Abstract
Background & objectives:
Little is known about the prevalence of Chlamydia trachomatis infection in Indian women with infertility. To improve the diagnosis of C. trachomatis infection in developing countries, there is an urgent need to establish cost-effective molecular test with high sensitivity and specificity. This study was conducted to determine the diagnostic utility of a real time-PCR assay for detention of C. trachomatis infection in infertile women attending an infertility clinic in north India. The in house real time-PCR assay was also compared with a commercial real-time PCR based detection system.
Method:
Endocervical swabs, collected from 200 infertile women were tested for C. trachomatis by three different PCR assays viz. in-house real time-PCR targeting the cryptic plasmid using published primers, along with omp1 gene and cryptic plasmid based conventional PCR assays. Specimens were also subjected to direct fluorescence assay (DFA) and enzyme immunoassay (EIA) Performance of in-house real time-PCR was compared with that of COBAS Taqman C. trachomatis Test, version 2.0 on all in-house real time-PCR positive sample and 30 consecutive negative samples.
Results:
C. trachomatis infection was found in 13.5 per cent (27/200) infertile women by in-house real time-PCR, 11.5 per cent (23/200) by cryptic plasmid and/or omp1 gene based conventional PCR, 9 per cent (18/200) by DFA and 6.5 per cent (7/200) by EIA. The in-house real time-PCR exhibited a sensitivity and specificity of 100 per cent, considering COBAS Taqman CT Test as the gold standard. The negative and positive predictive values of the in-house real time-PCR were 100 per cent. The in-house real time-PCR could detect as low as 10 copies of C. trachomatis DNA per reaction.
Interpretation & conclusions:
In-house real time-PCR targeting the cryptic plasmid of C. trachomatis exhibited an excellent sensitivity and specificity similar to that of COBAS Taqman CT Test, v2.0 for detection of C. trachomatis infection in women attending an infertility clinic. In an effort to prevent Chlamydia infection associated infertility, we recommend screening of women with infertility due to C. trachomatis infection by in-house molecular method as a cost-effective solution in resource limited settings.
Keywords: Chlamydia trachomatis, endocervical swabs, infertility, real time-PCR
Sexually transmitted infections (STIs) are generally considered the leading preventable cause of infertility worldwide. In developing countries, STIs cause 70 per cent of all pelvic inflammatory diseases (PIDs), which can lead to infertility1. The obligate intracellular pathogen Chlamydia trachomatis is the most common sexually transmitted bacterial organism. Approximately 60 per cent of C. trachomatis genital infections in women can be asymptomatic and thus, remain undiagnosed and untreated2. Untreated chlamydia infection may ascend to the upper genital tract leading to severe reproductive complications like tubal factor infertility and ectopic pregnancy. High prevalence rates of C. trachomatis and the asymptomatic course of infection have led some industrialized countries to implement screening programmes to prevent the development of reproductive sequelae3,4. However, in India there are limited data for selective screening guidelines for C. trachomatis in infertile women. Hence, screening for C. trachomatis is not a routine component of infertility work-up.
Diagnosis of genital chlamydia infection had traditionally been by culture, rapid antigen detection methods and antibody testing. Conventional cell culture methods for C. trachomatis can vary in sensitivity between 60-80 per cent and are difficult to standardize, technically demanding and expensive5,6. Antigen detection tests like direct fluorescent antibody (DFA) assay requires expertise in fluorescent microscopy and is only suited for laboratories that test a limited number of specimens5,7. In addition, the overall sensitivity of antigen detection tests like enzyme immune assay (EIA) can be as low as 60 per cent8. Consequently, antigen detection assays have not been established as initial screening tests for C. trachomatis detection in clinical samples. Recently nucleic acid amplification techniques (NAATs), with the potential to offer improved sensitivity for diagnosing C. trachomatis infections, have become available9,10. Screening for C. trachomatis by polymerase chain reaction (PCR) assays performed on endocervical swab specimens provides the highest sensitivity, thereby leading to an early diagnosis which minimizes the risk of disease sequel and continued transmission of infections11,12. Real time-PCR has increasingly been used in recent years for detecting genital chlamydia infection, since it is easier to perform, provides faster results and being performed in a closed-tube format is less prone to contamination. Commercial real-time PCR assays like Amplicor PCR Test and COBAS Taqman Chlamydia trachomatis Test developed by Roche Diagnostics (Branchburg, New Jersey, USA) have been approved by the US Food and Drug Administration (FDA)13. These tests are widely used in western countries because of their high sensitivity and reliability in comparison with culture11. However, these systems are of limited use in routine diagnostics in resource-poor countries like India because of their high costs. Hence, the development of in-house real-time PCR assays with comparable sensitivity and specificity to commercial molecular amplification methods will offer an opportunity for lower cost tests for detecting genital chlamydia infection in developing countries.
The objectives of the present study were to evaluate the diagnostic utility of a real time-PCR assay for detection of C. trachomatis from endocervical swab specimen, determine the prevalence of C. trachomatis infection in women attending an infertility clinic in a north Indian tertiary care hospital and to identify factors that might be associated with increased frequency of genital chlamydia infection. In addition, the performance of the real time-PCR was evaluated in comparison with a conventional PCR detection system and a commercial real time-PCR system, the COBAS Taqman Chlamydia trachomatis Test, version 2.0 (Roche Diagnostics, Branchburg, New Jersey, USA).
Material & Methods
Study population: The study participants were consecutive women attending the Infertility Clinic of the Gynaecology and Obstetrics Outpatient Department of All India Institute of Medical Sciences, New Delhi, India between February 2010 and January 2012.
