Abstract
Formation of a eutherian mammal requires concurrent establishment of embryonic and extraembryonic lineages. The functions of the trophectoderm and primitive endoderm are to enable implantation in the maternal uterus, axis specification and delivery of nutrients. The pluripotent epiblast represents the founding cell population of the embryo proper, which is protected from ectopic and premature differentiation until it is required to respond to inductive cues to form the fetus. While positional information plays a major role in specifying the trophoblast lineage, segregation of primitive endoderm from epiblast depends upon gradual acquisition of transcriptional identity, directed but not initiated by fibroblast growth factor (FGF) signalling. Following early cleavage divisions and formation of the blastocyst, cells of the inner cell mass lose totipotency. Developing epiblast cells transiently attain the state of naive pluripotency and competence to self-renew in vitro as embryonic stem cells and in vivo by means of diapause. This property is lost after implantation as the epiblast epithelializes and becomes primed in preparation for gastrulation and subsequent organogenesis.
Keywords: cleavage, totipotency, trophoblast, epiblast, primitive endoderm, pluripotency
1. Background
Mammalian preimplantation development combines establishment of a small population of founder cells for the fetus with early differentiation of extraembryonic tissues required to facilitate implantation, patterning and nutrition. The transcriptional and translational machinery becomes activated to institute self-sufficient cell populations from the maternally dominated zygote. Once established, the embryonic lineage must be protected from premature differentiation to remain susceptible to subsequent positional and temporal patterning in order to orchestrate formation of all the tissues in the body. This property is known as naive pluripotency [1]. An interesting and biomedically relevant asset of the murine preimplantation epiblast is its ability to remain undifferentiated and proliferate when explanted into appropriate culture conditions in the form of embryonic stem (ES) cells. In this chapter, we review the current knowledge of how this intriguing state of ‘naive’ pluripotency is acquired in vivo.
2. Totipotency is a unique property of cleavage stages
The fertilized egg is capable of producing all embryonic as well as extraembryonic lineages. This distinctive ability is referred to as totipotency. However, preparation for totipotency in mammals begins long before fertilization. In mouse, the volume of the developing oocyte increases approximately 500-fold during intra-ovarian growth. Continuous transcription of the maternal genome yields around 100 pg messenger RNA in mature oocytes, with some transcripts remaining dormant in order to become activated after fertilization. By contrast, sperm has lost most of its organelles during spermatogenesis in exchange for motility and therefore depends on the egg to boot the embryonic genome. After fertilization, maternal proteins and transcripts pave the way to the first major wave of transcription at the 2-cell stage in mouse [2] and continue to play a role in the initial stages of development. The first five cell cycles, commonly referred as cleavage divisions, are characterized by a predominant S-phase, while G-phases are present but short and variable [3,4]. Cleavage occurs in the absence of cellular growth or increase in total cell mass [5] and strictly depends on the large cytosolic compartment of the fertilized egg (figure 1). Cells generated by cleavage divisions are referred to as blastomeres. At the 2-cell stage, blastomeres retain the ability to form an entire conceptus, evident from the formation of identical twins and demonstrated by the production of viable offspring in mice after destruction of one of the two blastomeres [6,7]. However, monozygotic twins are a rare phenomenon and recent work revealed that a minimum of four preimplantation epiblast cells has to be established for successful normal development [8]. Moreover, the efficiency for monozygotic twins can be increased by modulation of fibroblast growth factor (FGF) and Wnt signalling [8].
Figure 1.
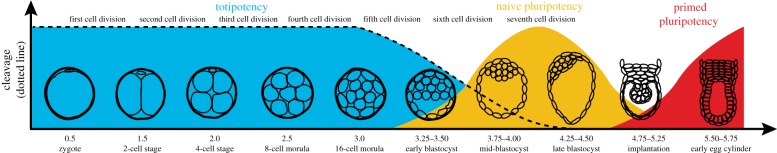
Overview of embryonic potential in relation to developmental stage from zygote to egg cylinder. Cleavage is indicated by the dotted line and correlates with totipotency (blue). Naive pluripotency (yellow) is established at the mid-blastocyst stage and persists until implantation. The terms totipotency, naive pluripotency and primed pluripotency (red) apply to the embryonic lineage only. (Online version in colour.)
Individual blastomeres of the 4- and 8-cell stage can also progress in development and form trophoblastic vesicles as well as small blastocysts [9], which can implant in the uterus when transferred into a synchronized recipient [10]. However, the resultant decidua mostly contained trophoblast giant cells and on only one occasion a retarded embryo [10], suggesting that single 1/4 and 1/8 blastomeres are not capable of producing an entire fetus on their own. Experiments in which isolated blastomeres from 4- and 8-cell stages were aggregated with host blastomeres from another embryo have shown that they are able to differentiate into both trophectoderm and inner cell mass (ICM) and yield viable pups [11]. Thus, their failure to form a normal fetus in isolation is most probably due to inadequate numbers of cells, rather than a restriction in developmental potential. The fact that all blastomeres derived from the 4- and 8-cell stage contribute to both extraembryonic and embryonic lineages demonstrates their principal equipotency.
3. Compaction controls the first lineage decision
One of the most intriguing questions in developmental biology is how lineage identity can be acquired from apparently uniform 8-cell blastomeres. A possible answer could be that early blastomeres might not be as identical as they appear. Several studies have highlighted differences between individual blastomeres, including differential methylation patterns [12], potency under the influence of certain conditions [13] and transcription factor kinetics [14]. However, the majority of blastomeres retain embryonic and extraembryonic potential and differentiate based on their position within the 8- to 16-cell embryo [15].
How do blastomeres ‘sense’ their position? A crucial event preceding the first lineage decision is compaction, which occurs at the late 8-cell stage, at around embryonic day (E) 2.75. During compaction, the blastomeres increase their intercellular interactions, thereby providing the essential spatial queues for the first lineage decision in the mammalian embryo. This allows the establishment of differential compartments. Initially formulated as the ‘inside–outside’ hypothesis [9], subsequent experiments have confirmed that the spatial location of individual blastomeres is instructive for their subsequent lineage allocation [11]. In normal development, the outer cells of the morula become biased towards the first extraembryonic lineage, the trophectoderm. Trophectoderm is required for implantation and subsequently will give rise to the placenta, an extraembryonic organ pivotal for nourishment, detoxification and patterning of the developing fetus [16]. By contrast, cells located in the inside tend to form the ICM of the early blastocyst. ICM cells maintain expression of the POU-domain transcription factor Oct4 (Pou5f1), which is downregulated in outside cells. In the absence of Oct4, the inside cells fail to maintain their identity and differentiate into trophectoderm [17]. Using ES cells, it has been shown that Oct4 acts cooperatively with Sox2 to induce expression of several pluripotency genes, including FGF4 [18] and Nanog [19]. In line with this, embryo profiling at single-cell resolution revealed Sox2 and Id2 as the earliest markers of inner and outer cells, respectively, specifically upregulated at the 16- and 32-cell stage [20].
During compaction, intercellular adhesion depends on E-cadherin [21], and outside cells acquire apical–basal polarity by asymmetric localization of the polarity proteins atypical protein kinase C [22], Par3 [23] and the actin-associated protein ezrin [24]. Interference with polarity regulators by RNAi microinjection perturbs trophectoderm development [23,25], placing polarization upstream of the first lineage decision in the embryo, but downstream of the ‘inside–outside’ spatial location of the individual blastomeres. This polarity is given particular consideration in the ‘polarity’ model of the first lineage decision during cleavage [26]. Key to the model is that the embryo becomes radially polarized at the compacted morula stage, originally discovered by the formation of an external microvillous pole on each blastomere. The model then suggests that this polarity can be inherited during the next (fourth) cleavage division, as most blastomeres will give rise to one polar cell, which inherits the outside surface, and one apolar cell, completely engulfed by other blastomeres. The remaining cells divide symmetrically by splitting the microvillous apical domain, thereby producing two polarized daughters, both of which harbour an outside surface. This model is consistent with the morphology of an average 16-cell embryo, which contains approximately 10–14 outer, polar and 2–6 inner, apolar cells [26–28]. It is worth pointing out that cell fates are not determined in the initial stages of blastocyst formation, as outside 16-cell blastomeres still retain the potential to become ICM at robust frequencies when put into an earlier stage. Moreover, aggregations of purely outer cells can form a new embryo, capable of development in the uterus [15], providing further evidence for the persistence of totipotency in a substantial proportion of blastomeres at this stage.
