Abstract
Background
Enterococcus is one of the major human pathogens able to acquire multiple antibiotic-resistant markers as well as virulence factors which also colonize remote ecosystems, including wild animals. In this work, we characterized the Enterococcus population colonizing the gut of Chilean Altiplano camelids without foreign human contact.
Material and methods
Rectal swabs from 40 llamas and 10 alpacas were seeded in M-Enterococcus agar, and we selected a total of 57 isolates. Species identification was performed by biochemical classical tests, semi-automated WIDER system, mass spectrometry analysis by MALDI-TOF (matrix-assisted laser desorption/ionization with a time-of-flight mass spectrometer), and, finally, nucleotide sequence of internal fragments of the 16S rRNA, rpoB, pheS, and aac(6)-I genes. Genetic diversity was measured by pulsed field gel electrophoresis (PFGE)-SmaI, whereas the antibiotic susceptibility was determined by the WIDER system. Carriage of virulence factors was explored by polymerase chain reaction (PCR).
Results
Our results demonstrated that the most prevalent specie was Enterococcus hirae (82%), followed by other non–Enterococcus faecalis and non–Enterococcus faecium species. Some discrepancies were detected among the identification methods used, and the most reliable were the rpoB, pheS, and aac(6)-I nucleotide sequencing. Selected isolates exhibited susceptibility to almost all studied antibiotics, and virulence factors were not detected by PCR. Finally, some predominant clones were characterized by PFGE into a diverse genetic background.
Conclusion
Enterococcus species from the Chilean camelids’ gut microbiota were different from those adapted to humans, and they remained free of antibiotic resistance mechanisms as well as virulence factors.
Keywords: Enterococcus, antibiotic resistance, molecular identification, camelids, PFGE, virulence factors
Latin American camelids are genetically related to those living in Asia or Africa, although they are much smaller, have no hump, and have adaptations that allow them to thrive at high elevations (1). Nowadays, they are divided into four species, two of them free-living (Lama guanicoe and Vicugna vicugna) and the other two domesticated (Lama glama and Lama pacos) (2, 3). The natural habitat of South American camelids is the Altiplano, the high Andean plateau extending through the countries of Bolivia, Peru, Argentina, and Chile. The Altiplano is a special ecosystem characterized by its high altitude and the scarcity of oxygen.
The main occupation of Altiplano natives is camelid stockbreeding, and they exploit both their meat and their fleece. The highest mortality and morbidity period of these animals is among the newborn; in industrial countries, this affects only 2–3% (4–7), whereas in the Altiplano the mortality is higher (8). In Chile, camelids inhabit the regions of Arica and Parinacota (about 3,800 m above sea level) and consist of a population of 19,066 alpacas and 17,392 llamas (9).
The major health diseases affecting camelids are the infectious diseases, including newborn diarrhea, acute respiratory processes, and bacteremia (10). Moreover, they have no access to veterinary control, and infrequent contact with foreign humans and antimicrobial drugs. In 1993, the first report of bacteremia in a llama by Enterococcus species (11) was described, and more recently neonatal sepsis was characterized by Dolente et al. (12). The aim of this work is to characterize the Enterococcus species of the gut microbiota of Chilean Altiplano camelids, focusing on their identification by phenotypic and molecular methods.
Material and methods
Samples collection
A total of 50 rectal swabs were collected from 40 alpacas and 10 llamas in different farms of Putre (Parinacota). Samples were collected during 2 different days, maintained at room temperature using Copan Transystem swabs (Copan, Brescia, Italy), and transported to the Antofagasta University. Samples were pre-enriched on brain–heart infusion broth at 37°C for 18 h, and then seeded on M-Enterococcus and incubated at 37°C for 48 h. All media were purchased from Difco (Detroit, MI).
Bacterial identification and antibiotic susceptibility
From each plate, at least five different colonies were reseeded and identified to the species level by MALDI-TOF mass spectrometry (MS). Complementary phenotypic analysis as well as antibiotic susceptibility were performed by the WIDER semiautomatic system (Fco. Soria Melguizo, Madrid, Spain). Clinical and Laboratory Standards Institute (CLSI) criteria for antibiotic susceptibility breakpoints were applied (13). Whole DNA of all enterococci strains was obtained by resuspending one colony in 200 µl of distilled water, heating at 95°C for 10 min, centrifuging for 5 min at 13,000 rpm, and collecting the supernatant. Five µl of the supernatant were used as a template in the polymerase chain reaction (PCR). Enterococcal species were assigned after PCR amplification of 16S rRNA, rpoB, rpoA, aac(6′)-I, and pheS genes; subsequent nucleotide sequencing in a ABI PRISM 3130 Genetic Analyzer (Applied Biosystems, San Mateo, CA); and comparison with the BLAST database tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Primers and conditions are shown in Table 1.
