Abstract
Importance in the Field
Phosphatases of Regenerating Liver (PRLs) are novel oncogenes that interact with many, well-established cell signaling pathways that are misregulated in cancer and is known to drive cancer metastasis when overexpressed.
Areas Covered in this Review
This review will cover basic information of the discovery and characteristics of the PRL family. We will also report findings on the role of PRL in cancer, cell functions, and cell signaling. Furthermore, PRL’s suitability as a novel drug target will be discussed along with current methods being developed to facilitate PRL inhibition.
Expert Opinion
PRLs show great potential as novel drug targets for anti-cancer therapeutics. Studies indicate that PRL can perturb major cancer pathways such as Src/ERK1/2 and PTEN/PI3K/Akt. Upregulation of PRLs has also been shown to drive cancer metastasis. However, in order to fully realize its therapeutic potential, a deeper understanding of the function of PRL in normal tissue and in cancer must be obtained. Novel and integrated biochemical, chemical biological and genetic approaches will be needed to identify PRL substrate(s) and to provide proof-of-concept data on the druggability of the PRL phosphatases.
Keywords: Adhesion Junctions, Adhesion molecules, Cell migration, Colorectal cancer, De-trimerization, ERK1/2, EMT, Epithelial to Mesenchymal Transition, Filamentous Actin Dynamics, Monoclonal Antibody targeting, phosphatase and tensin homologue deleted on chromosome 10, Small molecule inhibition, Phosphatase of Regenerating Liver, PI3K/Akt, PRL, PTEN, Receptor Tyrosine Kinases, Src kinase
1. INTRODUCTION
The addition and removal of phosphate groups is a highly conserved driving force in the regulation of cellular processes. A very precise balance between phosphorylation and dephosphorylation must be maintained in order for proper cell functions to be carried out. Protein kinases and phosphatases are the keepers of this balance with kinases adding phosphate groups and phosphatases removing them. As such, mutation and/or altered regulation of these proteins have long been associated with disease making them tempting therapeutic and preventative targets. For many years, kinases have dominated the drug development industry with phosphatases generally being ignored until recently. Phosphatases were once thought to be non-specific and very difficult to study (1,2). However, improved technology and novel substrate-binding techniques began to provide mounting evidence showing that phosphatases are equally implicated in disease including cancer, diabetes, neurological diseases, and more (1). Novel methods for drug discovery have since been implemented to increase drug specificity by targeting unique structural features near the catalytic domain and regulatory sites (1,2). Consequentially, phosphatases have been brought into the spotlight in academic research as potential therapeutic targets.
Protein phosphatases are a diverse group of enzymes that defy attempts at simple classification. Generally, phosphatases can be divided into two superfamilies: enzymes that show specificity towards dephosphorylating serine and threonine residues (Ser/Thr phosphatases) and those that specialize in the dephosphorylation of tyrosine residues (Protein Tyrosine Phosphatases (PTPs)) (1). These two broad categories contain numerous families and subfamilies of proteins all of which are structurally unique and many of which are thought to have evolved independently from one another (1). A majority of phosphatase genes encode protein tyrosine phosphatases which can be identified by their HCX5R active site motif where the active site cysteine acts as the essential nucleophile during dephosphorylation (1,2). One of the largest and most diverse classes of PTP includes classical PTPs, which can be divided into Receptor-like and Non-transmembrane PTPs, and Dual Specificity Phosphatases (DSPs) (1). Dual Specificity Phosphatases are unique from the classical PTPs due to their ability to dephosphorylate both tyrosine and serine/threonine residues (1). DSPs tend to be very diverse from each other functionally, being able to dephosphorylate a wide variety of substrates (1). This review will focus on Phosphatases of Regenerating Liver, or PRLs, a family of novel DSPs, as potential drug targets for cancer therapy.
2. PRL Structural Features
PRL-1 was first discovered as a strongly up-regulated, immediate-early gene in regenerating rat liver (3). Later, its family members, PRL-2 and PRL-3, were identified by searching the Murine Expressed Sequence Tags database and were found to be highly similar in both amino acid sequence and structure to each other and PRL-1 (4). PRL-1 and PRL-2 were most similar in sequence, exhibiting 87% identity, while PRL-3 exhibited 76% and 79% identity to PRL-1 and 2 respectively (4,5). Further structural and sequential analysis allowed the PRLs to be categorized within the PTP superfamily (5,6). All the PRLs share the CX5R active site, P-loop, and WDP loop motifs typical of PTPs (5,6) (Figure 1). Structural similarities to PTEN, Cdc14, MKP, and other dual specificity phosphatases (DSPs) presents strong evidence that the PRLs are able to dephosphorylate both tyrosine and serine/threonine residues (5,6). One feature that makes the PRLs unique from all other phosphatases is the presence of a CAAX prenylation motif next to a polybasic region in the C-terminal domain (6,7). Prenylation, as discovered in studies of proteins like Ras and Rab, is known to facilitate localization of proteins to the plasma membrane (8). Indeed, both PRL-1and 3 have been found to associate with the plasma membrane and early endosome in mammalian cell lines (9,10,11,12,13). PRL has also been shown to be regulated by the redox status of its active site and the formation of a disulfide bond when fully oxidized (14,15,16). Since its discovery, the PRL family has been studied extensively in cell culture and in several model animal systems.
Figure 1.
A. Ribbon Diagram of the Structure of PRL-1: The C-terminal end of PRL contains the CAAX prenylation domain and adjoining polybasic region and has been shown to be vital to PRL phosphatase activity. This structure shows the active site sequence, which is located on the P-loop, containing the catalytic Cysteine (6). The WPD-Loop contains Asp72 which acts as a general acid during catalysis (6). The WPD-Loop of PRL contains some variations from other PTPs that may account for low, in vitro activity (6). One difference is that the conserved serine or threonine placed after the invariant Arginine in most PTPs is replaced by an Alanine in PRL (6). PRL also lacks key hydrogen bonds that are known to stabilize the P-Loop of other PTPs for catalysis (6). The Q-Loop is important for hydrogen bonding with scissilie oxygen and active site water molecule (6). B. PRL Trimer: PRL crystalizes in a trimer that fixes the C-Terminal ends of each monomer such that they all face towards the plasma membrane. This trimer is oriented with the C-terminal ends facing out (into the “membrane”) with the active sites on the opposite face (in the “cytoplasm”). This is how the PRL trimer is hypothesized to dock onto the membrane. The trimer state has been shown to be physiologically relevant and targetable for disassociation.
3. Cellular Localization and Tissue Expression
In the cell, PRL will localize to the plasma membrane if the C-terminal CAAX motif is prenylated (7,8,13,). In HEK293 cells, PRL-1 constructs containing a mutated CAAX motif were found in the soluble cytoplasmic fraction whereas PRL-1 wild-type (WT) was found primarily in the membrane faction (7). This evidence showing that the CAAX domain is critical for membrane association coincides with studies with farnesyl-transferase inhibitor (FTI-277) based in NIH 3T3 cells stably expressing PRL-1,-2, or −3 (13). Immunofluorescent staining from both studies corroborated the conclusion drawn from the subcellular fractionation data and also revealed that a fraction of the PRL that was not prenylated had become localized to the nucleus (7,13). The polybasic region preceding the CAAX motif is hypothesized to be required for nuclear localization in the absence of prenylation and for recruitment to the membrane (8). When all six charged residues of the polybasic region were mutated to alanines, PRL did not localize to the plasma membrane or the nucleus but was found entirely in the cytosolic fraction (7).
Dumaual et al. conducted an extensive assessment of PRL-1 and −2 expression in human tissue samples. They found that PRL-1 and −2 were expressed almost ubiquitously across all tissues and organ systems with PRL-1 expression levels being slightly more restricted and less intense than PRL-2 (17). PRL-3 however, has a much more restricted expression pattern, primarily in the heart, skeletal muscle, vasculature, and brain, and is generally expressed at a lower level than PRL −1 and −2 (18). PRL-1 and −2 are also strongly expressed in the central nervous system (CNS) of developing Drosophila, amphioxus, and zebrafish (19). In mice, PRL-1 and −3 expression levels are much more restricted, showing expression in colon and intestine, but having very low or no expression in other organ systems (45,48).
