Abstract
Coxsackievirus A9 (CAV9), a member of the Enterovirus genus of Picornaviridae, is a common human pathogen and is one of a significant number of viruses containing a functional arginine-glycine-aspartic acid (RGD) motif in one of their capsid proteins. Previous studies identified the RGD-recognizing integrin αvβ3 as its cellular receptor. However, integrin αvβ6 has been shown to be an efficient receptor for another RGD-containing picornavirus, foot-and-mouth disease virus (FMDV). In view of the similarity in sequence context of the RGD motifs in CAV9 and FMDV, we investigated whether αvβ6 can also serve as a receptor for CAV9. We found that CAV9 can bind to purified αvβ6 and also to SW480 cells transfected with β6 cDNA, allowing expression of αvβ6 on their surface, but it cannot bind to mock-transfected cells. In addition, a higher yield of CAV9 was obtained in β6-expressing cells than in mock-transfected cells. There was no similar enhancement in infection with an RGD-less CAV9 mutant. We also found β6 on the surface of GMK cells, a cell line which CAV9 infects efficiently by an RGD-dependent mechanism. Significantly, this infection is blocked by an antibody to αvβ6, while this antibody did not block the low level of infection by the RGD-less mutant. Thus, integrin αvβ6 is an RGD-dependent receptor for CAV9 and may be important in natural CAV9 infections.
Coxsackievirus A9 (CAV9), a member of the Enterovirus genus of the Picornaviridae family, is a common human pathogen that causes central nervous system infections and myocarditis as well as a range of milder illnesses (11, 43). The picornavirus particle is about 28 nm in diameter and consists of a naked capsid with icosahedral symmetry, surrounding a positive sense RNA genome of around 7,100 to 8,500 nucleotides (41). The capsid is made up of 60 copies of each of four proteins, VP1 to VP4, and interacts with receptors during the early stages of infection. A number of picornavirus receptors have been identified, and they include several integrins (9). Integrins are named for their role in integrating the intracellular cytoskeleton with the extracellular matrix and participate in a number of cell-cell, cell-matrix interactions. They are heterodimeric proteins with one α and one β subunit, and each contains an exodomain, the site of interaction with ligands, a transmembrane region, and a cytoplasmic domain. The 8 known β subunits and 14 known α subunits combine to give at least 21 different heterodimeric combinations (16, 38, 40).
The arginine-glycine-aspartic acid (RGD) motif was the first integrin-binding sequence to be identified (38) and is recognized by several integrins (α5β1, α8β1, αIIbβ3, αvβ1, αvβ3, αvβ5, αvβ6 and αvβ8). Picornaviruses from three diverse genera are known to contain an RGD motif that is involved in cell entry: foot-and-mouth disease viruses (FMDV) (Aphthovirus) (26), human parechoviruses (Parechovirus) (5, 10, 17, 42); echovirus 9 (E9), and CAV9 (both Enterovirus) (6, 36, 51). In the case of FMDV it is located in the prominent GH loop of VP1, whereas in the other viruses it is located close to the VP1 C terminus (14). Essentially all natural isolates of these viruses contain the RGD motif (7, 21, 22, 32, 39, 52), although laboratory-adapted or genetically engineered strains lacking the motif can be recovered and may grow well in some cell lines, indicating that alternative, RGD-independent entry pathways are being used (2, 8, 15, 24, 50). For instance, a series of CAV9 deletion or substitution mutants lacking the RGD motif grow in several cell lines, such as GMK and A549, albeit inefficiently (13, 15). The mutants, including D4, which has a precise deletion of the RGD tripeptide, have unimpaired growth in RD cells, showing that this RGD-independent infection can be efficient in some cell lines. The RGD-independent receptor(s) has not been identified, and it is not known if the same molecule is used in these different cell lines.
One known RGD-recognizing integrin, αvβ3, is reported to be involved in cell entry of E9, CAV9, FMDV, and parechoviruses (3, 18, 23, 32-35, 37, 44-46), but other species of integrins have also been implicated in infection by the latter two (19-21, 23, 35). Researchers have previously identified amino acid sequence identity between the residues flanking the RGD motif of CAV9 and part of latent transforming growth factor β1 (TGF-β1), later shown to be a ligand for integrin αvβ6 (7, 31). Moreover, FMDV has been shown to recognize αvβ6, and this picornavirus shares the same consensus sequence, RGDM/LXXL, seen in CAV9 isolates (27).
These observations prompted us to investigate the potential use of integrin αvβ6 as a receptor by CAV9. Here we show that CAV9 binds to purified αvβ6 and to αv-containing cells transfected with the β6 subunit. In addition, the presence of the β6 chain greatly enhances susceptibility of the cells to CAV9 infection but not to infection by the RGD-less mutant D4, while antibody to αvβ6 efficiently blocks infection in GMK cells. Thus, αvβ6 is a functional RGD-dependent CAV9 receptor.
MATERIALS AND METHODS
Virus, cell lines, and media.
CAV9 (Griggs strain) was purified by sucrose gradient centrifugation as previously described (6). RD cells were maintained in minimal essential medium (MEM) containing 10% fetal calf serum (FCS), 2% vitamins, 2% nonessential amino acids, and 100 μg of gentamicin/ml. GMK cells were maintained in MEM containing 10% heat-inactivated FCS, 1% nonessential amino acids, and 100 μg of gentamicin/ml. The human colon carcinoma cell line, SW480, transfected to express either the full-length human β6- or human β3-integrin subunit and the mock-transfected cells (49), were cultivated in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% FCS (heat inactivated), 2 mM glutamine, penicillin (100 U/ml), streptomycin (100 μg/ml), and 1 mg of Geneticin (Invitrogen)/ml. CHO cells expressing soluble human αvβ6 (49) were grown in DMEM supplemented with 10% FCS, penicillin (100 U/ml), and streptomycin (100 μg/ml).
