Abstract
All fully sequenced baculovirus genomes, with the exception of the dipteran Culex nigripalpus nucleopolyhedrovirus (CuniNPV), have previously been from Lepidoptera. This study reports the sequencing and characterization of a hymenopteran baculovirus, Neodiprion lecontei nucleopolyhedrovirus (NeleNPV), from the redheaded pine sawfly. NeleNPV has the smallest genome so far published (81,755 bp) and has a GC content of only 33.3%. It contains 89 potential open reading frames, 43 with baculovirus homologues, 6 identified by conserved domains, and 1 with homology to a densovirus structural protein. Average amino acid identity of homologues ranged from 19.7% with CuniNPV to 24.9% with Spodoptera exigua nucleopolyhedrovirus. The conserved set of baculovirus genes has dropped to 29, since NeleNPV lacks an F protein homologue (ac23/ld130). NeleNPV contains 12 conserved lepidopteran baculovirus genes, including that for DNA binding protein, late expression factor 11 (lef-11), polyhedrin, occlusion derived virus envelope protein-18 (odv-e18), p40, and p45, but lacks 21 others, including lef-3, me53, immediate early gene-1, lef-6, pp31, odv-e66, few polyhedra 25k, odv-e25, protein kinase-1, fibroblast growth factor, and ubiquitin. The lack of identified baculovirus homologues may be due to difficulties in identification, differences in host-virus interactions, or other genes performing similar functions. Gene parity plots showed limited colinearity of NeleNPV with other baculoviruses, and phylogenetic analysis indicates that NeleNPV may have existed before the lepidopteran nucleopolyhedrovirus and granulovirus divergence. The creation of two new Baculoviridae genera to fit hymenopteran and dipteran baculoviruses may be necessary.
Several lepidopteran baculovirus genomes and one dipteran baculovirus genome have been fully sequenced, but none so far have been reported from Hymenoptera. Hymenopterans are ancient insects that have existed since the early to mid-Mesozoic era (41, 54). Sawflies (Symphyta) are primitive hymenopterans (22) that have existed since the Triassic period (206 to 248) × 106 years ago (54). Baculoviruses infecting the Hymenoptera likely represent more ancient viruses than those infecting Lepidoptera, which first appeared in the Cretaceous period (65 to 144) × 106 years ago during the late Mesozoic to Cenozoic eras (41, 54). Analysis of the polyhedrin gene from various baculoviruses, including that from Neodiprion sertifer (NeseNPV), suggested that hymenopteran nucleopolyhedroviruses (NPVs) may have diverged from the lepidopteran baculoviruses before the separation of the lepidopteran NPVs and the granuloviruses (GVs) (55, 71). With the radiation of both Lepidoptera and Hymenoptera, their respective baculoviruses may have undergone host-dependent evolution with their hosts (55, 71).
This study was undertaken to sequence and characterize the genome of the hymenopteran nucleopolyhedrovirus NeleNPV, from the redheaded pine sawfly, Neodiprion lecontei. This insect is a pest of young, natural pine stands, plantations, and greenhouse cultures and may cause complete defoliation and death of small trees, reduced growth, branch mortality and tree deformity (18). NeleNPV was first identified from the redheaded pine sawfly in Ontario in 1950 and proved to be highly infectious (10). Sawfly NPVs replicate in the nuclei of midgut epithelium cells, causing infectious diarrhea and sloughing off of infected cells during the advanced stage of the disease. The gregarious nature of sawflies leads to the rapid spread of the virus through a population, with insects dying 4 to 7 days after infection (22). NeleNPV is available as a registered product called Lecontvirus and is an effective control agent for N. lecontei infestations (19). NeleNPV is now one of the first two fully sequenced hymenopteran baculoviruses, the second being NeseNPV (24).
Baculoviruses are divided into NPVs or GVs based on their occlusion body (OB) formation. NPVs are found mainly in lepidopterans but have been identified in other insect orders, including Hymenoptera, Diptera, Coleoptera, Thysanura, and Trichoptera, and contain multiple virions with either single or multiple nucleocapsids (22, 66). All hymenopteran NPVs contain single nucleocapsids (22). GVs are occluded within granulin, each OB contains a single virion, and GVs have been found only in lepidopterans (22).
Baculoviruses contain a set of conserved genes that are involved in essential functions, such as viral replication, transactivation, production of structural proteins, assembly, and release of progeny viruses. The evolution of large genomes, particularly in lepidopteran NPVs, has reduced viral dependency on the host cell machinery and led to an increased number of auxiliary genes that are not essential but provide a selective advantage (49). To date, the complete genomes of 23 baculoviruses are available in GenBank. As the number of sequenced genomes has increased, the number of conserved genes has decreased. Previous reports list 62 to 67 conserved baculovirus genes (13, 31, 32, 36). With the publication of the Culex nigripalpus nucleopolyhedrovirus (CuniNPV) genome, that number decreased to 30 genes conserved in 13 baculoviruses (2, 33). The number of conserved baculovirus genes remained at 30 with the sequencing of Mamestra configurata NPV isolate 90/2 (MacoNPV A) (44), M. configurata NPV-96B (MacoNPV B) (43), Phthorimaea operculella GV (PhopGV) (GenBank accession number AF499596), Rachiplusia ou multiple NPV (RoMNPV) (27), Adoxophyes honmai NPV (AdhoNPV) (GenBank accession number AP006270), Adoxophyes orana granulovirus (AdorGV) (69), Choristoneura fumiferana MNPV (GenBank accession number NC_004778), Cryptophlebia leucotreta GV (CrleGV) (42), Helicoverpa armigera NPV (GenBank accession number NC_003094), and C. fumiferana defective NPV (GenBank accession number AY327402).
Our early work on NeleNPV estimated its size to be in the order of 82,000 bp based on restriction enzyme digestion, making it the smallest known baculovirus genome. We hypothesized that its small genome would consist mainly of essential baculovirus genes, that it would contain a smaller core of conserved genes, and that its sequence would provide useful information on the evolution of baculoviruses. Here we report on the complete sequence and gene organization of NeleNPV and compare it with other baculovirus genomes.
MATERIALS AND METHODS
Virus production and DNA preparation.
N. lecontei larvae infected with NeleNPV were collected between 1975 and 1980 from areas throughout Ontario or from Christian Island in southern Georgian Bay in 1995. Insects were freeze-dried, ground to a fine powder, lyophilized, and stored at 4°C as previously described (18). The lyophilized powder was diluted in 0.5% sodium dodecyl sulfate (final concentration), stirred overnight, filtered, and centrifuged (2,500 × g, 30 min). The OB-containing pellets were washed with double-distilled H2O, centrifuged three times (2,500 × g, 30 min), and then passed through two 60, 50, 10% discontinuous sucrose gradients (40,000 × g, 90 min, 20°C). The OBs between the 50 and 60% sucrose cushions were removed, passed through 10 to 45% continuous sucrose gradients (40,000 × g, 90 min, 4°C), diluted with Tris-EDTA (TE), and centrifuged (87,000 × g, 90 min). The pellets were resuspended in 0.5 ml of TE, and DNA was extracted from the purified OBs using the Qiagen genomic tip 20/G DNA extraction kit using 1 ml of general lysis buffer (G2) supplemented with 100 μl of proteinase K (20 mg/ml) with overnight incubation (50°C). The standard Qiagen protocol was then followed. Yields of virus were low, since NeleNPV infection is restricted to the midgut, nucleocapsids contain only a single virion, and the freeze-dried samples were heavily contaminated, with insect debris trapping much of the virus. Purified DNA was checked by restriction analysis on agarose gels for purity and concentration.
DNA cloning and sequencing.
Viral DNA was sheared into small fragments by nebulization, cloned, and sequenced by Qiagen Sequencing Services using a combination of shotgun cloning, primer walking, and PCR to generate gap-spanning fragments, for an average 12× coverage. An ABI PRISM 377XL or ABI PRISM 3700 sequencer was used with Qiagen modified ABI sequencing chemistries and Qiagen purified DNA. The sequence data were manually edited and automatically assembled by using LASERGENE's DNAStar Seqman program, version 4.06, for sequence assembly and contig management. Some polymorphisms were found and were attributed to the virus being from field isolates. These ambiguities have been left and given standard codes as determined by program analysis (M = A,C; R = A,G; W = A,T; S = C,G; Y = C,T; K = G,T; V = A,C,G; H = A,C,T; D = A,G,T; B = C,G,T; N = A,C,G,T).
Sequence analysis.
DNA sequence data were analyzed using DNAStar LASERGENE programs, version 4.06 or 5.05, and MacVector sequence analysis software, version 4.1.4. Open reading frames (ORFs) encoding more than 50 amino acids (aa) with minimal overlap were accepted as putative genes; otherwise, the largest ORF was selected. Sequence data were submitted to GenBank, and database searches were performed using the National Center for Biotechnology Information (NCBI) ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) and BLAST (4). If difficulties were encountered identifying homologues with standard protein-protein BLAST (blastp) searches, further analysis was done using blastp for short, almost exact matches, PSI BLAST (5) using E values of 10 and 0.01 for iterations, analysis using the Simple Modular Architecture Research Tool (SMART), version 3.4 (58, 59), and Smith Waterman similarity searches using DeCypher with default settings (coe02.ucalgary.ca). An ORF was considered clearly identified if any of the following criteria were met. (i) A blastp search showed a baculovirus match with an E value of 0.1 or less. (ii) Amino acid identity to a baculovirus homologue was 20% or greater based on DNAStar Megalign ClustalW analysis of complete ORFs. (iii) A conserved domain was found. (iv) Matches were close but did not meet the first three criteria, but ORFs showed a significant match to clearly identified ORFs in NeseNPV as determined by A Garcia-Maruniak, et al. (24). The sequence was analyzed for repeats using MacVector's Pustell DNA matrix, Emboss Palindrome (23), and Tandem Repeats Finder (9). Multiple alignments were performed using DNAStar's Megalign ClustalW alignment, and percent amino acid identity indicates the percentage of identical residues between complete ORFs. Phylogenetic trees were constructed using ClustalW protein alignments and PAUP 4.0b10 (62), using maximum parsimony analysis with heuristic search and stepwise addition options, and were confirmed using bootstrap analysis with heuristic search and 1,000 replicates. Gene parity plots were performed on the NeleNPV genome versus the genomes of Autographa californica MNPV (AcMNPV) (7), Helicoverpa armigera single-nucleocapsid NPV (SNPV) (HaSNPV) (13), CuniNPV (2), and Plutella xylostella GV (PxGV) (28), using established methods (13, 34, 37).
Nucleotide sequence accession number.
The NeleNPV genome sequence has been deposited in GenBank under accession number AY349019.
RESULTS AND DISCUSSION
Nucleotide sequence analysis.
The NeleNPV genome was 81,755 bp in size, making it the smallest baculovirus genome so far known, with others ranging from 99,657 bp for AdorGV (69) to 178,733 bp for Xestia c-nigrum GV (XcGV) (30). The G+C content was 33.3%, with the lowest so far published being that of CrleGV at 32.4% (42) and the highest being 57.5% for Lymantria dispar MNPV (LdMNPV) (40) (Table 1).
TABLE 1.