4. Hippo signalling conveys cellular polarity into lineage-specific gene expression
A key question in the context of embryonic lineage specification is how ‘inside’ or ‘outside’ spatial information is translated into transcriptional programmes. These are established by lineage-specific master regulators, including Cdx2 and Gata3 for trophectoderm versus Oct4, Sox2 and Nanog in the ICM [20,29–32]. Cdx2 null embryos are capable of trophectoderm specification but require Cdx2 for morphological integrity, subsequent development and implantation [33]. The discovery that loss of Tead4 leads to complete failure in blastocyst cavity formation places it upstream of the trophectoderm transcriptional network [34,35]. Intriguingly, Tead4 activity is not mediated by specific expression, but rather by intracellular localization regulated by the Hippo signalling cascade [36]. Hippo signalling is a highly evolutionarily conserved pathway, which, in the context of the mouse embryo, integrates positional information into lineage specification (figure 2a). In mammalian embryos, Hippo signalling is active in inside cells when Lats1/2 phosphorylates the Yorki homologues Yap1 and Wwtr1 [36]. Phosphorylated Yap1 is excluded from the nucleus and degraded. Consequently, Yap1 cannot act as co-activator for Tead4, resulting in failure to induce the trophectodermal programme via expression of Gata3 and Cdx2 [36,37]. In outside cells, Lats1/2 remains inactive, allowing Yap1 to enter the nucleus, and in combination with Tead4, to prime the cell towards trophectoderm. Consistent with this, reduction of Lats1/2 in the early preimplantation embryo prevents ICM lineage formation [38]. Recent work suggests that Lats1/2 activity is controlled by Nf2, which promotes interaction between the adherens junctions and Amot, another regulatory component of Hippo signalling in early mouse development [39,40].
Figure 2.
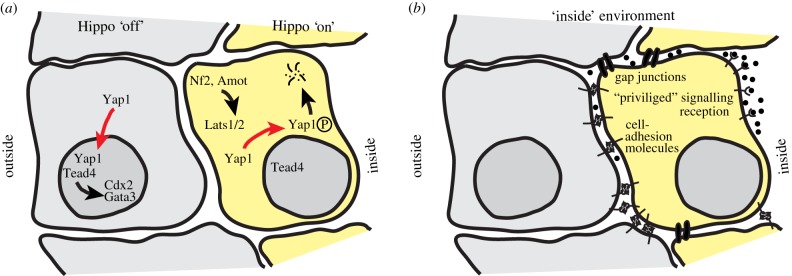
Hippo signalling and an ‘inside’ requirement for the establishment of the embryonic lineage. (a) Schematic of Hippo signalling activities in inside and outside cells of a 16-cell morula. (b) Hippo signalling alone is not sufficient for embryonic lineage formation. Potential signalling activities mediated by the inside environment are outlined. (Online version in colour.)
5. Inner cell mass specification requires an ‘inside’ environment
Hippo signalling alone is not sufficient to control entirely the first lineage decision. Nf2 overexpression fails to alter Yap localization, probably because of other missing components in outside cells [40]. Knockdown of Lats1/2 leads to ectopic Cdx2 expression in the ICM, but concurrent with persistent expression of Oct4 and Nanog, suggesting incomplete conversion of inner cells to bona fide trophectoderm [38]. Thus, additional information may be required to establish ICM fate [38], besides the lack of an apical domain. For instance, inside cells may use gap junction-mediated intercellular communication and adherens junctions, potentially leading to cytoskeletal alterations and signalling activities via focal adhesion kinases (figure 2b). Furthermore, inside cells may reside in a privileged position to receive signalling molecules. Considering the confined intercellular space, even small amounts of secreted ligand would be experienced at higher concentrations inside. Finally, inside cells may be exposed to a specific ‘basal’ environment as the result of asymmetrical protein localization in outside cells. Functional evidence for an ‘inside’ requirement in addition to Hippo signalling comes from blastomeres grown in isolation [41]. Blastomeres were separated after each of the first five cell divisions (1/32), subjected to lineage marker expression profiling, and compared to ICM and trophectoderm cells. Although their expression pattern was distinct from both, it was closer to trophectoderm than ICM [41], corroborating the requirement for an inside environment for ICM specification. This study also demonstrated that singled blastomeres preferentially contribute to trophectoderm in morula aggregations [41]. Interestingly, Hippo signalling is induced in singled blastomeres, suggesting that loss of apical–basal polarity is insufficient to adopt ICM fate (figure 3). In support of this, blastomeres have the ability to give rise to functional trophectoderm when transferred into a recipient female as single cells, but do not form embryonic tissues [10]. Embryos at the 4-cell stage denuded of the zona pellucida can rearrange their cells into various configurations during culture. Those adopting a linear configuration, where intercellular interactions are low, result in blastocysts with significantly fewer ICM cells [42] and exhibit inferior development when transferred into the uterus, compared with tetrahedral configurations, where intercellular interactions are maximized [43]. Single 1/4 and 1/8 blastomeres give rise to ‘blastocysts' at frequencies of 40% and 15%, respectively, while the number of empty trophoblastic vesicles increases [9]. Collectively, these data suggest that blastomeres grown in isolation, despite the loss of apical–basal polarity, become biased towards trophectoderm and fail to enter the embryonic lineage.
Figure 3.
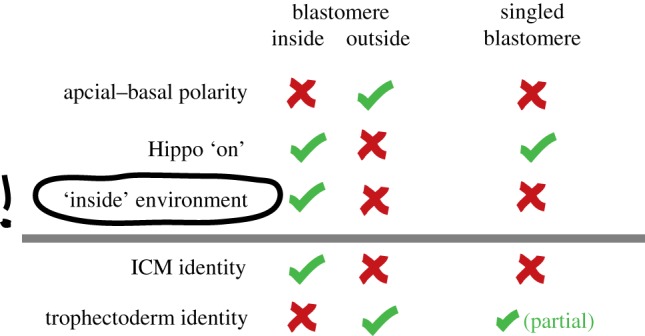
Summary of the cellular characteristics of blastomeres in the embryo and singled blastomeres grown in isolation [41]. Although singled blastomeres lack apical–basal polarity and active Hippo signalling (similar to ‘inside’ cells destined to become ICM), they partially recapitulate trophectoderm identity. This suggests an important role for the ‘inside’ environment in the establishment of the embryonic lineage, which is lacking in singled blastomeres. (Online version in colour.)