Table 1.
Primers and conditions used in this study
| Primer | Sequence (5′→3′) | Size | Reference |
|---|---|---|---|
| BAc08F | AGAGTTTGATCCTGGCTCAG | 1,398 | |
| Uni 1390R | GACGGGCGGTGTGTACAA | ||
| rpoA-21-F | GACAGACCCTCACGAATA | 533 | Naser et al. (14) |
| rpoA-23-R | AGTTCATCATGCTGTAGTA | ||
| aac-1 | GGATAGCGGATGATTATCA | 840 | Del Campo et al. (15) |
| aac-2 | TAAGAGTTTAATGAATAATTA | ||
| pheS-21-F | CAYCCNGCHCGYGAYATGC | 455 | Naser et al.(14) |
| pheS-22-R | CCWARVCCRAARGCAAARCC | ||
| ace-F | GTCTGTCTTTTCACTTGTTTCT | 1,003 | Rich et al.(16) |
| ace-R | GAGCAAAAGTTCAATCGTTGAC | ||
| efaA-F | GACAGACCCTCACGAATA | 705 | Eaton and Gasson (17) |
| efaA-R | AGTTCATCATGCTGTAGTA | ||
| cylA-F | TGGATGATAGTGATAGGAAGT | 517 | Eaton and Gasson (17) |
| cylA-R | TCTACAGTAAATCTTTCGTCA | ||
| agg-F | AAGAAAAAGAAGTAGACCAAC | 1,553 | Eaton and Gasson (17) |
| agg-R | AAACGGCAAGACAAGTAAATA | ||
| esp-F | TTGCTAATGCTAGTCCACGACC | 933 | Eaton and Gasson (17) |
| esp-R | GCGTCAACACTTGCATTGCCGA | ||
| gelE-F | ACGCATTGCTTTTCCATC | 419 | Eaton and Gasson (17) |
| gelE-R | ACCCCGTATCATTGGTTT |
Virulence genes detection
The presence of efaA, agg, gelE, esp, cylA, and ace genes was investigated by PCR using specific primers and conditions previously described (Table 1). Positive and negative control strains were included in each experiment.
Genetic relationship
Pulsed field gel electrophoresis (PFGE) genomic DNA was prepared in agarose plugs, digested with SmaI (New England BioLabs, Beverly, MA), and run on a CHEF-DR III (Bio-Rad Laboratories, Hercules, CA), as described previously (18). Electrophoresis conditions were 6 V/cm2, 14°C, and 22 h 5–35s. Band pattern analysis was performed constructing an UPGMA (unweighted pair group method with arithmetic mean) dendrogram based on Dice's coefficient by the Phoretrix 5.0 software (TotalLab, Newcastle upon Tyne, UK). Isolates were considered related if their PFGE banding patterns were ≥80% similar.
Results and discussion
After selective culture of the 50 rectal swabs from 40 llamas and 10 alpacas, a total of 57 unrelated colonies presenting compatible morphology with Enterococcus were finally selected. Initially, up to five morphologically different colonies were analyzed from each sample, but finally only one colony by species and pulsotype was further selected. The most frequently human-related species, Enterococcus faecalis and Enterococcus faecium, were not found, and some discrepancies were detected in their identification (Table 2).
Table 2.
Bacterial identification obtained by the different systems used in the 57 isolates
| Identification system | ||||||
|---|---|---|---|---|---|---|
| WIDER | MALDI-TOF | 16S rRNA | rpoB | pheS | aac(6′)-I | |
| E. hirae | 41 | 46 | 47 | 47 | 47 | 47 |
| E. mundtii | 12 | 7 | 6 | 6 | 6 | – |
| E. casseliflavus | 3 | 3 | 3 | 3 | 3 | – |
| E. gallinarum | 1 | 1 | 1 | 1 | 1 | – |
According to material previously described (14), the species was finally assigned by the 16S rRNA, rpoB, pheS, and aac(6′)-I genes, which were concordant in all of them. The worst result was obtained with the semiautomated WIDER system, based on phenotypical and biochemical tests. MALDI-TOF technology misidentified only the Enterococcus hirae isolates, which were assigned to Enterococcus mundtii. Nevertheless, MALDI-TOF could be actually considered as a reliable method for enterococcal identification (19–22).