Comparison of baculovirus genomes
| Size (bp) | % G+C | No. of ORFs | No. of hr'sa | No. of brob genes | No. of homo- logues in NeleNPVc | % Overall aa id with NeleNPVd | |
|---|---|---|---|---|---|---|---|
| NeleNPV | 81,755 | 33.3 | 89 | 0 | 0 | ||
| AcMNPV | 133,894 | 40.7 | 154 | 8 (9) | 1 | 41 | 23.2 |
| OpMNPV | 131,993 | 55.1 | 152 | 5 | 3 | 42 | 23.0 |
| BmNPV | 128,413 | 40.4 | 136 | 7 | 5 | 42 | 23.4 |
| LdMNPV | 161,046 | 57.5 | 163 | 13 | 16 | 42 | 23.7 |
| SeMNPV | 135,611 | 44.0 | 139 | 6 | 0 (1) | 42 | 24.9 |
| EppoMNPV | 118,584 | 41.0 | 135 | 5 | 1 | 42 | 24.0 |
| HaSNPV | 131,403 | 39.1 | 135 | 5 | 3 | 42 | 24.7 |
| SpltMNPV | 139,342 | 42.7 | 141 | 17 | 2 | 42 | 23.7 |
| CuniNPV | 108,252 | 50.9 | 109 | 4 | 6 | 29 | 19.7 |
| XcGV | 178,733 | 41.0 | 181 | 9 | 7 | 42 | 22.2 |
| PxGV | 100,999 | 40.7 | 120 | 4 | 0 | 42 | 23.6 |
| CpGV | 123,500 | 45.2 | 143 | 0 | 1 | 42 | 23.1 |
| AdorGV | 99,657 | 34.5 | 119 | 0 | 0 | 42 | 23.1 |
| CrleGV | 110,907 | 32.4 | 129 | 3 | 0 | 42 | 23.2 |
Only ORFs meeting criteria for clear identification included, ac27 iap not included.
Calculated using ClustalW results for complete ORFs; average based on shared ORFs. aa id, amino acid identity.
The genome contained 89 potential ORFs, accounting for 88.8% of the total sequence. Identifying homologues was difficult due to the low similarity of the NeleNPV genome to other baculovirus genomes. Only 50 ORFs met our criteria for clear identification with 43 identified as baculovirus homologues, 6 recognized by the presence of conserved domains, and 1 as a potential match with a structural protein from densoviruses (Fig. 1; Table 2).
FIG. 1.
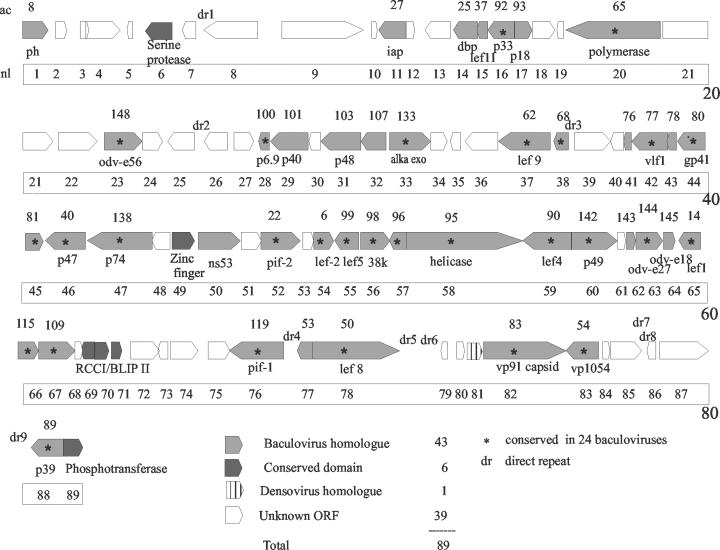
Linear map of the NeleNPV genome. Arrows indicate the position and direction of transcription for potential ORFs, with polyhedrin (ph) shown as ORF 1. NeleNPV ORF numbers and potential gene names are shown below the arrows, and AcMNPV ORF numbers (7) are shown above the arrows.
TABLE 2.
Characteristics of NeleNPV ORFse
| ORF no. | Name | Position | Size (aa) | AcMNPV ORF no. (% aa id)a | HaSNPV ORF no. (% aa id) | CuniNPV ORF no. (% aa id) | PxGV ORF no. (% aa id) | Comment(s)b |
|---|---|---|---|---|---|---|---|---|
| 1 | Polyhedrin | 1>744 | 247 | 8 (46.7) | 1 (45.9) | X | 4 (36.7) | NeabNPVpolyhedrin, e = e − 143, aa id 98% |
| 2 | 961>1287 | 108 | Internal repeats | |||||
| 3 | 1680>1922 | 80 | ||||||
| 4 | 1843>2841 | 332 | ||||||
| 5 | 3049<3207 | 52 | ||||||
| 6 | 3671<4450 | 259 | Trypsin-like serine protease, AAD21829 C. felis CfSP-2, e = 8e − 46, aa id 38.7% | |||||
| 7 | 4663<5058 | 131 | Internal repeat | |||||
| Direct repeat 1 | 5152-5194 | 2 43-bp direct repeats | ||||||
| 5223-5265 | ||||||||
| 8 | 5279<6859 | 526 | Internal repeat | |||||
| 9 | 7567>9951 | 794 | ||||||
| 10 | 10181>10345 | 54 | ||||||
| 11 | iap-3 | 10334<11116 | 260 | X | 103 (23.8) | X | 98 (17.6) | Sf iap AF186378, e = 3e − 33, aa id 28.4% |
| 12 | 11194>11436 | 80 | ||||||
| 13 | 11733<12476 | 247 | Transmembrane domain, coiled coil | |||||
| 14 | dbp | 12565<13275 | 236 | 25 (18.6) | 25 (17.3) | X | 61 (14.3) | HaSNPV ORF25 dbp, e = 0.94, aa id 17.3%c |
| 15 | lef-11 | 13256<13561 | 101 | 37 (16.7) | 32 (23.5) | X | 46 (12.4) | LsNPV lef 11, e = 0.014, aa id 23.5% |
| 16 | 13575<14345 | 256 | 92 (21.4) | 80 (23.1) | 14 (15.6) | 76 (20.3) | MacoNPV B ORF 92, e = 2e −22, aa id 22.5% | |
| 17 | 14347>14859 | 170 | 93 (14.8) | 81 (18.4) | X | 75 (15.9) | LsNPV ORF93, e = 0.037, aa id 14.0% | |
| 18 | 14868>15518 | 216 | Signal peptide | |||||
| 19 | 15584<15775 | 63 | Transmembrane domain | |||||
| 20 | dna pol | 15806<18577 | 923 | 65 (25.8) | 67 (27.3) | 91 (15) | 93 (23.7) | MacoNPV B ORF114 DNA polymerase, e = 2e − 61, aa id 28.1% |
| 21 | 18576>20762 | 728 | 2 coiled coils | |||||
| 22 | 20927>22069 | 380 | ||||||
| 23 | odv-e56 | 22321>23331 | 336 | 148 (32.6) | 15 (38) | 102 (20.8) | 16 (37.7) | PhopGV ORF 16 odv-e56, e = 1e − 54, aa id 37.7% |
| 24 | 23386>23964 | 192 | ||||||
| 25 | 24120<24905 | 261 | ||||||
| Direct repeat 2 | 24924-25029 | 2 53-bp direct tandem repeats | ||||||
| 26 | 25183<25863 | 226 | ||||||
| 27 | 26082>26654 | 190 | ||||||
| 28 | p6.9 | 26772<27080 | 102 | 100 (30.4) | 88 (23.3) | 23 (34.8) | 67 (40.4) | AdorGV ORF 72 p6.9, aa id 64.3%d |
| 29 | p40 | 27110<28210 | 366 | 101 (9.4) | 89 (10.4) | X | 66 (11.4) | XcGV ORF 93 p40, aa id 13.9% |
| 30 | 28231<28581 | 116 | ||||||
| 31 | p48 | 28578<29750 | 390 | 103 (11.1) | 91 (14.8) | X | 63 (12.7) | MacoNPV A ORF 83 p48, e = 0.026, aa id 15.1% |
| 32 | 29769<30485 | 238 | 106/107 (8.1) | 101 (18.4) | X | 40 (17.9) | LdMNPV ORF140, e = 6e − 10, aa id 16.7% | |
| 33 | alk-exo | 30588>31796 | 402 | 133 (24.3) | 114 (19.6) | 54 (16.8) | 106 (23.0) | PxGV ORF106 alk exo, e = 3e − 25, aa id 23.0% |
| 34 | 31793>32266 | 157 | Transmembrane domain, coiled coil | |||||
| 35 | 32364<32657 | 97 | Internal repeats | |||||
| 36 | 32787<33746 | 319 | ||||||
| 37 | lef-9 | 33771<35282 | 503 | 62 (31.5) | 55 (33.5) | 59 (17.1) | 99 (32.3) | PhopGV ORF 109 lef-9, e = 2e − 75, aa id 34.1% |
| 38 | 35308<35733 | 141 | 68 (15.5) | 64 (21.6) | 58 (18.1) | 96 (17.8) | SpltNPV ORF66, e = 2e − 04, aa id 21.8%, signal peptide, transmembrane domain | |
| Direct repeat 3 | 35829-35886 | 2 29-bp direct tandem repeats | ||||||
| 39 | 35984>37045 | 353 | ||||||
| 40 | 37042<37419 | 125 | ||||||
| 41 | 37416<37646 | 76 | 76 (18.2) | 70 (19.5) | X | 91 (22.1) | SeNPV ORF95, e = 1.4, aa id 24.7%d | |
| 42 | vlf-1 | 37649<38713 | 354 | 77 (22.8) | 71 (28.5) | 18 (17.5) | 89 (28.8) | SpltNPV ORF74, vlf-1, e = 1e − 41, aa id 29.6% |
| 43 | 38682<38966 | 94 | 78 (14.7) | 72 (20.0) | X | 88 (13.5) | AdorGV ORF 90, aa id 20.9%d | |
| 44 | gp41 | 38987<39799 | 270 | 80 (26.9) | 73 (30.6) | 33 (7.4) | 87 (20.3) | HaSNPV ORF 73 gp41, e = 2e − 19 |
| 45 | 39821>40348 | 175 | 81 (34.1) | 74 (35.2) | 106 (17.0) | 86 (34.7) | LdMNPV ORF 89, e = 5e − 23, aa id 34.1% | |
| 46 | p47 | 40601<41770 | 389 | 40 (21.3) | 35 (19.2) | 73 (8.5) | 51 (21.7) | CpGV ORF68 p47, e = 9e − 25, aa id 22.8% |
| 47 | p74 | 41772<43673 | 633 | 138 (38.3) | 20 (36.4) | 74 (34.4) | 49 (32.8) | RoMNPV ORF138 p74, e = e − 124, aa id 39.0% |
| 48 | 43651<44187 | 178 | ||||||
| 49 | 44247>44903 | 218 | Dm NP_609448, e = 1e − 13, aa id 17.4%, 4 C2H2 zinc fingers | |||||
| 50 | 45012>46235 | 407 | NeseNPV AF121349, e = 2e − 29, aa id 45.7%, internal repeat | |||||
| 51 | 46237>46815 | 192 | ||||||
| 52 | pif-2 | 46834>47988 | 384 | 22 (44.6) | 132 (43.8) | 38 (43.9) | 37 (47.2) | NeseNPV AF121349, e = e − 152, aa id 74.3% |
| 53 | 48058>48372 | 104 | ||||||
| 54 | lef-2 | 48374>48961 | 195 | 6 (13.8) | 117 (16.3) | 25 (14.8) | 32 (11.2) | NeseNPV AF121349 lef-2, e = 7e − 27, aa id 35.7% |
| 55 | lef-5 | 48989<49696 | 235 | 99 (25.4) | 87 (25.8) | 88 (9.3) | 69 (28.4) | NeseNPV AF121349 lef-5, e = 2e − 66, aa id 58% |
| 56 | 38k | 49687>50604 | 285 | 98 (30.1) | 86 (27.3) | 87 (25.5) | 70 (30.4) | OpMNPV ORF99 38k, e = 8e − 27, aa id 26.2% |
| 57 | 50590<51096 | 168 | 96 (25.4) | 85 (27.2) | 90 (26) | 71 (23.5) | CuniNPV ORF 90, e = 3e − 16, aa id 26.0% | |
| 58 | helicase | 51083>54487 | 1134 | 95 (16.8) | 84 (20.4) | 89 (11.9) | 72 (17.8) | PxGV ORF72 helicase, e = 7e − 45 |
| 59 | lef-4 | 54484<55890 | 468 | 90 (20.9) | 79 (22.1) | 96 (16) | 78 (25.9) | PxGV ORF78 lef-4, e = 2e − 28 |
| 60 | p49 | 55904>57232 | 442 | 142 (20.5) | 9 (22.1) | 30 (12.4) | 14 (16.9) | HaSNPV ORF 9 p49, e = 1e − 22 |
| 61 | 57244>57480 | 78 | transmembrane domain | |||||
| 62 | odv-e18 | 57503>57760 | 85 | 143 (14.3) | 10 (14.6) | X | 13 (24.4) | XcGV ORF 12 odv-e18, aa id 27.4%d |
| 63 | odv-ec27 | 57775>58563 | 262 | 144 (21.3) | 11 (18.3) | 32 (9.1) | 80 (16.0) | BmNPV ORF120 odv-e27, e = 1e − 05, aa id 21.7% |
