Abstract
Glutaminase is the enzyme that converts glutamine into glutamate, which serves as a key excitatory neurotransmitter and one of the energy providers for cellular metabolism. Previous studies have revealed that mice lacking glutaminase 1 (GLS1), the dominant isoform in the brain and kidney, died shortly after birth due to disrupted glutamatergic transmission, suggesting the critical role of GLS1 in the physiological functions of synaptic network. However, whether GLS1 regulates neurogenesis, a process by which neurons are generated from neural progenitor cells (NPCs), is unknown. Using a human NPC model, we found that both GLS1 isotypes, kidney-type glutaminase and glutaminase C, were upregulated during neuronal differentiation, which were correlated with the expression of neuronal marker microtubule-associated protein 2 (MAP-2). To study the functional impact of GLS1 on neurogenesis, we used small interference RNA targeting GLS1 and determined the expressions of neuronal genes by western blot, real-time polymerase chain reaction, and immunocytochemistry. siRNA silencing of GLS1 significantly reduced the expression of MAP-2, indicating that GLS1 is essential for neurogenesis. To unravel the specific process(es) of neurogenesis being affected, we further studied the proliferation and survival of NPCs in vitro. siRNA silencing of GLS1 significantly reduced the Ki67+ and increased the TUNEL+ cells, suggesting critical roles of GLS1 for the proliferation and survival of NPCs. Together, these data suggest that GLS1 is critical for proper functions of NPCs, including neuronal differentiation, proliferation, and survival.
Introduction
Neural stem/progenitor cells (NPCs) are multipotent self-renewing cells capable of differentiation into neurons, astrocytes, and oligodendrocytes [1]. Neurogenesis, by which new neurons are generated, is a coordination of NPC self-renewal, proliferation, migration, cell fate commitment, survival, neuronal differentiation, and eventual integration of the newborn neurons into the existing networks [2,3]. Neurogenesis is robust throughout the embryonic brain but limited to specific brain areas in adulthood [3–5]. Attenuated neurogenesis results in brain malformations, neurotransmission disruption, and cognition impairment [6–8], suggesting that neurogenesis constitutes important physiological functions for the brain homeostasis and activity. The limited neurogenesis in the adult brain has posed a challenge for the brain regeneration after injuries and diseases [9–14]. Therefore, vigorous research is being conducted to identify endogenous factor(s) involved in neurogenesis.
Glutaminase 1 (GLS1) is an enzyme that converts glutamine into glutamate [15]. In the central nervous system (CNS), GLS1 is expressed at high levels, particularly in neurons [16]. Based on the in situ hybridization data provided by “Allen Brain Atlas: Developing Mouse Brain” at http://developingmouse.brain-map.org/gene/show/14436, the GLS1 signal becomes detectable at embryonic day 11.5 (E11.5) with the expression level log (−1.5). The expression of GLS1 increases during brain development and reaches the plateau before birth (at E18.5) with an expression level log (3). The expression of GLS1 remains high in postnatal mouse CNS. Kidney-type glutaminase (KGA) and glutaminase C (GAC) are two splice isoforms of GLS1 [17]. The product of glutaminase-catalyzed reaction is glutamate, a classical and the most abundantly used excitatory neurotransmitter. Glutamate has long been implicated in the maturation of neurons [18–21]. Specifically, an in vitro study on NPCs has revealed the role of glutamate in neuronal differentiation through the activation of AMPA receptors [20]. However, the in vivo effect of glutamate on neurogenesis remains incompletely understood. Glutamate has been suggested to be beneficial for neurogenesis [22–24], but studies on different anatomical regions in the mammalian brain and with different experimental approaches yielded discrepant results [18]: a low dose of glutamate (50 μM) applied to mouse brain slice culture increased cell proliferation in the ventricular zone, but decreased cell proliferation in the subventricular zone [24]. However, a high dose of glutamate (300 μM) applied to brain slice culture decreased cell proliferation and survival in the ventricular zone [25].
GLS1 is critical for synaptic integration in the CNS. Gls1 knockout mice died at postnatal day 1 due to impairment in respiratory function that is controlled by glutamatergic synaptic network [26]. Although GLS1 is critical for CNS synaptic transmission and glutamate has been extensively studied for embryonic and adult CNS neurogenesis, the specific role of GLS1 in neurogenesis has not been identified. In this study, we report that GLS1 is an essential enzyme for the neuronal differentiation, proliferation, and survival of human NPCs in vitro.
Materials and Methods
Isolation and culture of human NPC
Human fetal brain tissue (12–16 weeks postconception) was obtained from elective abortions carried out at the University of Washington in full compliance with the National Institute of Health (NIH), University of Nebraska Medical Center (UNMC), and University of Washington ethical guidelines. Human cortical NPCs were isolated from human fetal brain tissue, as previously described [27]. Briefly, NPCs were cultured in substrate-free tissue culture flasks and grown as neurospheres in a neurosphere initiation medium (NPIM), which consisted of X-Vivo 15 (BioWhittaker, Walkersville, ME) with N2 supplement (Gibco BRL, Carlsbad, CA), neural cell survival factor-1 (NSF-1; BioWhittaker), basic fibroblast growth factor (20 ng/mL; Sigma-Aldrich, St. Louis, MO), epidermal growth factor (20 ng/mL; Sigma-Aldrich), leukemia inhibitory factor (10 ng/mL; EMD Millipore Corporation, Billerica, MA), and 60 ng/mL N-acetylcysteine (Sigma-Aldrich). Cells were passaged at 2-week intervals, as previously described [27].
