Abstract
The accessory Nef protein allows human immunodeficiency virus type 1 (HIV-1) to persist at high levels and to cause AIDS in infected humans. The function of HIV-1 group M subtype B nef alleles has been extensively studied, and a variety of in vitro activities believed to be important for viral pathogenesis have been established. However, the function of nef alleles derived from naturally simian immunodeficiency virus (SIV)-infected chimpanzees, the original host of HIV-1, or from the HIV-1 N and O groups resulting from independent zoonotic transmissions remains to be investigated. In the present study we demonstrate that SIVcpz and HIV-1 group N or O nef alleles down-modulate CD4, CD28, and class I or II MHC molecules and up-regulate surface expression of the invariant chain (Ii) associated with immature major histocompatibility complex (MHC) class II. Furthermore, the ability of Nef to interact with the p21-activated kinase 2 was generally conserved. The functional activity of HIV-1 group N and O nef genes did not differ significantly from group M nef alleles. However, SIVcpz nef genes as a group showed a 1.8- and 2.0-fold-higher activity in modulating CD28 (P = 0.0002) and Ii (P = 0.016) surface expression, respectively, but were 1.7-fold less active in down-regulating MHC class II molecules (P = 0.006) compared to HIV-1 M nef genes. Our finding that primary SIVcpz nef alleles derived from naturally infected chimpanzees modulate the surface expression of various human cellular receptors involved in T-cell activation and antigen presentation suggests that functional nef genes helped the chimpanzee virus to persist efficiently in infected humans immediately after zoonotic transmission.
Nef is a myristoylated regulatory protein of the primate lentiviruses. Early studies demonstrated that unlike other accessory viral factors, such as Vpr or Vpx (23), Nef is clearly important for the pathogenicity of simian immunodeficiency viruses (SIVmac) in infected rhesus macaques (37). Compared to wild-type SIVmac239, nef deletion mutants resulted in levels of viral RNA that were reduced by several orders of magnitude and an asymptomatic course of infection. In contrast, individual deletions in vpr or vpx had little effect on the efficiency of viral replication in infected macaques and did not prevent progression to simian AIDS (23). Subsequent studies revealed that forms of the virus with exclusive nef deletions can be detected in some long-term nonprogressors of human immunodeficiency type 1 (HIV-1) infection (17, 38). Similar to the results obtained with the SIV-macaque model, these human individuals showed very low levels of viral RNA and DNA and delayed or no disease progression.
The knowledge that Nef is a key factor of lentiviral pathogenesis has stimulated a great research interest, and a variety of Nef activities believed to contribute to AIDS progression have been established. Nef down-modulates cell surface expression of CD4, the primary receptor for HIV and SIV infection (1, 21). This Nef function could promote virion release, enhance incorporation of the Env protein into viral particles, prevent superinfection, alter T-cell receptor (TCR) signaling, and impair CD4+ helper T-cell functions (7, 40, 64). Nef also reduces major histocompatibility complex class I (MHC-I) cell surface expression (42, 73), likely allowing HIV-1 to escape from host immune surveillance (13). More recently, it has been shown that Nef also affects MHC-II antigen presentation by down-regulation of surface expression of mature MHC-II and up-regulation of the MHC-II-associated invariant chain (Ii) (72, 77). Furthermore, Nef might impair TCR signaling by down-modulating cell surface expression of CD28 (78) and expression or signaling of CD3 (6, 33, 34). Nef also increases virion infectivity and accelerates viral replication in primary human T cells and in human lymphoid tissue ex vivo (11, 24, 47, 76). Nef itself has no catalytic activity and seems to exert its activities by interactions with various components of the cellular signal transduction and endocytic machinery, such as serine-threonine or tyrosine kinases and adaptor protein complexes (reviewed in references 61 and 75).
The relative importance of these different Nef activities for the virulence of HIV and SIV remains to be clarified. However, experiments with rhesus macaques with SIVmac variants containing nef alleles that are selectively impaired in specific in vitro activities indicate that a variety of Nef functions, including T-cell activation (19) and down-modulation of CD4 (35), CD3 (50), or MHC-I (49) molecules, mediates a selective advantage for viral replication in vivo. Various Nef activities are genetically separable and can be modulated during the course of HIV-1 and SIV infection, likely to optimize viral spread in infected humans or macaques at different stages of disease progression (10, 54). Taken together, these studies imply that a combination of Nef activities allows pathogenic HIV-1 and SIVmac strains to persist efficiently at high levels and to cause immunodeficiency.
Humans are originally not the natural host of HIV-1 (4, 30). Current evidence suggests that the HIV-1 M, N, and O groups arose from three independent transmissions of SIV from the Pan troglodytes troglodytes subspecies of chimpanzees to humans (30). The descendants of one of these SIVcpz(P.t.t.) strains, now designated the HIV-1 main group (HIV-1 group M), has spread worldwide and infected about 60 million people (AIDS epidemic update, www.unaids.org). In comparison, infections with group O (outlier) viruses account only for a small fraction of AIDS cases and are mainly limited to Gabon, Cameroon, Equatorial Guinea, and surrounding countries (30). Group N (non-M, non-O) infection has only been documented in a very limited number of individuals from Cameroon (3, 74) SIVcpz is also found in the Pan troglodytes schweinfurthii subspecies of chimpanzees (55, 68, 69), but SIVcpz(P.t.s.) has not been found in humans (30). Interestingly, SIVcpz itself may have a relatively recent origin and may have arisen through cross-species transmission and recombination of SIVs from smaller hosts on which chimpanzees prey (5). Notably, there is only one report regarding AIDS-like symptoms in SIVcpz-infected chimpanzees (31), suggesting that they do usually not develop immunodeficiency.
Nef genes derived from pathogenic HIV-1 M subtype B and SIVmac strains have been intensively studied. However, the functions of nef alleles derived from members of the HIV-1 N and O groups or from SIVcpz are currently unknown. To address this, we investigated the ability of SIVcpz(P.t.t.) and SIVcpz(P.t.s.) as well as HIV-1 M, O, and N Nef proteins to modulate the cell surface expression of various cellular receptors and to interact with the p21-activated kinase 2 (PAK-2) (46, 60) in human cells. We wanted to clarify whether Nef activities might have been lost, altered, or acquired after cross-species transmission of the virus from chimpanzees to humans. We were also interested in elucidating whether differences in Nef function could explain the different spread of the HIV-1 M, N, and O groups in the human population and/or the distinct clinical outcome in humans and chimpanzees. Our results demonstrate that the ability of Nef to modulate the surface expression of various cellular receptors and to interact with PAK-2 is well conserved among all groups of HIV-1 and SIVcpz. However, we also found quantitative differences in the ability to modulate CD28, MHC-II, and Ii surface expression that might have implications for the virulence and spread of different groups of HIV-1 and SIVcpz.
MATERIALS AND METHODS
HIV-1 and SIVcpz nef alleles.
HIV-1 M subtype B nef alleles were amplified from uncultured peripheral blood mononuclear cells (PBMC) as described previously (10, 39). The 039 nm-94, 168mb-95, 032an93-93, and Priso nef alleles were derived from HIV-1-infected individuals from the United Kingdom, and the LT-87, FA-93, and AD-93 nef alleles are from patients from the United States (10, 39). Two consensus nef alleles obtained from nonprogressors (NPcon) or immunodeficient individuals (Pcon) infected with HIV-1 M subtype B strains were generated as described previously (10). The HIV-1 M subtype A (92UG029) and F (93BR029) nef alleles were amplified from human PBMC infected with the respective virus stocks obtained through the National Institutes of Health AIDS Reagent Program. The origins of the remaining nef alleles are summarized in Table 1. HIV-1 O Ca9, 13127, 8161, and 2171 were isolated and propagated in primary human PBMC as described previously (18). The HIV-1 N YBF30 and YBF116 (3, 74) nef alleles were also amplified from PBMC. The SIVcpz US (20) and GAB2 (B. H. Hahn, unpublished data) nef alleles were amplified from clones derived from spleen and uncultured PBMC DNA, respectively. SIVcpz TAN1 (69), TAN2.2, and TAN3.1 (M. L. Santiago and B. H. Hahn, unpublished data) were amplified from fecal viral RNA. The SIVcpz Cam DNA samples were obtained by short-term cocultivation of PBMC from the naturally infected chimpanzees Cam3 (14) and Cam5 (14, 48) with human PBMC. The remaining SIVcpz nef alleles were amplified from plasma RNA derived from Noah (ch-No) and an uncultured PBMC sample derived from Niko (ch-Ni), respectively. Both are infected with the highly divergent SIVcpz-ant strain (55, 80). ch-No was infected in Zaire, whereas ch-Ni was exposed to blood from cage mate ch-No (56, 79).