4. Role of PRL in Cancer
PRL-3 was first brought into the spotlight as a potential oncogene in 2001(20). The study used samples of metastatic liver lesions from colorectal cancer (CRC) patients and applied serial analysis of gene expression (SAGE) technology to create a gene expression profile to compare to non-metastatic libraries (20). Of the 144 genes that were found to be misregulated in the metastatic lesions, PRL-3 was the most consistently up-regulated (20). Interestingly, the expression level of PRL-3 in colorectal cancer primary tumors was much lower than in corresponding hepatic metastatic lesions suggesting that PRL-3 may play an important role in metastasis as opposed to carcinogenesis (20). Subsequent studies showed that, while PRL-3 was more consistently elevated in liver and lung metastasis, many other types of CRC metastasis exhibited high PRL-3 expression including brain, ovary, and lymph node lesions (11). Furthermore, it was shown that high levels of PRL-3 expression in the CRC primary tumor could predict the presence of distant metastasis (11). A Kaplan-Meier analysis for metastasis-free survival also revealed that patients with high levels of PRL-3 expression in resected CRC primary tumors were at a greater risk for liver/lung metastasis than those with low PRL-3 expression (11). Interestingly, the prognostic value of PRL-3 expression in CRC only correlates significantly in regards to risk of long distance metastasis and does not affect the presence of lymph node metastasis (11). Several more independent studies corroborate these findings (21,22). This suggests that PRL-3 may be useful for determining patient prognosis, allowing more judicious use of aggressive CRC therapies.
The original studies linking PRL-3 expression to CRC patient prognosis resulted in the discovery of similar findings in other types of cancer. Two studies show that PRL-3 overexpression can be found in breast cancer. Radke et al. found elevated PRL-3 expression in 116 out of 135 (85.9%) samples of ductal carcinoma in situ (DCIS), and 190 out of 246 (77.2%) samples of invasive carcinoma (12). Wang et al., on the other hand, found PRL-3 overexpression in breast cancer, but in a smaller portion of samples (133 out of 382 (34.8%)) (23). Radke et al. found that elevated PRL-3 in patients with node-positive breast cancer significantly impacted long term survival but did not significantly affect the survival of node-negative patients (12). Interestingly, Wang et al. found the opposite to be true, where PRL-3 overexpression was associated with reduced the life span of node-negative patients but not node-positive patients (23). Furthermore, a recent study by Hao et al. found PRL-3 overexpression in the primary tumor to be significantly correlated with breast cancer metastasis to the lymph node, while Wang et al. and Radke et al. reported that lymph node metastasis was independent of PRL-3 expression in the primary tumor (12,23,24). Further studies will be required to reconcile these conflicting results.
Along with the common colorectal cancer and breast cancer, PRL-3 has been shown to be up regulated in many other types of cancer. Up-regulated PRL-3 has been found in gastric carcinomas and was found to be significantly correlated to lymph node metastasis (25,26,27,28,29,30,31). Ovarian (32,33), liver (34,35), oral (36), cervical (37), esophageal (38), lung cancer (39,40), multiple myeloma (9,41), acute myeloid leukemia (AML) (42,43), and nasopharyngeal cancer (44) virtually all have been shown to exhibit high levels of PRL-3 expression. As with CRC, breast cancer, and gastric cancer, PRL-3 overexpression is overwhelmingly correlated with poor prognosis and progression to metastasis in all of the aforementioned cancer types (Table 1).
Table 1.
Summary of PRL dysregulations in cancer.
| Cancer Type | Primary Tumors with High PRL Expression |
Metastasis with High Levels of PRL Expression? |
High expression of PRL in Primary tumor predictive of Metastasis? |
Clinical Features Affected by PRL Overexpression |
Clinical Features NOT Affected by PRL Overexpression |
Reference |
|---|---|---|---|---|---|---|
| Colorectal | Yes | Yes: Especially in lung and liver |
Yes: Distant Metastasis |
Chance of Distant metastasis Sigificant reduction in Disease-free survival (DFS) |
Lymph- node metastasis Tumor size Tumor stage |
(20,11,21,22) |
| Breast | Yes: 116 out of 135 (85.9%) samples of DCIS and 190 out of 246 (77.2%) samples of invasive carcinoma |
Yes | Yes: Distant Metastasis |
Decrease in disease-free survival, overall trend. Significant decrease in DFS in node-positive samples (but not node- negative) |
Lymph-node metastasis. Tumor stage. |
(12) |
| Breast | Yes: 133 out of 382 (34.8%) samples |
Yes | No data. | Significant decrease in DFS in node-negative patients. (but not node- positive) |
Tumor stage Age |
(23) |
| Breast | Yes | Yes | Yes: increased lymph-node and distant metastasis |
Significantly correlated with lymph-node metastasis and distant metastasis. Decrease in DFS. |
Tumor stage | (24) |
| Gastric | Yes | Yes | Yes: Increased lymph-node and distant metastasis |
Significant decrease in DFS and distant metastasis. |
Tumor Stage Tumor Size |
(25,26,27,28,29, 30,31) |
| Ovarian | Yes | Yes | Yes: Increased metastasis |
Significant decrease in DFS and distant metastasis. |
No data | (32,33) |
| Liver | Yes | Yes | Yes: Increased metastasis |
Significant decrease in DFS and distant metastasis. |
No data | (34,35) |
| Oral | Yes | Yes | Yes: Increased metastasis |
Significant decrease in DFS and distant metastasis. |
No data | (36) |
| Cervical | Yes | Yes | Yes: Increased metastasis |
Significant decrease in DFS and distant metastasis. |
No data | (37) |
| Esophageal | Yes | Yes | Yes: Increased metastasis |
Significant decrease in DFS and distant metastasis. |
No data | (38) |
| Lung | Yes | Yes | Yes: Increased metastasis |
Significant decrease in DFS and distant metastasis. |
No data | (39,40) |
| Multiple Myeloma |
Yes | No Data | No Data | Significant decrease in DFS |
No data | (9,41) |
| Acute Myeloid Leukemia |
Yes | No Data | No Data | Significant decrease in DFS and possible driver for drug-resistance. |
No data | (42,43) |
| Nasopharyngeal | Yes | Yes | Yes: Increased metastasis |
Significant decrease in DFS and distant metastasis. |
No data | (44) |
| Prostate | Yes | No Data | No Data | No data. | No Data | (46) |
While there have been many studies investigating the role of PRL-3 in cancer, fewer have explored the relevance of PRL-1 and −2. Some of the earliest evidence of PRL-1 and −2 involvement in human cancer came from a study focusing on the generation of PRL-3 and −1 monoclonal antibodies to be used to diagnose cancer metastasis. Li et al. tested PRL-1 and PRL-3 specific antibodies on a human multiple cancer tissue array (45). The PRL-1 antibody hit on 10 cancer tissues including renal carcinoma and ovary lymphoma (45). Wang et al. provided evidence that PRL-2 can serve as an oncogene in prostate cancer (46). In this study, PRL-2 was shown to be overexpressed in prostate cancer cell lines LNCaP, PC3, and DU145 and regulated tumor cell migration and invasion (46). Additionally, PRL-2 transcription was found to be elevated in three samples of advanced prostate cancer in comparison to the corresponding normal prostate tissue (46). In situ hybridization of PRL-2 antisense oligonucleotide also showed PRL-2 elevation in four tumor samples in comparison to four samples of benign prostatic hyperplasia (46) (Table 1). Recently, a study by Dumaual et al. showed that both PRL mRNAs are highly to moderately expressed in all but six tumor tissue samples examined (47). Furthermore, PRL −1 and −2 mRNA was found to be more overexpressed in poorly differentiated tumors in comparison to well differentiated tumors (47).
The role of PRL in cancer has also been studied in animal models. A conditional PRL-3 KO mouse was created to study the effect of PRL-3 loss on azoxymethane (AOM)-dextran sodium sulphate (DSS) induced tumorigenesis (48). A one-time treatment with the carcinogen AOM followed by long term inflammation induced by DSS is a well-established protocol for inducing colorectal carcinogenesis for study in murine model systems (48). Cohorts of WT and PRL-3 KO mice were subjected to AOM-DSS treatment for 12 to 16 weeks (48). The tumors in the colons of the two cohorts were similar in size but PRL-3 KO mice had significantly fewer tumors at 16 weeks treatment than WT mice (48). These results provide evidence implicating PRL-3 in colon carcinogenesis. The involvement of PRL-2 in cancer was evaluated in breast cancer–prone MMTV-ErbB2 transgenic mice (49). MMTV-PRL-2 transgenic mice engineered to overexpress PRL-2 in mammary tissue did not exhibit spontaneous tumorigenesis, but they exhibited an accelerated development of mammary tumors initiated by introduction of an MMTV-ErbB2 transgene (49). The results support that PRL-2 plays a role in breast cancer progression.