Human NPC differentiation
Differentiation of NPCs to neurons was performed as previously described [27]. Briefly, trypsin- and mechanically dissociated NPCs were plated on poly-d-lysine-coated 24-well plates in the NPIM for 24 h. Cells were subsequently changed to a serum-free Neurobasal medium (Gibco BRL) supplemented with B27 (NB27 medium; Gibco BRL). Cells were collected for RNA or protein analyses, or fixed for immunocytochemistry at indicated days after neuronal differentiation.
siRNA knockdown of glutaminase
siRNA knockdown in NPCs was performed, as previously described [28]. Briefly, siRNA targeting GLS1 (ID# s5840) was purchased from Applied Biosystems, Inc. (Foster City, CA). Before they were changed into the serum-free Neurobasal medium for neuronal differentiation, NPCs were transfected with 100 nM siRNA duplexes for 24 h in the presence of siIMPORTER (Upstate Cell Signaling Solutions, Charlottesville, VA) according to the manufacturer's instructions. Nonspecific control siRNA from Applied Biosystems, Inc. (ID# AM4635) was transfected with the same concentration as controls to GLS1 siRNA.
Protein extraction and western blot
Cells were rinsed twice with 1× phosphate-buffered saline (PBS) and lysed by M-PER Protein Extraction Buffer (Pierce, Rockford, IL) containing 1× protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN). Protein concentrations were determined using a BCA Protein Assay Kit (Pierce). Proteins (20–30 μg) from cell lysates were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis. After electrophoretic transfer to polyvinyldifluoridene membranes (Millipore Corporation and Bio-Rad, Hercules, CA), proteins were treated with purified primary antibodies for microtubule-associated protein 2 (MAP-2) (mouse, cat#MAB3418, 1:1,000; Millipore Corporation), KGA (rabbit, 1:1,000; Dr. N. Curthoys, Colorado State University, Fort Collins, CO), GAC (rabbit, 1:500; Dr. N. Curthoys, Colorado State University), GAD67 (mouse, cat#MAB5406, 1:500; Millipore Corporation), poly ADP ribose polymerase (PARP) (rabbit, cat# 9542s, 1:1,000; Cell Signaling Technologies, Beverly, MA), or β-actin (Sigma-Aldrich) overnight at 4°C followed by a horseradish peroxidase-linked secondary anti-rabbit or anti-mouse antibody (1:10,000; Cell Signaling Technologies). Antigen–antibody complexes were visualized by the Pierce ECL Western Blotting Substrate (Pierce). For data quantification, films were scanned with a CanonScan 9950F scanner; the acquired images were then analyzed on a Macintosh computer using the public domain NIH ImageJ program (at http://rsb.info.nih.gov/nih-image/).
RNA extraction and TaqMan real-time RT-PCR
RNA was isolated with the TRIzol Reagent (Invitrogen Corp, Carlsbad, CA) and RNeasy Kit according to the manufacturer's protocol (Qiagen, Inc., Valencia, CA). Primers used for real-time reverse-transcription–polymerase chain reaction (RT-PCR) were GAC (ID# 528445: forward sequence 5-TATGGAAAAAAGTGTCACCTGAGTCA-3, reverse sequence 5-GCTTTTCTCTCCCAGACTTTCCATT-3, probe sequence 5-AATGAGGACATCTCTACAACTGTA-3); KGA (ID# 489954: forward sequence 5-CGAAGATTTGCTTTGTCAGCTATGG-3, reverse sequence 5-CTCTGCAGCAGCTACATGGA-3, probe sequence 5-CAGCGGGACTATGATTC-3); MAP-2 (ID# hs00258900) and GAPDH (ID# 4310884E) were commercial products from Applied Biosystems, Inc. Real-time RT-PCR was carried out using the one-step quantitative TaqMan assay in a StepOne™ Real-Time PCR system (Applied Biosystems, Inc.). Relative KGA and GAC mRNA levels were determined and standardized with a GAPDH internal control using the comparative ΔΔCT method. All primers used in the study were tested for amplification efficiencies and the results were similar.
Immunocytochemistry
Cells were fixed in 4% PFA and washed in PBS, as previously described [27]. Cells were then incubated overnight at 4°C with primary antibodies, followed by goat anti-mouse IgG Alexa Fluor 488 secondary antibodies (1:1,000; Molecular Probes, Eugene, OR) for 1 h at 25°C. Primary antibodies included mouse MAP-2 (1:500; Millipore Corporation), rabbit GFAP (1:2,000; Dako, Carpinteria CA), and mouse anti-Ki67 (1:500; BD Biosciences, San Diego, CA). All antibodies were diluted in 0.1% Triton X-100 and 2% bovine serum albumin in PBS. Cells were counterstained with DAPI (1:1,000; Sigma-Aldrich) to identify nuclei. Morphological changes were visualized and captured with a Nikon Eclipse E800 microscope equipped with a digital imaging system or a Zeiss META 510 confocal microscope (Carl Zeiss MicroImaging, LLC, Thornwood, NY). Images were imported into Image-Pro Plus, version 7.0 (Media Cybernetics, Silver Spring, MD), for quantification. A total of 500–1,000 immunostained cells from 10 random fields per culture were manually counted using magnifications of 20× objective lens.
Analyses of glutamate by Amplex Red Glutamic acid/Glutamate oxidase Assay Kit
Intracellular glutamate levels in the cells were determined by Amplex Red Glutamic acid/Gutamate oxidase Assay Kit (Invitrogen Corp) based on the manufacturer's instruction. Cell lysates were diluted to the same protein concentration before entering the assay.
In situ TUNEL assays
Primary human NPCs were transfected with siRNA for control siRNA or GLS1 siRNA. Three days after transfection, cells were stained with an in situ TUNEL assay (Roche Diagnostics, Indianapolis, IN). TUNEL+ NPCs and total cells were counted after acquiring random images (10) from immunostained fields using a Nikon Eclipse TE2000E microscope. A minimum of 10 fields was counted for each treatment condition.
Statistical analysis
Data were analyzed as mean±SEM. The data were evaluated statistically by the analysis of variance followed by the Tukey test for paired observations. The two-tailed Student's t-test was used to compare two groups. Significance was considered when P<0.05. All experiments were performed with at least three donors to account for any donor-specific differences. Assays were performed at least three times in triplicate or quadruplicate.
