Abstract
Exposed epitopes of the spike protein may be recognized by neutralizing antibodies against severe acute respiratory syndrome (SARS) coronavirus (CoV). A protein fragment (S-II) containing predicted epitopes of the spike protein was expressed in Escherichia coli. The properly refolded protein fragment specifically bound to the surface of Vero cells. Monoclonal antibodies raised against this fragment recognized the native spike protein of SARS CoV in both monomeric and trimeric forms. These monoclonal antibodies were capable of blocking S-II attachment to Vero cells and exhibited in vitro antiviral activity. These neutralizing antibodies mapped to epitopes in two peptides, each comprising 20 amino acids. Thus, this region of the spike protein might be a target for generation of therapeutic neutralizing antibodies against SARS CoV and for vaccine development to elicit protective humoral immunity.
The genome sequence of severe acute respiratory syndrome (SARS) coronavirus (CoV) provided direct evidence that a new CoV is responsible for the recent SARS epidemic in Asia and Canada (11, 15). This new CoV appears to have host specificities different from those of previously known CoVs. One of the potential cell tropism determinants is the spike (S) protein that recognizes the host receptor (2, 4, 21). The genome sequence of the SARS CoV predicts an S protein of 1,255 amino acids (aa) (GenBank accession no. 29836496) with 23 potential N-linked glycosylation sites (11, 15), and the N terminus of the S protein contains a signal peptide that is presumably removed in the mature virion (11, 15). Unlike some CoVs, such as human CoV (HCoV) 229E, in which the S protein is cleaved into S1 and S2 subunits, the S protein in SARS CoV is not cleaved. Alignment of the SARS CoV S protein with the S proteins of other CoVs, including HCoV 229E, showed low homology. However, the SARS CoV S protein could be aligned in the S2 region with higher homology than with the S1 region. S2 contains the membrane-anchoring and fusion region, which plays a key role in virus assembly and entry (8). Despite the lack of significant homology with the S1 protein of HCoV 229E, transmissible gastroenteritis virus (TGEV), or mouse hepatitis virus (MHV), which contains the receptor binding site (1, 3, 10, 16), it is likely that the receptor binding site of the SARS CoV S protein is located in the N-terminal part corresponding to the S1 region.
The host receptor for group I CoVs, such as HCoV 229E and TGEV, is aminopeptidase N (also known as CD13) (4, 21). MHV, a group II CoV (5, 6), recognizes carcinoembryonic antigen-related cell adhesion molecules as another group of CoV receptors. Viral entry is initiated by attachment of the S protein to the specific host receptor, which triggers a conformational change in the S protein. This receptor-induced conformational change exposes the fusogenic region embedded in the S2 region that inserts into the cellular membrane (12, 22). In HCoV 229E, the receptor binding domain has been narrowed to a region comprising residues 407 to 547 in S1. Protein fragments with these residues retain the ability to bind the host receptor, aminopeptidase N, and are recognized by monoclonal antibody (MAb) 4-9H.5 (1). Similar results were obtained for TGEV, where the receptor binding region is located between residues 506 and 655 (7, 18). Neutralizing determinants are also located in this region. The receptor binding domain of MHV also maps to the N-terminal end (330 aa) of the S protein (10). The definitive region that binds the host receptor has not been clearly mapped on the S protein of SARS CoV.
In this report, we describe a protein fragment of the SARS CoV S protein (residues 485 to 625). When this fragment is produced as a refolded recombinant protein from E. coli, it attaches specifically to the surface of Vero E6 cells. The MAbs against this fragment recognize the native S protein in SARS CoV and exhibit neutralizing activity against SARS CoV infection in cell culture. Thus, this region of the S protein may be related to receptor recognition and contains neutralizing epitopes of the SARS CoV S protein.
MATERIALS AND METHODS
Expression of S fragments.
Nineteen oligonucleotides of both sense and antisense sequences were synthesized (IDT, Coralville, Iowa) for the S-II fragment (aa 485 to 625). Four picomoles of each oligonucleotide was mixed in 1× PCR buffer and incubated with 2.5 U of Taq polymerase in a final volume of 50 μl at 72°C for 20 min. Five microliters of the annealed and ligated product was used as a template for a PCR with 5′ and 3′ primers for 30 cycles. The 438-bp PCR product was purified and cloned into the pET100/D/TOPO expression vector (Invitrogen). In addition, the cDNA fragments encoding aa 145 to 480 (S-I) and 601 to 973 (S-III) of the SARS CoV S protein were obtained by reverse transcription-PCR with the RNA genome of SARS CoV (Toronto-2 isolate) as the template and cloned into the pET32a vector. All proteins were expressed in transformed BL21(DE3) host cells induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 h. The inclusion bodies containing the recombinant proteins were dissolved in 8 M urea and refolded in 20 mM Tris buffer.
Flow cytometry analysis of the binding activity of S fragments.
The purified, refolded recombinant protein fragments were dialyzed against phosphate-buffered saline (PBS), and then 2 mg of protein per ml was conjugated with LC-Biotin (Pierce) at a 1:25 protein/biotin molar ratio in accordance with the manufacturer's instructions. After conjugation, free biotin was removed by dialysis. For flow cytometry analysis, 106 Vero E6 cells (American Type Culture Collection) were incubated with the diluted biotinylated S-II or control protein in 20 μl of fluorescence-activated cell sorter (FACS) buffer (5% fetal calf serum, 0.01% NaN3 in PBS) at room temperature (RT) for 30 min. After washing with FACS buffer, cells were further incubated with 20 μl of phycoerythrin (PE)-conjugated streptavidin (Southern Biotechnology Associates). Ten thousand viable cells were analyzed with a FACScan flow cytometer (BD). Mean fluorescence intensity (MFI) was analyzed with WinMdi software.
Preparation of viral stock and whole-cell lysates from SARS CoV-infected Vero cells.
