Abstract
We have constructed a human immunodeficiency virus type 1 (HIV-1)-based lentiviral vector expressing a 937-base antisense sequence against the HIV-1 envelope gene. Transduction of CD4+ T lymphocytes with this vector results in expression of the therapeutic antisense sequence and subsequent inhibition of productive HIV-1 replication. In this report, we examined the effect of antisense-mediated suppression on the potential development of virus escape mutants using a permissive T-cell line cultured under conditions that over serial passages specifically allowed for generation and amplification of mutants selected for by antisense pressure. In the resulting virus clones, we found a significant increase in the number of deletions at the envelope target region (91% compared to 27.5% in wild-type HIV). Deletions were most often greater than 1 kb in length. These data demonstrate for the first time that during antisense-mediated suppression of HIV, mutants develop as a direct result of selective pressure on the HIV genomic RNA. Interestingly, in clones where deletions were not observed, there was a high rate of A-G transitions in mutants at the antisense target region but not outside this region, which is consistent with those mutations that are predicted as a result of antisense-mediated modification of double-stranded RNA by the enzyme double-stranded RNA-specific adenosine deaminase. These clones were not found to be escape mutants, as their replicative ability was severely attenuated, and they did not replicate in the presence of vector.
Human immunodeficiency virus (HIV) is estimated to infect 34 to 46 million people worldwide and almost 1 million people in the United States. The mortality due to AIDS is estimated to be approximately 3 million per year worldwide and 15,000 per year in the United States (5, 46). Although treatment and prevention efforts have increased, infection and death rates continue to rise in most countries. Treatment for HIV infection is mostly limited to the highly active antiretroviral therapy, which consists of a triple cocktail of anti-HIV drugs. These drugs have been successful in reducing viral loads and extending life expectancy, but they require life-long drug regimens that lead to a series of adverse effects, in particular a range of metabolic disorders (3, 34, 48). These drugs are expensive and require strict adherence to be effective and prevent resistance and, therefore, cannot be used worldwide to successfully fight the epidemic (28, 45). Furthermore, transmission of drug-resistant strains of HIV has been on the rise since 1995 (23) and is as high as 18.5% in the United States (M. Mascolini, 11th Int. Workshop HIV Drug Resist., 2002).
Several groups have investigated using RNA-based technology in order to inhibit HIV infection or replication in cells. RNA-mediated inhibition of HIV is an attractive alternative to drug therapy, since RNA is not immunogenic, nor would RNA-based therapy carry with it the extensive adverse effects associated with drug therapy or require a scheduled dosing regimen. Alternative to antisense, the RNA technology examined to date includes RNA interference (RNAi) (7, 18, 22, 29, 35) and gene therapy using ribozymes (11, 12, 36). These technologies are derived from natural processes that participate in posttranscriptional gene silencing, or in cleavage of viral RNA in the case of ribozymes, but have been engineered to inhibit HIV receptors (29) or HIV transcripts (7, 18, 22, 29, 35). Each of these approaches requires recognition of the cognate RNA sequence in order to mediate their suppression.
It has been shown that introduction of short interfering RNAs (siRNAs) into a cell results in sequence-specific degradation of genomic HIV type 1 (HIV-1) RNA (18; M. Mascolini, 11th Int. Workshop HIV Drug Resist., 2002). RNAi has also been used to inhibit HIV entry and replication in CD4-positive cells via the targeting of the CD4 molecule and the gag gene. Recently, a method for long-term expression of siRNAs has been demonstrated which allows for the long-term production of anti-HIV genes that previously had only exerted a 3- to 5-day effect (33).
Others have investigated by using ribozymes the suppression of HIV-1 replication. Anti-HIV ribozymes recognize their target sequences through short recognition motifs of about 20 bases flanking the ribozyme catalytic unit and then cleave the bound RNA at a target cutting sequence that also contains a short specific nucleotide sequence (39). This approach has been successful, and testing of these constructs has led to phase I clinical trials for safety.
HIV has an unusually high mutation rate of 3 × 10−5 per replication cycle, a rate 2- to 10-fold higher than that of other retroviruses (13). The short length of siRNA molecules, about 20 to 30 bases, allows HIV to mutate and evade siRNA suppression (4, 8). Likewise, ribozyme recognition sequences are short in length and the cutting site requires a specific upstream nucleotide, and so a single mutation at the ribozyme cleavage site will result in a transcript that is resistant to cleavage. Hence, the remarkable mutability of HIV reduces the impacts of these approaches.
In contrast to RNAi and ribozymes, long antisense sequences targeting HIV require significantly greater levels of mutation on the part of HIV to allow escape from replication inhibition. Double-stranded RNAs are targeted by the enzyme double-stranded RNA-specific adenosine deaminase (dsRAD/DRADA), which is constitutively expressed in the nucleus and may be induced in the cytoplasm by type I and II interferons (31, 32). dsRAD/DRADA converts adenosines to inosines in double-stranded RNAs and causes partial unwinding of the helix, which is subsequently destroyed.
Antisense targeting of HIV transcripts or receptors has been previously described (6, 9, 10, 19, 21, 25-27, 40, 44, 47) and has established the feasibility of using antisense against HIV. However, the data from these studies have not yet led to efficacy in the clinic, in part due to low transduction efficiencies and persistence of modified cells. New vector and cell processing technology available today provide a new opportunity to achieve therapeutic efficacy.
We have developed an HIV-1-based lentivirus vector, VRX496, containing a 937-base antisense payload to HIV. This vector has been shown to efficiently and stably transduce patient T cells in vitro and inhibit HIV replication and CD4 downregulation after HIV challenge in normal CD4+ T lymphocytes and in lymphocytes isolated from early- and late-stage HIV-infected patients (17a). VRX496 is currently being evaluated for safety and tolerability in a phase I clinical trial initiated in 2003 in which VRX496-transduced autologous T-cell transplantation is intended for the treatment of HIV infection (http://www.fda.gov/ohrms/dockets/ac/01/briefing/3794b3.htm).
Gene therapy allows for long-term production of therapeutic RNA molecules. The use of retroviral gene therapy vectors in particular is especially useful, since they are capable of inserting the gene of interest stably into target cells, allowing for permanent expression. Lentivirus-based vectors may be safer than the commonly used Moloney murine leukemia virus (MoMuLV)-based retroviral vector, since MoMuLV has been shown to cause leukemia in mice (14, 43) and recently in two children transplanted with stem cells modified with the virus (2, 15, 16). Lentiviruses are not recognized for their ability to cause oncogenesis, which is reflected in the fact that millions of people are infected with HIV but a link between HIV and T-cell lymphomas is exceptionally rare, even though memory T cells are known to harbor integrated virus for years (41). Approximately one-third of these proviruses are defective and are unable to kill the cell upon activation and, therefore, would permit the outgrowth of a transformed cell were it present (37).
In the present study, we examined the effect of antisense suppression on HIV by sequencing viruses grown under conditions that specifically allowed for generation and amplification of HIV mutants escaping from vector-mediated suppression in order to gain insight into the mechanism of action of antisense and to determine the efficacy of such a therapy in the context of HIV, given the high mutation rate of the virus. We found that antisense-mediated selective pressure led to deletions of large regions in the antisense target region of the envelope gene at a significantly higher rate in the selected viruses than in the controls. Most interestingly, selected viruses that contained point mutations harbored significantly higher numbers of A-G transitions in the antisense target region than wild-type (wt) HIV, which correlates with that predicted using the dsRAD/DRADA model for antisense-mediated destruction of target RNAs. Finally, we examined the ability of the antisense to prevent evolution of virus escape mutants that are resistant to the antisense antiviral actions.
