Abstract
HepG2-conditioned medium (CM) facilitates early differentiation of murine embryonic stem cells (mESCs) into hematopoietic cells in two-dimensional cultures through formation of embryoid-like colonies (ELCs), bypassing embryoid body (EB) formation. We now demonstrate that three-dimensional (3D) cultures of alginate-encapsulated mESCs cultured in a rotating wall vessel bioreactor can be differentially driven toward definitive erythropoiesis and cardiomyogenesis in the absence of ELC formation. Three groups were evaluated: mESCs in maintenance medium with leukemia inhibitory factor (LIF, control) and mESCs cultured with HepG2 CM (CM1 and CM2). Control and CM1 groups were cultivated for 8 days in early differentiation medium with murine stem cell factor (mSCF) followed by 10 days in hematopoietic differentiation medium (HDM) containing human erythropoietin, m-interleukin (mIL)-3, and mSCF. CM2 cells were cultured for 18 days in HDM, bypassing early differentiation. In CM1, a fivefold expansion of hematopoietic colonies was observed at day 14, with enhancement of erythroid progenitors, hematopoietic genes (Gata-2 and SCL), erythroid genes (EKLF and β-major globin), and proteins (Gata-1 and β-globin), although ζ-globin was not expressed. In contrast, CM2 primarily produced beating colonies in standard hematopoietic colony assay and expressed early cardiomyogenic markers, anti-sarcomeric α-actinin and Gata-4. In conclusion, a scalable, automatable, integrated, 3D bioprocess for the differentiation of mESC toward definitive erythroblasts has been established. Interestingly, cardiomyogenesis was also directed in a specific protocol with HepG2 CM and hematopoietic cytokines making this platform a useful tool for the study of erythroid and cardiomyogenic development.
Introduction
Hematopoiesis in the early mouse embryo has been recapitulated by in vitro differentiation culture of human and murine embryonic stem cells (mESCs) by which mesodermal cells commit to the hematopoietic lineage before giving rise to other blood cells [1–7]. During embryogenesis, hematopoietic cells originate from the lateral plate mesoderm in an ordered program of development occurring in conjunction with cells of the cardiovascular system and share a common precursor, the hemangioblast [8–11]. The study of the hemangioblast, hemogenic endothelial cells and endothelial-to-hematopoietic transition [12–14] is critical to the understanding of related pathologies and for the development of a robust and reliable in vitro method of ESC differentiation toward cells, such as red blood cells (RBCs), which may be used in future clinical translational protocols [15–17].
Current protocols for in vitro production of RBCs from ESCs are fragmented and involve multiple (three) stages: (1) maintenance/expansion of undifferentiated ESCs, (2) spontaneous differentiation through formation of embryoid bodies (EBs) incorporating all three germ layers, and (3) dissociation of EBs with re-plating for terminal differentiation to desired lineages [16,18]. Ideal conditions for such cultures are not fully identified but result in sub-optimal control of differentiation and heterogeneity [19,20]. To date, the most efficient protocols for generation of oxygen-transporting, enucleated RBCs from ESCs require co-culture with feeder cells and a complex, fragmented multi-step process requiring at least 1 month of culture, which renders such protocols impractical and difficult to scale up [16,17]. EB cavitation during the process also results in cell loss and further reduces the yield and quality of cellular products [21,22]. In contrast to these two-dimensional (2D) cultures, three-dimensional (3D) systems can produce a controllable and scalable high cell-density culture of ESCs with a reduction in shear stress to cells that still maintain the capacity to terminally differentiate [23–26]. However, these methodologies for directed differentiation involved EB formation [25,26] and generated heterogeneous cell types [20] that were unsuitable for potential therapeutic applications. Furthermore, 3D cultures with increased cell density face mass transport limitations in static conditions resulting in an inhomogeneous environment, and a decrease in total cell output [27]. To overcome these limitations, we and others use bioreactors to accommodate dynamic ESC culture conditions and enable large-scale production [26,28–32].
Our previous work demonstrated that HepG2 conditioned medium (CM), obtained from cultures of the human hepatocarcinoma cell-line HepG2, enhanced mesoderm formation from mESCs [33,34] in the absence of EB formation [31,35,36], and facilitated early differentiation of hematopoietic cells in standard, monolayer 2D cultures [36]. We have recently developed an integrated bioprocess for the expansion and differentiation of mESCs using encapsulation and a rotating wall vessel (RWV) bioreactor system in a single-step process that does not require formation of EBs or the disruption of cell aggregates, and results in 3D tissue formation [31,32]. We now extend these studies and describe an efficient, integrated bioprocess for in vitro erythropoiesis from mESCs that (1) facilitates 3D culture through encapsulation of undifferentiated mESCs in alginate hydrogels, (2) uses CM to efficiently stimulate mesoderm formation, (3) bypasses EB formation, and (4) involves culture in an RWV bioreactor that requires minimal handling (cell passaging is not required) and which is scalable and automatable in a cost-effective manner. Our results demonstrate a viable platform for the convenient study and application of definitive erythroid development from mESCs based on early exposure to murine stem cell factor (mSCF) and, interestingly, also enables the study of the competitive developmental pathway used in hematopoiesis and cardiomyogenesis in the presence of hematopoietic cytokines.
Materials and Methods
Cell cultures
Cell lines and encapsulation of mESCs
Undifferentiated mESCs (E14/TG2-a cell line; ATCC; P<20) were cultivated according to standard mESC culture protocol [31,33] before the encapsulation process. Maintenance of HepG2 cells (ATCC, HB-8605; P<80) and preparation of CM was done as previously described [33]. Undifferentiated mESCs (10×106 cells) were suspended in 4 mL sterile-filtered (0.2 μm) alginic acid (w/v) and 0.1% (v/v) gelatin solution [both from Sigma-Aldrich; diluted in phosphate-buffered saline (PBS), pH 7.4] [31]. The cell-gel solution was passed through a peristaltic pump (Model P-1; Amersham Biosciences) and dropped using a 25-gauge needle into sterile alginate gelation solution (100 mM CaCl2; Sigma) with 10 mM HEPES (Sigma) and 0.01% (v/v) Tween (Sigma) at pH 7.4 with magnetic stirring. The alginate-encapsulated cells remained in gently stirred CaCl2 solution for 6–10 min, and were then washed twice with PBS.
Bioprocessing of 3D mESCs
A total number of 1×107 undifferentiated mESCs were encapsulated in alginate hydrogel to produce 500 cell beads (approximately 2×104 cells per bead; bead diameter=2.5 mm), and the beads were cultured in a 50 mL RWV HARV bioreactor (Synthecon), with daily replenishment of medium. Three groups were assessed simultaneously in RWV culture (Table 1). The control group consisted of undifferentiated mESCs encapsulated and cultured for 3 days in standard maintenance medium with leukemia inhibitory factor (LIF) as previously described [31,33], followed by 8 days in early differentiation medium (EDM) containing Iscove's modified Dulbecco's medium (IMDM; Invitrogen), 15% FBS (Gibco), 2 mM l-glutamine (Gibco), 0.45 mM α-monothio-glycerol (MTG; Sigma), 10 mg/mL bovine insulin, 5.5 mg/mL human transferrin, 5 ng/mL sodium selenite (ITS liquid media supplement; Sigma), and 50 mg/mL ascorbic acid (Sigma), and supplemented 40 ng/mL mSCF (R&D Systems). Thereafter, the cells were cultured for 10 days in hematopoietic differentiation medium (HDM) directed toward erythropoiesis consisting of EDM with mSCF supplemented with 10 ng/mL mIL3 and 3 U/mL human erythropoietin (hEPO) (all from R&D Systems; modified from previous studies [36,37]). The first experimental group (CM1) consisted of undifferentiated mESCs encapsulated and cultured for 3 days in a 1:1 (v/v) mixture of HepG2 CM and standard maintenance basal medium (BM) [33,36], followed by 8 days of culture in EDM with mSCF and 10 days in HDM, described earlier. The second experimental group (CM2) was similarly cultured for 3 days in a 1:1 (v/v) mixture of HepG2 CM and BM, followed by direct culture in HDM (bypassing the early differentiation step) for 18 days (Table 1). In preliminary experiments, 3D-encapsulated mESCs exposed only to CM throughout the 21 day RWV culture period (with LIF added in the first 3 days only) was also evaluated as an additional control group.