Results
GLS1 splice variants KGA and GAC were upregulated during neuronal differentiation
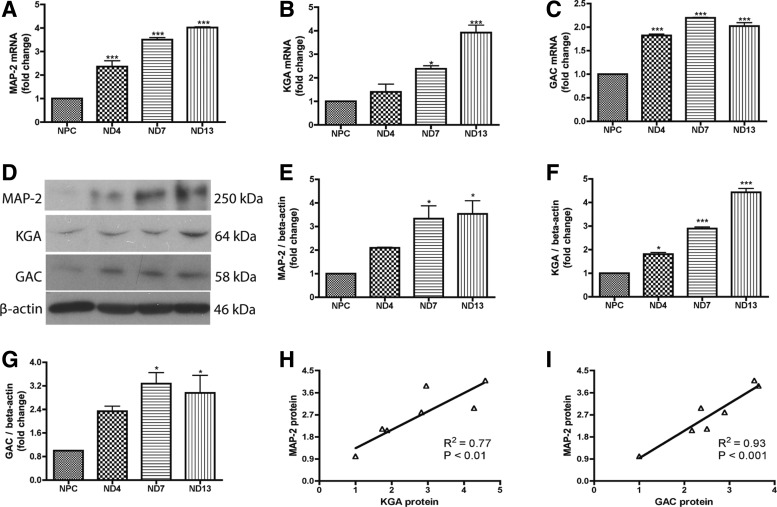
Human NPCs were differentiated to neurons in adherent cultures for 2 weeks. Total RNA was isolated at various time points, including day 0, 4, 7, and 13 after differentiation. The gene expression of neuronal marker MAP-2 continued to increase throughout this period of time (Fig. 1A), suggesting successful and continuous neuronal differentiation of the culture. Interestingly, both KGA and GAC mRNA were upregulated during neuronal differentiation (Fig. 1B, C). To confirm the upregulation of these genes after neuronal differentiation, we collected parallel protein samples and determined protein levels of MAP-2, KGA, and GAC through western blot (Fig. 1D–G). The neuronal differentiation resulted in a sharp increase of MAP-2 (Fig. 1D, E). Similarly, the KGA protein had 3- and 4.5-fold upregulation at 7 and 13 days after differentiation, respectively (Fig. 1D, F). The GAC protein had 2- and 3.5-fold upregulation at 4 and 7 days after differentiation, respectively (Fig. 1D, G). Both mRNA and protein levels of KGA were positively correlated with the MAP-2 levels (Fig. 1H and Supplementary Fig. S1A; Supplementary Data are available online at www.liebertpub.com/scd). Similarly, there was a significant correlation between both mRNA and protein levels of GAC and MAP-2 (Fig. 1I and Supplementary Fig. S1B). Together, the upregulation of KGA and GAC and their correlation with MAP-2 during neuronal differentiation suggests that KGA and GAC may be required for NPCs to differentiate into neurons.
FIG. 1.
Glutaminase 1 (GLS1) isotypes, kidney-type glutaminase (KGA), and glutaminase C (GAC) were upregulated during neural progenitor cell (NPC) differentiation to neurons. (A–G) Human NPCs were exposed to the neuron differentiation medium for differentiation. (A–C) At 0, 4, 7, and 13 days after differentiation, mRNA was collected and expressions of microtubule-associated protein 2 (MAP-2) (A), KGA (B), and GAC (C) were analyzed using real-time reverse transcription–polymerase chain reaction (RT-PCR). Data were normalized to GAPDH and presented as fold change compared to NPCs. (D–G) In parallel, protein lysates were collected and analyzed by western blot for the expressions of MAP-2 (D, E), KGA (D, F), and GAC (D, G). β-Actin was used as loading control. Protein levels were normalized as a ratio to beta-actin after densitometric quantification and presented as fold change relative to NPCs. Data are shown as the mean±SEM of three independent experiments with three different donors. (H, I) Correlation of the gene expression levels of KGA (H) and GAC (I) with MAP-2 was determined by Spearman correlation. *P<0.05, ***P<0.001 compared with NPCs, n=3.
siRNA knockdown of GLS1 impaired neuronal differentiation
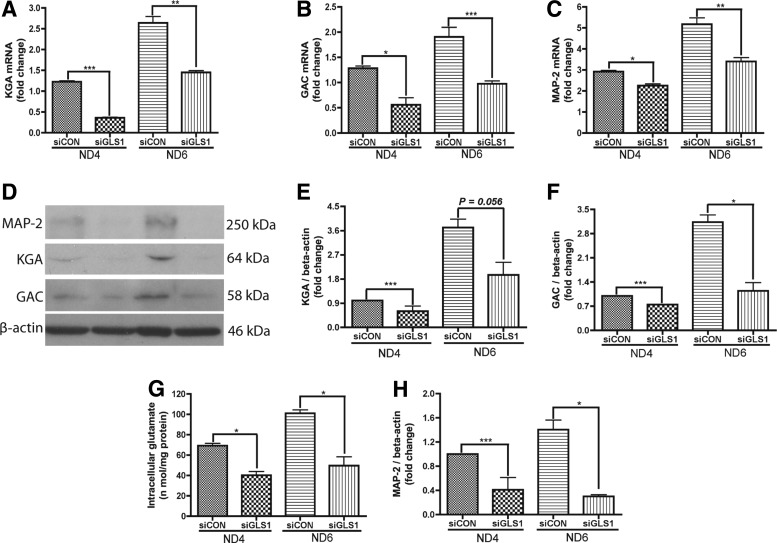
To test whether KGA and GAC were required for neuronal differentiation, we used siRNA to specifically silence GLS1 expression in NPCs. The GLS1 siRNA-transfected NPCs were induced to differentiate in neuronal differentiation media for 6 days. The knockdown of GLS1 resulted in significant decreases of both KGA (Fig. 2A, D, E) and GAC (Fig. 2B, D, F) mRNA and protein levels compared with the control siRNA group. The knockdown also resulted in a decrease of intracellular glutamate (Fig. 2G), indicating that the knockdown had reduced the enzymatic function of KGA and GAC. After siRNA knockdown of GLS1, MAP-2 protein and mRNA were reduced by more than 50% of that in control siRNA-transfected cells (Fig. 2C, D, H). The reduction of the mature neuronal marker MAP-2 in GLS1-deficient cells indicates that GLS1 is required for neuronal differentiation.