| 64 | 58590>58922 | 110 | 145 (24.4) | 12 (23.7) | X | 12 (23.2) | CpGV ORF9, e = 2e − 07, aa id 30.4% | |
| 65 | lef-1 | 58925<59560 | 211 | 14 (19.8) | 124 (29.2) | 45 (28.8) | 55 (29.2) | PhopGV ORF 66 lef-1, e = 9e − 16, aa id 30.7% |
| 66 | 59622>60203 | 193 | 115 (26.3) | 98 (30.4) | 46 (25.3) | 29 (28.0) | SpltNPV ORF107, e = 1e − 12, aa id 25.8% | |
| 67 | 60357>61421 | 354 | 109 (20.0) | 54 (18.0) | 69 (16.1) | 43 (17.7) | MacoNPV A ORF 80, e = 1e − 13, aa id 22.5% | |
| 68 | 61421>61630 | 69 | Signal peptide | |||||
| 69 | 61617>62027 | 136 | RCC1/BLIP-II | |||||
| 70 | 61993>62394 | 133 | RCC1/BLIP-II | |||||
| 71 | 62479>62778 | 99 | RCC1/BLIP-II | |||||
| 72 | 63034>63885 | 283 | ||||||
| 73 | 63835<64128 | 97 | Transmembrane domain | |||||
| 74 | 64127>64924 | 265 | ||||||
| 75 | 65298>65927 | 209 | ||||||
| 76 | pif | 65909<67501 | 530 | 119 (26.9) | 111 (28.5) | 29 (28.2) | 7 (28.1) | AcMNPV ORF119 pif, e = 2e − 59 |
| Direct repeat 4 | 67561-67606 | 3 46-bp direct repeats | ||||||
| 67652-67697 | ||||||||
| 67742-67787 | ||||||||
| 77 | 67897<68346 | 149 | 53 (13.6) | 43 (18.2) | X | 112 (11.6) | CpGV ORF 134, e = 2.4 aa id 15.7%c | |
| 78 | lef-8 | 68351>70882 | 843 | 50 (29.7) | 38 (30.6) | 26 (19.7) | 109 (32.1) | PhopGV ORF 121 lef-8, e = e − 113, aa id 33.9% |
| Direct repeat 5 | 71364-71562 | 3 67-bp direct tandem repeats | ||||||
| Direct repeat 6 | 72017-72105 | 2 45-bp direct tandem repeats | ||||||
| 79 | 72085<72279 | 64 | ||||||
| 80 | 72547>72783 | 78 | 2 transmembrane domains | |||||
| 81 | 72879>73325 | 148 | Casphalia extranea densovirus NP_694840, e = 1e − 11, aa id 30.2% | |||||
| 82 | vp91 capsid | 73334>75745 | 803 | 83 (22.8) | 76 (22.9) | 35 (21.6) | 84 (23.4) | HaSNPV ORF77 vp91 capsid, e = 3e − 56 |
| 83 | vp1054 | 75747<76688 | 313 | 54 (20.4) | 47 (17.5) | 8 (15.0) | 115 (15.4) | LdMNPV ORF57 vp1054, e = 2e − 09, aa id 19.1% |
| 84 | 76813>77022 | 69 | ||||||
| 85 | 77030>77947 | 305 | ||||||
| Direct repeat 7 | 77914-77943 | 2 30-bp repeats, one within nl85 | ||||||
| 78383-78412 | ||||||||
| Direct repeat 8 | 78002-78161 | 2 160-bp direct tandem repeats overlapping nl86 | ||||||
| 78162-78321 | ||||||||
| 86 | 78101<78361 | 86 | Signal peptide, two transmembrane domains | |||||
| 87 | 78468>79898 | 476 | Internal repeat | |||||
| Direct repeat 9 | 79967-80009 | 2 43-bp direct repeats | ||||||
| 80056-80098 | ||||||||
| 88 | vp39 capsid | 80133<81080 | 315 | 89 (18.7) | 78 (17.7) | 24 (11.0) | 79 (14.2) | SpltNPV ORF81 p39, e = 7e − 20, aa id 20.5% |
| 89 | 81089>81640 | 183 | Phosphotransferase | |||||
| Avg (23.2) | Avg (24.7) | Avg (19.7) | Avg (23.6) |
% Amino acid identity (aa id) calculated for complete ORFs using ClustalW.
Top NCBI blastp 2.2.5 or SMART WU-BLAST 2.0 match with e-value and % aa id. If top % aa id is listed in columns 5 to 8, %, it is not repeated. In some instances top blastp match and ORF with highest % aa id are not the same; if no baculovirus homologue was clearly identified, domains and features as identified with SMART or NCBI blast conserved domain database are given.
Accepted based on NeleNPV results.
Accepted based on aa id of >20%.
ORFS in bold are considered clearly identified using criteria given in Materials and Methods. < and > indicate direction of transcription. e values are given as follows: e = 8e − 46, e = 8 × 10−46.
Repeat regions.
Nine direct repeat regions were found in NeleNPV, but they showed little similarity to typical NPV homologous regions (hr's). NPV hr's are characterized by the presence of multiple tandemly repeated perfect or imperfect palindrome sequences within a direct repeat and have been implicated as origins of DNA replication (39) and as enhancers of transcription (26). Although NPV hr's typically show similarity to each other, differences have also been noted in CuniNPV (2), HaSNPV (13), and Spodoptera litura MNPV (SpltMNPV) (50).
NeleNPV direct repeat regions were up to 77% AT rich and contained two or three copies of direct repeat sequences, ranging in size from 29 to 160 bp, located either in tandem or separated by 30 to 439 bp (Fig. 2). Direct repeats within each region were not the same as those in other regions, with the closest match being 55.2% nucleotide identity between those in regions 3 and 5. Part of direct repeat 4 (31 bp), however, was repeated four times in nl2 and three times in nl35. Repeat 1, located between ORFs 7 and 8, contained two 43-bp direct repeats (97.7% nucleotide identity) separated by 28 bp. Repeat 2, between ORFs 25 and 26, contained two 53-bp direct tandem repeats (100% nucleotide identity). Repeat 3, between ORFs 38 and 39, had two 29-bp direct tandem repeats (93.1% nucleotide identity), and repeat 4, between ORFs 76 and 77, contained three 46-bp direct repeats separated by 45 bp (84.8 to 80.0% nucleotide identity). Repeat 5 and 6 were both between ORFs 78 and 79, with repeat 5 having three 67-bp direct tandem repeats (88.1 to 90.8% nucleotide identity) and repeat 6 having two 45-bp direct tandem repeats (93.3% nucleotide identity) with one partially overlapping nl79. Repeat 7 had two 30-bp direct repeats (80% nucleotide identity), and repeat 8 had two 160-bp tandem direct repeats (95.6% nucleotide identity), with the left direct repeat of region 7 overlapping nl85 and separated from the right direct repeat by 439 bp. Both tandem repeats in region 8 overlapped nl86. Repeat 9, between ORFs 87 and 88, had two 43-bp direct repeats (100% nucleotide identity) located 46 bp apart (Fig. 2).
FIG. 2.

Alignment of NeleNPV direct repeat sequences. Direct repeat regions are numbered according to their order in the genome. The repeat sequences are aligned to obtain maximum similarity, and their locations within the genome are indicated. Sequences within repeat 4 that are found within nl2 and nl36 are bolded and underlined. Arrows indicate palindromes.
Several ORFs (nl7, nl8, nl35, nl49, nl50, and nl87) contained internal repeats. Interestingly, nl7 and nl8 bordered repeat 1, and nl87 was between repeats 8 and 9. None of the ORFs with internal repeats showed homology to baculovirus repeat ORFs (bro's); however, similarities may exist that were at too low a level to identify. The ORFs containing internal repeats and found near repeat regions may not be functional but may be part of the repeat regions. Ambiguous bases did not appear to be randomly distributed; instead, repeat regions contained a higher number, with 12 of 162 or 7.4% of ambiguous bases being found in the direct repeats, while these regions accounted for only 1.4% of the genome.
Perfect and imperfect palindromes ranging in size from 11 to 40 bp were found in several of the repeat regions but were not necessarily embedded within the direct repeats. Those found within the direct repeats are shown in Fig. 2. The palindromes showed limited similarity to each other and no similarity to palindromes in other baculovirus hr's.
Repeat regions in GVs show more variability than those in NPVs, and most do not have a palindromic core (69). XcGV repeat regions are AT rich and contain direct imperfect repeats that are highly variable between each region and within each region (30), similar to those in NeleNPV. Little sequence similarity, however, exists between the NeleNPV and XcGV repeat regions. Typical NPV hr sequences are also not found in Cydia pomonella GV (CpGV) (46); instead, one major repeat region and 13 copies of a single repeated imperfect palindrome are found, with six of the repeats found within potential ORFs (46). Similarly, NeleNPV has a longer repeat region (repeat 8), and some of the NeleNPV direct repeats and palindromes overlap or are within ORFs, but none exhibit sequence similarity to CpGV repeats.