5. Cell Function
PRL-1 was originally identified as an immediate early gene, whose expression was induced during liver regeneration after partial hepatectomy (50). Subsequently, PRLs were found to be elevated in many tumor cell lines, and cells expressing high levels of PRLs exhibited enhanced proliferation and anchorage-independent growth (8, 50, 51, 52, 53, 54).
It became clear from accumulated studies in human metastatic lesions that PRL-1, −2, and −3 also play an important role in cancer progression. The first insights to the molecular basis for PRL involvement in metastasis and cancer was from a study published in 2003 (22). Using a Chinese Hamster Ovary (CHO) cell line stably expressing Myc-tagged-PRL-1, −3, or β-Gal (control), the authors ascertained the effect of PRL-1 and −3 overexpression on cell mobility and cell invasiveness (22). PRL expressing cells were found to be significantly more mobile and invasive than the controls (22). Additionally, cells expressing a catalytically inactive mutant PRL showed significantly less activity and movement than WT controls, confirming that PRL-3 played an important role in cellular migration and that its ability to induce migration is dependent on its phosphatase activity (22). To assess whether or not cells overexpressing PRL could induce metastatic lesions, Myc-PRL-1, −3, and β-Gal CHO cells were injected into the tail vein of ten-week-old female nude mice (22). All mice that received injections of either myc tagged PRL-1 or PRL-3 developed numerous lung metastases while none of the control mice injected with cells expressing β-Gal cells developed tumors (22). These results were the first to show that PRL overexpression could induce metastasis in vivo. Several papers were published later with similar results in both mouse melanoma and HEK293 cells lines (5,9,50,55,56,57,58,59). All studies showed evidence that increasing PRL-3 expression in cells resulted in increased cell adhesion, migration, invasiveness, and proliferation both in vivo and in vitro. The oncogenic effect was also shown to be dependent on PRL-3 phosphatase activity, as no phenotype was observed when cell were transfected with PRL-3 constructs whose active site was mutated (60). Sun et al. extended this further by showing that PRL trimerization and the presence of the C-terminal polybasic regions were also required to incite the oncogenic phenotype (7). For some time after the discovery of the PRLs, most of the interest had been focused on PRL-3 and −1. Recently, however, a study by Wang et al. revealed that PRL-2 can also affect cell migration and invasion (46). When PRL-2 was knocked down by short hairpin RNA (shRNA) or siRNA, A549 cells (a lung cancer cell line that naturally has high PRL expression) migrated slower through Transwell migration and Matrigel invasion chambers than control cells did (46). This effect could be rescued by transfecting a siRNA-resistant mutant PRL-2 construct into cells with (siRNA) silenced PRL-2 (46). Also, like PRL-1 and −3, PRL-2 constructs with mutated catalytic sites and/or CAAX domains could not induce increased cell migration and invasion (46).
Along with cell migration, invasion, adhesion, and proliferation PRLs have been shown to affect cell apoptosis and angiogenesis. Three papers reveal a novel connection between PRLs and p53. Basak et al. show that PRL-3 is up-regulated in a p53-dependent manner after submitting control and Doxorubicin (DNA damaging agent)-treated murine embryonic fibroblast (MEF) cells to a microarray analysis (61). A scan of the PRL-3 locus revealed two p53 binding sites, one of which p53 was shown to bind to directly via chromatin immunoprecipitation (61). PRL-3 expression in MEFs result in fewer BrdU positive cells which is indicative of cell cycle arrest (61). PRL-3 overexpression was also able to arrest HT1080 cells but failed to arrest RKO and U2OS cancer cells (61). The molecular mechanism for cell cycle arrest was shown to be dependent on increased Akt activation via overexpression of PRL-3, followed by negative feedback regulation of the PI3K/Akt pathway and subsequent transcription of growth arresting genes (specifically Foxo, p21, and p27) (61) (Figure 2). Surprisingly, Basak et al. found that knocking down PRL-3 did not produce the opposite effect, but instead resulted again in cell cycle arrest (61). Since down-regulating PRL-3 in p53-null MEF cells did not reproduce this effect it was concluded cell cycle arrest in PRL-3 ablated cells was due to increased p53 activity (61). Furthermore, p19Arf was found to be up-regulated in PRL-3 ablated MEF cells providing a potential molecular mechanism for p53-dependent cell cycle arrest via p19Arf sequestration of MDM2 (negative regulator of p53) (61) (Figure 2).
Figure 2.
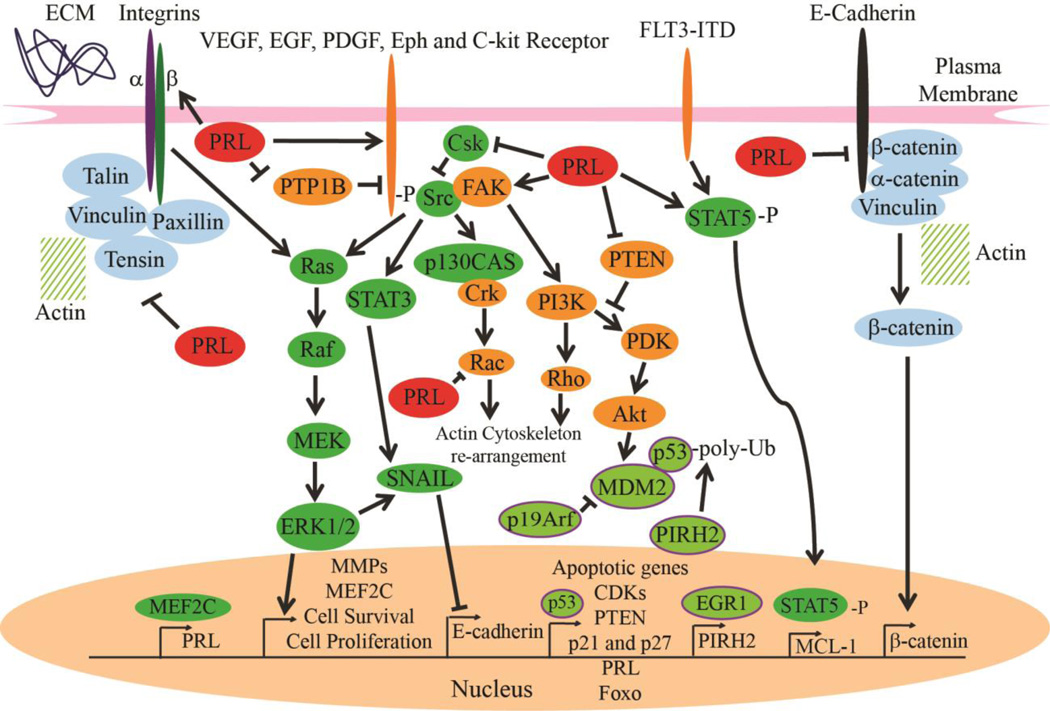
Cell Signaling Pathways Regulated by PRLs: A summarization of the most important oncogenic and tumor suppressive pathways that PRL has been shown to effect including Src/ERK1/2, p53, PTEN/PI3K/Akt, Adhesion proteins, and growth factor receptors.
Two papers by Min et al. showed that p53 could also be regulated by PRL-1 and −3 in cancer cells (Table 2). Up-regulation of PRL-1 and −3 can both inhibit p53 and p53-mediated apoptosis individually with the opposite being true when PRL-1 or −3 is knocked down (62,63). Western blot analysis of PRL-1 or −3 overexpressing cells reveals that apoptosis inhibition can be caused by PRL-mediated activation of MDM2 via PI3K activation and PIRH2 (p53 ubiquitinase) transcription via EGR1 activation (62,63) (Figure 2). Min et al. did not encounter the same cell cycle arrest phenotype that was discovered by Basak et al., however, this may be explained by carcinogenic defects in cancer cell lines used by Min et al (62,63).
Table 2.