The original Toronto-2 isolate received from Heinz Feldmann (Winnipeg, Canada) was diluted 1:1,000 in Dulbecco modified Eagle medium (DMEM), and 5 ml was added to 90 to 95% confluent Vero E6 cells in 162-cm2 tissue culture flasks (n = 4). After 1 h of incubation at 37°C in 5% CO2, 25 ml of DMEM and 1% bovine serum albumin (BSA) were added to the flasks, which were then incubated for 72 h at 37°C in 5% CO2. Cells were scraped, and flask contents were pooled. Samples were centrifuged for ∼10 min at 300 × g, and cells were resuspended in 8 ml of PBS, vortexed, and frozen at −70°C until used. Whole-cell lysates were prepared by mixing an equal volume frozen cell lysate with 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer and boiling the mixture for 5 min. Clarified supernatant was stored at −70°C in 1-ml aliquots for viral stock. Numbers of viral PFU per milliliter were determined by standard plaque assay on Vero E6 cells.
Generation of polyclonal antibodies and MAbs against S-II.
For polyclonal antibody production, 6- to 8-month-old goats were subcutaneously immunized with 5 mg of S-II in Freund's complete adjuvant and boosted with 5 mg of S-II in PBS every 2 weeks. After 8 weeks of primary immunization, antisera were collected 1 week after each boost and affinity purified with S-II-conjugated Sepharose 4B. The purified anti-S-II polyclonal antibody was horseradish peroxidase (HRP) conjugated by a standard method. For generation of an anti-S-II MAb, 6- to 8-week-old female BALB/c mice were primarily immunized with 200 μg of S-II in Freund's complete adjuvant in the footpads and boosted four times with the same amount of S-II in PBS weekly. Three days after the last immunization, the lymphocytes from the draining lymph nodes were fused with NS1 myeloma cells and cultured in hypoxanthine-aminopterin-thymidine medium. The hybridoma clones were first screened by enzyme-linked immunosorbent assay (ELISA) with a second recombinant S-II protein as the antigen. All ELISA-positive clones were further confirmed by Western blot analysis with whole-cell lysates of SARS CoV-infected Vero cells. The selected hybridoma clones were subcloned three times by limiting dilution. The MAbs were generated in ascites of inoculated BALB/c mice and purified by (NH4)2SO4 precipitation, followed by ion-exchange chromatography.
Generation of MAbs against N.
Details of the experiment in which MAbs against N were generated will be presented elsewhere (T.Z. et al., submitted for publication). Briefly, for generation of anti-Nn, -Nc, and -M MAbs, 6- to 8-week-old female BALB/c mice were primarily immunized with 200 μg each of two recombinant N antigens (Nn and Nc) in Freund's complete adjuvant in the footpads and boosted four times with the same amount of antigens in PBS weekly. Nn corresponds to aa 1 to 108, and Nc corresponds to aa 253 to 422. The hybridoma clones producing anti-N antibodies were first screened by ELISA with the full-length N protein as the antigen. All ELISA-positive clones were subsequently screened by Western blot analysis with whole-cell lysates of SARS CoV-infected Vero cells. The selected hybridoma clones were subcloned three times by limiting dilution. The MAbs were generated in ascites of inoculated BALB/c mice and purified by (NH4)2SO4 precipitation, followed by ion-exchange chromatography. MAbs 5E10 (anti-Nn) and 13-3 (anti-Nc) were used in this study.
Immunohistochemistry staining of SARS CoV-infected cells.
Ninety to 95% confluent monolayers of WI38 cell were infected with a 1:1,000 dilution of HCoV 229E, B-SC-1 cells were infected with OC-43, and Vero E6 cells were infected with SARS HCoV (Toronto-2). At 72 h postinfection, the 229E-infected WI38 and OC43-infected B-SC-1 cell monolayers were washed with PBS and scraped and 25 μl of cells was spotted onto heavy Teflon-coated slides. The slides were air dried in a biosafety cabinet and fixed in cold acetone for 10 min. SARS CoV-infected Vero cells were fixed in 24-well plates with acetone and air dried. Fixed cells were stored at −20°C until staining. For immunohistochemical staining, the fixed cells were rehydrated with PBS and blocked with 20% goat serum in PBS at RT for 40 min. Cells were then incubated with 0.2 μg of HRP-conjugated anti-S-II MAb per ml at RT for 30 min, and the reaction was developed with 3,3′-diaminobenzidine (DAB) substrate buffer (Sigma). The staining was examined under an inverted fluorescence microscope and photographed at a magnification of ×400.
Sandwich ELISA for quantitative measurement of SARS CoV S protein.
High-binding ELISA plates (Costar) were coated with 4 μg of purified anti-S-II MAbs per ml in PBS at 4°C overnight and blocked with 3% BSA-PBS at RT for 1 h. The whole-cell lysates from SARS CoV-infected Vero cells, prepared as described above, were serially diluted in 3% BSA-PBS, depending on the viral titer, and 100 μl of diluted lysates was added and the mixture was incubated at RT for 40 min. The captured S protein was detected by incubation with 1 μg of HRP-conjugated polyclonal anti-S-II antibody per ml at RT for another 40 min. The reaction was developed with TMB buffer 3,3′,5,5′-tetramethylbenzidine and the optical density (OD) values were recorded with an ELISA plate reader (Bio-Rad) at wavelengths of 450 and 650 nm.
Western blot and mass spectrum analyses of SARS CoV S protein.
Whole-cell lysates were prepared in SDS-PAGE sample buffer under nonreducing (0.5% SDS) and reducing (0.5% SDS with 2 mM β-mercaptoethanol) conditions, subjected to SDS-7% PAGE, and transferred onto a nitrocellulose membrane. The blots were blocked with 5% milk-PBS and probed with 1 μg of HRP-conjugated anti-S-RBS MAb per ml at 4°C overnight. The reaction was developed with enhanced chemiluminescence buffer. The samples prepared under nonreducing and reducing conditions were also digested with trypsin, and the tryptic peptides were analyzed on a PE-Sciex (Concord, Ontario, Canada) API III triple-quadrupole mass spectrometer in the University of Alabama Comprehensive Cancer Center (P30 CA-13148). The molecular mass pattern of the nonreducing sample was the same as that of the reducing sample. The nonreducing ∼600-kDa species could therefore only be interpreted as the trimeric complex of the ∼180-kDa monomeric S protein.
