Abstract
Monolayers of cystic fibrosis transmembrane conductance regulator (CFTR)-deficient human tracheal glandular cells (CF-KM4) were subjected to phage biopanning, and cell-internalized phages were isolated and sequenced, in order to identify CF-KM4-specific peptide ligands that would confer upon adenovirus type 5 (Ad5) vector a novel cell target specificity and/or higher efficiency of gene delivery into airway cells of patients with cystic fibrosis (CF). Three different ligands, corresponding to prototypes of the most represented families of phagotopes recovered from intracellular phages, were designed and individually inserted into Ad5-green fluorescent protein (GFP) (AdGFP) vectors at the extremities of short fiber shafts (seven repeats [R7]) terminated by scissile knobs. Only one vector, carrying the decapeptide GHPRQMSHVY (abbreviated as QM10), showed an enhanced gene transduction of CF-KM4 cells compared to control nonliganded vector with fibers of the same length (AdGFP-R7-knob). The enhancement in gene transfer efficiency was not specific to CF-KM4 cells but was observed in other mammalian cell lines tested. The QM10-liganded vector was referred to as AdGFP-QM10-knob in its knobbed version and as AdGFP-QM10 in its proteolytically deknobbed version. AdGFP-QM10 was found to transduce cells with a higher efficiency than its knob-bearing version, AdGFP-QM10-knob. Consistent with this, competition experiments indicated that the presence of knob domains was not an absolute requirement for cell attachment of the QM10-liganded vector and that the knobless AdGFP-QM10 used alternative cell-binding domains on its capsid, including penton base capsomer, via a site(s) different from its RGD motifs. The QM10-mediated effect on gene transduction seemed to take place at the step of endocytosis in both quantitative and qualitative manners. Virions of AdGFP-QM10 were endocytosed in higher numbers than virions of the control vector and were directed to a compartment different from the early endosomes targeted by members of species C Ad. AdGFP-QM10 was found to accumulate in late endosomal and low-pH compartments, suggesting that QM10 acted as an endocytic ligand of the lysosomal pathway. These results validated the concept of detargeting and retargeting Ad vectors via our deknobbing system and redirecting Ad vectors to an alternative endocytic pathway via a peptide ligand inserted in the fiber shaft domain.
Viruses have developed natural strategies to infect and grow in specific tissues of particular hosts, and their specificity and restricted host range make them theoretically the best-adapted vectors to transfer therapeutic genes into recipient cells that are their natural targets. However, when desired cell targets are not part of the natural host range of the virus, genetic modifications are required to adapt the virus and its derived vectors to these novel hosts. Since viral tropism is, in large part, under the control of primary virus-cell interaction occurring at the cell surface, the most obvious modification consists of altering the viral outer capsid or envelope in order to alter the viral tropism and redirect the virus to new cell receptors. Human adenovirus (Ad), which is now commonly used as a gene vector, is a nonenveloped virus whose capsid is constituted of 240 hexons and 12 pentons located at the 12 apices of the icosahedral capsid. The penton capsomer is formed from two components, the fiber and the penton base. The fiber has three structural domains: the tail, which is anchored to the penton base; the shaft, constituted of multiple repeats of a 15-amino-acid motif; and a terminal globular structure, called the “knob” (reviewed in reference 55). Attachment of Ad of species C (e.g., Ad type 2 [Ad2] and Ad5) to the cell surface is mediated by a high-affinity interaction between the knob domain of the fiber and the CAR (coxsackievirus-adenovirus receptor) protein (4, 67). Following attachment, the interaction between the RGD motifs of the penton base and αvβ3/5 integrin molecules promotes endocytosis and internalization of the virus and inititates cell signaling (14, 31-34, 38, 66, 73, 74).
However, Ads of other species use different attachment receptors, recently identified as sialic acid residues for species D Ad37 (1, 2) and CD46 for species B members Ad11 and Ad35 (13, 57). In addition, the absence or low level of expression of CAR molecules and their location at the basolateral tight junctions of the airway epithelium cells are two parameters that are detrimental to the usage of unmodified Ad vectors in gene therapy protocols aiming at certain tissues, such as the airways or lung epithelia (71, 72, 79). In a previous study, it was shown that CF-KM4 cells, an immortalized human tracheal submucosal gland serous cell line from a cystic fibrosis (CF) patient carrying a ΔF508:ΔF508 mutation, and its counterpart, MM39, a tracheal submucosal gland serous cell line from a healthy subject (24, 39), were poorly permissive for Ad5 infection and Ad5-mediated gene transduction (12). The limiting factor for the viral infection was shown to reside at the steps of virus-cell attachment and entry: CF-KM4 and MM39 cells do not express CAR at their surfaces or at the apical or basolateral poles, and Ad5 binds to α(2→6)-linked sialic acid residues of sialoglycoproteins (SA-GP) on CF-KM4 cells and to heparan sulfate glucosaminoglycans on MM39 cells (12). Although both CF-KM4 and MM39 cells express high levels of αvβ3/5 integrins, Ad5 endocytosis was found to occur via a penton base-RGD- and integrin-independent pathway. However, the addition of a stretch of seven lysine residues at the C terminus of the fiber knob domain significantly enhanced the gene transduction of MM39 cells, and replacement of the knobs by RGD motifs at the extremities of shortened shafts increased the Ad5-mediated gene transfer efficiencies in both cell lines (12). Considering the promiscuous nature of RGD-dependent integrins and the lack of recognition specificity of the oligolysine ligand, none of these fiber modifications could be envisaged to retarget Ad5 to any cell-specific surface molecule.
In the present study, we attempted to improve both the efficiency and specificity of Ad-mediated gene delivery into human airway cells, as previously performed using chimeric Ad (78), fiber-modified Ad5 vectors and, using CF transmembrane conductance regulator (CFTR)-deficient tracheal glandular CF-KM4 cells as a target model system in vitro. Since SAα(2→6)-GP were apparently low-efficiency receptors for Ad5 in CF-KM4 cells, we looked for surface molecules which could advantageously be used as alternative Ad receptors and whose ligands could be inserted into the fibers of Ad5 vectors. We therefore panned monolayers of living CF-KM4 cells by using a bacteriophage library and designed three different oligopeptides that each corresponded to one of the most represented families of phagotopes recovered from intracellular phages. Only one, the decapeptide GHPRQMSHVY (abbreviated as QM10), was found to enhance Ad-mediated gene transduction of CF-KM4 cells when inserted in the fiber shaft of an Ad5-GFP vector (AdGFP-QM10). Analysis of the early steps of vector-cell interaction indicated that QM10 acted as an endocytic ligand with retargeting properties. QM10 did not increase the cell-binding capacity of the recombinant virus but intervened at the step of endocytosis in both quantitative and qualitative manners by redirecting vector particles to an endocytic compartment different from the class of early endosomes used by species C members and already described as the residence of Ad7, a species B member (41). Targeting AdGFP-QM10 to this alternative entry pathway seemed to confer upon the vector a significant advantage in terms of gene transfer efficiency in a number of human and simian cell lines. In addition, data from cell internalization experiments using knobbed and knobless vectors suggested that the knob domain contributed to the function of endosomal escape of Ad virions.
MATERIALS AND METHODS
Cells.
The simian virus 40-immortalized human tracheal gland cell lines MM39 (normal) and CF-KM4 (CFTR deficient) were maintained as monolayers on collagen I-coated flasks (Biocoat; Becton-Dickinson, Bedford, Mass.) in Dulbecco's modified Eagle's medium (DMEM)-Ham's F12 supplemented with 1% Ultroser G (Gibco-Invitrogen, Rockville, Md.), penicillin (200 U/ml), streptomycin (200 μg/ml), and epinephrin (3 μM; Sigma, St. Louis, Mo.). The E1A-E1B-trans-complementing HEK-293 cell line (abbreviated as 293) (CRL 1573) was obtained from the American Type Culture Collection (Manassas, Va.), and E1A-E1B-fiber-trans-complementing 293 (293-Fiber) cells (30) were obtained from Transgene SA (Strasbourg, France). The cells were cultured as monolayers in DMEM supplemented with 10% fetal calf serum (FCS; Sigma), penicillin (200 U/ml), and streptomycin (200 μg/ml; Gibco-Invitrogen) at 37°C and 5% CO2, and for 293-Fiber cells, hygromycin was added at 350 μg/ml. The human cell lines HeLa (cervix epitheliod carcinoma; CCL 2), A549 (lung alveolar carcinoma; CCL 185), HEp2 (larynx epidermoid carcinoma; CCL 23), MRC-5 (fetal lung), HRT-18 (ileocecal adenocarcinoma; CCL 244), PLC/PRF/5 (hepatoma; CRL 8024), and RD (rhabdomyosarcoma; CCL 136); the simian cell lines Vero (African green monkey kidney; CCL 81), LLC-MK2 (rhesus monkey kidney; CCL 7), and BGM (buffalo green monkey kidney epithelial [28]); canine MDCK cells (dog kidney; CCL 34); hamster CHO-K1 cells (Chinese hamster ovary; CCL 61); and mouse L20B cells (a recombinant murine cell line expressing human poliovirus receptor CD155 on the cell surface [75]) were all generously provided by the Clinical Virology Laboratory (Hospices Civils de Lyon, Domaine Rockefeller, Lyon, France). Except for the MRC-5, BGM, and L20B cells, the cells were originally from the American Type Culture Collection. The cells were cultured as monolayers in DMEM as described above. Daudi cells (Epstein-Barr virus-transformed B lymphoma cells) were cultured in RPMI medium supplemented with 10% FCS (21). For the production of recombinant Ad proteins, Spodoptera frugiperda cells (Sf9 subclone) were propagated in TNM medium (Gibco-Invitrogen); cultured as monolayers with 10% FCS, penicillin, and streptomycin as mentioned above; and maintained at 28°C (25, 26, 47).
Identification of CF-KM4 cell ligands.
Confluent monolayers of CF-KM4 cells (4 × 105 cells) were incubated with an aliquot (3 × 1010 phage in 0.25 ml of Tris-buffered saline) of a phage-displayed hexapeptide library (61) for 20 min at 37°C and then rinsed twice in Tris-buffered saline at room temperature (RT). In order to detach cell surface-adsorbed phage, the cells were incubated with 0.25 ml of elution buffer (0.1 M glycine-HCl buffer, pH 3.0, 2 M urea) for 10 min at RT. The cells were then harvested by scraping them with a rubber policeman, pelleted at 2,000 × g for 3 min, and lysed by allowing them to swell and vortexing them in 0.1 ml of hypotonic buffer (5 mM Tris-HCl buffer, pH 7.5, 1 mM Na2 EDTA). Cell debris was eliminated by low-speed centrifugation (500 × g for 3 min), and the supernatant containing cell-internalized phages was recovered for phage isolation, amplification, and phagotope sequencing according to previously published methods (19, 21).
Ad and Ad vectors.
Wild-type (WT) Ad5 and the replication-competent vector Ad5Luc3 (40), harboring the firefly luciferase gene (luc) under the control of the simian virus 40 early promoter inserted in the E3 region of the Ad5 genome, were propagated in HeLa cells. Recombinant Ad5 vectors with E1 deleted and with WT fibers were propagated in 293 cells, whereas fiber-modified Ad5 vectors with E1 deleted were grown in 293-Fiber cells until the last amplification step of the vector stock, which was performed in 293 cells. Virus stocks were purified by CsCl gradient ultracentrifugation according to conventional methods (9). The construction of Ad5 vectors with modified fibers has been described in detail in previous studies (17, 22, 36, 37). In brief, the backbone vector AdGFP-R7-(Xa)-knob (abbreviated AdGFP-R7-knob) was an Ad5-based vector with its deleted E1 region replaced by a green fluorescent protein (GFP) expression cassette under the cytomegalovirus promoter and a genetically modified fiber that contained the following domains, from the N to the C terminus: (i) the tail, (ii) the N-terminal seven shaft repeats (R7), (iii) a trimerization signal (PDVASLRQQVAELQGQVQHLQAAFSQYKKVELFPNG) called the neck region peptide (NRP) from the human lung surfactant protein D, (iv) a tridecapeptide linker (AKKLNDAQAPKSD), (v) the desired cell ligand, (vi) the cleavage site for factor Xa protease, and (vii) the last shaft repeat and the terminal knob domain (Fig. 1). The cleavage site for factor Xa consisted of the tetrapeptide IEGR, and the cleavage occurred after the arginine residue. Recombinant fibers were rescued into the Ad5-GFP genome as previously described (36). The backbone vector AdGFP-R7-knob, which carried nonliganded, short-shafted fibers with NRP, the factor Xa site, and the terminal knob but no specific cell ligand (Fig. 1b and c), was used as the control vector in our experiments, with or without cleavage by factor Xa. For the construction of liganded AdGFP-R7-knob vector with new cell tropism, three peptide ligands potentially involved in phage endocytosis (abbreviated QM10, SY12, and LAP25, respectively) were individually inserted into the fiber shaft domain upstream of the factor Xa cleavage site. For proteolytic deknobbing of Ad5 vectors, virus samples were treated with 1 U of factor Xa per 109 physical virus particles in cleavage buffer (50 mM Tris, pH 8.0, 100 mM NaCl, 5 mM CaCl2) for 16 h at 22°C.
FIG. 1.
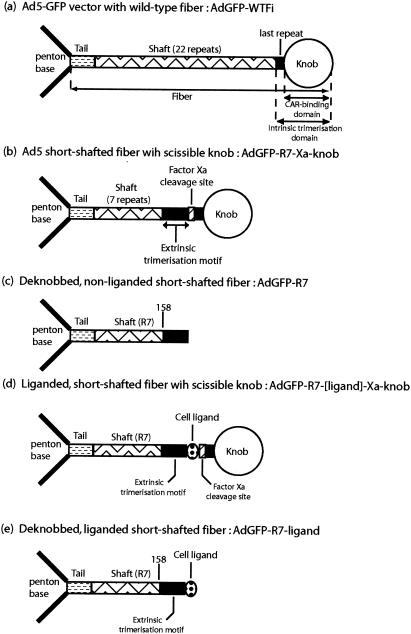
Schematic representation of genetic constructs of Ad5-based vectors (AdGFP) carrying WT fiber (a) or recombinant fibers (b to e). (a) In WT fiber, the tail domain, bound to a penton base capsomer (left), the shaft domain, the last shaft repeat, and the knob are shown. (b to e) In short-shafted recombinant fibers with seven-repeat shafts (R7), the different fiber domains are shown. The extrinsic trimerization motif, inserted downstream of amino acid residue 158 of the shaft, the factor Xa cleavage site, and the cell ligand are also shown.