αvβ6 purification.
Soluble human αvβ6 was produced from transfected CHO cells as described previously (49). Briefly, the cells were grown in roller bottles, the medium was changed to serum-less DMEM, and the cells were incubated for 24 h at 37°C. The medium was then collected, clarified by centrifugation, filtered, and concentrated by Centricon Plus-80 (Amicon). The integrin was bound to RGD-Sepharose, washed with Tris-buffered saline (TBS) (pH 7.4) supplemented with 1 mM MgCl2, 1 mM MnCl2, and 2 mM CaCl2, and eluted with TBS containing 10 mM EDTA by overnight incubation. The yield was usually 5 to 20 μg per roller bottle. The identity of the isolated material was confirmed by blotting, performed as described previously (49). The αv chain was recognized by using a polyclonal antibody to αvβ3 (AGK4; a kind gift from Merja Roivainen) and a specific αv monoclonal (L230; American Type Culture Collection), and the β6 chain was recognized by a β6-specific monoclonal antibody (clone CSβ6, MAB2076Z; Chemicon). No signal was seen when the blots were probed with specific β3 or αvβ3 monoclonal antibodies (clone B3A, MAB2023Z, and clone LM609, MAB1976Z, respectively; Chemicon) (data not shown). The αvβ6 preparation was ca. 50 to 70% pure with some contaminating smaller-molecular-size (ca. 60 to 50 kDa on nonreducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis) impurities (data not shown).
In vitro binding of CAV9 to purified integrins.
αvβ3 and αvβ6 binding assays were performed on Maxisorb 96-well plates (Nunc, Inc.). These were coated with approximately 300 ng of αvβ6/well, purified as described above (quantified by spectroscopy), or with αvβ3 (Chemicon) by incubating overnight at 4°C in 150 μl of TBS (pH 7.4) containing 1 mM MgCl2, 1 mM MnCl2, and 2 mM CaCl2. The wells were then blocked with 100 μl of the coating buffer supplemented with 2% bovine serum albumin (BSA) for 2 h at room temperature. For virus binding, different amounts of purified CAV9 were applied to the wells and were incubated for 1 h. All binding and inhibition experiments were done in 100 μl, and for binding the coating TBS buffer was supplemented with 0.1% BSA. Between steps the wells were washed three times with this buffer. Binding was detected by using a 1:500 dilution of rabbit antiserum against CAV9 and mouse anti-rabbit immunoglobulin G horseradish peroxidase-conjugated secondary antibody (1:1,000 dilution; DAKO). Substrate conversion by horseradish peroxidase was detected at 492 nm after 30 min of incubation in the dark at room temperature, and the reaction was stopped by the addition of 200 μl of 1 M H2SO4. To study peptide blocking of αvβ3-CAV9 binding, 90 μl of binding buffer containing the peptide was mixed with 10 μl of the virus and was incubated with the immobilized integrin. To enhance the poor peptide blocking effect on αvβ6-CAV9 binding, the integrin was preincubated for 30 min with peptide in a volume of 90 μl, followed by the addition of 10 μl of virus and 1 h of incubation.
Flow cytometry analysis.
Flow cytometry was performed as described previously (20). Briefly, cells were harvested with EDTA and were resuspended at ∼1 × 107 cells per ml in TBS, pH 7.4, containing 1 mM CaCl2, 0.5 mM MgCl2, 2% normal goat serum, and 3% BSA (buffer A). Cells (30 μl) were incubated sequentially with primary antibodies (10 μg of cell suspension/ml in buffer A) on ice for 30 min followed by secondary antibodies conjugated with R-phycoerythrin (Southern Biotechnology Associates). The primary antibodies were against β6, clone CSβ6 (MAB2076Z; Chemicon); αvβ3, clone LM609 (MAB1976Z; Chemicon); αvβ5, clone P1F6 (MAB1961Z; Chemicon); and α5β1, SAM-1 (Serotec). The cells were then washed and resuspended in 1% paraformaldehyde in phosphate-buffered saline. Background fluorescence was determined in the absence of the primary antibody. Fluorescent staining was analyzed by flow cytometry using a FACSCalibur (Becton Dickinson) counting 10,000 cells per sample.
Virus binding assay.
Cells, prepared in buffer A as described above (supplemented with 1 mM MnCl2), were incubated with purified CAV9 (5 μg/ml) for 1 h on ice, followed by addition of the anti-CAV9 monoclonal (MAB947; Chemicon) and a goat anti-mouse immunoglobulin G2b-specific, R-phycoerythrin conjugate. All steps were carried out in the presence of 1 mM manganese. Background fluorescence was determined under two conditions: in the absence of the virus and in the absence of the anti-CAV9 monoclonal. Both background conditions gave near-identical results.
Plaque reduction assay using purified soluble αvβ6 integrin.
GMK and RD cells, grown on 6-well plates, were infected with 100 μl of 2 × 102 PFU of CAV9 preincubated with serial dilutions of the soluble purified αvβ6 integrin for 30 min at 37°C. The cells were washed with Hank's solution and were covered with 0.5% carboxymethyl cellulose in MEM supplemented with 1% FCS, glutamine, penicillin, and streptomycin. The cells were incubated for 2 days and were stained with crystal violet prior to counting plaques.
Virus blocking assays using anti-αvβ6 monoclonal antibody.