Infertile women were initially screened at the outpatient department by transvaginal sonography done in follicular phase and pelvic ultrasound scans to exclude patients with polycystic ovarian syndrome (PCOS), uterine fibroid (> 5 cm in size or impinging on the uterine cavity), endometriosis and structural anomalies of the genital tract. Screening also included a basal hormone evaluation done between days 2 and 5 of the ovarian cycle to exclude women with abnormal levels of serum follicular stimulating hormone (FSH), luteinizing hormone (LH), prolactin and thyroid stimulating hormone (TSH). Women with a diagnosis of male factor infertility as determined by an abnormal semen analysis of the male partner were also excluded. The laboratory records of infertile women, in whom the above mentioned causes of infertility had been ruled out, were reviewed for the results of endometrial aspirate PCR for Mycobacterium tuberculosis and endocervical swab culture and Gram staining for Neisseria gonorrhoeae. Patients who tested positive for M. tuberculosis and N. gonorrhoeae were also excluded from the study. Eligible patients were enrolled in the study only after obtaining written informed consent. The study protocol was approved by the Ethical Committee of the Institute.
All participants received a clinical examination by a gynaecologist and interviewed following a structured questionnaire gathering socio-demographic characteristics and information on risk behaviour, genital tract symptoms, previous gynaecological problems and obstetric history. Hysterosalpingography, sonograpgy and/or laparoscopy findings were recorded from the case sheets of the enrolled patients. The cohabiting partners of the women were also invited to participate in the study. They were enrolled only after obtaining written informed consent.
Sample collection and processing: Four dacron-tipped endocervical swabs were collected from each participant after cleaning the exocervix with a sterile swab to remove mucus and exudates. Thirty milliliters (ml) of first-void urine (FVU) samples were collected from each of the enrolled male partners.
The first endocervical swab was transported to the laboratory in 0.2 M sucrose phosphate buffer chlamydial transport medium for C. trachomatis PCR assays. DNA was extracted from the first swab using QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer's instructions. The extracted DNA was stored at -20°C till further use. The second swab was collected for COBAS Taqman C. trachomatis Test, version 2.0 (Roche Diagnostics, Branchburg, New Jersey, USA). It was transported at room temperature in the M4-RT Microtest culture transport medium (Microtest, Remel Inc, USA), processed according to the manufacturer's instructions to obtain C. trachomatis DNA and stored at - 20˚C till use. The third and fourth swab specimens were subjected to direct fluorescence assay (DFA) for demonstration of elementary bodies (Microtrak, Syva Corp., California, USA) and C. trachomatis antigen detection by enzyme immunoassay (EIA) (Microtrak II, Syva Corp., California, USA), respectively.
FVU samples collected from the male participants were concentrated by centrifugation at 500×g for 30 min at 4°C (Cold Centrifuge, Hermle, Germany) and filtered through 0.45μm membrane filter (Millipore, USA). Nucleic acid extraction from the processed FVU samples was similarly performed using QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany).
Real time-PCR assay: Real time- PCR assay targeting a 71 base-pair (bp) DNA segment of the cryptic plasmid of C. trachomatis was performed following the protocol described by Jaton et al14. A forward primer Ctr_F (5’-CATGAAAACTCGTTCCGAAATAGAA-3’), a reverse primer Ctr_R (5’-TCAGAGCTTTAC CTAACAACGCATA-3’) and a minor-groove binder probe labelled with 5’-FAM (5’-TCGCATG- CAAGATATCGA-3’) were used. The primers and probe were prepared by Applied Biosystems (Foster City, California, USA). The reactions were performed in a final volume of 20 μl, including 0.2 μM of each primer, 0.1 μM of probe, 10 μl of 2X TaqMan Universal Master Mix (Applied Biosystems, USA) and 5 μl DNA sample. Cyclic conditions were 2 min at 50°C, 10 min at 95°C, followed by 45 cycles of 15 sec at 95°C and 1 min at 60°C. Amplification and PCR product detection were performed with the ABI prism 7500 real time-PCR System (Applied Biosystems, USA).
Determination of analytical sensitivity of real time-PCR assay for C. trachomatis: A plasmid containing the target gene was constructed. DNA was extracted from the plasmid and the target sequence was amplified with CTr_F and CTr_R primers. The final 50 μl reaction mixture contained 0.4 μM of each primer, 1.25 U of Taq DNA polymerase (GeneiTaq, Bangalore Genei, India), 5 μl of 10X PCR buffer [1× PCR buffer is 10 mmol/l Tris–HCl (pH 8.8 at 25 °C), 1.5 mmol/l MgCl2 50 mmol/l KCl, and 0.1% Triton X-100], 400 μM deoxynucleoside triphosphate mixture, 5 μl of DNA and ultrapure sterile water. The PCR conditions used were 1 cycle of initial denaturation at 95°C for 10 min, followed by 50 cycles of 95°C for 15 sec, 60°C for 1 min and 72°C for 30 sec in a thermocycler (MJ Research, Waltham, MA, USA).
PCR products were purified with QIAGEN gel extraction kit (QIAGEN, Hilden, Germany) and cloned into pGEMT easy vector (Promega, USA) according to the manufacturer's protocol. For transformation of the recombinant plasmid, DH5α strain of Escherichia coli was used. Isolation of recombinant plasmid DNA was performed with QIAprep Miniprep (QIAGEN, Hilden, Germany). For confirmation of the right clone, the plasmid was digested with EcoRI restriction enzyme (New England Biolabs, United Kingdom) and the restricted product was run on 1 per cent agarose gel. (Fig. 1). The plasmid concentration was determined as 108 copies/μl by spectrophotometric quantification (Nanodrop ND-1000 v3.3.1, Nanodrop Technologies, Inc., Wilmington, USA). The plasmid was ten-fold diluted up to 1 DNA copy per reaction. To determine the sensitivity of the assay, each dilution was used as template in triplicate for real-time PCR amplification.