Ad titration.
Since all our uncleaved Ad5 clones carried the fiber knob domain and bound to CAR, the concentration in infectious viral particles of CsCl-purified Ad stocks could be determined and compared using plaque assays in 293 cell monolayers according to conventional methods (36), and the infectivity titers were expressed as PFU per milliliter. The concentration in physical particles (PP) was deduced from the protein concentration determined in the same CsCl-purified Ad stocks, using the Bradford protein assay (Bio-Rad, Hercules, Calif.) with bovine serum albumin (BSA) (2× crystallized BSA; Bio-Rad) as the standard. The number of PP was calculated from the total protein content of the sample, taking the mass of 2.91 × 10−16 g for a single virion, i.e., 3.4 × 1012 virions per mg of protein (16). The infectivity index represented the ratio of infectious to physical particles (PFU/PP) and usually ranged between 1:25 and 1:50 for WT Ad5 (9). For the propagation of Ad clones, we used a multiplicity of infection (MOI) ranging from 2 to 10 PFU/cell, corresponding to 50 to 500 PP/cell. For probing cellular functions (e.g., attachment, endocytosis, and vesicular escape), the MOI used varied from 5 × 102 to 5 × 104 PP/cell.
Recombinant Ad proteins and competition assays.
Recombinant Ad penton base and fiber proteins were isolated in native and soluble forms from lysates of Ad5-infected HeLa cells or baculovirus-infected Sf9 cells and purified according to a previously published method (6) adapted to fast protein liquid chromatography (43, 44). The genetic constructions of recombinant Autographa californica multiple nuclear polyhedrosis viruses expressing full-length Ad5 fiber or fiber knob domain have been described in detail elsewhere (21, 47, 56). Since penton base proteins from Ad2 and -5 have 98% amino acid sequence identity (45), our recombinant Ad2 penton base proteins, WT and the RGD mutant R340E, isolated from A. californica multiple nuclear polyhedrosis virus-infected Sf9 cells (25, 26), were used in competition experiments with Ad5 virions. Protein samples were analyzed by conventional sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and immunoblotting using the required antibodies, as previously described (26, 47, 56). The Ad protein concentration was estimated by the intensity of Coomassie blue staining of protein bands in SDS gels, measured by scanning at 610 nm in an automatic densitometer (REP-EDC; Helena Laboratories, Beaumont, Tex.), using a range of known BSA concentrations for calibration.
Cell-binding competition assays were performed using purified hexon, penton base, full-length fiber, and knob proteins and the penton base mutant R340E. Each capsid protein was individually mixed with each vector, in large excess over its copy number in the Ad virion, and incubated at 4°C with cells to allow attachment but not entry. For solubility reasons, hexon protein was added in only 100-fold excess over the hexon content of the virions, whereas penton base, fiber, and knob proteins were added in 103-fold excess. Unadsorbed virus was rinsed off, and the cells were assayed for GFP expression 28 h postinfection (p.i.).
Immunological reagents and flow cytometry. (i) Antibodies.
1D6.14, an anti-Ad5 fiber knob monoclonal antibody (MAb) with CAR-blocking activity, was kindly supplied by D. T. Curiel (University of Alabama at Birmingham) (36, 54). MAb 4D2.5, directed toward the conserved fiber tail motif (18, 19), was obtained from J. A. Engler (University of Alabama at Birmingham). Other antibodies against Ad proteins were described in a previous study (12).
(ii) Flow cytometry.
Confluent or subconfluent cells were rinsed with phosphate-buffered saline (PBS), detached with PBS containing 1 mM Na2 EDTA and trypsin (0.1 mg/ml), fixed with paraformaldehyde (3% in PBS) for 10 min at RT, and resuspended in PBS containing BSA at 0.1 mg/ml. Cell samples (106 cells/0.1 ml) were analyzed by flow cytometry using a FACScalibur flow cytometer (Becton Dickinson).
Fluorescent probes and IF microscopy. (i) Labeling of cell ligands.
To generate fluorescent Cy2-Ad or Cy3-Ad vectors, Ad virions were covalently conjugated with two different fluorophores, the carbocyanine bisfunctional dyes Cy2 and Cy3 (Amersham Biosciences, Little Chalfont, United Kingdom), according to a published protocol (31) with some modifications. The main modification of the protocol consisted of replacing the dialysis step, which often provokes the precipitation of virions and macromolecules, or the gel exclusion chromatography step, which dilutes the virus inoculum, by blocking the excess of coupling reagent with 20 mM lysine, followed by dilution of the fluorescent-dye-conjugated ligands in culture medium. Cy2- or Cy3-labeled Ads were used within 2 h after coupling. Human alpha-2 macroglobulin (α2 M; Sigma) was conjugated with fluorescene isothiocyanate (FITC) using the EZ-label FITC protein-labeling kit (Pierce, Rockford, Ill.) and the protocol recommended by the manufacturer. Immunofluorescence (IF) microscopy of cell-bound virus was performed using an Axiovert 135 microscope (Zeiss) equipped with an AxioCam videocamera and a quantitative image analysis program.
(ii) Endosomal compartment markers.
Mouse MAb to transferrin receptor (anti-CD71) and rabbit antibodies against the Rab proteins Rab4 and Rab11 were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, Calif.). Mouse MAb to LAMP-1 (anti-CD107a) was from BD PharMingen (San Diego, Calif.), and rabbit antibody against the Rab5 protein was from Stressgen Biotechnologies (Victoria, British Columbia, Canada). FITC-conjugated anti-mouse immunoglobulin G was from Pierce, and FITC-conjugated anti-rabbit immunoglobulin G was from Santa Cruz Biotechnoloy. Internalized ligands and cell compartments were analyzed in IF microscopy, using a Zeiss LSM 510 META confocal microscope.
Assays for cell attachment, endocytosis, and vesicular escape of Ad5 vectors. (i) Cell attachment.
Confluent cell monolayers were incubated with aliquots of Cy3-Ad vectors ranging from 5 × 103 to 5 × 104 particles per cell at 0°C for 1 h. After being rinsed, cell-adsorbed fluorescent signal was quantitated by IF microscopy. The cell-binding index, expressed as arbitrary units (AU), was calculated using the following formula: number of positive fluorescent cells per field × (mean fluorescent intensity per cell − mean background fluorescent signal of control cells). The number of individual cells analyzed ranged between 100 and 200 in each separate experiment.
(ii) Endocytosis.
Confluent cell monolayers were incubated with aliquots of Cy2- or Cy3-Ad vectors ranging from 5 × 103 to 5 × 104 particles per cell at 37°C for 1 h and then treated with 0.05% trypsin and 0.53 mM Na4 EDTA in Hanks's balanced salt solution (Hank's balanced salt solution-trypsin-EDTA 1×; Gibco-Invitrogen) at 37°C for 15 min to detach possible surface-sequestered viruses. After fixation in 3% paraformaldehyde in PBS, the intensity of the internalized fluorescent signal was determined by fluorescence-activated cell sorter (FACS) analysis, as described above. The endocytic index, also expressed as AU, was calculated as described above, using the following equation: percentage of fluorescent cells × (mean fluorescence intensity per cell − mean background fluorescent signal of control cells).
(iii) Ad-mediated endosomolysis, vesicular escape, and virus internalization.
The Ad-mediated endosomolysis, vesicular escape, and virus internalization were determined using a toxin assay, as previously described (12). In brief, cell monolayers were preincubated with ricin agglutinin (RcA; 120,000 molecular weight; Sigma) at concentrations ranging from 0 to 10 μg of RcA per aliquot of 5 × 105 cells for 1 h at 37°C, in methionine- and cysteine-free culture medium, and in the presence (or absence) of Ad vector, at a constant input of 104 PP/cell. [35S]methionine and [35S]cysteine (≥1,000 Ci/mM; PRO-MIX; Amersham) were added at 15 μCi per 5 × 105-cell sample, and incubation proceeded for another 2 h at 37°C. The cells were then rinsed with culture medium, detached from the support, and dissolved in 0.2 N NaOH-1% SDS. Cellular proteins were precipitated by the addition of 10 volumes of trichloroacetic acid at 10%, and the precipitates were retained by filtration on GF/C glass filters. Ad-RcA-mediated inhibition of cell protein synthesis was evaluated from trichloroacetic acid-precipitable radioactivity, determined by scintillation counting in a liquid scintillation spectrometer (LS6500; Beckman Coulter Inc., Fullerton, Calif.).
Ad-mediated gene transduction assays.
Ad5 vector, carrying the reporter gene luc or gfp, was added to treated or mock-treated cells in a final volume of 200 μl in culture medium and at MOIs ranging from 0 to 30 PFU/cell. After incubation for 1 h at 0°C with intermittent rocking, the cell monolayers were washed with cold medium to remove unadsorbed virus. Prewarmed medium was then added, and the cells were transferred to 37°C for 24 to 36 h. The cells were then harvested, and the efficiency of Ad-mediated gene transfer was estimated by the level of reporter gene product expression, luciferase or GFP. Luciferase activity was assayed in cell lysates using luciferase substrate solution (Promega) in a Lumat LB-9501 luminometer (Berthold Bioanalytical, Bad Wildbad, Germany). The results were expressed in relative light units per milligram of whole protein present in cell lysates (20). GFP activity was determined using FACS analysis or IF microscopy with a quantitative image analysis program, as described above. The index of gene transfer efficiency (GTE) (12), expressed as AU, was measured by the following equation: GTE index = percentage of GFP-positive cells × (mean fluorescence intensity − background fluorescent signal of control cells).
Electron microscopy (EM).
Mock- or AdGFP vector-transduced cells were harvested at early times after infection (0.5 to 1 h p.i.), pelleted, fixed with 2% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.4, and postfixed with osmium tetroxide (1% in 0.1 M cacodylate buffer, pH 7.4). Cell specimens were dehydrated and embedded in Epon-812 (Fulham, Latham, N.Y.). Sections were stained with 7% uranyl acetate in methanol and poststained with 2.6% alkaline lead citrate in H2O. Specimens were examined under a Jeol 1200-EX electron microscope equipped with a MegaView II high-resolution transmission EM camera and a Soft Imaging system of analysis (Eloïse, Roissy, France).
RESULTS
Screening for peptide ligands of cell internalization receptors in CF-KM4 cells.
Since CF-KM4 cells do not express CAR, the high-affinity receptor for Ad5, we looked for other surface molecules that could act as substitute receptors for Ad5 and be targeted by recombinant Ad5 vectors. As there was no direct screening method available for identifying ligands of CF-KM4 cells, we applied the indirect approach of biopanning monolayers of living cells (11, 62), using a phage-displayed hexapeptide library (19, 21). Two classes of phage populations were isolated. One consisted of extracellular phages, eluted from the surfaces of intact cells and referred to as “cell binders.” The cell binders carried peptide aptamers corresponding to ligands of surface-exposed domains of plasma membrane molecules which would not necessarily lead to phage endocytosis and entry. The other population of phages recovered after cell lysis corresponded to intracellular phages. It has been shown that recombinant bacteriophages carrying a portion of the Ad penton base sequence overlapping the RGD peptide motif were capable of binding to αv integrins of the mammalian-cell plasma membrane and were found to be endocytosed into endosomal vesicles (10). We then postulated that intracellular phage recovered from CF-KM4 cells were competent for cell entry and that this novel competency, conferred upon the bacteriophages by the extrinsic peptides displayed on their pIII attachment proteins, could also be conferred upon recombinant Ad vectors by insertion of the same peptides into their fibers.
The intracellular phage were thus isolated, amplified, and sequenced, and the most represented phagotopes were grouped in families according to their sequence homologies. Alignment of overlapping peptides sharing common motifs allowed us to design consensus oligopeptides with longer sequences than the original hexapeptide phagotopes, e.g., 10- or 12-mers (19, 21). Three major ligand families emerged from this analysis. The first family of intracellular phage contained recurrent peptide motifs like LLTV, RMQ, and QPPG. A search in data banks revealed the same motifs in a lysosomial acid phosphatase (LAP), which has the property of recirculating between the plasma membrane and the lysosomal membrane (48, 49). Since the phagotopes overlapped the LAP sequence between residues 399 and 423 (LLTVLFRMQAQPPGYRHVADGQDHA), it is referred to as LAP25 in the present study. The two other phagotope families were represented by the decapeptide GHPRQMSHVY and the dodecapeptide TAYSSYMKGGKF, designated QM10 and SY12, respectively. Sequence comparison with proteins in data banks did not elicit any plasma membrane protein or protein domain with any known receptor activity or potentiality.
Construction and nomenclature of AdGFP vectors carrying liganded fibers with scissile knobs (Fig. 1).
Each of the three ligands LAP25, QM10, and SY12 was inserted into the fiber of the backbone vector AdGFP-R7-knob (22), upstream of the factor Xa cleavage site, to generate the recombinant vectors AdGFP-LAP25-knob, AdGFP-QM10-knob, and AdGFP-SY12-knob, respectively. trans-complementing HEK-293-Fiber cells were transfected with each of the Ad DNA genomes recombined with liganded fiber genes, and plaques were isolated and amplified. However, we were not able to amplify the plaques obtained after transfection with AdGFP-LAP25-knob DNA, likely due to the cytotoxic effect provoked by the LAP25-liganded recombinant fiber. Normal virus growth and infectious virus stocks with high titers were obtained in trans-complementing 293 cells with AdGFP-QM10-knob and AdGFP-SY12-knob vectors (Table 1). The infectivity indexes (estimated from the PP/PFU ratio per milliliter) of AdGFP-SY12-knob and AdGFP-QM10-knob were 5- to 10-fold lower than that of the normal, long-shafted fiber-bearing vector AdGFP-WTFi but 6- to 3-fold higher than the corresponding control vector, AdGFP-R7-knob, which carried nonliganded fibers of the same length (Table 1). After ablation of the knob by factor Xa digestion, the two liganded vectors were designated AdGFP-QM10 and AdGFP-SY12, and the nonliganded control vector was designated AdGFP-R7 (Fig. 1c and e). When analyzed by SDS-polyacrylamide gel electrophoresis and immunoblotting, the three recombinant vectors showed fiber protein and fiber cleavage products migrating with the apparent molecular mass expected before and after cleavage by factor Xa (reference 22 and data not shown).
TABLE 1.