6. Totipotency is gradually lost in the inner cell mass
A widely understated characteristic of the early ICM is its totipotency (figure 1). In this context, we define totipotency as the ability of a cell to contribute to all embryonic and extraembryonic lineages. Clearly, this totipotency is different from the ‘absolute’ totipotency of the zygote, which is capable of forming an entire organism from one cell. However, ICMs isolated from early blastocysts have the ability to regenerate trophectoderm, resulting in miniature blastocysts [44,45], and can differentiate into trophectoderm when explanted in vitro [46,47]. Furthermore, they can contribute to trophectoderm in ICM–morula aggregations [48]. Aggregation of several isolated ICMs can compensate for cell numbers and regulate their combined size to produce apparently normal blastocysts. Strikingly, more than one-third of these aggregates give rise to complete egg cylinders upon transfer into recipient female mice [48]. A recent study tested the developmental potential of ICM cells at various blastocyst stages and found that early ICM cells frequently contribute to trophectoderm when injected into a morula, confirming the previously observed developmental plasticity [49]. This ability is gradually lost after E3.5 when the ICM cell number exceeds approximately 16–19 cells [48,49], concomitant with the second lineage decision in the mouse embryo: the segregation of pluripotent epiblast and primitive endoderm (PrE).
7. The second lineage decision: partitioning the inner cell mass into preimplantation epiblast and primitive endoderm
With the advent of accessible custom-made antibodies and fluorescent lineage reporters, the process of PrE and epiblast segregation has been interrogated and is reviewed in great detail elsewhere [50–54]. Here, we outline the differences of the second lineage decision compared to the position-dependent induction of trophectoderm discussed above.
The early PrE marker, Gata6, is initially co-expressed with the pluripotent epiblast marker, Nanog, in the early ICM [55]. Consistent with this, a recent study has shown that at the early blastocyst stage (32-cell), the transcriptome of individual ICM cells is indistinguishable [56]. However, within the next couple of hours of development, small transcriptional changes become progressively manifested and the cells subsequently segregate into two discrete populations [20,56]. In mouse, this process is mainly driven by FGF signalling [57,58]. A cardinal feature of epiblast cells is their temporal unresponsiveness to FGF signalling during the segregation process. Transcriptome analysis of early ICM and epiblast cells has shown that FGFR2, FGFR3 and FGFR4 are specific to the PrE lineage, while FGFR1 is expressed in all cells [56]. Loss of FGF4, FGFR2 or its downstream mediator, Grb2, ablates PrE formation [57,59,60], whereas loss of the other FGF receptors exhibits phenotypes at later stages of development. Therefore, FGFR2 is the essential receptor for PrE specification. However, initiation of the PrE transcriptional programme does not exclusively depend on FGF signalling; embryos completely devoid of FGF4 exhibit mosaic expression of early markers of PrE, such as Gata6 and Sox17 [61].
In line with the genetic evidence, exogenous modulation of FGF signalling in culture from the mid-blastocyst stage or earlier influences ICM cell fate [62–64]. Inhibition of the FGF/Erk pathway with synthetic inhibitors directs ICM cells to become epiblast, whereas supplementation with exogenous FGF4 or FGF2 leads preferentially to PrE. The high concentrations of ligand required to accomplish this lineage switch seem somewhat perplexing, but these may approximate in real terms to the high expression levels of FGF4 secreted by epiblast progenitors [56,65] acting over a comparatively short range within the ICM. Evidence that physiological levels of FGF4 can direct immature ICM cells to become PrE is provided by formation of chimaeras between ES cells and cleavage stage embryos. During the aggregation process, ES cells will preferentially occupy the inside compartment of the embryo, displacing the host cells. The resulting fetus is frequently composed entirely of ES cell derivatives [66], whereas the extraembryonic endoderm almost exclusively originates from the host embryo [67] (figure 4). Once initiated, the inverse correlation of FGF4 in presumptive epiblast cells and its cognate receptor, FGFR2, in PrE precursors increases in order to reinforce the differential identity of the two lineages [20]. By the time the embryo is ready to implant in the uterus, the cells are irreversibly committed to their respective lineages [49,68].
Figure 4.
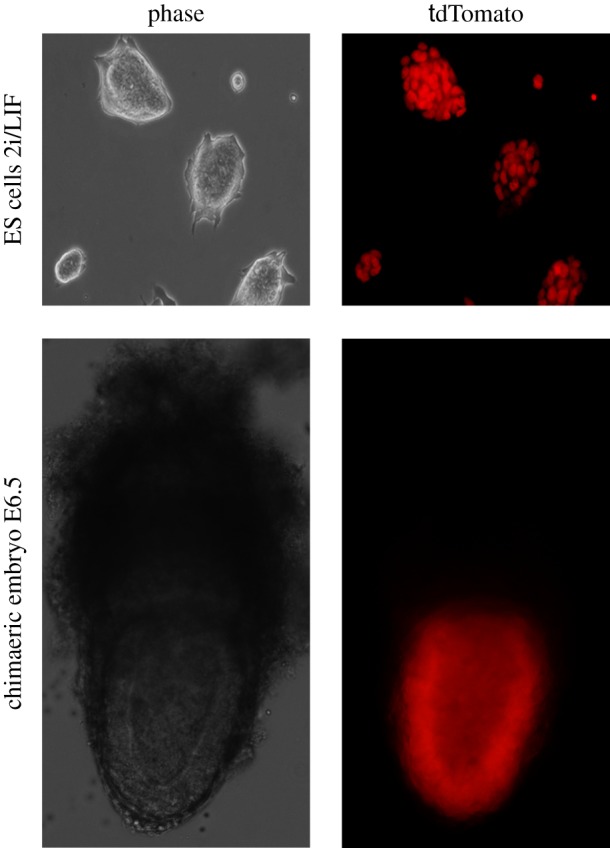
ES cells taking over the host embryo. Fluorescently labelled (tdTomato) mouse ES cells, grown under serum- and feeder-free 2i/LIF culture conditions (upper panel), were injected into non-labelled host morulae. The embryonic compartment (postimplantation epiblast) of the resulting chimaeras apparently consists entirely of donor-derived cells (lower panel). Left images: bright field; right images: fluorescence. (Online version in colour.)
The important question of how the symmetry of transcriptional regulators is broken in the early ICM is still debated. It has been suggested that stochastic fluctuations in gene expression, followed by signal re-enforcement, are sufficient to explain the second lineage decision [56]. Alternatively, it has been proposed that the origin of ICM cells influences their subsequent allocation to epiblast or PrE [28,69]. Live image tracing of embryos from early cleavage stages revealed a trend for the majority of cells becoming internalized during the fourth cell cycle to contribute to the epiblast, whereas those entering the ICM during the fifth or sixth cell cycle tended to generate PrE [28]. In another study, which used retrospective lineage tracing of fluorescent markers, no significant difference was observed between early and late entering cells [63]. The apparent controversy was resolved, as most discrepancies in the outcome were interpreted to originate from different experimental set-ups with both authors agreeing that ‘embryogenesis is a highly dynamic and regulative process with subtle trends that influence cell fate’ [70,71]. We support the notion that certain biases are most likely present in normal embryos, however any of these reported molecular lineage biases in mouse preimplantation development can readily be overridden by topological rearrangements for the first, and modulation of FGF signalling for the second lineage decision [49,62,63].
8. Naive pluripotency is acquired during epiblast specification and captured in embryonic stem cells
Naive pluripotency is the ability of a cell to self-renew while retaining the potential for unbiased differentiation and germline contribution in the context of normal development. Compelling evidence that ES cells are derived from the preimplantation epiblast was provided by Brook & Gardner [72], by means of micro-dissection of periimplantation embryos. Almost half of the epiblasts disaggregated and scattered over the culture well gave rise to one, two or occasionally three clonal ES cell lines. The fact that only a maximum of three clonal lines could be derived from a single preimplantation epiblast led to the speculation that maybe only a subpopulation of cells can give rise to an ES cell colony [72,73], suggesting that the property of naive pluripotency is not epiblast-wide. More recently, the use of two inhibitors (2i) in combination with leukaemia inhibitory factor (LIF) has allowed the derivation of ES cells from ‘recalcitrant’ mouse strains and rats [74–76]. PD0325901 mediates mitogen-activated protein kinase signalling inhibition, thereby eliminating auto-induced differentiation [77,78], while the glycogen synthase kinase 3 inhibitor CHIR99021 positively stimulates the biosynthetic capacity of ES cells and stabilizes β-catenin [79]. β-catenin has been shown to sequester a repressor of pluripotency genes, Tcf3 (Tcf7l1), from the nucleus, which stimulates expression of the naive pluripotency factors Esrrb, Nanog and Klf2 [80,81]. In 2i/LIF, ES cell derivation from the E4.5 blastocyst is very efficient. Dissociated ICMs at this stage have been shown to produce ES cell colonies from all embryos analysed with numbers of clones ranging from two to 12 [62], throwing into question the hypothesis that naive pluripotency is restricted to privileged cells within the epiblast [73].