Immunoprecipitation of SARS CoV with anti-S-II MAb.
One milliliter of 108 PFU of the Toronto-2 virus was added to anti-S MAbs, which were directly conjugated to Sepharose 4B and incubated for 30 min at 4°C in a shaker. After washing, the immunoprecipitated viruses were dissolved in 100 μl of SDS-PAGE buffer. The dissolved proteins were separated by SDS-7% PAGE and blotted. The S and N proteins were detected by Western blot analysis with HRP-conjugated anti-S and anti-N MAbs.
Neutralizing activity of MAbs and antisera in SARS CoV-infected Vero cells.
Ten-fold-diluted antisera or MAbs were added to duplicate 24-well plates containing confluent monolayers of Vero cells in Iscove's modified Eagle medium-2% fetal bovine serum-2 mM l-glutamine-antibiotics. SARS HCoV (Toronto-2 isolate) was added to each well at a final concentration of 0.5 PFU/ml, and plates were incubated for 72 h at 37°C in 5% CO2. Medium was removed from the wells, one set of plates was fixed with 80% acetone in PBS for immunostaining, and 0.1% SDS was added to the other plates for analysis by ELISA.
Neutralizing activity of MAbs by plaque assays.
Equal volumes of MAb (100, 10, and 1 μg/100 μl) and SARS HCoV (103 PFU of Toronto-2 virus per 100 μl in DMEM) were mixed and incubated at 37°C in 5% CO2 for 1 h. Two hundred microliters of the MAb-virus mixture was added to confluent monolayers of Vero cells in six-well plates in duplicate. Plates were incubated for 1 h at 37°C in 5% CO2. Cells were overlaid with agar and incubated for 72 h at 37°C in 5% CO2 Wells were fixed, overlays were removed, and cells were stained with crystal violet. Plaques were counted and recorded.
Detection of SARS CoV N protein in infected Vero cells.
For sandwich ELISA, high-binding ELISA plates (Costar) were coated with two purified anti-SARS N protein MAbs, reactive with the N or C terminus of the N protein, at 4 μg/ml in PBS at 4°C overnight and blocked with 3% BSA-PBS at RT for 1 h. The whole-cell lysates from SARS CoV-infected Vero cells, prepared as described above, were serially diluted in 3% BSA-PBS (1:10, 1:100, and 1:1,000), 100-μl volumes of diluted lysates were added, and the mixtures were incubated at RT for 40 min. The captured N protein was detected by incubation with 1 μg of HRP-conjugated polyclonal anti-SARS N protein antibody per ml at RT for another 40 min. The reaction was developed with TMB buffer, and the OD values were recorded with an ELISA plate reader (Bio-Rad) at wavelengths of 450 and 650 nm. A standard curve with the full-length recombinant N protein was generated, and the N protein concentration in the infected cells was calculated. For Western blot analysis, whole-cell lysates were prepared in the reducing SDS-PAGE sample buffer, subjected to SDS-10% PAGE, and transferred onto a nitrocellulose membrane. The blots were blocked with 5% milk-PBS and probed with 1 μg of HRP-conjugated anti-SARS N MAb per ml at 4°C overnight. The reaction was developed with enhanced chemiluminescence buffer. For immunohistochemistry staining of SARS CoV-infected cells, SARS CoV-infected Vero cells were fixed in 24-well plates with acetone and air dried. The fixed cells were rehydrated with PBS and blocked with 20% goat serum PBS at RT for 40 min. Cells were then incubated with 0.2 μg of HRP-conjugated anti-SARS N MAb per ml at RT for 30 min, and the reaction was developed with DBA substrate buffer (Sigma). The staining was examined under an inverted fluorescence microscope and photographed at a magnification of ×400.
Mapping of neutralizing epitopes with anti-S-II MAbs.
Six truncated S-II peptides were expressed in E. coli with the pET32a vector. Six peptides were truncated at the N or C terminus of S-II and made 20, 40, and 60 residues shorter, respectively. The purified peptides were used to coat an ELISA plate, which was incubated with each of the HRP-conjugated anti-S-II MAbs. OD values greater than 2.0 were judged positive, and those less than 0.1 were judged negative.
RESULTS
Expression and characterization of S protein fragments.
Amino acid sequence alignment showed a low homology of the SARS CoV S protein to those of other CoVs. However, it is likely that the exposed epitopes and the receptor binding regions are located in the S protein corresponding to the S1 subunit on the basis of sequence analysis and modeling (11, 15, 17). With the EMBOSS:Antigenic program (9, 14), the top five predicted antigenic sites could be found in the S1 region (aa 1 to 690) of the SARS CoV S protein (Fig. 1A). A protein fragment of residues 145 to 480 (S-I) could present two peptides, and a protein fragment of residues 485 to 625 (S-II) could present another two peptides. Residues 4 to 11 are not considered because a short peptide may not form a stable conformation. Interestingly, S-II has partial homology to the identified receptor binding domain (residues 407 to 547) of the S protein of HCoV 229E (GenBank accession no. 13604338) (Fig. 1B). S-I has homology to the receptor binding N-terminal region of MHV (data not shown), but it did not bind the surface of Vero cells, as shown below. The codons in the coding region for the selected S-II fragment were optimized for usage in E. coli (13), and oligonucleotides grouped with sticky overlap ends were annealed together as nicked double-stranded DNA and ligated before cloning into the expression vector pET100/D-TOPO (Invitrogen). Protein expression was induced with IPTG, and S-II was expressed as an insoluble protein in E. coli. The inclusion bodies were dissolved in 8 M Urea, refolded, and purified by Ni affinity chromatography (Fig. 1C). To determine whether the designed S protein fragments contained any Vero cell binding activities, the purified S-I and refolded S-II fragments were biotinylated and used as a probe for specific binding to the surface of Vero E6 cells. Flow cytometry analysis demonstrates that S-II, but not a control protein, bound to the surface of Vero E6 cells (Fig. 1D). There was a dose-dependent increase in the staining intensity of bound S-II but not the control protein, as shown by the increasing MFI (Fig. 1E). S-I and S-III (aa 601 to 973) were also expressed and tested for the ability to bind to Vero E6 cells. There was no significant binding of the other two fragments (Fig. 1D). These results indicate that the designated S-II peptide could be involved in specific binding to the Vero cell surface, perhaps the receptor(s). Therefore, S-II was selected for further studies for its potential to generate neutralizing antibodies.