Summary of potential PRL effector molecules.
| PRL effector molecule |
Effect of PRL Overexpression |
Reference |
|---|---|---|
| p53 | Inhibition | (62,63) |
| Src | Activation | (54,67,68) |
| ERK1/2 | Activation | (54,67,68) |
| STAT3 | Activation | (54,67,68) |
| p130Cas | Activation | (54,67,68) |
| Csk | Inhibition | (54,67) |
| MMPs | Activation | (68) |
| FAK | Activation (PRL1) | (68) |
| Rho A and C | Activation | (10) |
| Rac | Inhibition | (10) |
| Integrin α | Inhibition | (71,72,73) |
| Integrin β | Enhance Src interaction |
(73,74, 75) |
| c-fos | Inhibition | (71) |
| Actin Cytoskeleton |
Uncoupling from adhesive proteins |
(69) |
| E-Cadherin | Inhibition | (70) |
| γ-catenin | Inhibition | (70) |
| Vinculin | Inhibition | (70) |
| Fibronectin | Protein level Up- regulation |
(70) |
| Snail | Protein Level Up- regulation |
(70) |
| PTEN | Inhibition | (70,76,77) |
| Akt | Activation | (70,76,77) |
| PDGFR | Activation | (75) |
| Eph | Activation | (75) |
| EGFR | Activation | (78) |
| PTP1B | Inhibition | (78) |
| KCNN4 | Activation | (79) |
| Phosphoinositides | Dephosphorylation | (80) |
| miR 21,17,19a | Induction | (81) |
| miR 495, 551a | Induction | (82) |
| STAT5 | Activation | (43) |
| Histone Deacetylase 4 |
Activation | (43) |
| Mcl-1 | Induction | (43) |
PRL-3 has been found to be strongly expressed in tumor vasculature and noted by many early PRL studies that it may play a role in induction of angiogenesis (5,35,60,64,65). Early studies have shown that PRL-3 overexpression in human umbilical vascular endothelial cells (HUVEC) can induce tube formation and a recent paper by Xu et al. explored the molecular mechanism in detail (66). In this study, vascular endothelial growth factor (VEGF) is shown to induce the transcription of PRL-3 in HUVEC cells via MEF2C transcription factor (Figure 2), which was shown to bind to the PRL-3 promoter both in vitro and in vivo (66). Furthermore, VEGF could not induce PRL-3 expression when MEF2C was knocked down by siRNA and PRL-3 knockdown in HUVEC resulted in compromised tube formation (66).
6. Signaling mediated by PRL
Since its discovery as an oncogene in colorectal cancer, PRL-3 has been exhaustively researched in cell culture. The most confounding factor in many of these studies is the lack of a putative substrate. Despite this PRL-3 has been shown to alter several major cell signaling pathways, including PTEN and Src, making it an alluring target for study and cancer treatment (Summarized in Table 2). One of the first mechanistic pathways to be associated with PRL is Src and its downstream targets (Figure 2.). A study by Liang et al. showed that when PRL-3 was overexpressed in HEK293 cells Src kinase activity was increased by 180% in comparison to control and PRL-3 C104S mutant cells (54). Downstream targets ERK1/2, STAT3, and p130Cas also experienced increased phosphorylation, which is consistent with increased Scr activity (54) (Figure 2). Purified PRL-3 was not able to phosphorylate Src directly; however, Csk protein (negative regulator of Src) expression was shown to be significantly reduced in cells overexpressing PRL-3 (54) (Figure 2). Indeed, when Csk was reconstituted to normal levels, via tetracycline expression in cells overexpressing PRL-3, Src phosphorylation and the oncogenic phenotype of PRL-3 was negated (54). A follow up study on the activity of PRL-3 on Csk revealed that PRL-3 could inhibit Csk translation due to increased phosphorylation of Ser-51 of eIF2α (67). Subsequent studies indicated that PRL-1 can also activate Src and its downstream targets (67,68). Unlike PRL-3, PRL-1 activation increased Tyrosine 416 phosphorylation as opposed to Tyr527 and increased FAK activation along with pre-established p130Cas, ERK1/2, and STAT3 (68) (Figure 2). Furthermore, active forms of matrix metalloproteinases 2 and 9 (MMP 2 and 9) were expressed at higher levels than in control cells (68). MMP2 and 9 are regulated by ERK1/2, can degrade collagen of the basement membrane, and are often found overexpressed in cancer, providing another mechanism by which PRL can induce cell migration/invasion (68) (Figure 2).
A study by Fiordalisi et al. shows that PRLs can regulate Rho and Rac activity in SW480 cells (10) (Figure 2.). Rho A, C, and Rac are known to promote actin polymerization associated with cell mobility (10). In cells that have been transfected with PRL-1 and −3 overexpression vectors, RhoA, and RhoC expression levels increased by 4- to 7-fold while Rac was reduced 60 to 70 percent (10). Further testing showed that, in order to promote cell invasion and mobility in a Rho-dependent manner, the presence of Rho effector ROCK and PRL phosphatase activity is required (10). This report correlates with other studies that assert that PRL can affect actin cytoskeleton dynamics (69).
PRL is also known for interacting with adhesive proteins to modulate cell migration and invasiveness (69,70). It has been shown that PRL can interact with integrin proteins (71,72,73,74,75) (Figure 2). When up-regulated, PRL can dephosphorylate integrin β, enhancing integrin β binding with Src kinases (which, in turn, decreases the coupling of integrins to actin and impairs integrin-mediated adhesion) (73, 74, 75). Furthermore, PRL overexpression has been shown to suppress integrin α (but not integrin β) expression levels (71,72,73,74). The down-regulation of integrin α by PRL-3 is hypothesized to be carried out via inhibition of c-fos and that integrin α binding to PRL can inhibit its interaction with the β subunit (71,72,73,74). E-cadherin, γ-catenin, and vinculin are also down-regulated with PRL-3 over-expression, while mesenchymal markers fibronectin and Snail are up-regulated (70). The adhesive proteins are upstream of PI3K/Akt, which has been shown to be up-regulated in DLD-1 cells overexpressing PRL-3 (70) (Figure 2). The mechanism for this action may be the result of PTEN downregulation (70). PTEN is a negative regulator of the PI3K pathway and is down-regulated in DLD-1 cells over-expressing PRL-3 (70). PRL-2’s ability to suppress PTEN level and promote PI3K/Akt pathway activation is also documented in PRL-2 deficient murine models (76,77).
PRL-mediated Src and PTEN/Akt activation can also be explained by upstream activity with receptor tyrosine kinases (RTKs) (Figure 2). PRL-3 has been shown to up-regulate PDGFR, Eph, and integrin receptor array in a proteomic analysis of HEK293 cells overexpressing PRL-3 (75). Phosphoproteomic data support intracellular activation of an extensive signaling network normally governed by extracellular ligand-activated transmembrane growth factor, cytokine, and integrin receptors in the PRL3 cells (75). Another study also revealed evidence of PRL-3 induced activation of EGFR in A431 epidermoid carcinoma cells (78). PRL-3 overexpression induced a state of EGFR addiction in both cell lines and in patient tumor sample, causing hypersensitivity to EGFR inhibition (78). It was concluded that PRL-3 regulates EGFR by transcriptionally down-regulating PTP1B, causing EGFR hyperphosphorylation and activation (78) (Figure 2). These studies are of particular note due to the fact that the effect of PRL-3 on receptor tyrosine kinases is able to explain and corroborate established PRL literature that PRL alters Src/ERK, PI3K/Akt, and actin/adhesive protein dynamics (75,78).
Several independent studies also show that PRL effects some additional pathways including upregulation of KCNN4 potassium channels (79), activity toward phosphoinositides (80), induction of micro RNAs (miR) 21, 17, 19a (81), and interaction with miR-495 and miR551a (82). One study proposes a novel role of PRL-3 downstream of an internal tandem duplication mutant of fms-like tyrosine kinase 3 (FLT3-ITD) present in approximately 25% of AML patients (43). In an attempt to rectify FLT3-ITD-positive AML drug resistance, Zhou et al. treated AML cell lines with both FLT3 inhibitor ABT-869 and a histone deacetylase inhibitor suberoylanilide hydroxamic acid (SAHA) and were successful in reducing AML colony formation and inducing apoptosis in tumor cells (43). Further studies revealed that PRL-3 was strongly downregulated in cells that received combined ABT-869 and SAHA treatment and showed evidence that PRL-3 may be responsible for FTL3 drug resistance through activation of the Stat-pathway, interaction with histone deacetylase 4, and upregulation of Mcl-1 (a protein that has been shown to incite tumor development and maintenance in many different cancers including hematological malignancies) (Figure 2.) (43). In light of this evidence, PRL-3 may prove to be a potent target for treatment of drug-resistant AML.