FIG. 2.
Lack of GLS1 impaired the expression of MAP-2 during neuronal differentiation. (A–H) Human NPCs were transfected by control siRNA or GLS1 siRNA and then exposed to the neuron differentiation medium for 4 days and 6 days. RNA was collected and the expression levels of KGA (A), GAC (B), and MAP-2 (C) at 4 and 6 days were analyzed using real-time RT-PCR. Data were normalized to GAPDH and presented as fold changes. In parallel, protein samples were analyzed by western blot for the expressions of KGA (D, E), GAC (D, F), and MAP-2 (D, H). β-Actin was used as loading control. Protein levels were normalized as a ratio to beta-actin after densitometric quantification and presented as fold change relative to the control siRNA group. Intracellular glutamate levels were tested using the Amplex Red Glutamic acid/Gutamate oxidase Assay Kit (G). Data are shown as the mean±SEM of three different donors. *P<0.05, **P<0.01, ***P<0.001 compared with the control siRNA group, n=3.
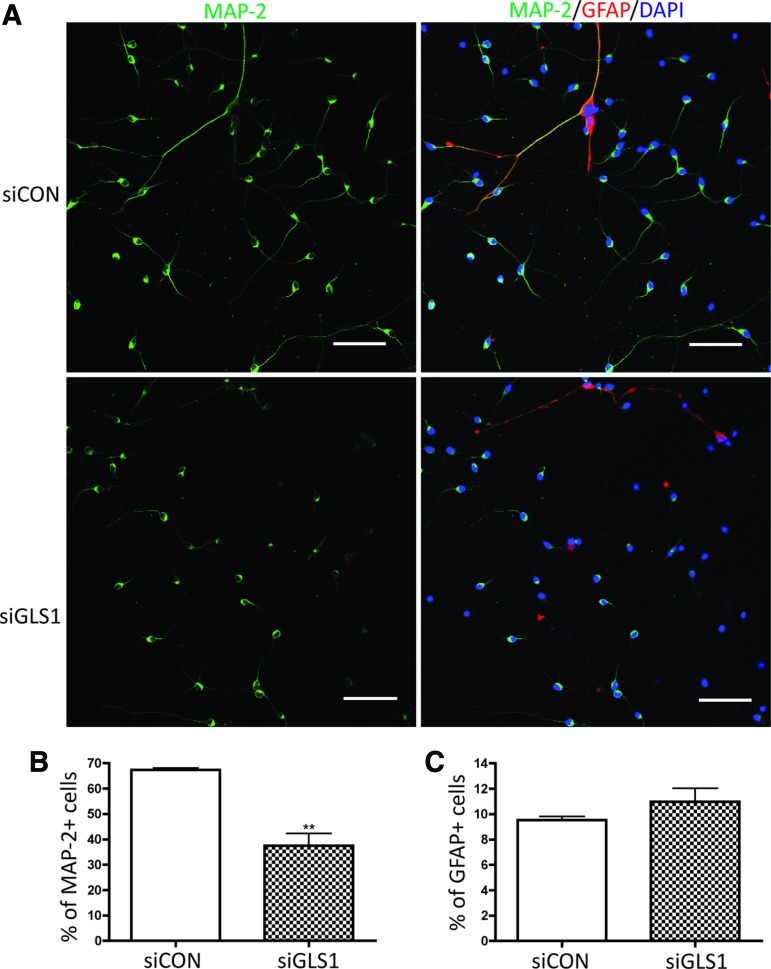
To further characterize the morphological changes of neurons after GLS1 knockdown, we labeled MAP-2 in the differentiated cultures through immunocytochemistry (Fig. 3A). Human NPCs transfected with control siRNA had ∼70% of the cells positive for MAP-2 (Fig. 3B) at 6 days after differentiation. In contrast, NPCs transfected with GLS1 siRNA had a significantly lower percentage (40%) of cells positive for MAP-2 (Fig. 3B). The impairment was specific to neuronal differentiation since the astrocyte differentiation, determined by GFAP immunostaining, did not change after GLS1 knockdown when compared with the control siRNA group (Fig. 3C). The number of the total counted cells is provided in Supplementary Table S1. These data suggest that GLS1 is required for the formation of mature neurons.
FIG. 3.