Fig. 1.
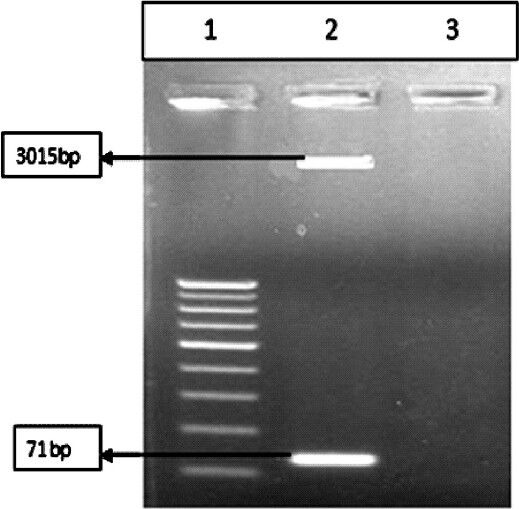
Restriction digestion of PCR product by EcoRI enzyme. Lane 1: 50 bp ladderk, Lane 2: Digested product, Lane 3: Blank.
Controls for the real-time PCR: The plasmid containing the target gene was constructed and used as positive control at a concentration of 100 copies per reaction in each run. A negative control in the form of template-free master mix solution was included in each run of amplification. An inhibition control was tested for each sample by adding to the PCR master mixture 4 μl of DNA sample and 1 μl of 100 copies of positive control plasmid. Amplification was considered efficient for a specimen if the corresponding inhibition control demonstrated the same threshold cycle (Ct) (±1) as 100 copies per reaction of positive control. A sample was considered negative if the fluorescent signal did not increase within 35 cycles. An amplification plot of the real-time PCR is shown in Fig. 2.
Fig. 2.
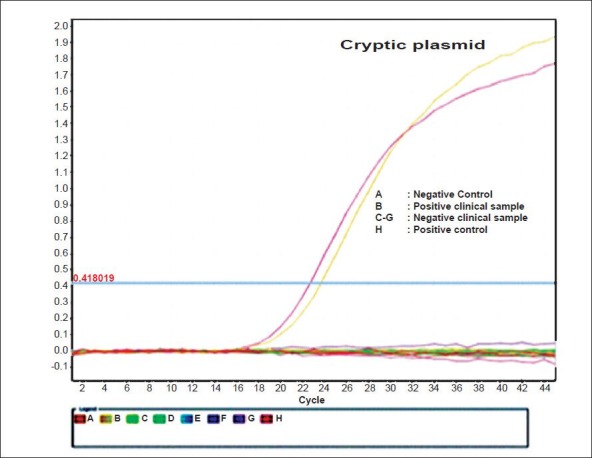
TaqMan real time-PCR amplification plot (ΔRn vs Cycle) targetting cryptic plasmid.
Specificity of real time-PCR assay: To assess the specificity, the real time-PCR assay was performed with DNA extracted from the positive cultures of organisms viz. Ureaplasma urealyticum NCTC 10177, Mycoplasma hominis NCTC 10111, Neisseria gonorrhoeae ATCC 49226, Candida albicans ATCC 10231, Lactobacillus spp. (clinical isolate), Escherichia coli ATCC 25922, Klebsiella pneumoniae (ATCC 13883) and Enterococcus faecalis ATCC 29212. An inhibition control (DNA of each organism spiked with positive control) was simultaneously tested for each organism.
Conventional PCR assay for amplification of 243 bp region of C. trachomatis cryptic plasmid: PCR targeting a sequence of the cryptic plasmid was done using primers KL-1 (5’TCCGGAGCGAGTTACGAAGA3’) and KL-2 (5’AATCAATGCCCGGGATTGGT3’) as described previously15.
Conventional PCR assay for amplification of 1071 bp region of C. trachomatis omp1 gene: PCR amplification of omp1 gene was performed using primers NLO (5’-ATG AAAAAACTCTTGAAATCG-3’) and NRO (5’-CTCAACTGTAACTGCGTATTT-3’) as described by Gao et al16.
COBAS Taqman CT Test, v 2.0: To evaluate the sensitivity, specificity, positive and negative predictive values of the real-time PCR for C. trachomatis, all samples tested positive for C. trachomatis by real time-PCR were analyzed by COBAS Taqman CT Test, v2.0. In addition, 30 consecutive samples negative by real-time PCR, collected between December 2011 and February 2012, were also subjected to COBAS Taqman CT test.
Simultaneous PCR amplification of target DNA using C. trachomatis specific complimentary primers and detection of cleaved dual fluorescent dye-labelled oligonucleotide probes were performed using the automated COBAS Taqman 48 analyzer system (Roche Diagnostics, Branchburg, New Jersey, USA) as recommended by the manufacturer. The C. trachomatis internal control (a plasmid containing the primer sequences used for the amplification of C. trachomatis) was always run in parallel for each sample to ensure the validity of each negative result.
A sample was considered true positive when it tested positive by two different PCR assays i.e. (i) real-time PCR positive and cryptic plasmid/omp1 gene conventional PCR positive, (ii) real-time PCR positive and COBAS Taqman CT Test positive, or (iii) cryptic plasmid/omp1 gene conventional PCR positive and COBAS Taqman CT Test positive.