RD and GMK cell monolayers in 24-well plates were washed with serum-free medium twice and then were incubated with 1.0 to 10 μg/ml of mouse anti-human αvβ6 antibody (10D5; Chemicon) on a rocking plate at room temperature for 50 min. After this time, 100 μl of a 104-PFU/ml stock of virus was added to the wells. The monolayers were incubated under the same conditions for a further 50 min. Two milliliters of plaque overlay medium (0.5% carboxymethyl cellulose in growth medium) was added to each well, and the plates were incubated at 37°C with 6% CO2 for 2 to 3 days. The monolayers were then washed with phosphate-buffered saline and were stained with 0.1% crystal violet.
Growth of CAV9 on SW480 cells expressing integrin β6 and β3 subunits.
CAV9 and the RGD deletion mutant D4 (105 PFU/ml) were adsorbed to confluent monolayers of mock-, β3-, and β6-transfected SW480 cells on a rocking plate at room temperature for 1 h before they were incubated at 37°C. Seventy-two hours postinfection cells were freeze-thawed three times, and plaque assays were carried out on the freeze-thawed suspensions.
RESULTS
CAV9 binds to purified integrin αvβ6.
To investigate the potential of αvβ6 as a CAV9 receptor, we used a solid-phase assay to analyze the binding of CAV9 to the purified integrin. We included αvβ3 in these experiments because this integrin has previously been shown to act as a CAV9 receptor (37). The results (Fig. 1) showed that CAV9 binds efficiently to both integrins in vitro. When 10 mM EDTA was added to the binding buffer, CAV9 did not bind to either integrin, in agreement with the expected metal ion dependence of RGD-integrin receptors.
FIG. 1.
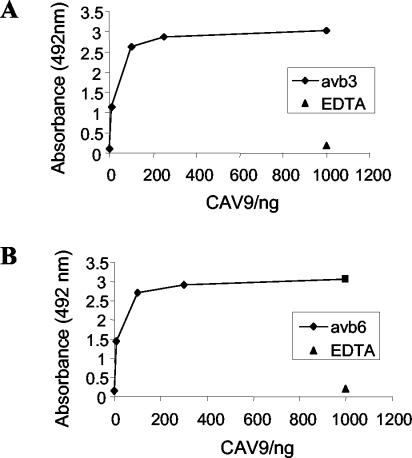
In vitro binding of CAV9 to integrins αvβ3 and αvβ6. (A) CAV9 binding to integrin αvβ3. (B) CAV9 binding to integrin αvβ6. In each case 10 to 1,000 ng of virus was applied to wells coated with approximately 300 ng of receptor. Each data point is the average of duplicate measurements. There is no binding in the presence of EDTA (1,000 ng of virus tested).
The specificity of the CAV9-integrin interaction was further studied in blocking experiments by using synthetic peptides (Fig. 2). Four different peptides were used: an RGD-containing peptide (GRGDSP) and its RGE-containing derivative (GRGESP); an RGD-containing peptide based on the CAV9 VP1 C terminus (QSRRRGDMST); and an unrelated control peptide (NGKKKNWKKIM) based on part of the human parechovirus 1 (HPeV1) VP3 sequence. Figure 2A shows that both of the RGD-containing peptides inhibit binding of CAV9 to αvβ3 in a dose-dependent manner and that GRGDSP is a more potent inhibitor than the CAV9-derived peptide. The inhibitory effect of these peptides was specific, because the control peptides had no effect on virus binding. Similarly, both of the RGD peptides were found to inhibit CAV9 binding to αvβ6, although these peptides were less effective than αvβ3 because a higher peptide concentration was required to inhibit virus binding (Fig. 2B). In the case of αvβ6, to demonstrate a blocking effect it was necessary to preincubate the integrin and peptide prior to adding the virus. In contrast to αvβ3, the CAV9 VP1-derived peptide was more efficient than GRGDSP as an inhibitor of virus binding to αvβ6 and the VP3 control peptide had a partial effect on virus binding.
FIG. 2.
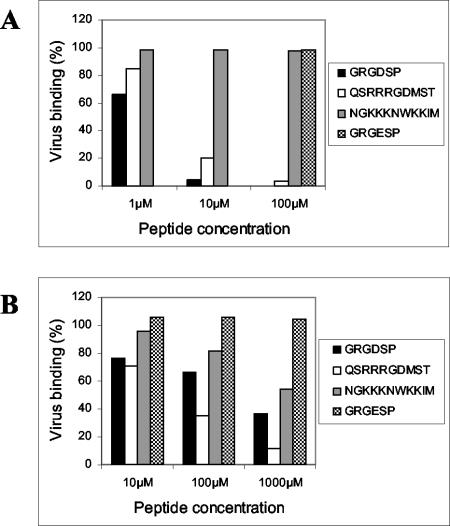
Inhibition of CAV9 binding to purified integrins αvβ3 and αvβ6 by RGD peptides. (A) CAV9 binding to integrin αvβ3. (B) CAV9 binding to integrin αvβ6. The peptides used were GRGDSP, GRGESP*, QSRRRGDMST (based on the CAV9 VP1 C terminus), and NGKKKNWKKIM (a control peptide based on part of the HPeV1 VP3 protein). The y axis represents binding in the presence of the appropriate peptide as a percentage of binding without added peptide. The GRGESP peptide was used only at a concentration of 100 μM in the αvβ3 experiment.
CAV9 binds to cells transfected with the integrin β6-subunit gene.