Biological characteristics of Ad5 vectors carrying unliganded or liganded fibersa
| Ad5 vector | Virion titerb (PP/ml) | Infectious titerc (PFU/ml) | PP/PFU ratio |
|---|---|---|---|
| AdGFP-WTFid | 1.00 × 1012 | 1.50 × 1011 | 7 |
| AdGFP-R7-knobe | 1.00 × 1012 | 3.75 × 109 | 266 |
| AdGFP-QM10-knob | 3.20 × 1011 | 4.00 × 109 | 80 |
| AdGFP-SY12-knob | 1.10 × 1012 | 2.88 × 1010 | 38 |
| AdGFP-LAP25-knob | NAf | NA | NA |
Samples of Ad5 vector, purified by ultracentrifugation in CsCl gradient, were assayed for infectivity and virion content.
The number of virions present in each sample, comprising infectious and uninfectious virus particles, was calculated from the penton base protein content of the samples, determined by Western blot analysis (37).
The titer in infectious particles was determined by plaque titration in 293 cells.
Control vector with WT nonliganded, long-shafted fibers.
Control vector with nonliganded, short-shafted fibers.
NA, not applicable.
Efficiency and cell specificity of gene transduction mediated by liganded AdGFP vectors.
The functionality of the fiber-inserted ligands was evaluated for AdGFP-QM10-knob and AdGFP-SY12-knob in transduction assays of CF-KM4 cells, control MM39 cells, and a variety of other cell targets from different species in comparison with their nonliganded counterpart, AdGFP-R7-knob, and with AdGFP-WTFi. The GTE was expressed as the percentage of GFP-positive cells (Fig. 2a), but also as the GTE index (12), as shown in Fig. 2b. Since some cell populations were found to be heterogeneous in their levels of GFP expression, which was particularly high in a limited number of cells, the parameter of mean fluorescence intensity was taken into account and included in the calculations of the GTE index.
FIG. 2.
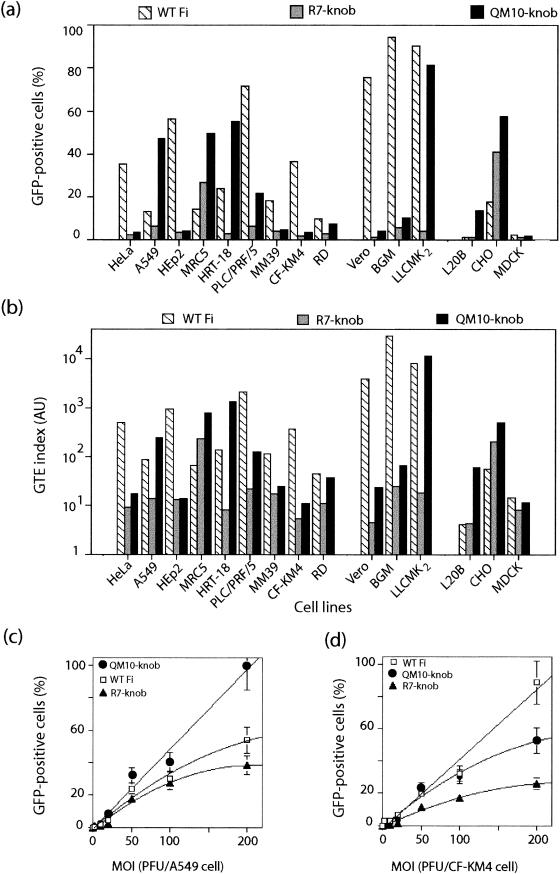
Comparison of GTEs in the QM10-liganded vector AdGFP-QM10-knob (QM10-knob) and the nonliganded control vectors AdGFP-WTFi (long-shafted fibers) (WT Fi) and AdGFP-R7-knob (short-shafted fibers) (R7-knob) in various cell lines at an MOI of 20 PFU/cell. (a and b) GFP expression was analyzed by FACS, and the results are expressed as the percentage of GFP-positive cells (a) and the GTE index (b), as described in Materials and Methods. (c and d) Dose dependence of gene transduction mediated by AdGFP-WTFi, AdGFP-R7-knob, and AdGFP-QM10-knob in A549 (c) and CF-KM4 (d) cells. The cells were transduced at various MOIs, as indicated on the x axis, and GFP expression was determined by counting GFP-positive cells by IF microscopy.
In most cells, gene transduction was 4- to 60-fold lower with the nonliganded, short-shafted vector AdGFP-R7-knob than with AdGFP-WTFi, as expressed by the percentage of GFP-positive cells (Fig. 2a), and the difference was even more pronounced as measured by the GTE index, ranging from 1 to 3 orders of magnitude (Fig. 2b). These results indicated that the length of the fiber shaft greatly influenced the mechanism of Ad cell recognition. This was consistent with previous observations and with a recent model of the interaction between the cell receptor and the Ad capsid in the case of long and flexible versus short and rigid fibers, showing that the length and flexibility of the fiber shaft are critical parameters for the usage of Ad receptors, such as integrins (52, 53) or CAR (76), as well as for virus infectivity (60). However, the GTE was higher with AdGFP-R7-knob than with AdGFP-WTFi in MRC5 and CHO cells and equivalent with both vectors in L20B cells (Fig. 2a and b).
The level of GFP expression in cells transduced by AdGFP-QM10-knob, compared to its nonliganded version, AdGFP-R7-knob, showed that the QM10 ligand consistently and significantly augmented the GTE in all the cell lines tested, but to various degrees (Fig. 2a and b). This enhancement ranged from 2- to 40-fold, depending on the MOI and the cell type. In LLC-MK2 and RD cells, the GFP level was equivalent to that mediated by AdGFP-WTFi, whereas in other cell lines (A549, MRC5, HRT-18, L20B, and CHO), it was 3- to 10-fold higher. In tracheal glandular cells, the difference between AdGFP-QM10-knob and AdGFP-R7-knob was not significant for MM39 cells and was <3-fold for CF-KM4 cells. AdGFP-SY12-knob gave levels of gene transfer similar to those of AdGFP-R7-knob (data not shown), indicating that the effect observed with the decapeptide ligand QM10 was not simply a property of any foreign peptide inserted at that site in Ad5 vectors but was sequence related. However, our results also implied that QM10, although isolated from CF-KM4 cell-internalized phages, was not tissue specific or even species specific. In fact, QM10 was more efficient in promoting gene transduction of A549 (terminal bronchial cells from human lung carcinoma), MRC5 (primary fibroblastic cells from human fetal lung), HRT-18 (human ileocecal adenocarcinoma cells), and RD cells (from human rhabdomyosarcoma), as well as LLC-MK2 cells (simian kidney cells), than the CF-KM4 human tracheal glandular cells from which it issued.
Dose dependence of gene transfer by different vectors in airway cells.
Since the possible low density of some cell surface receptors could mask the usefulness of the modifications of fibers at low vector doses, gene transduction was assayed at various MOIs of AdGFP-WTFi, AdGFP-R7-knob, and AdGFP-QM10-knob in cell lines with different QM10 ligand responsiveness, A549 (high) and CF-KM4 (low) cells, respectively (Fig. 2a and b). No difference in the levels of gene transfer was detectable at low infectious doses of the different vectors. At higher doses, however, gene transduction of A549 cells was found to increase in a dose-dependent manner. A nearly linear dose-response was observed with AdGFP-QM10-knob between 20 and 200 PFU/cell, and almost 100% of cells were found to be GFP positive at the highest MOI, whereas GFP expression reached a plateau with AdGFP-WTFi (50%) and AdGFP-R7-knob (35%) for MOIs higher than 100 PFU/cell (Fig. 2c). In CF-KM4 cells, a plateau was also reached at an MOI of 100 with AdGFP-R7-knob (25%) and AdGFP-QM10-knob (50%), whereas AdGFP-WTFi-mediated gene transduction increased linearly between 20 and 200 PFU/cell, with 90% transduced cells at an MOI of 200 (Fig. 2d). Thus, the difference in the efficiency of gene transduction between WT and fiber-modified AdGFP vectors was better detected at relatively high vector doses. These results also suggested that limiting factors other than the number of cell receptors could exist in the CAR-integrin entry pathway, as for AdGFP-WTFi in A549 cells, as well as with vectors using alternative pathways, as for AdGFP-QM10-knob in CF-KM4 cells.
Absence of knob requirement for gene transfer mediated by AdGFP vectors carrying short-shafted liganded fibers.
We then determined whether the terminal knob domain of the fiber was required for gene transduction by our liganded vectors and how efficient knobless vectors would be in this process. A549, HeLa, CF-KM4, and MM39 cells were incubated at 37°C for 1 h with each vector, and the GTE indices of AdGFP-R7-knob and AdGFP-R7, AdGFP-QM10-knob and AdGFP-QM10, and AdGFP-SY12-knob and AdGFP-SY12 were compared. The ratios of the GTE indices of the knobbed versus deknobbed versions of each vector provided insight into the role of the knob domain in the interaction of liganded versus nonliganded vectors with the different cell targets. As a control for nonspecific proteolytic effect, AdGFP-WTFi vector carrying nonscissible WT fibers was subjected to factor Xa digestion under the same conditions. Except for MM39 cells and the control vector AdGFP-R7, for which deknobbing drastically reduced gene transduction efficiency, the other cell-vector pairs involving AdGFP-R7±knob or AdGFP-SY12±knob seemed to be relatively unaffected by the presence or absence of the knob domain (GTE ratio, ∼1) (Fig. 3b, d, and i). In contrast, ablation of the knob domain in AdGFP-QM10 resulted in a significant enhancement of gene transduction of the four cell lines, and in particular A549, HeLa, and MM39 cells (five- to eightfold) (Fig. 3f, h, and i). This implied that the knob domain was dispensable for achieving the early steps of the virus cycle, which led to gene delivery by the AdGFP-QM10 vector into CAR+ and CAR− cells.
FIG. 3.
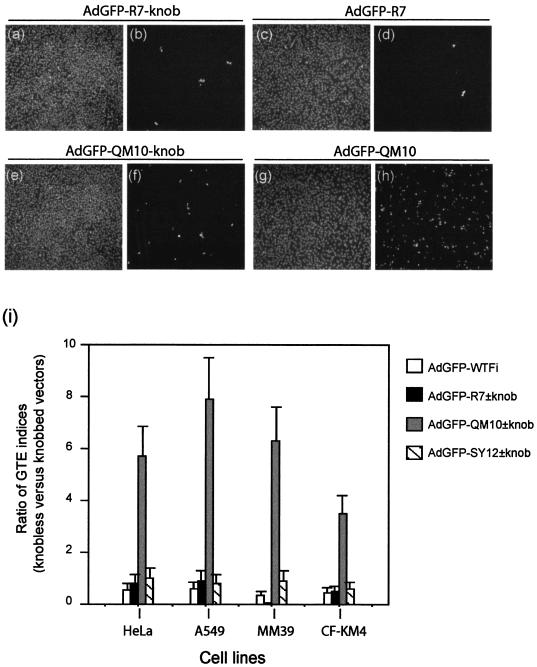
Effects of vector deknobbing on gene transduction of various cell lines mediated by knobless or knob-carrying versions of AdGFP vectors. HeLa and A549 cells (CAR+ CFTR+), MM39 glandular cells (CAR− CFTR+), and CF-KM4 cells (CAR− CFTR−) were infected by AdGFP-R7-knob (a, b, and i) or its knobless version, AdGFP-R7 (c, d, and i); AdGFP-QM10-knob (e, f, and i) or AdGFP-QM10 (g, h, and i); or AdGFP-SY12-knob or AdGFP-QMSY12 (i) at a constant MOI (30 PFU/cell). GFP expression was estimated 36 h p.i. by IF microscopy (b, d, f, and h) and quantitated by FACS analysis (i). (a, c, e, and g) DAPI staining of cell samples shown in panels b, d, f, and h. In panel i, the GTE is expressed as the ratios of GTE indices, i.e., AdGFP-R7 versus AdGFP-R7-knob, AdGFP-QM10 versus AdGFP-QM10-knob, and AdGFP-SY12 versus AdGFP-SY12-knob. The data shown are from three separate experiments. The error bars indicate standard deviations.
These results were conceivable for liganded vectors, such as AdGFP-SY12 and AdGFP-QM10, considering that the proteolytic removal of the knob had exposed the peptide ligands of the fiber shafts and made them directly accessible to their specific receptor molecules at the cell surface. However, the finding that the control nonliganded vector AdGFP-R7 still transduced cells in its deknobbed version indicated that our short-shafted AdGFP vectors, regardless of the occurrence of the knob or a cell ligand at the extremities of their short-shafted fibers, bound to the cell surface via domains of capsid components other than the fiber knob. The next set of experiments were designed to analyze the mechanism of the absence of the knob requirement for gene transduction by our nonliganded control vector and also to determine at which step(s) of the viral infection the QM10 ligand enhanced the efficiency of Ad-mediated gene transfer, attachment, endocytosis, endosomal release, nuclear addressing, or several of these four steps successively.
Role of cell ligands and knob domains in AdGFP cell interaction. (i) Cell binding of liganded versus nonliganded vectors.
Virus attachment to the cell surface was assayed at 4°C on CF-KM4, MM-39, and A549 cells, using Cy3-labeled virions of AdGFP-R7±knob, AdGFP-QM10±knob, and AdGFP-SY12±knob. Cell-bound fluorescence was determined by quantitative imaging in IF microscopy and compared in knobbed and deknobbed versions of each vector. The intensity of the signal of cell-bound viruses was very similar for the three knob-bearing vectors in each cell line, and the variations were within the range of experimental error (data not shown). In no case could the level of cell binding of AdGFP-QM10-knob account for the 40-fold-higer level of gene transfer observed with the vector in A549 cells compared to that of the nonliganded vector AdGFP-R7-knob or for its 3-fold-higher level in CF-KM4 cells (Fig. 2). This suggested that the QM10 and SY12 peptides did not function as ligands of virus attachment receptors in A549, CF-KM4, and MM39 cells but acted at later steps, at least in the presence of the knob domain. This was consistent with the fact that QM10 and SY12 are peptides derived from cell-internalized bacteriophages and not from extracellular, plasma membrane-bound phages.
(ii) Cell binding of knobbed versus deknobbed vectors.
After ablation of the knob, a slight augmentation of the cell-binding level (ca. twofold) was observed for the AdGFP-QM10 vector in the three cell lines compared to AdGFP-QM10-knob, but no significant variations were observed with nonliganded or SY12-liganded vectors, with or without their knobs (data not shown). These results suggested that the mechanism of increase in gene transfer efficiency shown by AdGFP-QM10-knob did not take place at the cell attachment step, at least in a quantitative manner. They also suggested that the knob domain was dispensable for the step of cell attachment of the vectors, as already observed for the overall gene transduction process (Fig. 3).