TABLE 1.
nef alleles from the HIV-1 N and O and SIVcpz groups analyzeda
| Clone | Group | Country of origin | Size of nef ORF (bp) | Source | Comment(s) | Reference(s) |
|---|---|---|---|---|---|---|
| YBF30 | HIV-1 N | Cameroon | 639 | Cultured human PBMC DNA | F, 40 yr, AIDS | 3, 74 |
| YBF106 | HIV-1 N | Cameroon | 639 | Cultured human PBMC DNA | M, 51 yr, AIDS | 3, 74 |
| Ca-9k7 | HIV-1 O | Cameroon | 636 | Cultured human PBMC DNA | Symptomatic individual | 18 |
| 13127k4 | HIV-1 O | Cameroon | 657 | Cultured human PBMC DNA | F, 22 yr, ARC | 18 |
| 2171k1 | HIV-1 O | Cameroon | 636 | Cultured human PBMC DNA | F, 35 yr, AIDS | 18 |
| 8161k9 | HIV-1 O | Cameroon | 636 | Cultured human PBMC DNA | F, 52 yr, AIDS | 18 |
| US | SIVcpz(P.t.t.) | Unknown | 630 | Uncultured P. t. troglodytes spleen DNA | Wild-caught animal | 20 |
| GAB2c148 | SIVcpz(P.t.t.) | Gabon | 618 | Uncultured P. t. troglodytes PBMC DNA | Wild-caught animal | Unpublished data |
| Cam3k1/k5 | SIVcpz(P.t.t.) | Cameroon | 624 | Cultured human PBMC DNA | Wild-caught (as an infant) animal | 14 |
| Cam5k2 | SIVcpz(P.t.t.) | Cameroon | 645 | Cultured human PBMC DNA | Wild-caught (as an infant) animal | 14, 48 |
| TAN1 | SIVcpz(P.t.s.) | Tanzania | 588 | Fecal RNA | Wild animal | 69 |
| TAN2.2 | SIVcpz(P.t.s.) | Tanzania | 588 | Fecal RNA | Wild animal | 70 |
| TAN3.1 | SIVcpz(P.t.s.) | Tanzania | 588 | Fecal RNA | Wild animal | 70 |
| ch-Nok5 | SIVcpz(P.t.s.) | DRC | 597 | P.t. schweinfurthii plasma RNA | Wild-caught animal | 55, 80 |
| ch-Nik4 | SIVcpz(P.t.s.) | 597 | Uncultured P.t. schweinfurthii PBMC DNA | Infected by blood from ch-No | 56, 79 |
Abbreviations: F, female; M, male; ARC, AIDS-related complex; DRC, Democratic Republic of Congo.
Cloning and sequencing.
Sequence analysis and cloning of HIV-1 M subtype B nef alleles into the bicistronic pCG expression vector coexpressing the green fluorescent protein (GFP) has been described previously (10). The SIVcpz US, GAB2, and TAN1 nef alleles were amplified from cloned DNA by a single round of PCR with primers introducing XbaI and MluI restriction sites just up- and downstream of the nef open reading frame (ORF), respectively.
TAN2.2 and TAN3.1 nef ORFs were initially amplified from fecal virion RNA from two wild chimpanzees essentially as described previously (69, 70) and subsequently subjected to another round of PCR amplification with primers introducing flanking XbaI and MluI sites. The 93BR029, 92UG029, HIV-1 N YBF30 and YBF116, SIVcpz Cam3, Cam5, Ch-Ni and HIV-1 O Ca9, 13127, 8161, and 2171 nef alleles were amplified from genomic DNA extracted from PBMC by standard methods. Viral RNA was extracted from a plasma sample derived from Ch-No with the QIAamp viral RNA mini kit (QIAGEN Inc., Hilden, Germany), and cDNA was prepared by using an avian myeloblastosis virus-reverse transcription-based cDNA synthesis kit (Boehringer Mannheim) following the protocols of the manufacturers. We employed nested PCR methods with oligonucleotides flanking the nef ORF because proviral DNA or genomic viral RNA is often present at low copy numbers. The outer primers were as follows: for HIV-1 M 93BR029 and 92UG029, pF1 (5′-GCAGTAGCTGAGGGGACAGATAGG-3′) and pF2 (5′-CCAGTACAGGCAAAAAGCAGCTGC-3′); for SIVcpz Cam, pCPZ-NEF1 (5′-CTTATTAGATACAACAGCAATTGC-3′) and pCPZ-NEF2 (5′-AAACCACGCCCAGTCCCG-3′); for SIVcpz Ant (Ch-No, Ch-Ni), pSIVcpzant5 (5′-GAAGTACCTAGGCGCATCAG-3′) and pSIVcpzant3 (5′-CCATAGTCACTCCCATAGTG-3′); for HIV-1 O, pHIV-1O5l (5′-GCAGTGGCAGTTGCCAATTGGACTG-3′) or pHIV-1O5r (5′-CTTAACATCCAAGAAGAATTAGA-3′) and pHIV-1O3 (5′-CACACTGGAAAGTCCCCGCCGTGTCAGC-3); for HIV-1 N, HIVNNEF1 (5′-CTTAATACAACAGCTATTGTAGTAG-3′) and HIVNNEF2 (5′-CAGCTCTGAGGGCAAGCCACTCC-3′). The inner primers introduced flanking XbaI and MluI sites (underlined) and were as follows: pHIVMXba5 (5′-GAATCTAGATGTAGATGGGGCAAGTGGTCA-3′) and p93BR029Mlu3 (5′-TCCGACGCGTTCAGCAGTCCAATAGTA-3′) or p92UG029Mlu (5′-TCCGACGCGTTCAGCAGTCTTTGTA-3′); pSIVcpzCam5Xba5 (5′-GAATCTAGATATAACATGGGTAACAAGTGGTCAAAAAG-3′) and pSIVcpzCam5Mlu3 (5′-GTCAACGCGTCTACTGCGCAGGCGCTGGGTTGTGGTC-3′); pSIVcpzCam3Xba5 (5′-GAATCTAGATATAACATGGGCAACAGGTGGTC-3′) and pSIVcpzCam3Mlu3 (5′-CGCACCGTTCAGTTCTGGTAATACTCCGGATG-3′); pSIVcpzantXba5 (5′-TGCTCTAGATATAAGATGGGTTCTGCATGGTCT-3′) and pSIVcpzantMlu3 (5′-CGGTTTTATACGCGTCTGTTAGCAGTCTTAGT-3′); HIV-1OXba5 (5′-AAAGAGTCTAGATGTAACATGGGAAACGTAT-3′) and pHIV-1OMlu3 (5′-TCCCTTACGCGTCTTCAGGTCAGCAGTTTTA-3′); HIVNNEF3Xba (5′-GAATCTAGAGGAAGGGGAATATTACACATAC-3′) and HIVNNEF4Mlu (5′-CGACGCGTCGGAAAGTCCCTGGCGGAAAGT-3′). PCR fragments were purified from agarose gels with the GeneClean kit (QIAGEN Inc.) and cloned with the TA cloning kit (Invitrogen Corp., San Diego, Calif.) as recommended by the manufacturers. Per sample, 5 to 10 individual TA clones containing inserts of the expected size were sequenced on both strands. For functional characterization, one or two intact representative nef alleles that were identical or closely related to the sample-specific consensus sequence were inserted into the bicistronic pCG vector as described above. For Western blot analysis, we also amplified all nef alleles with primers, resulting in the fusion of the AU-1 peptide tag to the C terminus of Nef. All PCR-derived inserts were sequenced to confirm that no undesired nucleotide changes were present.
Transfections and cell culture.
Transfection of Jurkat T cells was performed with the DMRIE-C reagent (GibcoBRL, Karlsruhe, Germany) according to the manufacturer's instructions. HeLa CIITA cells were transfected with Metafectene (Biontex, Munich, Germany). Briefly, 2.5 μg of DNA in 100 μl of Dulbecco’s modified Eagle medium (DMEM) (Invitrogen) were mixed with 12 μl of Metafectene in 100 μl of DMEM and incubated for 30 min at room temperature. Subsequently, the mixture was added to 2 × 105 cells and incubated for 6 h at 37°C. Thereafter, the medium was changed, and cells were analyzed by fluorescence-activated cell sorting on the following day. Transfection efficiencies varied between 20 and 35%. HeLa CIITA and Jurkat T cells were cultured as described previously (10, 72).
Flow cytometry.
CD4, MHC-I, CD28, CD3, and GFP reporter molecules in Jurkat T cells transfected with a bicistronic vector coexpressing Nef and GFP were measured as described previously (10). Down-modulation of MHC-II and up-regulation of Ii was measured on transfected HeLa CIITA cells (72, 77). The following phycoerythrin-conjugated antibodies were used: anti-human CD4, anti-human CD3, and anti-LeuTM-28 (BD Biosciences, Pharmingen), anti-CD74/R-PE M-B741 (Ancell), anti-HLA-ABC Antigen/RPE (DAKO), mouse anti-human HLA-DR TÜ36 (Caltag laboratories), and L243 (BD Biosciences). Staining with both TÜ36 or L243 gave similar results. For the quantitation of Nef-mediated down- or up-regulation, the mean channel numbers of red fluorescence were determined for cells expressing no, low, medium, or high levels of GFP as described previously (10, 72). The mean channel numbers of red fluorescence obtained for cells transfected with a control construct expressing GFP only were divided by the corresponding numbers obtained for cells coexpressing Nef and GFP to calculate the values for down- or up-modulation, respectively. The same ranges of green fluorescence were used in all calculations.
IVKAs.
In vitro kinase assays (IVKAs) were performed as described previously (46, 60). Briefly, 293T HEK cells were cotransfected with pEBB-PAK2-HA, pEBG-Cdc42V12, and pEBB-lacZ and expression vectors for the different AU1-tagged nef alleles by using the Lipofectamine transfection agent (Gibco BRL) according to the manufacturer's instructions. Forty-eight hours after transfection, cells were washed with phosphate-buffered saline and lysed in IVKA lysis buffer (50 mM HEPES [pH 7.4], 150 mM NaCl, 10% glycerol, 1% Triton X-100, 1 mM EGTA, 1.5 mM MgCl2, 1 mM Na orthovanadate, 1 mM phenylmethylsulfonyl fluoride, 10 μg of aprotinin/ml). Lysates were cleared and corrected over β-galactosidase activity by an o-nitrophenyl-β-d-galactopyranoside assay. The corrected lysates were used for Western blot analysis with a mix of anti-hemagglutinin (anti-HA) (for PAK-2) and anti-AU1 (for Nef) antibodies (both from BABCO, Richmond, Calif.). Immunoprecipitations were performed with anti-AU1-coupled protein G-Sepharose beads. After washing the immunoprecipitations three times with IVKA lysis buffer and twice with IVKA buffer (50 mM HEPES, [pH 7.4], 5 mM MgCl2), the beads were subjected to an IVKA for 30 min at 30°C. Proteins were separated on sodium dodecyl sulfate (SDS)-8% polyacrylamide gel electrophoresis (PAGE) gels for IVKAs and on 12% gels for Western blot analysis.