Antigen detection assays: Detection of C. trachomatis antigen in the endocervical swabs was performed by DFA and EIA. The sensitivity, specificity, positive and negative predictive values of the real-time PCR was compared with that of the antigen detection assays.
Direct fluorescence assay (DFA): The endocervical swabs collected for chlamydia antigen detection by DFA were directly smeared onto wells of the MicroTrak slides (Syva Corp., California, USA). The slides were fixed with cold methanol and stained by fluorescin isothiocyanate (FITC) conjugated anti-Chlamydia monoclonal antibodies (Syva Corp., California, USA) for 30 min at 37°C in a humid chamber. The slides were examined for typical apple-green fluorescent elementary bodies (EBs) at x 1,000 magnification of fluorescent microscope. The positive and negative control slides provided by the manufacturer (Syva Corp, California, USA) were included in each batch of the test. The positive criterion was set at three distinct fluorescent EBs per smear.
Enzyme immunoassay for Chlamydia antigen detection: Detection of C. trachomatis antigen in the fourth endocervical swabs was performed using Microtrak II Chlamydia EIA kit (Syva Microtrak, California, USA) according to the manufacturer's instructions. Patient specimens with absorbance values greater than or equal to the cut-off value measured spectrophotometrically at 450 nm were considered presumptively positive for Chlamydia.
Statistical analysis: All statistical analyses were performed using STATA version 11.2 software (STATA Corp LP, TX, USA). Sensitivities, specificities, positive predictive values (PPVs) and negative predictive values (NPVs) were calculated with 95% confidence interval.
Results
During the two year study period, case sheets of 250 infertile women, in whom male factor infertility, hormonal and ovulatory disorders, PCOS, uterine fibroid, endometriosis and structural anomalies had been ruled out by initial screening, were reviewed. Of these 250 women, 49 (19.6%) women with diagnosis of genital tract tuberculosis and one with gonococcal infection were excluded. Two hundred women were found eligible and enrolled. The mean age of these 200 women was 30.09 ± 2.12 yr. Male partners of 34 per cent of the infertile women (68 males) agreed to participate in the study. The mean age of the male participants was 40.25 ± 2.32 yr.
Of the 200 women, 91 (45.5%) had primary infertility. The remaining 109 (54.5%) women belonged to the secondary infertility group. The most frequent cause of infertility was tubal blockade in 156 (78.5%) of women, followed by the presence of ovarian pathology like ovarian adhesions and oophoritis in 21 (10.5%) patients. In 23 (11.5%) of cases, cause of infertility remained unexplained. At the time of enrollment, 20 (10%) women were symptomatic for mucopurulent vaginal discharge, while 21 (10.5%) had complaints of lower abdominal pain. History of abortion and ectopic pregnancy were present in 78 (39%) and 26 (13%) of women, respectively.
Analytical sensitivity and specificity of real-time PCR: To determine the analytical sensitivity of the PCR, the plasmid that was constructed containing the target sequence was used as positive control. Ten-fold serial dilutions (1000 to 1 copies) of the positive control plasmid were run with the real-time PCR. The level of sensitivity of the real-time PCR was found to be 10 copies of target DNA per reaction. When 10-fold dilutions were tested in duplicate in five different runs, 100 per cent of replicates were positive at a concentration of 10 copies of C. trachomatis DNA per reaction, showing an excellent reproducibility with 10 copies.
No amplification was observed when DNA of eight bacteria, which may be present in urogenital samples, was tested with the real-time PCR, which indicated the high specificity of this PCR assay. In addition, presence of these bacterial DNA did not inhibit the amplification of the positive control in the real-time PCR cycle.
Presence of C. trachomatis infection by PCR assays: Of the 268 samples received at the laboratory during the two years study period, 200 (74.6%) were endocervical specimens and 68 (25.5%) were urine samples, collected from 200 women and 68 men, respectively. All samples were tested by real-time PCR and conventional PCR for C. trachomatis cryptic plasmid and omp1 gene.
The rates of C. trachomatis infection in the 200 infertile women as detected by conventional cryptic plasmid and/or omp1 gene PCR and real-time PCR were 11.5 per cent (23/200) and 13.5 per cent (27/200), respectively. There were no samples, which tested positive by conventional cryptic plasmid and/or omp1 gene PCR but negative by real-time PCR. Thus real-time PCR detected four (2%) extra samples positive for C. trachomatis compared to conventional PCR.
Three per cent (2/68) of the male partners tested positive for C. trachomatis by both cryptic plasmid and/or omp1 gene PCR and real-time PCR assays. Of the 68 couples enrolled in the study, two had both partners infected and two couples had only the female partner infected with C. trachomatis.
Comparison of real time-PCR and conventional PCR with COBAS Taqman CT Test, v2.0: To test the reliability of the real time-PCR assay, all the 27 endocervical swab samples previously tested positive by real time-PCR, were also tested by COBAS Taqman CT Test, v2.0. These 27 samples were found to be positive by COBAS Taqman CT Test, v2.0. Thus, the four endocervical swabs with discrepant results i.e. negative by cryptic plasmid and/or omp1 gene PCR and positive by real-time PCR and were considered true positives.
Of the 30 consecutive swab specimens collected between December 2011 and February 2012 that were negative by real-time PCR, 27 specimens were also found to be negative by COBAS Taqman CT Test. The remaining three specimens were found to be inhibited in the first run. When retested, all three were found negative. Thus, the real-time PCR exhibited 100 per cent sensitivity and specificity when considering COBAS Taqman CT Test as the gold standard. The positive and negative predictive values of the real-time PCR was 100 per cent. The diagnostic accuracy of the real-time PCR was also 100 per cent (Table I).