(iii) Antibody-mediated competition for cellular attachment of vectors via the knob domain.
In a previous study, experiments using anti-knob 1D6.14 MAb have suggested that the fiber knob domain was involved in the binding of Ad5 to surface SA-GP molecules on CF-KM4 and to heparan sulfate glucosaminoglycans on MM39 cells (12). This was confirmed in the present study using AdGFP-WTFi vector carrying its natural long-shafted fibers: a 98% inhibition of attachment to CF-KM4 cells was observed in the presence of a large excess of 1D6.14 MAb (Table 2). However, under the same conditions, the knob-blocking antibody 1D6.14 had a much smaller inhibitory effect on the cell attachment of the short-shafted fiber vectors AdGFP-R7-knob and AdGFP-SY12-knob (∼20 and 50% inhibition, respectively) and an even smaller effect (only ∼10% inhibition) on AdGFP-QM10-knob (Table 2). This suggested that recombinant Ad5 carrying short-shafted fibers bound to cell surface molecules via sites of the knob domain different from the 1D6.14 epitope. Alternatively, but not exclusively, the virus-cell binding could be mediated by viral capsid proteins or domains different from the fiber knob, e.g., penton base, as for the nonliganded vector, or the new cellular ligand inserted into the fiber, as for AdGFP-QM10-knob, or several binding sites simultaneously. In any case, this confirmed the major role played by the fiber shaft in controlling the interaction of Ad virions with cell surface receptors (52, 53, 60, 76). The subsequent experiments were aimed at determining which viral capsid protein(s), or specific domain(s) thereof, could compensate for the absence of the knob or its blockade by MAb and acted as an alternative or/and auxiliary cell attachment component(s) in the knobbed or deknobbed versions of our AdGFP vectors.
TABLE 2.
Effects of blocking anti-knob MAb 1D6.14 on the attachment of nonliganded and liganded AdGFP vectors to CF-KM4 cellsa
| Ad5 vector | GFP expressionb
|
Inhibition of cell binding (%) | |
|---|---|---|---|
| Without 1D6.14 | With 1D6.14 | ||
| AdGFP-WTFi | 100 | 2 ± 1 | 98 |
| AdGFP-R7-knob | 100 | 81 ± 12 | 19 |
| AdGFP-QM10-knob | 100 | 89 ± 11 | 11 |
| AdGFP-SY12-knob | 100 | 45 ± 8 | 55 |
Cell attachment of Ad vectors was assayed by gene transduction following virus-cell binding experiments conducted at low temperature to prevent virus entry. Cell attachment was performed at an MOI of 100 for 2 h at 4°C in the absence (control samples) or presence of aliquots of 1D6.14 ascites fluid corresponding to 15 μg of monoclonal immunoglobulin G (a ca. 103-fold excess over the fiber content of the vectors). The inoculum was then removed, and the cells were postincubated for 24 h at 37°C.
GFP expression was quantitatively determined by FACS analysis, and its levels are expressed as percentages of the values in corresponding control samples. The data presented are the mean of three separate experiments ± standard deviation.
Receptor-binding domains in the capsid of knobbed and deknobbed AdGFP vectors.
Cell-binding competition assays were performed in the presence of purified Ad proteins, hexon, full-length fiber, knob, or penton base (WT or the RGD mutant R340E), individually mixed with each vector and added in large excess over their copy numbers in the Ad virion. Vector and competitor were coincubated at 4°C with CF-KM4 or A549 cells to allow attachment but not entry. Unadsorbed virus was rinsed off, and the cells were assayed for GFP expression at 28 h p.i. The attachment of AdGFP-QM10-knob to CF-KM4 cells was not influenced by an excess of hexon or penton base proteins (Fig. 4a). However, it was competed by knob protein to significant levels (∼50%) and to a lesser degree (25 to 30%) by full-length fiber, a difference which might reflect variations in the accessibilities of different sites on the viral capsid proteins. In its knobless version, AdGFP-QM10 was not competed by knob or fiber, as expected, but was competed to ∼50% by penton base, WT and the R340E mutant. Similar patterns of competition for CF-KM4 cell binding were observed with the control vectors AdGFP-R7-knob and AdGFP-R7 (Fig. 4a). Only slight differences in cell-binding competition assays were observed between CF-KM4 and A549 cells for the different vectors (Fig. 4b): full-length fiber was apparently more efficient in competing with the knobbed vectors AdGFP-R7-knob and AdGFP-QM10-knob for attachment to A549 cells, a result which might be due to the high accessibility of CAR to its fiber ligand. Taken together, our results suggested that the lack of knob in our short-shafted AdGFP vectors could be compensated for by other components of the viral capsid, mainly the penton base. The data obtained with penton base R340E mutant implied that the binding site(s) on the penton base capsomer did not involve the RGD motif. This was reminiscent of a previous observation that in CF-KM4 cells, cell endocytosis and entry of the Ad5Luc3 vector were apparently independent of the penton base and RGD ligand (12).
FIG. 4.
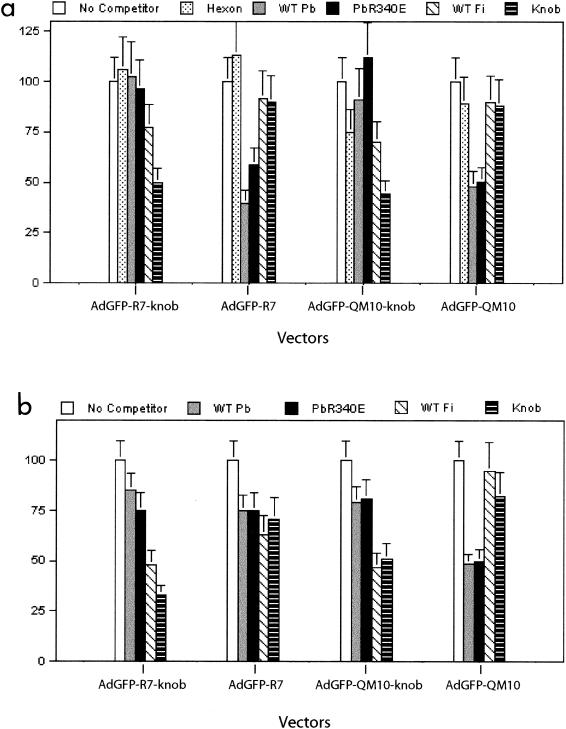
Cell-binding competition assays between Ad5 capsid components and knobbed or deknobbed versions of nonliganded or liganded AdGFP vectors. CF-KM4 (a) and A549 (b) cell monolayers were incubated with a mixture of AdGFP vector and a large excess of capsid protein under conditions allowing cell attachment but not cell entry. After the removal of unadsorbed virus, the cells were transferred to 37°C for 24 h, and the amount of cell-bound virus was indirectly assayed by the level of GFP expression, determined by FACS analysis. Data from two separate experiments are expressed as percentages of the values in corresponding control samples. The error bars indicate standard deviations. WT Pb, wild-type penton base; WT Fi, wild-type fiber; PbR340E, penton base RGD mutant.
In A549 cell-binding assays, however, the penton base competed as poorly as fiber protein with the control knobless vector AdGFP-R7 (Fig. 4b). This pattern, and the fact that a maximum level of only 50% inhibition was reached in other cell-binding competition assays using a 3-log-unit excess of knob protein over knob-carrying vectors, as well as using a 3-log-unit excess of the penton base protein over knobless vectors, strongly suggested that several different domains on the viral capsid of short-shafted fiber vectors could serve alternatively or even simultaneously as cell-binding sites. Of note, cellular binding through hexon capsomer has been reported (3), although no significant competition effect was observed with hexon protein in our experiments (Fig. 4a). Likewise, the fiber shaft could participate in the cell-binding process to some extent. In particular, the fiber shaft domain of our AdGFP vectors still carried the conserved basic peptide KKTK at positions 91 to 94, a motif which has been proposed to be responsible for the binding of Ad fiber to heparan sulfate proteoglycans and acidic carbohydrates (8). Moreover, some cell-binding activity for the nonviral trimerization peptide from the neck region of the human lung surfactant protein D inserted at the C-terminal end of the fiber shaft in our vectors could not be excluded. This would explain the absence of significant change in the nonliganded AdGFP-R7 vector infectivity after deknobbing, as shown in Fig. 3.
Cellular uptake of knobbed versus deknobbed AdGFP vectors.
The mechanism of gene transfer enhancement mediated by the QM10 ligand in different cell lines was then investigated at the steps of endocytosis and entry of Ad virions.
(i) Endocytosis.
Endocytosis of AdGFP-QM10-knob, AdGFP-SY12-knob, and AdGFP-R7-knob in their knobbed and deknobbed versions was then assayed in A549, MM39, and CF-KM4 cells, using flow cytometry analysis of cells rendered fluorescent by intracellular Cy3-labeled virions. In the three cell lines, the numbers of internalized vector particles were not significantly different for AdGFP-R7-knob and AdGFP-SY12-knob, and deknobbing of the two vectors resulted in a slight increase in endocytosis in MM39 and CF-KM4 cells and a decrease for AdGFP-R7 in A549 cells (Fig. 5a). However, the difference between the numbers of endocytosed virions in AdGFP-QM10-knob and AdGFP-QM10 was significant in A549 cells (twofold) (Fig. 5a) and CF-KM4 cells (fivefold) (Fig. 5c). This suggested that deknobbing of QM10 fibers resulted in a better exposure of the QM10 motifs, which in turn increased the efficacy of endocytosis and/or possibly further steps.
FIG. 5.
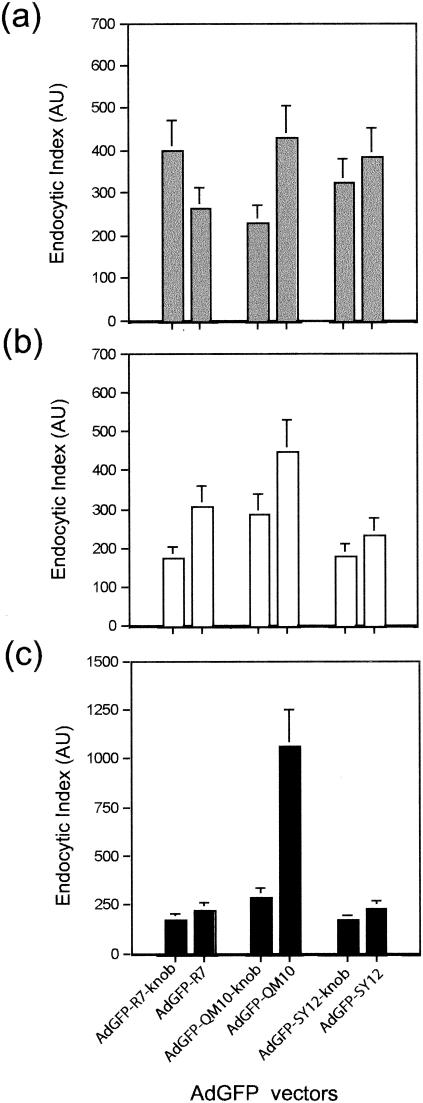
Endocytosis assays of knobbed versus deknobbed versions of nonliganded vector AdGFP-R7±knob and liganded vectors AdGFP-QM10±knob and AdGFP-SY12±knob in A549 (a), MM39 (b), and CF-KM4 (c) cells. Cy3-labeled vector particles were incubated with cell monolayers at 37°C for 1 h, cell surface-bound virus was detached by EDTA-trypsin treatment, and the cells were assayed for intracellular fluorescent signal by FACS analysis. Data from three separate experiments are expressed as endocytic indices in AU, as described in Materials and Methods. The error bars indicate standard deviations.
(ii) Endosomolysis.
Ad-mediated endosomolysis and vesicular escape of Ad virions was determined from the degree of Ad-mediated augmentation of cell protein synthesis inhibition by the toxin RcA as a result of their coendocytosis and subsequent vesicular release. RcA agglutinin blocks cell protein synthesis, and its effect is enhanced by the endosomal escape of coendocytosed Ad virions into the cytosol (12, 20, 58). The kinetics of protein synthesis inhibition was analyzed after incubation of cells with RcA and each of the three Ad vectors for 2 h at 37°C, at different RcA concentrations and constant input of Ad vector particles, and the 50% inhibitory concentration (IC50) was determined. In their knobbed versions, all three vectors showed similar curves of RcA dose response, with 10 to 20% residual protein synthesis at 20 pg of RcA and 104 virions per cell, and the IC50 values ranged between 0.8 and 4.0 pg of RcA and 104 virions per cell, depending on the cell type (Fig. 6).
FIG. 6.
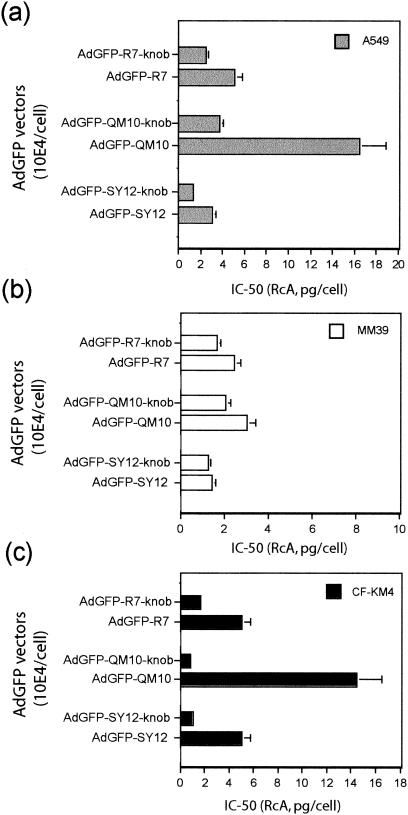
Endosomolysis assays. Cells were coincubated with toxin (RcA) and AdGFP vectors for 1 h at 37°C with different toxin-to-cell ratios and a constant vector input (104 PP/cell). The vesicular escape and cell internalization of the coendocytosed RcA and AdGFP vector were assayed in A549 (a), MM39 (b), and CF-KM4 (c) cells using toxin-induced inhibition of host-cell protein synthesis. The data indicated on the x axis represent the values of RcA concentration (in picograms per cell) which provoked a 50% inhibition of cell protein synthesis. The data presented are the means of four separate experiments ± standard deviations.