Phylogenetic analysis.
For phylogenetic analysis, a protein sequence alignment (235 positions in length) was constructed, including the SIVgsn sequence (15) which was used as an out-group to root the tree. An ambiguous region including indels was deleted from the analysis (positions 23 to 40). Phylogenetic tree construction was done by using the 4.0b10 version of the PAUP* package (D. L. Swofford, Sinauer Assoc., Inc., Sunderland, Mass.) with the factory default settings. Trees were constructed by using the neighbor-joining and maximum-parsimony methods. The reliability of the branching order was analyzed by bootstrap analysis with 1,000 replicates.
Statistical methods.
The mean activities of HIV-1 and SIVcpz nef alleles were compared by using Student's t test. Similar results were obtained with the Mann-Whitney test. The software package StatView, version 4.0 (Abacus Concepts, Berkeley, Calif.), was used for all calculations.
GenBank accession numbers.
The HIV-1 M subtype B 039 nm-94 (AF129351), 168mb-95 (AF129382), 032an93-93 (AF129349), Priso (1) (AF129394), LT-87 (AF129391), FA-93 (AF129388), and AD-93 (AF129383); HIV-1 N YBF30 (CAA06817); and SIVcpz US (AAD17911) and TAN1 (AAO13967) nef sequences have been previously deposited in the GenBank database and can be retrieved by using the accession numbers given in parentheses.
Nucleotide sequence accession number.
Nef sequences derived from HIV-1 M 92UG029 and 93Br029; HIV-1 O Ca9, 13127, 8161, and 2171; HIV-1 N 116; and SIVcpz US, Cam5, Cam3, GAB2, TAN2.2, TAN3.1, Ch-Ni, and Ch-No have been submitted to the GenBank sequence database and have been assigned the accession numbers AY536901 to AY536916.
RESULTS
Characteristics of HIV-1 and SIVcpz Nef sequences.
The origins of the HIV-1 and SIVcpz Nef sequences analyzed are summarized in Table 1. Briefly, nef alleles were derived from 9 SIVcpz-infected chimpanzees, 4 of the P. t. troglodytes subspecies and 5 of the P. t. schweinfurthii subspecies. Nine of the 10 SIVcpz nef alleles were derived from chimpanzees who had been infected in the wild in Cameroon, Gabon, Tanzania, or the Democratic Republic of Congo (Table 1). Our samples represent the majority of cloned SIVcpz strains described to date. Importantly, most SIVcpz nef alleles were obtained directly from uncultured chimpanzee material, such as the spleen (US), PBMC (GAB2 and Ch-Ni), plasma (Ch-No), and fecal material (TAN1, TAN2, and TAN3). Thus, they do not contain changes representing adaptation to human cell culture. Additionally, we characterized the functional activity of 13 HIV-1 M (10, 39), 4 HIV-1 O (18), and 2 HIV-1 N (3, 74) nef alleles. The group M and O nef alleles were each representative of one patient at a single time point. Only nef alleles encoding full-length Nef proteins were included in the analysis. The size of the nef ORFs ranged from 588 bp [SIVcpz(P.t.s.) TAN clones] to 657 bp (HIV-1 O 13127k2) due to length variations near the 5′ end of the nef ORF and a 15- to 18-bp deletion close to the 3′ end of the SIVcpz(P.t.s.) nef. The newly derived Nef sequences from the HIV-1 M, N, and O or SIVcpz (P.t.t.) and SIVcpz(P.t.s.) groups fell into the expected clusters in phylogenetic trees, confirming their authenticities (Fig. 1). This result also excludes the possibility of cross-contamination during PCR amplification.
FIG. 1.
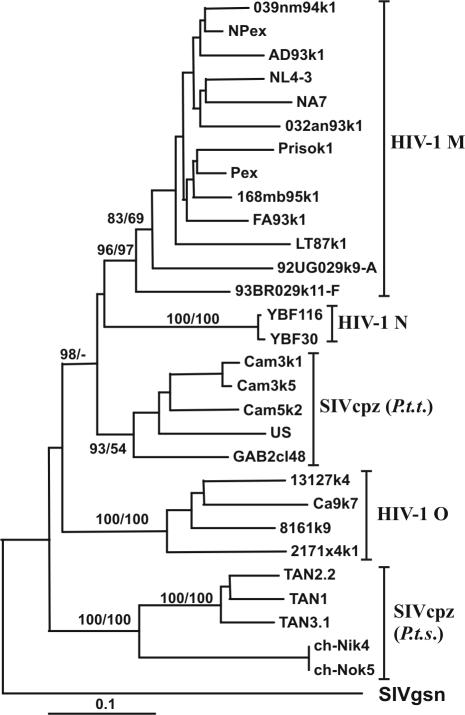
Evolutionary relationships among HIV-1 and SIVcpz Nef sequences. A phylogenetic tree was constructed based on Nef amino acid sequences by the neighbor-joining method. The tree was rooted with the SIVgsn Nef sequence as an out-group. Confidence in tree nodes was determined with 1,000 bootstrap resampling replicates in parallel with neighbor joining and maximum parsimony (confidence values before and after the slash, respectively).
Inspection of an alignment of Nef amino acid sequences revealed some group-specific sequence variations in domains with putative functional relevance (Fig. 2). N-myristoylation of Nef is critical for virtually all in vitro activities and a consensus sequence for the N-myristoyl transferase (MGXXXS6) (62) together with a basic residue at position 7 is almost always conserved in HIV-1 M Nef proteins (22). Unexpectedly, however, Nef proteins obtained from all four HIV-1 O strains and from the three SIVcpz(P.t.s.) TAN strains, derived from three different Tanzanian chimpanzee communities in Gombe, Kalande, and Mitumba, contained changes of S6 to R, T, G, or A (Fig. 2). In contrast, an N-terminal MGXXWSK/R motif in Nef was conserved among the remaining HIV-1 M and N and SIVcpz strains. In comparison, an M residue at position 20 (numbering corresponds to the position in the NL4-3 Nef amino acid sequence), which has been implicated in MHC-I down-modulation (8), was present in all but one (93BR029) HIV-1 Nef but absent in all SIVcpz sequences (Fig. 2). Furthermore, V194 or M194 in HIV-1 Nef sequences was replaced by I or L in most SIVcpz Nef sequences. Thus, the M20 and V/M194 sequence variations in Nef might represent adaptations to the human host.
FIG. 2.
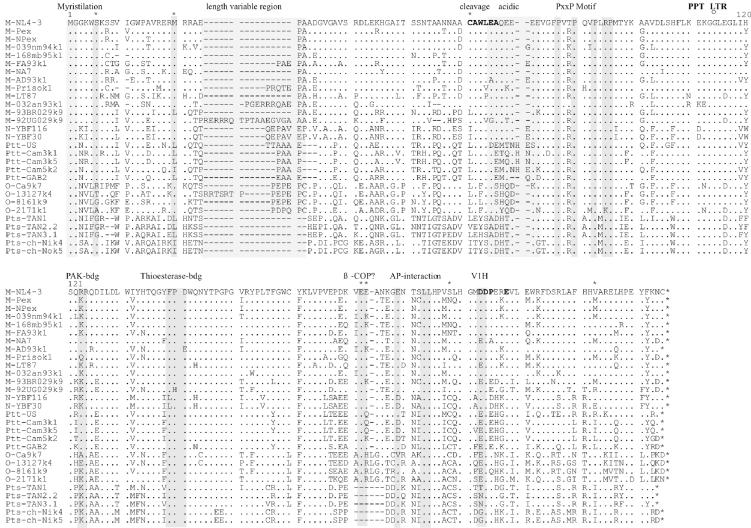
Alignment of HIV-1 and SIVcpz Nef sequences. The NL4-3 sequence is shown in the upper panel for comparison. Some conserved sequence elements in Nef, the position of the polypurine tract (PPT), and the start of the 3′ long terminal repeat (LTR) are indicated schematically. Asterisks above the alignment indicate positions where amino acid variations between the different groups are observed; the number gives the corresponding amino acid position in the NL4-3 Nef. Dots indicate identity with the consensus sequence, and dashes indicate gaps introduced to optimize the alignment. bdg, binding; VIH, catalytic subunit of vacuolar ATPase.
The HIV-1 M Nef protein can be cleaved at CAW↓LEA by the viral protease, although the biological relevance of this process remains unclear (53, 81). Notably, the C50 residue was only conserved in HIV-1 M Nef sequences, and the putative cleavage site was absent in the SIVcpz(P.t.s.) Nef sequences. The acidic region (EEEE65), described to represent a PACS-1 binding site (58), was conserved in HIV-1 M and N Nef sequences but variable in group O and SIVcpz Nef sequences (Fig. 2). In contrast, the proline-rich motif (PXXP)3, representing an SH3 binding site (41, 67), was generally conserved (Fig. 2). Two positively charged residues (K/R105R), which are critical for PAK-2 binding (71), were altered to AR or ER in the HIV-1 O Ca9k7 and 13127k4 Nef sequences (Fig. 2). A previously proposed thioesterase binding site (FPD123) (12, 43) was changed to LFP in the HIV-1 N Nef proteins but was otherwise well conserved. In contrast, a diacidic motif (EE155), which has a controversial role in β-COP binding (36, 57), was frequently changed to EK, ER, or EQ and deleted in the SIVcpz(P.t.s.) Nef proteins (Fig. 2). In comparison, with the single exception of HIV-1 O Ca9, an E/DXXXLL165 motif in Nef that interacts with adaptor protein complexes and is critical for CD4 down-regulation (9, 16, 28) was conserved in all Nef sequences analyzed. All six Nef sequences derived from patient Ca9, however, contained a V instead of an E or D residue in this domain (Fig. 2 and data not shown). A diacidic DD175 or ED175 motif, proposed to bind V1H (44), was present in all but the SIVcpz(P.t.s.) Nef sequences. Interestingly, the C-terminal flexible loop of all SIVcpz(P.t.s.) Nef sequences, which is known to be of great functional relevance (22), contained a deletion of 5 to 6 amino acid residues. Some features distinguished the HIV-1 M from the HIV-1 N and O and SIVcpz Nef sequences. For example, G3 in group M was replaced by K, N, or S residues. Furthermore, the C55 residue in group M Nef sequences was replaced by L or I in the remaining Nef sequences. Conversely, the HIV-1 N and O and SIVcpz Nef sequences contained a C instead of a S or N residue at position 169 (Fig. 2). In summary, the N-terminal myristoylation site, the proline-rich region, and the dileucine motif were well conserved among HIV-1 M, N, and O and SIVcpz Nef sequences, whereas other domains proposed to be important for HIV-1 M Nef function were more variable.