The above data show that CAV9 binds to αvβ6 in an authentic cation- and RGD-dependent manner. We next tested whether it can also recognize this integrin on the cell surface. For these studies we used the human colon carcinoma cell line SW480, stably transfected to express either human αvβ6 or αvβ3 (Fig. 3A). Because manganese (Mn) ions are known to enhance ligand binding to several integrins, including αvβ3 (18, 20, 21, 28, 40), these experiments were carried out in the presence of 1 mM Mn ions (see Materials and Methods). These studies showed that CAV9 binds only to SW480 cells expressing αvβ6 (SW480-αvβ6) and not to the mock-transfected cells (SW480-mock) or to cells expressing αvβ3 (SW480-αvβ3).
FIG. 3.
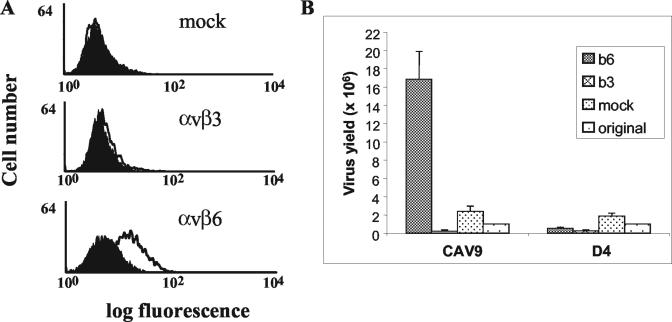
Flow cytometric analysis of CAV9 binding to SW480 cells expressing αvβ3 or αvβ6 and enhanced growth of CAV9 in cells expressing αvβ6. (A) Control cell samples were processed in the absence of virus (filled histogram). Virus binding (open histogram) was detected on SW480-αvβ6. Virus binding was not detected with SW480-mock or SW480-αvβ3 cells. (B) Levels (in PFU) of CAV9 and the RGD-less derivative D4 obtained when grown on monolayers of SW480-αvβ3 (b3), SW480-αvβ6 (b6), and SW480-mock (mock) cells for 72 h. The input virus inoculum is also shown (labeled “original”). The results are an average of duplicate measurements.
Infection of SW480-αvβ6 and SW480-αvβ3 cells by CAV9.
We next determined whether the increase in CAV9 binding to SW480 cells expressing αvβ6 results in a productive infection. SW480-mock, SW480-αvβ6, and SW480-αvβ3 cells were infected with CAV9, and the level of virus present at 72 h postinfection was determined by plaque assay. Figure 3B shows that infection of SW480-αvβ6 cells results in an increased level of CAV9 over the input virus, but this is not seen in mock- and β3-transfected cells. To confirm the role of the RGD motif in infection of SW480-αvβ6, we also infected these cells with the RGD-less CAV9 mutant, D4. No significant increase in the level of D4 was seen following infection of any of the cell lines.
GMK cells express αvβ6.
GMK cells have been used extensively in studies on CAV9, as this virus grows efficiently on these cells in an RGD-dependent manner. Therefore, we used specific antibodies to determine the RGD-binding integrins expressed on GMK cells. Flow cytometric analysis indicated that GMK cells express the RGD-binding integrins, α5β1, αvβ3, αvβ5, and the β6 subunit. Because the β6 subunit is normally expressed at the cell surface only when associated with the αv chain, these data indicate that GMK cells express αvβ6 (Fig. 4).
FIG. 4.
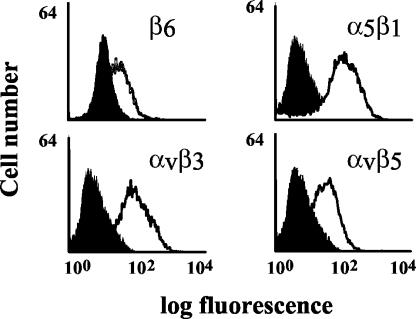
Flow cytometric analysis of integrin expression on GMK cells. Cells were stained with integrin-specific monoclonal antibodies (open histogram) as indicated on the figure. Control cell samples were processed in the absence of the primary antibody (filled histogram). Antibodies: β6, CSβ6; αvβ3, LM609; αvβ5, P1F6; and α5β1, SAM-1.
Blocking of infection of susceptible cells by antibody to αvβ6.
We then investigated whether αvβ6 is used as a receptor to initiate CAV9 infection of GMK cells. Infection-blocking assays with an anti-αvβ6 monoclonal antibody (10D5) revealed that CAV9 infection of GMK cells (Fig. 5A) is efficiently inhibited by this monoclonal antibody. By contrast, the antibody did not affect infection of GMK cells by D4 (Fig. 5C) or of RD cells by CAV9 (Fig. 5B). An interaction between CAV9 and αvβ6 was further confirmed by an experiment using the purified preparation of soluble αvβ6 to inhibit CAV9 infection in a plaque reduction assay. The preparation efficiently blocked CAV9 infection in a dose-dependent manner (Fig. 5D).
FIG. 5.
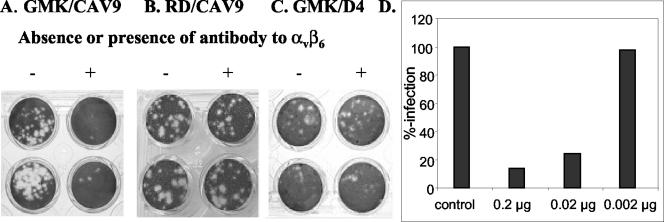
Plaque reduction assays. (A to C) Blocking effect of an antibody to αvβ6 (10D5) on the infection of cells by CAV9 and the RGD-less mutant D4. −, no antibody; +, 1.0 μg of antibody. (A) CAV9 on GMK cells; (B) CAV9 on RD cells; (C) D4 on GMK cells. (D) Blocking of CAV9 infection in GMK cells by purified soluble αvβ6. Inhibition was measured by plaque reduction. Bars represent different amounts of αvβ6 incubated with CAV9 prior to addition to the cell monolayer.