Although ES cells are commonly derived from the blastocyst stage, they can be established from various preimplantation stages and even from single blastomeres [82–85]. The resultant ES cells have very similar characteristics, suggesting that, during derivation, they progress to a common developmental stage from which in vitro self-renewal can ensue. Single cell ES cell derivation from dissociated embryos from 8-cell to the early postimplantation egg cylinder stage in 2i/LIF on gelatin demonstrates that clonal ES cell lines can be derived efficiently only from mid- and late blastocyst stages [86]. This study further showed that during derivation, epiblast cells do not traverse through distinct developmental states at a transcriptional level and cluster with the preimplantation epiblast at all times [86]. Thus, the window of opportunity to capture the epiblast state in vitro coincides with the initiation of ICM heterogeneity and epiblast specification. This is further supported by the observation that clonal ES cell colony numbers strictly correlate with preimplantation epiblast cell numbers, which can be modulated by activation and inhibition of FGF signalling [62,86]. Collectively, this demonstrates that naive pluripotency is a state acquired during preimplantation development, rather than representing a refined derivative of totipotency.
Epiblast cells can self-renew in vitro and the foundation for this property may be rooted in their self-renewal ability in vivo. Diapause is a facultative condition of embryonic arrest in rodents and other species [87–89], which occurs when implantation is prevented by oestrogen deprivation caused by persistent suckling of a previous litter. This phenomenon can be mimicked experimentally by ovariectomy or administration of an oestrogen antagonist. In diapause, the embryo develops until the late blastocyst stage and segregates epiblast and PrE. Interestingly, diapause embryos were originally used to derive mouse ES cells [90] and have been shown to facilitate ES cell derivation in conventional culture conditions on feeders and in the presence of serum [72]. Quantification of inner cells from diapause embryos revealed a small but significant increase in ICM cell number, implying that the cells continue to proliferate [91]. The fact that diapause embryos retain their developmental potential suggests that mouse epiblast cells can undergo self-renewal in vivo.
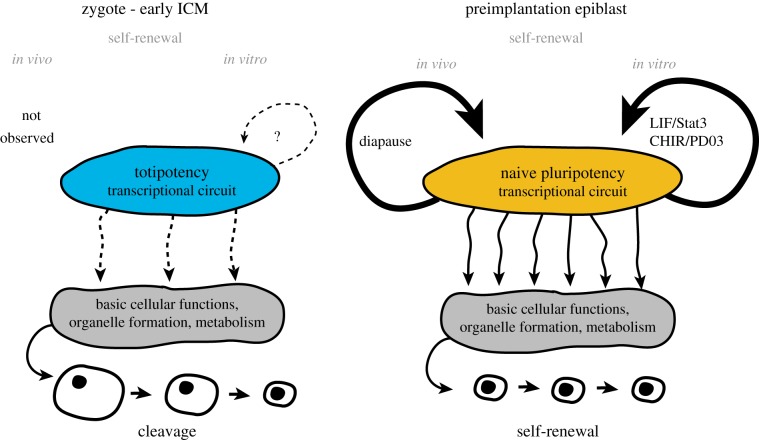
The transcriptional network of the initially totipotent developmental stages changes drastically after almost every cell division [20,92] and the common features of totipotency in vivo therefore remain ill defined. Developmentally, the closest totipotent state to naive pluripotency would be the early ICM, when the embryo is still undergoing cleavage [5]. We propose that the transcriptional networks operating during totipotent stages in vivo are incompletely connected to the basic cellular housekeeping machinery, including cell cycle checkpoints, and are thus incompatible with self-sufficiency and autonomous proliferation. By contrast, naive pluripotent epiblast cells have developed the capacity for cell-autonomous self-renewal in vitro and, during diapause, in vivo (figure 5). Currently, there are no culture conditions established to capture pure populations of authentic, self-renewing blastomeres or early ICM cells. Such totipotent cell lines would have to co-express both early epiblast and extraembryonic markers, readily differentiate into extraembryonic lineages in vitro within 48 h and efficiently contribute to both embryonic and extraembryonic tissues in chimaera assays. The establishment of self-renewing totipotent cells in vitro will strongly depend upon artificial integration of the totipotent transcriptional circuit to the housekeeping machinery. Moreover, it is likely that the temporal presence of maternal genes substantially contributes to a totipotent transcriptional network. Such key factors would have to be identified and expressed in a dosage and time-controlled manner in genetically engineered cells. In contrast to ES cells, self-renewing totipotent cells would lack a genuine embryonic counterpart and therefore it might be challenging, although theoretically possible, to generate such lines in the future.
Figure 5.
Representation of hypothetical totipotent and naive pluripotent transcriptional circuitries. Early embryonic cells from zygote to the early ICM stage strictly undergo cleavage and are unable to support self-sufficient proliferation. This may be caused by incomplete transcriptional interactions of totipotent circuitries with the basic housekeeping machinery. By contrast, cleavage ends at around the time of epiblast specification, thus rendering the preimplantation epiblast capable of cell-autonomous self-renewal. (Online version in colour.)
9. Prerequisites for acquisition of epiblast identity
Early ICM cells co-express epiblast markers, such as Klf2, Sox2 and Nanog, and early PrE markers, including Gata6, Pdgfra and FGFR2 [20,56,86]. This delicate balance of opposing lineage specifiers sets the scene for complete lineage segregation within 24 h. Notably, this timing differs substantially from PrE-like differentiation from ES cells in both embryoid body [93] and monolayer [94] based protocols, in which robust PrE marker induction typically takes around 5 days or longer [94–96]. In presumptive epiblast cells, Nanog and Sox2 become upregulated and repress the sequential activation of the PrE specifiers [64,97,98].
Transcriptional differences during development would predict certain associated epigenetic motifs. Genome wide erasure of DNA-methylation is associated with naive pluripotency [99,100]. This resetting of epigenetic signatures is potentially crucial for unrestricted germ-layer differentiation. In females, the paternally inherited X-chromosome is silenced during the first round of cleavage divisions. Reactivation occurs transiently and exclusively in the embryonic lineage just before implantation [101]. Moreover, there is a correlation of the epigenetic status in epiblast cells in the embryo and ES cells in vitro. Electron spectroscopic imaging of early mouse development has shown that in morula and epiblast the chromatin is distributed as an extended meshwork of uncompacted fibres, indistinguishable from that of ES cells. By contrast, the chromosomes of extraembryonic lineages were found to be denser and more compacted [102]. This supports the notion that naive pluripotency is associated with an open chromatin state.
Another potential factor involved in epiblast specification may be the duration of occupation of an internal position and/or the exposure to extracellular matrix within the ICM. The early ICM expresses a very specific pattern of Laminin511 (Lama5, Lamb1, Lamc1), integrins and fibronectin [65,86]. Isolated early ICM cells can develop the properties of functional epiblast in vitro, when cultured on an attachment matrix consisting of Laminin511 and fibronectin in the presence of 2i/LIF [86]. The history of cell divisions in the preimplantation embryo may similarly contribute to the maturation of a self-sufficient, pluripotent founder cell population. Acquisition of epiblast or PrE fate is a gradual process [20,56,103]. The ability of isolated ICM cells to give rise to ES cell colonies in vitro appears to coincide with the departure of potential for inter-lineage conversion [86]. An intriguing possibility is that each ICM cell becomes irreversibly committed to either PrE or epiblast within a single cell cycle, most likely the seventh (figure 1). This may also coincide with the end of cleavage and the initiation of embryonic growth.