Table I.
Comparison of real time-PCR, conventional cryptic plasmid and omp1 gene PCR, direct fluorescence assay (DFA) and enzyme immunoassay (EIA) assays for C. trachomatis with COBAS Taqman CT Test, v 2.0 (n=57)
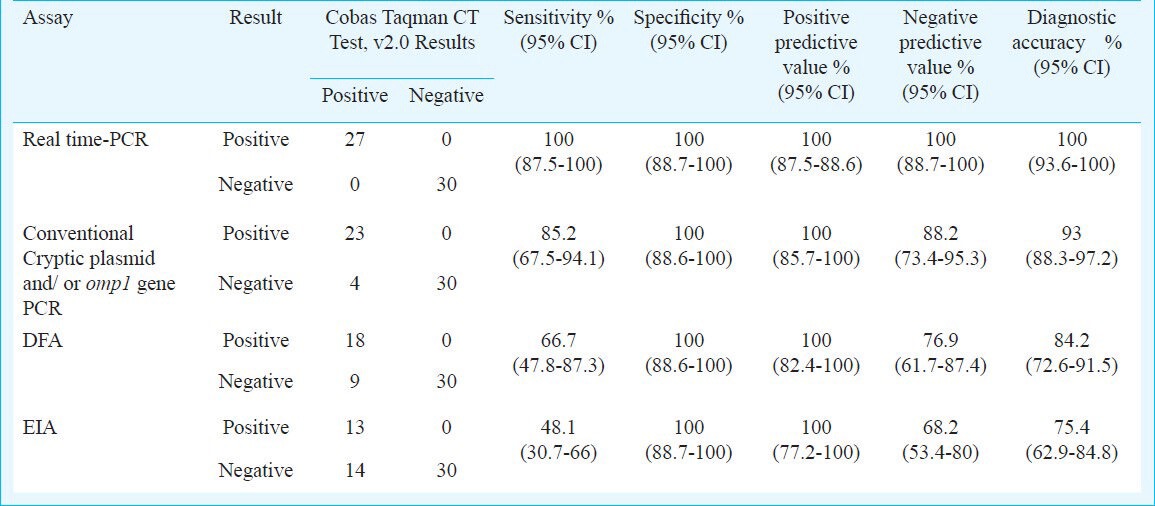
Compared with COBAS Taqman CT Test, conventional PCR for C. trachomatis demonstrated a sensitivity of 85.2 per cent, specificity of 100 per cent, positive predictive value of 100 per cent, negative predictive value of 88.2 per cent and a diagnostic accuracy of 93 per cent. Thus, real-time PCR was approximately 15 per cent more sensitive compared to conventional PCR along with a 12 per cent higher negative predictive value (Table I).
Results of DFA and EIA for detection of C. trachomatis: DFA and EIA detected 9 per cent (18/200) and 6.5 per cent (13/200) of infertile women as being positive for C. trachomatis, respectively. All the samples positive for C. trachomatis by DFA or EIA were identified as positive by both cryptic plasmid/omp1 gene PCR and real-time PCR assays. Compared with COBAS Taqman CT Test, DFA for C. trachomatis demonstrated a sensitivity of 66.7 per cent, specificity of 100 per cent, positive predictive value of 100 per cent and negative predictive value of 76.9 per cent. The sensitivity, specificity, positive and negative predictive values of ELISA for detection of C. trachomatis antigen from swab specimens were 48.1, 100, 100 and 68.2 per cent, respectively (Table I).
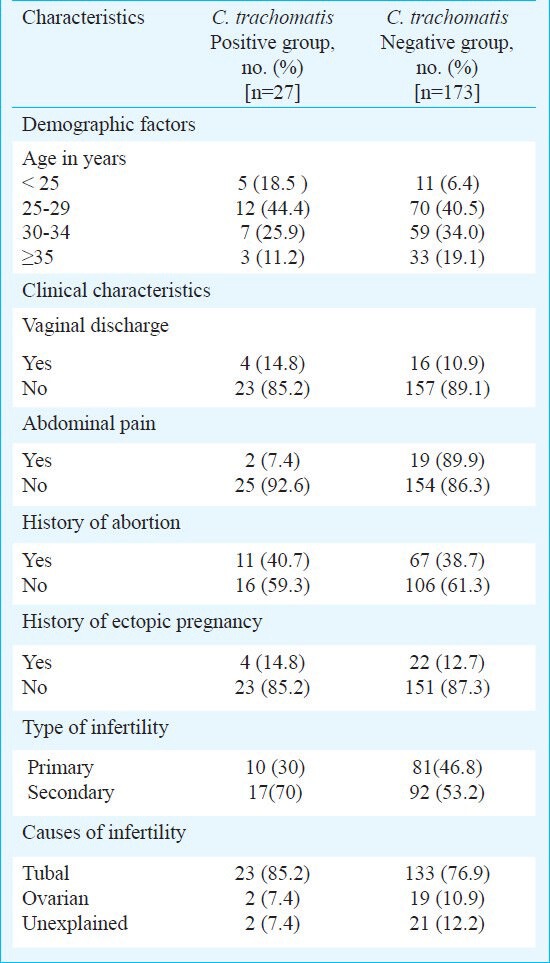
Association of C. trachomatis with demographic and clinical variables: Presenting symptoms and demographic characteristics of women with infertility in relation to C. trachomatis status are given in Table II. However, the rates of infection did not differ significantly between the different age groups. The presence of C. trachomatis infection was slightly higher in women with secondary infertility (15.6%) than in women with primary infertility (10.9%). The frequency of C. trachomatis infection was also found to be higher in women with tubal factor infertility (14.7%, 23/156). C. trachomatis infection was detected in 9.5 per cent (2/21) of infertile women with ovarian causes like ovarian adhesions and oophoritis. However, we found no association between detection of C. trachomatis in infertile women and the causes of infertility. No signs and symptoms could be identified as significantly associated with C. trachomatis infection in infertile women.