DISCUSSION
Infection by CAV9 appears to be mediated through at least two pathways, one dependent on and the other independent of its RGD motif (15, 36). To date, the RGD-recognizing integrin αvβ3 is the only integrin implicated as a receptor for CAV9 (37, 44). In contrast, although FMDV has also been reported to recognize αvβ3 (3), it additionally interacts with several other integrins, including αvβ6 (18-21). The amino acid sequence immediately following the RGD tripeptide in CAV9 and FMDV is similar (see Introduction), which prompted us to investigate whether there is an interaction between αvβ6 and CAV9. In the present study we have shown that αvβ6 is a receptor for CAV9. Several main findings support this conclusion. (i) CAV9 binds to purified αvβ6 in an authentic cation- and RGD-dependent interaction. (ii) CAV9 binding to SW480 cells is greatly enhanced following transfection with β6 cDNA, which allows expression of αvβ6 at the cell surface. (iii) Infection of SW480 cells is also enhanced by expression of β6. This infection is also dependent on the VP1 RGD, because infection of SW480 cells by an RGD-deleted CAV9 mutant, D4, was not enhanced by αvβ6 expression. (iv) GMK cells express αvβ6, and infection of these cells by CAV9 is inhibited by a function-blocking monoclonal antibody to αvβ6.
Our in vitro studies indicated that CAV9 can interact with purified αvβ3 and αvβ6 in a solid-phase assay (Fig. 1), and hence both can potentially act as a receptor for virus attachment to the host cell. In each case, binding is specific as it is reduced substantially by the addition of RGD-containing peptides (Fig. 2). These peptides were less effective in blocking CAV9 binding to αvβ6, possibly suggesting that the CAV9-αvβ6 interaction is of higher affinity. The generic peptide, GRGDSP, is a poor inhibitor of FMDV binding to αvβ6 expressed on CHO cells (19), consistent with the weak effect this peptide had on the interaction between purified αvβ6 and CAV9 (Fig. 2B).
In addition to binding to purified αvβ6, we showed by flow cytometry that CAV9 can also bind to SW480 cells expressing this integrin (Fig. 3A) (49). In contrast, CAV9 did not bind to mock-transfected SW480 cells lacking the β6 subunit or to SW480 cells expressing αvβ3. Binding differences of CAV9 to SW480-αvβ6, SW480-αvβ3, and SW480-mock cells were also reflected in the infection of these cells (Fig. 3B). CAV9 growth in SW480-αvβ6 but not in SW480-αvβ3 or SW480-mock cells gave a significant increase in yield over the input level. In contrast, the RGD-deleted mutant D4 did not show discrimination between these cells, indicating that infection of SW480-αvβ6 is RGD-dependent. Thus, CAV9 binds to αvβ6 in vitro, while expression of this integrin on the SW480 cell surface confers the ability to bind and be infected by CAV9. αvβ6 can therefore act as a functional RGD-dependent CAV9 receptor.
It has been reported that infection of SW480-αvβ6 cells by another enterovirus, coxsackievirus B1 (CBV1), is enhanced relative to that of mock-transfected cells (1). This observation is enigmatic, as CBV1 does not contain an RGD motif and therefore would not be expected to interact with αvβ6 in the usual way. It is known that β6-transfected cells have an increased growth rate, and this, or another effect on signaling, could alter the cellular environment and affect CBV1 replication (49). However, in our experiments D4 did not show increased infectivity of SW480-αvβ6 cells (Fig. 3B), so it seems more likely that enhanced infectivity of CAV9 is a direct effect arising from the use of αvβ6 as an RGD-dependent receptor.
The simian cell line GMK has been used extensively to study CAV9 infection (15, 36, 37, 44, 47). We therefore investigated whether infection of this cell line could also be mediated by αvβ6. Flow cytometry showed that several RGD-recognizing integrins are present on GMK cells (Fig. 4). The level of αvβ6 appears to be low but is significant, because a function-blocking αvβ6 antibody efficiently inhibited infection of GMK cells by CAV9 (Fig. 5A). It is, however, known that ligation and/or cross-linking of an integrin at the cell surface may activate intracellular signaling pathways that modulate the function of other integrin species expressed on the same cell in a process termed integrin cross-talk (4). Although we believe it unlikely, it is therefore possible that the effect of the anti-αvβ6 monoclonal could be to downregulate the activity of other integrin molecules used by CAV9 on GMK cells rather than simple blocking of the αvβ6-RGD interaction. The fact that purified αvβ6 blocked infection in a dose-dependent manner (Fig. 5D) argues against this nonspecific effect.
The almost complete blocking of infection by anti-αvβ6 suggests that αvβ6 is the major receptor used by CAV9 to initiate infection of GMK cells. Other RGD-recognizing integrins expressed on these cells, including αvβ3, may not be as important in entry, although the residual levels of infectivity may be due to interaction with other integrins. In this connection, it is interesting that the previously reported levels of blocking of CAV9 infection by αvβ3 antibodies (around 50%) (44, 47) are much less than that observed here for anti-αvβ6. Despite this relatively weak effect on infection, αvβ3 antibodies can compete with CAV9 for binding to GMK cells, while expression of αvβ3 on CHO cells makes these cells susceptible to CAV9 binding and infection (44), suggesting that αvβ3 can act as a functional receptor in both these cell lines. The failure to infect SW480-αvβ3 cells is therefore surprising, given that SW480-αvβ6 cells were infected. It is likely that more than one molecule is required for CAV9 entry, and GRP78, β2-microglobulin, and major histocompatibility complex class I have been implicated in other cells (44, 47, 48), while we have observed that some CAV9 strains can utilize heparan sulfate (unpublished data). One possibility is that SW480-αvβ3 cells do not possess the full repertoire of cofactors required for a productive infection mediated by αvβ3 entry, while these cofactors are present on GMK and CHO cells. An absence of a critical factor could potentially prevent virus binding if it were to lead to a subtle alteration in protein conformation, and this may explain the disparity between the in vitro binding of CAV9 to αvβ3 and the lack of binding to SW480-αvβ3 cells. Surprisingly, given that αvβ3 is considered an RGD-dependent integrin, binding of CAV9 to CHO cells expressing αvβ3 is not dependent on the RGD motif, because an RGD-less mutant can still bind to these cells (46). This RGD independence contrasts with the RGD-dependent infectivity of SW480-αvβ6 cells by CAV9 (Fig. 3) and may contribute to the apparent difference in functionality of αvβ3 expressed on SW480 and CHO cells.