FIG.1.
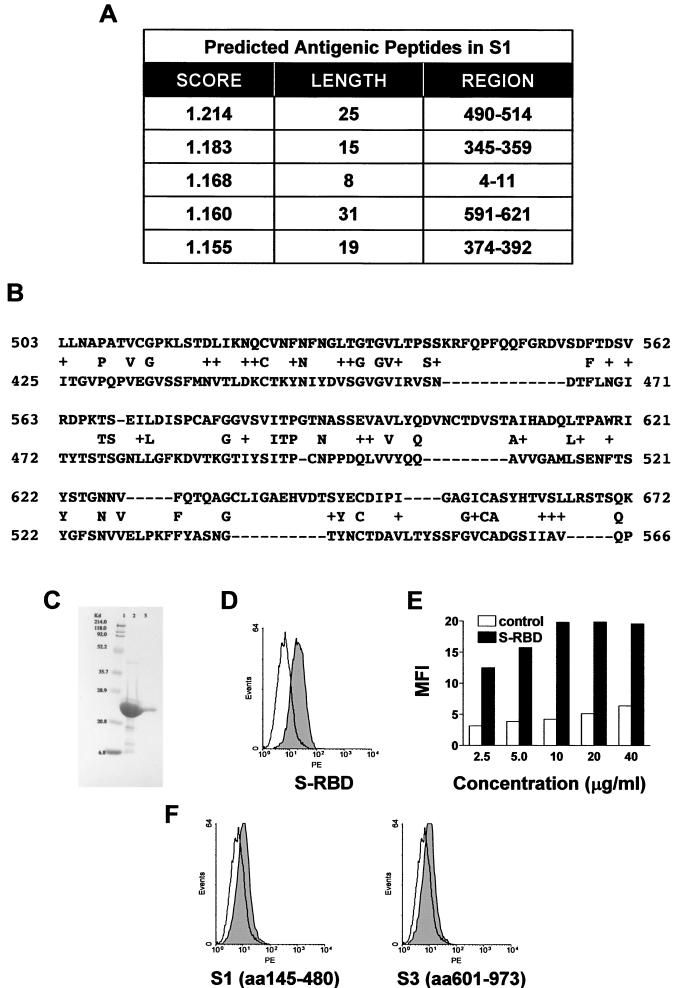
Antigenic analysis of the S protein. (A) Potential antigenic peptides predicted by EMBOSS:Antigenic. The scores are related to the probability that the amino acid sequence is an antigenic determinant on the basis of empirical data and the distribution of amino acids (9). (B) BLAST analysis of the S protein sequences of SARS CoV and HCoV 229E was performed with the full-length amino acid sequences of both proteins at the National Center for Biotechnology Information website. The letters between the two sequences are conserved residues, and the plus signs indicate homologous changes. The starting point corresponds to the first amino acid aligned by BLAST, and only the beginning portion of the alignment is shown here. The shaded region showed significant homology (60%) in the receptor binding region of the HCoV 229E S protein. (C) The codons for S-II were optimized for usage in E. coli, and 18 oligonucleotides grouped with sticky overlap ends were synthesized and annealed together as nicked double-stranded DNA and ligase treated. The synthesized cDNA was cloned into the expression vector pET100/D-TOPO with a His6 tag and an EK recognition site at the N terminus (Invitrogen), and protein expression was induced with IPTG in BL21/ED3 host cells. The inclusion bodies containing S-II were dissolved in 8 M urea, refolded, and purified by Ni affinity chromatography. The yield of purified, refolded, soluble S-II was >2 mg/liter. Lanes: 1, molecular mass marker; 2, denatured S-II expressed in E. coli; 3, purified, refolded S-II. (D) The purified, refolded S-II was biotinylated and used as a probe for specific binding to the surface of Vero cells. Vero E6 cells (106; American Type Culture Collection) were first incubated with a 20-μl volume of 10 μg of biotinylated S-II or a control His6-tagged S.Tag protein (Novagen) per ml at RT for 30 min and then further incubated with 20 μl of PE-conjugated streptavidin. The binding of S-II to Vero cells was demonstrated by flow cytometry analysis. S-II, solid; control, open. S-II showed selective binding to Vero cells compared to two recombinant protein fragments covering aa 145 to 480 (S-I) or 601 to 973 (S-III). S-RBD, S protein receptor binding domain. (E) To demonstrate dose-dependent binding of S-II to Vero cells, Vero cells were incubated with the indicated concentration of the S-II fragment (solid) or a control (open) and MFI was determined. (F) The same flow cytometry analysis as in panel D, using fragments S-I and S-III.
Characterization of MAbs against S-II.