Table 1.
Experimental Plan
| Experimental conditions | ||||
|---|---|---|---|---|
| 2D | 3D | |||
| Experimental groups | Maintenance | Maintenance | Early differentiation | Late differentiation |
| Control | BM+LIF (3 days) | BM+LIF (3 days) | EDM+mSCF (8 days) | HDM (10 days) |
| CM1 | BM+LIF (3 days) | BM+HepG2 CM (3 days) | EDM+mSCF (8 days) | HDM (10 days) |
| CM2 | BM+LIF (3 days) | BM+HepG2 CM (3 days) | HDM (18 days) | |
Three mESCs groups were assessed: (1) control (3 days' standard maintenance medium and then 8 days' early differentiation medium), (2) CM1 (3 days' CM and then 8 days' early differentiation medium), and (3) CM2 (3 days' CM, bypassing early differentiation). All underwent hematopoietic differentiation with hEPO, mIL-3, and mSCF to 21 culture days.
2D, two-dimensional; 3D, three-dimensional; BM, basal medium; EDM, early differentiation medium; HDM, hematopoietic differentiation medium; LIF, leukemia inhibitory factor; mESC, murine embryonic stem cell; mSCF, murine stem cell factor.
Live and dead assay
Viability of cells within the alginate hydrogels was observed using two-colour fluorescent Live/Dead Assay (Molecular Probes) as per manufacturer's instructions. Briefly, 4 mM EthD-1 and 2 mM calcein AM solution was prepared in PBS and added to the alginate-encapsulated cells, with 30 min incubation at room temperature in the dark. The cell-hydrogel beads were washed twice with PBS and visualized under an Olympus BX51 microscope (Olympus). Images were captured using the F-view unit (Soft Imaging System GmbH) with U-MNIBA3 (FITC) and U-MWIY2 (Texas Red) filters. Image staining intensity was quantified with Image J analysis software [38] (National Institute of Health).
MTS assay
Growth kinetics of encapsulated cells was evaluated by the Cell Titer 96 Aqueous One Solution Reagent MTS assay (Promega). MTS solution (20 μL) in 100 μL fresh medium was added to five hydrogel beads and incubated for 3 h before assessment of the resultant color change (absorbance at 490 nm) using an enzyme-linked immunosorbent assay reader (ELx808; BIO-TEK).
Metabolic analysis
Culture supernatant (1.5 mL) was collected every 3 days and concentrations of glucose, glutamine, ammonia, lactate, and pH level were assessed using a Bio-Profile Analyzer 400 (Nova-Biomedical). Fresh medium in culture, without cells, served as the control.
Cell immunophenotyping
Fifty alginate cell-hydrogel beads were collected and dissolved in a sterile depolymerization buffer consisting of 50 mM tri-sodium citrate dehydrate (Fluka), 77 mM sodium chloride (Fisher Scientific), and 10 mM HEPES (Sigma) for 15–20 min by gentle stirring. After dissolution, de-capsulated cells were washed twice with PBS. A cell suspension (100 μL) of 1×106 cells/mL was prepared in PBS, loaded into a cuvette well of a cytocentrifuge (Hettich Rotina 46R), and spun for 3 min at 130 g. Cells were incubated with directly conjugated anti-mouse CD-71 fluorescein isothiocyanate-immunoglobin (FITC; clone RI7217) and anti-mouse TER119 phycoerythrin (PE; clone TER-119) antibodies (all from Biolegend) in PBS buffer containing 2% (v/v) FBS and 0.1% (w/v) sodium azide (Sigma). Isotype controls (all from Biolegend) IgG2ak-FITC (Clone RTK2758) and IgG2b-PE (Clone RTK4530) were used.
Immunohistochemistry of beating colonies formed from the colony-forming unit (CFU) assay of the CM2 group was performed after a gentle washing step to remove methylcellulose, followed by fixing with paraformaldehyde for 1 h. Primary antibody, rabbit anti-mouse sarcomeric α-actinin (1:100 polyclonal) and secondary antibody, goat-anti-rabbit IgG-FITC (1:150) were used as per manufacturer's instructions (all from Abcam). Samples were washed twice in PBS (Gibco) and mounted using Vectashield TM with 1.5 μg/mL 4′,6′ diamidino-2-phenylindole (DAPI) (Vector Laboratories). Cells and colonies were visualized on an Olympus BX51 microscope, and images were captured using the F-view unit (GmbH) with U-MNIBA3 (FITC) and U-MWIY3 (PE) filters. Image staining intensity was quantified with Image J analysis software [38] (NIH). Images were not manipulated.
Hematopoietic CFU assay
After 21 days culture in the RWV, a cell suspension containing 2×105 cells/mL was collected by dissolving alginate cell hydrogels in a sterile depolymerization buffer as outlined earlier. The cells were then plated in standard methylcellulose hematopoietic colony-forming medium containing IMDM supplemented with 15% FBS, 2 mM l-glutamine, 150 μM MTG, 1% BIT (1% BSA, 10 μg/mL Insulin, and 200 μg/mL Transferrin; Stem Cell Technologies), 150 ng/mL mSCF, 30 ng/mL mIL-3, 30 ng/mL hIL-6, 3 U/mL hEPO (all from R&D), and 1% basic methylcellulose medium (Stem Cell Technologies). Cell suspensions were placed on low-adherence 35 mm Petri dishes (VWR International Ltd.) and cultivated for an additional 14 days at 37°C with 5% CO2 in a humidified incubator. Resultant colonies were scored and counted using standard criteria for burst-forming unit-erythroid (BFU-E), CFU-erythroid (CFU-E), CFU-granulocyte, macrophage (CFU-GM), and CFU-granulocyte, erythrocyte, monocyte, megakaryocyte (CFU-GEMM). Colonies were individually plucked for further analysis.
Wright–Giemsa staining
Cytospins (1×106 cells/mL) from CFUs of control and CM1 groups were prepared, stained with Wright–Giemsa (Sigma; 10 s), and subsequently washed in deionized (DI) water for 2 min. Cells were visualized with an Olympus BX51 microscope; images were captured using the DP50 camera (Olympus) and not manipulated after capture.
Reverse transcriptase-polymerase chain reaction
Cell pellets from de-capsulated beads and CFUs were stored at −80°C before RNA extraction using the total RNA isolation kit (Qiagen Ltd.). Single-stranded cDNA synthesis was performed with 1 mg total RNA using a random primer and AMV reverse transcriptase with an RNase inhibitor (Promega). The subsequent polymerase chain reaction was performed as previously described [36] with primers detailed in Table 2.
