Abstract
Inflammasomes are large multiprotein complexes that have the ability to sense intracellular danger signals through special NOD-like receptors or NLRs. They include NLRP3, NLRC4, AIM2 and NLRP6. They are involved in recognizing diverse microbial (bacteria, viruses, fungi and parasites), stress and damage signals, which result in direct activation of caspase-1, leading to secretion of potent pro-inflammatory cytokines and pyroptosis. NLRP3 is the most studied antimicrobial immune response inflammasome. Recent studies reveal expression of inflammasomes in innate immune response cells including monocytes, macrophages, neutrophils, and dendritic cells. Inflammasome deficiency has been linked to alterations in the gastrointestinal microflora. Alterations in the microbiome population and/or changes in gut permeability promote microbial translocation into the portal circulation and thus directly to the liver. Gut derived lipopolysaccharides (LPS) play a significant role in several liver diseases. Recent advancements in the sequencing technologies along with improved methods in metagenomics and bioinformatics have provided effective tools for investigating the 1014 microorganisms of the human microbiome that inhabit the human gut. In this review, we examine the significance of inflammasomes in relation to the gut microflora and liver. This review also highlights the emerging functions of human microbiota in health and liver diseases.
Keywords: inflammasomes, microbiota, liver disease, inflammation
Abbreviations: LPS, lipopolysaccharides; NOD, nucleotide-binding oligomerization domain; NLR, NOD-like receptor; PAMPs, pathogen associated molecular patterns; DAMP, damage associated molecular patterns; AIM2, absent in melanoma 2; IL, interleukin; IFN, interferon; TNF-α, tumor necrosis factor-α; NACHT, domain present in NAIP, CIITA, HET-E (Podospora anserina incompatibility, locus protein) and telomerase associated protein; LRR, leucine-rich repeat; MDP, muramyl dipeptide; ATP, adenosine triphosphate; ROS, reactive oxygen species; NAIP, neuronal apoptosis inhibitor protein; HMGB1, high-mobility group box1; BMDMs, bone marrow-derived macrophages; CTB, Cholera toxin B; TLR, toll-like receptor; NK/NKT, natural killer/natural killer T cells; mCMV, mouse cytomegalovirus; NAFLD, non-alcoholic fatty liver disease; CARD, caspase activation and recruitment domain
Inflammation is the defense mechanism of the body following tissue injury. In health, this defense mechanism is smoothly switched on and off following initiation and cessation of injury. However, continuous exposure to the noxious stimulus leads to chronic inflammation, parenchymal cell loss, healing by fibrosis and, in the case of the liver, eventually to liver cirrhosis. This is the common pathogenetic process underpinning many chronic liver diseases. The process of inflammation involves innate immune cells and production of pro-inflammatory cytokines IL-1α, IL-1β, and TNF-α.1
Macrophages are important cells of the innate immune response and play a crucial role in the initiation and resolution of inflammation. They perform three key functions, namely phagocytosis and destruction of infectious agents, antigen presentation and immune modulation, and initiating the release of various cytokines and growth factors.2–4 Two types of macrophages have been described. M1 macrophages or immune effector cells engulf and digest damaged cells. They are activated by pro-inflammatory mediators such as lipopolysaccharides (LPS), IL-1β and IFN-γ. In turn, these cytokines produce other pro-inflammatory cytokines (TNF-α, IFN-γ, IL-6 and IL-12) which generate reactive oxygen species.5,6 On the other hand, M2 macrophages function in wound healing and tissue repair and produce the anti-inflammatory cytokine IL-10, which turns off immune system activation.
Disturbances of steady state or acute damage lead to the initiation of inflammation.7 Several receptors have been studied that distinguish between homeostasis and agents harmful to the host. Some receptors recognize pathogen associated molecular patterns (PAMPs) which are important for the survival of microbes. Thus, pathogens that are devoid of these structures can be explicitly sensed in the tissues.8 Alteration in tissue homeostasis due to microbial or non-microbial agents causes the release of damage associated molecular patterns (DAMPs). These molecular patterns help in sensing stressed tissue.9
Inflammasomes
The term inflammasome was first introduced by Martinon and co-workers.10 They are large multiprotein complexes which have the ability to sense intracellular danger signals through NOD-like receptors or NLRs.11 NOD-like receptors are members of the pattern recognition receptor family. Two domains that play a key role in the activation of the inflammasome are the C-terminal leucine rich repeat (LRR) domain and the N-terminal domain present in NAIP, CIITA, HET-E (Podospora anserine incompatibility locus protein) and telomerase associated protein (NACHT) domain. Recognition of the ligand depends on the leucine rich-repeat (LRR) domain whereas oligomerization and dNTPase activity are functions of the central NACHT domain.6 A complex is formed between the NLR sensor and the effector molecule, pro-caspase-1, which may or may not require an adaptor molecule such as apoptosis-associated speck-like caspase activation and recruitment domain (CARD) domain containing protein (ASC).10–12 Activation of the inflammasome is a two-step process. Signal 1, due to toll-like receptor (TLR) activation, helps in up-regulating expression of the inflammasome while signal 2 triggers its functional activation with the help of inflammasome ligand. Although, many inflammasomes have been discovered till date, such as: NLRP1, 2, 3, 6, 10, 12, NLRC4 and AIM2, we will elaborate the main inflammasome prototypes in this review namely: NLRP1 (NALP1), NLRP3 (NALP3, cryopyrin), NLRC4 (IPAF) and AIM2.11,12 Although differing in ligand recognition sites and utilization of adaptor molecules, the core function of different inflammasomes is activation of caspase-1.