BALB/c mice were immunized with the refolded S-II fragment in Freund's complete adjuvant and boosted with S-II in PBS four times. Lymphocytes from the draining lymph nodes were fused with NS1 myeloma cells. Four hybridoma clones producing high-affinity anti-S-II antibody, S26, S34, S78, and S84, were selected. HRP-conjugated S34 and S78 reacted with acetone-fixed SARS CoV (Toronto-2 strain)-infected Vero cells but not with noninfected Vero cells or cells infected with two common HCoVs, 229E and OC43 (Fig. 2A). All anti-S-II MAbs, but not a control MAb, were able to capture the native form of the S protein from whole-cell lysates of SARS CoV-infected Vero cells. In an ELISA format with the MAbs used individually as a capture antibody and an HRP-conjugated polyclonal goat anti-S-II antibody used as the detecting antibody, the content of the S protein in SARS CoV-infected Vero cells could be quantitatively measured. At 72 h after infection, the content of the S protein in whole-cell lysates was well correlated with the number of viral PFU (Fig. 2B). Neither ELISAs with the control capture antibody nor ELISAs with cell lysates from noninfected Vero cells showed a significant reaction (data not shown). In a Western blot paradigm, the whole-cell lysate of SARS CoV-infected Vero cells was prepared under both reducing and nonreducing conditions, electrophoresed by SDS-PAGE, and probed with each of the four MAbs against S-II. All of the antibodies recognized the S protein prepared under both reducing and nonreducing conditions. Under reducing conditions, the antibodies recognized an ∼180-kDa protein as predicted (Fig. 2C, lane 1). However, under nonreducing conditions, an ∼600-kDa protein predominated (Fig. 2C, lane 2). The reactivity of the MAb was specific for SARS CoV, as the lysates of the control Vero cells did not show any reaction under either reducing or nonreducing conditions (Fig. 2C, lanes 3 and 4). The S protein in the whole-cell lysate of infected Vero cells could be immunoprecipitated with MAb against S-II, and SDS-PAGE analysis of the affinity-purified S protein demonstrated that the ∼600-kDa protein was homotrimeric, as identified by mass spectrometry (see Materials and Methods), indicating that the SARS S protein might exist as a trimer in its native form. Conjugated to Sepharose 4B, each of the anti-S-II MAbs was able to immunoprecipitate viral particles in the supernatants from SARS CoV-infected Vero cells. After immunoprecipitation of viral particles, both the S and N proteins could be detected in a Western blot assay with HRP-conjugated anti-S and anti-N MAbs (Fig. 2D). These results indicate that MAbs raised against the designated S-II peptide specifically recognize the native S protein of SARS CoV.
FIG.2.
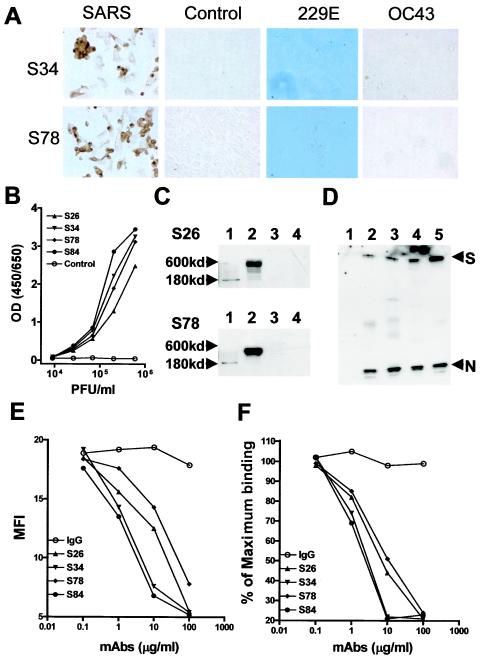
Characterization of MAbs against S-II. (A) Vero cells either uninfected as controls or infected with 0.5 PFU of SARS CoV (Toronto-2 isolate) per ml per well for 24 h, WI38 cells infected with 229E, and B-SC-1 cells infected with OC43 for 72 h were cytospun onto slides, acetone fixed, and stained with 0.2 μg of HRP-conjugated MAbs (S34 and S78) per ml at RT for 40 min. The reaction was developed with DAB substrate. (B) To measure S protein in SARS CoV-infected Vero cells, whole-cell lysates were prepared from Vero cells infected with SARS CoV (Toronto-2 isolate). Numbers of viral PFU per milliliter of the supernatant from the cell lysates were determined at 6 × 105 PFU/ml by standard plaque assay on Vero E6 cells. Cell lysates were generated by mixing equal volumes of frozen cell lysate and 2× SDS sample buffer and boiling the mixture for 5 min. Cell lysates were dialyzed against PBS overnight prior to ELISA. ELISA plates were coated with 4 μg of each of the indicated MAbs per ml and blocked with 3% BSA. The cell lysate was diluted to the indicated concentrations and incubated at RT for 1 h. The captured S protein was detected with HRP-conjugated polyclonal goat anti-S antibody. (C) Western blot analysis of the S protein of SARS CoV. Whole-cell lysate from either SARS CoV-infected Vero cells obtained as described above (lane 1 and 2) or control noninfected Vero cells (lane 3 and 4) was separated by SDS-7% PAGE under both reducing (lane 1 and 3) and nonreducing (lane 2 and 4) conditions and blotted. The blots were probed with HRP-conjugated S26 or S78 at 4°C overnight and developed by chemiluminescence. (D) To immunoprecipitate SARS CoV, anti-S-II MAbs were conjugated to Sepharose 4B and incubated with 2 × 108 PFU of virus in a 1-ml total volume at RT for 30 min and then washed twice with PBS. The immunoprecipitated samples were dissolved in SDS denaturing buffer and separated by SDS-7% PAGE under nonreducing conditions. After transfer, the blot was probed with a mixture of HRP-conjugated anti-N and anti-S MAbs (lanes: 1, control; 2, S26; 3, S34; 4, S78; 5, S84). (E) Flow cytometry analysis of blocking activity of anti-S-II MAbs. Vero cells (106) were incubated with 10 μg of biotinylated S-II per ml in the presence of various concentrations of each MAb, followed by PE-conjugated streptavidin and determination of MFI by quantitative flow cytometry. (F) ELISA of blocking activity of anti-S-II MAbs. The membrane fraction of Vero E6 cells was lysed in 1% Triton-PBS and passed through an S-II-conjugated affinity column. The bound proteins were biotinylated on the column and eluted with pH 3.0 glycine-HCl buffer for the detecting agents. ELISA plate was coated with 5 μg of S-II per ml in PBS and incubated with 1 μg of biotinylated binding proteins of Vero cells per ml in the presence of various concentrations of anti-S-II MAbs. The plate-bound binding proteins were then detected with HRP-conjugated streptavidin. Maximum binding was determined as the OD values in the absence of MAb. IgG, immunoglobulin G.