Table 2.
Description of Polymerase Chain Reaction Primers
| Reverse transcriptase polymerase chain reaction primers | ||||
|---|---|---|---|---|
| Genes | Forward 5′-3′ | Reverse 5′-3′ | Length (bp) | PCR cycles |
| Pluripotency | ||||
| Oct-4 | TGTGGACCTCAGGTTGACT | CTTCTGCAGGGCTTTCATGT | 181 | 27 |
| Cardiomyogenic progenitors | ||||
| Gata-4 | CTCCTACTCCAG CCCCTACC | GTGGCATTGCTGGAGTTACC | 571 | 30 |
| α-MHC | ACCGTGGACTACAACAT | CTTTCGCTCGTTGGGA | 285 | 33 |
| Mesoderm | ||||
| Brac-T | AAGGAACCACCGGTCATCG | CGTGTGCGTCAGTGGTGTGTAATG | 321 | 27 |
| Flk-1 | CCTGGTCAAACAGCTCATCA | AAGCGTCTGCCTCAATCACT | 599 | 30 |
| Hematopoietic progenitors | ||||
| Gata-2 | TGCAACACACCACCCGATACC | CAATTTGGACAACAGGTGCCC | 336 | 35 |
| SCL | TAGCCTTAGCCAGCCGCTCG | GCGGAGGATCTCATTCTTGC | 277 | 35 |
| Erythroid | ||||
| EKLF | ACCACCCTGGGACAGTTTCT | GAAGGGTCCTCCGATTTCAG | 519 | 35 |
| Gata-1 | ATGCCTGTAATCCCAGCACT | TCATGGTGGTAGCTGGTAGC | 581 | 35 |
| βH1-globin | TTTGCACACTTGAGATCATCTC | GGTCTCCTTGAGGTTGTTCCATG | 610 | 30 |
| β-globin | ATGGTGCACCTGACTGATGCTG | GGTTTAGTGGTACTTGTGAGCC | 447 | 30 |
| House-keeping | ||||
| GAPDH | CATCACCATCTTCCAGGAGC | ATGCCAGTGAGCTTCCCGTC | 500 | 27 |
Oct-4, octamer-binding transcription factor-4; Gata-1, -2, and -4, glutamyl-tRNA(Gln) amidotransferase subunit A -1, −2, and -4; α-MHC, alpha myosin heavy chain; Brac-T, brachyury transcription factor T-gene; Flk-1, fetal liver kinase-1; SCL/Tal-1, stem cell leukemia/T-cell acute lymphocytic protein 1; EKLF,- erythroid-like kruppel factor; βH1-globin, hemoglobin beta-H1 chain; β-globin, beta-major globin; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; PCR, polymerase chain reaction.
Western blot analysis
Total protein extracted from CFUs of control mESCs, CM1, and CM2 groups was quantified using the BCA™ Assay Kit (Thermo Fisher Scientific). BSA (Sigma) was used as a known protein standard. Protein blotting reactions were performed as previously described [36]. Primary antibodies used were rabbit anti-mGata-1 polyclonal (1:2,000), rabbit anti-mζ-globin polyclonal (1:2,000), chicken anti-mβ-major globin polyclonal (1:2,000), and rabbit anti-mGAPDH polyclonal (1:20,000). Secondary antibodies, anti-rabbit, and anti-chicken polyclonal IgG were conjugated to Horseradish peroxide (HRP; 1:10,000 was used for each antibody, all antibodies were from Santa Cruz Biotechnology).
Statistical analysis
All experiments were done in quadruplicate (n=4) on three separate occasions (n=3), and each value represents mean±standard deviation. Statistical significance was evaluated using two-way analysis of variance (ANOVA) with P values ≤0.05 considered significant.
Results
Alginate-encapsulated mESCs exposed to HepG2 CM and hematopoietic cytokines have enhanced viability and proliferation yet similar metabolism
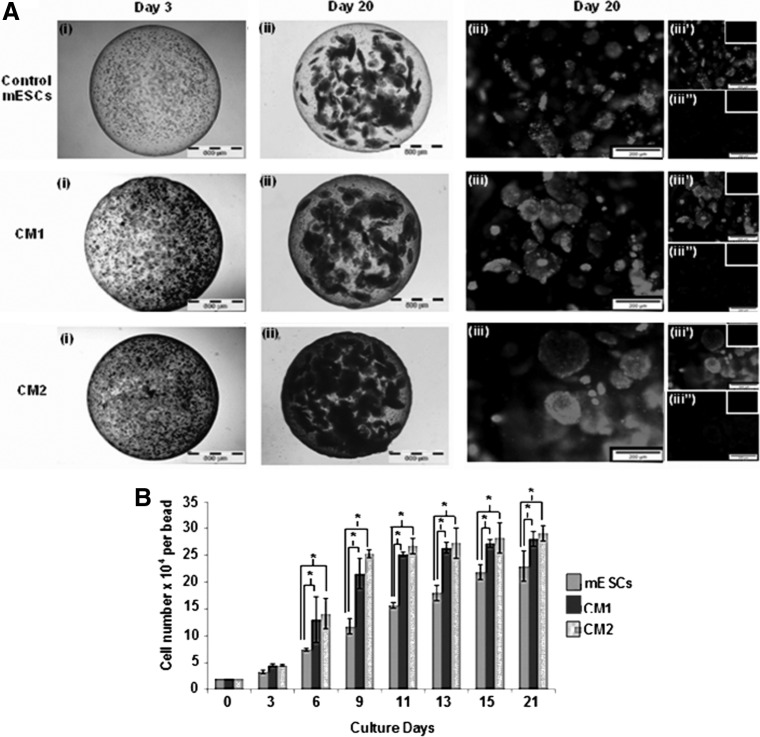
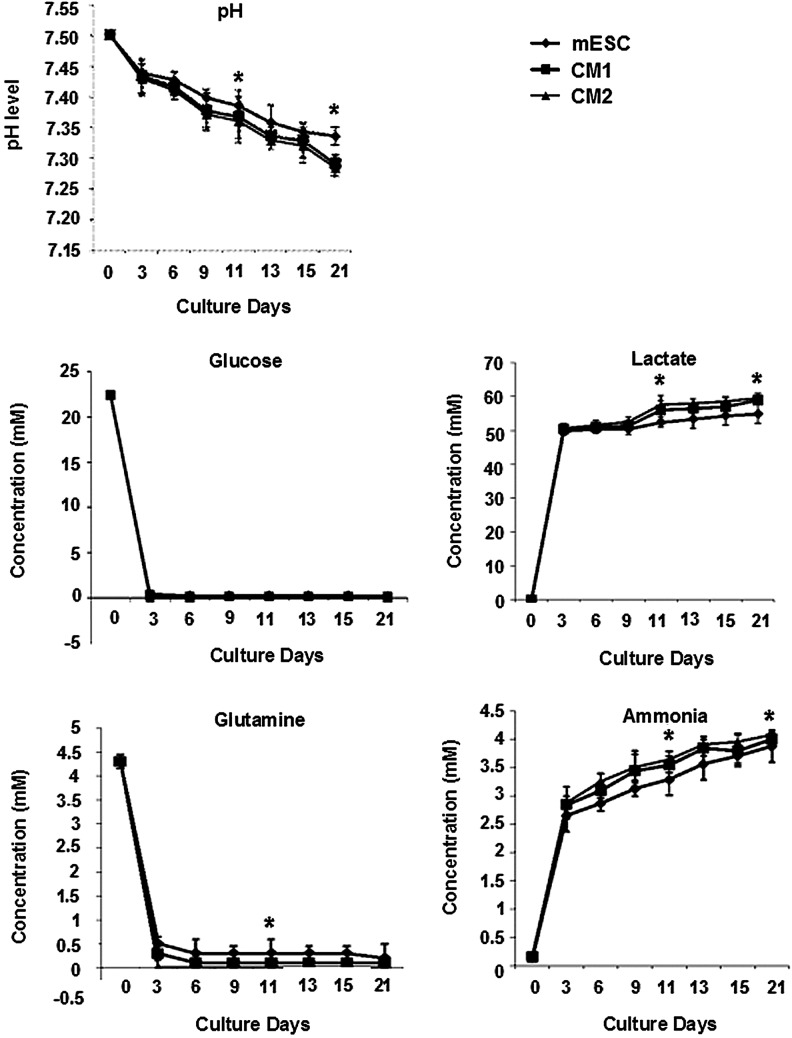
Viability of encapsulated mESCs within the alginate hydrogels was determined using a two-color fluorescence live/dead assay that measures intracellular esterase activity and plasma membrane integrity. At day 3, encapsulated undifferentiated mESCs were observed in small cell aggregates of 20 μm (Fig. 1). However, during early differentiation and terminal hematopoietic differentiation, more and larger viable cell aggregates were formed after 20 days of culture with the largest observed in CM2 (200 μm)>CM1>Control (100 μm), indicating that nutrient mass transport limitations were not experienced within the hydrogels. Cells of the CM2 group had the highest viability (>95%), 2.3-fold higher than that of the control group (1×; CM1 1.5-fold). A higher proliferation rate was observed for CM2 and CM1 when compared with that of the control from 6 days until 15 days of culture before reaching a plateau phase; CM2 had the highest number of cells at the end of culture with an approximately 15-fold expansion (Fig. 1B). Correlating with these higher cell densities later in the culture, the pH level initially was 7.5 and gradually declined to 7.3 (Fig. 2). In contrast, nutrient consumption kinetics showed a