Interaction with PAK-2.
The Nef sequence comparison revealed that some functional protein motifs previously identified in HIV-1 M Nef proteins (22) are also conserved in HIV-1 N and O and SIVcpz(P.t.t.) or SIVcpz(P.t.s.) Nef sequences. However, as described above, sequence variations in the N-terminal myristoylation site, the acidic region, and the flexible C-loop were also detected, suggesting possible functional differences between the different groups of HIV-1 and SIVcpz nef alleles. For further analysis, all nef genes were cloned into a bicistronic vector coexpressing Nef and the GFP from a single bicistronic RNA (27, 72). To study the ability of the nef alleles to associate with the active PAK-2, 293T cells were cotransfected with constructs expressing AU-1-tagged Nef proteins and HA-tagged PAK2-HA. To ensure the presence of active PAK-2, a dominant-active mutant of the p21 GTPase Cdc42 (V12 mutant) was cotransfected. Western blot analysis revealed similar quantities of PAK-2 in all cellular extracts (Fig. 3, upper panel). In comparison, some Nef proteins, e.g., 2171x4k1 (Fig. 3, lane 6) and TAN1 (Fig. 3, lane 11), were only expressed at low levels. The electrophoretic mobility of the different Nef proteins varied between approximately 25 and 30 kDa. Up to three forms of Nef could be detected in some cellular extracts, likely resulting from differences in posttranslational modifications. Next, anti-Nef immune complexes were subjected to IVKAs and separated by SDS-PAGE. All Nef proteins interacted with PAK-2, albeit with different efficiencies (Fig. 3, lower panel). Notably, the quantity of PAK-2 detected did not correlate with the expression levels of the respective Nef proteins. For example, the NL4-3 (lane 2) and GAB2cl48 (lane 8) Nef proteins precipitated less PAK-2 than the 2171x4k1 (lane 6) and ch-Ni2k4 (lane 13) or ch-No5k1 (lane 14) Nef proteins, although they were expressed with higher efficiencies (Fig. 3). The alteration of RR106 to ER in the dibasic motif of the 13127k4 Nef did not impair PAK-2 binding (Fig. 3, lane 5). These results demonstrate that the interaction of Nef with PAK-2 is highly conserved within the SIVcpz-HIV-1 group.
FIG. 3.
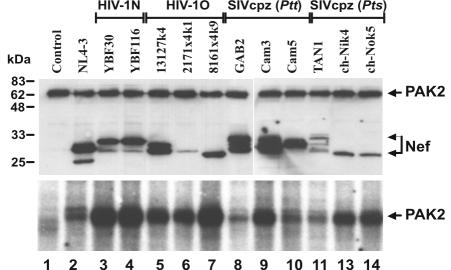
Interaction of HIV-1 and SIVcpz Nef proteins with PAK-2. 293T cells were cotransfected with expression plasmids for HA-tagged PAK-2, Cdc42V12, and the AU1-tagged Nef proteins indicated. An aliquot of the cell lysates was directly separated by SDS-PAGE and analyzed for Nef and PAK-2 expression by Western blot analysis with a mixture of AU-1- and HA-specific antibodies (upper panel). To detect PAK-2 interaction, Nef immunocomplexes were precipitated with anti-AU1 antibody, subjected to IVKAs, and separated by SDS-PAGE (lower panel).
Functional activity of HIV-1 and SIVcpz nef alleles.
Next, we investigated the ability of the HIV-1 and SIVcpz nef alleles to modulate the surface expression of various human cellular receptors. Flow cytometric analysis demonstrated that nef alleles derived from both groups of SIVcpz and all three groups of HIV-1 efficiently down-modulate cell surface expression of CD4 and MHC-I and up-regulate expression of Ii associated with immature MHC-II complexes (examples are shown in Fig. 4). In comparison, the effects of Nef on CD28 and MHC-II surface expression were relatively weak. Unlike SIVmac or HIV-2 nef genes (6, 33, 34), none of the HIV-1 and SIVcpz nef alleles down-modulated CD3 cell surface expression (data not shown). In our experimental system, both Nef and GFP are expressed in transiently transfected cells at a constant ratio, allowing them to readily quantitate the effect of Nef on the surface expression levels of cellular receptors and to assess whether the different groups of SIVcpz and HIV-1 nef alleles show more subtle functional differences. As described previously (10, 72) and indicated in Fig. 4, we defined four areas of green fluorescence representing no, low, medium, and high levels of GFP and, hence, Nef expression. For quantitative fluorescence-activated cell sorting analysis, the mean fluorescence intensity obtained on Jurkat or HeLa CIITA cells transiently transfected with the different nef alleles was divided by the mean fluorescence obtained for cells transfected with a control construct containing inactivating point mutations in nef. As expected from previous studies on HIV-1 M subtype B nef (10), the functional activity varied considerably between different nef alleles (Fig. 5). On average, the HIV-1 M, N, and O and SIVcpz(P.t.t.) groups of nef alleles down-modulated CD4 cell surface expression levels about 16-fold (Fig. 5). The SIVcpz(P.t.s.) nef alleles were significantly less active (10.7 ± 1.2; n = 5; P = 0.023; values give average x-fold down-modulation of CD4 ± standard error of the mean) than the SIVcpz(P.t.t.) nef alleles (16.6 ± 1.2; n = 5). However, three of the four SIVcpz(P.t.t.) nef alleles which efficiently down-modulated CD4 were obtained after short-term passage in human PBMC (Fig. 5). Thus, it remains to be clarified whether primary P. t. troglodyte nef alleles are also usually more active than P. t. schweinfurthii nef alleles in down-modulating CD4. The 1.5-fold functional difference between the SIVcpz(P.t.s.) and HIV-1 M, N, and O nef alleles (15.8 ± 1.7; n = 17) failed to reach significance (P = 0.126) because several HIV-1 nef alleles showed only moderate activity in CD4 down-regulation (Fig. 5). More-detailed analysis revealed, however, that the unpassaged SIVcpz(P.t.t.) and SIVcpz(P.t.s.) nef alleles were significantly less efficient in CD4 down-modulation (11.8 ± 1.3; n = 7; P = 0.005) than those derived from human AIDS patients (18.6 ± 1.4; n = 11) but showed an activity comparable to HIV-1 nef alleles obtained during asymptomatic infection (10.9 ± 2.1; n = 6). This finding is consistent with previous reports showing that Nef-mediated CD4 down-modulation is increased after progression to immunodeficiency (10, 54). Unexpectedly, of the four HIV-1 O Nef proteins analyzed, the Ca9k7 Nef showed the highest activity in down-modulation of CD4 cell surface expression (Fig. 5), although it contained a mutation of E/DXXXLL to VXXXLL in the interaction site with adaptor protein complexes (Fig. 2).
FIG. 4.
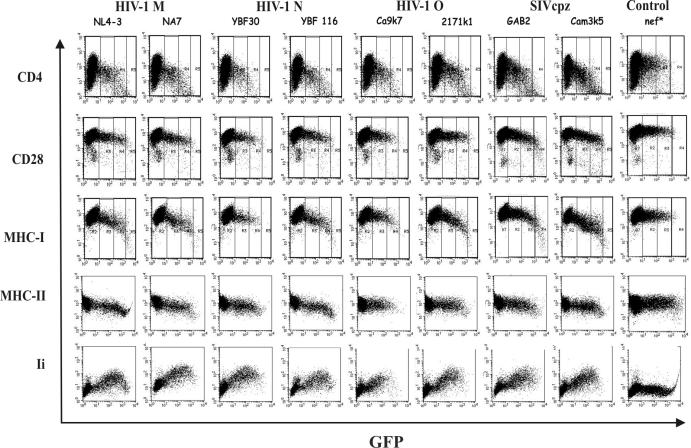
Modulation of human cell surface receptors by nef alleles derived from the HIV-1 M, N, and O groups or SIVcpz(P.t.t.) and SIVcpz(P.t.s.) groups. Jurkat T or HeLa-CIITA cells were transfected with the indicated HIV-1 and SIVcpz Nef expression plasmids and assayed for surface expression of CD4, CD28, MHC-I, MHC-II, and Ii. Quantification was performed as described in Materials and Methods. In the upper three panels, the areas of no, low, medium, and high levels of GFP and, hence, Nef expression (72) are indicated. The results were confirmed in two independent experiments.
FIG. 5.