NLRP1 Inflammasome
The first inflammasome described consists of NACHT, PYD (pyrin domain) and LRR domains. The C-terminal CARD domain interacts directly with caspase-1. Importantly, activity of NLRP1 inflammasome is further enhanced by the presence of ASC.13 Activation of NLRP1 is triggered by (muramyl dipeptide) and the lethal Bacillus anthracis toxin.14–17 A unique feature of NLRP1 is that it can localize in the nucleus unlike other inflammasomes which are distributed in the cytoplasm.18
NLRP3 Inflammasome
Hoffman and co-workers (2001) first described the NLRP3, the most elaborately characterized inflammasome, containing NACHT, LRR and PYD domains containing protein 3 or cryoporin.19. Absence of CARD domain makes it important for the ASC molecule to be present for complex formation.13 Three main pathways have been described for activation of this inflammasome. The first is potassium efflux, due to the presence of P2X7 purinergic receptors that sense extracellular ATP, which recruits pannexin and causes activation of NLRP3.20–22 Secondly, crystals or large particles such as silica, asbestos, aluminum, amyloid, monosodium urate and cholesterol also lead to activation of NLRP3 inflammasome.23–31 Thirdly, activation of NLRP3 inflammasome also depends on reactive oxygen species (ROS), as suggested by a study in which blocking of priming by ROS inhibitors prevented activation of the NLRP3 inflammasome.12
NLRC4 Inflammasome
NLRC4 inflammasome contains protein 4 and is activated by flagellin of Gram-negative and Gram-positive bacteria as well as type III secretion system (T3SS) of Gram-negative bacteria.32–34 However, the mechanism involving activation of this inflammasome is not yet fully understood. Although other NLR proteins, such as murine 143 NAIP5 and NAIP2 also interact with bacterial flagellin or type III secretion system (T3SS), it is the rod components that cause the assembly and activation of NLRC4 inflammasome.35 Recently, it has been shown that Salmonella typhimurium infection of NLRP4 macrophages causes phosphorylation of NLRC4 S533. This is followed by the conformational changes necessary for NLRC4 inflammasome activity and host innate immunity.36
AIM2 Inflammasome
AIM2 is a cytosolic inflammasome which senses dsDNA. It is activated by bacterial, viral and mammalian host DNA, causing caspase-1 activation.37–39 AIM2 can bind directly to its ligand. Strikingly, it recognizes the mammalian DNA which acts as a contributory factor toward the pathogenesis of autoimmune diseases.40 The association of helicase receptor RIG of dsRNA with the inflammasome adaptor molecule ASC leads to inflammasome activation which triggers caspase-1 activation.41
Inflammasome Activates Caspase-1
Inflammatory processes involve pro-inflammatory cytokine IL-1β.42 Though synthesis of IL-1β does not require any signal sequence, interestingly, its activation and release from cellular compartment is dependent on the cysteine protease, caspase-1. Following the protein secretion pathway, caspase-1 also contributes to the processing and secretion of IL-18.
Moreover, IL-1α and fibroblast growth factor-2 are also secreted by the caspase-1 mechanism.43 Bergsbaken and group studied the role of caspase-1 in pyroptosis, which is programmed cell death involving both apoptosis (DNA fragmentation) and necrosis (inflammation and releasing cytokines).44 A recent study supported that pyroptosis occurs due to altered secretion and release of IL-1β.45 Basically, caspase-1 becomes proteolytically active by controlled dimerization in the inflammasome where it is synthesized as the inactive zymogen, pro-caspase-1. Figure 1 depicts the different inflammatory caspases in both humans and mice. Caspase-1 activating complex has been called the inflammasome.10 The sequence of caspase-1 activation and IL-1β secretion results in the activated inflammasome. Although studies by Andrei et al have found caspase-1 in secretory lysosomes along with other lysosomal proteins and pro-IL-1β, much more light needs to be shed on the molecular mechanism associated with secretion of IL-1β. Release of IL-1β depends on ATP-triggered potassium efflux which causes calcium-dependent phospholipase C activation. This in turn triggers the activation of phospholipase A2 and the process of exocytosis releases IL-1β.46
Figure 1.
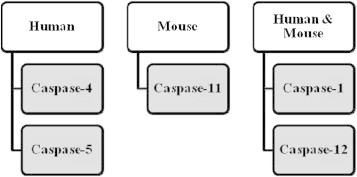
Inflammatory caspases.
The activation of caspase-1 by inflammasome complexes due to PAMPs and DAMPs leads to the maturation and secretion of IL-1β and IL-18. Recently, certain effector mechanisms like induction of pyroptosis, restricted bacterial replication, cellular repair by activation of lipid metabolic pathways and the secretion of DAMPs and cytokines have been reported to be activated independent of IL-1β and IL-18. Thus, non-canonical functions of caspase-1 demonstrate various mechanisms which elaborate the role of inflammasomes that might help in innate immunity, repair process and host defense.47
Caspase-11 belongs to caspase-1 subfamily of proteases and is known to be homologous to the human caspase-4. Not yet fully explored, caspase-11 is believed to be an inflammatory caspase like caspase-1 with 46% similarity between the two.48 Also, caspase-11 is involved in secretion of IL-1α and high-mobility group box1 (HMGB1) which is an important inflammatory mediator.49 Kayagaki et al investigated the role of caspase-11. Casp11 −/− mice was generated by targeting Casp11 exon 5 which contain the critical catalytic residue cysteine 254. Similar amount of IL-1β secretion was observed from both wild type and Casp11 −/− bone marrow-derived macrophages (BMDMs) by canonical stimuli (ATP, Francisella tularensis, Pseudomonas aeruginosa, flagellin, monosodium urate, nigericin and calcium pyrophosphate). However, Cholera toxin B (CTB), Escherichia coli, Citrobacter rodentium and Vibrio cholera stimuli showed that caspase-11 is indispensible. Thus, caspase-11 dependent inflammasomes are called non-canonical inflammasomes. Caspase-1 is important for proteolytic processing and release of IL-1β, IL-18, IL-1α, HMGB1 and pyroptosis following canonical inflammasome activation. However, in the case of non-canonical inflammasomes, stimulators CTB, E. coli, C. rodentium and V. cholera lead to secretion of IL-1α, HMGB1 and can trigger a death signal independent of caspase-1. CTB and E. coli cause secretion of caspase-11 independent of NLRP3 and ASC. Two important pathways followed by caspase-11 are: caspase-1 dependant production of IL-1β and IL-18 and caspase-1 independent macrophage death in response to non-canonical activators. Kayagaki and co-workers showed that caspase-11 may prove to be the important effector of inflammatory response instead of caspase-1. They further suggested that caspase-4 and caspase-5 may be better therapeutic targets as compared to caspase-1 for the patients suffering from sepsis.49
Inflammasomes Initiate Antimicrobial Host Response
The role of inflammasome has been extensively studied in vivo by Elinav and group revealing its association with microbial innate immune response.50 NLRP3, with its involvement in antibacterial, viral, fungal and parasitic immune responses, is the most studied inflammasome in regard to the antimicrobial response. An example which depicts the activation of NLRP3 is the influenza A virus acting as an indirect activator of inflammasome. Viral RNA recognition through toll-like receptor 7 (TLR7) induces transcription of NLRP3 inflammasome components.51 TLRs play a pivotal role in host cell recognition and responses to microbial pathogens by acting as germline-encoded pattern recognition receptors (PRRs).52