The four anti-S-II MAbs were able to inhibit the binding of S-II to the Vero E6 cell surface in a dose-dependent fashion (Fig. 2E). Among the four antibodies, S34 and S84 showed more blocking activity than either S26 or S78. The blocking activity of MAbs was further confirmed in a competitive ELISA in which the S-II affinity-purified cell membrane proteins of Vero E6 cells were biotinylated and incubated with the plate-coated S-II peptide in the presence of various concentrations of MAbs. A similar inhibition of binding of S-II to the Vero cell surface was observed (Fig. 2F). These results indicate that anti-S-II MAbs are capable of blocking the surface binding activities of S-II.
In vitro antiviral activity of anti-S-II MAbs.
If the S-II region of the S protein is involved in the attachment of the SARS CoV to its host cells, we would expect the blocking activity of MAbs against S-II to have a parallel neutralizing activity. To test this hypothesis, we examined the neutralizing activity of MAbs by assessing the inhibition of productive SARS CoV infection of Vero E6 cells in culture. As the expression of the N protein in SARS CoV-infected cells is a sensitive marker of infection, we have developed a sensitive and high-throughput ELISA for quantitative measurement of SARS CoV N protein (T.Z. et al., submitted). The antiviral activity of each MAb was evaluated by measuring the N protein with a quantitative ELISA, Western blotting, and in situ immunohistochemistry. By ELISA, control immunoglobulin G had no effect on the infection of Vero cells compared to that of cells incubated with virus alone. In contrast, all four anti-S-II MAbs inhibited SARS CoV infection (Fig. 3A). Two MAbs, S34 and S84, at 10 μg/ml completely inhibited viral infection at a multiplicity of infection of 0.5 and showed no detectable N protein comparable to that in noninfected control cells. The other two MAbs produced a >80% reduction in N protein production at 100 μg/ml. Western blot analysis of whole-cell lysates confirmed the absence of detectable viral protein in cell cultures incubated with 10 μg of S34 and S84 per ml (Fig. 3B). In situ immunohistochemistry staining demonstrated that there was no significant staining of SARS CoV N protein in cells incubated with antibodies at concentrations of ≥10 μg/ml. The neutralizing activity of some MAbs was also confirmed by the plaque reduction assay (1). The four antibodies showed similar activities at 50 μg/ml when a total of 103 PFU of virus per well was added to a six-well plate.
FIG. 3.
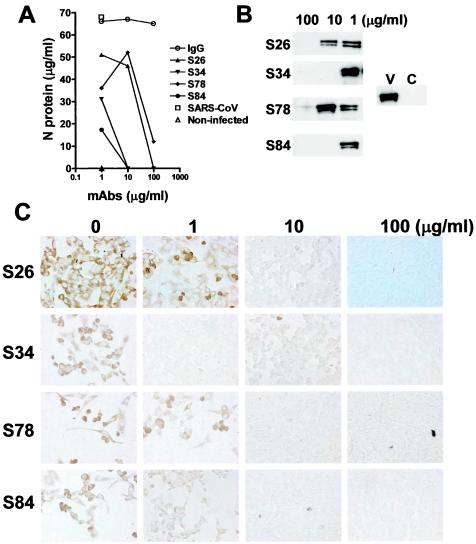
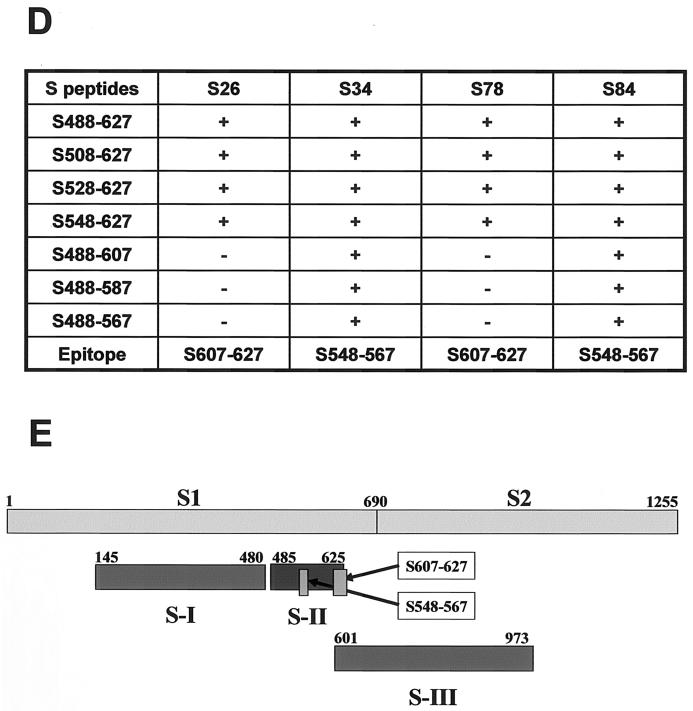
Neutralizing activity of anti-S-II MAbs. Various concentrations of diluted MAbs were added to duplicate 24-well plates containing confluent monolayers of Vero76 cells in Iscove's modified Eagle medium-2% fetal bovine serum-2 mM l-glutamine-antibiotics. SARS CoV (Toronto-2 isolate) was added to each well at a final concentration of 0.5 PFU/ml, and plates were incubated for 72 h at 37°C in 5% CO2. Medium was removed from the wells, one set of plates was fixed with 80% acetone in PBS for immunostaining, and 0.1% SDS was added to the other set of plates for analysis by ELISA. (A) Quantitative measurement of the SARS CoV N protein for neutralizing activity. A quantitative ELISA of SARS CoV N protein was developed for measurement of viral infection (T.Z. et al., submitted). Briefly, an ELISA plate was coated with two MAbs against the N and C termini of the SARS CoV N protein. Whole-cell lysates (0.1% SDS) of infected cells were diluted 1:100 and 1:1,000 in 3% BSA-PBS and incubated in a MAb-coated plate. The captured N protein was then detected with HRP-conjugated polyclonal goat anti-N antibody. The N protein content was determined with the full-length of recombinant N protein as the standard. IgG, immunoglobulin G. (B) Western blot analysis of the N protein in MAb-treated Vero cells infected with SARS CoV. Five microliters of whole-cell lysate was separated by SDS-10% PAGE and blotted. The N protein was probed with an HRP-conjugated MAb against N protein. (C) Immunohistochemistry detection of SARS CoV-infected cells. Acetone-fixed cells were stained with an HRP-conjugated anti-N MAb and revealed by DBA substrate buffer. Photos were taken at a magnification of ×400. The values at the top are the concentrations of the MAbs used for neutralization. (D) Neutralizing epitope mapping of anti-S-II MAbs. A series of the truncated S-II peptides were expressed in E. coli with a His6 tag at the N terminus. The truncated peptides were used to coat ELISA plates, which were incubated with 1 μg of HRP-conjugated anti-S-II MAbs per ml. (E) Location of the neutralizing epitopes relative to the S-I, S-II, and S-III fragments within the S protein.