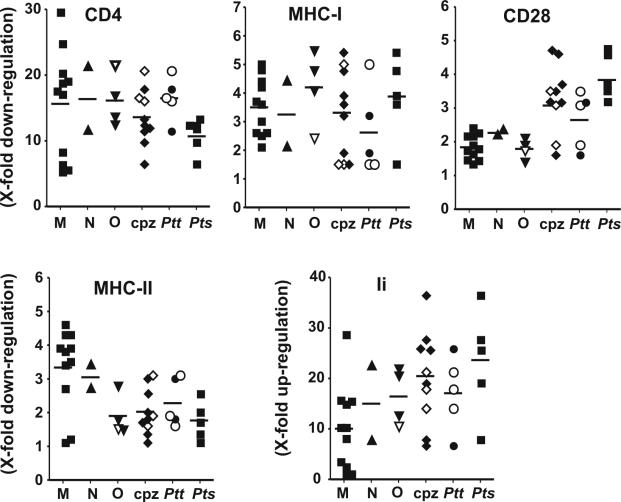
Functional activity of nef alleles derived from different groups of HIV-1 and SIVcpz. For the quantitation of Nef-mediated CD4, CD28, Ii, and MHC-I or -II downregulation, the mean channel numbers of red fluorescence were determined for cells with high levels of GFP and, hence, Nef (72). The numbers obtained for cells transfected with vector expressing GFP only were divided by the corresponding numbers obtained for cells coexpressing Nef and GFP, to calculate the values for down- or up-modulation (n-fold), respectively. Each symbol represents the functional activity of one individual nef allele. The results were confirmed in at least three independent experiments. The horizontal bars indicate average activities. The open triangle in the HIV-1 O panel specifies the Ca9k7 nef allele which contains an E/DXXXLL to VXXXLL mutation in the C-proximal endocytosis signal. The open symbols in the SIVcpz panels specify the three Cam nef alleles that have been obtained after short-term passage of SIVcpz in human PBMC. Please note that different scales are used on the y axes.
The SIVcpz(P.t.t.) and SIVcpz(P.t.s.) and HIV-1 M, N, and O groups of nef alleles did not differ significantly in their ability to down-regulate MHC-I (Fig. 5). This is remarkable because in SIVcpz(P.t.t.) and SIVcpz(P.t.s.) Nef proteins, the M20 residue is changed to L or I and the acidic domain is poorly conserved (Fig. 2). Both M20 and the acidic cluster have recently been implicated in Nef-mediated MHC-I down-regulation (8). Unexpectedly, the SIVcpz nef alleles (3.3 ± 0.3; n = 10) were significantly more active than HIV-1 nef genes (1.9 ± 0.1; n = 17) in down-modulating CD28 (Cpz versus HIV-1, P < 0.0001; Cpz versus HIV-1 M, P = 0.0002) (Fig. 5). CD28 is an important costimulatory factor in TCR-dependent activation of T cells by antigen-presenting cells (APCs). Nef alleles derived from the P. t. troglodytes subspecies of chimpanzees (2.6 ± 0.4; n = 5) were less active than those derived from the P. t. schweinfurthii subspecies (3.9 ± 0.3; n = 5; P = 0.03) but still more effective than HIV-1 M nef alleles (1.8 ± 0.1; n = 11; P = 0.016). It has been previously suggested that down-modulation of CD4 and CD28 are mediated by similar domains in Nef (78). Thus, it is noteworthy that the SIVcpz(P.t.s.) nef alleles showed the lowest functional activity in CD4 down-modulation but the highest activity in down-regulation of CD28 (Fig. 5). Overall, the SIVcpz (2.0 ± 0.2; n = 10) and HIV-1 O (1.9 ± 0.3; n = 4) Nef proteins were 1.7-fold less active than the group M Nef proteins (3.3 ± 0.4; n = 11) in down-modulating MHC-II molecules (P = 0.006 and 0.04, respectively). However, many HIV-1 and SIVcpz nef alleles showed only marginal activity in this assay (Fig. 4 and 5). In contrast, nef alleles from all groups of HIV-1 and SIVcpz dramatically up-regulated Ii cell surface expression (Fig. 5). On average, the HIV-1 M nef genes (10.1 ± 2.5; n = 11) were 2.0-fold less active than the SIVcpz nef alleles (20.2 ± 2.9; n = 10; P = 0.016). Notably, the strength of PAK-2 binding did not correlate with other in vitro Nef functions investigated. We are currently investigating whether this conserved interaction may be critical for other activities, e.g., the ability of Nef to stimulate viral replication in T cells.
Taken together, our findings suggest that the different groups of SIVcpz and HIV-1 nef alleles show subtle but significant functional differences in the ability to modulate CD4, CD28, MHC-II, and Ii cell surface expression. More importantly, however, our results demonstrate that primary SIVcpz nef genes efficiently modulate the surface expression of various human receptors without any adaptation to human cells.
DISCUSSION
In this study, we demonstrated that the ability of Nef to modulate cell surface expression of human CD4, CD28, MHC-I, MHC-II, and Ii molecules and to interact with PAK-2 is conserved among the HIV-1 M, N, and O and SIVcpz(P.t.t.) and SIVcpz(P.t.s.) groups of primate lentiviruses. We have only begun to understand the relevance of these nef activities for viral replication in vivo and for the ability of primate lentiviruses to cause disease in their respective hosts. Studies in long-term nonprogressors of HIV-1 infection and in SIVmac-infected macaques have demonstrated that grossly defective nef genes result in several-log-reduced viral loads and an attenuated clinical course of infection (17, 37, 38). More-recent findings with human individuals infected with HIV-1 subtype B strains and in the SIVmac-macaque model indicate that a combination of Nef activities is relevant for the maintenance of high viral loads and the development of AIDS (10, 19, 35, 49, 50, 54). We found that nef alleles derived from SIVcpz(P.t.t.) and SIVcpz(P.t.s.) strains that have never been passaged in human cells modulate cell surface expression of human CD4, CD28, MHC-I, and Ii molecules with an efficiency comparable to nef genes obtained from pathogenic HIV-1 strains (Fig. 5). These data suggest that SIVcpz nef alleles are capable of reducing cytotoxic T lymphocyte lysis of infected human cells, altering TCR signaling, and affecting MHC-II-restricted antigen presentation without adaptive changes. The finding that SIVcpz and HIV-1 Nef proteins perform similar activities in vitro suggests that both might also have related effects on viral replication in vivo. Concordant with this, preliminary findings indicate that SIVcpz may persist in infected chimpanzees at levels comparable to HIV-1 infection of human individuals (66, 70, 79).
HIV-1 infection causes AIDS in the great majority of infected individuals, whereas HIV-1 or SIVcpz infection of chimpanzees usually does not result in CD4+-T-cell depletion or disease (51, 52, 56, 66). Host, viral, or environmental factors could explain the different outcome of HIV-1 and SIVcpz infection of humans and chimpanzees, respectively. The functional activity of SIVcpz nef alleles in chimpanzee cells and the effect of an intact nef gene on the efficiency of viral replication in this primate species remain to be investigated. Nevertheless, the cellular receptors investigated in our study are well conserved, and it seems highly likely that SIVcpz nef alleles are also capable of manipulating the surface expression of chimpanzee-derived molecules. In conclusion, Nef-mediated modulation of various cellular receptors involved in T-cell activation and antigen presentation most likely contributes to optimal viral fitness in infected apes but is usually not associated with induction of disease. Differences in other Nef functions or other viral genes might exist. Nonetheless, our finding that HIV-1 and SIVcpz nef alleles perform similar activities is concordant with previous studies indicating that viral determinants alone are not sufficient for pathogenicity in vivo but that host factors are also determinative for the absence of clinical symptoms in primates naturally infected with SIV (32).
Consistent with the idea that the host environment is implicated in the clinical outcome, HIV-1 O and N, which infect the same host and for which express nef alleles modulating CD4, MHC-I, CD28, MHC-II, and Ii surface expression comparable to those derived from the HIV-1 M group (Fig. 5), are pathogenic in humans (3, 18, 74). Our preliminary data indicate that group O and N nef alleles are also capable of enhancing virion infectivity and stimulating HIV-1 replication in PBMC cultures (J. Münch, F. Bailer, and N. Kirchhoff, unpublished data), although a larger number of HIV-1 O and, particularly, N nef alleles needs to be analyzed to assess possible quantitative differences. Nevertheless, concordant with previously published results (3, 18, 74), our study on nef functions supports that HIV-1 M, O, and N strains may not reveal major differences in their pathogenic properties.
An interesting aspect of this study is that several sequence motifs thought to be important for Nef function, including domains involved in N-myristoylation, Nef cleavage, trafficking, kinase interaction, or signaling, were mutated in functionally active SIVcpz and HIV-1 O or N Nef proteins. For example, the HIV-1 O Nef proteins missed the S residue at position 6, which has been implicated in membrane targeting and phosphorylation of Nef (22), and the two group N Nef proteins had a mutation in the thioesterase binding site implicated in CD4 down-modulation (12, 43) (Fig. 2). Furthermore, the HIV-1 O Ca9 Nef contains a mutation of E/DXXXLL to VXXXLL in the interaction site of Nef with adaptor protein complexes (9, 16, 28) but was highly active in down-modulating CD4. This result is consistent with the observation that mutation of EXXXLL to AXXXL does not significantly affect Nef-mediated CD4 down-modulation (16). Thus, at least in the context of group N or O Nef proteins, S6 near the N terminus, F121 in the thioesterase binding motif, and the E/D residue in the adaptor protein interaction site are not required for the ability of Nef to modulate surface expression of CD4. Similarly, of the three motifs in Nef that have recently been implicated in MHC-I down-modulation, M20, EEEE65, and P72XXP75 (8, 27, 58), only the SH3 binding site was highly conserved. In all group N, O, and Cpz Nef proteins, the M20 residue was changed to L or I, and frequently, only two negatively charged residues were present in the acidic domain. Nevertheless, these Nef proteins were as active in down-modulating MHC-I as the HIV-1 M Nef proteins containing intact M20, EEEE65, and P72XXP75 residues. It remains to be clarified whether these motifs can still perform their proposed functions and the changes are simply tolerated, whether changes elsewhere in Nef compensate for inactivating mutations in these domains, or whether they are just not crucial for Nef function.