It would appear that NeleNPV repeat regions show little sequence similarity to baculovirus hr's but that they share a closer similarity to the organization of repeat regions in GVs than to those in NPVs. The lack of typical baculovirus hr's in NeleNPV may reflect a different genome replication strategy for the virus or its replication in a nonlepidopteran host.
Conserved ORFs.
The number of conserved core baculovirus genes has decreased from 30 (2, 33) to 29, since an F protein homologue (ac23/ld130) was not found in NeleNPV. Of the 62 conserved lepidopteran baculovirus genes (32), only 41 were clearly identified in NeleNPV. Three of these, nl14 (ac25/dna binding protein [dbp]), nl29 (ac101/p40), and nl77 (ac53) met our identification criteria using earlier blastp versions but did not with blastp 2.2.5. They have been accepted, however, since they were strong homologues of ORFs found independently in NeseNPV that have been identified as baculovirus homologues (24). NeleNPV contained 12 genes conserved in the lepidopteran baculoviruses but not found in the dipteran virus CuniNPV, including homologues to ac8 (polyhedrin), ac25 (dbp), ac37 (lef-11), ac53, ac76, ac78, ac93, ac101 (p40), ac103 (p45), ac106, ac143 (odv-e18), and ac145. Conserved lepidopteran baculovirus genes not clearly identified in NeleNPV include homologues to ac10 (protein kinase 1 [pk1]), ac13, ac23, ac28 (lef-6), ac29, ac32 (fibroblast growth factor [fgf]), ac35 (ubiquitin), ac36 (pp31), ac38, ac46 (odv-e66), ac61 (few polyhedrin 25K), ac66, ac67 (lef-3), ac75, ac82, ac94 (odv-e25), ac102, ac110, ac139 (me35), ac146, and ac147 (immediate early gene-1 [ie-1]) (Table 3).
TABLE 3.
Conserved genes in baculovirus genomesd
| Gene function | Genes present in all baculovirusesa | Genes present in all lepidopteran baculoviruses and NeleNPV | Genes present in all lepidopteran baculoviruses but not in NeleNPV |
|---|---|---|---|
| Replication | lef-2 (ac6), lef-1 (ac14), dnapol (ac65), helicase (ac95) | dbp1 (ac25) | lef-3 (ac67), me53 (ac139), ie-1 (ac147) |
| Transcription | p47 (ac40), lef-8 (ac50), lef-9 (ac62), vlf-1 (ac77), lef-4 (ac90), lef-5 (ac99) | lef-11 (ac37) | lef-6 (ac28), pp31/39K (ac36) |
| Structural proteins | vp1054 (ac54), gp41 (ac80), vp91/p95 (ac83), vp39 (ac89), p6.9 (ac100), p74 (ac138), odv-e27 (ac144), odv-e56 (ac148), | polh (ac8), odv-e18 (ac143) | ac23/ld130b, odv-e66 (ac46), fp25K (ac61), odv-e25 (ac94), pk1 (ac10) |
| Per os infectivity factorsc | pif (ac119) | ||
| pif-2(ac22) | |||
| Auxiliary | alk-exo (ac133) | fgf (ac32), ubiquitin (ac35) | |
| Unknown | ac68, ac81, ac92, ac96, 38K (ac98), ac109, ac115, ac142 | ac53, ac76, ac78, ac93, p40 (ac101), ac106, p45 (ac103), ac145 | 38.7K (ac13), ac29, ac38, ac66, ac75, ac82, p12 (ac102), ac110, ac146, |
Based on the genomes of 22 completely sequenced baculoviruses as listed in the text.
CuniNPV, a dipteran baculovirus, also has a potential ld130 homologue.
P74 is listed as a structural protein but is also necessary for per os infectivity.
Replication genes.
All the previously considered conserved baculovirus replication genes, lef-2 (nl54/ac6), lef-1 (nl65/ac14), DNA-polymerase (nl20/ac65), and helicase (nl58/ac95), were found in NeleNPV. The conserved lepidopteran baculovirus replication gene dbp (nl14/ac25) was present, but lef-3 (ac67), me35 (ac139), and ie-1 (ac147) were absent. nl25 showed a very low blastp match to lef-3 but was not close enough to meet our criteria. NeleNPV was also missing several variable DNA replication genes or those involved in replication in some but not all baculoviruses, including proliferating cell nuclear antigen (ac49), lef-7 (ac125), p35 (ac135) (45), ie-2 (ac151), and pe38 (ac153) (Table 4), as well as helicase-2, dna-ligase, RNase reductase-1 (op32), RNase reductase-2 (op34), and dutpase (op31) (31).
TABLE 4.
Variable lepidopteran baculovirus genes found in AcMNPV but not in NeleNPV
| Gene function | Genes in AcMNPV but not NeleNPVa,b |
|---|---|
| Replication | pcna (ac49), lef-7 (ac125), p35 (ac135), ie-2 (ac151), pe38 (ac153) |
| Transcription | 39k (ac36), lef-12 (ac41), lef-10 (ac53a) |
| Structural | ptp (ac1), orf1629 (ac9), p80/p87 capsid (ac104), gp67 (ac128), p24 (ac129), pp34-calyx (ac131), p10 (ac137) |
| Auxiliary | conotoxin (ac3), egt (ac15), iap-1 (ac27), sod (ac31), chitinase (ac126), vcath (ac127) |
| Unknown | ac4, ac5, ac7, ac11, ac12, ac16, ac17, ac18, ac19, ac26, ac30, ac33, ac34, ac39, ac43, ac44, ac45, ac51, ac52, ac55, ac56, ac57, ac58, ac59, ac60, ac63, ac70, ac72, ac73, ac74, ac84, ac85, ac87, ac88, ac91, ac97, ac106, ac107, ac108, ac111, ac112, ac113, ac114, ac116, ac117, ac118, ac120, ac121, ac122, ac124, ac125, ac140, ac149, ac150, ac152, ac154 |
| bro (ac2), arif-1 (ac20/21), pkip-1 (ac24), gta (ac42), ets (ac47), etm (ac48), gp37 (ac64), met (ac69), iap-2 (ac71), pnk/pnl (ac86), he65 (ac105), pk2 (ac123), gp16 (ac130), p25 (ac132), p94 (ac134), p26 (ac136), exon0/ie0 (ac141) |
Variable genes are those found in some but not all baculovirus genomes.
Some missing genes may be present but due to low homology were not identified.
Transcription-specific genes.
All conserved transcription-specific genes, lef-5 (nl55/ac99), lef-4 (nl59/ac90), lef-8 (nl78/ac50), lef-9 (nl37/ac62), very late factor-1 (nl42/ac77), and p47 (nl46/ac40) were found. Lef-11 (nl15/ac37) was present but is no longer considered a conserved baculovirus gene, since it is absent from CuniNPV. Lef-6 (ac28) and pp31/39k (ac36) were absent, as in CuniNPV. Variable transcription-specific genes, including ac36, lef-12 (ac41), and lef-10 (ac53a), were not found (Table 4).
Structural protein genes.
Conserved baculovirus structural genes identified include odv-e56 (nl23/ac148), p6.9 (nl28/ac100), gp41 (nl44/ac80), p74 (nl47/ac138), odv-ec27 (nl63/ac144), vp91/p95 capsid (nl82/ac83), vp1054 capsid-associated protein (nl83/ac54), and vp39 capsid (nl88/ac89). Polyhedrin and odv-e18, present in all sequenced lepidopteran baculoviruses but not in CuniNPV, were also found (Table 3).
Neither a GP64/67 homologue nor an F protein homologue (ac23/ld130) was found in NeleNPV. Membrane fusion proteins mediate the fusion of budded virus to cell membranes and the release of nucleocapsids (51). Previously, GP64/67 homologues have been found in all group I NPVs, and F protein homologues (ac23/ld130) have been found in group II NPVs (56). Ld130 homologues are thought to be the primordial baculovirus envelope fusion protein (51). When blastp searches failed to identify a GP64/67 or F protein (ac23/ld130) homologue in NeleNPV, all unidentified ORFs were examined more thoroughly as listed in Materials and Methods.
F proteins are acid pH dependent and contain a predicted signal peptide at the amino terminus, a transmembrane domain near the carboxyl terminus, up to 11 conserved cysteines, and a furin cleavage site (40, 51, 56). nl38 (141 aa) and nl86 (86 aa) contained signal peptides and transmembrane domains but were much shorter than ld130 (676 aa) and lacked the conserved cysteines. Nl38 met our identification criteria as a homologue to SpltMNPV ORF 66, further suggesting that it is not an F protein homologue. nl18, nl62, and nl68 showed signal peptides with possible overlapping transmembrane domains, but due to large size differences, the conserved cysteines were difficult to assess. Nl87 contained five potential conserved cysteines but did not have a signal peptide or transmembrane domain. Although ld130 homologues generally show low conservation (56), the fact that we were unable to find matches with either BLAST searches, Smith Waterman searches, or searches for conserved features suggests that NeleNPV may be the first NPV found without an envelope fusion protein. Similar results have recently been found with another hymenopteran baculovirus, NeseNPV (24).
Analysis of AcMNPV exon0 (ac141) has shown that exon0 is expressed as a late gene and all early transcripts from this gene region are spliced to form ie-0. exon0 knockouts produced with AcMNPV BACmids have shown that exon0 is essential for budded virus production (X. Dai and D. Theilmann, personal communication). The NeleNPV genome lacks exon0 and ie-0 homologues. The absence of both ld130 and exon0 homologues in NeleNPV suggests that the budded virus phenotype may not play a role in the biology of hymenopteran NPVs or that other unidentified proteins may be involved in budded virus production. It is also possible, however, that these homologues are present but their similarity was too low for clear identification.
Questions have been raised on how sawfly viruses might spread from cell to cell, whether they would produce a budded virus or have a GP64 homologue and, if not, what other mechanism they might use for cell-to-cell transmission (22). The lack of an insect colony or a cell culture for NeleNPV has made the presence or absence of budded virus in vivo difficult to determine.
Conserved lepidopteran structural genes pk-1 (ac10) (31, 33) and odv-e66 (ac46) were missing. Genes for two potential proteins, weakly matching protein kinases, were seen. Nl9 showed a low SMART match to a phosphotransferase with possible dual specificity as a Ser/Thr/Tyr kinase (STYKc domain; SMART accession number SM0221). Smith Waterman searches with nl9 showed level 2 evidence for a cAMP- and cGMP-dependent protein kinase phosphorylation site (CAMP PHOSPHO site) (PROSITE: PDOC00004) or a tyrosine kinase phosphorylation site (PROSITE: PDOC00007). By using a search for short nearly exact matches, nl25 showed a low blastp match to a possible vaccinia virus protein kinase (AAA48288; e = 0.50), and Smith Waterman searches showed level 2 evidence for a CAMP PHOSPHO site. Compared to ac10 (pk1), however, nl9 and nl25 showed very low amino acid identity and neither was accepted as a pk1 homologue. The presence of such a homologue, however, cannot be discounted at the present time.
Variable structural genes few polyhedra 25K (ac61) and odv-e25 (ac94), as well as protein tyrosine phosphatase (ptp) (ac1), orf1629 (ac9), p80/p87 capsid (ac104), gp64/67 (ac128), p24 (ac129), pp34 calyx (ac131), p10 (ac137), and enhancin (31) were not seen (Table 4).