TABLE 1.
Neutralization of MAbs by plaque assay
| MAb | Activity (PFU) at indicated concn (μg/ml):
|
||
|---|---|---|---|
| 5 | 50 | 500 | |
| S26 | 54 | 12 | 4 |
| S34 | 300 | 15 | 3 |
| S84 | 300 | 12 | 1 |
| S78 | 69 | 14 | 1 |
| Positive control | 320 | ||
To map the S-II epitopes recognized by these neutralizing antibodies, we truncated the S-II peptide at both the N and C termini and expressed a panel of shorter peptides in E. coli. The profile of MAb binding to these truncated peptides (Fig. 3D) indicates that S26 and S78 recognize an epitope within the 20 aa at the C terminus (aa 607 to 627), whereas S34 and S84 recognize an epitope (aa 548 to 567) in the middle of the S-II peptide immediately adjacent to a region significantly homologous to the receptor binding domain of HCoV 229E (Fig. 1B and 3E). Therefore, although S34 and S84 had the strongest neutralizing activity, there are at least two epitopes in the S protein that could elicit protective immunity against SARS CoV infection.
S-II elicits a protective antibody response.
BALB/c mice were subcutaneously immunized with 100 μg of S-II in Freund's complete adjuvant and boosted twice at weekly intervals. The titer of specific anti-S-II reactivity and of neutralizing activity for sera obtained at 2, 4, and 8 weeks after initial immunization showed a time-dependent increase (Fig. 4A and B). Compared to preimmune sera, significant neutralizing activity was observed as early as 2 weeks after primary immunization although the titers were low. However, high titers and maximal protection were observed after 4 weeks and were maintained at 8 weeks. Thus, S-II, a small part of the S protein of SARS CoV, is sufficient to elicit an endogenous, protective antiviral antibody response.
FIG. 4.
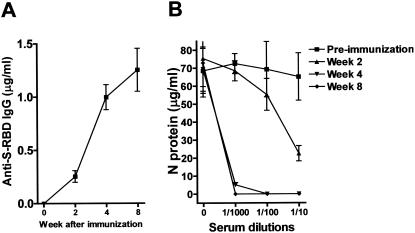
S-II elicits a protective antibody response against SARS CoV. Six- to 8-week-old female BALB/c mice were subcutaneously immunized with 100 μg of S-II in Freund's complete adjuvant and boosted twice with S-II in PBS weekly. Serum samples were collected at the indicated time points after primary immunization. (A) Serum levels of anti-S-II antibody were measured by indirect ELISA with an anti-S-II MAb as the standard. IgG, immunoglobulin G. (B) The 10-fold-diluted sera were added to Vero E6 cell cultures 1 h prior to infection of each well with 0.5 PFU of SARS CoV per ml. The neutralizing activity of antisera was determined by quantitative measurement of N protein by ELISA as described above. The results are presented as the mean ± the standard deviation of two pools of six mice at each time point.
DISCUSSION
Selecting from regions of the S protein of SARS CoV that are predicted to potentially present antigenic determinants, a small region (S-II) comprising residues 485 to 625 was expressed as a recombinant protein in E. coli. After proper refolding, the solubilized S-II fragment was shown to specifically bind the surface of Vero cells by FACS analysis. This suggests that antibodies against this region could give rise to neutralizing activities. A panel of MAbs was raised with the recombinant S-II protein. A series of tests showed that the MAbs could recognize the native S protein, block the attachment of S-II to Vero cells, and neutralize SARS CoV infection in cell culture.
Virus entry involves attachment of the S1 region to the host receptor and exposure of the fusion peptide that results from the conformational change induced by the interaction of the S1 region with the receptor. However, studies with HCoV 229E, MHV, and TGEV further defined polypeptide regions that can bind the host receptor. A receptor binding domain of HCoV 229E was identified as aa 407 to 547 (3), and that of TGEV was identified as aa 506 to 655 (7, 18), which corresponds to aa 279 to 417 by amino acid sequence alignment (3). A receptor binding domain was mapped to the N-terminal 330 aa of MHV (10). These observations suggest that a defined receptor binding domain could be located in various regions of the S protein, depending on the host receptor. The entire S1 region may be required for the full function while a local region is responsible for attachment to the receptor. S-II was shown to bind the host cell surface in this study, which is most closely related to the receptor binding domain identified for HCoV 229E with moderate homology in the local region. This is consistent with the studies of Wong et al. (19) and Xiao et al. (20) in which the more C-terminal region of the SARS CoV S protein was shown to bind the isolated receptor angiotensin-converting enzyme 2. The minimum overlapping region between S-II and other angiotensin-converting enzyme 2 binding fragments is aa 485 to 510.