MATERIALS AND METHODS
Vectors.
The pVRX494 plasmid was derived from a pUC18 backbone with distinctive cis elements of HIVNL4-3 (derived from the plasmid pNL4-3) (1) sequentially cloned into its multicloning site. The vector contains a 5′ long terminal repeat (LTR) and partial gag sequence (a ClaI site was introduced after the gag initiation codon, creating a frameshift in order to generate a stop codon immediately downstream); central polypurine track (cPPT) and central termination sequence; antisense HIV envelope; rev response element (RRE); enhanced green fluorescent protein (EGFP); and a PPT and 3′ LTR (Fig. 1A).
FIG. 1.
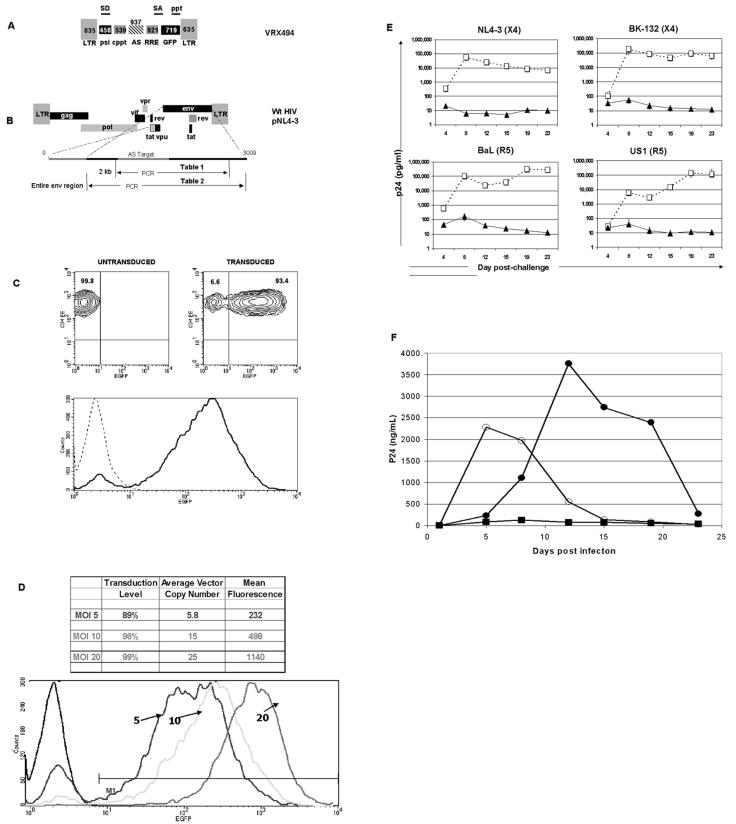
Schematic representation of HIV-1-based vector expressing antisense to the HIV-1 envelope region. (A) The antisense-expressing vector contains a 5′ and 3′ LTR, splice donor site (SD), splice acceptor site (SA), packaging signal (PSI), a cPPT (CPT) and central termination sequence (CTS), RRE, EGFP as a marker gene, a 937-base antisense sequence to the HIV envelope gene, and a 3′ PPT. (B) PCR primers were designed to amplify both the antisense target region and part of the envelope (Table 1; 1,936 bases), or the entire envelope region (Table 2; 2,544 bases). (C) Flow cytometry of untransduced and VRX494-transduced primary CD4+ T lymphocytes at 20 TU per cell. Upper panels show a density blot of untransduced (left) and transduced (right) lymphocytes, and relative percentages are shown. The bottom panel shows the mean fluorescent intensity (MFI) from the upper panels for untransduced (dotted line) and transduced (solid line) cultures. (D) Flow cytometric analysis of primary CD4 T lymphocytes transduced with VRX494 at 5, 10, and 20 TU per cell. Shown at the top is the transduction level, average vector copy number, and MFI in cells at each level of transduction. The lower panel shows the shift in MFI in graph format, in comparison to untransduced cells (dark black line). (E) Challenge of transduced (closed circles) and untransduced (open squares) primary CD4+ T-lymphocyte cultures with various HIV strains at an MOI of 0.001. Top and bottom panels show challenge with X4 and R5 strains of HIV, respectively. Virus strains are shown above each panel, and virus replication was measured by ELISA for p24 production. Bars represent standard deviations (n = 3). (F) Challenge with HIV at an MOI of 0.3 in untransduced SupT1 cells (open circles), SupT1 cells transduced at 20 TU per cell with an empty vector without antisense to the HIV env gene (closed circles), or SupT1 cells transduced at 20 TU per cell with the same vector containing a 937-base antisense sequence to the HIV env gene (squares). Transduction rates and MFI in Sup T1 cells for antisense and control vectors were 97% and 586 and 96% and 887, respectively. n = 3.
The VRX494 vector was made via cotransfection of 293T cells (BioReliance, Rockville, Md.) with the pVRX494 plasmid and a helper plasmid containing gag, pol, rev, and tat for amplification and the envelope G protein of vesicular stomatitis virus for pseudotyping. The titers of these vectors were determined on HeLa-tat cells (NIH AIDS Reagent Program; www.aidsreagent.org), and the titers were on average 5 × 106 transducing units (TU)/ml (unconcentrated supernatant).
VRX494 is the research analogue for our clinical vector, VRX496, and differs only in that VRX494 expresses a functional GFP gene instead of a molecular tag. VRX496 is currently being evaluated for safety and tolerability in a phase I clinical trial. Preclinical studies have shown in vitro and in mice that the vector does not significantly mobilize, if at all (24). Lack of mobilization is likely due to the antisense product targeting the HIV envelope gene, resulting in packaged vector particles incapable of subsequent transduction.
Virus and cell lines.
HIVNL4-3 was the wt virus strain used. It is derived from a clone constructed from two North American strains of HIV, and its sequence and characterization are described in reference 1. Viral stocks were made by transfection of 293T cells with pNL4-3 plasmid and subsequent collection of supernatant at 48 h, and then the supernatant was filtered through a 45-μm-pore-size filter. Virus titers were quantified by 50% tissue culture infective dose (TCID50) assay on SupT1 cells (NIH AIDS Reagent Program; www.aidsreagent.org) and were calculated to be 1.3 × 106 infectious particles/ml of supernatant. HIV strains BK132 (X4), Ba-L (R5), and US1 (R5) were obtained from the Center for AIDS Research at the University of Pennsylvania.
Primary cell isolation, culture, transduction, and challenge.
Peripheral blood from healthy donors was obtained from AllCells (Berkeley, Calif.), and lymphocytes were isolated by Ficoll-Hypaque density gradient separation. Cells were resuspended in blocking buffer (phosphate-buffered saline with Ca2+ and Mg2+, 0.5% bovine serum albumin, 5 mM EDTA, and human antibody). CD4+ T lymphocytes were subsequently purified by positive selection by magnetic cell sorter (Miltenyi Biotec, Auburn, Calif.). Purity was determined to be above 95% by flow cytometric staining for CD4/CD3.