E. hirae was the most frequent species detected (82.5%), followed by E. mundtii (10.5%), Enterococcus casseliflavus (5.2%), and, finally, Enterococcus gallinarum (1.7%). Previous reports of gut microbiota from wild animals, including geese, free-living raptors, and other animals, confirmed the presence of these species (23–25).
Antibiotic resistance for penicillin, ampicillin, amoxicillin/clavulanate, daptomycin, levofloxacin, erythromycin, linezolid, minocycline, and nitrofurantoin was not detected in any enterococcal isolate, and glycopeptide resistance was confirmed in three E. casseliflavus isolates (CMI=8 µg/ml for vancomycin and susceptibility to teicoplanin). Five E. hirae isolates and one E. casseliflavus isolate exhibited resistance to quinupristin/dalfopristin. The E. gallinarum isolate exhibited resistance to fosfomycin and rifampin. Virulence genes were not amplified in any enterococcal strains. Dissemination of antibiotic resistance mechanisms and virulence genes has been demonstrated in enterococcal isolates from wild animals (25–28), although in our case, the Altiplano remains a remote region with scarce antibiotic exposition.
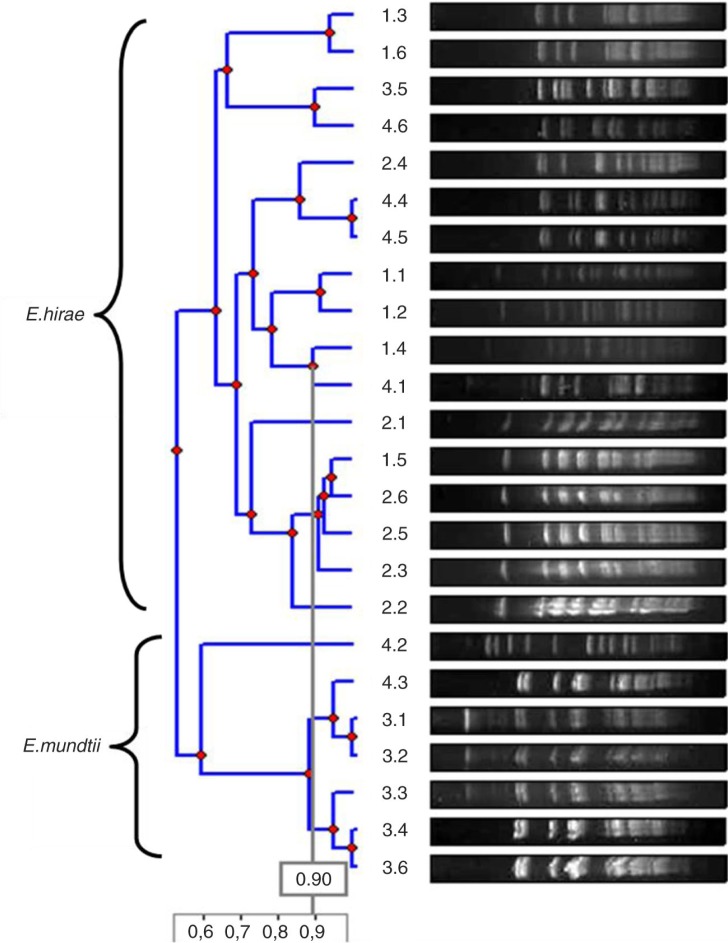
As to the genetic variability of these isolates by the PFGE analysis, eight unrelated band patterns for the E. hirae isolates and three for the E. mundtii isolates were identified (Fig. 1). Pulsotype F was the most frequent, representing 57% of all E. hirae isolates, suggesting a common source of this clone or animal-to-animal transmission. Although the animals belonged to different farms and are separated at night, during the day they roam freely and can have contact.
Fig. 1.
Dendrogram of the genetic relationship of the most frequent E. hirae and E. mundtii pulsotypes.
