosis virus enhances delayed-early gene expression. J. Virol. 60:215-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guarino, L. A., B. Xu, J. Jin, and W. Dong. 1998. A virus-encoded RNA polymerase purified from baculovirus-infected cells. J. Virol. 72:7985-7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harrison, R. L., and B. C. Bonning. 2003. Comparative analysis of the genomes of Rachiplusia ou and Autographa californica multiple nucleopolyhedroviruses. J. Gen. Virol. 84:1827-1842. [DOI] [PubMed] [Google Scholar]
- 29.Hashimoto, Y., T. Hayakawa, Y. Ueno, T. Fujita, Y. Sano, and T. Matsumoto. 2000. Sequence analysis of the Plutella xylostella granulovirus genome. Virology 275:358-372. [DOI] [PubMed] [Google Scholar]
- 30.Hayakawa, T., R. Ko, K. Okano, S. Seong, C. Goto, and S. Maeda. 1999. Sequence analysis of the Xestia c-nigrum granulovirus genome. Virology 262:277-297. [DOI] [PubMed] [Google Scholar]
- 31.Herniou, E. A., T. Luque, X. Chen, J. M. Vlak, D. Winstanley, J. S. Cory, and D. R. O'Reilly. 2001. Use of whole genome sequence data to infer baculovirus phylogeny. J. Virol. 75:8117-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herniou, E. A., J. A. Olszewski, J. S. Cory, and D. R. O'Reilly. 2003. The genome sequence and evolution of baculoviruses. Annu. Rev. Entomol. 48:211-234. [DOI] [PubMed] [Google Scholar]
- 33.Hu, Z. H., B. M. Arif, F. Jin, J. W. M. Martens, X. W. Chen, J. S. Sun, D. Zuidema, R. W. Goldbach, and J. M. Vlak. 1998. Distinct gene arrangement in the Buzura suppressaria single-nucleocapsid nucleopolyhedrovirus genome. J. Gen. Virol. 79:2841-2851. [DOI] [PubMed] [Google Scholar]
- 34.Huber, J. 1986. Use of baculoviruses in pest management programs, p. 182-197. In R. R. Granados, and B. A. Federici (ed.), The biology of baculoviruses, vol. II, Practical application for insect control. CRC Press, Boca Raton, Fla. [Google Scholar]
- 35.Hyink O., R. A. Dellow, M. J. Olsen, K. M. Caradoc-Davies, K. Drake, E. A. Herniou, J. S. Cory, D. R. O'Reilly, and V. K. Ward. 2002. Whole genome analysis of the Epiphyas postvittana nucleopolyhedrovirus. J. Gen. Virol. 83:957-971. [DOI] [PubMed] [Google Scholar]
- 36.Ijkel, W. F. J., E. A. van Strien, J. G. M. Heldens, R. Broer, D. Zuidema, R. W. Goldbach, and J. M. Vlak. 1999. Sequence and organization of the Spodoptera exigua multicapsid nucleopolyhedrovirus genome. J. Gen. Virol. 80:3289-3304. [DOI] [PubMed] [Google Scholar]
- 37.Ijkel, W. F. J., M. Westenberg, R. W. Goldbach, G. W. Blissard, J. M. Vlak, and D. Zuidema. 2000. A novel baculovirus envelope fusion protein with a proprotein convertase cleavage site. Virology 275:30-41. [DOI] [PubMed] [Google Scholar]
- 38.Kang, W., M. Suzuki, E. Zemskov, K. Okano, and S. Maeda. 1999. Characterization of baculovirus repeated open reading frames (bro) in Bombyx mori nucleopolyhedrovirus. J. Virol. 73:10339-10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kikhno, I., S. Gutierrez, L. Croizier, G. Croizier, and M. Lopez Ferber. 2002. Characterization of pif, a gene required for the per os infectivity of Spodoptera littoralis nucleopolyhedrovirus. J. Gen. Virol. 83:3013-3022. [DOI] [PubMed] [Google Scholar]
- 40.Kogan, P. H., and G. W. Blissard. 1994. A baculovirus gp64 early promoter is activated by host transcription factor binding to CACGTG and GATA elements. J. Virol. 68:813-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kolomiets, N. G., G. V. Stadnitskii, and A. I. Vorontsov. 1979. The European pine sawfly: distribution, biology, economic importance, natural enemies and control, p. 138. Amerind Publishing, New Delhi, India.