significant reduction from 23 to 0 mM and from 4.5 to 0.15 mM, for glucose and glutamine, respectively, even at day 3 of culture with a commensurate accumulation of lactate and ammonia. The kinetics was indicative of the high metabolic activity of alginate-encapsulated cells within the RWV bioreactor cultures. Interestingly, despite these drastic changes in nutrients and metabolites, the viability and proliferation of cells in culture (Fig. 1) was adequately supported by medium exchange every 3 days. Although the CM2 group had a similar proliferation profile to that of CM1 (both were higher than the control), there was a significant difference in pH level, glutamine consumption, lactate and ammonia production between CM2 and both CM1 and control on day 11 (when mESCs and CM1 were exposed to HDM) and at the end of the culture, indicative of the lower metabolic requirements of maturing cells within CM2 at these time points. In summary, encapsulation and culture in a RWV bioreactor facilitated high viable cell densities at 21 days, with a total output of 1.5×108 cells total in 500 hydrogel beads (15-fold expansion), minimal mass-transport effects, and the potential for control and optimization of metabolic parameters.
FIG. 1.
Morphology, viability, and proliferation of three-dimensional (3D)-murine embryonic stem cells (mESCs). (A) Representative mESC-beads on (i) day 3 and (ii) day 20. Magnification: 4×; Scale: 500 μm. (iii) day 20 viability in composite images of Calcien AM (iii’) and nonviable cells (Ethidium homodimer-1;) (iii’’); negative controls in insets. Magnification: 20×. Scale: 200 μm. (B) Cell proliferation over 21 days [mean number of cells±standard deviation (SD); n=3, *P≤0.05].
FIG. 2.
pH, nutrient, and metabolite concentrations in supernatants. pH, glucose, and glutamine levels decreased during 21 days of culture in all groups, concomitant with lactate and ammonia accumulation by day 3 when cells were exposed to differentiation medium. Differences were observed in CM2 on day 11 and at the end of culture, day 21. *P≤0.05.
Directed hematopoietic differentiation is dependent on early exposure to mSCF, whereas cardiomyogenic differentiation occurs with early exposure to mSCF, mIL-3, and hEPO
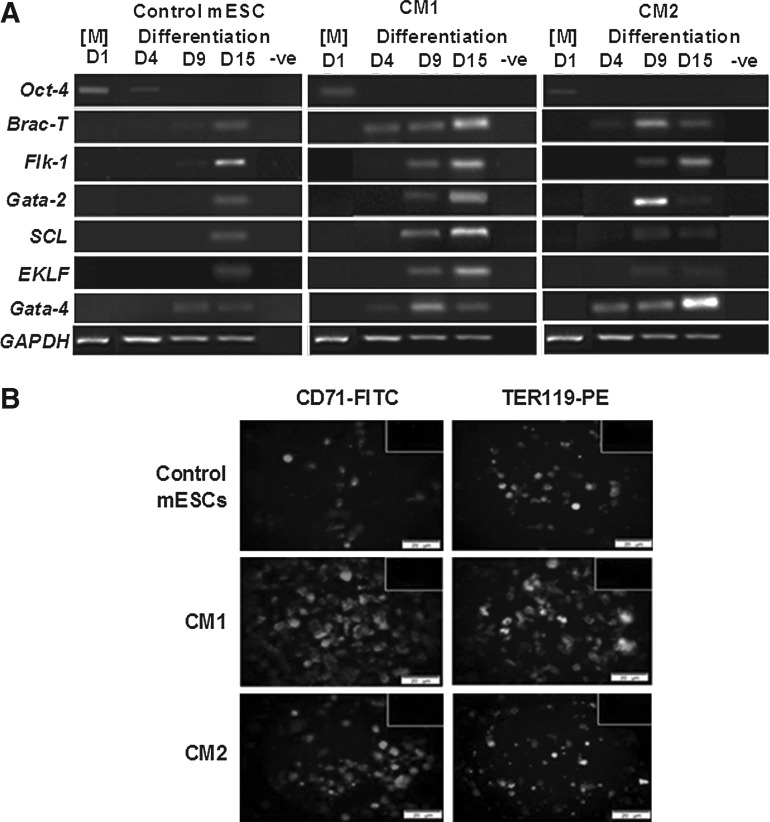
Gene expression analysis was performed on the de-capsulated cells at days 1, 4, 9, and 15 of culture (Fig. 3A). Expression of the pluripotency gene, Oct-4, was maintained in the control group until at least day 4, indicating that at least some pluripotent capacity was retained, which was in contrast to the CM1 and CM2 groups exposed to the HepG2 CM. During differentiation, the mesoderm genes, Brachyury (Brac-T) and Flk-1, were clearly expressed early at day 4 and 9 in the CM1 and CM2 groups with delayed expression at day 15 in the control group, consistent with the known effects of HepG2 CM in enhancing mesoderm formation [33,36]. Expression of the early hematopoietic and erythroid genes Gata-2, SCL, and EKLF was earlier in CM1 and CM2 compared with that of the control, with the highest consistent expression in CM1. Interestingly, although Gata-4 increased from day 4 in all conditions, a distinct high intensity band was observed in CM2 at day 15 of culture in conjunction with maintained expression of Flk-1 and coincident with a fall in all hematopoiesis-specific genes, in particular Gata-2, which peaked at day 9; this expression pattern was suggestive of cardiomyogenic gene upregulation under the influence of hematopoietic cytokines within the differentiation medium. The enhanced hematopoietic maturation in CM1 by day 15 was supported by the increased expression of erythroid antigens CD71 (2.6-fold higher) and TER119 (1.8-fold higher) compared with that of CM2 (1.4-fold CD71, 1.1-fold TER119) and control (1×) groups (Fig. 3B). These results suggested that the conditions of CM1 were most conducive to directed erythroid differentiation and that of CM2, which bypassed the early differentiation stage, expressed a more prominent cardiomyogenic signature on direct exposure to hematopoietic cytokines.