Interestingly, the inhibitory effect of RcA on host cell protein synthesis was consistently lower in the presence of the knobless vectors AdGFP-R7, AdGFP-QM10, and AdGFP-SY12 than with their knobbed versions, irrespective of the cell ligand. However, the difference was particularly striking with AdGFP-QM10 vector in CF-KM4 cells, in which a 15- to 20-fold augmentation of the IC50 value was observed, and in A549 cells, showing a 4- to 5-fold increase (Fig. 6). Since deknobbing did not negatively affect the cell attachment of AdGFP-QM10 vector and positively influenced its endocytosis, these results suggested that the fiber knob domain played a role in the endosomal release of coendocytosed Ad virions and toxin. The association of RcA plus AdGFP-QM10-knob was the pair of coendocytosed macromolecules which was apparently the most sensitive to the deknobbing process in CF-KM4 and A549 cells, suggesting that the QM10 ligand acted mainly at the step of Ad endocytosis and cell entry. This could theoretically be explained by at least two mechanisms. (i) QM10 ligand could have decreased the level of endocytosis of the Ad vector, but this hypothesis could be excluded, since it was contradicted by the FACS analysis of internalized virions (Fig. 5) and by the positive effect of QM10 on the efficiency of Ad-mediated gene transfer (Fig. 2). (ii) A significant proportion of AdGFP-QM10±knob could be endocytosed in an endocytic compartment different from the one in which most RcA and AdGFP-R7-knob (or RcA and AdGFP-SY12-knob) were coendocytosed.
Endocytic compartments of nonliganded and liganded AdGFP vectors.
The cellular compartments in which AdGFP-R7-knob, AdGFP-SY12-knob, and AdGFP-QM10-knob were endocytosed were investigated in CF-KM4 and A549 cells. The cells were coincubated with aliquots of Cy3- and Cy2-labeled particles of each vector pairwise and analyzed by confocal IF microscopy. Only partial colocalization was observed for AdGFP-QM10-knob and AdGFP-SY12-knob (Fig. 7a to c), whereas the majority of AdGFP-R7-knob and AdGFP-SY12-knob virions were found to colocalize in the same vesicular compartment (Fig. 7d to f). Colocalization events were very rare with AdGFP-QM10-knob and the control vector AdGFP-R7-knob (Fig. 7g to i) and even more rarely observed in cells coinfected with knobless AdGFP-QM10 and AdGFP-R7-knob, where most of the Cy2 and Cy3 signals emanated from separate compartments (Fig. 7j to l). These patterns suggested that our vectors used different endocytic pathways, one common to AdGFP-R7-knob and AdGFP-SY12-knob and another independent one used by AdGFP-QM10-knob and its knobless version.
FIG. 7.
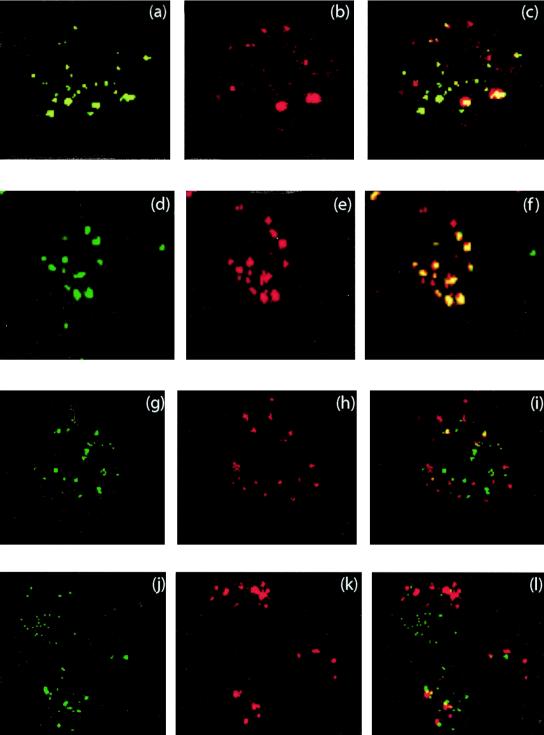
Intracellular localization of liganded vectors AdGFP-QM10-knob and AdGFP-SY12-knob in comparison with that of the control nonliganded vector AdGFP-R7-knob. The vectors were coincubated pairwise with CF-KM4 cells and analyzed by confocal IF microscopy 45 min after input. (a to c) Cy2-labeled AdGFP-QM10-knob and Cy3-labeled AdGFP-SY12-knob; (d to f) Cy2-labeled AdGFP-R7-knob and Cy3-labeled AdGFP-SY12-knob; (g to i) Cy2-labeled AdGFP-QM10-knob and Cy3-labeled AdGFP-R7-knob; (j to l) Cy2-labeled knobless AdGFP-QM10 and Cy3-labeled AdGFP-R7-knob. Overlay pictures are shown in panels c, f, i, and l.
The nature of the compartments in which vectors resided was then analyzed using fluorescent probes of endosomal vesicles. Cells incubated at 37°C for 45 min with Cy3-labeled vectors were harvested, fixed, and permeabilized and then reacted with FITC-labeled antibodies specific for endosomal-compartment markers and examined by confocal IF microscopy. Transferrin receptor (TfR) and the Rab protein Rab5 were both used as markers of clathrin-coated vesicles and early endosomes, Rab4 was used for early and recycling endosomes, Rab11 was used for recycling endosomes of epithelial cells (51), and LAMP-1 was used for late endosomes and lysosomes (48). Both AdGFP-R7-knob and AdGFP-SY12-knob colocalized with the IF signals of TfR (Fig. 8a to c) and Rab5 (not shown), but not with LAMP-1 (Fig. 8g to i), confirming the roles of clathrin-coated vesicles and early endosomes in early steps of endocytosis and entry of Ad5, a prototype member of species C Ad (15, 41, 42). By contrast, no significant occurrence of AdGFP-QM10-knob within the TfR/Rab5 endosomal compartment could be detected at 45 min (Fig. 8d to f). Instead, colocalization of AdGFP-QM10-knob and LAMP-1 signals was observed with a high frequency (Fig. 8j to l). This indicated that a significant proportion of AdGFP-QM10-knob vector particles followed an endocytic pathway different from that of AdGFP-R7-knob and AdGFP-SY12-knob and were preferentially endocytosed into and/or addressed to the late, acidic endosomal compartment.
FIG. 8.
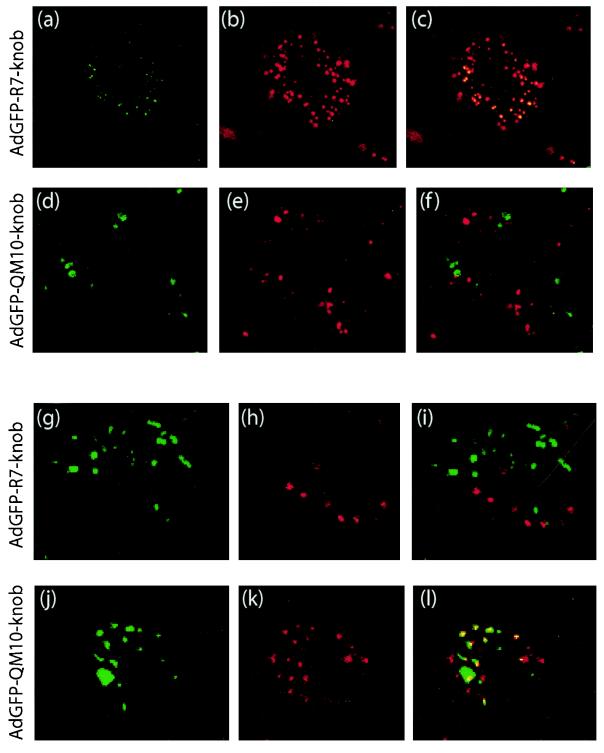
Cell compartmentalization of nonliganded vector AdGFP-R7-knob and liganded vector AdGFP-QM10-knob in CF-KM4 cells analyzed by confocal IF microscopy 45 min after input. After fixation and permeabilization, the cells were reacted with FITC-labeled anti-TfR (a to f) or anti-LAMP-1 (g to l) antibody. (a, d, g, and j) Signal of FITC-labeled antibodies. (b, e, h, and k) Signal of Cy3-labeled vector particles. (c, f, i, and l) Merging of the two signals. (a to c) FITC-labeled anti-TfR antibody and Cy3-labeled AdGFP-R7-knob. (d to f) FITC-labeled anti-TfR antibody and Cy3-labeled AdGFP-QM10-knob. (g to i) FITC-labeled anti-LAMP-1 antibody and Cy3-labeled AdGFP-R7-knob. (j to l) FITC-labeled anti-LAMP-1 antibody and Cy3-labeled AdGFP-QM10-knob.
This was confirmed by the results of coincubation of our vectors with human α2 M, a ligand of cell receptors targeted to one of the two well-characterized endosomal pathways (5). Tf ligands have been shown to be directed to early and recycling endosomes that are relatively alkaline (pH ∼6.0 to 6.8), whereas α2 M ligands do not recycle to the cell surface but progress rapidly through the endosomal pathway to late, relatively acidic (pH ∼5) endosomes (63, 68, 77, 80). FITC-labeled α2 M and each of our three Cy3-labeled AdGFP vectors were therefore incubated with CF-KM4 and A549 cells, and their possible colocalization was analyzed by confocal IF microscopy. AdGFP-R7-knob and AdGFP-SY12-knob were very rarely found to colocalize with α2 M (not shown). However, a majority of AdGFP-QM10-knob vector particles were found to reside in the same vesicular compartment as α2 M after 45 to 60 min of incubation at 37°C (Fig. 9a to c). In its knobless version, the events of colocalization of AdGFP-QM10 with α2 M also occurred with high frequency (Fig. 9d to f).
FIG. 9.
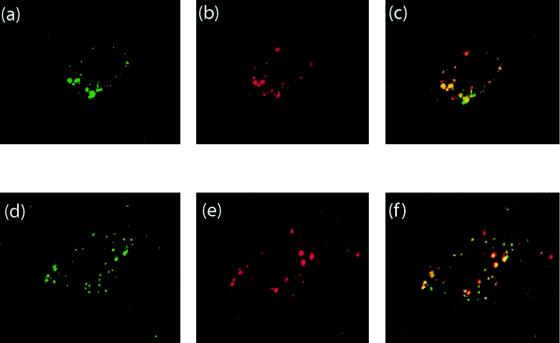
Colocalization of human α2 M and QM10-liganded AdGFP vector in its knob-bearing version, AdGFP-QM10-knob (a to c), and its knobless version, AdGFP-QM10 (d to f), in A549 cells 45 min after input, analyzed by confocal IF microscopy. (a and d) Signal of FITC-labeled α2 M; (b and e) signal of Cy3-labeled vector particles; (c and f) signal overlay.
EM analysis of intracellular vector particles.
Samples of A549 and CF-KM4 cells were incubated at 37°C for 1 h with AdGFP-R7-knob and AdGFP-QM10-knob at 104 virions per cell and processed for EM. The average number of cell-associated virions, determined from at least 30 sections of individual cells, was ∼10-fold higher for AdGFP-QM10-knob than for nonliganded AdGFP-R7-knob, a result consistent with our FACS analysis. As expected, most of the cell-associated virions of AdGFP-R7-knob were found in typical endosomal vesicles, and rare particles were visible at the cell surface (not shown). In contrast, AdGFP-QM10-knob virions were found in greater numbers both at the cell surface (Fig. 10a to c) and in intracellular compartments (Fig. 10d and 11). Enlargement of plasma membrane-bound AdGFP-QM10-knob virions often revealed thin filaments linking one or more vertices of the vector capsid to the cell surface, as depicted in Fig. 10b and c. The average length of these links, 145 ± 6 Å (n = 12), was compatible with the dimensions of the short-shafted fibers of our vectors (R7), calculated from experimental determinations (7). Without taking into account the 36 residues of our trimerization motif and the 13 residues of the linker, the knob diameter of 49 Å and the length increment of 13.4 to 13.5 Å per shaft repeat gave a theoretical value of 94 Å for the R7 shaft and a value of 143 Å for the whole R7 knob fiber.
FIG. 10.
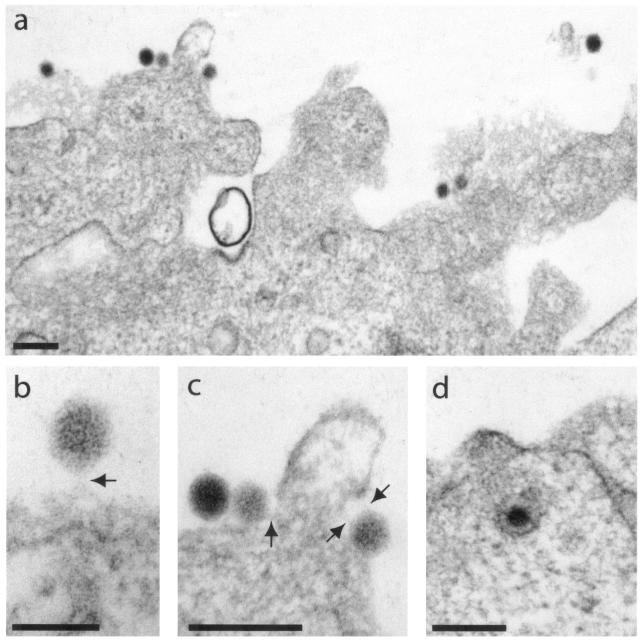
EM analysis of early events in the interaction of AdGFP-QM10-knob vector with A549 cells. The cells were incubated with an input of 104 virions per cell for 1 h at 37°C. (a) General view of cellular apical surface. (b and c) enlargements of plasma membrane-bound virions. The arrows indicate filaments linking the virus to the cell surface, whose lengths were compatible with that of a short-shafted fiber of seven repeats. (d) Endocytosed virion. Bars, 200 (a, c, and d) and 100 (b) nm.
FIG. 11.