- 42.Kuzio, J., M. N. Pearson, S. H. Harwood, C. J. Funk, J. T. Evans, J. M. Slavicek, and G. F. Rohrmann. 1999. Sequence and analysis of the genome of a baculovirus pathogenic for Lymantria dispar. Virology 253:17-34. [DOI] [PubMed] [Google Scholar]
- 43.Li, L., C. Donly, Q. Li, L. G. Willis, B. A. Keddie, M. A. Erlandson, and D. A. Theilmann. 2002. Identification and genomic analysis of a second species of nucleopolyhedrovirus isolated from Mamestra configurata. Virology 297:226-244. [DOI] [PubMed] [Google Scholar]
- 44.Li, L., S. H. Harwood, and G. F. Rohrmann. 1999. Identification of additional genes that influence baculovirus late gene expression. Virology 255:9-19. [DOI] [PubMed] [Google Scholar]
- 45.Li, L., and G. F. Rohrmann. 2000. Characterization of a baculovirus alkaline nuclease. J. Virol. 74:6401-6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li, Q., C. Donly, L. Li, L. G. Willis, D. A. Theilmann, and M. Erlandson. 2002. Sequence and organization of the Mamestra configurata nucleopolyhedrovirus genome. Virology 294:106-121. [DOI] [PubMed] [Google Scholar]
- 47.Lin, G., and G. W. Blissard. 2002. Analysis of an Autographa californica multicapsid nucleopolyhedrovirus lef-6 null virus: LEF-6 is not essential for viral replication but appears to accelerate late gene transcription. J. Virol. 76:5503-5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu, A., and E. B. Carstens. 1993. Immediate-early baculovirus genes transactivate the p143 gene promoter of Autographa californica nuclear polyhedrosis virus. Virology 195:710-718. [DOI] [PubMed] [Google Scholar]
- 49.Lu, A., P. J. Krell, J. M. Vlak, and G. F. Rohrmann. 1997. Baculovirus DNA replication, p. 171-186. In L. K. Miller (ed.), The baculoviruses. Plenum Press, New York, N.Y.
- 50.Lu, A., and L. K. Miller. 1995. The roles of eighteen baculovirus late expression factor genes in transcription and DNA replication. J. Virol. 69:975-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu, A., and L. K. Miller. 1997. Regulation of baculovirus late and very late gene expression, p. 193-211. In L. K. Miller (ed.), The baculoviruses. Plenum Press, New York, N.Y.
- 52.Lung, O., M. Westenberg, J. M. Vlak, D. Zuidema, and G. W. Blissard. 2002. Pseudotyping Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV): F proteins from group II NPVs are functionally analogous to AcMNPV GP64. J. Virol. 76:5729-5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luque, T., R. Finch, N. Crook, D. R. O'Reilly, and D. Winstanley. 2001. The complete sequence of the Cydia pomonella granulovirus genome. J. Gen. Virol. 82:2531-2547. [DOI] [PubMed] [Google Scholar]
- 54.Maguire, T., P. Harrison, O. Hyink, J. Kalmakoff, and V. K. Ward. 2000. The inhibitors of apoptosis of Epiphyas postvittana nucleopolyhedrovirus. J. Gen. Virol. 81:2803-2811. [DOI] [PubMed] [Google Scholar]
- 55.Maruniak, J. E. 1986. Baculovirus structural proteins and proteins synthesis, p. 29-146. In R. R. Granados, and B. R. Federici (ed.), The biology of baculoviruses, vol. I. Biological properties and molecular biology. CRC Press, Boca Raton, Fla.
- 56.McLachlin, J. R., and L. K. Miller. 1994. Identification and characterization of vlf-1, a baculovirus gene involved in very late gene expression. J. Virol. 68:7746-7756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mikhailov, V. S., A. L. Mikhailova, M. Iwanaga, S. Gomi, and S. Maeda. 1998. Bombyx mori nucleopolyhedrovirus encodes a DNA-binding protein capable of destabilizing duplex DNA. J. Virol. 72:3107-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Monsma, S. A., A. G. P. Oomens, and G. W. Blissard. 1996. The gp64 envelope fusion protein is an essential baculovirus protein required for cell-to-cell transmission of infection. J. Virol. 70:4607-4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moraes, R. R., and J. E. Maruniak. 1996. Detection and identification of multiple baculoviruses using the polymerase chain reaction (PCR) and restriction endonuclease analysis. J. Virol. Methods 63:207-217. [DOI] [PubMed] [Google Scholar]
- 60.Nakai, M., C. Goto, W. Kang, M. Shikata, T. Luque, and Y. Kunimi. 2003. Genome sequence and organization of a nucleopolyhedrovirus isolated from the smaller tea tortrix, Adoxophyes honmai. Virology 316:171-183. [DOI] [PubMed] [Google Scholar]