In conclusion, Enterococcus spp. from the Chilean camelids’ gut microbiota were different from those adapted to humans (E. faecalis and E. faecium), and remained free of antibiotic resistance mechanisms as well as virulence factors. Genetic studies demonstrated the existence of several lineages in each species, suggesting a non-clonal situation. Although Enterococcus might be a major cause of bacteremia in newborn animals (11, 12), our results pointed out that potentially pathogenic isolates are not present in the gut of these animals.
Conflict of interest and funding
The authors have not received any funding or benefits from industry or elsewhere to conduct this study. Part of this work has been finnanced by the European Commission Project EvoTAR-282004 and the Proyecto CODEI N°5393, DINV, Universidad de Antofagasta.
References
- 1.Wheeler JC. Evolution and present situation of the South-American camelidae. Biol J Linn Soc. 1995;54:271–95. [Google Scholar]
- 2.Kadwell M, Fernandez M, Stanley HF, Baldi R. Genetic analysis reveals the wild ancestors of the llama and the alpaca. Proc R Soc Lond. 2001;268:2575–84. doi: 10.1098/rspb.2001.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marín JC, Zapata B, González BA, Bonacic C, Wheeler JC, Casey C, et al. Sistemática, taxonomía y domesticación de alpacas y llamas: nueva evidencia cromosómica y molecular. Rev Chil Hist Nat. 2007;80:121–40. [Google Scholar]
- 4.D'Alterio GL, Knowles TG, Eknaes EI, Loevland IE, Foster AP. Postal survey of the population of South American camelids in the United Kingdom in 2000/01. Vet Rec. 2006;158:86–90. doi: 10.1136/vr.158.3.86. [DOI] [PubMed] [Google Scholar]
- 5.Davis R, Keeble E, Wright A, Morgan KL. South American camelids in the United Kingdom: population statistics, mortality rates and causes of death. Vet Rec. 1998;142:162–6. doi: 10.1136/vr.142.7.162. [DOI] [PubMed] [Google Scholar]
- 6.Sharpe MS, Lord LK, Wittum TE, Anderson DE. Pre-weaning morbidity and mortality of llamas and alpacas. Aust Vet J. 2009;87:56–60. doi: 10.1111/j.1751-0813.2008.00377.x. [DOI] [PubMed] [Google Scholar]
- 7.Wright A, Davis R, Keeble E, Morgan KL. South American camelids in the United Kingdom: reproductive failure, pregnancy diagnosis and neonatal care. Vet Rec. 1998;142:214–15. [PubMed] [Google Scholar]
- 8.Ameghino E, De Martini J. Boletín de divulgación del Instituto Veterinario de Investigaciones Tropicales y de Altura (IVITA) Lima, Perú: Universidad de San Marcos; 1991. Mortalidad de crías de alpacas; pp. 71–80. [Google Scholar]
- 9.Instituto Nacional de Estadística (INE) Santiago, Chile: Instituto Nacional de Estadística; 2007. VII Censo Nacional Agropecuario y Forestal. [Google Scholar]
- 10.Martín C, Pinto J, Chris E, Vázquez C, Dolores M. Camélidos Sudamericanos: estado sanitario de sus crías. RCCV. 2010;4:37–50. [Google Scholar]
- 11.Burkhardt J, Janovitz E, Bowersock T, Higgins R. Septicemic Enterococcus infection in an adult llama. J Vet Diagn Invest. 1993;5:106. doi: 10.1177/104063879300500126. [DOI] [PubMed] [Google Scholar]
- 12.Dolente BA, Lindborg S, Palmer JE, Wilkins PA. Culture-positive sepsis in neonatal camelids: 21 cases. J Vet Intern Med. 2007;21:519. doi: 10.1892/0891-6640(2007)21[519:csincc]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 13.Clinical Laboratory Standards Institute. Wayne, PA: CLSI; 2012. Performance standards for antimicrobial susceptibility testing: twenty-second informational. Supplement M100–S22. [Google Scholar]
- 14.Naser SM, Dawyndt P, Hoste B, Gevers D, Vandemeulebroecke K, Cleenwerck I, et al. Identification of lactobacilli by pheS and rpoA gene sequence analyses. Int J Syst Evol Microbiol. 2005;57:2777–89. doi: 10.1099/ijs.0.64711-0. [DOI] [PubMed] [Google Scholar]