Interestingly, activation of mouse NLRP1 inflammasome plays a pivotal role in initiation of antimicrobial response to B. anthracis infection. This involves caspase-1 triggered pyroptosis of infected macrophages, limiting infection and initiating antimicrobial neutrophilic reaction.53 Hsu et al showed that the formation of NLRP1 inflammasome was ASC-dependent in humans whereas mouse NLRP1b caused the activation of caspase-1 in an ASC-independent manner.17 Indeed, muramyl dipeptide can be sensed by NLRP1, like NOD2 and NLRP3.54 Microbial infection elicits effector mechanisms during the process of host defense.7 Pyroptosis plays a key role during in vivo infection by Salmonella enterica Typhimurium, Legionella pneumophila or Burkholderia thailandensis through its caspase-1 mediated effects, although there have been studies on the role of IL-1β and IL-18 induced by inflammasome during infection by influenza virus and Shigella.55 The cooperation of distinct inflammasomes plays a key role in host defense and in clearing infection by eliciting a protective immune response. Infection by Salmonella typhimurium is an example of an overlapping effect of inflammasomes, where deficiency in vivo of either NLRP3 or NLRC4 alone does not lead to increased bacterial infection. Surprisingly, NLRC4 and NLRP3 deficient mice had increased chances of infection when compared to mice with caspase-1 deficiency.56
Liver and the Inflammasomes
While the liver contains a preponderance of parenchymal cells or hepatocytes, it also has large numbers of immune cells including macrophages, neutrophil leukocytes, dendritic cells, T cells, NK/NKT cells and B lymphocytes.6 Recent studies have revealed the expression of inflammasomes in innate immune cells including monocytes, macrophages, neutrophils, and dendritic cells. Furthermore, non-immune cells including hepatocytes,57–59 stellate cells,60 endothelial cells,61,62 and myofibroblasts63 also contain the functionally active inflammasome complex. Studies at the mRNA levels support the expression of NLRP1, 2, 3, 6, 10, 12, and NLRC4 in the liver.59 Comparing the human spleen and liver, Lech et al concluded that levels of NLRP10 inflammasome expression were higher in the liver, whereas murine liver has shown high level expression of NLRP6 inflammasome.64
Intracellular Pathogens and the Inflammasomes
Studies by two prominent groups of researchers have shown that, though phenotypically normal, AIM2 deficient macrophages were not able to generate inflammasomes following infection by Francisella species, vaccinia virus or mouse cytomegalovirus (mCMV).65,66 In vivo studies have highlighted low levels of serum IL-18 and less secretion of interferon-γ (IFN-γ) by splenic NK cells in AIM2 deficient mice infected with mCMV.66 Likewise, AIM2 deficient mice infected with F. tularensis novicida have high bacterial loads, low levels of serum IL-18 and lower survival than do wild type mice.65
Mice deficient in ASC or caspase-1 show similar phenotype as described for the AIM2 deficient mice. This evidence suggests that AIM2, ASC and caspase-1 function in a single signaling pathway that helps in detecting infection with Francisella, vaccinia virus or mCMV and provokes protective host responses.67 A significant relationship between the interferon response to the infection and the inflammasome response can be inferred. Deficiency of AIM2 has been found to increase the interferon response to dsDNA transfection or Francisella infection but elicits a normal response in the case of mCMV infection.65,66 A recent study by Liu and group68 established that the production of IL-1β and IL-18 in NLRP3 and NLRC4 inflammasomes plays a critical role in host defense against enteric infection caused by C. rodentium. An enteric bacterial pathogen in the intestinal tract of the mouse, C. rodentium triggers inflammatory responses similar to those seen in humans infected with enteropathogenic and enterohemorrhagic E. coli. Mice deficient in inflammasome components NLRP3, NLRC4, and caspase-1 were hyper-susceptible to C. rodentium-induced gastrointestinal inflammation. This can be attributed to defective IL-1β and IL-18 production.68
Gut microbiota and Inflammation
Gut microbiota is composed of strict anaerobes, facultative anaerobes and aerobes.69 In human, the gut microbiota is dominated by the Bacteroidetes and Firmicutes while Proteobacteria, Verrucomicrobia, Actinobacteria, Fusobacteria, and Cyanobacteria are present in trace amounts.70 Recent reports suggest the existence of over 35000 bacterial species in the human gut microbiota.71 An important characteristic of gut microbes is their heterogeneity.69 Frank and co-workers studied biopsy samples from the small intestine and colon of healthy individuals and found different bacterial groups at different sites in the gastrointestinal tract. The small intestine was enriched with Firmicutes and Actinobacteria whereas prevalence of Bacteroidetes and the Lachnospiraceae family of the Firmicutes was more in colonic samples.71 A thick and complex mucus layer separated the intestinal epithelium from the luminal cavity. Studies by Swidsinski et al demonstrated the inability of many bacterial species to access the mucus layer and the epithelial crypts from the intestinal lumen.72 Colonization of the human gut by microbes starts at the time of birth.69 Interestingly, different gut microbial populations develop in infants born by caesarean section compared with those born by vaginal delivery.73 The composition of GALT (gut associated lymphoid tissue) is affected by presence of microbial colonies in the gastrointestinal tract. There is steep rise in the number of intraepithelial lymphocytes immediately after exposure to luminal microorganisms.74,75 The intestinal immune system and the resident gut microbiota have a complex relationship and, at times, it is difficult for the epithelial cells and the mucosal immune system to distinguish pathogens from non-pathogens. In addition, bacterial antigens are detected by the intestinal epithelial cells and may play a role in initiation and regulation of innate and adaptive immune responses. Major histo-compatibility complex I and II molecules and toll-like receptors (TLRs) help in signal transmission from bacteria to adjacent immune cells such as macrophages, dendritic cells and lymphocytes.76,77 TLRs were so named due to their similarity to the receptor encoded by the Toll-gene that was first studied in the fruit fly, Drosophila melanogaster (toll meaning ‘fantastic’ in German). More than ten types of TLRs are known in humans. Sites where TLRs can be easily studied in myelomonocytic, dendritic, endothelial and epithelial cells. TLRs are capable of recognizing PAMPs and their interaction triggers processes activating complex intracellular signaling cascades and up-regulating inflammatory genes. Secretion of pro-inflammatory cytokines, interferon production and myeloid cell recruitment also depends on communication between TLRs and PAMPs. TLRs–PAMPs association can also stimulate expression of co-stimulatory molecules leading to the activation of adaptive immune response by antigen presenting cells.78
NLRs are other important membrane-bound receptors besides the TLRs for the detection of proteins. More than twenty NLRs have been known till date, although the best characterized are NOD1 and NOD2. The presence of NLRs matter more where there is low concentration of TLRs for example on the epithelial cells of the gastrointestinal tract.79
Inflammasomes Sense Microflora