P10 (ac137), considered either a structural protein (31) or an auxiliary protein (33, 49), was not clearly identified in NeleNPV. P10 is a very late protein that is generally poorly conserved at the amino acid level, but its size, hydrophilicity distribution, and secondary structure are conserved (72). Four structural domains implicated in various functions are usually found, including a coiled-coil domain at the amino-terminal end, followed by a proline-rich domain, a variable region not found in all P10s, and a positively charged carboxy terminus often containing serine or threonine residues (63, 64, 65, 68). Known P10 proteins range in size from 70 aa for Bombyx mori NPV (BmNPV) (25) to 105 aa for SpltMNPV (50) and are usually located between p26 and p74 (65). Nl53 showed a weak blastp match with P10 from Buzura suppressaria NPV (BusuNPV) (34), showing 47% identity over 26 of 54 amino acids, but amino acid identity over the entire ORF was much lower. Nl53 had two potential start sites, giving a size of either 104 or 80 amino acids, both within the expected P10 size range. A putative late baculovirus transcriptional start site, TAAG (12), was found at position −85 to −82 with respect to the translational start codon of the smaller transcript and −10 to −7 of the putative larger transcript. The conserved A at −3 (65) was seen only with the larger transcript. Nl53 was located near the p74 homologue (nl47) and a weak match to a potential RNA binding protein (nl48) that could be related to p26. SMART analysis did not indicate a coiled-coil domain, but by using MacVector's combined Chou-Fasman and Robson-Garnier method for determining secondary structure, nl53 was determined to have two areas of helix-forming residues. Only one proline was found in the expected proline-rich domain. Hydrophilicity profiles (Kyte-Doolittle scale) showed that nl53 had a positively charged carboxy terminus and had a serine in this area, as expected for P10 proteins. With ClustalW, nl53 showed the highest amino acid identity to the Epiphyas postvittana MNPV (36) P10, but this was only 13.8%. Amino acid identity, however, is usually low for P10 proteins, and AcMNPV P10, for example, shares only 20.1% amino acid identity with SpltMNPV P10 (65). We cannot, therefore, rule out the possibility that nl53 may be a P10 homologue.
Per os infectivity factors.
To date, the products of three genes have been identified as essential for per os infectivity of baculoviruses. These include structural protein P74 (ac138) (20), PIF (ac119) (38), and PIF-2 (ac22) (52), all of which have been identified in NeleNPV, as nl47, nl76, and nl52, respectively (Table 3). The presence of genes involved in per os infectivity in an ancient virus such as NeleNPV strengthens the idea of these genes being essential (20, 38, 52) and that per os infectivity is a conserved process in baculoviruses. While early baculoviruses may not have needed budded viruses, they could not have survived without an efficient means of insect-to-insect transmission.
Auxiliary genes.
Analysis of the NeleNPV genome has supported the hypothesis that small baculovirus genomes would carry few auxiliary genes (49), since most baculovirus genes in the auxiliary class (2, 22, 31, 33) were not found in NeleNPV (Table 4). Only alkaline exonuclease (alk-exo) remained as a conserved baculovirus auxiliary gene (nl33/ac133). Conserved lepidopteran auxiliary genes, fgf (ac32) and ubiquitin (ac35), were not found (Table 3).
Inhibitors of apoptosis proteins.
NeleNPV contained one iap-like gene (nl11) but its top eight blastp matches were to insect iap genes, including those from Spodoptera frugiperda (GenBank accession number AAF35285) (e = 3 × 10−33), Trichoplusia ni (accession number AAF19819) (e = 4 × 10−32), Anopheles gambiae (accession number EAA04007) (e = 5 × 10−32), and B. mori (accession number AAK5760) (e = 10−31). Top blastp insect virus matches included BusuNPV iap-1 (e = 10−28), Amsacta moorei entomopoxvirus (EPV) iap (AmEPV) (8) ORF 21 (e = 5 × 10−6), and MacoNPV A iap-3 (e = 2 × 10−24). BusuNPV iap-1 is designated by its order in the viral genome and not by homology and is actually an iap-3 (op35) homologue (34). ClustalW alignments with nl11 showed the highest amino acid identity to an iap from A. gambiae (accession number EAA04007) (32.2%), with the closest baculovirus match being CpGV ORF 17 (iap-3) and LdMNPV ORF 139 (iap-3) at 28.7% amino acid identity.
Nl11 contained two baculovirus inhibitor of apoptosis protein repeats (BIRs), an internal repeat between the BIRs and a transmembrane domain at the C termini, but did not have a RING finger. It is not yet known if nl11 is a functional iap gene. While BIRs are critical for iap activity (11, 16, 17) and may be sufficient for antiapoptotic activity in mammalian cells (57), the RING finger may also play an important role in inhibition of apoptosis, especially in baculoviruses (47).
It has been suggested that viral iap genes might have originated from their hosts, since S. frugiperda iap genes share sequence and functional similarity with their baculovirus counterparts (35). This hypothesis is supported by the fact that nl11 shows a closer identity to insect iap genes than to baculovirus iap genes.
Conserved baculovirus ORFs of unknown function.
Conserved baculovirus ORFs of unknown function found in NeleNPV included homologues to ac68, ac81, ac92, ac96, ac98 (38K), ac109, ac115, and ac142. Homologues of conserved lepidopteran baculovirus ORFs of unknown function, including ac53, ac76, ac78, ac93, ac101 (p40), ac103 (p45), ac106, and ac145, were also found in NeleNPV, but homologues to ac13 (38.7K), ac29, ac38, ac66, ac75, ac82, ac102 (p12), ac110, and ac146 could not be identified by our criteria (Table 3).
Other clearly identified ORFs.
Blastp searches showed that nl50 was similar to a protein found in NeseNPV (accession number AAF24987) (e = 3 ×10−29; amino acid identity, 45.7%). No baculovirus matches were found for these ORFs, suggesting that hymenopteran baculoviruses may contain ORFs unique to that group.
Six ORFs were accepted as clearly identified based on the presence of conserved domains but did not show baculovirus matches. Nl6 had a conserved trypsin-like serine protease domain identified with the NCBI conserved domain search (e = 2 × 10−61) and with SMART (e = 8.4 × 10−83). Trypsin-like serine proteases are enzymes that exploit serine in their catalytic site, and they include a wide range of peptidase activities, such as exopeptidase, endopeptidase, oligopeptidase and omega-peptidase activities (SMART accession number SM0020). The top blastp match for nl6 was a Ctenocephalides felis (cat flea) trypsin-like serine protease (GenBank accession number AAD21829) (e = 8 × 10−46), and their amino acid identity (38.7%) was much higher than the average amino acid identity of NeleNPV ORFs to NPV homologues. Other top matches were to trypsin-like serine proteases from B. mori (accession number VDP_BOMMO) (e = 4 × 10−45) and Drosophila melanogaster (GenBank accession number NP_523518) (e = 10−43).
Nl49 contained four clearly identified C2H2 zinc finger domains. Zinc finger domains are nucleic acid binding protein structures composed of 25 to 30 amino acid residues in a C-X2-C-X12-H-X3-H-type motif in which zinc binds in a tetrahedral array to yield a finger-like projection that interacts with nucleotides in the major groove of the nucleic acid (pfam00096). No insect virus matches were found for nl49; instead, the highest blastp match was to a zinc finger protein from D. melanogaster (GenBank accession number NP_609448) (e = 10−13; amino acid identity, 17.4%).
BLAST analysis showed that nl69, nl70, and nl71 contained a conserved α-tubulin suppressor domain (ATSI) and related regulator of chromosome condensation factor (RCC1). SMART analysis also showed the presence of RCC1/β-lactamase inhibitor protein II domains. There were no significant blastp matches to insect viruses, but all three ORFs showed significant matches to proteins in A. gambiae (nl69 [accession number EAA06079; e = 10−20; amino acid identity, 36.5%], nl70 [accession number EAA06079; e = 4 × 10−11; amino acid identity, 27.6%] and nl71 [accession number A04764; e = 2 × 10−4; amino acid identity, 23.0%]). Nl70 also showed a match to chromatin-binding protein BJ1 in D. melanogaster (accession number S15028; e = 10−10; amino acid identity, 23.9%), a homologue of the vertebrate RCC1 gene.
Nl89 appeared to be a phosphotransferase, since BLAST searches showed two potential phosphotransferase domains, DUF60 (pfam01885) and KptA (COG1859), and SMART analysis also showed a DUF60 conserved domain. Two members of the DUF60 family of proteins have been annotated as phosphotransferases, although this has not been supported experimentally, and KptA is a probable RNA 2′-phosphotransferase.
Nl81 showed significant blastp matches to proteins from densoviruses, including viral protein 1-4 from Casphalia extranea densovirus (accession number NP_694840; e = 10−11; amino acid identity, 30.2%), a structural protein from Periplaneta fuliginosa densovirus (accession number BAA82965; e = 5 × 10−11; amino acid identity, 24.2%), and a capsid protein from Bombyx mori densovirus (accession number NP_694837; e = 10−9; amino acid identity, 24.8%). Neither nl81 nor its potential densovirus homologues showed blastp matches to any other baculoviruses. A potential densovirus homologue has recently been reported for CrleGV (42), but Crle9 showed little similarity to nl81. Densoviruses replicate in the nucleus, and it is conceivable that a horizontal gene transfer to NeleNPV could have taken place via recombination or transposition. It is not known if the potential densovirus homologue encodes a functional protein in NeleNPV.
Other possible homologues.
Sequence analysis provided clues to other possible homologues or functions, but results were often inconclusive, and 39 ORFs remained unidentified (Fig. 1; Table 2). Potential features, such as transmembrane domains, signal peptides, coiled-coil domains, and internal repeats, are listed in Table 2.
Several unattributed ORFs showed acceptable matches to ORFs in EPVs by using earlier blastp analysis but did not with blastp 2.2.5. Others had acceptable E values with blastp 2.2.5 for short, nearly exact matches, had amino acid identities greater than 20%, or had protein domains similar to those of potential homologues. Nl68, for example, had a blastp E value of 0.008 with Melanoplus sanguinipes EPV ORF 186, a member of the leucine-rich-repeat family (1), when blastp was used for short, nearly exact matches; nl80 had an E value of 2.2 with AmEPV ORF 208, but their sizes were similar and both contained transmembrane domains; nl84 had an E value of 0.036 with AmEPV ORF 051 with use of blastp for short, nearly exact matches; nl86 had a blastp E value of 0.55 with AmEPV ORF 196, and both contained transmembrane domains, were similar in size, and shared 23.0% amino acid identity. Smith Waterman searches showed that nl21 was a potential homologue of MSV156, with an E value of 0.000004.
NeleNPV DNA had a high A+T content (66.5%), and the A+T content of ORFs with potential EPV matches was even higher, ranging from 68.0 to 79.7%. The possible homology of NeleNPV ORFs with those from EPVs may be due to random similarities, since MsEPV and AmEPV also have a very high A+T content (1, 8). NeleNPV ORFs with low identity to EPV ORFs, however, were clustered in two areas. NeleNPV is not the only baculovirus that may contain ORFs similar to those found in EPVs. Spindlin or gp37 (ac64), a gene rich in A+T residues and found in many NPVs, is a homologue of fusolin from EPVs (6, 70).