Table II.
Demographic and clinical characteristics of infertile women with and without C. trachomatis infection (n=200)
Discussion
Genital C. trachomatis infections are an extreme socio-economic and public health concern due to the potential for severe long-term consequences in women including infertility. This warrants mandatory screening of women attending infertility clinics17,18. Early and accurate diagnosis of C. trachomatis infection requires the use of highly sensitive and specific laboratory techniques.
The present results showed presence of C. trachomatis among 13.5 per cent infertile women as determined by the highly sensitive real-time PCR assay. Similar detection rates of C. trachomatis in infertile women by PCR have been reported in previous studies from developing countries19,20. El Qouqa et al19 detected C. trachomatis in 20.2 per cent of endocervical swabs from women attending gynaecology and infertility centres in Ghaza, Palestine by plasmid-based PCR. Similarly, frequency of C. trachomatis infection in infertile Iranian women by PCR was found to be 13.7 per cent20.
A low frequency of C. trachomatis (2.7%) infection using NAATs in subfertile women undergoing in vitro fertilization has been reported by de Barbeyrac et al21 from Bourdeaux, France. The low prevalence was attributed to the selection of a low-risk population for STI with respect to age (32 yr in women) and asymptomatic individuals, who apart from their infertility problem, were healthy individuals. In contrast, approximately 50 per cent of women enrolled in our study were below 30 yr of age and around 42 per cent were either symptomatic for vaginal discharge/low abdominal pain suggestive of pelvic inflammatory disease (PID) or had a history of abortion/ectopic pregnancy. Low detection rates (2.4%) of C. trachomatis in infertile women have also been reported from Ghana, where first-void urine FVU samples were investigated by PCR22. A higher sensitivity of endocervical specimens for diagnosing genital C. trachomatis infection in women by nucleic-acid amplification methods as compared to samples like FVU has been shown, which could contribute to the enhanced rates of C. trachomatis detection by our PCR assays. However, FVU has been found to be the most sensitive diagnostic specimen for C. trachomatis detection by NAATs in male patients23.
High seroprevalence of anti-chlamydia antibodies ranging from 40 to 65 per cent among infertile women had been observed24. In a study from India by Malik et al25. Chlamydia immunoglobin G (IgG) antibodies were detected in 55 per cent of women with secondary infertility. However, the same group of researchers in a previous study26 had reported a comparatively lower prevalence of C. trachomatis infection in infertile women by culture and antigen detection assays (27% in primary infertility patients and 30 per cent in secondary infertility patients). Similar discrepancies were observed in a study from Rwanda27 in which the overall positivity of C. trachomatis in infertile women as determined by PCR was 3.6 per cent compared to an IgG seroprevalence ranging between 17.3 to 18.8 per cent. A higher frequency of C. trachomatis infection in infertile women by serological testing was observed in these studies as compared to the detection rates by culture, antigen detection and PCR assays. This can be attributed to the persistence of Chlamydia antibodies in serum long after treatment and resolution of C. trachomatis infection. Thus, presence of antibodies cannot differentiate between an acute, chronic or a resolved C. trachomatis infection which is the major limitation of serological testing. In addition, cross-reactive antibodies can be induced in response to lipopolysaccharides of organisms of the genus Chlamydia including C. pneumoniae and Gram-negative bacterial lipopolysacchradies28.
The COBAS Taqman CT Test v2.0 (Roche Diagnostics, Branchburg, New Jersey, USA) had shown a high sensitivity (92.9%) and specificity (100%) on urine and endocervical specimens with positive and negative predictive values of 99.7 and 100 per cent, respectively29. The real-time PCR targeting the cryptic plasmid had sensitivity and specificity of which were equivalent to that of COBAS Taqman CT test. The real-time PCR used in the present study was able to detect a very low copy number of DNA (10 copies) per reaction, which probably explains the higher sensitivity of this assay compared to that of our conventional cryptic plasmid and omp1 gene PCR. Although isolates of C. trachomatis that lack the cryptic plasmid with the risk of producing false negative results have been described, plasmidless strains are extremely rare. Hence, primers and probes were designed targeting the cryptic plasmid, which is present in 7 to 10 copies in C. trachomatis.
Compared with NAATs, the C. trachomatis antigen detection by DFA and EIA in our study demonstrated low sensitivities of 66.7 and 48.2 per cent, respectively. It was further observed that 4.5 per cent samples tested positive by real-time PCR and COBAS Taqman CT Test v2.0 and hence considered true positives, were negative by DFA. Estimated sensitivity of DFA compared to culture and non-culture methods in different laboratory settings can vary from 61-92 per cent13. This can be explained by the fact that the diagnostic performance of the DFA test is highly dependent on the number of chlamydial elementary bodies present in a sample. Similarly, sensitivity of EIA applied to endocervical swabs can be as low as 60 per cent, using culture as the reference standard13. Thus, direct antigen detection by DFA and EIA cannot be recommended as appropriate screening tests for diagnosing C. trachomatis infection in infertile women.