CD4+ T cells were cultured in X-Vivo-15 (Biowhittaker) containing 10% human serum (NABI, Boca Raton, Fla.). For transduction, T lymphocytes were plated at 106 cells per well in a 24-well plate, and then VRX494 vector was added at 20 TU per ml concomitantly with immobilized CD3/28 beads, using anti-human CD3 (clone BB11; Diaclone, Besancon, France) and anti-human CD28 (clone BL8; Diaclone) antibodies on M450 epoxy beads (Dynal, Lake Success, N.Y.) at a ratio of three beads per cell and with 100 U of interleukin-2 (Chiron, Emeryville, Calif.)/ml. T lymphocytes were cultured for 3 days, centrifuged, washed three times to remove vector, and replated in 2 ml of medium, and 4 days later the beads were removed. At each passage after bead removal, cells were replated at 0.5 × 106 cells per ml. Upon bead depletion, 2 × 106 transduced or untransduced cells from healthy donors were challenged with wt HIV at a multiplicity of infection (MOI) of 0.001 overnight, and then washed four times. HIV-containing supernatants were recovered twice a week, and cells were counted and replated at 2 × 106 in 4 ml of T-cell medium.
SupT1 transduction and selection of mutants.
SupT1 cells were transduced with VRX494 vector overnight at an MOI of 5, 10, or 20. Cells were washed the following day. Flow cytometric analysis for EGFP expression was performed to determine the transduction rate (percentage of cells transduced).
Naïve SupT1 cells and transduced SupT1 cells were challenged with HIVNL4-3 at an MOI of 0.1 overnight. The next day, cells were washed three times. Time points were taken twice weekly during passaging of the cells. At each time point, supernatants were taken for p24 enzyme-linked immunosorbent assay (ELISA), which was performed according to the manufacturer's specifications (Beckman Coulter, Inc., Brea, Calif.). Reinfections were performed in the same manner by using virus supernatants normalized for p24 concentration from the challenge experiment instead of the viral stock.
Isolation of HIV episomal DNA and cloning.
Selected and wt HIV was harvested from culture supernatants and used to infect naïve SupT1 cells at the maximum concentrations allowed. Cells were collected when cytopathic effect was observed (about 5 days later). Episomal HIV DNA was isolated by mini-prep (QIAGEN, Inc., Valencia, Calif.). It is of interest that isolation of whole genomic DNA did not yield virus product after PCR, which is why episomal viral DNA was isolated instead. Virus envelope was amplified by PCR using either the forward pNL4-3 (6567-6590) (5′-TAAAGCCATGTGTAAAATTAACCC-3′) and reverse pNL4-3 (8503-8485) (5′-CAGGCTCCGCAGATCGTCC-3′) primers, which encompass the entire antisense region and part of the nontarget envelope region (see Table 1), or the forward pNL4-3 (6221-6240) (5′-CTACAGTACTTGGCACTAGC-3′) and reverse pNL4-3 (8785-8762) (5′-CTTCCAGTCCCCCCTTTTCTTTT-3′) primers, which encompass the entire envelope region (see Table 2). Amplified sequences were then cloned into the TOPO construct backbone as part of the TOPO TA cloning kit (Invitrogen Corporation, Carlsbad, Calif.). Individual clones were digested for deletion analysis. All intact clones and some deletion clones were sequenced for mutation analysis and detailed deletion analysis (Elim Biopharmaceuticals, Inc., Hayward, Calif.).
TABLE 1.
Deletions at the antisense target region
| Virus | No. of clones | No. of deletions | % with deletions |
|---|---|---|---|
| Wild type | 40 | 11 | 27.5 |
| Breakthrough | 290 | 264 | 91 |
TABLE 2.
Type and percentage of mutations in HIV selected for by serial passaging through Sup T1 cells
| Clonea | No. of A-G transitions | No. of mutations | Mutation rate (%)
|
||
|---|---|---|---|---|---|
| Entire ENV | ASb region | Non-AS region | |||
| WT | 0 | 0 | 0 | 0 | |
| BT90/BT-P1 | 108 | 109 | NAc | 11.66 | 0.1 |
| BT1 | 1 | 3 | 0.12 | 0 | 0.18 |
| BT2 | 3 | 8 | 0.31 | 0.53 | 0.12 |
| BT3 | 2 | 7 | 0.31 | 0.53 | 0.18 |
| BT4 | 4 | 9 | 0.35 | 0.53 | 0.25 |
| BT5 | 1 | 0.04 | 0 | 0.06 | |
| BT6 | 4 | 0.16 | 0.11 | 0.18 | |
| BT7 | 2 | 0.08 | 0.11 | 0.06 | |
| BT9 | 2 | 7 | 0.31 | 0.32 | 0.31 |
| BT11 | 2 | 3 | 0.12 | 0.11 | 0.12 |
| BT12 | 1 | 7 | 0.27 | 0.21 | 0.31 |
| BT14 | 4 | 0.16 | 0.11 | 0.18 | |
| BT15 | 2 | 4 | 0.16 | 0.21 | 0.12 |
| BT17 | 1 | 5 | 0.19 | 0.21 | 0.18 |
| BT19 | 2 | 4 | 0.16 | 0 | 0.25 |
| BT22 | 1 | 3 | 0.16 | 0.21 | 0.12 |
| BT24 | 1 | 2 | 0.08 | 0.11 | 0.06 |
| BT25 | 3 | 5 | 0.19 | 0.21 | 0.18 |
| BT26 | 1 | 2 | 0.12 | 0.11 | 0.12 |
| BT27 | 1 | 6 | 0.23 | 0.32 | 0.18 |
| BT31 | 1 | 3 | 0.16 | 0.21 | 0.12 |
| BT33 | 2 | 4 | 0.16 | 0.11 | 0.18 |
| BT34 | 2 | 0.16 | 0.11 | 0.18 | |
| BT35 | 2 | 0.08 | 0 | 0.12 | |
| BT36 | 2 | 0.08 | 0 | 0.12 | |
| BT37 | 4 | 8 | 0.27 | 0.32 | 0.25 |
| BT38 | 2 | 4 | 0.16 | 0.11 | 0.18 |
| BT39 | 2 | 4 | 0.16 | 0.21 | 0.12 |
| BT40 | 4 | 7 | 0.23 | 0.32 | 0.18 |
| BT41 | 2 | 0.08 | 0.11 | 0.06 | |
| BT43 | 4 | 0.16 | 0.11 | 0.18 | |
| BT44 | 2 | 4 | 0.16 | 0.32 | 0.06 |
| BT45 | 2 | 5 | 0.19 | 0.11 | 0.25 |
| BT46 | 1 | 0.04 | 0 | 0.06 | |
| BT47 | 1 | 0.04 | 0 | 0.06 | |
| BT48 | 5 | 7 | 0.27 | 0.32 | 0.25 |
| BT50 | 2 | 0.08 | 0.11 | 0.06 | |
| BT51 | 1 | 0.04 | 0 | 0.06 | |
| BT52/BT-P2 | 13 | 17 | 0.66 | 1.5 | 0.18 |
| BT53/BT-P3 | 15 | 17 | 0.66 | 1.6 | 0.12 |
| BT55 | 2 | 0.08 | 0 | 0.12 | |
| BT56 | 3 | 0.12 | 0 | 0.18 | |
| BT57 | 1 | 3 | 0.12 | 0.32 | 0.06 |
| BT58 | 1 | 5 | 0.19 | 0.32 | 0.12 |
| BT60 | 3 | 5 | 0.19 | 0.32 | 0.12 |
| BT61 | 4 | 8 | 0.31 | 0.32 | 0.31 |
| BT65 | 2 | 0.08 | 0 | 0.12 | |
| BT73 | 1 | 5 | 0.19 | 0.21 | 0.18 |
| BT74 | 1 | 4 | 0.16 | 0.21 | 0.12 |
| BT75 | 2 | 7 | 0.27 | 0.32 | 0.25 |
The three virus clones with significantly higher mutation rates are shown in bold.
AS, antisense.
NA, not available (only 1,247 of 1,627 nucleotides of BT-P1 were sequenced).