An elaborate study of alterations of weight, metabolism and inflammation due to regulation of the gut flora by inflammasomes would be the next major step in understanding innate immunity and host response.7 A recent study has shown the association of NLRP6 inflammasome with the autoinflammation-promoting microbiota expansion. The microbiota includes bacterial taxa such as Prevotellaceae and TM7.80 There are certain unknown mechanisms about the sensing of microflora by inflammasomes and regulating tissue repair and regeneration. Coordination of the mucosal immune response during the process of inflammation needs to be fully understood. The linking of inflammasome deficiency to alterations in the gastrointestinal microflora has been done in different studies and has helped in understanding the metabolic syndrome, obesity and atherosclerosis, both in mice and human.81,82 Additionally, a study on rural African children demonstrated the presence of Prevotellaceae in the faecal matter, whereas it was absent in the European population. This gives an indication that the genetic and the environmental factors also contribute toward the regulation of the microflora composition.83
Connecting Gut and Liver
Gut microbes play a significant role in various body functions such as digestion of food and vitamins. In addition, they also help in shaping immunity in the host.84,85 Alterations in the microbiome population and/or changes in gut permeability promote microbial translocation into the portal circulation that delivers blood directly to the liver. Danger signals derived from the microbiome trigger the inflammatory cascade and activate immune cells.86 Liver and spleen are the major organs that help in removing bacterial toxins and their lipopolysaccharides (LPS) from the blood.87 Mechanisms that balance the barrier function of the gut and the ability of the liver to detoxify have been studied.86,88 It was found that gut derived LPS played a significant role in several liver diseases.89,90 Lu et al observed an increase in inflammatory responses as there was attrition of the protective ability of the liver to detoxify LPS derived from the gut.91 A positive correlation between liver dysfunction and bacterial translocation has been reported by others also.92,93
This close interplay between the gut and the liver has been termed the gut–liver axis.94 Liver is the organ most exposed to gut derived toxins (bacteria and bacterial products), due to the portal circulation. Ethanol, ammonia and acetaldehyde produced in the intestine are metabolized in the liver. Importantly, these toxins keep a check on the Kupffer cell activity and cytokine production.95 While it has been evident that there is an increased level of bacterial lipopolysaccharide (LPS) in the portal and/or systemic circulation, further studies are needed to reveal the complete picture of the variety and quantity of gut-derived microbial products being transferred to the liver or reaching the systemic circulation after bypassing the liver through porto-systemic shunts. This factor has been implicated in several types of chronic liver diseases.96 Liver macrophages or Kupffer cells are activated by the bacteria and bacterial products. This activation is dependent on an NFkB-mediated mechanism which releases cytokines such as tumor necrosis factor alpha (TNF-α). As soon as cytokines are released, specific intracellular pathways are activated including pro-apoptotic signals via the caspase cascade.97 LPS behaves as a hepatotoxin causing morphological and functional changes. Accumulation of polymorphonuclear cells induces an acute inflammatory response, releasing reactive oxygen metabolites, proteases and other enzymes. The whole process culminates in worsening of liver damage.98 An inter-relation between the gut microbiota with its altered composition and the liver is depicted in Figure 2 where the activation of inflammasome complex causes the release of IL-1β (Figure 2).
Figure 2.
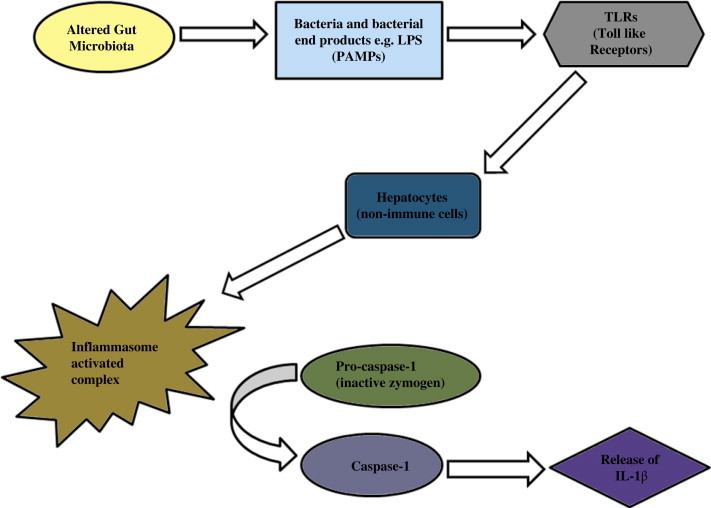
Activated caspase-1 causes the release of IL-1β with the alterations in gut microbiota aggravating liver diseases.
About 80 years ago it was shown that the patients with chronic liver disease had altered gut microbiota. About 20–75% of patients suffering from the chronic liver disease have demonstrated changes in microbiome, in particular small intestine bacterial overgrowth i.e. SIBO.94 Patients suffering from non-alcoholic fatty liver disease (NAFLD) have reported SIBO correlating with severity of steatosis.95,99 While many more studies need to be done for understanding the link between SIBO and NASH, intestinal permeability is the key toward better insight.94 The development of insulin resistance and related metabolic disorders is influenced by the gut microbiota which plays a pivotal role in fat storage and energy harvesting.100–102 Strikingly, fewer Bacteroidetes and more Firmicutes in the obese as compared to the lean controls demonstrate the affect of gut microbiota on calorie proportions obtained from the intestine.103
Study by Flavell and co-workers have demonstrated the link between inflammasomes, gut microbiota and NAFLD.104 As already discussed, inflammasomes are multiprotein complexes which induce production of pro-inflammatory cytokines IL-1β and IL-18. The group showed that dysbiotic and colitogenic microflora in the mice due to deficiency in the components of the NLRP6 inflammasome could be transmitted to co-housed wild-type mice which led to IBD in them. A pivotal finding in their studies was that NLRP6 and NLRP3 inflammasomes (through activation of the effector protien IL-18) negatively regulated NAFLD/NASH progression and caused metabolic syndrome via a cell-extrinsic effect of modulation of the gut microbiota through activation of the effector protein IL-18.105 It was shown that changes in the composition of the gut microbiota caused the portal circulation to be flooded with Toll-like receptor TLR4 and TLR9 agonists. This enhanced hepatic expression of tumor necrosis factor (TNF) and aggravated steatosis and inflammation, resulting in the progression of NAFLD/NASH. Flavell and group have described inflammasomes as steady-state sensors and regulators of colonic microbiota. Astonishingly, it was discovered that the deficiency in components of two inflammasomes, NLRP6 and NLRP3 (both of which include ASC and caspase-1) resulted in an altered transmissible, colitogenic intestinal microbial community.104
Human Microbiome Project
The human microbiome project (HMP) is working toward complete characterization of the human microbiome which will help researchers and scientists to understand variations in human microbiome due to factors like population, genotype, disease, age, nutrition, medication and environmental factors.106 This would help in understanding the influence of the microbiome on human health and disease. Recent advancement in sequencing technologies along with development of metagenomic and bioinformatics methods have increased insight into investigating the 1014 microorganisms that inhabit the human gut.107 Handelsman and Rodon were the first to describe metagenomics.108,109 Metagenomics gives an understanding of the functional features of non-cultivated bacteria by analyzing the collective genomes available in a defined environment or ecosystem.107 Two large insert libraries were the first among the human gut metagenomic libraries that were generated after analyzing faecal samples from six healthy people and six Crohn's disease patients.110,111
With the upcoming metagenomics and high throughput technologies, Bajaj and group made an interesting observation where they studied the sigmoid mucosal microbiome of cirrhotic patients and control group, comparing the patients with hepatic encephalopathy (HE) and no-HE and further analyzing the cognition and inflammation in these patients.112 This study revealed a vast difference between the mucosal microbiome and the stool microbiome. Patients with cirrhosis especially HE were lacking potentially beneficial autochthonous bacteria and an increase in potentially pathogenic bacteria was observed. This gives a brighter view leading to the association of HE patients with poor cognitive performance and inflammation.