Since women aged < 25 yr are more likely to have genital infections with C. trachomatis, selective screening of younger women in resource limited setting has been recommended28. In the present study also, the proportion of C. trachomatis infected infertile women was higher in the younger age group (<25 yr) although not statistically significant. Similar to seroprevalence studies26,28 in which genital chlamydia infection was associated with secondary infertility, the secondary infertility group in our study had a higher proportion of C. trachomatis infection. However, we could not find any significant difference in detection rates of C. trachomatis by PCR between women with primary and secondary infertility. Hence, all women attending an infertility clinic need to be screened for genital chlamydia infection, irrespective of their infertility status.
In our study the cryptic plasmid and omp1 gene PCR were performed on all samples as a part of routine diagnostic services. The real time PCR was also performed on all samples in parallel. However, the Roche PCR was done only on the samples, which tested positive by real-time PCR and on a limited number of consecutive swab samples that were negative by real-time PCR. The justification for this shortcoming was to save the cost of an expensive closed system commercial kit. In addition, our study was limited to 200 infertile women attending the outpatient department of a tertiary care hospital. Larger study population with multi-centric patient recruitment or spanning over a longer time period can be useful for a thorough evaluation of the diagnostic utility of in-house real-time PCR assays for C. trachomatis in infertile women in developing countries.
In conclusion, the real time-PCR targeting the cryptic plasmid of C. trachomatis used in this study had sensitivity and specificity comparable to that of COBAS Taqman CT Test v2.0. Thus, the real time-PCR has the potential to contribute in the initial screening for infertility in diagnostic laboratories in India.
Acknowledgment
Authors thank the Indian Council of Medical Research, New Delhi, India, for financially supporting the project. Authors also thank Dr Sarman Singh, Professor, Faculty-in Charge, Clinical Microbiology, Department of Laboratory Medicine, All India Institute of Medical Sciences, New Delhi, for allowing to use the Roche thermal cycler and COBAS Taqman CT Test v2.0.
References
- 1.Paavonen J, Eggert-Kruse W. Chlamydia trachomatis: impact on human reproduction. Hum Reprod Update. 1999;5:433–7. doi: 10.1093/humupd/5.5.433. [DOI] [PubMed] [Google Scholar]
- 2.Mania-Pramanik J, Kerkar S, Sonawane S, Mehta P, Salvi V. Current Chlamydia trachomatis infection, a major cause of infertility. J Reprod Infertil. 2012;13:204–10. [PMC free article] [PubMed] [Google Scholar]
- 3.Low N, Bender N, Nartey L, Shang A, Stephenson JM. Effectiveness of chlamydia screening: systematic review. Int J Epidemiol. 2009;38:435–48. doi: 10.1093/ije/dyn222. [DOI] [PubMed] [Google Scholar]
- 4.Low N, Hocking J. The POPI trial: what does it mean for chlamydia control now? Sex Transm Infect. 2010;86:158–9. doi: 10.1136/sti.2010.043737. [DOI] [PubMed] [Google Scholar]
- 5.Black CM. Current methods of laboratory diagnosis of Chlamydia trachomatis infections. Clin Microbiol Rev. 1997;10:160–84. doi: 10.1128/cmr.10.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watson EJ, Templeton A, Russell I, Paavonen J, Mardh PA, Stary A, et al. The accuracy and efficacy of screening tests for Chlamydia trachomatis: a systematic review. J Med Microbiol. 2002;51:1021–31. doi: 10.1099/0022-1317-51-12-1021. [DOI] [PubMed] [Google Scholar]
- 7.Schachter J, Stamm WE, Quinn TC. Discrepant analysis and screening for Chlamydia trachomatis. Lancet. 1996;348:1308–9. doi: 10.1016/S0140-6736(05)65783-2. [DOI] [PubMed] [Google Scholar]
- 8.Mylonas I. Female genital Chlamydia trachomatis infection: where are we heading? Arch Gynecol Obstet. 2012;285:1271–85. doi: 10.1007/s00404-012-2240-7. [DOI] [PubMed] [Google Scholar]
- 9.Mushanski LM, Brandt K, Coffin N, Levett PN, Horsman GB, Rank EL. Comparison of the BD Viper System with XTR Technology to the Gen-Probe APTIMA COMBO 2 assay using the TIGRIS DTS system for the detection of Chlamydia trachomatis and Neisseria gonorrhoeae in urine specimens. Sex Transm Dis. 2012;39:514–7. doi: 10.1097/OLQ.0b013e31824f2f5b. [DOI] [PubMed] [Google Scholar]
- 10.Van Der Pol B, Liesenfeld O, Williams JA, Taylor SN, Lillis RA, Body BA, et al. Performance of the Cobas CT/NG test compared to the Aptima AC2 and Viper CTQ/GCQ assays for detection of Chlamydia trachomatis and Neisseria gonorrhoeae. J Clin Microbiol. 2012;50:2244–9. doi: 10.1128/JCM.06481-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michel CE, Solomon AW, Magbanua JP, Massae PA, Huang L, Mosha J, et al. Field evaluation of a rapid point-of-care assay for targeting antibiotic treatment for trachoma control: a comparative study. Lancet. 2006;367:1585–90. doi: 10.1016/S0140-6736(06)68695-9. [DOI] [PubMed] [Google Scholar]