RESULTS
Transduction and HIV challenge in vector-transduced primary lymphocytes.
VRX494 is a fully gutted, HIV-1-derived cppt-containing vector, which expresses an unspliced mRNA containing a 937-base antisense sequence targeted to the HIV envelope gene under the control of the HIV LTR (Fig. 1A and B). For purposes of analysis, GFP was also driven off of the LTR in a splice-dependent manner. VRX494 is identical to our clinical-grade vector currently being evaluated in humans for safety, VRX496, except that VRX496 does not contain GFP but instead contains a 186-base molecular tag for in vivo marking.
VRX494 efficiently transduces primary CD4+ T lymphocytes with a single addition of 20 TU per cell upon culture initiation (see Materials and Methods). Over 90% of lymphocytes in culture were positive for GFP expression within 1 week posttransduction, and GFP expression was evenly distributed among the positive cells (Fig. 1C). Lower levels of transduction still offered high levels of gene transfer, but at lower copy numbers (Fig. 1D). The number of vector copies per cell correlated to the level of gene expression from the vector, as observed with the higher mean fluorescence activity at higher copy numbers. The correlation between copy number and concentration of vector addition is different for different culture systems and cell types. Therefore, this relationship must be determined separately for each culture system
To examine the antiviral efficacy of the expressed anti-Env antisense region, we challenged transduced and untransduced primary CD4+ T-lymphocyte cultures in triplicate with primary (NL4-3) and laboratory isolates (BaL, US1, and BK-132) at an MOI of 0.001. The infecting HIV strains represented both the macrophage-tropic (CCR5) and T-cell-tropic (CXCR4) virus tropisms. Virus replication was measured by p24 ELISA, which quantitates the level of HIV capsid protein in culture supernatant and thus serves as a surrogate marker for HIV replication. Cultures were followed for a total of 23 days postchallenge. All untransduced cultures produced p24 at maximum quantities of around 100,000 pg/ml. Transduced cultures inhibited replication by three logs or greater, regardless of the strain or tropism of the challenge virus (Fig. 1E). Suppression of virus was continuous, and towards the end of culture no increase in p24 was detected. These data demonstrate long-term efficient inhibition of HIV strains with different tropisms. An in-depth study of the efficacy of VRX494 against endogenous virus in primary T lymphocytes isolated from HIV patients has also been conducted (17a).
We then investigated the relative role of the antisense payload to the “empty” vector alone. The empty vector was anticipated to have a partial effect on virus replication, since it serves as a TAR and RRE decoy. SupT1 cells were transduced with vector containing antisense (VRX494) or an identical but empty vector at 20 TU per cell to yield a culture that was >99% transduced as measured by GFP fluorescence. Cultures were then challenged with HIVNL4-3 at an MOI of 0.3. Culture supernatants were collected twice weekly for 3 weeks, and replication was measured by ELISA for p24 production. Empty vector delayed peak virus replication by 1 week and demonstrated peak p24 levels at day 12, compared to that at day 5 after challenge in untransduced cells (Fig. 1F). Despite the delay, the empty vector was unable to control virus replication, and cultures were destroyed by the end of the experiment at day 24. In contrast, cultures transduced with antisense-containing vector did not produce measurable amounts of p24 at any time, despite the high-MOI challenge. These data clearly demonstrate the important role of the antisense payload in control of HIV replication, and they are supported by data presented later in this paper reporting virus mutations above baseline specifically within the antisense-targeted region.
Selection of HIV mutants by vector-mediated suppression.
Although viral breakthrough is not normally observed in vector-transduced primary human CD4 T lymphocytes, we were able to define conditions in the permissive SupT1 T-cell line to facilitate viral breakthrough. This was possible since SupT1 cells are more permissible to HIV replication than primary cells, in part due to their high level of division. Thus, these cells could replicate HIV to higher titers, improving the chance of development of potential escape mutant viruses. In addition, SupT1 cells could be cultured for a longer time, which allowed detection of selected viruses after several weeks. Since wt HIV replicates but vector does not, low levels of transduction allow for marginal initial wt HIV replication that eventually overwhelms the vector and replicate under conditions of selective pressure during long-term culture. For SupT1 cells, we have found that transduction at 20 TU per cell is required for complete virus inhibition and, therefore, transduction at 10 or 5 TU per cell results in incomplete inhibition and therefore may be considered suboptimal in this specific in vitro system. SupT1 cells were transduced with suboptimal levels of the HIV antisense-expressing vector described schematically in Fig. 1A. Virus could be selected from cultures transduced at 5 TU per cell, which inefficiently transduces the culture, but not at 20 TU per cell (Fig. 2A). As a result of the difference in HIV replication kinetics in SupT1 cells compared to primary T lymphocytes, the level of transduction required for protection cannot be translated from SupT 1 cells to T lymphocytes. In this paper, in Sup T1 cells followed by a strong HIV challenge of 0.1, the breakpoint transduction level was between 5 and 10 TU per cell. However, we have found in primary T cells challenged at a more clinically relevant MOI of 0.01 or 0.001 that the breakpoint transduction level is much lower.
FIG. 2.
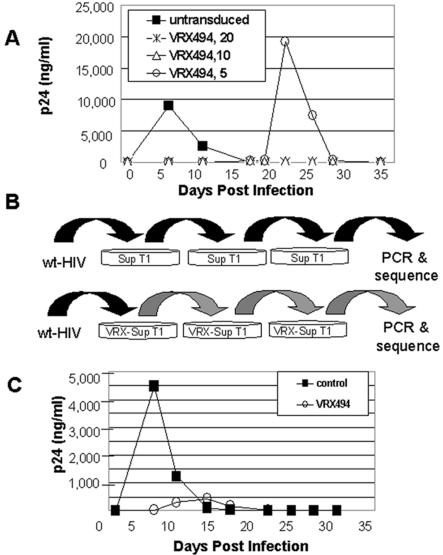
Selection of HIV by antisense in SupT1 cells. (A) Isolation of breakthrough wt HIV from antisense selective pressure can only be measured after challenge of SupT1 cells transduced at suboptimal doses of vector. SupT1 cells were transduced with vector at an MOI of 5, 10, or 20 TU/ml and then challenged with HIVNL4-3 at an MOI of 0.1. Virus production was measured by a p24 ELISA. (B) Schematic representation of selection procedure. SupT1 cells were mock transduced or transduced with vector at an MOI of 5 and then challenged with HIVNL4-3 at an MOI of 0.1. Culture supernatants were collected twice weekly and were tested for p24 protein levels in order to determine the point of peak replication. Culture supernatants from each group were collected during peak replication, normalized for p24, and subsequently used to infect naïve and transduced SupT1 cells for a total of three rounds of selection reflecting a selection period of 56 days. (C) After three rounds of selection, naïve SupT1 cells were infected with p24-normalized supernatants collected from the final passage of the third selection during peak replication. Supernatants were collected at the indicated times postinfection and assayed for p24 production by ELISA.
After infection of nontransduced SupT1 cells with HIV, peak p24 levels were observed 5 days later. However, in suboptimally transduced SupT1 cells, p24 levels did not peak until 20 to 25 days postinfection (Fig. 2A). Supernatants normalized for p24 and taken from normal and selected cultures were passaged serially three times for a total culture time of 56 days through transduced and nontransduced SupT1 cells (Fig. 2B). With each passage, selected virus was detected later than wt HIV. After serial selection, p24 was produced earlier in selected virus than before selection, but at lower levels (Fig. 2C, compare to A). This shift of the curve down and to the left suggests that a significant level of mutation may be occurring in response to antisense pressure.