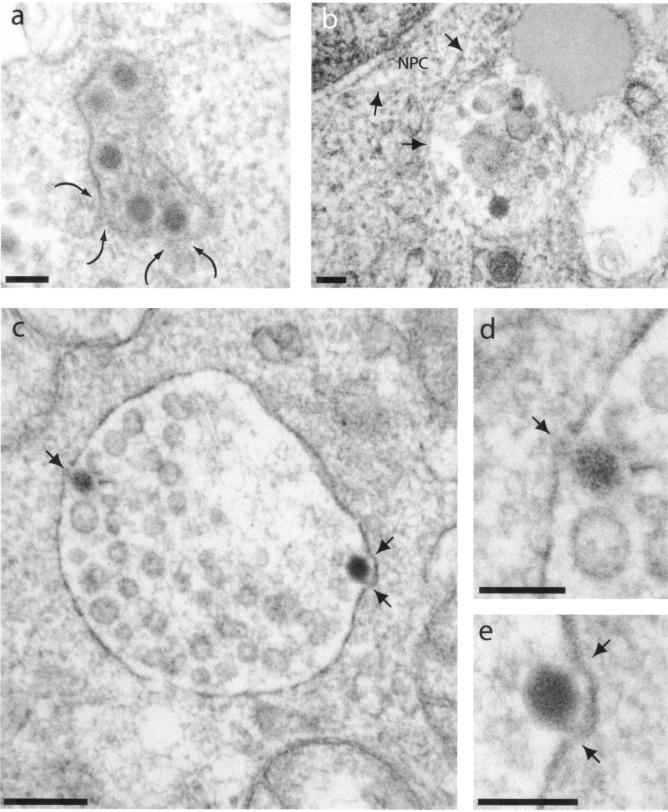
EM analysis of intracellular virions of AdGFP-QM10-knob in A549 cells. The cells were incubated with vector for 1 h at 37°C and at an input multiplicity of 104 virus particles per cell. (a) The curved arrows indicate two small intracytoplasmic vesicles opening into a larger one containing multiple virions and heterogeneous electron-dense material. (b) Vesicle containing one virion and heterogeneous and amorphous material. The arrows indicate microtubules apparently connecting endosomal vesicle to the nuclear envelope; NPC, nuclear pore complex. (c to e) The arrows point to junctions of intravesicular virions with the endosomal leaflet. Panels d and e are enlargements of areas with intravesicular virions shown in panel c. Bars, 100 (a, b, d, and e) and 200 (c) nm.
The intracellular virions of AdGFP-QM10-knob were seen within A549 cells in two unequally represented types of intravesicular compartments: on rare occasions, single virions occurred in small vesicles with discontinuities in the vesicular membrane leaflet (Fig. 10d) resembling the vesicles in which the control vector AdGFP-R7-knob was endocytosed. However, the most frequent events consisted of several AdGFP-QM10-knob virions present in the same vesicles containing electron-dense heterogeneous material (Fig. 11a), amorphous inclusions, or both (Fig. 11b), or in large vesicles containing multiple vesicles of smaller diameter reminiscent of multivesicular bodies (Fig. 11c). The morphological aspect of these vesicular contents was characteristic of the lysosomal compartment. Higher magnifications of the vector-vesicular membrane junctions showed fuzzy material interrupting the dark line of the membrane leaflet (Fig. 11d and e). These gaps might represent the first stages of endosomolysis. A similar pattern of cellular compartmentalization was observed in A549 cells with knobless AdGFP-QM10 vector, most of whose virions were found associated with multivesicular bodies (Fig. 12a and b) or with multilamellar inclusions of phospholipids already described in A549 cells (Fig. 12c). These observations suggested that AdGFP-QM10-knob and AdGFP-QM10 could follow the same endocytic pathway as Ad5WT through early endosomes, leading to their endosomal escape, but that late endosomes and lysosomes represented the preferred endocytic compartment to which QM10-liganded vectors were rapidly redirected and where they accumulated.
FIG. 12.
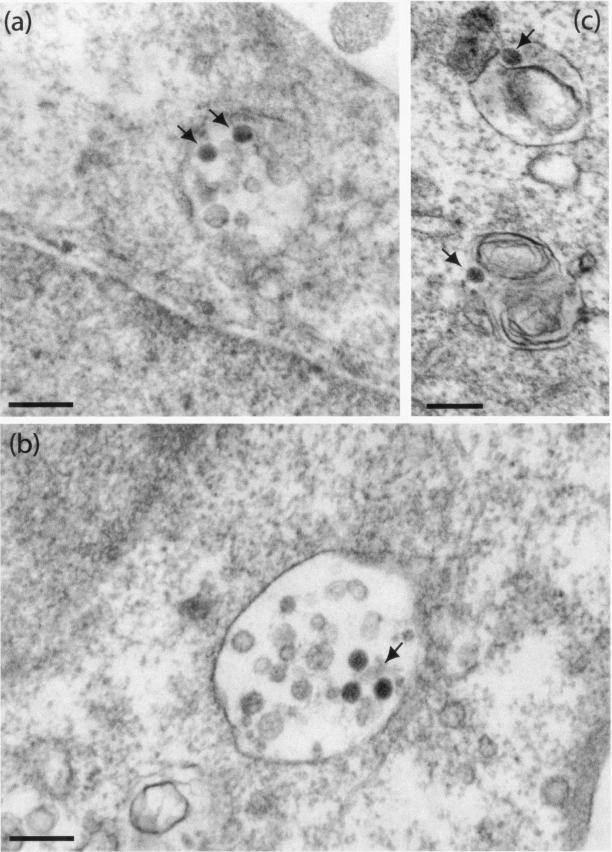
EM analysis of intracellular virions of knobless AdGFP-QM10 in A549 cells. The cells were incubated with vector for 1 h at 37°C and at an input multiplicity of 104 virus particles per cell. Virions (arrows) were seen associated with multivesicular bodies (a and b) or with multilamellar inclusion bodies (c) characteristic of A549 cells. Bars, 200 nm.
The cytoplasm of CF-KM4 cells has been shown to contain a very rich vesicular network (12), and this made it difficult to distinguish between early and late endosomal compartments in that cell line by using conventional EM. However, virions of the control vectors AdGFP-R7-knob and AdGFP-WTFi were more frequently found within vesicles with characteristics of clathrin-coated pits (Fig. 13a, b, d, and e) than were those of QM10-liganded vectors, a majority of which were found within large vesicles reminiscent of late endosomes or lysosomes (Fig. 13f and g). No obvious difference was observed between AdGFP-QM10-knob and its knobless version, AdGFP-QM10, in terms of preferred vesicular residence (compare Fig. 13f and g). Careful examination of intravesicular virions under EM showed that most of the particles of AdGFP-QM10-knob and AdGFP-QM10 presented a blurred aspect and/or irregular morphology, whereas AdGFP-WTFi and AdGFP-R7-knob virions had retained their sharp contour and regular, icosahedral shape (compare Fig. 13a to e with f to g). This suggested that at 1 h p.i. at 37°C in CF-KM4 cells, endocytosed virions of AdGFP-QM10-knob or AdGFP-QM10 were at a more advanced stage of uncoating than those of nonliganded vectors, a process which might contribute to the higher efficiency of transduction of CF-KM4 cells by QM10-liganded vectors.
FIG. 13.
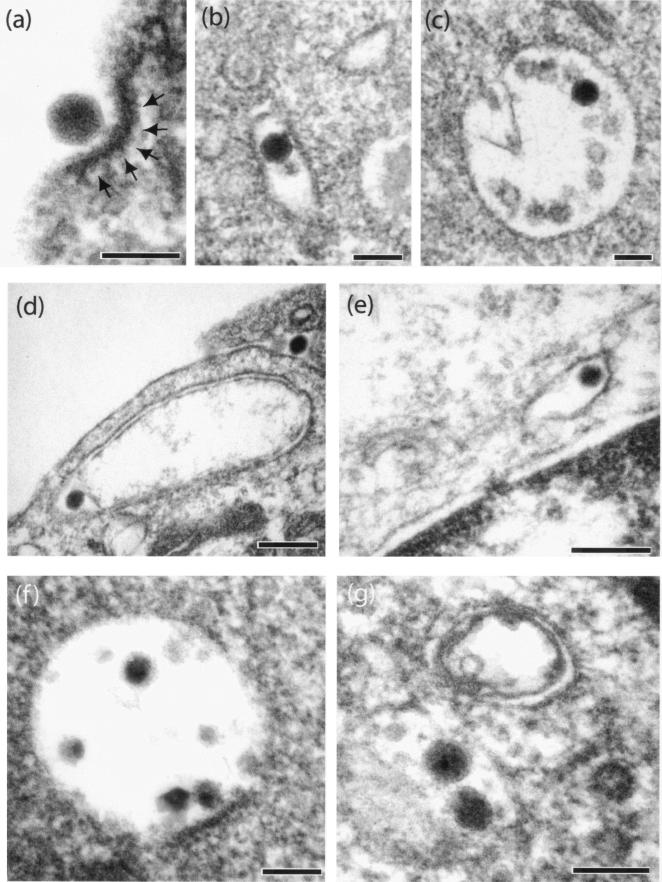
Early virus-cell interaction between CF-KM4 cells and QM10-liganded or control vectors analyzed under EM. The cells were incubated with vector particles for 1 h at 37°C and at an input multiplicity of 104 virions per cell. (a to c) AdGFP-R7-knob; (d and e) AdGFP-WTFi; (f) AdGFP-QM10-knob; (g) AdGFP-QM10. The arrows in panel a indicate an electron-dense layer of proteinic material, most likely clathrin, at the virus attachment site on the plasma membrane. Bars, 100 (a to c, f, and g) and 200 (d and e) nm.
DISCUSSION
The original goal of the present study was to improve the Ad-mediated gene delivery into airway cells of CF patients, using fiber-modified Ad5 vectors and CFTR-deficient tracheal glandular cells (CF-KM4) as the model system. Since SAα(2->6)-GP were apparently low-efficiency receptors for Ad5 in CF-KM4 cells and were largely responsible for their poor permissivity for this virus compared to HeLa or A549 cells (12), we looked for other surface molecules which could advantageously be used as alternative receptors by Ad5. When no specific cell surface markers are known in target tissues, biopanning of desired living cells has been proposed to select cell-specific ligands (62). Using this phage display strategy, it has been found that the insertion of the hexapeptide SIGLYP in the HI loop of the knob, the flexible region between β barrels H and I, has resulted in an endothelial cell-selective Ad vector (46). We therefore biopanned confluent monolayers of living CF-KM4 cells to isolate CF-KM4-specific peptide ligands that could be incorporated into the fibers of recombinant Ad5 to confer upon the resulting Ad5 vector a novel cell target specificity and/or higher efficiency of gene delivery. Three cell ligands, LAP25, QM10, and SY12, were designed from phagotopes of intracellular phages. Each ligand was inserted into the shaft domain of Ad5 fiber containing a factor Xa cleavage site and a proteolytically removable knob domain (Fig. 1) and recombined with the genome of an Ad5-based GFP-expressing vector (AdGFP). AdGFP vectors carried knob-terminated fibers during the reproductive cycle and were grown in the conventional HEK-293 cell line. After isolation of the vectors by conventional methods, proteolytic “stripping” by factor Xa released the knobs, and three cell-targeting ligands (one per fiber monomer) were then displayed at the C-terminal ends of their fiber shafts (22). In terms of gene therapy, proteolytic deknobbing of Ad fibers has the double advantage of (i) detargeting Ad vectors prior to their retargeting via specific ligands and (ii) decreasing the immune response against Ad vectors by preventing the knob-mediated activation of dendritic cells (44). The results of the present study confirmed the feasibility of redirecting proteolytically deknobbed Ad5 vectors to an alternative cell entry pathway via fiber shaft-inserted ligands.
We found that, out of the three AdGFP constructs, only two, the vectors carrying the QM10 and SY12 ligands (AdGFP-QM10-knob and AdGFP-SY12-knob, respectively), yielded viable viruses in the trans-complementing cell line. AdGFP-QM10-knob and AdGFP-SY12-knob were then assayed for gene transfer efficiency and the cell entry pathways into various cell lines, with or without proteolytic removal of their fiber knobs. AdGFP-QM10-knob was the only liganded vector that transduced CF-KM4 cells with a higher efficiency than the control nonliganded vector AdGFP-R7-knob. However, QM10 targeting was promiscuous, and the QM10 peptide conveyed targeting to several other cell types. The AdGFP-QM10-knob-mediated gene transfer enhancement occurred in a non-cell-specific manner, as most of the cell lines tested, and particularly A549, were transduced with significantly higher efficiency (Fig. 2). Since the QM10 peptide was selected on CF-KM4 cells, this lack of cell selectivity showed the limitations of the strategy of phage biopanning living cells to identify cell-specific ligands. No negative preselection step was included in our strategy, i.e., exposing the phage library to other cell types and collecting the nonbinding phages for further cell-specific binding, internalization, and characterization. The rationale for this strategic option was based on the consideration of two opposite and nonnegligible risks: (i) amplifying a number of phages displaying nonspecific or/and biologically inactive peptides versus (ii) loosing a significant proportion of individual phage specific for the desired cells during this step of phage exhaustion, due to their nonspecific binding to irrelevant cell types.
The knobless version of the QM10-liganded vector, AdGFP-QM10, was found to transduce cells with a similar or higher efficiency than its knob-bearing version, indicating that the gene transduction occurred in a knob-independent manner (Fig. 3). A lesser knob dependence and a higher penton base dependence have been reported for short-shafted fiber Ad vectors than for long-shafted fiber vectors (52, 53, 60, 76). Consistent with these observations, we found that our three short-shafted vectors, AdGFP-QM10-knob, AdGFP-SY12-knob, and AdGFP-R7-knob, could still bind to the cell surface and be efficiently endocytosed in their knobless versions (Table 2 and Fig. 3 and 5), confirming that they were apparently less dependent on the fiber knob domain than the WT fiber-bearing AdGFP vector for cell binding. Results from competition experiments indicated that their cell binding occurred via alternative sites on the viral capsid and that the penton base capsomers played a significant role in the vector-cell attachment, via a site(s) different from their RGD motifs (Fig. 4).
Analysis of the early steps of virus-cell interaction provided data suggesting that the enhancement of gene transfer efficiency observed with QM10-liganded AdGFP vector, with or without its knobs, occurred at the step of endocytosis in both quantitative and qualitative manners. Quantitatively, QM10-liganded vector particles were endocytosed in higher numbers than the control nonliganded vector. Qualitatively, AdGFP-QM10±knob was found to localize in the endocytic compartment corresponding to late endosomal and low-pH compartments that contain the endocytic ligand α2 M (Fig. 9). This compartment has been described as the one preferentially targeted by species B Ad7 (41), and it differs from the one used by our nonliganded Ad5 vector and by species C members, such as Ad2 or Ad5, consisting of early and recycling endosomes which are relatively alkaline and contain Tf ligands (Fig. 8). The effect of QM10 on the routing of Ad suggests that the decapeptide QM10 selected on CF-KM4 cells is an endocytic ligand of the lysosomal pathway. Our results therefore imply that it is possible, using a relatively short peptide ligand inserted into the fiber shaft, to redirect an Ad5 vector to an endocytic pathway unusual for this serotype, i.e., the lysosomal pathway specifically used by members of another Ad species.