The data presented here provide the first evidence that integrin αvβ6 is a receptor for CAV9. This is interesting in light of the observed sequence similarity between the regions surrounding the RGD motifs of CAV9 and latent TGF-β1, a ligand for αvβ6 (31), although TGF-β1 has also been reported to interact with αvβ1, αvβ8, and α8β1 (25, 29, 30). This may suggest that αvβ6 is an important receptor for CAV9 in natural infections as well as in the cell lines tested here. The RGD motif is a feature of all natural isolates of CAV9, although specific mutants lacking it can be generated in the laboratory (7, 15, 39). These grow less well than their RGD-containing parent in many cell lines and show reduced pathogenicity in a mouse model (12, 13, 15). While previous studies have related the function of the RGD motif only to interactions with the integrin αvβ3, we have shown here that CAV9 can also interact with αvβ6 and that this interaction is biologically relevant. This is consistent with the expression of αvβ6 on epithelial cells, the initial site of enterovirus infection.
Acknowledgments
The studies were supported by grants from the Wellcome Trust, the Academy of Finland, and the Sigrid Juselius Foundation.
We thank Maaria Vainio for excellent technical assistance and Merja Roivainen (KTL, Helsinki, Finland) and Jyrki Heino (University of Jyväskylä, Finland) for the kind gift of integrin antibodies.
REFERENCES
- 1.Agrez, M. V., D. R. Shafren, X. Gu, K. Cox, D. Sheppard, and R. D. Barry. 1997. Integrin alpha v beta 6 enhances coxsackievirus B1 lytic infection of human colon carcinoma cells. Virology 239:71-77. [DOI] [PubMed] [Google Scholar]
- 2.Baranowski, E., C. M. Ruiz-Jarabo, N. Sevilla, D. Andreu, E. Beck, and E. Domingo. 2000. Cell recognition by foot-and-mouth disease virus that lacks the RGD integrin-binding motif: flexibility in aphthovirus receptor usage. J. Virol. 74:1641-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berinstein, A., M. Roivainen, T. Hovi, P. W. Mason, and B. Baxt. 1995. Antibodies to the vitronectin receptor (integrin αvβ3) inhibit binding and infection of foot-and-mouth-disease virus to cultured-cells. J. Virol. 69:2664-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blystone, S. D., S. E. Slater, M. P. Williams, M. T. Crow, and E. J. Brown. 1999. A molecular mechanism of integrin crosstalk: αvβ3 suppression of calcium/calmodulin-dependent protein kinase II regulates α5β1 function. J. Cell Biol. 145:889-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boonyakiat, Y., P. J. Hughes, F. Ghazi, and G. Stanway. 2001. Arginine-glycine-aspartic acid motif is critical for human parechovirus 1 entry. J. Virol. 75:10000-10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, K. H., P. Auvinen, T. Hyypiä, and G. Stanway. 1989. The nucleotide sequence of coxsackievirus A9—implications for receptor binding and enterovirus classification. J. Gen. Virol. 70:3269-3280. [DOI] [PubMed] [Google Scholar]
- 7.Chang, K. H., C. Day, J. Walker, T. Hyypiä, and G. Stanway. 1992. The nucleotide sequences of wild-type coxsackievirus A9 strains imply that an RGD motif in VP1 protein is functionally significant. J. Gen. Virol. 73:621-626. [DOI] [PubMed] [Google Scholar]
- 8.Escarmis, C., E. C. Carrillo, M. Ferrer, J. F. G. Arriaza, N. Lopez, C. Tami, N. Verdaguer, E. Domingo, and M. T. Franze-Fernandez. 1998. Rapid selection in modified BHK-21 cells of a foot-and-mouth disease virus variant showing alterations in cell tropism. J. Virol. 72:10171-10179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans, D. J., and J. W. Almond. 1998. Cell receptors for picornaviruses as determinants of cell tropism and pathogenesis. Trends Microbiol. 6:198-202. [DOI] [PubMed] [Google Scholar]
- 10.Ghazi, F., P. J. Hughes, T. Hyypiä, and G. Stanway. 1998. Molecular analysis of human parechovirus type 2 (formerly echovirus 23). J. Gen. Virol. 79:2641-2650. [DOI] [PubMed] [Google Scholar]
- 11.Grist, N. R., and D. Reid. 1988. General pathogenicity and epidemiology, p. 221-239. In M. Berdinelli and H. Friedman (ed.), Coxsackieviruses: a general update, Plenum Press, New York, N.Y.