Lack of GLS1 specifically impaired neuronal differentiation. Human NPCs were transfected by control siRNA or GLS1 siRNA and then exposed to the neuron differentiation medium for 6 days. The cells were fixed and stained with MAP-2 to determine the levels of neuronal differentiation. (A) Representative pictures of MAP-2+ (green) and GFAP+ (red) cells were shown. Nuclei were labeled with DAPI (blue). (B, C) The percentage of MAP-2+ cells (B) and GFAP+ cells (C) was determined by counting the number of MAP-2+ or GFAP+ cells over the number of DAPI+ cells in each microscope field. Images were acquired from a Zeiss META 510 confocal microscope. Magnifications: 20× objective lens. Scale bar=50 μm. Data are shown as mean±SEM of 10 fields in each experimental group for the three donors. **P<0.01 compared with control siRNA group, n=3. Color images available online at www.liebertpub.com/scd
Knockdown of GLS1 impaired NPC proliferation and survival
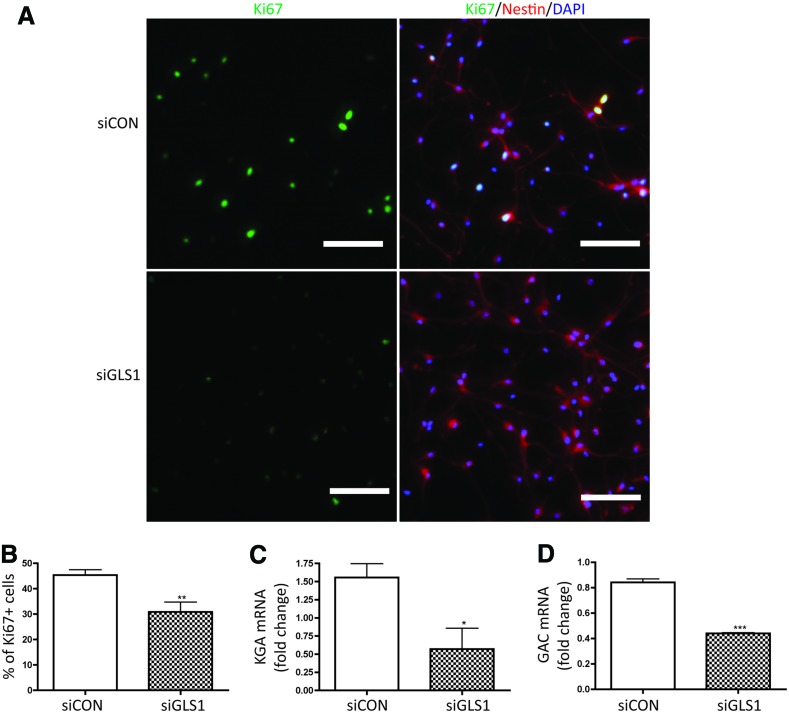
Neuronal differentiation is a key step of neurogenesis. However, proper NPC functions also include proliferation and survival. To determine whether lack of GLS1 affected NPC proliferation, we transfected NPCs with siRNA targeting GLS1 and maintained the culture under growth media for 3 days. Through immunocytochemistry that labeled the Ki67+ cells, we were able to identify the levels of NPC proliferation in the cultures. The percentage of Ki67+ cells was significantly decreased when GLS1 was silenced (Fig. 4A, B), suggesting a critical role of GLS1 in the NPC pool expansion. The number of the total counted cells is provided in Supplementary Table S1. The decrease of KGA and GAC mRNA levels by GLS1 siRNA transfection in NPCs was confirmed by real-time RT-PCR (Fig. 4C, D).
FIG. 4.
GLS1 silencing reduced NPC proliferation. Human NPCs were transfected by control siRNA or GLS1 siRNA, then maintained in growth media for 3 days. NPC proliferation was determined by immunolabeling the Ki67 of proliferating cells in culture. (A) Representative pictures of Ki67+ (green) cells in control siRNA- and siGLS1-transfected NPC culture were shown. Nestin (red) was used to label NPCs. Nuclei (blue) were labeled with DAPI. (B) The percentage of Ki67+ cells was determined by counting the number of Ki67+ cells over the number of DAPI+ cells in each microscope field. Images were acquired from a Nikon Eclipse TE2000E fluorescent microscope. Magnifications: 20× objective lens. Scale bar=50 μm. (C, D) KGA and GAC mRNA levels were confirmed to be reduced in NPCs transfected by siGLS1 kept in growth media. Data are shown as mean±SEM of 10 fields in each experimental group for the three donors. *P<0.05, **P<0.01, ***P<0.001 compared with the control siRNA group, n=3. Color images available online at www.liebertpub.com/scd
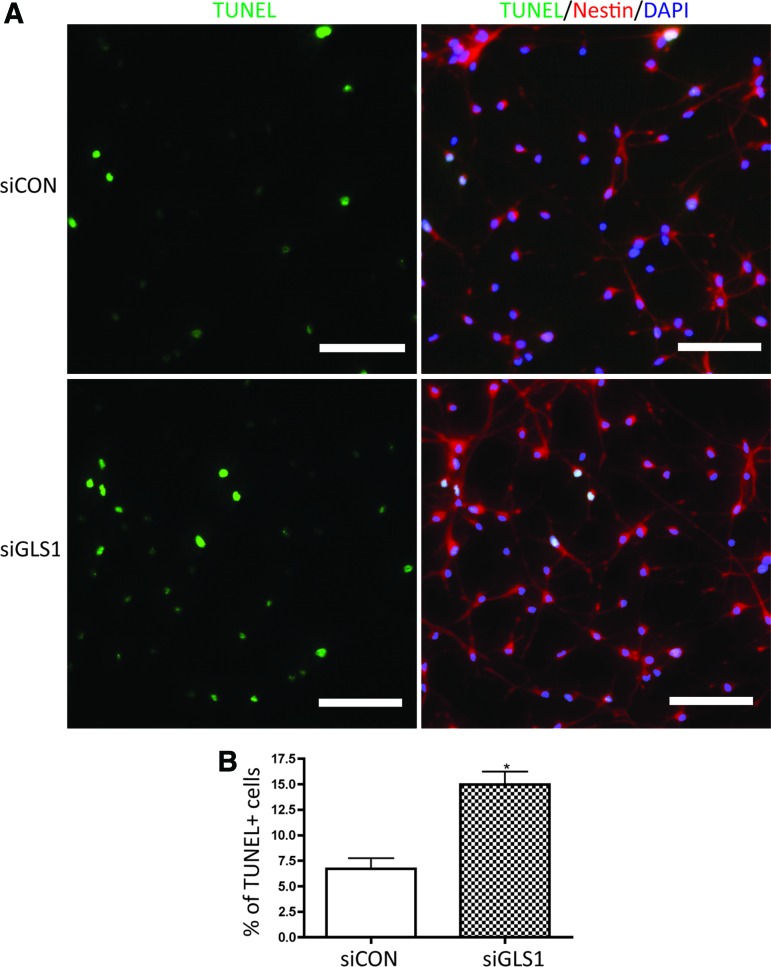
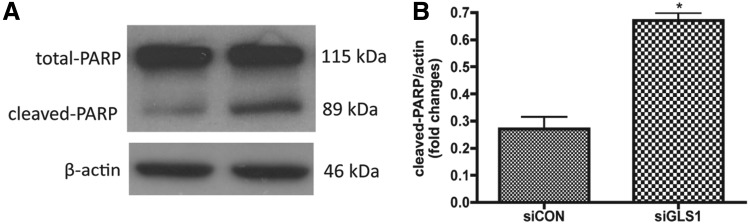
Next, we determined NPC survival after GLS1 was silenced by siRNA. The levels of apoptosis were identified by terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay. TUNEL detects DNA fragmentation by labeling the terminal end of nucleic acids. We applied TUNEL assay to undifferentiated NPCs kept in growth media. siRNA targeting GLS1 significantly increased the percentage of TUNEL+ cells in the human NPC culture compared with the siRNA control group (Fig. 5). The number of the total counted cells is provided in Supplementary Table S1. Therefore, the TUNEL assay revealed the occurrence of more cellular apoptosis when GLS1 was deficient. To further confirm the apoptosis, we determined PARP cleavage in the NPCs that lack GLS1. PARP is deactivated and cleaved by active caspase 3 (cleaved caspase 3) in the apoptosis cascade; thus, cleaved PARP represents ongoing apoptosis. Western blot analysis revealed the presence of cleaved PARP in both control and GLS1 siRNA groups. However, cleaved PARP was at significantly higher levels in the GSL1 siRNA group compared with the control siRNA group (Fig. 6). These data demonstrate the occurrence of cellular apoptosis after GLS1 was knocked down.