The SIVcpz(P.t.s.) nef alleles differed by several structural and functional aspects from the remaining SIVcpz(P.t.t.) and HIV-1 nef alleles. On average, they showed the lowest activity in CD4 down-regulation but were highly active in down-modulating CD28 and in enhancing surface expression of Ii (Fig. 5). For HIV-1 M, it has been demonstrated that Nef-mediated CD4 down-regulation correlates with the efficiency of viral replication in primary T cells and in human lymphoid tissue ex vivo (2, 25, 40, 45). We are currently investigating whether the relatively low activity of SIVcpz(P.t.s.) nef alleles in CD4 down-modulation also correlates with reduced replicative capacity. The high activity of most SIVcpz nef alleles in CD28 down-modulation and in enhancing Ii expression suggests that they might be particularly active in blocking T-cell activation by APCs. T-cell activation requires two sets of signals: (i) the interaction of B7 on APCs with the CD28 molecule on T-cells and (ii) the interaction of mature peptide MHC-II complexes with the TCR-CD4 complex. Increased Ii surface expression prevents efficient MHC-II-restricted antigen presentation (63). Notably, it has been reported that infected chimpanzees do not develop the high levels of generalized immune activation that infected humans acquire (66). Our ongoing experiments aim to clarify whether SIVcpz Nef proteins are usually more efficient in blocking T-cell activation than HIV-1 Nef proteins. The results might provide new insights into the pathogenesis of SIVcpz and HIV-1 infection.
Sexual contacts are responsible for >95% of all new HIV-1 infections worldwide. However, for a mainly sexually transmitted agent, the efficiency of HIV-1 transmission per intercourse is low (about 0.1% to 0.5%) and seems to correlate with the virus load in the infected individual (26, 59, 65). Nef-defective HIV-1 strains can be transmitted by contaminated blood products (17). However, the low viral loads make sexual transmission highly unlikely. The ability of primary SIVcpz nef alleles to manipulate key factors of human T cells and APCs, as demonstrated in this study, likely enabled the virus to maintain high viral loads in the new host, thereby accelerating the spread of HIV-1 in the human population.
In summary, our results suggest that the nef gene, known to be a key factor of viral pathogenesis, performs similar functions in apathogenic infection of chimpanzees with SIVcpz and in HIV-1-infected AIDS patients. SIV infection has been documented in about 30 African nonhuman primate species (30), and several of these SIV isolates can replicate in human cells (29). Nevertheless, these viruses were apparently only efficiently transmitted into humans from chimpanzees and sooty mangabeys (30). It will be interesting to see whether nef alleles derived from other SIV-infected primates are also capable of manipulating human cells in such a complex way or whether differences in Nef function might represent a barrier for successful cross-species transmission for most of these primate lentiviruses.
Acknowledgments
We thank Thomas Mertens for support and encouragement and Nadja Auer for excellent technical assistance. We are also grateful to Ingrid Bennett for critical reading of the manuscript, Thomas Schmid for help with statistical analysis, and Philippe Benaroch for providing HeLa CIITA cells. We are also indebted to all the patients who participated in the various research cohorts.
This work was supported by the Deutsche Forschungsgemeinschaft (DFG), the Wilhelm-Sander-Stiftung, and the National Institutes of Health (RO1 AI 44596 and RO1 AI50529).
REFERENCES
- 1.Aiken, C., J. Konner, N. R. Landau, M. E. Lenburg, and D. Trono. 1994. Nef induces CD4 endocytosis: requirement for a critical dileucine motif in the membrane-proximal CD4 cytoplasmic domain. Cell 76:853-864. [DOI] [PubMed] [Google Scholar]
- 2.Arganaraz, E. R., M. Schindler, F. Kirchhoff, M. J. Cortes, and J. Lama. 2003. Enhanced CD4 down-modulation by late-stage HIV-1 nef alleles is associated with increased Env incorporation and viral replication. J. Biol. Chem. 278:33912-33919. [DOI] [PubMed] [Google Scholar]
- 3.Ayouba, A., S. Souquieres, B. Njinku, P. M. Martin, M. C. Muller-Trutwin, P. Roques, F. Barre-Sinoussi, P. Mauclere, F. Simon, and E. Nerrienet. 2000. HIV-1 group N among HIV-1-seropositive individuals in Cameroon. AIDS 14:2623-2625. [DOI] [PubMed] [Google Scholar]
- 4.Bailes, E., R. R. Chaudhuri, M. L. Santiago, F. Bibollet-Ruche, B. H. Hahn, and P. M. Sharp. 2002. The evolution of primate lentiviruses and the origins of AIDS, p. 65-96. In T. Leitner (ed.), The molecular epidemiology of human viruses. Kluwer Academic Publishers, Norwell, Mass.
- 5.Bailes, E., F. Gao, F. Bibollet-Ruche, V. Courgnaud, M. Peeters, P. A. Marx, B. H. Hahn, and P. M. Sharp. 2003. Hybrid origin of SIV in chimpanzees. Science 300:1713. [DOI] [PubMed] [Google Scholar]
- 6.Bell, I., C. Ashman, J. Maughan, E. Hooker, F. Cook, and T. A. Reinhart. 1998. Association of simian immunodeficiency virus Nef with the T-cell receptor (TCR) zeta chain leads to TCR down-modulation. J. Gen. Virol. 79:2717-2727. [DOI] [PubMed] [Google Scholar]
- 7.Benson, R. E., A. Sanfridson, J. S. Ottinger, C. Doyle, and B. R. Cullen. 1993. Downregulation of cell-surface CD4 expression by simian immunodeficiency virus nef prevents viral super infection. J. Exp. Med. 177:1561-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blagoveshchenskaya, A. D., L. Thomas, S. F. Feliciangeli, C. H. Hung, and G. Thomas. 2002. HIV-1 Nef downregulates MHC-I by a PACS-1- and PI3K-regulated ARF6 endocytic pathway. Cell 111:853-866. [DOI] [PubMed] [Google Scholar]
- 9.Bresnahan, P. A., W. Yonemoto, and W. C. Greene. 1999. Cutting edge: SIV Nef protein utilizes both leucine- and tyrosine-based protein sorting pathways for down-regulation of CD4. J. Immunol. 163:2977-2981. [PubMed] [Google Scholar]
- 10.Carl, S., T. C. Greenough, M. Krumbiegel, M. Greenberg, J. Skowronski, J. L. Sullivan, and F. Kirchhoff. 2001. Modulation of different human immunodeficiency virus type 1 Nef functions during progression to AIDS. J. Virol. 75:3657-3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chowers, M. Y., C. A. Spina, T. J. Kwoh, N. J. Fitch, D. D. Richman, and J. C. Guatelli. 1994. Optimal infectivity in vitro of human immunodeficiency virus type 1 requires an intact nef gene. J. Virol. 68:2906-2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen, G. B., V. S. Rangan, B. K. Chen, S. Smith, and D. Baltimore. 2000. The human thioesterase II protein binds to a site on HIV-1 Nef critical for CD4 down-regulation. J. Biol. Chem. 275:23097-23105. [DOI] [PubMed] [Google Scholar]
- 13.Collins, K. L., B. K. Chen, S. A. Kalams, B. D. Walker, and D. Baltimore. 1998. HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lympocytes. Nature 391:397-401. [DOI] [PubMed] [Google Scholar]
- 14.Corbet, S., M. C. M. Nuller-Trutwin, P. Versmisse, S. Delarue, A. Ayouba, J. Lewis, S. Brunak, P. Martin, F. Brun-Vezinet, F. Simon, F. Barre-Sinoussi, and P. Mauclere. 2000. Env sequences of simian immunodeficiency viruses from chimpanzees in Cameroon are strongly related to those of human immunodeficiency virus group N from the same geographic area. J. Virol. 74:529-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Courgnaud, V., M. Salemi, X. Pourrut, E. Mpoudi-Ngole, B. Abela, P. Auzel, F. Bibollet-Ruche, B. Hahn, A. M. Vandamme, E. Delaporte, and M. Peeters. 2002. Characterization of a novel simian immunodeficiency virus with a vpu gene from greater spot-nosed monkeys (Cercopithecus nictitans) provides new insights into simian/human immunodeficiency virus phylogeny. J. Virol. 76:8298-8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Craig, H. M., M. W. Pandori, and J. C. Guatelli. 1998. Interaction of HIV-1 Nef with the cellular dileucine-based sorting pathway is required for CD4 down-regulation and optimal viral infectivity. Proc. Natl. Acad. Sci. USA 95:11229-11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deacon, N. J., A. Tsykin, A. Solomon, K. Smith, M. Ludford-Menting, D. J. Hooker, D. A. Mc Phee, A. L. Greenway, A. Ellett, and C. Chatfield. 1995. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science 270:988-991. [DOI] [PubMed] [Google Scholar]
- 18.Dittmar, M. T., L. Zekeng, L. Kaptue, J. Eberle, H. G. Krausslich, and L. Gurtler. 1999. Coreceptor requirements of primary HIV type 1 group O isolates from Cameroon. AIDS Res. Hum. Retrovir. 15:707-712. [DOI] [PubMed] [Google Scholar]