- 12.Harvala, H., H. Kalimo, L. Dahllund, J. Santti, P. J. Hughes, T. Hyypiä, and G. Stanway. 2002. Mapping of tissue tropism determinants in coxsackievirus genomes. J. Gen. Virol. 83:1697-1706. [DOI] [PubMed] [Google Scholar]
- 13.Harvala, H., H. Kalimo, G. Stanway, and T. Hyypiä. 2003. Pathogenesis of coxsackievirus A9 in mice: role of the viral arginine-glycine-aspartic acid motif. J. Gen. Virol. 84:2375-2379. [DOI] [PubMed] [Google Scholar]
- 14.Hendry, E., H. Hatanaka, E. Fry, M. Smyth, J. Tate, G. Stanway, J. Santti, M. Maaronen, T. Hyypiä, and D. Stuart. 1999. The crystal structure of coxsackievirus A9: new insights into the uncoating mechanisms of enteroviruses. Structure 7:1527-1538. [DOI] [PubMed] [Google Scholar]
- 15.Hughes, P. J., C. Horsnell, T. Hyypiä, and G. Stanway. 1995. The coxsackievirus A9 RGD motif is not essential for virus viability. J. Virol. 69:8035-8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hynes, R. O. 1992. Integrins: versatility, modulation, and signaling in cell adhesion. Cell 69:11-25. [DOI] [PubMed] [Google Scholar]
- 17.Hyypiä, T., C. Horsnell, M. Maaronen, M. Khan, N. Kalkkinen, P. Auvinen, L. Kinnunen, and G. Stanway. 1992. A distinct picornavirus group identified by sequence analysis. Proc. Natl. Acad. Sci. USA 89:8847-8851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson, T., A. Sharma, R. Abu-Ghazaleh, W. E. Blakemore, F. M. Ellard, D. L. Simmons, J. W. I. Newman, D. I. Stuart, and A. M. Q. King. 1997. Arginine-glycine-aspartic acid-specific binding by foot-and-mouth disease viruses to the purified integrin αvβ3 in vitro. J. Virol. 71:8357-8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson, T., A. P. Mould, D. Sheppard, and A. M. Q. King. 2002. Integrin αvβ1 is a receptor for foot-and-mouth disease virus. J. Virol. 76:935-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson, T., D. Sheppard, M. Denyer, W. Blakemore, and A. M. Q. King. 2000. The epithelial integrin αvβ6 is a receptor for foot-and-mouth disease virus. J. Virol. 74:4949-4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson, T., W. Blakemore, J. W. I. Newman, N. J. Knowles, A. P. Mould, M. J. Humphries, and A. M. Q. King. 2000. Foot-and-mouth disease virus is a ligand for the high-affinity binding conformation of integrin α5β1: influence of the leucine residue within the RGDL motif on selectivity of integrin binding. J. Gen. Virol. 81:1383-1391. [DOI] [PubMed] [Google Scholar]
- 22.Joki-Korpela, P., M. Roivainen, H. Lankinen, T. Pöyry, and T. Hyypiä. 2000. Antigenic properties of human parechovirus 1. J. Gen. Virol. 81:1709-1718. [DOI] [PubMed] [Google Scholar]
- 23.Joki-Korpela, P., V. Marjomäki, C. Krogerus, J. Heino, and T. Hyypiä. 2001. Entry of human parechovirus 1. J. Virol. 75:1958-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leippert, M., E. Beck, F. Weiland, and E. Pfaff. 1997. Point mutations within the βG-βH loop of foot-and-mouth disease virus O1K affect virus attachment to target cells. J. Virol. 71:1046-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu, M., J. S. Munger, M. Steadele, C. Busald, M. Tellier, and L. M. Schnapp. 2002. Integrin α8β1 mediates adhesion to LAP-TGF β1. J. Cell Sci. 115:4641-4648. [DOI] [PubMed] [Google Scholar]
- 26.Mason, P. W., E. Rieder, and B. Baxt. 1994. RGD sequence of foot-and-mouth disease virus is essential for infecting cells via the natural receptor but can be bypassed by an antibody-dependent enhancement pathway. Proc. Natl. Acad. Sci. USA 91:1932-1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, L. C., W. Blakemore, D. Sheppard, A. Atakilit, A. M. Q. King, and T. Jackson. 2001. Role of the cytoplasmic domain of the β-subunit of integrin αvβ6 in infection by foot-and-mouth disease virus. J. Virol. 75:4158-4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mould, A. P., S. K. Akiyama, and M. J. Humphries. 1995. Regulation of integrin α5β1-fibronectin interactions by divalent cations. J. Biol. Chem. 270:26270-26277. [DOI] [PubMed] [Google Scholar]
- 29.Mu, D. Z., S. Cambier, L. Fjellbirkeland, J. L. Baron, J. S. Munger, H. Kawakatsu, D. Sheppard, V. C. Broaddus, and S. L. Nishimura. 2002. The integrin αvβ8 mediates epithelial homeostasis through MT1-MMP-dependent activation of TGF-b1. J. Cell Biol. 157:493-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Munger, J. S., J. G. Harpel, F. G. Giancotti, and D. B. Rifkin. 1998. Interactions between growth factors and integrins: latent forms of transforming growth factor-β are ligands for the integrin αvβ1. Mol. Biol. Cell. 9:2627-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Munger, J. S., X. Z. Huang, H. Kawakatsu, M. J. D. Griffiths, S. L. Dalton, J. F. Wu, J. F. Pittet, N. Kaminski, C. Garat, M. A. Matthay, D. B. Rifkin, and D. Sheppard. 1999. The integrin αvβ6 binds and activates latent TGFβ1: a mechanism for regulating pulmonary inflammation and fibrosis. Cell 96:319-328. [DOI] [PubMed] [Google Scholar]