Future perspectives
Strong evidence relates the gut microbiome to the pathogenesis of liver diseases. Understanding molecular mechanisms involved in the production of gut bacterial toxins and their detoxification by liver leading to the activation of inflammasomal complex would open a new field toward the better understanding of the specific liver diseases. Although, innate immune system has been explored to understand the multiprotein complexes- inflammasomes, certain genetic and biochemical mechanisms that involve activation of the inflammasomes and their modulatory affects need to be answered. It would be interesting to explore certain inhibitory pathways or molecules that can act upon these NLR protein platforms to understand the pathogenesis of diseases and work toward therapeutic interventions.
Conflicts of interest
All authors have none to declare.
References
- 1.Szabo G., Csak T. Inflammasomes in liver diseases. J Hepatol. 2012;57:642–654. doi: 10.1016/j.jhep.2012.03.035. [DOI] [PubMed] [Google Scholar]
- 2.Hakansson A., Molin G. Gut microbiota and inflammation. Nutrients. 2011;3:637–682. doi: 10.3390/nu3060637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palmblad J. The role of granulocytes in inflammation. Scand J Rheumatol. 1984;13:163–172. doi: 10.3109/03009748409100381. [DOI] [PubMed] [Google Scholar]
- 4.Fujiwara N., Kobayashi K. Macrophages in inflammation. Curr Drug Targets Inflamm Allergy. 2005;4:281–286. doi: 10.2174/1568010054022024. [DOI] [PubMed] [Google Scholar]
- 5.Anderson C.F., Mosser D.M. A novel phenotype for an activated macrophage: the type 2 activated macrophage. J Leukoc Biol. 2002;72:101–106. [PubMed] [Google Scholar]
- 6.Gordon S. Alternative activation of macrophages. Nat Rev. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 7.Strowig T., Henao-Mejia J., Elinav E., Flavell R. Inflammasomes in health and disease. Nature. 2012;481:278–286. doi: 10.1038/nature10759. [DOI] [PubMed] [Google Scholar]
- 8.Medzhitov R., Janeway C.A., Jr. Innate immunity: the virtues of a nonclonal system of recognition. Cell. 1997;91:295–298. doi: 10.1016/s0092-8674(00)80412-2. [DOI] [PubMed] [Google Scholar]
- 9.Matzinger P. Tolerance, danger, and the extended family. Annu Rev Immunol. 1994;12:991–1045. doi: 10.1146/annurev.iy.12.040194.005015. [DOI] [PubMed] [Google Scholar]
- 10.Martinon F., Burns K., Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of pro-IL-beta. Mol Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 11.Martinon F., Mayor A., Tschopp J. The inflammasomes: guardians of the body. Annu Rev Immunol. 2009;27:229–265. doi: 10.1146/annurev.immunol.021908.132715. [DOI] [PubMed] [Google Scholar]
- 12.Bauernfeind F., Ablasser A., Bartok E. Inflammasomes: current understanding and open questions. Cell Mol Life Sci. 2011;68:765–783. doi: 10.1007/s00018-010-0567-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schroder K., Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 14.Boyden E.D., Dietrich W.F. NALP1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat Genet. 2006;38:240–244. doi: 10.1038/ng1724. [DOI] [PubMed] [Google Scholar]
- 15.Wickliffe K.E., Leppla S.H., Moayeri M. Anthrax lethal toxin-induced inflammasome formation and caspase-1 activation are late events dependent on ion fluxes and the proteasome. Cell Microbiol. 2008;10:332–343. doi: 10.1111/j.1462-5822.2007.01044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bruey J.M., Bruey-Sedano M., Luciano F. Bcl-2 and Bcl-XL regulate proinflammatory caspase-1 activation by interaction with NALP1. Cell. 2007;129:45–46. doi: 10.1016/j.cell.2007.01.045. [DOI] [PubMed] [Google Scholar]
- 17.Hsu L.C., Ali S.R., Mcgillivray S. A NOD2-NALP1 complex mediates caspase-1-dependent IL-1b secretion in response to Bacillus anthracis infection and muramyl dipeptide. Proc Natl Acad Sci U S A. 2008;105:7803–7808. doi: 10.1073/pnas.0802726105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kummer J.A., Broekhuizen R., Everett H. Inflammasome components NALP1 and 3 show distinct but separate expression profiles in human tissues suggesting a site-specific role in the inflammatory response. J Histochem Cytochem. 2007;55:443–452. doi: 10.1369/jhc.6A7101.2006. [DOI] [PubMed] [Google Scholar]
- 19.Hoffman H.M., Mueller J.L., Broide D.H., Wanderer A.A., Kolodner R.D. Mutation of a new gene encoding a putative pyrin-like protein causes familial cold autoinflammatory syndrome and Muckle–Wells syndrome. Nat Genet. 2001;29:301–305. doi: 10.1038/ng756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mariathasan S., Weiss D.S., Newton K. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- 21.Kanneganti T.D., Lamkanfi M., Kim Y.G. Pannexin-1 mediated recognition of bacterial molecules activates the cryoporin inflammasome independent of toll-like receptor signalling. Immunity. 2007;26:433–443. doi: 10.1016/j.immuni.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 22.Petrilli V., Papin S., Dostert C., Mayor A., Martinon F., Tschopp J. Activation of the NALP3 inflammasome is triggered by low intracellular potassium concentration. Cell Death Differ. 2007;14:1583–1589. doi: 10.1038/sj.cdd.4402195. [DOI] [PubMed] [Google Scholar]