- 15.Del Campo R, Galán JC, Tenorio C, Ruiz-Garbajosa P, Zarazaga M, Torres C, et al. New aac(6′)-I genes in Enterococcus hirae and Enterococcus durans: effect on {beta}-lactam/aminoglycoside synergy. J Antimicrob Chemother. 2005;55:1053–5. doi: 10.1093/jac/dki138. [DOI] [PubMed] [Google Scholar]
- 16.Rich RL, Kreikemeyer B, Owens RT, LaBrenz S, Narayana SV, Weinstock GM, et al. Ace is a collagen-binding MSCRAMM from Enterococcus faecalis . J Biol Chem. 1999;274:26939–45. doi: 10.1074/jbc.274.38.26939. [DOI] [PubMed] [Google Scholar]
- 17.Eaton TJ, Gasson MJ. Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl Environ Microbiol. 2001;67:1628–35. doi: 10.1128/AEM.67.4.1628-1635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murray BE, Singh KV, Heath JD, Sharma BR, Weinstock GM. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J Clin Microbiol. 1990;28:2059–63. doi: 10.1128/jcm.28.9.2059-2063.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Facklam RR, Collins MD. Identification of Enterococcus species isolated from human infections by a conventional test scheme. J Clin Microbiol. 1989;27:731–4. doi: 10.1128/jcm.27.4.731-734.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goh SH, Facklam RR, Chang M, Hill JE, Tyrrell GJ, Burns EC, et al. Identification of Enterococcus species and phenotypically similar Lactococcus and Vagococcus species by reverse checkerboard hybridization to chaperonin 60 gene sequences. J Clin Microbiol. 2000;38:3953–9. doi: 10.1128/jcm.38.11.3953-3959.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benagli C, Rossi V, Dolina M, Tonolla M, Petrini O. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for the identification of clinically relevant bacteria. PLoS One. 2011;6:e16424. doi: 10.1371/journal.pone.0016424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang H, Ohlsson AK, Ullberg M, Ozenci V. Evaluation of species-specific PCR, Bruker MS, VITEK MS and the VITEK 2 system for the identification of clinical Enterococcus isolates. Eur J Clin Microbiol Infect Dis. 2012;31:3073–7. doi: 10.1007/s10096-012-1667-x. [DOI] [PubMed] [Google Scholar]
- 23.Han D, Unno T, Jang J, Lim K, Lee SN, Ko G, et al. The occurrence of virulence traits among high-level aminoglycosides resistant Enterococcus isolates obtained from feces of humans, animals, and birds in South Korea. Int J Food Microbiol. 2011;144:387–92. doi: 10.1016/j.ijfoodmicro.2010.10.024. [DOI] [PubMed] [Google Scholar]
- 24.Marrow J, Whittington JK, Mitchell M, Hoyer LL, Maddox C. Prevalence and antibiotic-resistance characteristics of Enterococcus spp. isolated from free-living and captive raptors in Central Illinois. J Wildl Dis. 2009;45:302–13. doi: 10.7589/0090-3558-45.2.302. [DOI] [PubMed] [Google Scholar]
- 25.Radhouani H, Silva N, Poeta P, Torres C, Correia S, Igrejas G. Potential impact of antimicrobial resistance in wildlife, environment and human health. Front Microbiol. 2014;5:23. doi: 10.3389/fmicb.2014.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lanthier M, Scott A, Lapen DR, Zhang Y, Topp E. Frequency of virulence genes and antibiotic resistances in Enterococcus spp. isolates from wastewater and feces of domesticated mammals and birds, and wildlife. Can J Microbiol. 2010;56:715–29. doi: 10.1139/w10-046. [DOI] [PubMed] [Google Scholar]
- 27.Poeta P, Costa D, Igrejas G, Rodrigues J, Torres C. Phenotypic and genotypic characterization of antimicrobial resistance in faecal enterococci from wild boars (Sus scrofa) Vet Microbiol. 2007;125:368–74. doi: 10.1016/j.vetmic.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 28.Radhouani H, Igrejas G, Carvalho C, Pinto L, Gonçalves A, Lopez M, et al. Clonal lineages, antibiotic resistance and virulence factors in vancomycin-resistant enterococci isolated from fecal samples of red foxes (Vulpes vulpes) J Wildl Dis. 2011;47:769–73. doi: 10.7589/0090-3558-47.3.769. [DOI] [PubMed] [Google Scholar]