- 61.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 62.Nissen, M. S., and P. D. Friesen. 1989. Molecular analysis of the transcriptional regulatory region of an early baculovirus gene. J. Virol. 63:493-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Olszewski, J., and L. K. Miller. 1997. Identification and characterization of a baculovirus structural protein, VP1054, required for nucleocapsid formation. J. Virol. 71:5040-5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Olszewski, J., and L. K. Miller. 1997. The role for baculovirus GP41 in budded virus production. Virology 233:92-301. [DOI] [PubMed] [Google Scholar]
- 65.Pang, Y., J. Yu, L. Wang, X. Hu, W. Bao, G. Li, C. Chen, H. Han, S. Hu, and H. Yang. 2001. Sequence analysis of the Spodoptera litura multicapsid nucleopolyhedrovirus genome. Virology 287:391-404. [DOI] [PubMed] [Google Scholar]
- 66.Pearson, M. N., R. M. Bjornson, C. Ahrens, and G. F. Rohrmann. 1993. Identification and characterization of a putative origin of DNA replication in the genome of a baculovirus pathogenic for Orgyia pseudotsugata. Virology 197:715-725. [DOI] [PubMed] [Google Scholar]
- 67.Pearson, M., R. Bjornson, G. Pearson, and G. Rohrmann. 1992. The Autographa californica baculovirus genome: evidence for multiple replication origins. Science 257:1382-1384. [DOI] [PubMed] [Google Scholar]
- 68.Pearson, M. N., R. L. Russell, and G. F. Rohrmann. 2001. Characterization of a baculovirus-encoded protein that is associated with infected-cell membranes and budded virions. Virology 291:22-31. [DOI] [PubMed] [Google Scholar]
- 69.Pei, Z., G. Reske, Q. Huang, B. D. Hammock, Y. Qi, and N. Chejanovsky. 2002. Characterization of the apoptosis suppressor protein P49 from the Spodoptera littoralis nucleopolyhedrovirus. J. Biol. Chem. 277:48677-48684. [DOI] [PubMed] [Google Scholar]
- 70.Pijlman, G. P., A. J. Pruijssers, and J. M. Vlak. 2003. Identification of pif-2, a third conserved baculovirus gene required for per os infection of insects. J. Gen. Virol. 84:2041-2049. [DOI] [PubMed] [Google Scholar]
- 71.Possee, R. D. and G. F. Rohrmann. 1997. Baculovirus genome organization and evolution, p. 109-133. In L. K. Miller (ed.), The baculoviruses. Plenum Press, New York, N.Y.
- 72.Rohrmann, G. F., M. N. Pearson, T. J. Bailey, R. R. Becker, and G. S. Beaudreau. 1981. N-terminal polyhedrin sequences and occluded baculovirus evolution. J. Mol. Evol. 17:329-333. [DOI] [PubMed] [Google Scholar]
- 73.Rypniewski, W. R., A. Perrakis, C. E. Vorgias, and K. S. Wilson. 1994. Evolutionary divergence and conservation of trypsin. Protein Eng. 7:55-64. [DOI] [PubMed] [Google Scholar]
- 74.Tettelin, H., D. Radune, S. Kasif, H. Khouri, and S. L. Salzberg. 1999. Optimized multiplex PCR: efficiently closing a whole-genome shotgun sequencing project. Genomics 62:500-507. [DOI] [PubMed] [Google Scholar]
- 75.Vlak, J. M., and G. E. Smith. 1982. Orientation of the genome of Autographa californica nuclear polyhedrosis virus: a proposal. J. Virol. 41:1118-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang, H., X. Chen, H. Wang, B. M. Arif, J. M. Vlak, and Z. Hu. 2001. Nucleotide sequence and transcriptional analysis of a putative basic DNA-binding protein of Helicoverpa armigera nucleopolyhedrovirus. Virus Genes 22:113-120. [DOI] [PubMed] [Google Scholar]
- 77.Wheeler, D. L., D. M. Church, S. Federhen, A. E. Lash, T. L. Madden, J. U. Pontious, G. D. Schuler, L. M. Schriml, E. Sequeira, T. A. Tatusova, and L. Wagner. 2003. Database resources of the National Center for Biotechnology. Nucleic Acids Res. 31:28-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wormleaton, S., J. Kuzio, and D. Winstanley. 2003. The complete sequence of the Adoxophyes orana granulovirus genome. Virology 311:350-365. [DOI] [PubMed] [Google Scholar]
- 79.Wu, X., and L. A. Guarino. 2003. Autographa californica nucleopolyhedrovirus orf69 encodes an RNA cap (nucleoside-2′-O)-methyltransferase. J. Virol. 77:3430-3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang, S., and L. K. Miller. 1998. Expression and mutation analysis of the baculovirus very late factor 1 (vlf-1) gene. Virology 245:99-109. [DOI] [PubMed] [Google Scholar]
- 81.Zanotto, P. M. A., B. D. Kessing, and J. E. Maruniak. 1993. Phylogenetic interrelationships among baculoviruses: evolutionary rates and host associations. J. Invertebr. Pathol. 62:147-164. [DOI] [PubMed] [Google Scholar]
- 82.Zoog, S. J., J. J. Schiller, J. A. Wetter, N. Chejanovsky, and P. D. Friesen. 2002. Baculovirus apoptotic suppressor P49 is a substrate inhibitor of initiator caspases resistant to P35 in vivo. EMBO J. 21:5130-5140. [DOI] [PMC free article] [PubMed] [Google Scholar]