10. Exit from naive pluripotency in vivo
A major rearrangement of the epiblast occurs following implantation. From a loosely adherent ball of cells, a single-layered cup-shaped epithelium emerges. This process was long believed to occur as a result of apoptosis in the cells not in contact with the visceral endoderm in a BMP-dependent manner [104]. Recently, this hypothesis has been elegantly refuted and alternatively attributed to self-organizational behaviour of the epiblast [105]. During implantation, epiblast cells rearrange to form a rosette, probably due to basal membrane-stimulated integrin signalling. This establishment of apical–basal polarity is a prerequisite for lumenogenesis and subsequent gastrulation [105]. The transcriptional signature specific to the primed state of pluripotency includes downregulation of naive pluripotency markers such as Rex1, Klf2, Klf4, Tbx3 and Tfcp2l1 as well as upregulation of Pou3f1, Otx2 and FGF5 [86,106–108].
One of the key drivers of exit from naive pluripotency is FGF signalling. Preimplantation epiblast cells, and ES cells, their in vitro equivalent, autonomously drive progression of development by FGF4 expression [57,78]. Activation of the Erk-cascade directs transition to the early postimplantation epiblast, a tissue responsive to inductive cues for germ-layer specification and subsequent development. Furthermore, preimplantation epiblast cells express Nodal and upregulate Acvr2b upon implantation [86], which may facilitate the specification process. By contrast, the Wnt/Gsk3b signalling pathway has been implicated in maintenance of naive pluripotency [79–81,109]. Downregulation of Wnt/Gsk3b signalling is required for the transition from a naive to a primed state in vitro [79,109]. Interestingly, PrE cells express high levels of the Wnt inhibitor Dkk1 [86], potentially facilitating the pre- to postimplantation epiblast transition. However, recent work demonstrated that mice lacking the porcupine homologue Porcn (a protein required for acetylation and function of Wnt ligands) develop normally until gastrulation [110]. Further studies will be required to elucidate fully the complex role of Wnts, Gsk3b and β-catenin in preimplantation development.
Changes in signalling pathway activities between pre- and early postimplantation development are reflected in pluripotent stem cell lines derived from postimplantation epiblasts (EpiSCs), which exhibit distinct culture requirements from those of ES cells [106,107]. EpiSCs self-renew in the presence of FGF and Activin A, whereas ES cells differentiate upon activation of these pathways. Conversely, 2i-based culture conditions are detrimental for EpiSCs, suggesting that the ability to thrive in the absence of FGF signalling is a distinctive feature of mouse ES cells. In corroboration of this observation, the capacity for isolated epiblast cells to generate naive pluripotent cell lines in feeder-free 2i/LIF culture conditions is rapidly lost in the early postimplantation embryo [86], an event which functionally marks the exit from naive pluripotency in vivo.
11. Concluding remarks
The establishment of a pool of cells poised to respond to positional and signalling cues to form a highly complex organism is an elegant achievement of mammalian development. The first cell fates are specified by means of positional information, with an ‘inside’ requirement for the embryonic lineage. Cleavage continues and the inner cells set aside another extraembryonic lineage, subsequently required for patterning of the embryo. Towards the end of preimplantation development, the embryonic cells exit cleavage, a fundamental prerequisite for embryonic growth. At this time, the epiblast acquires the intriguing state of naive pluripotency, which can then be captured in vitro as self-renewing ES cells.
Funding statement
We wish to thank the MRC, Wellcome Trust and University of Cambridge for our funding.
References
- 1.Nichols J, Smith A. 2009. Naive and primed pluripotent states. Cell Stem Cell 4, 487–492. ( 10.1016/j.stem.2009.05.015) [DOI] [PubMed] [Google Scholar]
- 2.Schultz RM. 1993. Regulation of zygotic gene activation in the mouse. BioEssays 15, 531–538. ( 10.1002/bies.950150806) [DOI] [PubMed] [Google Scholar]
- 3.Streffer C, van Beuningen D, Molls M, Zamboglou N, Schulz S. 1980. Kinetics of cell proliferation in the pre-implanted mouse embryo in vivo and in vitro. Cell Tissue Kinetics 13, 135–143. [DOI] [PubMed] [Google Scholar]
- 4.Howlett SK, Webb M, Maro B, Johnson MH. 1985. Meiosis II, mitosis I and the linking interphase: a study of the cytoskeleton in the fertilised mouse egg. Cytobios 43, 295–305. [PubMed] [Google Scholar]
- 5.Aiken CE, Swoboda PP, Skepper JN, Johnson MH. 2004. The direct measurement of embryogenic volume and nucleo-cytoplasmic ratio during mouse pre-implantation development. Reproduction 128, 527–535. ( 10.1530/rep.1.00281) [DOI] [PubMed] [Google Scholar]
- 6.Tarkowski AK. 1959. Experiments on the development of isolated blastomers of mouse eggs. Nature 184, 1286–1287. ( 10.1038/1841286a0) [DOI] [PubMed] [Google Scholar]
- 7.Hoppe PC, Whitten WK. 1972. Does X chromosome inactivation occur during mitosis of first cleavage? Nature 239, 520 ( 10.1038/239520a0) [DOI] [PubMed] [Google Scholar]
- 8.Morris SA, Guo Y, Zernicka-Goetz M. 2012. Developmental plasticity is bound by pluripotency and the FGF and Wnt signaling pathways. Cell Rep. 2, 756–765. ( 10.1016/j.celrep.2012.08.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tarkowski AK, Wroblewska J. 1967. Development of blastomeres of mouse eggs isolated at the 4- and 8-cell stage. J. Embryol. Exp. Morphol. 18, 155–180. [PubMed] [Google Scholar]
- 10.Rossant J. 1976. Postimplantation development of blastomeres isolated from 4- and 8-cell mouse eggs. J. Embryol. Exp. Morphol. 36, 283–290. [PubMed] [Google Scholar]
- 11.Hillman N, Sherman MI, Graham C. 1972. The effect of spatial arrangement on cell determination during mouse development. J. Embryol. Exp. Morphol. 28, 263–278. [PubMed] [Google Scholar]
- 12.Torres-Padilla ME, Parfitt DE, Kouzarides T, Zernicka-Goetz M. 2007. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature 445, 214–218. ( 10.1038/nature05458) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piotrowska-Nitsche K, Perea-Gomez A, Haraguchi S, Zernicka-Goetz M. 2005. Four-cell stage mouse blastomeres have different developmental properties. Development 132, 479–490. ( 10.1242/dev.01602) [DOI] [PubMed] [Google Scholar]
- 14.Plachta N, Bollenbach T, Pease S, Fraser SE, Pantazis P. 2011. Oct4 kinetics predict cell lineage patterning in the early mammalian embryo. Nat. Cell Biol. 13, 117–123. ( 10.1038/ncb2154) [DOI] [PubMed] [Google Scholar]
- 15.Rossant J, Vijh KM. 1980. Ability of outside cells from preimplantation mouse embryos to form inner cell mass derivatives. Dev. Biol. 76, 475–482. ( 10.1016/0012-1606(80)90395-4) [DOI] [PubMed] [Google Scholar]