- 12.Semeniuk H, Zentner A, Read R, Church D. Evaluation of sequential testing strategies using non-amplified and amplified methods for detection of Chlamydia trachomatis in endocervical and urine specimens from women. Diagn Microbiol Infect Dis. 2002;42:43–51. doi: 10.1016/s0732-8893(01)00315-7. [DOI] [PubMed] [Google Scholar]
- 13.Sachdeva P, Patel AL, Sachdev D, Ali M, Mittal A, Saluja M. Comparison of an in-house PCR assay, direct fluorescence assay and the Roche AMPLICOR Chlamydia trachomatis kit for detection of C. trachomatis. J Med Microbiol. 2009;58:867–73. doi: 10.1099/jmm.0.008698-0. [DOI] [PubMed] [Google Scholar]
- 14.Jaton K, Bille J, Greub G. A novel real-time PCR to detect Chlamydia trachomatis in first-void urine or genital swabs. J Med Microbiol. 2006;55(pt 7):1667–74. doi: 10.1099/jmm.0.46675-0. [DOI] [PubMed] [Google Scholar]
- 15.Mahony J, Chong S, Jang D, Luinstra K, Faught M, Dalby D, et al. Urine specimens from pregnant and nonpregnant women inhibitory to amplification of Chlamydia trachomatis nucleic acid by PCR, ligase chain reaction, and transcription-mediated amplification: identification of urinary substances associated with inhibition and removal of inhibitory activity. J Clin Microbiol. 1998;36:3122–6. doi: 10.1128/jcm.36.11.3122-3126.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao X, Chen XS, Yin YP, Zhong MY, Shi MQ, Wei WH, et al. Distribution study of Chlamydia trachomatis serovars among high-risk women in China performed using PCR-restriction fragment length polymorphism genotyping. J Clin Microbiol. 2007;45:1185–9. doi: 10.1128/JCM.02076-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oakeshott P, Kerry S, Atherton H, Aghaizu A, Hay S, Taylor-Robinson D, et al. Community-based trial of screening for Chlamydia trachomatis to prevent pelvic inflammatory disease: the POPI (prevention of pelvic infection) trial. Trials. 2008;9:73. doi: 10.1186/1745-6215-9-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oakeshott P, Kerry S, Aghaizu A, Atherton H, Hay S, Taylor-Robinson D, et al. Randomised controlled trial of screening for Chlamydia trachomatis to prevent pelvic inflammatory disease: the POPI (prevention of pelvic infection) trial. BMJ. 2010;340:c1642. doi: 10.1136/bmj.c1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.El Qouqa IA, Shubair ME, Al Jarousha AM, Sharif FA. Prevalence of Chlamydia trachomatis among women attending gynecology and infertility clinics in Gaza, Palestine. Int J Infect Dis. 2009;13:334–41. doi: 10.1016/j.ijid.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 20.Hossein Rashidi B, Chamani Tabriz L, Haghollahi F, Ramezangadeh F, Shariat M, Rahimi Foroushani A, et al. Prevalence of Chlamydia trachomatis infection in fertile and infertile women: a molecular and serological study. J Reprod Infertil. 2009;10:32–41. [Google Scholar]
- 21.de Barbeyrac B, Papaxanthos-Roche A, Mathieu C, Germain C, Brun JL, Gachet M, et al. Chlamydia trachomatis in subfertile couples undergoing an in vitro fertilization program: a prospective study. Eur J Obstet Gynecol Reprod Biol. 2006;129:46–53. doi: 10.1016/j.ejogrb.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 22.Siemer J, Theile O, Larbi Y, Fasching PA, Danso KA, Kreienberg R, et al. Chlamydia trachomatis infection as a risk factor for infertility among women in Ghana, West Africa. Am J Trop Med Hyg. 2008;78:323–7. [PubMed] [Google Scholar]
- 23.Falk L, Coble BI, Mjörnberg PA, Fredlund H. Sampling for Chlamydia trachomatis infection - a comparison of vaginal, first-catch urine, combined vaginal and first-catch urine and endocervical sampling. Int J STD AIDS. 2010;21:283–7. doi: 10.1258/ijsa.2009.009440. [DOI] [PubMed] [Google Scholar]
- 24.Thomas K, Coughlin L, Mannion PT, Haddad NG. The value of Chlamydia trachomatis antibody testing as part of routine infertility investigations. Hum Reprod. 2000;15:1079–82. doi: 10.1093/humrep/15.5.1079. [DOI] [PubMed] [Google Scholar]
- 25.Malik A, Jain S, Rizvi M, Shukla I, Hakim S. Chlamydia trachomatis infection in women with secondary infertility. Fertil Steril. 2009;91:91–5. doi: 10.1016/j.fertnstert.2007.05.070. [DOI] [PubMed] [Google Scholar]
- 26.Malik A, Jain S, Hakim S, Shukla I, Rizvi M. Chlamydia trachomatis infection & female infertility. Indian J Med Res. 2006;123:770–5. [PubMed] [Google Scholar]
- 27.Muvunyi CM, Dhont N, Verhelst R, Temmerman M, Claeys G, Padalko E. Chlamydia trachomatis infection in fertile and subfertile women in Rwanda: prevalence and diagnostic significance of IgG and IgA antibodies testing. Hum Reprod. 2011;26:3319–26. doi: 10.1093/humrep/der350. [DOI] [PubMed] [Google Scholar]
- 28.Mårdh PA. Tubal factor infertility, with special regard to chlamydial salpingitis. Curr Opin Infect Dis. 2004;17:49–52. doi: 10.1097/00001432-200402000-00010. [DOI] [PubMed] [Google Scholar]
- 29.Rockett R, Goire N, Limnios A, Turra M, Higgens G, Lambert SB, et al. Evaluation of the cobas 4800 CT/NG test for detecting Chlamydia trachomatis and Neisseria gonorrhoeae. Sex Transm Infect. 2010;86:470–3. doi: 10.1136/sti.2010.042812. [DOI] [PubMed] [Google Scholar]