Anti-HIV antisense results in a high frequency of large deletions in the target envelope region.
To characterize the mutations that led to the change in HIV phenotype after vector-mediated suppression, a 2-kb region was amplified via PCR using wt HIV-specific primers and then subcloned for sequencing (Fig. 1B). The mutations primarily fell into two categories: (i) those with large deletions in the envelope region, and (ii) those with only point mutations (discussed in the following section). The percentage of deletions in the breakthrough virus was much higher than in wt HIV (Table 1; 91% compared to 27.5%), and the deletions always encompassed the antisense target region (Fig. 3). The higher rate of deletion in virus from transduced cells indicated that antisense was exerting significant selective pressure on HIV to delete the region that binds the antisense region in the vector. Many of these deletions are quite large, which would result in severe debilitation of the virus. The ability of these mutants to persist with such defects likely resulted from copackaging and/or pseudotyping with wt HIV in cells. Alternatively, coinfection of complementing defective viruses in a cell could allow propagation of otherwise-replication-incompetent viral species.
FIG. 3.
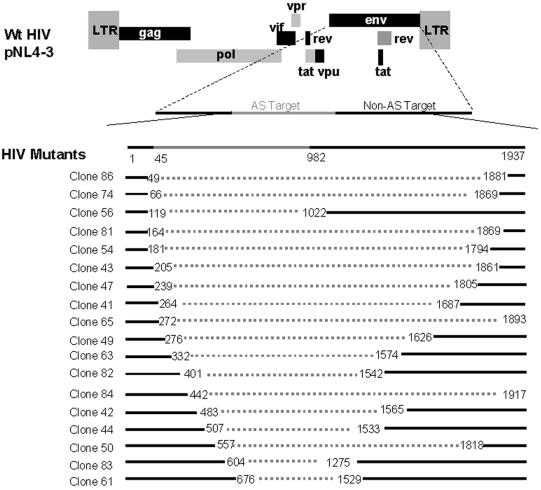
Deletion analysis of selected viruses. Viruses obtained from serial selection in SupT1 cells as described in the legend for Fig. 2 were subcloned and sequenced. Representative clones that contain large deletions are shown. Deletions are represented by the dotted lines, and the antisense target region is indicated at the top of the figure in grey. Please note that the figure is not drawn to scale. Most deletions observed were larger than 1 kb in length, and all deletions overlapped with the target region of the envelope.
High levels of A-G transitions in point mutations in selected viruses.
Viruses that contained point mutations ranged from having background levels of mutations similar to that of wt HIV to having high levels of mutations resulting in replication-defective viruses. The mutation frequency was examined in the antisense-targeted region, the nontargeted region, and in the entire envelope region, using the primers presented in Fig. 1B. The sequence changes from the entire wt HIV envelope in Table 2 are presented as follows: the number of A-G transitions are shown in the second column for comparison to the total number of point mutations within the antisense target region of the envelope; the mutation rates for the entire envelope or antisense region only are also shown. The vast majority of the clones displayed similar mutation rates (ranging from 0.04 to 0.35%) to the wt HIV control (ranging from 0 to 0.312%) (data not shown) over the entire region of the envelope. This suggests that the mutations in these clones may be equivalent to the spontaneous variations of wt HIV. In addition, we conducted a t test on the mutation rate within the antisense target region compared to that outside the antisense target region and found that for those viruses exhibiting overall spontaneous mutation rates, there was no statistical difference between the regions (P > 0.05). Therefore, the selected viruses are predicted to behave similarly to wt HIV, which is controlled by the antisense region expressed by our vector. However, three virus clones did have a significantly higher mutation rate in the envelope, BTP1, BTP2, and BTP3 (Table 2). In these clones, we noted that in the antisense target region, A-to-G transitions were more common than any other kinds of base changes, representing 99, 76, and 88% of the changes, respectively (Table 2, columns 2 and 3). Antisense-mediated selection of viruses containing these mutations was consistent with the known mechanism of antisense degradation of target transcripts by dsRAD/DRADA. Significantly, these clones only have increased rates of mutation within the antisense region and not outside the region complementary to the antisense, strongly suggesting that the mutations are a direct result of antisense-mediated selective pressure on the HIV genome.
Replication competence of viruses with high levels of A-G transitions.
As mentioned above, most of the viruses isolated from the transduced cells exhibited mutation rates similar to that found in virus isolated from mock-transduced cells. The reason for this is because most transcripts targeted by antisense are degraded instead of released from the cell. However, we were able to isolate three clones that exhibited significantly more point mutations in the antisense target region of the envelope gene but not outside the target region, BT-P1, BT-P2, and BT-P3 (Table 2). The overall mutation rates for these clones were 0.66, 0.66, and 4.25%, respectively (note that only 1,247 of 1,627 nucleotides of BT-P1 were sequenced, and therefore Table 2 shows no value for the “entire Env” data for BT-P1). These rates significantly exceeded the upper limit of the basal mutation rate of 0.35%. This is more striking when looking at mutation rates in the target region alone: BT-P1 (11.66%), BT-P2 (1.5%), and BT-P3 (1.6%). The location and frequency of these mutations are displayed in greater detail in the sequence map alignment presented in Fig. 4.
FIG. 4.

Sequence of the antisense target region of three clones with a high mutation rate. Three clones, BT-P1, BT-P2, and BT-P3 (Table 2) that had significantly higher mutation rates than wt HIV were sequenced. The nucleotide changes are shown and are predominantly A-G transitions. The antisense target region is shown in bold black, and the underlined region represents the V3 loop. Mutations were concentrated within the antisense target region.
Due to the high percentage of base changes in these mutants, it was possible that the viruses would no longer be sensitive to antisense vector inhibition. To address this question, the functionality of the mutant envelopes was tested with respect to their ability to support viral replication and subsequent resistance to vector inhibition. To that end, the envelope regions from the mutant viruses were cloned back into the HIVNL4-3 backbone. The resultant viruses were tested for their replication ability by measuring the viral titers in terms of TCID50 in nontransduced cells (Fig. 5A). Two of the three mutants were incapable of replicating in nontransduced SupT1 cells (BT-P1 and BT-P3). One mutant was able to replicate (BT-P2), but not to wt HIV levels. These data indicate that the selected viruses are not increasingly virulent, but instead demonstrate a decrease in fitness or lost replicative ability.
FIG. 5.
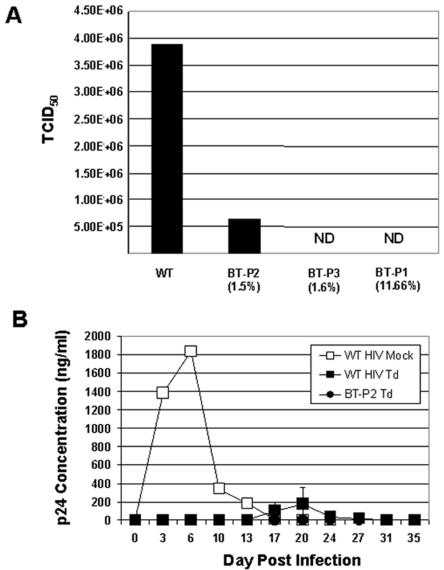
Replicative ability of selected mutants. (A) Virus variants selected for by antisense pressure are not fit. Viruses were produced by transfecting 293T cells with the mutant virus plasmids. Supernatants were collected at 48 h posttransfection for determination of virus titer by TCID50 in SupT1 cells. Actual values were as follows: 3.88 × 106 for wt HIV, 6.56 × 105 for BT-P2, and 0 for both BT-P3 and BT-P1 (ND is shown on the graph to indicate not detected). Mutation rates in the antisense region are shown in parentheses. BT-P2 and BT-P3 were full-length envelopes that were inserted back into HIVNL4-3. (B) Selected viruses cannot replicate in SupT1 cells transduced with vector expressing antisense. SupT1 cells were transduced with vector at an MOI of 11 and then challenged with either wt HIV or BT-P2 at an MOI of 0.01 (WT HIV Td and PT-P2 Td, respectively). Virus replication was measured by p24 ELISA. wt HIV mock-infected cultures (WT HIV Mock) were terminated after day 27 due to virus-induced cell death. Cells transduced with vector showed no evidence of HIV replication. Standard error is shown for WT HIV Td, but not for WT HIV Mock, since only one culture was used as a replication control. BT-P1 and BT-P3 were not tested because they do not replicate.