Ad serotypes of species B present an apparent intrinsic contradiction: their infectivity indices, ranging from 1:300 to 1:8,000 for serotypes 3 and 7 in various cells, are much lower than those of species C members (9), but they have similar prevalences in human Ad infections. Since a low infectivity index (PFU/PP ratio) reflects low efficiency in late steps in the virus cycle, e.g., virus assembly and/or DNA encapsidation, this must be compensated for in species B Ad by higher efficiency in another step(s) of their replication cycle than in species C members. One of these might be endocytosis, uncoating, or intracellular routing of incoming virions. Several types of experimental evidence, including data with our QM10-liganded vector, support this hypothesis. (i) Replacement of Ad5 knob with Ad3 knob has been found to enhance several steps in virus replication (27). (ii) Construction of chimeric Ad5 carrying fibers with serotype 35 knobs has revealed that the fiber knob is a major determinant of the cell trafficking of incoming Ad virions (59). (iii) Likewise, using chimeric Ad5 virions carrying Ad7 fibers (Ad5fiber7), it has been shown that targeting to the lysosomal pathway is under the control of Ad7 fiber and that this pathway and the delayed escape of Ad7 from this compartment may advantageously be used by this serotype and other serotypes of species B to be transported within these organelles through the cytoplasm to the nucleus (41). It has been suggested that engineering gene therapy vectors, in particular nonviral vectors, to direct them to the lysosomal pathway might be beneficial to their transduction efficiency (41). (iv) Our results with the QM10 peptide demonstrate the feasibility of this concept, at least using fiber-modified serotype 5, and suggest that the QM10-mediated retargeting of Ad5 to this alternative pathway confers upon QM10-liganded Ad5 a significant advantage as a vector in enhancing its gene transfer efficiency. The observation that addressing to the late, acidic endosome compartment was knob independent and that the same enhancement effect in gene transfer was obtained with knobbed and knobless versions of QM10-liganded Ad5 vector would be another advantage in ablating the CAR recognition of undesired CAR+ cells.
The cellular molecule(s) which binds the peptide ligand QM10, acts as an alternative endocytic receptor for Ad5, and modifies its intracellular routing remains to be identified. Since our QM10-liganded AdGFP vector resided in the same lysosomal compartment as α2 M, it is possible that it bound to α2 M receptor (70). Of note, a portion of the QM10 peptide, the tetrapeptide motif SHVY, is found at position 1118 in the C-terminal moiety of α2 M (64), which includes its receptor binding domain (residues 1313 to 1450 [23, 29]). Whatever their nature, these QM10 receptors seemed to be well represented and conserved in most mammalian-cell species, as indicated by the gene transfer enhancement shown by AdGFP-QM10-knob in the different cell lines tested (Fig. 2). The identification and functional characterization of such a molecule(s) would not only help improve cell trafficking and gene delivery by Ad, and possibly nonviral, vectors but would also provide valuable information on the endocytic machinery of mammalian cells. It still needs to be determined whether QM10 would be functional only in the context of the fiber shaft or would also be active in the context of another capsid protein, such as penton base, hexon, or protein IX, or as a decapeptide incorporated in a nonviral vector.
The fiber knob domain carries several biological functions that are essential in the Ad life cycle: (i) cell binding and tissue tropism of the virus (9, 35, 65); (ii) activation of dendritic cells (44); (iii) trimerization signal for the fiber protein, a function that is critical for Ad, since fiber monomers fail to bind legitimately to penton base to form penton capsomer and to be encapsidated (36, 37); (iv) determinants of Ad assembly and cellular release, as fiberless Ads are 4 orders of magnitude less infectious and have severely reduced progeny yields compared to WT Ad (30, 50, 69); (v) binding to CAR in the tight junctions to facilitate the release of progeny virions into the lumen (71); and, in addition, (vi) the results of studies of cell trafficking of chimeric Ad5fiber7 suggest that the penton base capsomers are not directly responsible for the endosomal escape of Ad virions into the cytosol but that this function is carried by the fibers (41). The results of our experiments with Ad vectors carrying scissile knobs supported the hypothesis that fiber could exert a control on the endosomal escape of Ad virions: our control nonliganded vector and SY12-liganded vector, both of which used the WTAd5 endocytic pathway, were apparently less efficient in vesicular escape in their knobless than in their knobbed versions when assayed in coendocytosis with the RcA toxin (Fig. 6). The data with our deknobbed vectors would therefore assign this novel function of endosomolysis to the knob domain of the fiber.
A detailed analysis of Ad-dependent cell membrane lysis has shown that the most effective lytic activity occurred at pH 5.5 for both Ad7 and chimeric Ad5fiber7 virions, versus 6.0 for Ad5, and it has been proposed that fiber would be responsible for endosomolysis, either (i) by interfering directly in this process or (ii) rather indirectly, by defining the optimal pH for membrane lysis and acting as a pH sensor that would trigger the endosomal lytic activity (41). The second hypothesis would be consistent with our data with AdGFP-QM10±knob. If the fiber knob domain functions as a pH sensor, endosomolysis would be more dependent upon the presence of the knob in an unusually acidic environment than in the relatively alkaline compartment of early endosomes used by species C Ad and nonliganded Ad5 vectors. This is the case for AdGFP-QM10±knob: it localized to the acidic lysosomal compartment, and the endosomal release of coendocytosed RcA, and AdGFP-QM10±knob vector, was the most negatively affected by deknobbing (Fig. 6). Further studies, using knob protein provided in cis or trans to incoming Ad virions following one or the other endocytic pathway, would be necessary to understand the mechanism of knob-mediated endosomal escape of Ad at the molecular level.
Acknowledgments
This work was financially supported by the French Cystic Fibrosis Association Vaincre la Mucoviscidose (VLM; AO.2002.TG-0203), the Programme Hospitalier de Recherche Clinique (PHRC 94-97.017), the Association Française contre les Myopathies (AFM; 2003.1477-9481), and Got-A-Gene AB, Göteborg, Sweden. F.G. was the recipient of a VLM fellowship.
We are deeply grateful to Simone Peyrol and Sandra Balvay-Garvi (Centre Commun d'Imagerie de la Faculté de Médecine Laennec) for their help in specimen processing and EM analyses, to David Curiel (University of Alabama at Birmingham) for his generous gift of 1D6.14 MAb, to Monika Lusky (Transgene SA, Strasbourg, France) for providing us with the 293-Fiber cell line, and to Emmanuelle Groscarret and Yvette Jaquet (Laboratoire de Virologie Médicale des Hospices Civils de Lyon, Domaine Rockefeller, Lyon, France) for kindly supplying us with a dozen different mammalian cell lines. We also thank Catherine Figarella (Groupe de Recherches sur les Glandes Exocrines, Faculté de Médecine de Marseille, Marseille, France) for her valuable advice about CF-KM4 and MM39 cell culture, Béatrice Burdin (Service Commun d'Imagerie de l'UCBL, Lyon, France) for her precious help in confocal IF microscopy, and Cathy Berthet for her expert secretarial aid.
REFERENCES
- 1.Arnberg, N., K. Edlund, A. H. Kidd, and G. Wadell. 2000. Adenovirus type 37 uses sialic acid as a cellular receptor. J. Virol. 74:42-48. [PMC free article] [PubMed] [Google Scholar]
- 2.Arnberg, N., A. H. Kidd, K. Edlund, F. Olfat, and G. Wadell. 2000. Initial interactions of subgenus D adenoviruses with A549 cellular receptors: sialic acid versus alpha(v) integrins. J. Virol. 74:7691-7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balakireva, L., G. Schoen, E. Thouvenin, and J. Chroboczek. 2003. Binding of adenovirus capsid to dipalmitoylcholine provides a novel pathway for virus entry. J. Virol. 77:4858-4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergelson, J. M., J. A. Cunningham, G. Droguett, E. A. Kurt-Jones, A. Krithivas, J. S. Hong, M. S. Horwitz, R. L. Crowell, and R. W. Finberg. 1997. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320-1323. [DOI] [PubMed] [Google Scholar]
- 5.Borth, W. 1992. Alpha 2-macroglobulin, a multifunctional binding protein with targeting characteristics. FASEB J. 6:3345-3353. [DOI] [PubMed] [Google Scholar]
- 6.Boulanger, P., and F. Puvion. 1973. Large-scale preparation of soluble adenovirus hexon, penton and fiber antigens in highly purified form. Eur. J. Biochem. 39:37-42. [DOI] [PubMed] [Google Scholar]
- 7.Chroboczek, J., R. W. Ruigrok, and S. Cusack. 1995. Adenovirus fiber. Curr. Top. Microbiol. Immunol. 199:163-200. [DOI] [PubMed] [Google Scholar]
- 8.Dechecchi, M. C., P. Melotti, A. Bonizzato, M. Santacatterina, M. Chilosi, and G. Cabrini. 2001. Heparan sulfate glycosaminoglycans are receptors sufficient to mediate the initial binding of adenovirus types 2 and 5. J. Virol. 75:8772-8780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Defer, C., M. T. Belin, M. L. Caillet-Boudin, and P. Boulanger. 1990. Human adenovirus-host cell interactions: comparative study with members of subgroups B and C. J. Virol. 64:3661-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Giovine, M., B. Salone, Y. Martina, V. Amati, G. Zambruno, E. Cundari, C. M. Failla, and I. Saggio. 2001. Binding properties, cell delivery, and gene transfer of adenoviral penton base displaying bacteriophage. Virology 282:102-112. [DOI] [PubMed] [Google Scholar]
- 11.Fontana, L., M. Nuzzo, L. Urbanelli, and P. Monaci. 2003. General strategy for broadening adenovirus tropism. J. Virol. 77:11094-11104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaden, F., L. Franqueville, S. S. Hong, V. Legrand, C. Figarella, and P. Boulanger. 2002. Mechanism of restriction of normal and cystic fibrosis transmembrane conductance regulator-deficient human tracheal gland cells to adenovirus (Ad) infection and Ad-mediated gene transfer. Am. J. Respir. Cell Mol. Biol. 27:628-640. [DOI] [PubMed] [Google Scholar]
- 13.Gaggar, A., D. M. Shayakhmetov, and A. Lieber. 2003. CD46 is a cellular receptor for group B adenoviruses. Nat. Med. 8:746-755. [DOI] [PubMed] [Google Scholar]
- 14.Greber, U. F., P. Webster, J. Weber, and A. Helenius. 1996. The role of the adenovirus protease on virus entry into cells. EMBO J. 15:1766-1777. [PMC free article] [PubMed] [Google Scholar]
- 15.Greber, U. F., M. Willetts, P. Webster, and A. Helenius. 1993. Stepwise dismantling of adenovirus 2 during entry into cells. Cell 75:477-486. [DOI] [PubMed] [Google Scholar]
- 16.Green, M., M. Piña, R. Kimes, P. C. Wensink, L. A. MacHattie, and C. A. Thomas, Jr. 1967. Adenovirus DNA. I. Molecular weight and conformation. Proc. Natl. Acad. Sci. USA 57:1302-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henning, P., M. K. Magnusson, E. Gunneriusson, S. S. Hong, P. Boulanger, and L. Lindholm. 2002. Genetic modification of adenovirus 5 tropism by a novel class of ligands based on a three-helix bundle scaffold derived from staphylococcal protein A. Hum. Gene Ther. 13:1427-1439. [DOI] [PubMed] [Google Scholar]
- 18.Hong, J. S., and J. A. Engler. 1991. The amino terminus of the adenovirus fiber protein encodes the nuclear localization signal. Virology 185:758-767. [DOI] [PubMed] [Google Scholar]
- 19.Hong, S. S., and P. Boulanger. 1995. Protein ligands of the human adenovirus type 2 outer capsid identified by biopanning of a phage-displayed peptide library on separate domains of wild-type and mutant penton capsomers. EMBO J. 14:4714-4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hong, S. S., B. Gay, L. Karayan, M. C. Dabauvalle, and P. Boulanger. 1999. Cellular uptake and nuclear delivery of recombinant adenovirus penton base. Virology 262:163-177. [DOI] [PubMed] [Google Scholar]
- 21.Hong, S. S., L. Karayan, J. Tournier, D. T. Curiel, and P. Boulanger. 1997. Adenovirus type 5 fiber knob binds to MHC class I alpha2 domain at the surface of human epithelial and B lymphoblastoid cells. EMBO J. 16:2294-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong, S. S., M. K. Magnusson, P. Henning, L. Lindholm, and P. Boulanger. 2003. Adenovirus stripping: a versatile method to generate adenovirus vectors with new cell target specificity. Mol. Ther. 7:692-699. [DOI] [PubMed] [Google Scholar]
- 23.Jenner, L., L. Husted, S. Thirup, L. Sottrup-Jensen, and J. Nyborg. 1998. Crystal structure of the receptor binding domain of alpha 2-macroglobulin. Structure 6:595-604. [DOI] [PubMed] [Google Scholar]
- 24.Kammouni, W., B. Moreau, F. Becq, A. Saleh, A. Pavirani, C. Figarella, and M. D. Merten. 1999. A cystic fibrosis tracheal gland cell line, CF-KM4. Correction by adenovirus-mediated CFTR gene transfer. Am. J. Respir. Cell Mol. Biol. 20:684-691. [DOI] [PubMed] [Google Scholar]
- 25.Karayan, L., B. Gay, J. Gerfaux, and P. Boulanger. 1994. Oligomerization of recombinant penton base of adenovirus type 2 and its assembly with fiber in baculovirus-infected cells. Virology 202:782-795. [DOI] [PubMed] [Google Scholar]