- 19.Du, Z., S. M. Lang, V. G. Sasseville, A. A. Lackner, P. O. Ilyinskii, M. D. Daniel, J. U. Jung, and R. C. Desrosiers. 1995. Identification of a nef allele that causes lymphocyte activation and acute disease in macaque monkeys. Cell 82:665-674. [DOI] [PubMed] [Google Scholar]
- 20.Gao, F., E. Bailes, D. L. Robertson, Y. Chen, C. M. Rodenburg, S. F. Michael, L. B. Cummins, L. O. Arthur, M. Peeters, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436-441. [DOI] [PubMed] [Google Scholar]
- 21.Garcia, J. V., and A. D. Miller. 1991. Serine phosphorylation-independent downregulation of cell-surface CD4 by nef. Nature 350:508-511. [DOI] [PubMed] [Google Scholar]
- 22.Geyer, M., O. T. Fackler, and B. M. Peterlin. 2001. Structure-function relationships in HIV-1 Nef. EMBO Rep. 2:580-585.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gibbs, J. S., A. A. Lackner, S. M. Lang, M. A. Simon, P. K. Sehgal, M. D. Daniel, and R. C. Desrosiers. 1995. Progression to AIDS in the absence of genes for vpr or vpx. J. Virol. 69:2378-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glushakova, S., J. C. Grivel, K. Suryanarayana, P. Meylan, J. D. Lifson, R. C. Desrosiers, and L. Margolis. 1999. Nef enhances human immunodeficiency virus replication and responsiveness to interleukin-2 in human lymphoid tissue ex vivo. J. Virol. 73:3968-3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glushakova, S., J. Münch, S. Carl, T C. Greenough, J. L. Sullivan, L. Margolis, and F. Kirchhoff. 2001. CD4 down-modulation by human immunodeficiency virus type 1 Nef correlates with the efficiency of viral replication and with CD4(+) T-cell depletion in human lymphoid tissue ex vivo. J. Virol. 75:10113-10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gray, R. H., M. J. Wawer, R. Brookmeyer, N. K. Sewankambo, D. Serwadda, F. Wabwire-Mangen, T. Lutalo, X. Li, T. vanCott, T. C. Quinn, and the Rakai Project Team. 2001. Probability of HIV-1 transmission per coital act in monogamous, heterosexual, HIV-1-discordant couples in Rakai, Uganda. Lancet 357:1149-1153. [DOI] [PubMed] [Google Scholar]
- 27.Greenberg, M. E., A. J. Iafrate, and J. Skowronski. 1998. The SH3 domain-binding surface and an acidic motif in HIV-1 Nef regulate trafficking of class I MHC complexes. EMBO J. 17:2777-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenberg, M. E., L. DeTulleo, I. Rapoport, J. Skowronski, and T. Kirchhausen. 1998. A dileucine motif in HIV-1 Nef is essential for sorting into clathrin-coated pits and for downregulation of CD4. Curr. Biol. 8:1239-1242. [DOI] [PubMed] [Google Scholar]
- 29.Grimm, T. A., B. E. Beer, V. M. Hirsch, and K. A. Clouse. 2003. Simian immunodeficiency viruses from multiple lineages infect human macrophages: implications for cross-species transmission. J. Acquir. Immune Defic. Syndr. 32:362-369. [DOI] [PubMed] [Google Scholar]
- 30.Hahn, B. H., G. M. Shaw, K. M. De Cock, and P. M. Sharp. 2000. AIDS as a zoonosis: scientific and public health implications. Science 287:607-614. [DOI] [PubMed] [Google Scholar]
- 31.Heeney, J. L. 1995. AIDS: a disease of impaired Th-cell renewal? Immunol. Today 16:515-520. [DOI] [PubMed] [Google Scholar]
- 32.Hirsch, V. M., G. Dapolito, P. R. Johnson, W. R. Elkins, W. T. London, R. J. Montali, S. Goldstein, and C. Brown. 1995. Induction of AIDS by simian immunodeficiency virus from an African green monkey: species-specific variation in pathogenicity correlates with the extent of in vivo replication. J. Virol. 69:955-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Howe, A. Y., J. U. Jung, and R. C. Desrosiers. 1998. Zeta chain of the T-cell receptor interacts with nef of simian immunodeficiency virus and human immunodeficiency virus type 2. J. Virol. 72:9827-9834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iafrate, A. J., S. Bronson, and J. Skowronski. 1997. Separable functions of Nef disrupt two aspects of T cell receptor machinery: CD4 expression and CD3 signaling. EMBO J. 16:673-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iafrate, A. J., S. Carl, S. Bronson, C. Stahl-Hennig, T. Swigut, J. Skowronski, and F. Kirchhoff. 2000. Disrupting surfaces of Nef required for down-regulation of CD4 and for enhancement of virion infectivity attenuates simian immunodeficiency virus replication in vivo. J. Virol. 74:9836-9844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janvier, K., H. Craig, S. Le Gall, R. Benarous, J. Guatelli, O. Schwartz, and S. Benichou. 2001. Nef-induced CD4 downregulation: a diacidic sequence in human immunodeficiency virus type 1 Nef does not function as a protein sorting motif through direct binding to beta-COP. J. Virol. 75:3971-3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kestler, H. W., D. J. Ringler, K. Mori, D. L. Panicali, P. K. Sehgal, M. D. Daniel, and R. C. Desrosiers. 1991. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell 65:651-662. [DOI] [PubMed] [Google Scholar]
- 38.Kirchhoff, F., T. C. Greenough, D. B. Brettler, J. L. Sullivan, and R. C. Desrosiers. 1995. Absence of intact nef sequences in a long-term, nonprogressing survivor of HIV-1 infection. N. Engl. J. Med. 332:228-232. [DOI] [PubMed] [Google Scholar]
- 39.Kirchhoff, F., P. J. Easterbrook, N. Douglas, M. Troop, T. C. Greenough, J. Weber, S. Carl, J. L. Sullivan, and R. D. Daniels. 1999. Sequence variations in human immunodeficiency virus type 1 Nef are associated with different stages of disease. J. Virol. 73:5497-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lama, J., A. Mangasarian, and D. Trono. 1999. Cell-surface expression of CD4 reduces HIV-1 infectivity by blocking env incorporation in a Nef- and Vpu-inhibitable manner. Curr. Opin. Cell Biol. 9:622-631. [DOI] [PubMed] [Google Scholar]
- 41.Lee, C. H., K. Saksela, U. A. Mirza, B. T. Chait, and J. Kuriyan. 1996. Crystal structure of the conserved core of HIV 1 Nef complexed with a Src family SH3 domain. Cell 85:931-942. [DOI] [PubMed] [Google Scholar]
- 42.Le Gall, S., M. C. Prevost, J. M. Heard, and O. Schwartz. 1997. Human immunodeficiency virus type I Nef independently affects virion incorporation of major histocompatibility complex class I molecules and virus infectivity. Virology 229:295-301. [DOI] [PubMed] [Google Scholar]
- 43.Liu, L. X., N. Heveker, O. T. Fackler, S. Arold, S. Le Gall, K. Janvier, P. M. Peterlin, C. Dumas, O. Schwartz, S. Benichou, and R. Benarous. 2000. Mutation of a conserved residue (D123) required for oligomerization of human immunodeficiency virus type 1 Nef protein abolishes interaction with human thioesterase and results in impairment of Nef biological functions. J. Virol. 74:5310-5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu, X., H. Yu, S. H. Liu, F. M. Brodsky, and B. M. Peterlin. 1998. Interactions between HIV1 Nef and vacuolar ATPase facilitate the internalization of CD4. Immunity 8:647-656. [DOI] [PubMed] [Google Scholar]
- 45.Lundquist, C. A., M. Tobiume, J. Zhou, D. Unutmaz, and C. Aiken. 2002. Nef-mediated downregulation of CD4 enhances human immunodeficiency virus type 1 replication in primary T lymphocytes. J. Virol. 76:4625-4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manninen, A., M. Hiipakka, M. Vihinen, W. Lu, B. J. Mayer, and K. Saksela. 1998. SH3-domain binding function of HIV-1 Nef is required for association with a PAK-related kinase. Virology 250:273-282. [DOI] [PubMed] [Google Scholar]
- 47.Miller, M. D., M. T. Warmerdam, I. Gaston, W. C. Greene, and M. B. Feinberg. 1994. The human immunodeficiency virus-1 nef gene product: a positive factor for viral infection and replication in primary lymphocytes and macrophages. J. Exp. Med. 179:101-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muller-Trutwin, M. C., S. Corbet, S. Souquiere, P. Roques, P. Versmisse, A. Ayouba, S. Delarue, E. Nerrienet, J. Lewis, P. Martin, F. Simon, F. Barre-Sinoussi, and P. Mauclere. 2000. SIVcpz from a naturally infected Cameroonian chimpanzee: biological and genetic comparison with HIV-1 N. J. Med. Primatol. 29:166-172. [DOI] [PubMed] [Google Scholar]
- 49.Münch, J., N. Stolte, D. Fuchs, C. Stahl-Hennig, and F. Kirchhoff. 2001. Efficient class I MHC down-regulation by simian immunodeficiency virus Nef is associated with a strong selective advantage in infected rhesus macaques. J. Virol. 75:10532-10536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Münch, J., A. Janardhan, N. Stolte, C. Stahl-Hennig, P. Ten Haaft, J. L. Heeney, T. Swigut, F. Kirchhoff, and J. Skowronski. 2002. T-cell receptor:CD3 down-regulation is a selected in vivo function of simian immunodeficiency virus Nef but is not sufficient for effective viral replication in rhesus macaques. J. Virol. 76:12360-12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Novembre, F. J., J. de Rosayro, S. Nidtha, S. P. O'Neil, T. R. Gibson, T. Evans-Strickfaden, C. E. Hart, and H. M. McClure. 2001. Rapid CD4(+) T-cell loss induced by HIV-1 (NC) in uninfected and previously infected chimpanzees. J. Virol. 75:1533-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O'Neil, S. P., F. J. Novembre, A. B. Hill, C. Suwyn, C. E. Hart, T. Evans-Strickfaden, D. C. Anderson, J. deRosayro, J. G. Herndon, M. Saucier, and H. M. McClure. 2000. Progressive infection in a subset of HIV-1-positive chimpanzees. J. Infect. Dis. 182:1051-1062. [DOI] [PubMed] [Google Scholar]