FIG. 3.
Gene and protein assessment of 3D alginate-encapsulated mESCs. (A) Time course of gene expression by reverse transcriptase-polymerase chain reaction. Negative controls consisted of sample without cDNA. [M]=maintenance stage. (B) Immunohistochemistry of erythroid markers CD71 and TER119 at day 15 of culture. Negative controls in insets. Magnification: 20×. Scale: 200 μm.
Early exposure to mSCF favored definitive erythroid colony development, whereas early exposure to mSCF, mIL-3, and hEPO resulted in cardiomyogenic beating colonies (“CFU-Beat”)
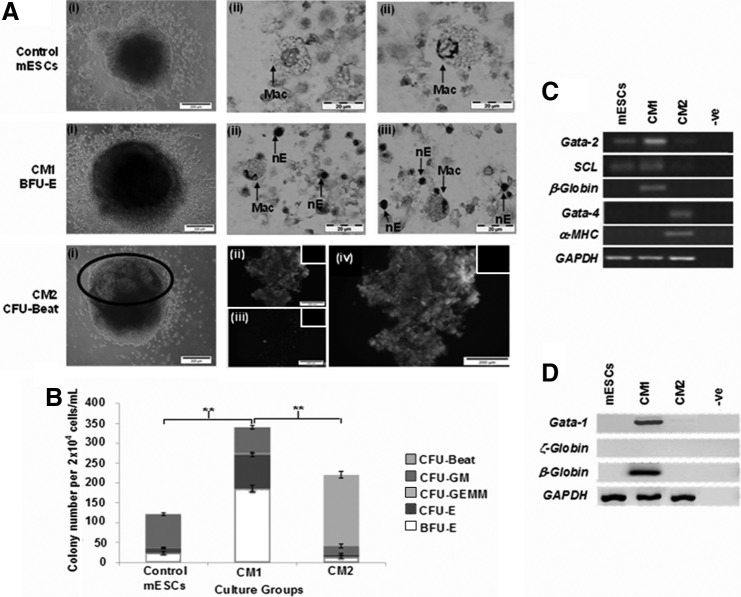
At the end of the 21-day culture period, de-capsulated terminally differentiated cells were re-plated in methylcellulose with a hematopoietic cytokine cocktail in CFU assay to assess functionality. The control group formed mostly myeloid progenitors, identified by formation of CFU-GM colonies, confirmed by Wright–Giemsa staining showing macrophages (Fig. 4A). In contrast, a higher number of hematopoietic progenitor colonies, consisting mostly of erythroid colonies, BFU-E, and CFU-E, was achieved from the CM1 group after 14-day methylcellulose culture (Fig. 4B; P≤0.01) with a fivefold enhancement compared with that of control and CM2 groups. Macrophages with nucleated erythrocytes (nE) were observed in cytospins of CM1, indicative of erythroid islands needed to support BFU-Es [39] (Fig. 4A). Interestingly, despite the early exposure to mSCF, mIL-3, and hEPO and the hematopoietic cytokine cocktail inherent in standard methylcellulose CFU assay, beating colonies (“CFU-Beat”) dominated the CFU assay of CM2 (Supplementary Video S1; 22 beats per minute observed; Supplementary Data are available online at www.liebertpub.com/scd). CFU-Beats were only observed in the CM2 group, with a small population of mixed hematopoietic colonies formed (16% of total). Encapsulated mESCs exposed only to HepG2 CM for 21 days in the RWV bioreactor did not form CFU-Beats (data not shown). Fluorescence staining for anti-sarcomeric α-actinin confirmed cardiomyogenic differentiation in the CFU-Beat (Fig. 4A). Further gene (Fig. 4C) and protein analysis (Fig. 4D) of CFUs from CM1 showed higher Gata-2, SCL, and β-major globin gene expression as well as protein expression of the early erythroid marker Gata-1 and β-globin with lack of ζ-globin, confirming that hematopoietic differentiation toward the definitive erythroid lineage was successfully achieved. In contrast, expression of Gata-4 and α−MHC genes was only observed from colonies collected from CM2, indicative of the dominant cardiomyogenic differentiation pathway in CFU-Beats on exposure to standard hematopoietic cytokines of the CFU assay. CFUs from the control group expressed early hematopoietic markers, Gata-2 and SCL, without expression of erythroid genes or proteins, confirming that the hematopoietic differentiation program was committed toward hematopoietic-myeloid lineage. These results indicated that, in this 3D system, early exposure of mESCs to mSCF directed definitive erythroid maturation, whereas early exposure to mSCF, mIL-3, and hEPO resulted in functional cardiomyogenic differentiation preferentially even on direct exposure to hematopoietic cytokines in standard hematopoietic CFU assay.
FIG. 4.
Clonogenic capacity. (A)(i) CFU morphology (beating area in black circle). (ii&iii) Wright–Giemsa (Mac=macrophages; nE=nucleated erythrocytes) and immunostaining of plucked colonies [(ii) anti-sarcomeric α-actinin-FITC, (iii) DAPI, (iv) composite image]. Negative control insets. Magnification: 20×. Scale: 200 μm. (B) Summary of CFUs (mean cell number±SD, n=3; **P<0.01). (C) CFU gene expression. (D) Western blot of CFUs. (Negative controls=no antibody/cDNA/protein loaded).
Discussion
We have described an integrated single-step 3D bioprocess for the efficient and directed generation of definitive erythroid and cardiomyogenic cells from alginate-encapsulated mESCs in the absence of standard EB or embryoid-like colony-formation. The formation of erythroid progenitors and beating colonies based on early exposure to hematopoietic cytokines was most interesting and indicated a differential effect of the hematopoietic cytokines IL-3 and EPO (in combination with SCF) toward cardiomyogenesis in the absence of specific cardiomyogenic factors, whereas SCF alone was most conducive to erythropoiesis from the mesoderm stage of development. This 3D system, more reflective of in vivo events than 2D systems, will be instructive as an in vitro model of erythropoiesis, in the study of the hemangioblast and in the competitive hematopoietic-cardiomyogenic developmental pathways. It could also provide a viable, robust, automatable, and scalable platform for large-scale production of stem cell-derived RBCs, and other ESC-derived cellular products, in translational protocols.