- 23.Martinon F., Petrilli V., Mayor A., Tardivel A., Tshopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. 2006;440:237–241. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- 24.Eisenbarth S.C., Colegio O.R., O‘Connor W., Sutterwala F.S., Flavell R.A. Crucial role for the Nalp3 inflammasome in the immunstimulatory properties of aluminium adjuvants. Nature. 2008;453:1122–1126. doi: 10.1038/nature06939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hornung V., Bauernfeind F., Halle A. Silica crystals and aluminium salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol. 2008;9:847–856. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dostert C., Petrilli V., Van Bruggen R., Steele C., Mossman B.T., Tschopp J. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science. 2008;320:674–677. doi: 10.1126/science.1156995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Halle A., Hornung V., Petzold G.C. The Nalp3 inflammasome is involved in the innate immune response to amyloid-beta. Nat Immunol. 2008;9:857–865. doi: 10.1038/ni.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamasaki K., Muto J., Taylor K.R. NLRP3/cryoporin is necessary for interleukin-1beta (IL-1beta) release in response to hyaluronan, an endogenous trigger of inflammation in response to injury. J Biol Chem. 2009;284:12762–12771. doi: 10.1074/jbc.M806084200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dostert C., Guarda G., Romero Malarial hemozoin is a Nalp3 inflammasome activating danger signal. PloS One. 2009;4:e6510. doi: 10.1371/journal.pone.0006510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharp F.A., Ruane D., Claass B. Uptake of particulate vaccine adjuvants by dendritic cells activate the NALP3 inflammasome. Proc Natl Acad Sci U S A. 2009;106:870–875. doi: 10.1073/pnas.0804897106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duewell P., Kono H., Rayner K.J. NLRP3 inflammasome are required for atherogenesis and activated by cholesterol crystals. Nature. 2010;464:1357–1361. doi: 10.1038/nature08938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miao E.A., Mao D.P., Yudkosky N. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc Natl Acad Sci U S A. 2010;107:3076–3080. doi: 10.1073/pnas.0913087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mariathasan S., Newton K., Monack D.M. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430:213–218. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- 34.Vinzing M., Eitel J., Lippmann J. NAIP and Ipaf control Legionella pneumophila replication in human cells. J Immunol. 2008;180:6808–6815. doi: 10.4049/jimmunol.180.10.6808. [DOI] [PubMed] [Google Scholar]
- 35.Zhao Y., Yang J., Shi J. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 2011;477:596–600. doi: 10.1038/nature10510. [DOI] [PubMed] [Google Scholar]
- 36.Qu Y., Misaghi S., Izrael-Tomasevic A. Phosphorylation of NLRC4 is critical for inflammasome activation. Nature. 2012 doi: 10.1038/nature11429. [DOI] [PubMed] [Google Scholar]
- 37.Muruve D.A., Pétrilli V., Zaiss A.K. The inflammasome recognizes cytosolic microbial and host DNA and triggers innate immune response. Nature. 2008;452:103–108. doi: 10.1038/nature06664. [DOI] [PubMed] [Google Scholar]
- 38.Hornung V., Ablasser A., Charrel-Dennis M. AIM2 recognizes cytosolic dsDNA and forms a caspase-1 activating inflammasome with ASC. Nature. 2009;26:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakahira K., Haspel J.A., Rathinam V.A. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rathinam V.A.K., Jiang Z., Waggoner S.N. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poeck H., Bscheider M., Gross O. Recognition of RNA virus by RIG-I results in activation of CARD9 and inflammasome signaling for interleukin-1b production. Nat Immunol. 2009;11:63–69. doi: 10.1038/ni.1824. [DOI] [PubMed] [Google Scholar]
- 42.Dinarello C.A. IL-1: discoveries, controversies and future directions. Eur J Immunol. 2010;40:599–606. doi: 10.1002/eji.201040319. [DOI] [PubMed] [Google Scholar]
- 43.Keller M., Ruegg A., Werner S., Beer H.D. Active caspase-1 is a regulator of unconventional protein secretion. Cell. 2008;132:818–831. doi: 10.1016/j.cell.2007.12.040. [DOI] [PubMed] [Google Scholar]
- 44.Bergsbaken T., Fink S.L., Cookson B.T. Pyroptosis: host cell death and inflammation. Nat Rev Microbiol. 2009;7:99–109. doi: 10.1038/nrmicro2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andrei C., Margiocco P., Poggi A., Lotti L.V., Torrisi M.R., Rubartelli A. Phospholipases C and A2 control lysosome-mediated IL-1 beta secretion: implications for inflammatory processes. Proc Natl Acad Sci. 2004;101:9745–9750. doi: 10.1073/pnas.0308558101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dinarello C.A. Interleukin-1 in the pathogenesis and treatment of inflammatory diseases. Blood. 2011;117:3720–3732. doi: 10.1182/blood-2010-07-273417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lamkanfi M. Emerging inflammasome effector mechanisms. Nat Rev Immunol. 2011:213–220. doi: 10.1038/nri2936. [DOI] [PubMed] [Google Scholar]
- 48.Wang S., Miura M., Jung Y.K., Zhu H., Li E., Yuan J. Murine caspase-11, an ICE-interacting protease, is essential for the activation of ICE. Cell. 1998;92:501–509. doi: 10.1016/s0092-8674(00)80943-5. [DOI] [PubMed] [Google Scholar]
- 49.Kayagaki N., Warming S., Lamkanfi M. Non-canonical inflammasome activation targets caspase-11. Nature. 2011;479:117–121. doi: 10.1038/nature10558. [DOI] [PubMed] [Google Scholar]
- 50.Elinav E., Strowig T., Henao-Mejia J., Flavell R.A. Regulation of the antimicrobial response by NLR proteins. Immunity. 2011;34:665–679. doi: 10.1016/j.immuni.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 51.Ichinohe T., Pang I.K., Iwasaki A. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat Immunol. 2010;11:404–410. doi: 10.1038/ni.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kawai T., Akira S. Toll-like receptors and their Crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34(5):637–640. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 53.Boyden E.D., Dietrich W.F. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat Genet. 2006;38:240–244. doi: 10.1038/ng1724. [DOI] [PubMed] [Google Scholar]
- 54.Faustin B., Lartique L., Bruey J.M. Reconstituted NALP1 inflammasome reveals two-step mechanism of caspase-1 activation. Mol Cell. 2007;25:713–724. doi: 10.1016/j.molcel.2007.01.032. [DOI] [PubMed] [Google Scholar]