Gene content.
It has been suggested from the analysis of up to 13 baculovirus genomes that 27 genes specific to GVs and 14 genes specific to lepidopteran NPVs might distinguish these two groups (32, 33). None of the unique NPV or GV genes was clearly identified in NeleNPV, making it distinct from either group. Low potential matches to the NPV-specific genes pkip (ac24) and p26 (ac136) were possible for NeleNPV, as was a low potential match with a metalloproteinase gene unique to GVs, but they were not considered clearly identified. An iap gene was found but it appeared closer to iap genes present in insects and to iap-3 in baculoviruses than to iap-5, found uniquely in GVs and iap-2 in NPVs. A list of 20 genes distinguishes group I from group II NPVs, 17 of which are found in group I (32, 33). NeleNPV did not appear to contain any of the group I- or II-specific NPV genes, which again suggests that it may not be a typical NPV.
It is possible that NeleNPV may contain other baculovirus genes not yet identified or that NeleNPV may not require the missing genes due to differences in host-virus interactions. The lepidopteran conserved genes and some auxiliary genes may have been acquired when baculoviruses evolved to infect tissues beyond the midgut and may not be necessary in hymenopterans. Cathepsin and chitinase, for example, are needed to break down the host cuticle after death, liquefy the cadaver, and release occlusion bodies (29, 60). Neither gene was found in NeleNPV, and they may not be necessary, since the virus is spread via sloughed-off intestinal cells, and infectious diarrhea and infected insects do not liquefy (22). Another gene missing in NeleNPV but conserved in all lepidopteran baculoviruses is fgf, a homologue of an insect fgf gene involved in tracheal development (61). Such an FGF protein might be involved in spread of NPVs through the trachea but would not be needed by NeleNPV, whose replication is restricted to the midgut. It is also possible that some unique hymenopteran baculovirus genes are functionally equivalent but do not show close sequence identity. No ie-1 homologue was found in NeleNPV, for example, but perhaps one or more other unidentified genes may be involved in the transactivation of early and late genes as a substitute to ie-1, or NeleNPV may depend on host factors for early transcription, as suggested for CuniNPV (2).
Factors affecting genome size.
NeleNPV is the smallest baculovirus genome so far reported. Despite its small size, however, there were still intergenic areas in the genome in which ORFs or repeat regions were not found. The lack of many baculovirus ORFs is an important factor contributing to the small size of NeleNPV, but another is the lack of repeated genes that account for the large size of many baculovirus genomes. The XcGV genome, for example, contains 30 repeated genes, accounting for 37.5 kb or 20% of the genome, and LdMNPV contains 32, accounting for 27.5 kb or 17% of the genome (30, 40). The type of repeated gene varies but may include bro genes, among others. NeleNPV had no clearly identified bro genes and contained no repeated baculovirus ORFs.
Size differences can also be attributed to numerous insertions or deletions in hr's. The Helicoverpa zea SNPV genome, for example, is larger than that of HaSNPV due mainly to insertions in hr's (14). SpltNPV contains the largest number of hr's with 17, each containing 2 to 29 palindromic repeats with an average length of 534 bp (50), greatly adding to its size. NeleNPV lacked typical NPV hr's, and its repeat regions were short, containing a maximum of three copies of direct repeat units, also contributing to its small size.
Comparison of NeleNPV with other baculoviruses.
Polyhedrin is generally considered one of the most highly conserved of baculovirus proteins (55), but nl1 (polyhedrin) shared only 35.5 to 49.2% amino acid identity with lepidopteran baculovirus polyhedrins versus 98% with Neodiprion abietis NPV polyhedrin and 82.1% with NeseNPV polyhedrin. Overall, polyhedrin was the most conserved baculovirus gene in NeleNPV, followed by pif-2 (nl52/ac22), DNA polymerase (nl20/ac65), and p6.9 (nl28/ac100).
The average overall amino acid identity of clearly identified NeleNPV homologues with other baculoviruses ranged from 19.7% with CuniNPV (29 common ORFs) to 24.9% with Spodoptera exigua MNPV (SeMNPV) (37) (42 common ORFs). The closest GV was PxGV, with 42 common ORFs averaging 23.6% amino acid identity, followed by CrleGV at 23.2% (Tables 1 and 2).
Although the majority of ORFS were closer to those in NPVs, many, including nl23 (odv-e56), nl28 (p6.9), nl29 (p40), nl33 (alk-exo), nl37 (lef-9), nl46 (p47), nl58 (helicase), nl59 (lef-4), nl62 (odv-e18), nl64, nl65 (lef-1), nl77, and nl78 (lef-8), showed a closer blastp match to GV homologues. The highest amino acid identity difference for a NeleNPV ORF compared to an NPV and GV homologue was seen with p6.9, with nl28 showing 30.4% amino acid identity with p6.9 in AcMNPV (ac100) and 64.3% amino acid identity with p6.9 in AdorGV (Ador72).
If NeleNPV ORFs were recently acquired from GVs, one would expect them to be clustered in both genomes and their amino acid identities to be close. It is unlikely that recent recombination events have occurred between NeleNPV and GVs, since NeleNPV homologues with higher identity to GV ORFs were spread throughout the genome, and in most cases there was little difference in amino acid identities between NPV and GV homologues. This may mean that NeleNPV is indeed an older baculovirus that existed before the GV/NPV spilt and thus shows similarity to both types of virus.
Gene parity plots.
Gene parity plots are used to display the gene order of genomes from any two viruses. Closely related baculoviruses generally show colinear arrangements of genes, and colinearity decreases with increased divergence between baculoviruses (34). Gene parity plots showed that gene order was not highly conserved between NeleNPV and a representative type I NPV (AcMNPV), type II NPV (HaSNPV), a GV (PxGV), or CuniNPV, except for the central portion of the genome containing lef-5 (ac99), 38k (ac98), ac96, and helicase (ac95), as has been reported with other baculoviruses (13) (Fig. 3). This region may be highly conserved as a result of transcriptional or regulatory constraints and may have been maintained over long evolutionary periods (13, 33). Three other clusters in the central region appeared to be conserved between NeleNPV and the lepidopteran NPVs, with some containing a single insertion or deletion, including p6.9 (ac100), p40 (ac101), and p48 (ac103) in one grouping, ac76, vlf (ac77), ac78, and gp41 (ac80) in a second, and p49 (ac142), odve-18 (ac143), odv-ec27 (ac144), and ac145 in a third (Fig. 3).
FIG. 3.
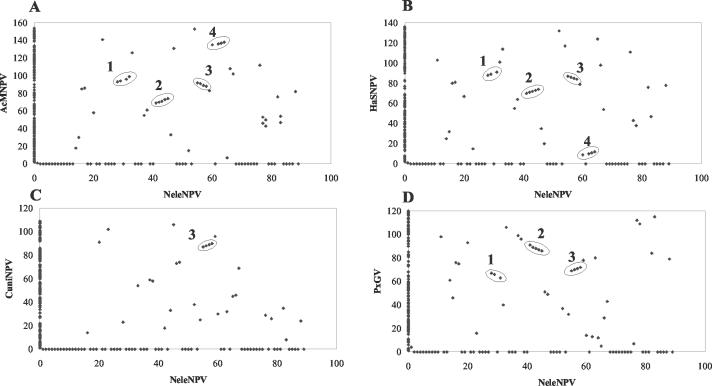
Gene parity plots. Comparison of NeleNPV with AcMNPV (A), HaSNPV (B), CuniNPV (C), and PxGV (D). AcMNPV ORFs are renumbered, with polyhedrin as ORF 1. The CuniNPV gene order is as listed in GenBank and not by ORF number for lef-5, 38k, ac96, and helicase. The graphic representations display the colinearity of gene arrangement between two genomes. ORFs unique to each virus are shown on the x axis and y axis, respectively. Most conserved clusters are circled: cluster 1, p6.9 (ac100), p40 (ac101), and p48 (ac103); cluster 2, ac76, vlf-1 (ac77), ac78, and gp41 (ac80); cluster 3, lef-5 (ac99), 38k (ac98), ac96, and helicase (ac95); cluster 4, p49 (ac142), odv-ec27 (ac143), odv-ec27 (ac144), and ac145.
Phylogeny of NeleNPV.
Baculoviruses may have had a common evolutionary origin in an ancestral virus that attacked the midgut or hepatopancreas of ancient arthropods and coevolved with their hosts, eventually spreading to Lepidoptera (22). Midgut cell sloughing may have been an important host-mediated selection pressure influencing NPV evolution, with virion production in the midgut epithelium being selected against due to its devastating effects on gregarious host populations. The success of lepidopteran baculoviruses might be due to their selection for ability to invade internal tissues so that a larger inoculum of virus could be produced (21, 22, 67).
NeleNPV replicates only in the midgut and so may represent an early stage in baculovirus evolution. A GV isolated from the Western grapeleaf skeletonizer Harrisina brillians (HbGV), a member of a primitive lepidopteran family (Zygaedidae), is the only GV restricted to midgut cells and may similarly represent an early GV. H. brillians larvae also lead a gregarious lifestyle, feed in groups, and transmit virus via infectious diarrhea and sloughed-off midgut epithelium cells (21). It would be interesting to compare the gene content and phylogeny of HbGV with NeleNPV, but only a few HbGV genes have been sequenced.
Phylogenies based on the combined sequence of shared genes have been found to be more robust than those based on the sequences of individual genes (32, 33). The most parsimonious tree produced by using the combined data set of 29 conserved baculovirus proteins from 24 baculovirus genomes placed NeleNPV and CuniNPV as separate branches that existed before the split of the lepidopteran GVs and NPVs (Fig. 4). This suggests that both NeleNPV and CuniNPV do not fit in the current NPV designation of group I or II NPVs. The separation of NeleNPV and CuniNPV into separate genera was corroborated with trees constructed by using individual genes and with a combined concatemer of 10 genes conserved in all fully sequenced baculoviruses and the Hz-1 virus, which infects Helicoverpa zea (15), although the divisions within group II NPVs varied in some cases (data not shown). It has already been proposed that CuniNPV is a baculovirus with unusual characteristics that may represent a new genus within the family Baculoviridae (2, 33, 48). Our phylogenetic analysis suggests that NeleNPV may also belong to a genus separate from the dipteran and lepidopteran baculoviruses.
FIG. 4.