The ability to monitor and modulate pH, glucose/glutamine consumption, lactate and ammonia production is instrumental in controlling and standardizing cell growth and differentiation outputs [40]. Although a drastic decline in glucose/glutamine concentrations and an accumulation of metabolic by-products was observed, the pH level (7.3) was acceptable [41] during the culture period and indicated that the feeding schedule was practical. Indeed, low-glucose conditions may be more suitable for ESC differentiation and growth [42–45], and are conducive to myeloid and erythroid growth [46] with glutamine-deficient conditions inducing erythroid differentiation [47,48]. Although glucose uptake increases during cardiomyogenesis [49], low glucose conditions can also support cardiomyogenic differentiation [50]. The effect of pH on cardiomyogenesis is not elucidated, but lower pH levels may induce formation of the three germ layers during ESC culture [51], and erythroid differentiation accelerates with increasing extracellular pH (range 7.1–7.6) [52]. The day 11 and 21 time points of the culture indicated altered metabolism of cells in the CM2 condition, leading to cardiomyogenesis and a future study of metabolic pathways in this model system may, therefore, be informative. The degree to which these parameters affect speed, type, quality, and homogeneity of cell differentiation, especially in 3D tissue culture, needs to be explored.
Early exposure of encapsulated mESCs to SCF in CM1 followed by continuous SCF and addition of EPO and IL-3 directed definitive erythropoiesis and is consistent with what is already known about the actions of these cytokines on hematopoietic stem and progenitor cells and erythroid development. SCF increases cell survival and proliferation of early erythroid progenitors and precursors toward CFU-E, and at the proerythroblast stage [53]. At later maturational stages, its main role is to prevent apoptosis and induce proliferation [54]. In synergy with EPO and SCF, more primitive erythroid progenitors require IL-3 to survive and proliferate in vitro [55]. EPO dramatically increases the number of CFU-E and erythroid precursors up to the stage of basophilic erythroblasts, mainly by preventing apoptosis [56]. The observation that early exposure to all three cytokines was most conducive to cardiomyocyte development rather than erythropoiesis may provide further insights into morphogens and patterning in response to these cytokines during embryogenesis [57] as well as the impact they have on combined pathological states of cardiac and bone marrow tissues in the adult.
One of the first key events that precedes the emergence of hematopoietic cells in in vitro studies of mESCs is the expression of the early mesoderm marker, Brac-T, and the coincident appearance of vascular endothelial growth factor (VEGF)-independent cells which form transitional mesoderm colonies of Brac-T+Flk1+ cells [58]. Flk-1 (VEGFR2) -positive cells from the primitive streak in early embryogenesis represent one of the earliest mesoderm precursors that are capable of forming cardiac [9], endothelial, and primitive erythroid cells [59]. The next stage is a Brac-T+Flk1+ VEGF-responsive blast (BL)-colony forming cell (CFC) that forms colonies of Brac-T-Flk1+SCL+ cells containing the hemangioblast, generating both endothelial and hematopoietic (either primitive and/or definitive) progeny [1,3]. Our current findings of Brac-T and Flk-1 upregulation, with variable SCL expression, in CM1 and CM2 from day 9 to 15 suggests hemangioblast development in the 3D culture [60]. The spontaneous formation of CFU-Beats after early and continued exposure to hematopoietic cytokines and without further adaptation, apart from standard hematopoietic methylcellulose CFU assay, was unexpected and suggested that cardioprogenitor commitment had occurred in 3D RWV by day 21 in CM2. Previously described cardiovascular-CFCs (CV-CFCs) from day 4.25 Flk-1+ multipotential cardiovascular progenitors were likely generated by a different progenitor population, had cardiac and vascular potential (rather than cardiac and hematopoietic potential as observed here), and occurred only after primary culture in cardio-conductive medium with subsequent secondary liquid culture in order to expand cells and induce contractions [9]. Although it is possible that other factors in HepG2 CM were instrumental in cardiomyogenesis (eg, fibronectin and myosin, heavy chain 3 [61]), there was differential maturation toward erythropoiesis in the absence of IL-3 and EPO, and on exposure of mESCs to CM alone for 21 days in the RWV bioreactor, no CFU-Beats were observed (data not shown), indicating that HepG2 CM mainly played a role in inducing earlier onset mesoderm formation [33,36] rather than having a critical role in the cardiomyogenic developmental pathway. Hematopoietic cytokines affect cardiomyogenic differentiation. In murine embryos, EPO triggers cardiomyocyte proliferation [62]; its receptor is detected by E10.5 until day E14.5 [62,63], and IL-3 can induce endothelial cell formation as well as stimulate and chemo-attract smooth muscle cell proliferation and migration [64]. The formation of mixed lineage hematopoietic cells in the same cultures as CFU-Beats suggests that this may be a good system for future studies on common progenitor cells involved in cardiomyocyte and hematopoietic development in the early embryo, the role of the hemangioblast, the interaction of different cytokines and morphogens at specific timepoints, and the influence of the 3D microenvironment and metabolism.
In summary, we have established a 3D erythropoietic and cardiomyogenic differentiation culture system, bypassing standard EB formation using alginate-encapsulated mESCs cultivated in a RWV bioreactor. This 3D culture platform has demonstrated the importance of SCF, EPO, and IL-3 in erythropoietic and cardiomyogenic maturation and should be a useful tool for the study of these differential and competitive pathways from ESCs in a relevant in vitro system with potential applications in future cellular therapies.
Supplementary Material
Acknowledgments
This work was funded by the Higher Ministry of Education Malaysia, Universiti Malaysia Terengganu, the Richard Thomas Leukaemia Fund, and the Northwick Park Hospital Leukaemia Research Trust Fund.
Author Disclosure Statement
The authors have no conflicts of interest.