Finally, we examined whether these mutants had evolved to evade vector-mediated antisense inhibition. SupT1 cells were transduced with vector at 11 TU per cell, a suboptimal level that allows delayed replication of wt HIV, and then subsequently challenged with wt HIV or BT-P2 as a representative mutant capable of replicating (Fig. 5A). As a control, mock-transduced cells were infected with wt HIV. As previously observed in Fig. 2A, in the absence of antisense, wt HIV replicated to high titers between days 6 and 9 postchallenge. In transduced cells, wt HIV p24 levels peaked at days 20 to 23, but BT-P2 was not able to replicate (Fig. 5B). These data support that the mutations acquired in the target envelope region of a selected virus capable of replication do not result in true escape mutants capable of evading the antisense therapy.
DISCUSSION
This report describes the use of an HIV-1-based lentiviral vector, VRX494, which was engineered to express a 937-base antisense sequence against the envelope region of HIV. The clinical analogue to VRX494, VRX496, is identical to VRX494 except that it expresses a molecular tag in place of GFP. VRX496 is currently being evaluated for safety in humans in an ongoing clinical trial. We demonstrated efficient gene delivery to primary lymphocytes and subsequent inhibition of various HIV strains at a clinically relevant MOI (clinically relevant MOIs range from 0.0001 and 0.02 according to the Current Protocols in Immunology and rates of infected cells in HIV-positive individuals).
Consideration of antisense gene therapy for treatment of HIV infection has been extensively investigated over the last decade. Several strategies have been employed, including targeting of the HIV coreceptor to prevent infection and spread (21), inhibition of regulatory genes like the packaging sequence, tar, and primer binding site (6), or targeting of structural or nonstructural viral gene products like rev, envelope, virulence factors, etc. (6, 10, 19, 26, 27, 40, 44, 47). Importantly, the ability of antisense to inhibit HIV replication in vivo was demonstrated in a monkey model given autologous T cells modified with a vector containing antisense against the tat/rev gene prior to challenge (10). Challenges to bringing this therapy to the clinic include a difficulty in reaching efficacy, and the first HIV gene therapy clinical trial using modified T-cell transfer demonstrated the safety of the approach but did not achieve efficacy (9). Expression of antisense from lentivirus-derived vectors offers increased transduction and efficacy in inhibiting HIV (25). Our vector is an HIV-based lentiviral vector expressing a long antisense for increased transduction and anti-HIV efficacy. Efficacy can also be improved after dosing of the patient by using a selection gene to increase the number of vector-modified cells in vivo (9). An alternative approach would be to transduce stem cells that have the ability to mature into virus-resistant lymphocytes (9, 26).
In order to determine whether suppression of virus would lead to escape mutants or resistance to virus, we studied the mechanism of virus suppression by defining conditions that allowed virus production during antisense-mediated inhibition and then analyzing the resultant viruses for mutations in the target envelope region and for any ability to escape from antisense. We found that antisense exhibited a strong selective pressure against HIV, resulting in mutants incapable of escaping the antisense antiviral under standard transduction and culture conditions, which have been translated to the clinical scale.
There was a significantly higher percentage of deletions in the envelope target region in virus selected for by serial passage through transduced cells (91% compared to 27.5%). The deletions were of a significant size (most frequently over 1 kb in length), which would result in a nonfunctional envelope that severely debilitates the virus. These mutants could continue to propagate either due to help from wt HIV virions that could copackage with (pseudotype) the mutated genome or, alternatively, due to coinfection with complementing helper virus. This is reflected in the pattern of replication of virus mutants after selection, in which the virus is detected earlier after infection of suboptimally transduced cells, but at much lower levels. The magnitude and frequency of the deletions in the envelope target region demonstrated a high level of inhibition mediated by even suboptimal levels of vector transduction in SupT1 cells.
Antisense-mediated inhibition of RNA transcripts was first described in prokaryotes and eukaryotes in the mid 1980s (42, 49). It is generally believed that antisense-mediated inhibition occurs primarily in the nucleus of eukaryotes, where high levels of the enzyme dsRAD/DRADA are constitutively expressed. This enzyme recognizes double-stranded RNA only and catalyzes the partial unwinding of double-stranded RNA by converting adenosine to inosine. It is proposed that this unwinding is the cause for nuclear retention and degradation of target transcripts, which then are unable to be exported to the cytoplasm for translation into proteins. An excellent in-depth study examining this mechanism of nuclear retention using the polyomavirus model, which downregulates its early genes through the production of antisense during late gene transcription, is presented in reference 20. dsRAD/DRADA can also be induced in the cytoplasm by interferon, which is produced during virus infections like HIV. Therefore, in addition to nuclear modification of transcripts, HIV genomes might be modified in the cytoplasm after antisense transcripts expressed from our vector are transported there, forming double-stranded RNA with wt HIV RNA.
A significantly increased number of A-G transitions occurred in the mutagenized HIV genomes produced during selection. Such hypermutations have been previously reported in a retrovirus model and in respiratory syncytial virus and vesicular stomatitis virus (17, 30, 38). One possible explanation for the hypermutation in the present model is that neutral or random mutations incorporated into the HIV genome during reverse transcription favor selection of A-G transitions due to an enzyme-induced bias. However, given the very high number of A-G transitions, a second intriguing possibility of enzyme-directed mutagenesis by A-I conversions that select for A-G transitions is presented. The pattern of mutations observed in these studies supports the hypothesis that during HIV infection, dsRAD/DRADA changes adenosine to inosine, and some of the modified HIV genomic RNAs escape nuclear retention and are packaged into progeny virions. Since inosine base pairs with cytosine during reverse transcription (which occurs inside the virion after packaging), the modified RNAs would result in mutant HIV genomes containing A-G transitions. These results demonstrate that antisense-mediated suppression is occurring through the dsRAD/DRADA pathway.
Finally, and perhaps most importantly for use of our therapeutic antisense vector for treatment of HIV-infected patients, breakthrough viruses do not occur in cells transduced at normal levels. An advantage to the lentiviral vector antisense delivery system is that it affords the ability to express longer antisense transcripts than the liposome delivery system, and the lentiviral delivery system may be safer than the MoMuLV-based system in terms of insertional oncogenic potential. The 937-base length of the antisense sequence is sufficiently long so that viral deletions large enough to escape suppression are unlikely to replicate. Under specific conditions designed to allow for emergence of breakthrough viruses for the purposes of analysis, selection of viruses from antisense pressure does not allow for true escape mutants to evolve. For those viruses that mutated significantly greater percentages of the target region of the envelope gene compared to wt HIV, their ability to escape antisense was examined by cloning the mutated regions into the wt HIV backbone and then measuring the sensitivity of these clones to antisense inhibition. Two of these three viruses that contained 109 and 17 mutations in the entire envelope region (108 and 15 of which were A-G transitions, respectively) were unable to replicate, and a third also containing 17 mutations in this region (13 of which were A-G transitions) did not replicate to wt HIV levels. The replication-competent mutant, however, could not replicate when added to vector-transduced cells, thus demonstrating that viruses that mutate significantly in response to antisense inhibition do not result in true escape mutants. The A-G transitions seen in the mutated HIV likely result in dysfunctional envelope proteins that persist through pseudotyping of mutated genomes with wt HIV genomes that express functional envelope proteins. However, the mutated genomes are not replication competent on their own.