FIG. 5.
GLS1 silencing increased NPC apoptosis. Human NPCs were transfected by control siRNA or GLS1 siRNA, then maintained in growth media for 3 days. NPC apoptosis was determined by terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay. (A) Representative pictures of TUNEL+ (green) cells in control siRNA- and siGLS1-transfected NPC culture were shown. Nestin (red) was used to label NPCs. Nuclei (blue) were labeled with DAPI. (B) The percentage of TUNEL+ cells was determined by counting the number of TUNEL+ cells over the number of DAPI+ cells in each microscope field. Images were acquired from a Nikon Eclipse TE2000E fluorescent microscope. Magnifications: 20× objective lens. Scale bar=50 μm. Data are shown as mean±SEM of 10 fields in each experimental group for the three donors. *P<0.05 compared with the control siRNA group, n=3. Color images available online at www.liebertpub.com/scd
FIG. 6.
Lack of GLS1 increased poly ADP ribose polymerase (PARP) cleavage in NPCs. Human NPCs were transfected by control siRNA or GLS1 siRNA, then maintained in growth media for 3 days. (A) Whole cell lysates were collected and NPC apoptosis was analyzed by western blot for PARP cleavage. β-Actin was used as loading control. (B) PARP cleavage levels were normalized as a ratio of cleaved PARP to actin after densitometric quantification of (A) and shown as fold change relative to the control siRNA group. *P<0.05 compared with the control siRNA group, n=3.
Discussion
Neurogenesis is a highly orchestrated process that includes maintenance and expansion of NPCs, NPC migration, and differentiation to neurons, neuronal survival, and integration of new neurons to the existing synaptic networks [6]. As a result, impairment in neurogenesis often causes brain malformations, neurotransmission disruptions, and cognitive difficulties and deterioration, as reviewed by Jessel and Sanes [2,6–8]. Despite the importance of neurogenesis to the health and diseases, factors involved in this process remain to be fully elucidated. In the current study, we show that GLS1 isoforms were upregulated and correlated with MAP-2 during neuronal differentiation (Fig. 1). siRNA silencing of GLS1 in NPCs impaired their subsequent neuronal differentiation, suggesting that GLS1 is critical for neuronal differentiation (Figs. 2 and 3). Furthermore, we showed reduced proliferation and increased apoptosis in GLS1-deficient cells, suggesting that GLS1 is required for NPC proliferation and survival (Figs. 4–6). Collectively, we characterized GLS1 regulation in NPCs and identified GLS1 as one of the essential factors for neurogenesis.
GLS1 is a critical mitochondrial enzyme for cellular metabolism. Its expression increases during mammalian neural development and remains high in postnatal CNS, indicating a functional relevance of GLS1 to brain development. We report for the first time that lack of GLS1 impairs NPC differentiation, proliferation, and survival. This finding is consistent with the report of a functional role of GLS2 on neuronal differentiation [29]. Since GLS1 expression is at a much higher level in mammalian CNS compared with GLS2, tight regulation of GLS1 is of critical importance to brain homeostasis.
The identification of GLS1 functions in neurogenesis has an important clinical implication. GLS1, particularly the GAC isotype, has been shown to regulate oncogenic transformation [30] and mediate excitotoxicity in HIV-1-associated neurocognitive disorders [31]. Therefore, inhibiting GLS1 may serve for therapeutic purposes in cancer treatment or neurodegenerative diseases. Indeed, inhibition of GLS1 by either a potent chemical inhibitor or siRNA silencing slowed the growth of glioma cells [32] or blocked oncogenic transformation of human breast cancer cells and B lymphoma cells [33]. Also, knocking down GAC in HIV-infected microglia or macrophages alleviated the damage to neurons in the in vitro cocultured settings [31]. Although targeting GLS1 clearly provides benefits in the treatment of cancer or neurodegenerative diseases, our studies with neurogenesis suggest that it may have unintended consequences on the NPCs, as the properties and functions of NPCs were impaired with deficient GLS1 expression. Therefore, genetic, immunological, or pharmacological inhibition of GLS1 needs to be cautious on the potential complications. Furthermore, Gls1 KO mice died at postnatal day 1 due to glutamatergic synaptic transmission disruptions [26], and Gls1+/− mice displayed deteriorated hippocampal activity and developed schizophrenia-like symptoms [34]. These studies on GLS1 knockout (Gls1 KO) mice are consistent with our evaluation of the critical role of GLS1 on neurogenesis, particularly on the formation of neuronal network.