References
- 1.Kennedy M, D'Souza SL, Lynch-Kattman M, Schwantz S. and Keller G. (2007). Development of the hemangioblast defines the onset of hematopoiesis in human ES cell differentiation cultures. Blood 109:2679–2687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kouskoff V, Lacaud G, Schwantz S, Fehling HJ. and Keller G. (2005). Sequential development of hematopoietic and cardiac mesoderm during embryonic stem cell differentiation. Proc Natl Acad Sci U S A 102:13170–13175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choi K, Kennedy M, Kazarov A, Papadimitriou JC. and Keller G. (1998). A common precursor for hematopoietic and endothelial cells. Development 125:725–732 [DOI] [PubMed] [Google Scholar]
- 4.Keller G, Kennedy M, Papayannopoulou T. and Wiles MV. (1993). Hematopoietic commitment during embryonic stem-cell differentiation in culture. Mol Cell Biol 13:473–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palis J, Mcgrath KE. and Kingsley PD. (1995). Initiation of hematopoiesis and vasculogenesis in murine yolk-sac explants. Blood 86:156–163 [PubMed] [Google Scholar]
- 6.Palis J, Robertson S, Kennedy M, Wall C. and Keller G. (1999). Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development 126:5073–5084 [DOI] [PubMed] [Google Scholar]
- 7.Lu SJ, Feng Q, Caballero S, Chen Y, Moore MAS, Grant MB. and Lanza R. (2007). Generation of functional hemangioblasts from human embryonic stem cells. Nat Methods 4:501–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Handel B, Montel-Hagen A, Sasidharan R, Nakano H, Ferrari R, Boogerd CJ, Schredelseker J, Wang YL, Hunter S, et al. (2012). Scl represses cardiomyogenesis in prospective hemogenic endothelium and endocardium. Cell 150:590–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kattman SJ, Huber TL. and Keller GM. (2006). Multipotent Flk-1(+) cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev Cell 11:723–732 [DOI] [PubMed] [Google Scholar]
- 10.Huber TL, Kouskoff V, Fehling HJ, Palis J. and Keller G. (2004). Haemangioblast commitment is initiated in the primitive streak of the mouse embryo. Nature 432:625–630 [DOI] [PubMed] [Google Scholar]
- 11.Fehling HJ, Lacaud G, Kubo A, Kennedy M, Robertson S, Keller G. and Kouskoff V. (2003). Tracking mesoderm induction and its specification to the hemangioblast during embryonic stem cell differentiation. Development 130:4217–4227 [DOI] [PubMed] [Google Scholar]
- 12.Lancrin C, Sroczynska P, Stephenson C, Allen T, Kouskoff V. and Lacaud G. (2009). The haemangioblast generates haematopoietic cells through a haemogenic endothelium stage. Nature 457:892–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eilken HM, Nishikawa SI. and Schroeder T. (2009). Continuous single-cell imaging of blood generation from haemogenic endothelium. Nature 457:896–900 [DOI] [PubMed] [Google Scholar]
- 14.Rhodes KE, Gekas C, Wang Y, Lux CT, Francis CS, Chan DN, Conway S, Orkin SH, Yoder MC. and Mikkola HKA. (2008). The emergence of hematopoietic stem cells is initiated in the placental vasculature in the absence of circulation. Cell Stem Cell 2:252–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Migliaccio AR, Whitsett C. and Migliaccio G. (2009). Erythroid cells in vitro: from developmental biology to blood transfusion products. Curr Opin Hematol 16:259–268 [DOI] [PubMed] [Google Scholar]
- 16.Lu SJ, Feng Q, Park JS, Vida L, Lee BS, Strausbauch M, Wettstein PJ, Honig GR. and Lanza R. (2008). Biologic properties and enucleation of red blood cells from human embryonic stem cells. Blood 112:4475–4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma F, Ebihara Y, Umeda K, Sakai H, Hanada S, Zhang H, Zaike Y, Tsuchida E, Nakahata T, Nakauchi H. and Tsuji K. (2008). Generation of functional erythrocytes from human embryonic stem cell-derived definitive hematopoiesis. Proc Natl Acad Sci U S A 105:13087–13092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keller GM. (1995). In-vitro differentiation of embryonic stem-cells. Curr Opin Cell Biol 7:862–869 [DOI] [PubMed] [Google Scholar]
- 19.Placzek MR, Chung IM, Macedo HM, Ismail S, Blanco TM, Lim M, Cha JM, Fauzi I, Kang Y, et al. (2009). Stem cell bioprocessing: fundamentals and principles. J R Soc Interface 6:209–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ungrin MD, Joshi C, Nica A, Bauwens C. and Zandstra PW. (2008). Reproducible, ultra high-throughput formation of multicellular organization from single cell suspension-derived human embryonic stem cell aggregates. PLoS One 3:e1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coucouvanis E. and Martin GR. (1999). BMP signaling plays a role in visceral endoderm differentiation and cavitation in the early mouse embryo. Development 126:535–546 [DOI] [PubMed] [Google Scholar]
- 22.Hernandez-Garcia D, Castro-Obregon S, Gomez-Lopez S, Valencia C. and Covarrubias L. (2008). Cell death activation during cavitation of embryoid bodies is mediated by hydrogen peroxide. Exp Cell Res 314:2090–2099 [DOI] [PubMed] [Google Scholar]
- 23.Bauwens C, Yin T, Dang S, Peerani R. and Zandstra PW. (2005). Development of a perfusion fed bioreactor for embryonic stem cell-derived cardiomyocyte generation: Oxygen-mediated enhancement of cardiomyocyte output. Biotechnol Bioeng 90:452–461 [DOI] [PubMed] [Google Scholar]
- 24.Bienaime C, Barbotin JN. and Nava-Saucedo JE. (2003). How to build an adapted and bioactive cell microenvironment?. A chemical interaction study of the structure of Ca-alginate matrices and their repercussion on confined cells. J Biomed Mater Res A 67A:376–388 [DOI] [PubMed] [Google Scholar]
- 25.Magyar JP, Nemir M, Ehler E, Suter N, Perriard JC. and Eppenberger HM. (2001). Mass production of embryoid bodies in microbeads. Ann N Y Acad Sci 944:135–143 [DOI] [PubMed] [Google Scholar]
- 26.Dang SM, Gerecht-Nir S, Chen J, Itskovitz-Eldor J. and Zandstra PW. (2004). Controlled, scalable embryonic stem cell differentiation culture. Stem Cells 22:275–282 [DOI] [PubMed] [Google Scholar]
- 27.Panoskaltsis N, Mantalaris A. and Wu JHD. (2005). Engineering a mimicry of bone marrow tissue ex vivo. J Biosci Bioeng 100:28–35 [DOI] [PubMed] [Google Scholar]
- 28.Fok EYL. and Zandstra PW. (2005). Shear-controlled single-step mouse embryonic stem cell expansion and embryoid body-based differentiation. Stem Cells 23:1333–1342 [DOI] [PubMed] [Google Scholar]
- 29.Fong WJ, Tan HL, Choo A. and Oh SKW. (2005). Perfusion cultures of human embryonic stem cells. Bioproc Biosyst Eng 27:381–387 [DOI] [PubMed] [Google Scholar]
- 30.Fridley KM, Fernandez I, Li MT, Kettlewell RB. and Roy K. (2010). Unique differentiation profile of mouse embryonic stem cells in rotary and stirred tank bioreactors. Tissue Eng 16:3285–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hwang YS, Cho J, Tay F, Heng JYY, Ho R, Kazarian SG, Williams DR, Boccaccini AR, Polak JM. and Mantalaris A. (2009). The use of murine embryonic stem cells, alginate encapsulation, and rotary microgravity bioreactor in bone tissue engineering. Biomaterials 30:499–507 [DOI] [PubMed] [Google Scholar]
- 32.Randle WL, Cha JM, Hwang YS, Chan KLA, Kazarian SG, Polak JM. and Mantalaris A. (2007). Integrated 3-dimensional expansion and osteogenic differentiation of murine embryonic stem cells. Tissue Eng 13:2957–2970 [DOI] [PubMed] [Google Scholar]