7. Physiological Roles of PRL
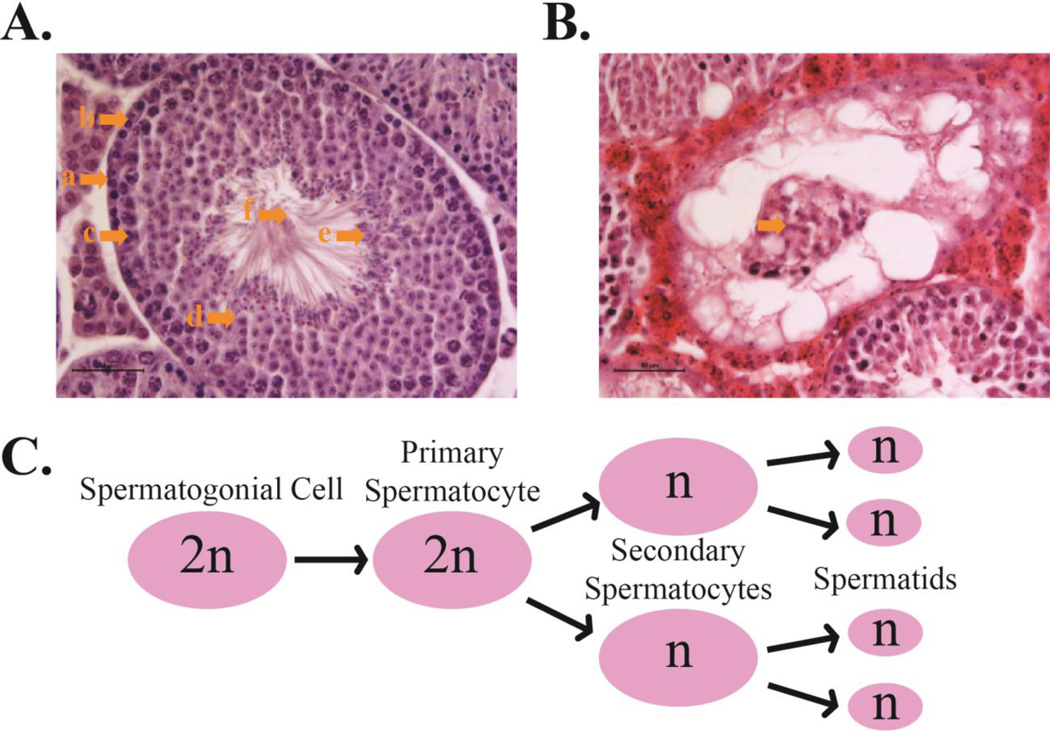
In order to study the physiological role of PRL in normal and cancerous tissue several genetically modified model organisms were created. Deletion of PRL2, the most ubiquitously expressed PRL family member, leads to retarded growth at both embryonic and adult stages. (76). This finding is congruent to evidence presented by previous studies that PRL plays a role in cell proliferation. Interestingly, another phenotype made itself evident in the PRL-2 KO lines in the form of placental hypotrophy in females (76). Immunohistochemistry staining of Ki67 revealed that placental hypotrophy was caused by a reduction of cell proliferation, especially in the spongiotrophoblast layer (76). Glycogen cells were also significantly reduced in the PRL-2 KO placenta raising the possibility of a nutrient defect in PRL-2 KO fetuses (76). Western Blot analysis of the whole placenta lysate showed a 70% decrease in Akt phosphorylation along with a 1.7-fold increase in PTEN in KO mice in comparison to WT (76). This data was corroborated in HEK 293 cells overexpressing PRL-2 (in this case the effect on PTEN and Akt phosphorylation was the reverse of that in placental tissue) (76). Furthermore, severe and progressive loss of fertility as well as hypotrophic testis were observed in male PRL-2 KO mice (77). Within the testis, spermatogenesis occurs in seminiferous tubules, which normally have a very distinct cellular structure with spermatogonia stem cells lining the basal lamina, followed by primary and secondary spermatocytes, and terminating with mature spermatids ready to be ejected into the lumen (Figure 3) (83). Mature sperm are then drained into the epididymis where they are stored until ejaculation (83). Histological cross-sections of PRL-2 KO seminiferous tubules show reduced cellularity in comparison to WT mice accounting for the small testis size (77). By the time the mice reach six months of age, male fertility and sperm count is severely reduced and testis cross-sections show large sections of seminiferous tubules that have completely shed their germ cells into the lumen (Figure 3) (77). Unlike the placenta however, the KO testis showed no decrease in cell proliferation (77). Instead, the cause of testicular atrophy in PRL-2 KO mice was due to a significant increase in apoptosis (77). This led to the hypothesis that the loss of fertility and hypo-cellularity of the testis was due to a failure to maintain spermatogonia stem cells as a direct result of the loss of PRL-2 (77). Furthermore, c-kit signaling, which is responsible for maintaining the self-renewal of spermatogonia (84), was also found to be attenuated in a post-transcriptional manner in PRL-2 null germ cells. PTEN, a negative regulator of c-kit signaling, was elevated in PRL-2 null germ cells (Figure 2) (84,77). More recent studies reveal that PRL-2 deficiency also impairs hematopoietic stem cell (HSC) self-renewal (85). Moreover, PRL-2 null hematopoietic stem and progenitor cells (HSPCs) are more quiescent and show reduced activation of the Akt and ERK1/2 signaling as a result of an increase in PTEN level (85). Furthermore, PRL-2 is found to be important for SCF-mediated HSPC proliferation and loss of PRL-2 decreased the ability of oncogenic KIT/D814V mutant to promote hematopoietic progenitor cell proliferation (85). Thus, PRL-2 plays critical roles in regulating HSC self-renewal and mediating SCF/Kit signaling (85,77). Together these findings establish that PRL-2 is required for a number of developmental processes (placenta formation, spermatogenesis, HSC self-renewal) and reveal a novel mechanistic connection between PRL-2 and PTEN (76,77,85). Given the strong cancer susceptibility to subtle variations in the level of PTEN, the ability of PRL-2 to repress PTEN expression qualifies it as an oncogene and a novel target for developing anti-cancer agents.
Figure 3.
Spermatogenesis and PRL-2 depletion: A. Healthy seminiferous tubule cross-section of a 3-month old male mouse. The seminiferous tubules sport a distinct cellular structure. a: Basal Lamina, b: Spermatogonial cells, c: Primary Spermatocytes, d: Secondary spermatocytes, e: Spermatids and mature sperm about to eject into the lumen, F: Lumen and maturing sperm tails. B. This is the seminiferous tubule of a 6-month old PRL-2 KO mouse. This cross-section shows complete shedding of the germ cells into the lumen (as indicated). C. Spermatogenesis procession.