- 16.Lawson KA, Dunn NR, Roelen BA, Zeinstra LM, Davis AM, Wright CV, Korving JPWFM, Hogan BLM. 1999. Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev. 13, 424–436. ( 10.1101/gad.13.4.424) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Smith A. 1998. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95, 379–391. ( 10.1016/S0092-8674(00)81769-9) [DOI] [PubMed] [Google Scholar]
- 18.Basilico C, Ambrosetti D, Fraidenraich D, Dailey L. 1997. Regulatory mechanisms governing FGF-4 gene expression during mouse development. J. Cell Physiol. 173, 227–232. () [DOI] [PubMed] [Google Scholar]
- 19.Boyer LA, et al. 2005. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956. ( 10.1016/j.cell.2005.08.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo G, Huss M, Tong GQ, Wang C, Li Sun L, Clarke ND, Robson P. 2010. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev. Cell. 18, 675–685. ( 10.1016/j.devcel.2010.02.012) [DOI] [PubMed] [Google Scholar]
- 21.Johnson MH, Maro B, Takeichi M. 1986. The role of cell adhesion in the synchronization and orientation of polarization in 8-cell mouse blastomeres. J. Embryol. Exp. Morphol. 93, 239–255. [PubMed] [Google Scholar]
- 22.Pauken CM, Capco DG. 2000. The expression and stage-specific localization of protein kinase C isotypes during mouse preimplantation development. Dev. Biol. 223, 411–421. ( 10.1006/dbio.2000.9763) [DOI] [PubMed] [Google Scholar]
- 23.Plusa B, et al. 2005. Downregulation of Par3 and aPKC function directs cells towards the ICM in the preimplantation mouse embryo. J. Cell Sci. 118, 505–515. ( 10.1242/jcs.01666) [DOI] [PubMed] [Google Scholar]
- 24.Louvet S, Aghion J, Santa-Maria A, Mangeat P, Maro B. 1996. Ezrin becomes restricted to outer cells following asymmetrical division in the preimplantation mouse embryo. Dev. Biol. 177, 568–579. ( 10.1006/dbio.1996.0186) [DOI] [PubMed] [Google Scholar]
- 25.Alarcon VB. 2010. Cell polarity regulator PARD6B is essential for trophectoderm formation in the preimplantation mouse embryo. Biol. Reprod. 83, 347–358. ( 10.1095/biolreprod.110.084400) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson MH, Ziomek CA. 1981. Induction of polarity in mouse 8-cell blastomeres: specificity, geometry, and stability. J. Cell Biol. 91, 303–308. ( 10.1083/jcb.91.1.303) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson MH, Ziomek CA. 1981. The foundation of two distinct cell lineages within the mouse morula. Cell 24, 71–80. ( 10.1016/0092-8674(81)90502-X) [DOI] [PubMed] [Google Scholar]
- 28.Morris SA, Teo RT, Li H, Robson P, Glover DM, Zernicka-Goetz M. 2010. Origin and formation of the first two distinct cell types of the inner cell mass in the mouse embryo. Proc. Natl Acad. Sci. USA 107, 6364–6369. ( 10.1073/pnas.0915063107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. 2003. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113, 643–655. ( 10.1016/S0092-8674(03)00392-1) [DOI] [PubMed] [Google Scholar]
- 30.Ralston A, Rossant J. 2008. Cdx2 acts downstream of cell polarization to cell-autonomously promote trophectoderm fate in the early mouse embryo. Dev. Biol. 313, 614–629. ( 10.1016/j.ydbio.2007.10.054) [DOI] [PubMed] [Google Scholar]
- 31.Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, Rossant J. 2005. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell 123, 917–929. ( 10.1016/j.cell.2005.08.040) [DOI] [PubMed] [Google Scholar]
- 32.Dietrich JE, Hiiragi T. 2007. Stochastic patterning in the mouse pre-implantation embryo. Development 134, 4219–4231. ( 10.1242/dev.003798) [DOI] [PubMed] [Google Scholar]
- 33.Strumpf D, et al. 2005. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 132, 2093–2102. ( 10.1242/dev.01801) [DOI] [PubMed] [Google Scholar]
- 34.Nishioka N, Yamamoto S, Kiyonari H, Sato H, Sawada A, Ota M, Nakao K, Sasaki H. 2008. Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mech. Dev. 125, 270–283. ( 10.1016/j.mod.2007.11.002) [DOI] [PubMed] [Google Scholar]
- 35.Yagi R, Kohn MJ, Karavanova I, Kaneko KJ, Vullhorst D, DePamphilis ML, Buonanno A. 2007. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development 134, 3827–3836. ( 10.1242/dev.010223) [DOI] [PubMed] [Google Scholar]
- 36.Nishioka N, et al. 2009. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev. Cell. 16, 398–410. ( 10.1016/j.devcel.2009.02.003) [DOI] [PubMed] [Google Scholar]
- 37.Ralston A, et al. 2010. Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development 137, 395–403. ( 10.1242/dev.038828) [DOI] [PubMed] [Google Scholar]
- 38.Lorthongpanich C, Messerschmidt DM, Chan SW, Hong W, Knowles BB, Solter D. 2013. Temporal reduction of LATS kinases in the early preimplantation embryo prevents ICM lineage differentiation. Genes Dev. 27, 1441–1446. ( 10.1101/gad.219618.113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hirate Y, et al. 2013. Polarity-dependent distribution of angiomotin localizes Hippo signaling in preimplantation embryos. Curr. Biol. 23, 1181–1194. ( 10.1016/j.cub.2013.05.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cockburn K, Biechele S, Garner J, Rossant J. 2013. The Hippo pathway member Nf2 is required for inner cell mass specification. Curr. Biol. 23, 1195–1201. ( 10.1016/j.cub.2013.05.044) [DOI] [PubMed] [Google Scholar]
- 41.Lorthongpanich C, Doris TP, Limviphuvadh V, Knowles BB, Solter D. 2012. Developmental fate and lineage commitment of singled mouse blastomeres. Development 139, 3722–3731. ( 10.1242/dev.086454) [DOI] [PubMed] [Google Scholar]
- 42.Graham CF, Lehtonen E. 1979. Formation and consequences of cell patterns in preimplantation mouse development. J. Embryol. Exp. Morphol. 49, 277–294. [PubMed] [Google Scholar]
- 43.Suzuki H, Togashi M, Adachi J, Toyoda Y. 1995. Developmental ability of zona-free mouse embryos is influenced by cell association at the 4-cell stage. Biol. Reprod. 53, 78–83. ( 10.1095/biolreprod53.1.78) [DOI] [PubMed] [Google Scholar]
- 44.Handyside AH. 1978. Time of commitment of inside cells isolated from preimplantation mouse embryos. J. Embryol. Exp. Morphol. 45, 37–53. [PubMed] [Google Scholar]
- 45.Spindle AI. 1978. Trophoblast regeneration by inner cell masses isolated from cultured mouse embryos. J. Exp. Zool. 203, 483–489. ( 10.1002/jez.1402030315) [DOI] [PubMed] [Google Scholar]
- 46.Nichols J, Gardner RL. 1984. Heterogeneous differentiation of external cells in individual isolated early mouse inner cell masses in culture. J. Embryol. Exp. Morphol. 80, 225–240. [PubMed] [Google Scholar]
- 47.Hogan B, Tilly R. 1978. In vitro development of inner cell masses isolated immunosurgically from mouse blastocysts. I. Inner cell masses from 3.5-day p.c. blastocysts incubated for 24 h before immunosurgery. J. Embryol. Exp. Morphol. 45, 93–105. [PubMed] [Google Scholar]
- 48.Rossant J, Lis WT. 1979. Potential of isolated mouse inner cell masses to form trophectoderm derivatives in vivo. Dev. Biol. 70, 255–261. ( 10.1016/0012-1606(79)90022-8) [DOI] [PubMed] [Google Scholar]