- 26.Karayan, L., S. S. Hong, B. Gay, J. Tournier, A. D. d'Angeac, and P. Boulanger. 1997. Structural and functional determinants in adenovirus type 2 penton base recombinant protein. J. Virol. 71:8678-8689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawakami, Y., H. Li, J. T. Lam, V. Krasnykh, D. T. Curiel, and J. L. Blackwell. 2003. Substitution of the adenovirus serotype 5 knob with a serotype 3 knob enhances multiple steps in virus replication. Cancer Res. 63:1262-1269. [PubMed] [Google Scholar]
- 28.Kok, T. W., T. Pryor, and L. Payne. 1998. Comparison of rhabdomyosarcoma, buffalo green monkey kidney epithelial, A549 (human lung epithelila) cells and human embryonic lung fibroblasts for isolation of enteroviruses from clinical samples. J. Clin. Virol. 11:61-65. [DOI] [PubMed] [Google Scholar]
- 29.Kolodziej, S. J., T. Wagenknecht, D. K. Strickland, and J. K. Stoops. 2002. The three-dimensional structure of the human alpha 2-macroglobulin dimer reveals its structural organization in the tetrameric native and chymotrypsin-alpha 2-macroglobulin complexes. J. Biol. Chem. 277:28031-28037. [DOI] [PubMed] [Google Scholar]
- 30.Legrand, V., D. Spehner, Y. Schlesinger, N. Settelen, A. Pavirani, and M. Mehtali. 1999. Fiberless recombinant adenoviruses: virus maturation and infectivity in the absence of fiber. J. Virol. 73:907-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leopold, P. L., B. Ferris, I. Grinberg, S. Worgall, N. R. Hackett, and R. G. Crystal. 1998. Fluorescent virions: dynamic tracking of the pathway of adenoviral gene transfer vectors in living cells. Hum. Gene Ther. 9:367-378. [DOI] [PubMed] [Google Scholar]
- 32.Leopold, P. L., G. Kreitzer, N. Miyazawa, S. Rempel, K. K. Pfister, E. Rodriguez-Boulan, and R. G. Crystal. 2000. Dynein- and microtubule-mediated translocation of adenovirus serotype 5 occurs after endosomal lysis. Hum. Gene Ther. 11:151-165. [DOI] [PubMed] [Google Scholar]
- 33.Li, E., D. Stupack, G. M. Bokoch, and G. R. Nemerow. 1998. Adenovirus endocytosis requires actin cytoskeleton reorganization mediated by Rho family GTPases. J. Virol. 72:8806-8812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, E., D. Stupack, R. Klemke, D. A. Cheresh, and G. R. Nemerow. 1998. Adenovirus endocytosis via alpha(v) integrins requires phosphoinositide-3-OH kinase. J. Virol. 72:2055-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Louis, N., P. Fender, A. Barge, P. Kitts, and J. Chroboczek. 1994. Cell-binding domain of adenovirus serotype 2 fiber. J. Virol. 68:4104-4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Magnusson, M. K., S. S. Hong, P. Boulanger, and L. Lindholm. 2001. Genetic retargeting of adenovirus: novel strategy employing “deknobbing” of the fiber. J. Virol. 75:7280-7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Magnusson, M. K., S. S. Hong, P. Henning, P. Boulanger, and L. Lindholm. 2002. Genetic retargeting of adenovirus vectors: functionality of targeting ligands and their influence on virus viability. J. Gene Med. 4:356-370. [DOI] [PubMed] [Google Scholar]
- 38.Meier, O., and U. F. Greber. 2003. Adenovirus endocytosis. J. Gene Med. 5:451-462. [DOI] [PubMed] [Google Scholar]
- 39.Merten, M. D., W. Kammouni, W. Renaud, F. Birg, M. G. Mattei, and C. Figarella. 1996. A transformed human tracheal gland cell line, MM-39, that retains serous secretory functions. Am. J. Respir. Cell Mol. Biol. 15:520-528. [DOI] [PubMed] [Google Scholar]
- 40.Mittal, S. K., M. R. McDermott, D. C. Johnson, L. Prevec, and F. L. Graham. 1993. Monitoring foreign gene expression by a human adenovirus-based vector using the firefly luciferase gene as a reporter. Virus Res. 28:67-90. [DOI] [PubMed] [Google Scholar]
- 41.Miyazawa, N., R. G. Crystal, and P. L. Leopold. 2001. Adenovirus serotype 7 retention in a late endosomal compartment prior to cytosol escape is modulated by fiber protein. J. Virol. 75:1387-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyazawa, N., P. L. Leopold, N. R. Hackett, B. Ferris, S. Worgall, E. Falck-Pedersen, and R. G. Crystal. 1999. Fiber swap between adenovirus subgroups B and C alters intracellular trafficking of adenovirus gene transfer vectors. J. Virol. 73:6056-6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Molinier-Frenkel, V., R. Lengagne, F. Gaden, S. S. Hong, J. Choppin, H. Gahery-Segard, P. Boulanger, and J.-G. Guillet. 2002. Adenovirus hexon protein is a potent adjuvant for activation of a cellular immune response. J. Virol. 76:127-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Molinier-Frenkel, V., A. Prévost-Blondel, S. S. Hong, R. Lengagne, S. Boudaly, M. K. Magnusson, P. Boulanger, and J.-G. Guillet. 2003. The maturation of murine dendritic cells induced by human adenovirus is mediated by the fiber knob domain. J. Biol. Chem. 278:37175-37182. [DOI] [PubMed] [Google Scholar]
- 45.Neumann, R., J. Chroboczek, and B. Jacrot. 1988. Determination of the nucleotide sequence for the penton base gene of human adenovirus type 5. Gene 69:153-157. [DOI] [PubMed] [Google Scholar]
- 46.Nicklin, S. A., D. J. Von Seggern, L. M. Work, D. C. Pek, A. F. Dominiczak, G. R. Nemerow, and A. H. Baker. 2001. Ablating adenovirus type 5 fiber-CAR binding and HI loop insertion of the SIGYPLP peptide generate an endothelial cell-selective adenovirus. Mol. Ther. 4:534-542. [DOI] [PubMed] [Google Scholar]
- 47.Novelli, A., and P. Boulanger. 1991. Deletion analysis of functional domains in baculovirus-expressed adenovirus type 2 fiber. Virology 185:365-376. [DOI] [PubMed] [Google Scholar]
- 48.Obermuller, S., C. Kiecke, K. von Figura, and S. Honing. 2002. The tyrosine motifs of Lamp 1 and LAP determine their direct and indirect targetting to lysosomes. J. Cell Sci. 115:185-194. [DOI] [PubMed] [Google Scholar]
- 49.Peters, C., M. Braun, B. Weber, M. Wendland, B. Schmidt, R. Polhlmann, A. Waheed, and K. von Figura. 1990. Targeting of a lysosomal membrane protein: a tyrosine-containing endocytosis signal in the cytoplasmic tail of lysosomal acid phosphatase is necessary and sufficient for targeting to lysosomes. EMBO J. 9:3497-3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Puvion-Dutilleul, F., V. Legrand, M. Mehtali, M. K. Chelbi-Alix, H. de Thé, and E. Puvion. 1999. Deletion of the fiber gene induces the storage of hexon and penton base proteins in PML/Sp100-containing inclusions during adenovirus infection. Biol. Cell 91:617-628. [PubMed] [Google Scholar]
- 51.Rodman, J. S., and A. Wandinger-Ness. 2000. Commentary: Rab GTPases coordinate endocytosis. J. Cell Sci. 113:183-192. [DOI] [PubMed] [Google Scholar]
- 52.Roelvink, P. W., I. Kovesdi, and T. J. Wickham. 1996. Comparative analysis of adenovirus fiber-cell interaction: adenovirus type 2 (Ad2) and Ad9 utilize the same cellular fiber receptor but use different binding strategies for attachment. J. Virol. 70:7614-7621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roelvink, P. W., A. Lizonova, J. G. Lee, Y. Li, J. M. Bergelson, R. W. Finberg, D. E. Brough, I. Kovesdi, and T. J. Wickham. 1998. The coxsackievirus-adenovirus receptor protein can function as a cellular attachment protein for adenovirus serotypes from subgroups A, C, D, E, and F. J. Virol. 72:7909-7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rogers, B. E., J. T. Douglas, C. Ahlem, D. J. Buchsbaum, J. Frincke, and D. T. Curiel. 1997. Use of a novel cross-linking method to modify adenovirus tropism. Gene Ther. 4:1387-1392. [DOI] [PubMed] [Google Scholar]
- 55.Russell, W. C. 2000. Update on adenovirus and its vectors. J. Gen. Virol. 81:2573-2604. [DOI] [PubMed] [Google Scholar]
- 56.Santis, G., V. Legrand, S. S. Hong, E. Davison, I. Kirby, J. L. Imler, R. W. Finberg, J. M. Bergelson, M. Mehtali, and P. Boulanger. 1999. Molecular determinants of adenovirus serotype 5 fibre binding to its cellular receptor CAR. J. Gen. Virol. 80:1519-1527. [DOI] [PubMed] [Google Scholar]
- 57.Segerman, A., J. P. Atkinson, M. Marttila, V. Dennerquist, G. Wadell, and N. Arnberg. 2003. Adenovirus type 11 uses CD46 as a cellular receptor. J. Virol. 77:9183-9191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seth, P. 1994. Mechanism of adenovirus-mediated endosome lysis: role of the intact adenovirus capsid structure. Biochem. Biophys. Res. Commun. 205:1318-1324. [DOI] [PubMed] [Google Scholar]
- 59.Shayakhmetov, D. M., Z. Y. Li, V. Ternovoi, A. Gaggar, H. Gharwan, and A. Lieber. 2003. The interaction between the fiber knob domain and the cellular attachment receptor determines the intracellular trafficking route of adenoviruses. J. Virol. 77:3712-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shayakhmetov, D. M., and A. Lieber. 2000. Dependence of adenovirus infectivity on length of the fiber shaft domain. J. Virol. 74:10274-10286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith, G. P., and J. K. Scott. 1993. Libraries of peptides and proteins displayed on filamentous phage. Methods Enzymol. 217:228-257. [DOI] [PubMed] [Google Scholar]
- 62.Smothers, J. F., S. Henikoff, and P. Carter. 2002. Affinity selection from biological libraries. Science 298:621-622. [DOI] [PubMed] [Google Scholar]
- 63.Sonowane, N. D., and A. S. Verkman. 2003. Determinants of [Cl−] in recycling and late endosomes and Golgi complex measured using fluorescent ligands. J. Cell Biol. 160:1129-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sottrup-Jensen, L., T. M. Stepanik, D. M. Wierzbicki, C. M. Jones, P. B. Lonblad, T. Kristensen, S. B. Mortensen, T. E. Petersen, and S. Magnusson. 1983. The primary structure of alpha 2-macroglobulin and localization of a factor XIIIa cross-linking site. Ann. N. Y. Acad. Sci. 421:41-60. [DOI] [PubMed] [Google Scholar]
- 65.Stevenson, S. C., M. Rollence, B. White, L. Weaver, and A. McClelland. 1995. Human adenovirus serotypes 3 and 5 bind to two different cellular receptors via the fiber head domain. J. Virol. 69:2850-2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Suomalainen, M., M. Y. Nakano, K. Boucke, S. Keller, and U. F. Greber. 2001. Adenovirus-activated PKA and p38/MAPK pathways boost microtubule-mediated nuclear targeting of virus. EMBO J. 20:1310-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tomko, R. P., R. Xu, and L. Philipson. 1997. HCAR and MCAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc. Natl. Acad. Sci. USA 94:3352-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tycko, B., and F. R. Maxfield. 1982. Rapid acidification of endocytic vesicles containing alpha 2-macroglobulin. Cell 28:643-651. [DOI] [PubMed] [Google Scholar]
- 69.van Beusechem, V. W., A. L. van Rijswijk, H. H. van Es, H. J. Haisma, H. M. Pinedo, and W. R. Gerritsen. 2000. Recombinant adenovirus vectors with knobless fibers for targeted gene transfer. Gene Ther. 7:1940-1946. [DOI] [PubMed] [Google Scholar]
- 70.Van Leuven, F., L. Stas, C. Hilliker, K. Lorent, L. Umans, L. Sterneels, L. Overbergh, S. Torrekens, D. Moechars, B. De Strooper, and H. van den Berghe. 1994. Structure of the gene (LRP1) coding for the human alpha 2-macroglobulin receptor lipoprotein receptor-related protein. Genomics 24:78-89. [DOI] [PubMed] [Google Scholar]
- 71.Walters, R. W., P. Freimuth, T. O. Moninger, I. Ganske, J. Zabner, and M. J. Welsh. 2002. Adenovirus fiber disrupts CAR-mediated intercellular adhesion allowing virus escape. Cell 110:789-799. [DOI] [PubMed] [Google Scholar]
- 72.Walters, R. W., T. Grunst, J. M. Bergelson, R. W. Finberg, M. J. Welsh, and J. Zabner. 1999. Basolateral localization of fiber receptors limits adenovirus infection from the apical surface of airway epithelia. J. Biol. Chem. 274:10219-10226. [DOI] [PubMed] [Google Scholar]
- 73.Wickham, T. J., E. J. Filardo, D. A. Cheresh, and G. R. Nemerow. 1994. Integrin αvβ5 selectively promotes adenovirus mediated cell membrane permeabilization. J. Cell Biol. 127:257-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wickham, T. J., P. Mathias, D. A. Cheresh, and G. R. Nemerow. 1993. Integrins αvβ3 and αvβ5 promote adenovirus internalization but not virus attachment. Cell 73:309-319. [DOI] [PubMed] [Google Scholar]
- 75.Wood, D. J., and B. Hull. 1999. L20B cells simplify culture of polioviruses from clinical samples. J. Med. Virol. 58:188-192. [PubMed] [Google Scholar]
- 76.Wu, E., L. Pache, D. J. von Seggern, T.-M. Mullen, Y. Mikyas, P. L. Stewart, and G. R. Nemerow. 2003. Flexibility of the adenovirus fiber is required for efficient receptor interaction. J. Virol. 77:7225-7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yamashiro, D. J., L. A. Borden, and F. R. Maxfield. 1989. Kinetics of alpha 2-macroglobulin endocytosis and degradation in mutant and wild-type Chinese hamster ovary cells. J. Cell Physiol. 139:377-382. [DOI] [PubMed] [Google Scholar]
- 78.Zabner, J., M. Chillon, T. Grunst, T. O. Moninger, B. L. Davidson, R. Gregory, and D. Armentano. 1999. A chimeric type 2 adenovirus vector with a type 17 fiber enhances gene transfer to human airway epithelia. J. Virol. 73:8689-8695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zabner, J., P. Freimuth, A. Puga, A. Fabrega, and M. J. Welsh. 1997. Lack of high-affinity fiber receptor activity explains the resistance of ciliated airway epithelia to adenovirus infection. J. Clin. Investig. 100:1144-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zen, K., J. Biwersi, N. Periasamy, and A. S. Verkman. 1992. Second messengers regulate endosomal acidification in Swiss 3T3 fibroblasts. J. Cell Biol. 119:99-110. [DOI] [PMC free article] [PubMed] [Google Scholar]