8. Strategies to target PRL
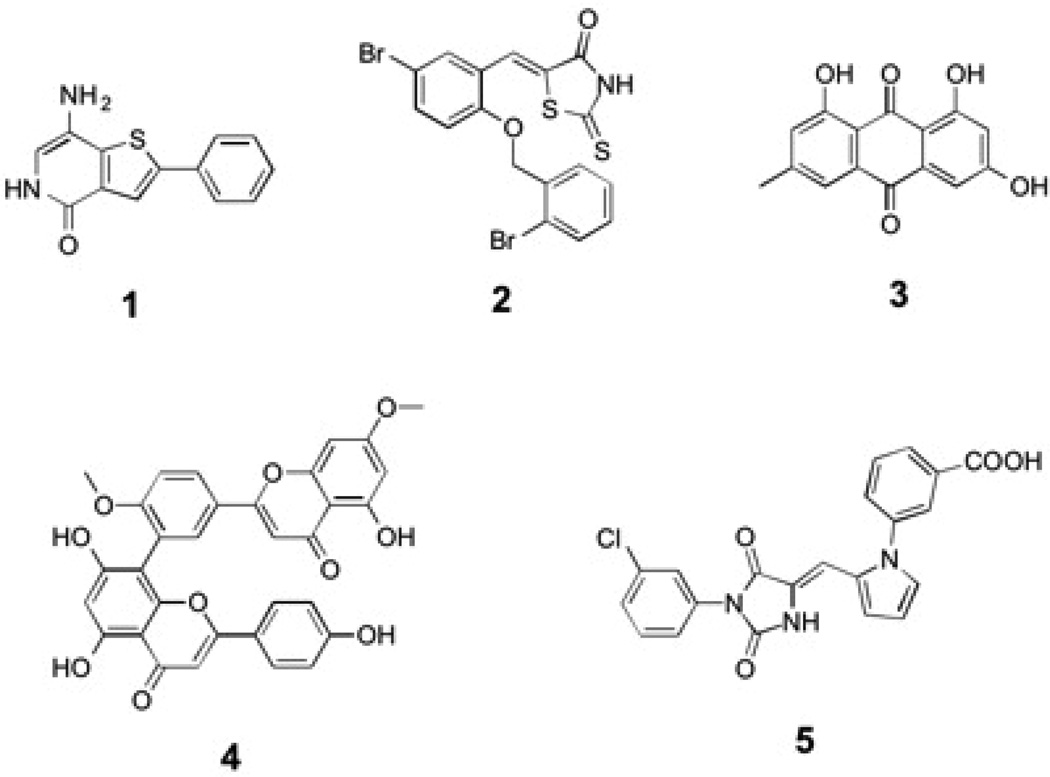
Given the roles of PRL in tumor progression and metastasis, there is increasing interest in targeting the PRLs for novel anti-cancer agents. Although PRL’s wide and shallow active site makes inhibitor specificity with small molecules very difficult, several small molecules have been reported to inhibit PRL activity. The most noteworthy is compound 1 (Figure 4), which was identified from a high-throughput screen of the Roche chemical library (86). Compound 1 inhibits all PRLs with IC50 values in the 100–300 nM range (86). It exhibits very high selectivity toward the PRLs, with minimal activity when tested in vitro against a panel of 11 other phosphatases (86). Furthermore, compound 1 is able to inhibit anchorage independent cell growth of colon cancer cells and suppress the migration of HUVEC cells, through a mechanism involving proteolytic cleavage of p130Cas (86). Compound 2 (Figure 4), a benzylidene rhodanine derivative, inhibits PRL-3 activity with an IC50 of 0.9 µM [2] (87). Compound 2 is able to block the invasion of B16F19 mouse melanoma cells, which express high levels of endogenous PRL-3 (87). An anthraquinone derivative, emodin (compound 3, Figure 3), inhibits PRL-3 and reduces cell migration and invasion of PRL-3-DLD-1 colon cancer cells (88). A biflavonoid (ginkgetin, compound 4, Figure 4) extracted from Taxus cuspidate also showed moderate PRL-3 inhibitory activity (89). Finally, compound 5 (Figure 4) was identified as a PRL-3 inhibitor using a structure-based virtual screening approach (90). Although some of these compounds can serve as promising leads for further development, further studies are required to correlate PRL inhibition with the anti-tumor activity.
Figure 4.
Small molecule PRL inhibitors. Compounds 1–5 as described in the text.
An alternative to inhibition of the phosphatase activity is interference with PRL trimerization. One interesting and unique feature of PRL-1 is that it forms trimers in crystalline state (7,91) (Figure 1). The domain on which trimerization occurs is away from the active site and on the structurally conserved C-terminal end (7,91). Crosslinking experiments using HA-tagged PRL-1 and PRL-3 showed that PRL does indeed form trimers inside the cells (7). Furthermore, trimerization was shown to be essential for PRL function (7). When PRL-1 proteins with mutated residues on the trimerization domain were stably overexpressed in cells lines the oncogenic increase in cell proliferation and migration due to PRL overexpression was rescued (7). Mutation of the trimer interface residues did not impede the ability of PRL-1 to associate with the membrane nor did the concentration of PRL-1 affect the catalytic rate constant in vitro (7). The notion that one can disrupt the function of a PTP by targeting trimerization and other unique regulatory domains instead of the catalytic site is a novel and exciting concept, since many of the active sites of PTPs are conserved and it can be difficult to obtain a drug that is specific for a single PTP (1,2).
Another method of therapeutically targeting PRL proteins is through the use of specific antibodies. For many years chemical inhibition or stimulation has dominated the medical field as the premier method for treating cancer and other diseases. Within the past decade, however, new and exciting methods of treatment have emerged in the form of gene therapy and antibody treatment (92). The use of antibodies as a powerful diagnostic tool and as a way to use the body’s own immune system to combat disease has become one of the hottest fields of therapeutic research (92). Three proof-of-concept papers show that injectable antibodies against intracellular PRL-1 and PRL-3 are able to prevent experimental metastases when mice are injected with cancer cells that express high PRL [92,93,94]. Mice with PRL-1 or −3 overexpressing cells had larger tumors in comparison to WT and vector controls (92,93,94). When treated with PRL-1 or −3 antibody, however, the tumor size became dramatically reduced (92,93,94).
9. Conclusion
The PRL family consists of novel DSPs found to be overexpressed in many cancers. The main cellular mechanisms of PRL in cancer is to induce cell proliferation, survival, invasion, and epithelial to mesenchymal transition. Due to the fact that PRL plays a causative role in cancer progression and metastasis and affects so many important oncogenic and tumorigenic pathways it makes for a very tempting therapeutic target for cancer treatment and possibly other conditions (i.e. male fertility). Some success has been made using small molecules to target PRL as well as two novel approaches. Overall, PRL shows much potential for development as a drug target.
10. Expert Opinion
Phosphatase of Regenerating Liver is a promising, novel oncogene in the field of cancer research. Increasing amounts of evidence indicate that PRLs interact with important oncogenes and tumor suppressors such as PTEN, p53, and Src and show much promise as a cancer therapy target. In addition to small molecules aimed at the active site, therapeutic targeting of the trimer interface for PRL inhibitor development and the use of PRL-specific antibodies are both novel and promising strategies for PRL-based anti-cancer agents. However, there is much work to be done before treatment in human cancer patients can be considered. The most significant hurdle that needs to be overcome before we can reach a genuine understanding of the function of PRL is the lack of a putative substrate. Many potential substrates and pathways have been proposed but none have been confirmed by dephosphorylation with PRL using traditional enzyme assays. The reason for this may be, as stated above, due in part to PRL’s relative inactivity in vitro. PRL is known to associate with the plasma membrane and that this association is necessary for its function as a protein phosphatase. The reason for its inactivity in vitro may indicate that PRL requires molecular interactions that can only be attained in a complex, by the formation of PRL trimers, or with an as of yet unidentified co-factor. However, in vivo substrate trapping is a viable method for discovering the substrate(s) of PRL when combined with mass spectrometry and cell-based assays (95). These trapping mutants are made by mutating key residues in the catalytic domain such that the enzyme can recognize and bind to its substrate but cannot dephosphorylate and release it (95). The in vivo method involves either a transient or stable transfection of tagged, substrate trapping mutant plasmids into a living cell to incubate before lysis and extraction of the mutant protein via immunoprecipitation (95). For the in vitro method, the mutated protein is tagged and conjugated to beads and then applied to cell lysate where it is incubated with free floating proteins (95). There are advantages and disadvantages to both methods. The in vivo method allows for the mutated protein to interact with substrates in a physiologically relevant cellular environment but can be difficult to scale up enough to achieve a novel and observable substrate band, especially if the substrate is not abundant in normal cellular conditions (95). Conversely, the in vitro method is easy to scale up for mass spectrometry or western blot analysis but may cause non-specific interactions (95). Practically, both methods are used, along with mass spectrometry and cell assays to maximize the chance of capturing and identifying a physiological substrate.
Another hurdle that confounds the ability to effectively study the PRLs is, in fact, their significant homology to each other. In terms of the effort to dissect the physiological role of PRL, no antibody exists as of yet that can completely differentiate between PRL-1 and 2 making it difficult to determine differential protein expression and function between the two. Also, since the PRLs may have similar and potentially overlapping roles, isoform specific knockout mice would be very useful to dissect PRL functions. Knockout mouse models for PRL-2 and PRL-3 have recently been reported (48,76) and it is likely that a PRL-1 knockout model will be available in the near future. Crossing these strains to generate multiple null mutations could add considerable insight into the in vivo function of this gene family.
In conclusion, there is a lack of detailed knowledge regarding the physiological function of PRLs that needs to be resolved before clinical drug development can be attempted. However, the ability to target and inhibit PRLs in vivo is invaluable in the effort to dissect the physiological function of these important oncogenes and to evaluate the therapeutic potential of targeting the PRLs. PRLs have been shown to greatly impact cellular growth, apoptosis, stem cell renewal, and can regulate several important oncogenes, tumor suppressors, and regulators of metabolism. Furthermore, there is evidence in murine models that PRL is critically important for proper embryonic development and cancer progression. The knowledge gained from studying the physiological and mechanistic aspects of PRL will be essential in facilitating the development of new therapeutic strategies for cancer and other diseases.
Highlights.
Phosphatases of Regenerating Liver are novel, dual specificity phosphatase and oncogenes
PRLs are found significantly overexpressed in many cancers including colorectal cancer, breast cancer, and gastric carcinoma.