- 53.Pandori, M. W., N. J. Fitch, H. M. Craig, D. D. Richman, C. A. Spina, and J. C. Guatelli. 1996. Producer-cell modification of human immunodeficiency virus type 1: Nef is a virion protein. J. Virol. 70:4283-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Patel, P. G., M. T. Yu Kimata, J. E. Biggins, J. M. Wilson, and J. T. Kimata. 2002. Highly pathogenic simian immunodeficiency virus mne variants that emerge during the course of infection evolve enhanced infectivity and the ability to downregulate CD4 but not class I major histocompatibility complex antigens. J. Virol. 76:6425-6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peeters, M., K. Fransen, E. Delaporte, M. Van den Haesevelde, G. M. Gershy-Damet, L. Kestens, G. van der Groen, and P. Piot. 1992. Isolation and characterization of a new chimpanzee lentivirus (simian immunodeficiency virus isolate cpz-ant) from a wild-captured chimpanzee. AIDS 6:447-451. [DOI] [PubMed] [Google Scholar]
- 56.Peeters, M., W. Janssens, M. Vanden Haesevelde, K. Fransen, B. Willems, L. Heyndrickx, L. Kestens, P. Piot, G. Van der Groen, and J. L. Heeney. 1995. Virologic and serologic characteristics of a natural chimpanzee lentivirus infection. Virology 211:312-315. [DOI] [PubMed] [Google Scholar]
- 57.Piguet, V., F. Gu, M. Foti, N. Demaurex, J. Gruenberg, J. L. Carpentier, and D. Trono. 1999. Nef-induced CD4 degradation: a diacidic-based motif in Nef functions as a lysosomal targeting signal through the binding of beta-COP in endosomes. Cell 97:63-73. [DOI] [PubMed] [Google Scholar]
- 58.Piguet, V., L. Wan, C. Borel, A. Mangasarian, N. Demaurex, G. Thomas, and D. Trono. 2000. HIV-1 Nef protein binds to the cellular protein PACS-1 to downregulate class I major histocompatibility complexes. Nat. Cell Biol. 2:163-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quinn, T. C., M. J. Wawer, N. Sewankambo, D. Serwadda, C. Li, F. Wabwire-Mangen, M. O. Meehan, T. Lutalo, and R. H. Gray. 2000. Viral load and heterosexual transmission of human immunodeficiency virus type 1. Rakai Project Study Group. N. Engl. J. Med. 342:921-929. [DOI] [PubMed] [Google Scholar]
- 60.Renkema, G. H., A. Manninen, D. A. Mann, M. Harris, and K. Saksela. 1999. Identification of the Nef-associated kinase as p21-activated kinase 2. Curr. Biol. 9:1407-1410. [DOI] [PubMed] [Google Scholar]
- 61.Renkema, G. H., and K. Saksela. 2000. Interactions of HIV-1 NEF with cellular signal transducing proteins. Front. Biosci. 5:268-283. [DOI] [PubMed] [Google Scholar]
- 62.Resh, M. D. 1999. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim. Biophys. Acta 1451:1-16. [DOI] [PubMed] [Google Scholar]
- 63.Roche, P. A., C. L. Teletski, D. R. Karp, V. Pinet, O. Bakke, and E. O. Long. 1992. Stable surface expression of invariant chain prevents peptide presentation by HLA-DR. EMBO J. 11:2841-2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ross, T. M., A. E. Oran, and B. R. Cullen. 1999. Inhibition of HIV-1 progeny virion release by cell-surface CD4 is relieved by expression of the viral Nef protein. Curr. Opin. Cell Biol. 9:613-621. [DOI] [PubMed] [Google Scholar]
- 65.Royce, R. A., A. Sena, W. Cates, Jr., and M. S. Cohen. 1997. Sexual transmission of HIV. N. Engl. J. Med. 336:1072-1078. [DOI] [PubMed] [Google Scholar]
- 66.Rutjens, E., S. Balla-Jhagjhoorsingh, E. Verschoor, W. Bogers, G. Koopman, and J. L. Heeney. 2003. Lentivirus infections and mechanisms of disease resistance in chimpanzees. Front. Biosci. 8:1134-1145. [DOI] [PubMed] [Google Scholar]
- 67.Saksela, K., G. Cheng, and D. Baltimore. 1995. Proline-rich (PxxP) motifs in HIV-1 Nef bind to SH3 domains of a subset of Src kinases and are required for the enhanced growth of Nef+ viruses but not for down-regulation of CD4. EMBO J. 14:484-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Santiago, M. L., C. M. Rodenburg, S. Kamenya, F. Bibollet-Ruche, F. Gao, E. Bailes, S. Meleth, S. J. Soong, J. M. Kilby, Z. Moldoveanu, B. Fahey, M. N. Muller, A. Ayouba, E. Nerrienet, H. M. McClure, J. L. Heeney, A. E. Pusey, D. A. Collins, C. Boesch, R. W. Wrangham, J. Goodall, P. M. Sharp, G. M. Shaw, and B. H. Hahn. 2002. SIVcpz in wild chimpanzees. Science 295:465. [DOI] [PubMed] [Google Scholar]
- 69.Santiago, M. L., F. Bibollet-Ruche, E. Bailes, S. Kamenya, M. N. Muller, M. Lukasik, A. E. Pusey, D. A. Collins, R. W. Wrangham, J. Goodall, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 2003. Amplification of a complete simian immunodeficiency virus genome from fecal RNA of a wild chimpanzee. J. Virol. 77:2233-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Santiago, M. L., M. Lukasik, S. Kamenya, Y. Li, F. Bibollet-Ruche, E. Bailes, M. N. Muller, M. Emery, D. A. Goldenberg, J. S. Lwanga, A. Ayouba, E. Nerrienet, H. M. McClure, J. L. Heeney, D. P. Watts, A. E. Pusey, D. A. Collins, R. W. Wrangham, J. Goodall, J. F. Brookfield, P. M. Sharp, G. M. Shaw, and B. H. Hahn. 2003. Foci of endemic simian immunodeficiency virus infection in wild-living eastern chimpanzees (Pan troglodytes schweinfurthii). J. Virol. 77:7545-7562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sawai, E. T., A. S. Baur, B. M. Peterlin, J. A. Levy, and C. Cheng-Mayer. 1995. A conserved domain and membrane targeting of Nef from HIV and SIV are required for association with a cellular serine kinase activity. J. Biol. Chem. 270:15307-15314. [DOI] [PubMed] [Google Scholar]
- 72.Schindler, M., S. Wuerfl, P. Benaroch, T. C. Greenough, R. Daniels, P. Easterbrook, M. Brenner, J. Münch, and F. Kirchhoff. 2003. Down-modulation of mature MHC class II and up-regulation of invariant chain cell surface expression are well conserved functions of human and simian immunodeficiency virus nef alleles. J. Virol. 77:10548-10556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schwartz, O., V. Marechal, S. Le Gall, F. Lemonnier, and J. M. Heard. 1996. Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 Nef protein. Nat. Med. 2:338-342. [DOI] [PubMed] [Google Scholar]
- 74.Simon, F., P. Mauclere, P. Roques, I. Loussert-Ajaka, M. C. Muller-Trutwin, S. Saragosti, M. C. Georges-Courbot, F. Barre-Sinoussi, and F. Brun-Vezinet. 1998. Identification of a new human immunodeficiency virus type 1 distinct from group M and group O. Nat. Med. 4:1032-1037. [DOI] [PubMed] [Google Scholar]
- 75.Skowronski, J., M. E. Greenberg, M. Lock, R. Mariani, S. Salghetti, T. Swigut, and A. J. Iafrate. 1999. HIV and SIV Nef modulate signal transduction and protein sorting in T cells. Cold Spring Harb. Symp. Quant. Biol. 64:453-663. [DOI] [PubMed] [Google Scholar]
- 76.Spina, C. A., T. J. Kwoh, M. Y. Chowers, J. C. Guatelli, and D. D. Richman. 1994. The importance of nef in the induction of human immunodeficiency virus type 1 replication from primary quiescent CD4 lymphocytes. J. Exp. Med. 179: 115-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stumptner-Cuvelette, P., S. Morchoisne, M. Dugast, S. Le Gall, G. Raposo, O. Schwartz, and P. Benaroch. 2001. HIV-1 Nef impairs MHC class II antigen presentation and surface expression. Proc. Natl. Acad. Sci. USA 98:12144-12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Swigut, T., N. Shody, and J. Skowronski. 2001. Mechanism for down-regulation of CD28 by Nef. EMBO J. 20:1593-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.ten Haaft, P., K. Murthy, M. Salas, H. McClure, R. Dubbes, W. Koornstra, H. Niphuis, D. Davis, G. van der Groen, and J. L. Heeney. 2001. Differences in early virus loads with different phenotypic variants of HIV-1 and SIV(cpz) in chimpanzees. AIDS 15:2085-2092. [DOI] [PubMed] [Google Scholar]
- 80.Vanden Haesevelde, M. M., M. Peeters, G. Jannes, W. Janssens, G. van der Groen, P. M. Sharp, and E. Saman. 1996. Sequence analysis of a highly divergent HIV-1-related lentivirus isolated from a wild captured chimpanzee. Virology 221:346.350. [DOI] [PubMed] [Google Scholar]
- 81.Welker, R., H. Kottler, H. R. Kalbitzer, and H. G. Krausslich. 1996. Human immunodeficiency virus type 1 Nef protein is incorporated into virus particles and specifically cleaved by the viral proteinase. Virology 219:228-236. [DOI] [PubMed] [Google Scholar]