- 49.Grabarek JB, Zyzynska K, Saiz N, Piliszek A, Frankenberg S, Nichols J, Hadjantonakis A-K, Plusa B. 2012. Differential plasticity of epiblast and primitive endoderm precursors within the ICM of the early mouse embryo. Development 139, 129–139. ( 10.1242/dev.067702) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xenopoulos P, Kang M, Hadjantonakis AK. 2012. Cell lineage allocation within the inner cell mass of the mouse blastocyst. Results Probl. Cell Differ. 55, 185–202. ( 10.1007/978-3-642-30406-4_10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schrode N, Xenopoulos P, Piliszek A, Frankenberg S, Plusa B, Hadjantonakis AK. 2013. Anatomy of a blastocyst: cell behaviors driving cell fate choice and morphogenesis in the early mouse embryo. Genesis 51, 219–233. ( 10.1002/dvg.22368) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamanaka Y, Ralston A, Stephenson RO, Rossant J. 2006. Cell and molecular regulation of the mouse blastocyst. Dev. Dyn. 235, 2301–2314. ( 10.1002/dvdy.20844) [DOI] [PubMed] [Google Scholar]
- 53.Oron E, Ivanova N. 2012. Cell fate regulation in early mammalian development. Phys. Biol. 9, 045002 ( 10.1088/1478-3975/9/4/045002) [DOI] [PubMed] [Google Scholar]
- 54.Saiz N, Plusa B. 2013. Early cell fate decisions in the mouse embryo. Reproduction 145, R65–R80. ( 10.1530/REP-12-0381) [DOI] [PubMed] [Google Scholar]
- 55.Plusa B, Piliszek A, Frankenberg S, Artus J, Hadjantonakis AK. 2008. Distinct sequential cell behaviours direct primitive endoderm formation in the mouse blastocyst. Development 135, 3081–3091. ( 10.1242/dev.021519) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ohnishi Y, et al. 2014. Cell-to-cell expression variability followed by signal reinforcement progressively segregates early mouse lineages. Nat. Cell Biol. 16, 27–37. ( 10.1038/ncb2881) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arman E, Haffner-Krausz R, Chen Y, Heath JK, Lonai P. 1998. Targeted disruption of fibroblast growth factor (FGF) receptor 2 suggests a role for FGF signaling in pregastrulation mammalian development. Proc. Natl Acad. Sci. USA 95, 5082–5087. ( 10.1073/pnas.95.9.5082) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rappolee DA, Basilico C, Patel Y, Werb Z. 1994. Expression and function of FGF-4 in peri-implantation development in mouse embryos. Development 120, 2259–2269. [DOI] [PubMed] [Google Scholar]
- 59.Feldman B, Poueymirou W, Papaioannou VE, DeChiara TM, Goldfarb M. 1995. Requirement of FGF-4 for postimplantation mouse development. Science 267, 246–249. ( 10.1126/science.7809630) [DOI] [PubMed] [Google Scholar]
- 60.Cheng AM, et al. 1998. Mammalian Grb2 regulates multiple steps in embryonic development and malignant transformation. Cell 95, 793–803. ( 10.1016/S0092-8674(00)81702-X) [DOI] [PubMed] [Google Scholar]
- 61.Kang M, Piliszek A, Artus J, Hadjantonakis AK. 2013. FGF4 is required for lineage restriction and salt-and-pepper distribution of primitive endoderm factors but not their initial expression in the mouse. Development 140, 267–279. ( 10.1242/dev.084996) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nichols J, Silva J, Roode M, Smith A. 2009. Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development 136, 3215–3222. ( 10.1242/dev.038893) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamanaka Y, Lanner F, Rossant J. 2010. FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst. Development 137, 715–724. ( 10.1242/dev.043471) [DOI] [PubMed] [Google Scholar]
- 64.Frankenberg S, Gerbe F, Bessonnard S, Belville C, Pouchin P, Bardot O, Chazaud C. 2011. Primitive endoderm differentiates via a three-step mechanism involving Nanog and RTK signaling. Dev. Cell. 21, 1005–1013. ( 10.1016/j.devcel.2011.10.019) [DOI] [PubMed] [Google Scholar]
- 65.Tang F, Barbacioru C, Bao S, Lee C, Nordman E, Wang X, Lao K, Surani MA. 2010. Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell Stem Cell 6, 468–478. ( 10.1016/j.stem.2010.03.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Poueymirou WT, et al. 2007. F0 generation mice fully derived from gene-targeted embryonic stem cells allowing immediate phenotypic analyses. Nat. Biotechnol. 25, 91–99. ( 10.1038/nbt1263) [DOI] [PubMed] [Google Scholar]
- 67.Beddington RS, Robertson EJ. 1989. An assessment of the developmental potential of embryonic stem cells in the midgestation mouse embryo. Development 105, 733–737. [DOI] [PubMed] [Google Scholar]
- 68.Gardner RL, Rossant J. 1979. Investigation of the fate of 4–5 day post-coitum mouse inner cell mass cells by blastocyst injection. J. Embryol. Exp. Morphol. 52, 141–152. [PubMed] [Google Scholar]
- 69.Krupa M, Mazur E, Szczepanska K, Filimonow K, Maleszewski M, Suwinska A. 2014. Allocation of inner cells to epiblast versus primitive endoderm in the mouse embryo is biased but not determined by the round of asymmetric divisions (8-->16- and 16-->32-cells). Dev. Biol. 385, 136–148. ( 10.1016/j.ydbio.2013.09.008) [DOI] [PubMed] [Google Scholar]
- 70.Yamanaka Y. 2011. Response: Cell fate in the early mouse embryo--sorting out the influence of developmental history on lineage choice. Reprod. Biomed. Online 22, 525–527; discussion 8 ( 10.1016/j.rbmo.2011.03.011) [DOI] [PubMed] [Google Scholar]
- 71.Morris SA. 2011. Cell fate in the early mouse embryo: sorting out the influence of developmental history on lineage choice. Reprod. Biomed. Online 22, 521–524. ( 10.1016/j.rbmo.2011.02.009) [DOI] [PubMed] [Google Scholar]
- 72.Brook FA, Gardner RL. 1997. The origin and efficient derivation of embryonic stem cells in the mouse. Proc. Natl Acad. Sci. USA 94, 5709–5712. ( 10.1073/pnas.94.11.5709) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chu LF, Surani MA, Jaenisch R, Zwaka TP. 2011. Blimp1 expression predicts embryonic stem cell development in vitro. Curr. Biol. 21, 1759–1765. ( 10.1016/j.cub.2011.09.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Buehr M, et al. 2008. Capture of authentic embryonic stem cells from rat blastocysts. Cell 135, 1287–1298. ( 10.1016/j.cell.2008.12.007) [DOI] [PubMed] [Google Scholar]
- 75.Hanna J, et al. 2010. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc. Natl Acad. Sci. USA 107, 9222–9227. ( 10.1073/pnas.1004584107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nichols J, Jones K, Phillips JM, Newland SA, Roode M, Mansfield W, Smith A, Cooke A. 2009. Validated germline-competent embryonic stem cell lines from nonobese diabetic mice. Nat. Med. 15, 814–818. ( 10.1038/nm.1996) [DOI] [PubMed] [Google Scholar]
- 77.Burdon T, Stracey C, Chambers I, Nichols J, Smith A. 1999. Suppression of SHP-2 and ERK signalling promotes self-renewal of mouse embryonic stem cells. Dev. Biol. 210, 30–43. ( 10.1006/dbio.1999.9265) [DOI] [PubMed] [Google Scholar]
- 78.Kunath T, Saba-El-Leil MK, Almousailleakh M, Wray J, Meloche S, Smith A. 2007. FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development 134, 2895–2902. ( 10.1242/dev.02880) [DOI] [PubMed] [Google Scholar]