- 32.Neff, S., D. Sa-Carvalho, E. Rieder, P. W. Mason, S. D. Blystone, E. J. Brown, and B. Baxt. 1998. Foot-and-mouth disease virus virulent for cattle utilizes the integrin αvβ3 as its receptor. J. Virol. 72:3587-3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neff, S., P. W. Mason, and B. Baxt. 2000. High-efficiency utilization of the bovine integrin αvβ3 as a receptor for foot-and-mouth disease virus is dependent on the bovine β3 subunit. J. Virol. 74:7298-7306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nelsen-Salz, B., H. J. Eggers, and H. Zimmermann. 1999. Integrin αvβ3 (vitronectin receptor) is a candidate receptor for the virulent echovirus 9 strain Barty. J. Gen. Virol. 80:2311-2313. [DOI] [PubMed] [Google Scholar]
- 35.Pulli, T., E. Koivunen, and T. Hyypiä. 1997. Cell-surface interactions of echovirus 22. J. Biol. Chem. 272:21176-21180. [DOI] [PubMed] [Google Scholar]
- 36.Roivainen, M., T. Hyypiä, L. Piirainen, N. Kalkkinen, G. Stanway, and T. Hovi. 1991. RGD-dependent entry of coxsackievirus A9 into host cells and its bypass after cleavage of VP1 protein by intestinal proteases. J. Virol. 65:4735-4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roivainen, M., L. Piirainen, T. Hovi, I. Virtanen, T. Riikonen, J. Heino, and T. Hyypiä. 1994. Entry of coxsackievirus A9 into host cells: specific interactions with αvβ3 integrin, the vitronectin receptor. Virology 203:357-365. [DOI] [PubMed] [Google Scholar]
- 38.Ruoslahti, E., and M. D. Pierschbacher. 1987. New perspectives in cell adhesion: RGD and integrins. Science 238:491-497. [DOI] [PubMed] [Google Scholar]
- 39.Santti, J., H. Harvala, L. Kinnunen, and T. Hyypiä. 2000. Molecular epidemiology and evolution of coxsackievirus A9. J. Gen. Virol. 81:1361-1372. [DOI] [PubMed] [Google Scholar]
- 40.Springer, T. A. 1990. Adhesion receptors of the immune system. Nature 346:425-434. [DOI] [PubMed] [Google Scholar]
- 41.Stanway, G. 1990. Structure, function and evolution of picornaviruses. J. Gen. Virol. 71:2483-2501. [DOI] [PubMed] [Google Scholar]
- 42.Stanway, G., N. Kalkkinen, M. Roivainen, F. Ghazi, M. Khan, M. Smyth, O. Meurman, and T. Hyypiä. 1994. Molecular and biological characteristics of echovirus 22—a representative of a new picornavirus group. J. Virol. 68:8232-8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stanway, G., T. Hovi, N. J. Knowles, and T. Hyypiä. 2002. Biological and molecular basis of picornavirus classification, p. 17-24. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses, ASM Press, Washington, D.C.
- 44.Triantafilou, M., K. Triantafilou, K. M. Wilson, Y. Takada, N. Fernandez, and G. Stanway. 1999. Involvement of β2-microglobulin and integrin αvβ3 molecules in the coxsackievirus A9 infectious cycle. J. Gen. Virol. 80:2591-2600. [DOI] [PubMed] [Google Scholar]
- 45.Triantafilou, K., M. Triantafilou, Y. Takada, and N. Fernandez. 2000. Human parechovirus 1 utilizes integrins αvβ3 and αvβ1 as receptors. J. Virol. 74:5856-5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Triantafilou, M., K. Triantafilou, K. M. Wilson, Y. Takada, and N. Fernandez. 2000. High affinity interactions of coxsackievirus A9 with integrin alpha v beta 3 (CD51/61) require the CYDMKTTC sequence of beta 3, but do not require the RGD sequence of the CAV-9 VP1 protein. Hum. Immunol. 61:453-459. [DOI] [PubMed] [Google Scholar]
- 47.Triantafilou, K., D. Fradelizi, K. Wilson, and M. Triantafilou. 2002. GRP78, a coreceptor for coxsackievirus A9, interacts with major histocompatibility complex class I molecules which mediate virus internalization. J. Virol. 76:633-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ward, T., R. M. Powell, P. A. Pipkin, D. J. Evans, P. D. Minor, and J. W. Almond. 1998. Role for β2-microglobulin in echovirus infection of rhabdomyosarcoma cells. J. Virol. 72:5360-5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weinacker, A., A. Chen, M. Agrez, R. I. Cone, S. Nishimura, E. Wayner, R. Pytela, and D. Sheppard. 1994. Role of the integrin alpha v beta 6 in cell attachment to fibronectin-heterologous expression of intact and secreted forms of the receptor. J. Biol. Chem. 269:6940-6948. [PubMed] [Google Scholar]
- 50.Zimmermann, H., H. J. Eggers, A. Zimmermann, W. Kraus, and B. Nelsen-Salz. 1995. Complete nucleotide sequence and biological properties of an infectious clone of prototype echovirus 9. Virus Res. 39:311-319. [DOI] [PubMed] [Google Scholar]
- 51.Zimmermann, H., H. J. Eggers, and B. Nelsen-Salz. 1996. Molecular cloning and sequence determination of the complete genome of the virulent echovirus 9 strain Barty. Virus Genes 12:149-154. [DOI] [PubMed] [Google Scholar]
- 52.Zimmermann, H., H. J. Eggers, and B. Nelsen-Salz. 1997. Cell attachment and mouse virulence of echovirus 9 correlate with an RGD motif in the capsid protein VP1. Virology 233:149-156. [DOI] [PubMed] [Google Scholar]