- 33.Hwang YS, Randle WL, Bielby RC, Polak JM. and Mantalaris A. (2006). Enhanced derivation of osteogenic cells from murine embryonic stem cells after treatment with HepG2-conditioned medium and modulation of the embryoid body formation period: application to skeletal tissue engineering. Tissue Eng 12:1381–1392 [DOI] [PubMed] [Google Scholar]
- 34.Hwang YS, Bishop AE, Polak JM. and Mantalaris A. (2007). Enhanced in vitro chondrogenic differentiation of murine embryonic stem cells. Biotechnol Bioproc E 12:696–706 [Google Scholar]
- 35.Hwang YS, Polak JM. and Mantalaris A. (2008). In vitro direct osteogenesis of murine embryonic stem cells without embryoid body formation. Stem Cells Dev 17:963–970 [DOI] [PubMed] [Google Scholar]
- 36.Fauzi I, Panoskaltsis N. and Mantalaris A. (2012). Enhanced hematopoietic differentiation towards erythrocytes from murine embryonic stem cells with HepG2-conditioned medium. Stem Cells Dev 21:3152–3161 [DOI] [PubMed] [Google Scholar]
- 37.Hiroyama T, Miharada K, Sudo K, Danjo I, Aoki N. and Nakamura Y. (2008). Establishment of mouse embryonic stem cell-derived erythroid progenitor cell lines able to produce functional red blood cells. PLoS One 3:e1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schneider CA, Rasband WS. and Eliceiri KW. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Narla A, Dutt S, McAuley JR, Al-Shahrour F, Hurst S, McConkey M, Neuberg D. and Ebert BL. (2011). Dexamethasone and lenalidomide have distinct functional effects on erythropoiesis. Blood 118:2296–2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Obradovic B, Carrier RL, Vunjak-Novakovic G. and Freed LE. (1999). Gas exchange is essential for bioreactor cultivation of tissue engineered cartilage. Biotechnol Bioeng 63:197–205 [DOI] [PubMed] [Google Scholar]
- 41.Zetterberg A. and Engstrom W. (1981). Mitogenic effect of alkaline-ph on quiescent, serum-starved cells. Proc Natl Acad Sci U S A 78:4334–4338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mochizuki H, Ohnuki Y. and Kurosawa H. (2011). Effect of glucose concentration during embryoid body (EB) formation from mouse embryonic stem cells on EB growth and cell differentiation. J Biosci Bioeng 111:92–97 [DOI] [PubMed] [Google Scholar]
- 43.Khoo MLM, McQuade LR, Smith MSR, Lees JG, Sidhu KS. and Tuch BE. (2005). Growth and differentiation of embryoid bodies derived from human embryonic stem cells: Effect of glucose and basic fibroblast growth factor. Biol Reprod 73:1147–1156 [DOI] [PubMed] [Google Scholar]
- 44.Conaghan J, Handyside AH, Winston RML. and Leese HJ. (1993). Effects of pyruvate and glucose on the development of human preimplantation embryos in-vitro. J Reprod Fertil 99:87–95 [DOI] [PubMed] [Google Scholar]
- 45.Quinn P. (1995). Enhanced results in mouse and human embryo culture using a modified human tubal fluid medium lacking glucose and phosphate. J Assist Reprod Gen 12:97–105 [DOI] [PubMed] [Google Scholar]
- 46.Tennant GB, Truran LN, Bailey-Wood R. and Burnett AK. (2000). Control of pH in human long-term bone marrow cultures with low-glucose medium containing zwitterion buffer lengthens the period of haemopoietic activity. Br J Haematol 109:785–787 [DOI] [PubMed] [Google Scholar]
- 47.Erard F, Dean A. and Schechter AN. (1981). Inhibitors of cell-division reversibly modify hemoglobin concentration in human erythroleukemia K562 cells. Blood 58:1236–1239 [PubMed] [Google Scholar]
- 48.Hiep NC, Kinohira S, Furuyama K. and Taketani S. (2012). Depletion of glutamine enhances sodium butyrate-induced erythroid differentiation of K562 cells. J Biochem 152:509–519 [DOI] [PubMed] [Google Scholar]
- 49.Florian M, Jankowski M. and Gutkowska J. (2010). Oxytocin increases glucose uptake in neonatal rat cardiomyocytes. Endocrinology 151:482–491 [DOI] [PubMed] [Google Scholar]
- 50.Crespo FL, Sobrado VR, Gomez L, Cervera AM. and McCreath KJ. (2010). Mitochondrial reactive oxygen species mediate cardiomyocyte formation from embryonic stem cells in high glucose. Stem Cells 28:1132–1142 [DOI] [PubMed] [Google Scholar]
- 51.Gerecht-Nir S, Cohen S. and Itskovitz-Eldor J. (2004). Bioreactor cultivation enhances the efficiency of human embryoid body (hEB) formation and differentiation. Biotechnol Bioeng 86:493–502 [DOI] [PubMed] [Google Scholar]
- 52.McAdams TA, Miller WM. and Papoutsakis ET. (1998). pH is a potent modulator of erythroid differentiation. Br J Haematol 103:317–325 [DOI] [PubMed] [Google Scholar]
- 53.Broudy VC. (1997). Stem cell factor and hematopoiesis. Blood 90:1345–1364 [PubMed] [Google Scholar]
- 54.Muta K, Krantz SB, Bondurant MC. and Dai CH. (1995). Stem-cell factor retards differentiation of normal human erythroid progenitor cells while stimulating proliferation. Blood 86:572–580 [PubMed] [Google Scholar]
- 55.Migliaccio G, Migliaccio AR. and Visser JWM. (1988). Synergism between erythropoietin and interleukin-3 in the induction of hematopoietic stem-cell proliferation and erythroid burst colony formation. Blood 72:944–951 [PubMed] [Google Scholar]
- 56.Ghezzi P. and Brines M. (2004). Erythropoietin as an antiapoptotic, tissue-protective cytokine. Cell Death Differ 11 (Suppl. 1):S37–S44 [DOI] [PubMed] [Google Scholar]
- 57.Ji RP, Phoon CKL, Aristizabal O, McGrath KE, Palis J. and Turnbull DH. (2003). Onset of cardiac function during early mouse embryogenesis coincides with entry of primitive erythroblasts into the embryo proper. Circ Res 92:133–135 [DOI] [PubMed] [Google Scholar]
- 58.Robertson SM, Kennedy M, Shannon JM. and Keller G. (2000). A transitional stage in the commitment of mesoderm to hematopoiesis requiring the transcription factor SCL/tal-l. Development 127:2447–2459 [DOI] [PubMed] [Google Scholar]
- 59.Nishikawa S, Nishikawa S, Hirashima M, Matsuyoshi N. and Kodama H. (1998). Progressive lineage analysis by cell sorting and culture identifies FLK1(+)VE-cadherin(+) cells at a diverging point of endothelial and hemopoietic lineages. Development 125:1747–1757 [DOI] [PubMed] [Google Scholar]
- 60.Yamaguchi TP, Dumont DJ, Conlon RA, Breitman ML. and Rossant J. (1993). Flk-1, an Flt-related receptor tyrosine kinase is an early marker for endothelial-cell precursors. Development 118:489–498 [DOI] [PubMed] [Google Scholar]
- 61.Kang YY, Nagy JM, Polak JM. and Mantalaris A. (2009). Proteomic characterization of the conditioned media produced by the visceral endoderm-like cell lines HepG2 and END2: toward a defined medium for the osteogenic/chondrogenic differentiation of embryonic stem cells. Stem Cells Dev 18:77–91 [DOI] [PubMed] [Google Scholar]
- 62.Wu XC, Richards NT. and Johns EJ. (1999). The influence of erythropoietin on the vascular responses of rat resistance arteries. Exp Physiol 84:917–927 [PubMed] [Google Scholar]
- 63.Wu H, Lee SH, Gao J, Liu X. and Iruela-Arispe ML. (1999). Inactivation of erythropoietin leads to defects in cardiac morphogenesis. Development 126:3597–3605 [DOI] [PubMed] [Google Scholar]
- 64.Brizzi MF, Formato L, Dentelli P, Rosso A, Pavan M, Garbarino G, Pegoraro M, Camussi G. and Pegoraro L. (2001). Interleukin-3 stimulates migration and proliferation of vascular smooth muscle cells—a potential role in atherogenesis. Circulation 103:549–554 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.