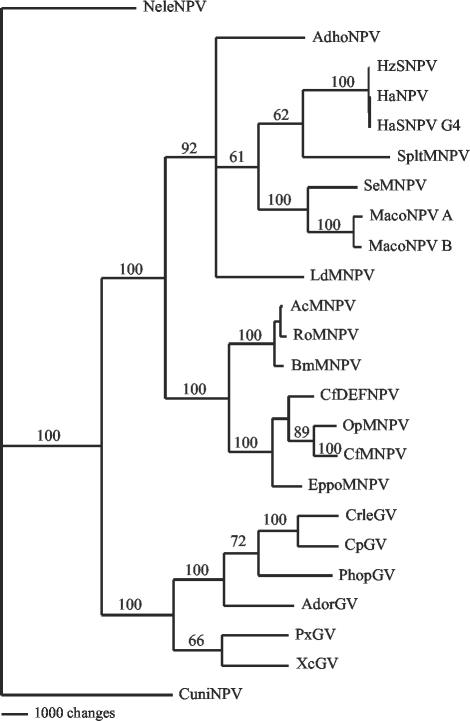
Baculovirus phylogeny based on complete genome data. Most-parsimonious tree based on analysis of the combined sequences of 29 conserved genes found in 24 fully sequenced baculovirus genomes. Bootstrap values for 1,000 replicates are given. Sequences include those for AcMNPV (7), Orgyia pseudotsugata MNPV (OpMNPV) (3), BmNPV (25), LdMNPV (40), SeMNPV (37), XcGV (30), PxGV (28), HaSNPV G4 (13), Helicoverpa armigera NPV (HaNPV) (GenBank accession number NC_003094), SpltMNPV (50), CpGV (46), CuniNPV (2), E. postvittana MNPV (EppoMNPV) (36), H. zea SNPV (HzSNPV) (14), MacoNPV A (44), MacoNPV B (43), PhopGV (GenBank accession number AF499596), RoMNPV (27), AdhoNPV (GenBank accession number AP006270), C. fumiferana defective NPV (CfDEFNPV) (GenBank accession number AY327402), Choristoneura fumiferana MNPV (CfMNPV) (GenBank accession number NC_004778), CrleGV (42), AdorGV (69), and NeleNPV (GenBank accession number AY349019).
There are many factors supporting the idea that NeleNPV represents a new baculovirus genus. Members of the Hymenoptera family are more ancient than Lepidoptera, and NeleNPV was isolated from a primitive hymenopteran. The virus showed low identity to all fully sequenced lepidopteran NPVs and GVs. Many genes conserved in lepidopteran baculoviruses, as well as genes identified as unique to lepidopteran NPVs and GVs, were not found in NeleNPV. Many ORFs showed no baculovirus matches, and some had significant matches to other insect viruses or to insect genes. With the exception of the lef-5-to-helicase region, gene order was not highly conserved in NeleNPV relative to other baculoviruses, and repeat regions lacked sequence homology to hr's found in NPVs or GVs. Phylogenetic trees suggest that NeleNPV, like CuniNPV, does not fit into the present NPV designation but may represent an early baculovirus that existed before the lepidopteran GVs and NPVs diverged from each other.
The present classification criteria encompassing only two genera are clearly insufficient to accommodate the hymenopteran and dipteran baculoviruses. Moreover, it is conceivable that baculoviruses from other insect orders will be discovered that also may not fit into the present genera. Other genera will have to be created and must be flexible enough to accommodate all baculoviruses, even ones yet to be discovered. We suggest adding two new genera to accommodate the hymenopteran and dipteran viruses and changing the names of the Baculoviridae genera to Alphabaculovirus, Betabaculovirus, Gammabaculovirus, and Deltabaculovirus. The proposed genera will accommodate not only the hymenopteran and dipteran baculoviruses but also the present groups of NPVs and GVs. This system has the flexibility for adding more genera in the future and is consistent with the nomenclature of viruses in other families, including the Herpesviridae, Retroviridae, and Entomopoxvirinae.
Acknowledgments
We thank David Theilmann for his help in the search for an ie-1 gene and Theilmann and Xiaojiang Dai for personal communications; Gary Blissard for his help in the search for an F-protein homologue; Elizabeth Herniou for her advice on phylogenetic analysis; and Alejandra Garcia-Maruniak and James Maruniak for sharing the NeseNPV sequence prior to publication. We also thank Kees VanFrankenhuyzen for the stock of NeleNPV and Lillian Pavlik for her excellent work in extracting viral DNA.
The research was supported by grants from Genome Canada, the Canadian Biotechnology Strategy Fund, and the NSERC Biocontrol Network.
REFERENCES
- 1.Afonso, C. L., E. R. Tulman, Z. Lu, E. Oma, G. F. Kutish, and D. L. Rock. 1999. The genome of Melanoplus sanguinipes Entomopoxvirus. J. Virol. 73:533-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Afonso, C. L., E. R. Tulman, Z. Lu, C. A. Balinsky, B. A. Moser, J. J. Becnel, D. L. Rock, and G. F. Kutish. 2001. The genome sequence of a baculovirus pathogenic for Culex nigripalpus. J. Virol. 75:11157-11165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahrens, C. H., R. L. Q. Russell, C. J. Funk, J. T. Evans, S. H. Harwood, and G. F. Rohrmann. 1997. The sequence of the Orgyia pseudotsugata multinucleocapsid nuclear polyhedrosis virus genome. Virology 229:381-399. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 5.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arif, B. M. 1995. Recent advances in the molecular biology of entomopoxviruses. J. Gen. Virol. 76:1-13. [DOI] [PubMed] [Google Scholar]
- 7.Ayres, M. D., S. C. Howard, J. Kuzio, M. Lopez-Ferber, and R. D. Possee. 1994. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology 202:586-605. [DOI] [PubMed] [Google Scholar]
- 8.Bawden, A. L., K. L. Glassberg, J. Diggans, R. Shaw, W. Farmerie, and R. W. Moyer. 2000. Complete genomic sequence of the Amsacta moorei entomopoxvirus: analysis and comparison with other poxviruses. Virology 274:120-139. [DOI] [PubMed] [Google Scholar]
- 9.Benson, G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bird, F. T. 1961. Transmission of some insect viruses with particular reference to ovarial transmission and its importance in the development of epizootics. J. Insect Pathol. 3:352-380. [Google Scholar]
- 11.Birnbaum, M. J., R. J. Clem, and L. K. Miller. 1994. An apoptosis-inhibiting gene from a nuclear polyhedrosis virus encoding a polypeptide with Cys/His sequence motifs. J. Virol. 68:2521-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blissard, G. W., and G. F. Rohrmann. 1990. Baculovirus diversity and molecular biology. Annu. Rev. Entomol. 35:127-155. [DOI] [PubMed] [Google Scholar]
- 13.Chen, X., W. F. J. Ijkel, R. Tarchini, X. Sun, H. Sandbrink, H. Wang, S. Peters, D. Zuidema, R. K. Lankhorst, J. M. Vlak, and Z. Hu. 2001. The sequence of the Helicoverpa armigera single nucleocapsid nucleopolyhedrovirus genome. J. Gen. Virol. 82:241-257. [DOI] [PubMed] [Google Scholar]
- 14.Chen, X., W. J. Zhang, J. Wong, G. Chun, A. Lu, B. F. McCutchen, J. K. Presnail, R. Hermann, M. Dolan, S. Tingey, Z. Hu, and J. M. Vlak. 2002. Comparative analysis of the complete genome sequences of Helicoverpa zea and Helicoverpa armigera single-nucleocapsid nucleopolyhedroviruses. J. Gen. Virol. 83:673-684. [DOI] [PubMed] [Google Scholar]
- 15.Cheng, C. H., S. M. Liu, T. Y. Chow, Y. Y. Hsiao, D. P. Wang, J. J. Huang, and H. H. Chen. 2002. Analysis of the complete genome sequence of the Hz-1 virus suggests that it is related to members of the Baculoviridae. J. Virol. 76:9024-9034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clem, R. J., and L. K. Miller. 1994. Control of programmed cell death by the baculovirus genes p35 and iap. Mol. Cell. Biol. 14:5212-5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crook, N. E., R. J. Clem, and L. K. Miller. 1993. An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. J. Virol. 67:2168-2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cunningham, J. C., P. DeGroot, and J. R. McPhee. 1984. Lecontvirus: a viral insecticide for control of redheaded pine sawfly, Neodiprion lecontei. Technical note no. 2, Biological control methods, ISSN 0826-0532. Canadian Forest Service, Sault Sainte Marie, Ontario, Canada.
- 19.De Groot, P., and J. C. Cunningham. 1983. Aerial spray trials with a baculovirus to control red headed pine sawfly in Ontario in 1979 and 1980. Information report FPM-X-63. Canadian Forest Service, Sault Sainte Marie, Ontario, Canada.
- 20.Faulkner, P., J. Kuzio, G. V. Williams, and J. A. Wilson. 1997. Analysis of p74, a PDV envelope protein of Autographa californica nucleopolyhedrovirus required for occlusion body infectivity in vivo. J. Gen. Virol. 78:3091-3100. [DOI] [PubMed] [Google Scholar]
- 21.Federici, B. A., and V. M. Stern. 1990. Replication and occlusion of a granulosis virus in larval and adult midgut epithelium of the western grapeleaf skelontizer, Harrisina brillians. J. Invertebr. Pathol. 56:401-414. [Google Scholar]
- 22.Federici. B. A. 1997. Baculovirus pathogenesis, p. 33-56. In L. K. Miller (ed.), The baculoviruses. Plenum Press, New York, N.Y.
- 23.Fuller, M. 1999. Emboss palindrome. Human Genome Mapping Project Resource Centre, Genome Campus, Cambridge, United Kingdom.