Neuronal differentiation, proliferation, and survival are three components of neurogenesis examined in the current study. How GLS1 functions to change each of those components is currently unclear. However, there are several potential pathways that GLS1 may take to assert its function. First, the natural product of GLS1 is glutamate, and glutamate is known to regulate neurogenesis. Interestingly, glutamate may have dichotomous effects on neurogenesis, depending on its concentrations. Low concentrations of exogenous glutamate (10 μM) introduced to cell or slice culture led to increased NPC proliferation and neurogenic potentials [19–21], whereas high concentrations of exogenous glutamate (300 μM) introduction resulted in impaired DNA synthesis and reduced cellular proliferation [25]. Second, glutamate has a direct neurotoxic effect on mature neurons [35–38]. Excess glutamate production by activated microglia and brain macrophages has been documented in several neurodegenerative diseases, including Parkinson's disease, Alzheimer's disease, stroke, and HIV-associated neurocognitive dementia [31,39]. Third, glutamate may serve as an alternative energy substrate for the citric acid cycle. Therefore, GLS1 deficiency may lead NPCs into a starvation-like state where normal activities like cellular proliferation and differentiation are attenuated. Furthermore, cells lacking energy are known to switch to the apoptotic signaling cascade. It is likely that more than one pathway play out in the NPCs that lack GLS1. More studies on glutamate signaling pathways leading to the impairment of NPC functions are needed.
In summary, our study demonstrated that GLS1 is required for neurogenesis in vitro because GLS1 deficiency impaired NPC differentiation, proliferation, and survival. These data shed light on a novel pathway to regulate neurogenesis and further suggest that great caution should be taken when targeting GLS1 in cancer or neurodegenerative disease therapy.
Supplementary Material
Acknowledgments
The authors thank Dr. Norman Curthoys for providing the KGA and GAC antibodies. They thank Dr. Hui Peng, Dr. Santhi Gorantla, and Ms. Li Wu for the technical support of this work. Julie Ditter, Lenal Bottoms, Myhanh Che, Johna Belling, and Robin Taylor provided outstanding administrative and secretarial support. This work was supported, in part, by research grants by the National Institutes of Health: R01 NS 41858-01, R01 NS 061642-01, P01 NS043985, and P20 RR15635-01 (J.Z.).
Author Disclosure Statement
No competing financial interests exist with the submitted manuscript.
References
- 1.McKay R. (1997). Stem cells in the central nervous system. Science 276:66–71 [DOI] [PubMed] [Google Scholar]
- 2.Jessell TM. and Sanes JR. (2000). Development. The decade of the developing brain. Curr Opin Neurobiol 10:599–611 [DOI] [PubMed] [Google Scholar]
- 3.Kintner C. (2002). Neurogenesis in embryos and in adult neural stem cells. J Neurosci 22:639–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lie DC, Song H, Colamarino SA, Ming GL. and Gage FH. (2004). Neurogenesis in the adult brain: new strategies for central nervous system diseases. Annu Rev Pharmacol Toxicol 44:399–421 [DOI] [PubMed] [Google Scholar]
- 5.Alvarez-Buylla A, Seri B. and Doetsch F. (2002). Identification of neural stem cells in the adult vertebrate brain. Brain Res Bull 57:751–758 [DOI] [PubMed] [Google Scholar]
- 6.Bergstrom T. and Forsberg-Nilsson K. (2012). Neural stem cells: brain building blocks and beyond. Ups J Med Sci 117:132–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nogues X, Corsini MM, Marighetto A. and Abrous DN. (2012). Functions for adult neurogenesis in memory: an introduction to the neurocomputational approach and to its contribution. Behav Brain Res 227:418–425 [DOI] [PubMed] [Google Scholar]
- 8.Ming GL. and Song H. (2005). Adult neurogenesis in the mammalian central nervous system. Annu Rev Neurosci 28:223–250 [DOI] [PubMed] [Google Scholar]
- 9.Parent JM. (2003). Injury-induced neurogenesis in the adult mammalian brain. Neuroscientist 9:261–272 [DOI] [PubMed] [Google Scholar]
- 10.Arvidsson A, Collin T, Kirik D, Kokaia Z. and Lindvall O. (2002). Neuronal replacement from endogenous precursors in the adult brain after stroke. Nat Med 8:963–970 [DOI] [PubMed] [Google Scholar]
- 11.Nakatomi H, Kuriu T, Okabe S, Yamamoto S, Hatano O, Kawahara N, Tamura A, Kirino T. and Nakafuku M. (2002). Regeneration of hippocampal pyramidal neurons after ischemic brain injury by recruitment of endogenous neural progenitors. Cell 110:429–441 [DOI] [PubMed] [Google Scholar]
- 12.Whitney NP, Eidem TM, Peng H, Huang Y. and Zheng JC. (2009). Inflammation mediates varying effects in neurogenesis: relevance to the pathogenesis of brain injury and neurodegenerative disorders. J Neurochem 108:1343–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doetsch F. and Scharff C. (2001). Challenges for brain repair: insights from adult neurogenesis in birds and mammals. Brain Behav Evol 58:306–322 [DOI] [PubMed] [Google Scholar]
- 14.Limke TL. and Rao MS. (2002). Neural stem cells in aging and disease. J Cell Mol Med 6:475–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kvamme E, Roberg B. and Torgner IA. (2000). Phosphate-activated glutaminase and mitochondrial glutamine transport in the brain. Neurochem Res 25:1407–1419 [DOI] [PubMed] [Google Scholar]
- 16.Mock B, Kozak C, Seldin MF, Ruff N, D'Hoostelaere L, Szpirer C, Levan G, Seuanez H, O'Brien S. and Banner C. (1989). A glutaminase (gis) gene maps to mouse chromosome 1, rat chromosome 9, and human chromosome 2. Genomics 5:291–297 [DOI] [PubMed] [Google Scholar]