- 55.Miao E.A., Leaf I.A., Treuting P.M. Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat Immunol. 2010;11:1136–1142. doi: 10.1038/ni.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Broz P., Newton K., Lamkanfi M., Mariathasan S., Dixit V.M., Monack D.M. Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella. J Exp Med. 2010;207:1745–1755. doi: 10.1084/jem.20100257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Csak T., Ganz M., Pespisa J., Kodys K., Dolganiuc A., Szabo G. Fatty acid and endotoxin activate inflammasomes in mouse hepatocytes that release danger signals to stimulate immune cells. Hepatology. 2011;54:133–144. doi: 10.1002/hep.24341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Burdette D., Haskett A., Presser L., McRae S., Iqbal J., Waris G. Hepatitis C virus activates interleukin-1 {beta} via caspase-1-inflammasome complex. J Gen Virol. 2012;93(Pt2):235–246. doi: 10.1099/vir.0.034033-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 59.Yan W., Chang Y., Liang X. High mobility group box 1 activates caspase-1 and promotes hepatocellular carcinoma invasiveness and metastases. Hepatology. 2012;55:1863–1875. doi: 10.1002/hep.25572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Watanabe A., Sohail M.A., Gomes D.A. Inflammasome-mediated regulation of hepatic stellate cells. Am J Physiol Gastrointest Liver Physiol. 2009;296:G1248–G1257. doi: 10.1152/ajpgi.90223.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kolly L., Busso N., Palmer G., Talabot-Ayer D., Chobaz V., So A. Expression and function of NALP3 inflammasome in rheumatoid synovium. Immunology. 2010;129:175–185. doi: 10.1111/j.1365-2567.2009.03174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Imaeda A.B., Watanabe A., Sohail M.A. Acetaminophen-induced hepatotoxicity in mice is dependent on Tlr9 and the Nalp3 inflammasome. J Clin Invest. 2009;119:305–314. doi: 10.1172/JCI35958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rawat R., Cohen T.V., Ampong B. Inflammasome up-regulation and activation in dysferlin-deficient skeletal muscle. Am J Pathol. 2010;176:2891–2900. doi: 10.2353/ajpath.2010.090058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lech M., Avila-Ferrufino A., Skuginna V., Susanti H.E., Anders H.J. Quantitative expression of RIG-like helicase, NOD-like receptor and inflammasomerelated mRNAs in humans and mice. Int Immunol. 2010;9:717–728. doi: 10.1093/intimm/dxq058. [DOI] [PubMed] [Google Scholar]
- 65.Fernandes-Alnemri T., Yu J.W., Juliana C. The AIM2 inflammasome is critical for innate immunity to Francisella Tularensis. Nat Immunol. 2010;11:385–393. doi: 10.1038/ni.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rathinam V.A.K., Jiang Z., Waggoner S.N. The AIM2 inflamma-some is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Krieg AM. AIMing 2 defend against intracellular pathogens. Nat Immunol. 2010;11(5):367–369. doi: 10.1038/ni0510-367. [DOI] [PubMed] [Google Scholar]
- 68.Liu Z., Zaki M.H., Vogel P. Role of inflammasomes in host defense against Citrobacter rodentium infection. J Biol Chem. 2012;287:16955–16964. doi: 10.1074/jbc.M112.358705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sekirov I., Russell S.L., Antunes L.C.M., Finlay B.B. Gut microbiota in health and disease. Physiol Rev. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 70.Eckburg P.B., Bik E.M., Bernstein C.N. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Frank D.N., St Amand A.L., Feldman R.A., Boedeker E.C., Harpaz N., Pace N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Swidsinski A., Loening-Baucke V., Lochs H., Hale L.P. Spatial organization of bacterial flora in normal and inflamed intestine: a fluorescence in situ hybridization study in mice. World J Gastroenterol. 2005;11:1131–1140. doi: 10.3748/wjg.v11.i8.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huurre A., Kalliomaki M., Rautava S., Rinne M., Salminen S., Isolauri E. Mode of delivery: effects on gut microbiota and humoral immunity. Neonatology. 2008;93:236–240. doi: 10.1159/000111102. [DOI] [PubMed] [Google Scholar]
- 74.Umesaki Y., Setoyama H., Matsumoto S., Okada Y. Expansion of alpha beta T cell receptor-bearing intestinal intraepithelial lymphocytes after microbial colonization in germ-free mice and its independence from thymus. Immunology. 1993;79:32–37. [PMC free article] [PubMed] [Google Scholar]
- 75.Helgeland L., Vaage J.T., Rolstad B., Midtvedt T., Brandtzaeg P. Microbial colonization influences composition and T cell receptor V beta repertoire of intraepithelial lymphocytes in rat intestine. Immunology. 1996;89:494–501. doi: 10.1046/j.1365-2567.1996.d01-783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cario E., Brown D., McKee M., Lynch-Devaney K., Gerken G., Podolsky D.K. Commensal associated molecular patterns induce selective toll-like receptor-trafficking from apical membrane to cytoplasmic compartments in polarized intestinal epithelium. Am J Pathol. 2002;160:165–173. doi: 10.1016/S0002-9440(10)64360-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hershberg R.M., Mayer L.F. Antigen processing and presentation by intestinal epithelial cells—polarity and complexity. Immunol Today. 2000;21:123–128. doi: 10.1016/s0167-5699(99)01575-3. [DOI] [PubMed] [Google Scholar]
- 78.Testro A.G., Visvanathan K. Toll-like receptors and their role in gastrointestinal disease. J Gastroenterol Hepatol. 2009;24:943–954. doi: 10.1111/j.1440-1746.2009.05854.x. [DOI] [PubMed] [Google Scholar]
- 79.Philpott D.J., Girardin S.E., Sansonetti P.J. Innate immune responses of epithelial cells following infection with bacterial pathogens. Curr Opin Immunol. 2001;13:410–416. doi: 10.1016/s0952-7915(00)00235-1. [DOI] [PubMed] [Google Scholar]