In conclusion, a long antisense vector results in mutations that cause sufficient debilitation of the virus. Other RNA-based approaches to HIV therapy that include ribozymes and RNAi use short sequences to identify the target RNA, which would quickly result in virus resistance. Theoretically, the use of several anti-HIV sequences in a single gene therapy construct may inhibit the evolution of resistant virus strains, although this has not yet been demonstrated over the long term. The length of the antisense message in the vector presented here is an important parameter to prevent virus resistance and ensure potential long-term suppression of HIV. These data support the safety of anti-HIV gene therapy mediated by antisense vector by demonstrating that it does not result in a population of viruses resistant to the gene therapy.
Acknowledgments
We thank Mari-Louise Hammarskjold and David Rekosh for helpful discussions.
REFERENCES
- 1.Adachi, A., H.-E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M.-A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baum, C., J. Dullmann, L. Zhixiong, B. Fehse, J. Meyer, D. A. Williams, and C. von Kalle. 2003. Side effects of retroviral gene transfer into hematopoietic stem cells. Blood 101:2099-2113. [DOI] [PubMed] [Google Scholar]
- 3.Behrens, G. M., M. Stoll, and R. E. Schmidt. 2000. Lipodystrophy syndrome in HIV infection. What is it, what causes it, and how can it be managed? Drug Safety 23:57-76. [DOI] [PubMed] [Google Scholar]
- 4.Boden, D., O. Pusch, F. Lee, L. Tucker, and B. Ramratnam. 2003. Human immunodeficiency virus type 1 escape from RNA interference. J. Virol. 77:11531-11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2002. Cases of HIV infection and AIDS in the United States, 2002. HIV/AIDS surveillance report, vol. 14. CDC Division of HIV/AIDS Prevention, Atlanta, Ga.
- 6.Chadwick, D. R., and A. M. Lever. 2000. Antisense RNA sequences targeting the 5′ leader packaging signal region of human immunodeficiency virus type-1 inhibits viral replication at post-transcriptional stages of the life cycle. Gene Ther. 7:1362-1368. [DOI] [PubMed] [Google Scholar]
- 7.Coburn, G. A., and B. R. Cullen. 2002. Potent and specific inhibition of human immunodeficiency virus type I replication by RNA interference. J. Virol. 76:9225-9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das, A. T., T. R. Brummelkamp, E. M. Westerhout, M. Vink, M. Madiredjo, R. Bernards, and B. Berkhout. 2004. Human immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition. J. Virol. 78:2601-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis, B., L. Humeau, and B. Dropulic. 2003. In vivo selection for human and murine hematopoietic cells transduced with a therapeutic MGMT lentiviral vector that inhibits HIV replication. Mol. Ther. 9:160-172. [DOI] [PubMed] [Google Scholar]
- 10.Donahue, R. E., B. A. Bunnell, M. C. Zink, M. E. Metzger, R. P. Westro, M. R. Kirby, T. Unangst, J. E. Clements, and R. A. Morgan. 1998. Reduction in SIV replication in rhesus macaques infused with autologous lymphocytes engineered with antiviral genes. Nat. Med. 4:181-186. [DOI] [PubMed] [Google Scholar]
- 11.Dropulic, B., D. A. Elkins, J. J. Rossi, and N. Sarver. 1993. Ribozymes: use as anti-HIV therapeutic molecules. Antisense Res. Dev. 3:87-94. [DOI] [PubMed] [Google Scholar]
- 12.Dropulic, B., M. Hermankova, and P. M. Pitha. 1996. A conditionally replicating HIV-1 vector interferes with wild type HIV-1 replication and spread. Proc. Natl. Acad. Sci. USA 93:11103-11108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freed, E. O, and M. A. Martin. 2001. HIVs and their replication, pp. 1971-2041. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 14.Gacon, G., S. Gisselbrecht, J. P. Piau, J. P. Boissel, J. Tolle, and S. Fischer. 1982. High level of tyrosine protein kinase in a murine lymphoma cell line induced by Moloney leukemia virus. EMBO J. 1:1579-1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hacein-Bey-Abina, S., C. von Kalle, M. Schmidt, M. P. McCormack, N. Wulffraat, P. Leboulch, A. Lim, C. S. Osborne, R. Pawlick, E. Morillon, R. Sorenson, A. Forster, P. Frase, J. I. Cohen, G. de Saint Basile, I. Alexander, U. Wintergerst, T. Frebourg, A. Aurias, D. Stoppa-Lyonnet, S. Romana, I. Radfort-Weiss, F. Gross, F. Valensi, E. Delabesse, E. Macintyre, F. Sigaux, J. Soulier, L. E. Leiva, M. Wissler, C. Prinz, T. H. Rabbitts, F. Le Deist, A. Fischer, and M. Cavazzana-Calvo. 2003. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 302:415-419. [DOI] [PubMed] [Google Scholar]
- 16.Hacein-Bey Abina, S., C. von Kalle, M. Schmidt, F. Le Deist, N. Wulffraat, E. McIntyre, I. Radford, J. L. Villeval, C. C. Fraser, M. Cavazzana-Calvo, and A. A. Fischer. 2003. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N. Engl. J. Med. 348:255-256. [DOI] [PubMed] [Google Scholar]
- 17.Hajjar, A. M., and M. L. Linial. 1995. Modification of retroviral RNA by double-stranded RNA adenosine deaminase. J. Virol. 69:5878-5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17a.Humeau, L. M., G. K. Binder, X. Lu, V. Slepuskin, R. Merling, P. Encheagaray, M. Pereira, T. Slepuskina, S. Barnett, L. K. Dropulic, R. Carrol, B. L. Levine, C. H. June, and B. Dropulic. Efficient lentiviral vector-mediated control of HIV-1 replication in lymphocytes from HIV+-infected individuals discordant for clinical status. Mol. Ther., in press. [DOI] [PubMed]
- 18.Jacque, J. M., K. Triques, and M. Stevenson. 2002. Modulation of HIV-1 replication by RNA interference. Nature 418:435-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, J. H., R. J. McLinden, J. D. Mosca, M. T. Vahey, W. C. Greene, and R. R. Redfield. 1996. Inhibition of HIV replication by sense and antisense rev response elements in HIV-based retroviral vectors. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 12:343-351. [DOI] [PubMed] [Google Scholar]
- 20.Kumar, M., and G. G. Carmichael. 1997. Nuclear antisense RNA induces extensive adenosine modifications and nuclear retention of target transcripts. Proc. Natl. Acad. Sci. USA 94:3542-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kusunoki, A., T. Saitou, N. Miyano-Kurosaki, and H. Takaku. 2001. Inhibition of the human chemokine receptor CXCR4 by antisense phosphorothioate oligodeoxyribonucleotides. FEBS Lett. 488:64-68. [DOI] [PubMed] [Google Scholar]