PRLs have been shown to interact with major oncogenic and tumor suppressor cell signaling pathways such as PTEN, PI3K/Akt, p53, and Src.
PRL plays a role in number of developmental processes (placenta, spermatogenesis, hematopoietic stem cell self-renewal).
PRL can be inhibited by active site-directed inhibitors as well as novel strategies that include vaccination by antibodies and disruption of PRL trimerization.
Acknowledgment
This work was supported in part by National Institutes of Health Grants CA69202 and CA152194.
Abbreviations
- PRL
Phosphatase of Regenerating Liver
- PTEN
phosphatase and tensin homologue deleted on chromosome 10
- RTK
Receptor Tyrosine Kinase
- EGFR
Epidermal Growth Factor Receptor
- PTPs
Protein Tyrosine Phosphatases
- DSPs
Dual Specificity Phosphatases
- PTP1B
Protein Phosphatase 1B
- CRC
colorectal cancer
- SAGE
serial analysis of gene expression
- DCIS
ductal carcinoma in situ
- MEF
Mouse Embryonic Fibroblasts
REFERENCES
- 1.Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nature Reviews Molecular Cell Biology. 2006;7.11:833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 2.Zhang ZY. Protein-tyrosine phosphatases: Biological function, structural characteristics, and mechanism of catalysis. Critical Reviews in Biochemistry and Molecular Biology. 1998;33:1–52. doi: 10.1080/10409239891204161. [DOI] [PubMed] [Google Scholar]
- 3. Mohn KL, Laz TM, Hsu JC, et al. The immediate-early growth response in regenerating liver and insulin-stimulated H-35 cells: comparison with serum-stimulated 3T3 cells and identification of 41 novel immediate-early genes. Molecular and Cellular Biology. 1991;11:381–390. doi: 10.1128/mcb.11.1.381. This paper records the discovery of PRL-1 in regenerating liver.
- 4. Zeng Q, Hong W, Tan YH. Mouse PRL-2 and PRL-3, two potentially prenylated protein tyrosine phosphatases homologous to PRL-1. Biochemical and Biophysical Research Communications. 1998;244:421–427. doi: 10.1006/bbrc.1998.8291. PRL 2 and 3 are discovered by Zeng et al.
- 5.Bessette DC, Qiu D, Pallen CJ. PRL PTPs: Mediators and markers of cancer progression. Cancer Metastasis Reviews. 2008;27(2):231–252. doi: 10.1007/s10555-008-9121-3. [DOI] [PubMed] [Google Scholar]
- 6. Sun JP, Wang WQ, Yang H, et al. Structure and biochemical properties of PRL-1, a phosphatase implicated in cell growth, differentiation, and tumor invasion. Biochemistry. 2005;44:12009–12021. doi: 10.1021/bi0509191. One of the first structural analyses of PRL-1
- 7. Sun JP, Luo Y, Yu X, et al. Phosphatase activity, trimerization, and the C-terminal polybasic region are all required for PRL1-mediated cell growth and migration. Journal of Biological Chemistry. 2007;282:29043–29051. doi: 10.1074/jbc.M703537200. Discovery of trimer form and the catalytic domain, CAAX motif and polybasic region found to be essential for PRL activity.
- 8.Cates CA, Michael RL, Stayrook KR, et al. Prenylation of oncogenic human PTP(CAAX) protein tyrosine phosphatases. Cancer Letters. 1996;110:49–55. doi: 10.1016/s0304-3835(96)04459-x. [DOI] [PubMed] [Google Scholar]
- 9.Fagerli UM, Holt RU, Holien T, et al. Overexpression and involvement in migration by the metastasis-associated phosphatase PRL-3 in human myeloma cells. Blood. 2008;111:806–815. doi: 10.1182/blood-2007-07-101139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fiordalisi JJ, Keller PJ, Cox AD. PRL tyrosine phosphatases regulate rho family GTPases to promote invasion and motility. Cancer Research. 2006;66:3153–3161. doi: 10.1158/0008-5472.CAN-05-3116. Records interaction with Rho family GTPases.
- 11.Kato H, Semba S, Miskad UA, et al. High expression of PRL-3 promotes cancer cell motility and liver metastasis in human colorectal cancer: A predictive molecular marker of metachronous liver and lung metastases. Clinical Cancer Research. 2004;10:7318–7328. doi: 10.1158/1078-0432.CCR-04-0485. [DOI] [PubMed] [Google Scholar]
- 12. Radke I, Gotte M, Kersting C, et al. Expression and prognostic impact of the protein tyrosine phosphatases PRL-1, PRL-2, and PRL-3 in breast cancer. British Journal of Cancer. 2006;95:347–354. doi: 10.1038/sj.bjc.6603261. PRL’s relevance in breast cancer.
- 13. Zeng Q, Si X, Horstmann H, et al. Prenylation-dependent association of protein-tyrosine phosphatases PRL-1, −2, and −3 with the plasma membrane and the early endosome. Journal of Biological Chemistry. 2000;275:21444–21452. doi: 10.1074/jbc.M000453200. PRL was found to associate to the membrane and early endosome.
- 14.Ishii T, Funato Y, Miki H. Thioredoxin-related protein 32 (TRP32) specifically reduces oxidized phosphatase of regenerating liver (PRL) Journal of Biological Chemistry. 2013;288(10):7263–7270. doi: 10.1074/jbc.M112.418004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Skinner AL, Vartia AA, Williams TD, Laurence JS. Enzyme activity of phosphatase of regenerating liver is controlled by the redox environment and its C-terminal residues. Biochemistry. 2009;48(20):4262–4272. doi: 10.1021/bi900241k. The enzymatic activity of PRL was first found to be regulated by redox environment.
- 16.Yu L, Kelly U, Ebright JN, et al. Oxidative stress-induced expression and modulation of Phosphatase of Regenerating Liver-1 (PRL-1) in mammalian retina. Biochimica et Biophysica Acta. 2007;1773:1473–1482. doi: 10.1016/j.bbamcr.2007.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dumaual CM, Sandusky GE, Crowell PL, Randall SK. Cellular localization of PRL-1 and PRL-2 gene expression in normal adult human tissues. Journal of Histochemistry and Cytochemistry. 2006;54:1401–1412. doi: 10.1369/jhc.6A7019.2006. First investigation into the expression pattern of PRL-1 and -2 in human tissue.
- 18. Matter WF, Estridge T, Zhang C, et al. Role of PRL-3, a human muscle-specific tyrosine phosphatase, in angiotensin-II signaling. Biochemical and Biophysical Research Communications. 2001;283:1061–1068. doi: 10.1006/bbrc.2001.4881. First investigation into the expression pattern of PRL-3 in human tissue
- 19. Lin MD, Lee HT, Wang SC, et al. Expression of Phosphatase of regenerating liver family genes during embryogenesis: an evolutionary developmental analysis among Drosophila, amphioxus, and zebrafish. BMC Developmental Biology. 2013;13(1):1–13. doi: 10.1186/1471-213X-13-18. One of the first reports of PRL-3 overexpression in colorectal cancer.
- 20.Saha S, Bardelli A, Buckhaults P, et al. A phosphatase associated with metastasis of colorectal cancer. Science. 2001;294:1343–1346. doi: 10.1126/science.1065817. [DOI] [PubMed] [Google Scholar]
- 21.Tamagawa H, Oshima T, Yoshihara K, et al. The expression of the phosphatase of regenerating liver 3 gene is associated with outcome in patients with colorectal cancer. Hepato-Gastroenterology. 2012;59(119):2122–2126. doi: 10.5754/hge11996. [DOI] [PubMed] [Google Scholar]
- 22. Zeng Q, Dong JM, Guo K, et al. PRL-3 and PRL-1 promote cell migration, invasion, and metastasis. Cancer Research. 2003;63:2716–2722. Early evidence linking PRL to cell migration and invasion.