- 80.Elinav E., Strowig T., Kau A.L. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang Z., Klipfell E., Bennett B.J. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Turnbaugh P.J., Hamady M., Yatsunenko T. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.De Filippo C., Cavalieri D., Di Paola M. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Robinson C.J., Bohannan B.J.M., Young V.B. From structure to function: the ecology of host-associated microbial communities. Microbiol Mol Biol Rev. 2010;74:453–476. doi: 10.1128/MMBR.00014-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gillevet P., Sikaroodi M., Keshavarzian A., Mutlu E.A. Quantitative assessment of the human gut microbiome using multitag pyrosequencing. Chem Biodivers. 2010;7:1065–1075. doi: 10.1002/cbdv.200900322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Son G., Kremer M., Hines I.N. Contribution of gut bacteria to liver pathobiology. Gastroenterol Res Pract. 2010;2010:453–563. doi: 10.1155/2010/453563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Szabo G., Bala S., Petrasek J., Gattu A. Gut-liver axis and sensing microbes. Dig Dis. 2010;28:737–744. doi: 10.1159/000324281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Szabo G., Bala S. Alcoholic liver disease and the gut-liver axis. World J Gastroenterol. 2010;16:1321–1329. doi: 10.3748/wjg.v16.i11.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Szabo G. LPS in liver disease. Falk Meet. 2008 [Google Scholar]
- 90.Wang H.J., Zakhari S., Jung M.K. Alcohol, inflammation, and gut-liver-brain interactions in tissue damage and disease development. World J Gastroenterol. 2010;16:1304–1313. doi: 10.3748/wjg.v16.i11.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lu M., Varley A.W., Ohta S., Hardwick J., Munford R.S. Host inactivation of bacterial lipopolysaccharide prevents prolonged tolerance following Gram-negative bacterial infection. Cell Host Microbe. 2008;4:293–302. doi: 10.1016/j.chom.2008.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bjarnason I., Peters T.J., Wise R.J. The leaky gut of alcoholism: possible route of entry for toxic compounds. Lancet. 1984;1:179–182. doi: 10.1016/s0140-6736(84)92109-3. [DOI] [PubMed] [Google Scholar]
- 93.Nanji A.A., Khettry U., Sadrzadeh S.M., Yamanaka T. Severity of liver injury in experimental alcoholic liver disease. Correlation with plasma endotoxin, prostaglandin E 2, leukotriene B 4, and thromboxane B 2. Am J Pathol. 1993;142:367–373. [PMC free article] [PubMed] [Google Scholar]
- 94.Compare D., Coccoli P., Rocco A., Nardone O.M., Maria S.D., Cartenı M., Nardone G Gut-liver axis: the impact of gut microbiota on non alcoholic fatty liver disease. Nutr Metab Cardiovasc Dis. 2012;22(6):471–476. doi: 10.1016/j.numecd.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 95.Nagata K., Suzuki H., Sakaguchi S. Common pathogenic mechanism in development progression of liver injury caused by non-alcoholic or alcoholic steatohepatitis. J Toxicol Sci. 2007;32(5):453–468. doi: 10.2131/jts.32.453. [DOI] [PubMed] [Google Scholar]
- 96.Han D.W. Intestinal endotoxemia as a pathogenetic mechanism in liver failure. World J Gastroenterol. 2002;8(6):961–965. doi: 10.3748/wjg.v8.i6.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bilzer M., Roggel F., Gerbes A.L. Role of Kupffer cells in host defense and liver disease. Liver Int. 2006;26(10):1175–1186. doi: 10.1111/j.1478-3231.2006.01342.x. [DOI] [PubMed] [Google Scholar]
- 98.Jirillo E., Caccavo D., Magrone T. The role f the liver in the response to LPS: experimental and clinical findings. J Endotoxin Res. 2002;8(5):319–327. doi: 10.1179/096805102125000641. [DOI] [PubMed] [Google Scholar]
- 99.Diamant M., Blaak E.E., de Vos W.M. Do nutrient-gut-microbiota interactions play a role in human obesity, insulin resistance and type 2 diabetes? Obes Rev. 2011;12(4):272–281. doi: 10.1111/j.1467-789X.2010.00797.x. [DOI] [PubMed] [Google Scholar]
- 100.Abu-Shanab A., Quigley E.M. The role of the gut microbiota in nonalcoholic fatty liver disease. Nat Rev Gastroenterol Hepatol. 2010;7(12):691–701. doi: 10.1038/nrgastro.2010.172. [DOI] [PubMed] [Google Scholar]
- 101.Turnbaugh P.J., Ley R.E., Mahowald M.A., Magrini V., Mardis E.R., Gordon J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 102.Backhed F., Ding H., Wang T. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004;101(44):15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ley R.E., Turnbaugh P.J., Klein S., Gordon J.I. Microbial ecology: human gut microbes associated with obesity. Nature. 2006 Dec 21;444(7122):1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 104.Wood Natalie J. Dysbiosis driven by inflammasome deficiency exacerbates hepatic steatosis and governs rate of NAFLD progression. Nat Rev Gastroenterol Hepatol. 2012;9:123. doi: 10.1038/nrgastro.2012.21. [DOI] [PubMed] [Google Scholar]
- 105.Henao-Mejia J., Elinav E., Jin C. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–186. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.The NIH HMP Working Group, Peterson J., Garges S. NIH Human Microbiome Project. Genome Res. 2009;19:2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lepage P., Leclerc M.C., Joossens M. A metagenomic insight into our gut’s microbiome. Gut. 2012 doi: 10.1136/gutjnl-2011-301805. [DOI] [PubMed] [Google Scholar]
- 108.Handelsman J., Rondon M.R., Brady S.F. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem Biol. 1998;5:R245–R249. doi: 10.1016/s1074-5521(98)90108-9. [DOI] [PubMed] [Google Scholar]
- 109.Rondon M.R., Raffel S.J., Goodman R.M. Toward functional genomics in bacteria: analysis of gene expression in Escherichia coli from a bacterial artificial chromosome library of Bacillus cereus. Proc Natl Acad Sci U S A. 1999;96:6451–6455. doi: 10.1073/pnas.96.11.6451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Manichanh C., Rigottier-Gois L., Bonnaud E. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Manichanh C., Chapple C.E., Frangeul L. A comparison of random Sequence reads versus 16S rDNA sequences for estimating the biodiversity of a metagenomic library. Nucleic Acids Res. 2008;36:5180–5188. doi: 10.1093/nar/gkn496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bajaj J.S., Hylemon P.B., Ridlon J.M. Colonic mucosal microbiome differs from stool microbiome in cirrhosis and hepatic encephalopathy and is linked to cognition and inflammation. Am J Physiol Gastrointest Liver Physiol. 2012;303(6):G675–G685. doi: 10.1152/ajpgi.00152.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]