- 17.Porter LD, Ibrahim H, Taylor L. and Curthoys NP. (2002). Complexity and species variation of the kidney-type glutaminase gene. Physiol Genomics 9:157–166 [DOI] [PubMed] [Google Scholar]
- 18.Schlett K. (2006). Glutamate as a modulator of embryonic and adult neurogenesis. Curr Top Med Chem 6:949–960 [DOI] [PubMed] [Google Scholar]
- 19.Suzuki M, Nelson AD, Eickstaedt JB, Wallace K, Wright LS. and Svendsen CN. (2006). Glutamate enhances proliferation and neurogenesis in human neural progenitor cell cultures derived from the fetal cortex. Eur J Neurosci 24:645–653 [DOI] [PubMed] [Google Scholar]
- 20.Whitney NP, Peng H, Erdmann NB, Tian C, Monaghan DT. and Zheng JC. (2008). Calcium-permeable AMPA receptors containing Q/R-unedited GluR2 direct human neural progenitor cell differentiation to neurons. FASEB J 22:2888–2900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luk KC, Kennedy TE. and Sadikot AF. (2003). Glutamate promotes proliferation of striatal neuronal progenitors by an NMDA receptor-mediated mechanism. J Neurosci 23:2239–2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cameron HA, McEwen BS. and Gould E. (1995). Regulation of adult neurogenesis by excitatory input and NMDA receptor activation in the dentate gyrus. J Neurosci 15:4687–4692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Di Giorgi-Gerevini V, Melchiorri D, Battaglia G, Ricci-Vitiani L, Ciceroni C, Busceti CL, Biagioni F, Iacovelli L, Canudas AM, et al. (2005). Endogenous activation of metabotropic glutamate receptors supports the proliferation and survival of neural progenitor cells. Cell Death Differ 12:1124–1133 [DOI] [PubMed] [Google Scholar]
- 24.Haydar TF, Wang F, Schwartz ML. and Rakic P. (2000). Differential modulation of proliferation in the neocortical ventricular and subventricular zones. J Neurosci 20:5764–5774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.LoTurco JJ, Owens DF, Heath MJ, Davis MB. and Kriegstein AR. (1995). GABA and glutamate depolarize cortical progenitor cells and inhibit DNA synthesis. Neuron 15:1287–1298 [DOI] [PubMed] [Google Scholar]
- 26.Masson J, Darmon M, Conjard A, Chuhma N, Ropert N, Thoby-Brisson M, Foutz AS, Parrot S, Miller GM, et al. (2006). Mice lacking brain/kidney phosphate-activated glutaminase have impaired glutamatergic synaptic transmission, altered breathing, disorganized goal-directed behavior and die shortly after birth. J Neurosci 26:4660–4671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng H, Huang Y, Rose J, Erichsen D, Herek S, Fujii N, Tamamura H. and Zheng J. (2004). Stromal cell-derived factor 1-mediated CXCR4 signaling in rat and human cortical neural progenitor cells. J Neurosci Res 76:35–50 [DOI] [PubMed] [Google Scholar]
- 28.Peng H, Erdmann N, Whitney N, Dou H, Gorantla S, Gendelman HE, Ghorpade A. and Zheng J. (2006). HIV-1-infected and/or immune activated macrophages regulate astrocyte SDF-1 production through IL-1beta. Glia 54:619–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Velletri T, Romeo F, Tucci P, Peschiaroli A, Annicchiarico-Petruzzelli M, Niklison-Chirou MV, Amelio I, Knight RA, Mak TW, Melino G. and Agostini M. (2013). GLS2 is transcriptionally regulated by p73 and contributes to neuronal differentiation. Cell Cycle 12:3564–3573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Erickson JW. and Cerione RA. (2010). Glutaminase: a hot spot for regulation of cancer cell metabolism?. Oncotarget 1:734–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang Y, Zhao L, Jia B, Wu L, Li Y, Curthoys N. and Zheng JC. (2011). Glutaminase dysregulation in HIV-1-infected human microglia mediates neurotoxicity: relevant to HIV-1-associated neurocognitive disorders. J Neurosci 31:15195–15204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seltzer MJ, Bennett BD, Joshi AD, Gao P, Thomas AG, Ferraris DV, Tsukamoto T, Rojas CJ, Slusher BS, et al. (2010). Inhibition of glutaminase preferentially slows growth of glioma cells with mutant IDH1. Cancer Res 70:8981–8987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang JB, Erickson JW, Fuji R, Ramachandran S, Gao P, Dinavahi R, Wilson KF, Ambrosio AL, Dias SM, Dang CV. and Cerione RA. (2010). Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell 18:207–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaisler-Salomon I, Miller GM, Chuhma N, Lee S, Zhang H, Ghoddoussi F, Lewandowski N, Fairhurst S, Wang Y, et al. (2009). Glutaminase-deficient mice display hippocampal hypoactivity, insensitivity to pro-psychotic drugs and potentiated latent inhibition: relevance to schizophrenia. Neuropsychopharmacology 34:2305–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olney JW. (1971). Glutamate-induced neuronal necrosis in the infant mouse hypothalamus. An electron microscopic study. J Neuropathol Exp Neurol 30:75–90 [DOI] [PubMed] [Google Scholar]
- 36.Choi DW. (1988). Glutamate neurotoxicity and diseases of the nervous system. Neuron 1:623–634 [DOI] [PubMed] [Google Scholar]
- 37.McCall A, Glaeser BS, Millington W. and Wurtman RJ. (1979). Monosodium glutamate neurotoxicity, hyperosmolarity, and blood-brain barrier dysfunction. Neurobehav Toxicol 1:279–283 [PubMed] [Google Scholar]
- 38.Newcomb R, Sun X, Taylor L, Curthoys N. and Giffard RG. (1997). Increased production of extracellular glutamate by the mitochondrial glutaminase following neuronal death. J Biol Chem 272:11276–11282 [DOI] [PubMed] [Google Scholar]
- 39.Erdmann N, Tian C, Huang Y, Zhao J, Herek S, Curthoys N. and Zheng J. (2009). In vitro glutaminase regulation and mechanisms of glutamate generation in HIV-1-infected macrophage. J Neurochem 109:551–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.