With the MAbs generated with S-II, we have identified the predicted 180-kDa S protein of SARS CoV. These MAbs also recognize a putatively trimerized form of the S protein, which may represent its native form, and recognition of the trimerized form may be important for antibody neutralizing activity. Our data indicate that at least one binding-related region of the SARS CoV S protein is located within amino acid residues 485 to 625. Epitope mapping demonstrates that two exposed regions, aa 607 to 627 and 548 to 567, are located in this peptide, which is separated from the suggested minimum binding region. Development of an effective antiviral antibody response in mice immunized with S-II suggests that S-II might be an appropriate target in vaccine development for generation of protective humoral immunity in humans. Moreover, identification of S-II as a critical antigenic determinant within the S protein will be helpful in the development of an immunoassay to evaluate effective, protective antibody responses in patients during SARS CoV infection. The in vitro antiviral activity of anti-S-II MAbs suggests strategies for the development of therapeutic neutralizing antibodies effective for passive immunization in the prevention and perhaps treatment of SARS CoV infection. In addition, paired MAbs specific for the SARS CoV S protein and able to immunoprecipitate intact viral particles will enable quantitative immunoassays for measurement of viral particles in specimens from SARS patients.
Acknowledgments
T.Z. is supported by a research grant from Sankyo Co., Ltd., Tokyo, Japan.
We thank William J. Koopman for critical comments.
REFERENCES
- 1.Bonavia, A., B. D. Zelus, D. E. Wentworth, P. J. Talbot, and K. V. Holmes. 2003. Identification of a receptor-binding domain of the spike glycoprotein of human coronavirus HCoV-229E. J. Virol. 77:2530-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyle, J. F., D. G. Weismiller, and K. V. Holmes. 1987. Genetic resistance to mouse hepatitis virus correlates with absence of virus-binding activity on target tissues. J. Virol. 61:185-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breslin, J. J., I. Mørk, M. K. Smith, L. K. Vogel, E. M. Hemmila, A. Bonavia, P. J. Talbot, H. Sjöstrom, O. Norén, and K. V. Holmes. 2003. Human coronavirus 229E: receptor binding domain and neutralization by soluble receptor at 37°C. J. Virol. 77:4435-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delmas, B., J. Gelfi, R. L'Haridon, L. K. Vogel, H. Sjostrom, O. Noren, and H. Laude. 1992. Aminopeptidase N is a major receptor for the enteropathogenic coronavirus TGEV. Nature 357:417-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dveksler, G. S., C. W. Dieffenbach, C. B. Cardellichio, K. McCuaig, M. N. Pensiero, G.-S. Jiang, N. Beauchemin, and K. V. Holmes. 1993. Several members of the mouse carcinoembryonic antigen-related glycoprotein family are functional receptors for the coronavirus mouse hepatitis virus-A59. J. Virol. 67:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dveksler, G. S., M. N. Pensiero, C. B. Cardellichio, R. K. Williams, G. S. Jiang, K. V. Holmes, and C. W. Dieffenbach. 1991. Cloning of the mouse hepatitis virus (MHV) receptor: expression in human and hamster cell lines confers susceptibility to MHV. J. Virol. 65:6881-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Godet, M., J. Grosclaude, B. Delmas, and H. Laude. 1994. Major receptor-binding and neutralization determinants are located within the same domain of the transmissible gastroenteritis virus (coronavirus) spike protein. J. Virol. 68:8008-8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holmes, K. V. 2003. SARS-associated coronavirus. N. Engl. J. Med. 348:1948-1951. [DOI] [PubMed] [Google Scholar]
- 9.Kolaskar, A. S., and P. C. Tongaonkar. 1990. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. 276:172-174. [DOI] [PubMed] [Google Scholar]
- 10.Kubo, H., Y. K. Yamada, and F. Taguchi. 1994. Localization of neutralizing epitopes and the receptor binding site within the amino-terminal 330 amino acids of the murine coronavirus spike protein. J. Virol. 68:5403-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marra, M. A., S. J. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 12.Matsuyama, S., and F. Taguchi. 2002. Receptor-induced conformational changes of murine coronavirus spike protein. J. Virol. 76:11819-11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakamura, Y., T. Gojobori, and T. Ikemura. 2000. Codon usage tabulated from the international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 28:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rice, P., I. Longden, and A. Bleasby. 2000. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 16:276-277. [DOI] [PubMed] [Google Scholar]
- 15.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. T. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rassmussen, R. Fouchier, S Gunther, A. D. M. E. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 16.Sanchez, C. M., A. Izeta, J. M. Sanchez-Morgado, S. Alonso, I. Sola, M. Balasch, J. Plana-Duran, J., and L. Enjuanes. 1999. Targeted recombination demonstrates that the spike gene of transmissible gastroenteritis coronavirus is a determinant of its enteric tropism and virulence. J. Virol. 73:7607-7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spiga, O., A. Bernini, A. Ciutti, S. Chiellini, N. Menciassi, F. Finetti, V. Causarono, F. Anselmi, F. Prischi, and N. Niccolai. 2003. Molecular modelling of S1 and S2 subunits of SARS coronavirus spike glycoprotein. Biochem. Biophys. Res. Commun. 310:78-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sune, C., G. Jimenez, I. Correa, M. J. Bullido, F. Gebauer, C. Smerdou, and L. Enjuanes. 1990. Mechanisms of transmissible gastroenteritis coronavirus neutralization. Virology 177:559-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong, S. K., W. Li, M. J. Moore, H. Choe, and M. Farzan. 2004. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 279:3197-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao, X., S. Chakraborti, A. S. Dimitrov, K. Gramatikoff, and D. S. Dimitrov. 2003. The SARS-CoV S glycoprotein: expression and functional characterization. Biochem. Biophys. Res. Commun. 312:1159-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yeager, C. L., R. A. Ashmun, R. K. Williams, C. B. Cardellichio, L. H. Shapiro, A. T. Look, and K. V. Holmes. 1992. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature 357:420-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zelus, B. D., J. H. Schickli, D. M. Blau, S. R. Weiss, and K. V. Holmes. 2003. Conformational changes in the spike glycoprotein of murine coronavirus are induced at 37°C either by soluble murine CEACAM1 receptors or by pH 8. J. Virol. 77:830-840. [DOI] [PMC free article] [PubMed] [Google Scholar]