- 24.Garcia-Maruniak, A., J. E. Maruniak, P. M. A. Zanotto, A. E. Doumbouya, J.-C. Liu, T. M. Merritt, and J. S. Lanoie. 2004. Sequence analysis of the genome of the Neodiprion sertifer nucleopolyhedrovirus. J. Virol. 78:7036-7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gomi, S., K. Majima, and S. Maeda. 1999. Sequence analysis of the genome of Bombyx mori nucleopolyhedrovirus. J. Gen. Virol. 80:1323-1337. [DOI] [PubMed] [Google Scholar]
- 26.Guarino, L. A., M. A. Gonzalez, and M. D. Summers. 1986. Complete sequence and enhancer function of the homologous DNA regions of Autographa californica nuclear polyhedrosis virus. J. Virol. 60:224-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrison, R. L., and B. C. Bonning. 2003. Comparative analysis of the genomes of Rachiplusia ou and Autographa californica multiple nucleopolyhedroviruses. J. Gen. Virol. 84:1827-1842. [DOI] [PubMed] [Google Scholar]
- 28.Hashimoto, Y., T. Hayakawa, Y. Ueno, T. Fujita, Y. Sano, and T. Matsumoto. 2000. Sequence analysis of the Plutella xylostella granulovirus genome. Virology 275:358-372. [DOI] [PubMed] [Google Scholar]
- 29.Hawtin, R. E., T. Zarkowska, K. Arnold, C. T. Thomas, G. W. Gooday, L. A. King, J. Kuzio, and R. D. Possee. 1997. Liquefaction of Autographa californica nucleopolyhedrovirus infected insects is dependent on the integrity of virus-encoded chitinase and cathepsin genes. Virology 238:243-253. [DOI] [PubMed] [Google Scholar]
- 30.Hayakawa, T., R. Ko, K. Okano, S. Seong, C. Goto, and S. Maeda. 1999. Sequence analysis of the Xestia c-nigrum granulovirus genome. Virology 262:277-297. [DOI] [PubMed] [Google Scholar]
- 31.Hayakawa, T., G. F. Rohrmann, and Y. Hashimoto. 2000. Patterns of genome organization and content in lepidopteran baculoviruses. Virology 278:1-12. [DOI] [PubMed] [Google Scholar]
- 32.Herniou, E. A., T. Luque, X. Chen, J. M. Vlak, D. Winstanley, J. Cory, and D. R. O'Reilly. 2001. Use of whole genome sequence data to infer baculovirus phylogeny. J. Virol. 75:8117-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herniou, E. A., J. A. Olszewski, J. S. Cory, and D. R. O'Reilly. 2003. The genome sequence and evolution of baculoviruses. Annu. Rev. Entomol. 48:211-234. [DOI] [PubMed] [Google Scholar]
- 34.Hu, Z. H., B. M. Arif, F. Jin, J. W. M. Martens, X. W. Chen, J. S. Sun, D. Zuidema, R. W. Goldbach, and J. M. Vlak. 1998. Distinct gene arrangement in the Buzura suppressaria single-nucleocapsid nucleopolyhedrovirus genome. J. Gen. Virol. 79:2841-2851. [DOI] [PubMed] [Google Scholar]
- 35.Huang, Q., Q. L. Deveraux, S. Maeda, G. S. Salvesen, H. R. Stennicke, B. D. Hammock, and J. C. Reed. 2000. Evolutionary conservation of apoptosis mechanisms: lepidopteran and baculoviral inhibitor of apoptosis proteins are inhibitors of mammalian caspase-9. Proc. Natl. Acad. Sci. USA 97:1427-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hyink, O., R. A. Dellow, M. J. Olsen, K. M. B. Caradoc-Davies, K. Drake, E. A. Herniou, J. S. Cory, D. R. O'Reilly, and V. K. Ward. 2002. Whole genome analysis of the Epiphyas postvittana nucleopolyhedrovirus. J. Gen. Virol. 83:957-971. [DOI] [PubMed] [Google Scholar]
- 37.Ijkel, W. F. J., E. A. van Strien, J. G. M. Heldens, R. Broer, R. W. Zuidema, and J. M. Vlak. 1999. Sequence and organization of the Spodoptera exigua multicapsid nucleopolyhedrovirus genome. J. Gen. Virol. 80:3289-3304. [DOI] [PubMed] [Google Scholar]
- 38.Kikhno, I., S. Gutierrez, L. Crozier, G. A. Crozier, and M. L. Ferber. 2002. Characterization of pif, a gene required for the per os infectivity of Spodoptera littoralis nucleopolyhedrovirus. J. Gen. Virol. 83:3013-3022. [DOI] [PubMed] [Google Scholar]
- 39.Kool, M., C. H. Ahrens, J. M. Vlak, and G. F. Rohrmann. 1995. Replication of baculovirus DNA. J. Gen. Virol. 76:2103-2118. [DOI] [PubMed] [Google Scholar]
- 40.Kuzio, J., M. N. Pearson, S. H. Harwood, J. Funk, J. T. Evans, J. M. Slavicek, and G. F. Rohrmann. 1999. Sequence and analysis of the genome of a baculovirus pathogenic for Lymantria dispar. Virology 253:17-34. [DOI] [PubMed] [Google Scholar]
- 41.Labandeira, C. C., and J. Sepkoski. 1993. Insect diversity in the fossil record. Science 261:310-314. [DOI] [PubMed] [Google Scholar]
- 42.Lange, M., and J. A. Jehle. 2003. The genome of the Cryptophlebia leucotrata granulovirus (CrleGV). Virology 317:220-236. [DOI] [PubMed] [Google Scholar]
- 43.Li, L., C. Donly, Q. Li, L. G. Willis, B. A. Keddie M. A. Erlandson, and D. A. Theilmann. 2002. Identification and genomic analysis of a second species of nucleopolyhedrovirus isolated from Mamestra configurata. Virology 297:226-244. [DOI] [PubMed] [Google Scholar]
- 44.Li, Q., C. Donly, L. Li, L. G. Willis, D. A. Theilmann, and M. Erlandson. 2002. Sequence and organization of the Mamestra configurata nucleopolyhedrovirus genome. Virology 294:106-121. [DOI] [PubMed] [Google Scholar]
- 45.Lu, A., P. J. Krell, J. M. Vlak, and G. F. Rohrmann. 1997. Baculovirus DNA replication, p. 171-191. In L. K. Miller (ed.), The baculoviruses. Plenum Press, New York, N.Y.
- 46.Luque, T., R. Finch, N. Crook, D. R. O'Reilly, and D. Winstanley. 2001. The complete sequence of Cydia pomonella granulovirus genome. J. Gen. Virol. 82:2531-2547. [DOI] [PubMed] [Google Scholar]
- 47.Maguire, T., P. Harrison, O. Hyink, J. Kalmakoff, and V. K. Ward. 2000. The inhibitors of apoptosis of Epiphyas postvittana nucleopolyhedrovirus. J. Gen. Virol. 81:2803-2811. [DOI] [PubMed] [Google Scholar]
- 48.Moser, B. A., J. J. Becnel, S. E. White, C. Afonso, G. Kutish, S. Shanker, and E. Almira. 2001. Morphological and molecular evidence that Culex nigripalpus baculovirus is an unusual member of the family Baculoviridae. J. Gen. Virol. 82:283-297. [DOI] [PubMed] [Google Scholar]
- 49.O'Reilly, D. R. 1997. Auxiliary genes of baculoviruses, p. 267-300. In L. K. Miller (ed.), The baculoviruses, Plenum Press, New York, N.Y.
- 50.Pang, Y., J. Yu, L. Wang, X. Hu, W. Bao, G. Li, C. Chen, H. Han, S. Hu, and H. Yang. 2001. Sequence analysis of the Spodoptera litura multicapsid nucleopolyhedrovirus genome. Virology 287:391-404. [DOI] [PubMed] [Google Scholar]
- 51.Pearson, M. N., C. Groten, and G. F. Rohrmann. 2000. Identification of the Lymantria dispar nucleopolyhedrovirus envelope fusion protein provides evidence for a phylogenetic division of the Baculoviridae. J. Virol. 74:6126-6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pijlman, G. P., A. J. P. Pruijssers, and J. M. Vlak. 2003. Identification of pif-2, a third baculovirus gene required for per os infection of insects. J. Gen. Virol. 84:2041-2049. [DOI] [PubMed] [Google Scholar]
- 53.Posse, D., and G. F. Rohrmann. 1997. Baculovirus genome organization and evolution, p. 109-134. In L. K. Miller (ed.), The baculoviruses. Plenum Press, New York, N.Y.
- 54.Reik, E. F. 1970. Fossil history, p. 168-186. In Reik E. F. (ed.), The insects of Australia. Melbourne University Press, Melbourne, Australia.
- 55.Rohrmann, G. F. 1992. Baculovirus structural proteins. J. Virol. 73:749-761. [DOI] [PubMed] [Google Scholar]
- 56.Rohrmann, G. F., and P. A. Karplus. 2001. Relatedness of baculovirus and gypsy retrotransposon envelope proteins. BMC Evol. Biol. 1:1. [Online.] http://www.biomedcentral.com/1471-2148/1/1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roy, N., Q. L. Deveraux, R. Takahashi, S. Guy, G. S. Salvesen, and J. C. Reed. 1997. The c-IAP-1 and c-IAP-2 proteins are direct inhibitors of specific caspase. EMBO J. 16:6914-6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schultz, J., F. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc. Natl. Acad. Sci. USA 95:5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schultz, J., R. R. Copley, T. Doerks. C. P. Ponting, and P. Bork. 2000. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 28:231-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Slack, J. M., J. Kuzio, and P. Faulkner. 1995. Characterization of v-cath, a cathepsin L-like proteinase expressed by the baculovirus Autographa californica multiple nuclear polyhedrosis virus. J. Gen. Virol. 76:1091-1098. [DOI] [PubMed] [Google Scholar]
- 61.Sutherland, D., C. Samakovlis, and M. A. Krasnow. 1996. Branchless encodes a Drosophila FGF homolog that controls tracheal cell migration and the pattern of branching. Cell 87:1091-1101. [DOI] [PubMed] [Google Scholar]
- 62.Swofford, D. L. 2003. PAUP*. Phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, Mass.
- 63.Van Oers, M. M., J. T. M. Flipsen, C. B. E. M. Reusken, E. L. Sliwinsky, R. W. Goldbach, and J. M. Vlak. 1993. Functional domains of the p10 protein of Autographa californica nuclear polyhedrosis virus. J. Gen. Virol. 74:563-574. [DOI] [PubMed] [Google Scholar]
- 64.Van Oers, M. M., J. T. M. Flipsen, C. B. E. Reusken, and J. M. Vlak. 1994. Specificity of baculovirus p10 functions. Virology 200:513-523. [DOI] [PubMed] [Google Scholar]
- 65.Van Oers, M. M., and J. M. Vlak. 1997. The baculovirus 10-kDA protein. J. Invertebr. Pathol. 70:1-17. [DOI] [PubMed] [Google Scholar]
- 66.Volkman, L. E., G. W. Blissard, P. Friesen, B. A. Keddie, R. Posse, and D. A. Theilmann. 1995. Family Baculoviridae, p. 104-113. In F. A. Murphy, C. M. Fauquet, D. H. L. Bishop, S. A. Ghabrial, A. W. Jarvis, G. P. Martelli, M. A. Mayo, and M. D. Summers (ed.), Virus taxonomy. Springer-Verlag, New York, N.Y.
- 67.Washburn, J. O., D. Trudeau, J. F. Wong, and L. E. Volkman. 2003. Early pathogenesis of Autographa californica multiple nucleopolyhedrovirus and Helicoverpa zea single nucleopolyhedrovirus in Heliothis virescens: a comparison of the ′M' and ′S' strategies for establishing fatal infection. J. Gen. Virol. 84:343-351. [DOI] [PubMed] [Google Scholar]
- 68.Wilson, J. A., J. E. Hill, J. Kuzio, and P. Faulkner. 1995. Characterization of the baculovirus Choristoneura fumiferana multicapsid nuclear polyhedrosis virus p10 gene indicates that the polypeptide contains a coiled-coil domain. J. Gen. Virol. 76:2923-2932. [DOI] [PubMed] [Google Scholar]
- 69.Wormleaton, S., J. Kuzio, and D. Winstanely. 2003. The complete sequence of the Adoxophyes orana granulovirus genome. Virology 311:350-365. [DOI] [PubMed] [Google Scholar]
- 70.Yuen, L., J. Dionne, B. Arif, and C. Richardson. 1990. Identification and sequencing of the spheroidin gene of Choristoneura biennis entomopoxvirus. Virology 175:427-433. [DOI] [PubMed] [Google Scholar]
- 71.Zanotto, P. M., B. D. Kessing, and J. E. Maruniak. 1993. Phylogenetic interrelationships among baculoviruses: evolutionary rates and host associations. J. Invertebr. Pathol. 62:147-164. . [DOI] [PubMed] [Google Scholar]
- 72.Zuidema, D., M. M. Van Oers, E. A. Van Strien, P. C. Caballero, E. J. Klok, R. W. Goldback, and J. M. Vlak. 1993. Nucleotide sequence and transcriptional analysis of the p10 gene of Spodoptera exigua nuclear polyhedrosis virus. J. Gen. Virol. 74:1017-1024. [DOI] [PubMed] [Google Scholar]