- 22.Lee, N. S., T. Dohjima, G. Bauer, H. Li, M.-J. Li, A. Ehsani, P. Salvaterra, and J. Rossi. 2002. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol. 19:500-505. [DOI] [PubMed] [Google Scholar]
- 23.Little, S. J., S. Holte, J.-P. Routy, E. S. Daar, M. Markowitz, A. C. Collier, R. A. Koup, J. W. Mellors, E. Connick, B. Conway, M. Kilby, L. Wang, J. M. Whitcomb, N. S. Hellmann, and D. D. Richman. 2002. Antiretroviral-drug resistance among patients recently infected with HIV. N. Engl. J. Med. 347:385-394. [DOI] [PubMed] [Google Scholar]
- 24.Lu, X., L. Humeau, V. Slepushkin, G. Binder, Q. Yu, T. Slepushkina, Z. Chen, R. Merling, B. Davis, Y.-N. Chang, and B. Dropulic. Safe two-plasmid production for the first clinical lentivirus vector that achieves >99% transduction in primary cells using a one-step protocol. J. Gen. Med., in press. [DOI] [PubMed]
- 25.Mautino, M. R., and R. A. Morgan. 2002. Inhibition of HIV-1 replication by novel lentiviral vectors expressing transdominant Rev and HIV-1 env antisense. Gene Ther. 9:421-431. [DOI] [PubMed] [Google Scholar]
- 26.Morel, F., S. Salimi, J. Markovits, T. W. Austin, and I. Plavec. 1999. Hematologic recovery in mice transplanted with bone marrow stem cells expressing anti-human immunodeficiency virus genes. Hum. Gene Ther. 10:2779-2787. [DOI] [PubMed] [Google Scholar]
- 27.Morgan, R. A., and R. Walker. 1996. Gene therapy for AIDS using retroviral mediated gene transfer to deliver HIV-1 antisense TAR and transdominant Rev protein genes to syngeneic lymphocytes in HIV-1 infected identical twins. Hum. Gene Ther. 7:1281-1306. [DOI] [PubMed] [Google Scholar]
- 28.Nijuis, M., S. Deeks, and C. Boucher. 2001. Implications of antiretroviral resistance on viral fitness. Curr. Opin. Infect. Dis. 14:23-28. [DOI] [PubMed] [Google Scholar]
- 29.Novina, C. D., M. F. Murray, D. M. Dykxhoorn, P. J. Beresford, J. Riess, S. K. Lee, R. G. Collamn, J. Lieberman, P. Shankar, and P. A. Sharp. 2002. siRNA-directed inhibition of HIV-1 infection. Nat. Med. 8:681-686. [DOI] [PubMed] [Google Scholar]
- 30.O'Hara, P. J., S. T. Nichol, F. M. Horodyski, and J. J. Holland. 1984. Vesicular stomatitis virus defective interfering particles can contain extensive genomic sequence rearrangements and base substitutions. Cell 36:915-924. [DOI] [PubMed] [Google Scholar]
- 31.Patterson, J. B., and C. E. Samuel. 1995. Expression and regulation by interferon of a double-stranded RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Mol. Cell. Biol. 15:5376-5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patterson, J. B., D. C. Thomis, S. L. Hans, and C. E. Samuel. 1995. Mechanism of interferon action: double-stranded RNA-specific adenosine deaminase from human cells is inducible by alpha and gamma interferons. Virology 210:508-511. [DOI] [PubMed] [Google Scholar]
- 33.Paul, C. P., P. D. Good, I. Winer, and D. R. Engelke. 2002. Effective expression of small interfering RNA in human cells. Nat. Biotechnol. 29:505-508. [DOI] [PubMed] [Google Scholar]
- 34.Powderly, W. G. 2002. Long-term exposure to lifelong therapies. J. Acquir. Immune Defic. Syndr. 29:S28-S40. [DOI] [PubMed] [Google Scholar]
- 35.Qin, X.-F., D. S. An, I. Chen, and D. Baltimore. 2003. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc. Natl. Acad. Sci. USA 100:183-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rigden, J. E., J. A. Ely, J. L. Macpherson, W. L. Gerlach, L. Q. Sun, and G. P. Symonds. 2000. The use of ribozyme gene therapy for the inhibition of HIV replication and its pathogenic sequelae. Curr. Iss. Mol. Biol. 2:61-69. [PubMed] [Google Scholar]
- 37.Rouzine, I. M., and J. M. Coffin. 1999. Search for the mechanism of genetic variation in the pro gene of human immunodeficiency virus. J. Virol. 73:8167-8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rueda, P., B. Garcia-Barreno, and J. A. Melero. 1994. Loss of conserved cysteine residues in the attachment (G) glycoprotein of two human respiratory syncytial virus escape mutants that contain multiple A-G substitutions (hypermutations). J. Virol. 198:653-662. [DOI] [PubMed] [Google Scholar]
- 39.Sarver, N., E. M. Cantin, P. S. Chang, J. A. Zaia, P. A. Ladne, D. A. Stephens, and J. J. Rossi. 1990. Ribozymes as potential anti-HIV-1 therapeutic agents. Science 247:1222-1225. [DOI] [PubMed] [Google Scholar]
- 40.Shahabuddin, M., and A. S. Khan. 2000. Inhibition of human immunodeficiency virus type 1 by packageable, multigenic antisense RNA. Antisense Nucleic Acid Drug Dev. 10:141-151. [DOI] [PubMed] [Google Scholar]
- 41.Siliciano, J. D., J. Kajdas, D. Finzi, T. C. Quinn, K. Chadwick, J. B. Margolick, C. Kovacs, S. J. Gange, and R. F. Siliciano. 2003. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat. Med. 9:727-728. [DOI] [PubMed] [Google Scholar]
- 42.Spencer, C. A., R. D. Gietz, and R. B. Hodgetts. 1986. Overlapping transcription units in the dopa decarboxylase region of Drosophila. Nature 322:279-281. [DOI] [PubMed] [Google Scholar]
- 43.Tsichlis, P. N. 1987. Oncogenesis by Moloney murine leukemia virus. Anticancer Res. 7:171-180. [PubMed] [Google Scholar]
- 44.Tung, F. Y., and M. H. Tung. 1996. Characterization of antisense RNA-mediated inhibition of SIV replication. J. Med. Virol. 48:321-325. [DOI] [PubMed] [Google Scholar]
- 45.Turner, B. J. 1993. Adherence to antiretroviral therapy by human immunodeficiency virus-infected patients. J. Infect. Dis. 185:S143-S151. [DOI] [PubMed] [Google Scholar]
- 46.UNAIDS Joint United Nations Programme on HIV/AIDS. 2003. AIDS epidemic update: December 2003. UNAIDS and World Health Organization, Geneva, Switzerland.
- 47.Veres, G., U. Junker, J. Baker, C. Barske, C. Kalfoglou, H. Ilves, S. Escaich, H. Kaneshima, and E. Bohnlein. 1998. Comparative analyses of intracellularly expressed antisense RNAs as inhibitors of human immunodeficiency virus type 1 replication. J. Virol. 72:1894-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vigouroux, C., S. Gharakhanian, Y. Salhi, T. H. Nguyen, N. Adda, W. Rozenbasum, and J. Capeau. 1999. Adverse metabolic disorders during highly active antiretroviral treatments. Diabetes Metab. 25:383-392. [PubMed] [Google Scholar]
- 49.Williams, T., and M. Fried. 1986. A mouse locus at which transcription from both DNA strands produces mRNAs complementary at their 3′ ends. Nature 322:275-279. [DOI] [PubMed] [Google Scholar]