- 79.Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. 2008. The ground state of embryonic stem cell self-renewal. Nature 453, 519–523. ( 10.1038/nature06968) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Martello G, Sugimoto T, Diamanti E, Joshi A, Hannah R, Ohtsuka S, Niwa H, Smith A. 2012. Esrrb is a pivotal target of the Gsk3/Tcf3 axis regulating embryonic stem cell self-renewal. Cell Stem Cell 11, 491–504. ( 10.1016/j.stem.2012.06.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wray J, Kalkan T, Gomez-Lopez S, Eckardt D, Cook A, Kemler R, Smith A. 2011. Inhibition of glycogen synthase kinase-3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nat. Cell Biol. 13, 838–845. ( 10.1038/ncb2267) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Delhaise F, Bralion V, Schuurbiers N, Dessy F. 1996. Establishment of an embryonic stem cell line from 8-cell stage mouse embryos. Eur. J. Morphol. 34, 237–243. ( 10.1076/ejom.34.4.0237) [DOI] [PubMed] [Google Scholar]
- 83.Tesar PJ. 2005. Derivation of germ-line-competent embryonic stem cell lines from preblastocyst mouse embryos. Proc. Natl Acad. Sci. USA 102, 8239–8244. ( 10.1073/pnas.0503231102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chung Y, Klimanskaya I, Becker S, Marh J, Lu SJ, Johnson J, Meisner L, Lanza R. 2006. Embryonic and extraembryonic stem cell lines derived from single mouse blastomeres. Nature 439, 216–219. ( 10.1038/nature04277) [DOI] [PubMed] [Google Scholar]
- 85.Wakayama S, Hikichi T, Suetsugu R, Sakaide Y, Bui HT, Mizutani E, Wakayama T. 2007. Efficient establishment of mouse embryonic stem cell lines from single blastomeres and polar bodies. Stem Cells 25, 986–993. ( 10.1634/stemcells.2006-0615) [DOI] [PubMed] [Google Scholar]
- 86.Boroviak TLR, Bertone P, Smith A, Nichols J. In press. The ability of inner cell mass cells to self-renew as embryonic stem cells is acquired upon epiblast specification. Nat. Cell Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hondo E, Stewart CL. 2005. Profiling gene expression in growth-arrested mouse embryos in diapause. Genome Biol. 6, 202 ( 10.1186/gb-2004-6-1-202) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ketchel MM, Banik UK, Mantalenakis SJ. 1966. A study of delayed implantation caused by parabiosis in pregnant rats. J. Reprod. Fertil. 11, 213–219. ( 10.1530/jrf.0.0110213) [DOI] [PubMed] [Google Scholar]
- 89.Mantalenakis SJ, Ketchel MM. 1966. Frequency and extent of delayed implantation in lactating rats and mice. J. Reprod. Fertil. 12, 391–394. ( 10.1530/jrf.0.0120391) [DOI] [PubMed] [Google Scholar]
- 90.Evans MJ, Kaufman MH. 1981. Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156. ( 10.1038/292154a0) [DOI] [PubMed] [Google Scholar]
- 91.Nichols J, Chambers I, Taga T, Smith A. 2001. Physiological rationale for responsiveness of mouse embryonic stem cells to gp130 cytokines. Development 128, 2333–2339. [DOI] [PubMed] [Google Scholar]
- 92.Xue Z, et al. 2013. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 500, 593–597. ( 10.1038/nature12364) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Martin GR. 1981. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl Acad. Sci. USA 78, 7634–7638. ( 10.1073/pnas.78.12.7634) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cho LT, Wamaitha SE, Tsai IJ, Artus J, Sherwood RI, Pedersen RA, Hadjantonakis A-K, Niakan KK. 2012. Conversion from mouse embryonic to extra-embryonic endoderm stem cells reveals distinct differentiation capacities of pluripotent stem cell states. Development 139, 2866–2877. ( 10.1242/dev.078519) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Doughton G, Wei J, Tapon N, Welham MJ, Chalmers AD. 2014. Formation of a polarised primitive endoderm layer in embryoid bodies requires fgfr/erk signalling. PLoS ONE 9, e95434 ( 10.1371/journal.pone.0095434) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang Y, Smedberg JL, Cai KQ, Capo-Chichi DC, Xu XX. 2011. Ectopic expression of GATA6 bypasses requirement for Grb2 in primitive endoderm formation. Dev. Dyn. 240, 566–576. ( 10.1002/dvdy.22447) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Artus J, Piliszek A, Hadjantonakis AK. 2011. The primitive endoderm lineage of the mouse blastocyst: sequential transcription factor activation and regulation of differentiation by Sox17. Dev. Biol. 350, 393–404. ( 10.1016/j.ydbio.2010.12.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chazaud C, Yamanaka Y, Pawson T, Rossant J. 2006. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Dev. Cell. 10, 615–624. ( 10.1016/j.devcel.2006.02.020) [DOI] [PubMed] [Google Scholar]
- 99.Leitch HG, et al. 2013. Naive pluripotency is associated with global DNA hypomethylation. Nat. Struct. Mol. Biol. 20, 311–316. ( 10.1038/nsmb.2510) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ficz G, et al. 2013. FGF signaling inhibition in ESCs drives rapid genome-wide demethylation to the epigenetic ground state of pluripotency. Cell Stem Cell 13, 351–359. ( 10.1016/j.stem.2013.06.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Heard E. 2004. Recent advances in X-chromosome inactivation. Curr. Opin. Cell Biol. 16, 247–255. ( 10.1016/j.ceb.2004.03.005) [DOI] [PubMed] [Google Scholar]
- 102.Rugg-Gunn PJ, Cox BJ, Ralston A, Rossant J. 2010. Distinct histone modifications in stem cell lines and tissue lineages from the early mouse embryo. Proc. Natl Acad. Sci. USA 107, 10 783–10 790. ( 10.1073/pnas.0914507107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kurimoto K, et al. 2006. An improved single-cell cDNA amplification method for efficient high-density oligonucleotide microarray analysis. Nucleic Acids Res. 34, e42 ( 10.1093/nar/gkl050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Coucouvanis E, Martin GR. 1999. BMP signaling plays a role in visceral endoderm differentiation and cavitation in the early mouse embryo. Development 126, 535–546. [DOI] [PubMed] [Google Scholar]
- 105.Bedzhov I, Zernicka-Goetz M. 2014. Self-organizing properties of mouse pluripotent cells initiate morphogenesis upon implantation. Cell 156, 1032–1044. ( 10.1016/j.cell.2014.01.023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RDG. 2007. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448, 196–199. ( 10.1038/nature05972) [DOI] [PubMed] [Google Scholar]
- 107.Brons IG, et al. 2007. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448, 191–195. ( 10.1038/nature05950) [DOI] [PubMed] [Google Scholar]
- 108.Iwafuchi-Doi M, Matsuda K, Murakami K, Niwa H, Tesar PJ, Aruga J, Matsuo I, Kondoh H. 2012. Transcriptional regulatory networks in epiblast cells and during anterior neural plate development as modeled in epiblast stem cells. Development 139, 3926–3937. ( 10.1242/dev.085936) [DOI] [PubMed] [Google Scholar]
- 109.ten Berge D, Kurek D, Blauwkamp T, Koole W, Maas A, Eroglu E, Siu RK, Nusse R. 2011. Embryonic stem cells require Wnt proteins to prevent differentiation to epiblast stem cells. Nat. Cell Biol. 13, 1070–1075. ( 10.1038/ncb2314) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Biechele S, Cockburn K, Lanner F, Cox BJ, Rossant J. 2013. Porcn-dependent Wnt signaling is not required prior to mouse gastrulation. Development 140, 2961–2971. ( 10.1242/dev.094458) [DOI] [PubMed] [Google Scholar]