- 23.Wang L, Peng L, Dong B, et al. Overexpression of phosphatase of regenerating liver-3 in breast cancer: Association with a poor clinical outcome. Annals of Oncology. 2006;17:1517–1522. doi: 10.1093/annonc/mdl159. [DOI] [PubMed] [Google Scholar]
- 24.Hao RT, Zhang XH, Pan YF, et al. Prognostic and metastatic value of phosphatase of regenerating liver-3 in invasive breast cancer. Journal of cancer research and clinical oncology. 2012;136.9:1349–1357. doi: 10.1007/s00432-010-0786-y. [DOI] [PubMed] [Google Scholar]
- 25.Bilici A, Ustaalioglu BB, Yavuzer D, et al. Prognastic significance of high phosphatase of regenerating Liver-3 expression in patients with gastric cancer who underwent curative gastrectomy. Digestive Diseases and Sciences. 2012;57(6):1568–1575. doi: 10.1007/s10620-012-2076-9. [DOI] [PubMed] [Google Scholar]
- 26.Dai N, Lu AP, Shou CC, Li JY. Expression of phosphatase of regenerating liver 3 is an independent prognostic indicator for gastric cancer. World Journal of Gastroenterology. 2009;15(12):1499–1505. doi: 10.3748/wjg.15.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li ZR, Wang Z, Zhu BH, et al. Association of tyrosine PRL-3 phosphatase protein expression with peritoneal metastasis of gastric carcinoma and prognosis. Surgery Today. 2007;37:646–651. doi: 10.1007/s00595-006-3437-9. [DOI] [PubMed] [Google Scholar]
- 28.Matsukawa Y, Semba S, Kato H, et al. Constitutive Suppression of PRL-3 inhibits invasion and proliferation of gastric cancer cell in vitro and in vivo. Pathobiology. 2010;77(3):155–162. doi: 10.1159/000292649. [DOI] [PubMed] [Google Scholar]
- 29.Miskad UA, Semba S, Kato H, Yokozaki H. Expression of PRL-3 phosphatase in human gastric carcinomas: close correlation with invasion and metastasis. Pathobiology. 2004;71:176–184. doi: 10.1159/000078671. [DOI] [PubMed] [Google Scholar]
- 30.Miskad UA, Semba S, Kato H, et al. High PRL-3 expression in human gastric cancer is a marker of metastasis and grades of malignancies: An in situ hybridization study. Virchows Archiv. 2007;450.3:303–310. doi: 10.1007/s00428-006-0361-8. [DOI] [PubMed] [Google Scholar]
- 31.Ooki A, Yamashita K, Kikuchi S, et al. Phosphatase of regenerating liver-3 as a prognostic biomarker in histologically node-negative gastric cancer. Oncology Reorts. 2009;21(6):1467–1475. doi: 10.3892/or_00000376. [DOI] [PubMed] [Google Scholar]
- 32.Polato F, Codegoni A, Fruscio R, et al. PRL-3 phosphatase is implicated in ovarian cancer growth. Clinical Cancer Research. 2005;11:6835–6839. doi: 10.1158/1078-0432.CCR-04-2357. [DOI] [PubMed] [Google Scholar]
- 33.Ren T, Jiang B, Xing X, et al. Prognastic significance of phosphatase of regenerating liver-3 expression in ovarian cancer. Pathology Oncology Research. 2009;15(4):555–560. doi: 10.1007/s12253-009-9153-1. [DOI] [PubMed] [Google Scholar]
- 34.Xu Y, Zhu M, Zhang S, et al. Expression and prognostic value of PRL-3 in human intrahepatic cholangiocarcinoma. Pathology Oncology Research. 2012;16(2):169–175. doi: 10.1007/s12253-009-9200-y. [DOI] [PubMed] [Google Scholar]
- 35.Zhao WB, Li Y, Liu X, et al. Evaluation of PRL-3 expression, and its correlation with angiogenesis and invasion in hepatocellular carcinoma. International Journal of Molecular Medicine. 2008;22(2):187–192. [PubMed] [Google Scholar]
- 36.Hassan NM, Hamada J, Kameyama T, et al. Increased expression of PRL-3 gene in human oral squamous cell carcinoma and dysplasia tissues. Asian Pacific Journal of Cancer Prevention. 2011;12(4):947–951. [PubMed] [Google Scholar]
- 37.Ma Y, Li B. Expression of phosphatase of regenerating liver-3 in squamous cell carcinoma of the cervix. Medical Oncology. 2011;28(3):775–780. doi: 10.1007/s12032-010-9514-3. [DOI] [PubMed] [Google Scholar]
- 38.Liu YQ, Li HX, Lou X, Lei JY. Expression of phosphatase of regenerating liver 1 and 3 mRNA in esophageal squamous cell carcinoma. 2008;132(8):1307–1312. doi: 10.5858/2008-132-1307-EOPORL. [DOI] [PubMed] [Google Scholar]
- 39.Achiwa H, Lazo JS. PRL-1 tyrosine phosphatase regulates c-Src levels, adherence, and invasion in human lung cancer cells. Cancer Research. 2007;67:643–650. doi: 10.1158/0008-5472.CAN-06-2436. [DOI] [PubMed] [Google Scholar]
- 40.Yamashita S, Masuda Y, Matsumoto K, et al. Down-regulation of the human PRL-3 gene is associated with the metastasis of primary non-small cell lung cancer. Annals of Thoracic and Cardiovascular Surgery. 2007;13:236–239. [PubMed] [Google Scholar]
- 41.Broyl A, Hose D, Lokhorst H, et al. Gene expression profiling for molecular classification of multiple myeloma in newly diagnosed patients. Blood. 2010;116:2543–2553. doi: 10.1182/blood-2009-12-261032. [DOI] [PubMed] [Google Scholar]
- 42.Yagi T, Morimoto A, Eguchi M, et al. Identification of a gene expression signature associated with pediatric AML prognosis. Blood. 2003;102:1849–1856. doi: 10.1182/blood-2003-02-0578. [DOI] [PubMed] [Google Scholar]
- 43. Zhou J, Bi C, Chng WJ, et al. “PRL-3, a metastasis associated tyrosine phosphatase, is involved in FLT3-ITD signaling and implicated in anti-AML therapy”. PloS one. 2011;6.5:e19798. doi: 10.1371/journal.pone.0019798. PRL may be a potent target for cancer therapy in patients with drug resistant FLT3-ITD-positive AML.
- 44.Zhou J, Wang S, Lu J, et al. Over-expression of phosphatase of regenerating liver-3 correlates with tumor progression and poor prognosis in nasopharyngeal carcinoma. International Journal of Cancer. 2009;124(8):1879–1886. doi: 10.1002/ijc.24096. [DOI] [PubMed] [Google Scholar]
- 45. Li J, Guo K, Koh VW, et al. Generation of PRL-3- and PRL-1-specific monoclonal antibodies as potential diagnostic markers for cancer metastases. Clinical Cancer Research. 2005;11:2195–2204. doi: 10.1158/1078-0432.CCR-04-1984. Creation of PRL-1 and -3 monoclonal antibodies and first recorded relevance of PRL-1 in cancer.
- 46. Wang Y, Lazo JS. Metastasis-associated phosphatase PRL-2 regulates tumor cell migration and invasion. Oncogene. 2012;31(7):818–827. doi: 10.1038/onc.2011.281. First recorded evidence of PRL-2 activity in cancer.
- 47.Dumaual CM, Sandusky GE, Soo HW, et al. Tissue-specific alterations of PRL-1 and PRL-2 expression in cancer. American Journal of Translational Research. 2012;4(1):83–101. [PMC free article] [PubMed] [Google Scholar]
- 48. Zimmerman MW, Homanics GE, Lazo JS. Targeted Deletion of the Metastasis-Associated Phosphatase Ptp4a3 (PRL-3) Suppresses Murine Colon Cancer. PloS one. 2013;8.3:e58300. doi: 10.1371/journal.pone.0058300. Study of impact of PRL-3 ablation in CRC mouse model
- 49.Hardy S, Wong NN, Muller WJ, Park M, Tremblay ML. Overexpression of the protein tyrosine phosphatase PRL-2 correlates with breast tumor formation and progression. Cancer Res. 2010;70(21):8959–8967. doi: 10.1158/0008-5472.CAN-10-2041. [DOI] [PubMed] [Google Scholar]
- 50. Diamond RH, Cressman DE, Laz TM, Abrams CS, Taub R. PRL-1, a unique nuclear protein tyrosine phosphatase, affects cell growth. Molecular and Cellular Biology. 1994;14:3752–3762. doi: 10.1128/mcb.14.6.3752. Early evidence of PRL-1 connection to cell growth, metastasis and cell invasion.
- 51.Wang J, Kirby C, Herbst R. The tyrosine phosphatase PRL-1 localizes to the endoplasmic reticulum and the mitotic spindle and is required for normal mitosis. Journal of Biological Chemistry. 2002;277(48):46659–46668. doi: 10.1074/jbc.M206407200. [DOI] [PubMed] [Google Scholar]
- 52.Wang Q, Holmes DI, Powell SM, Lu QL, Waxman J. Analysis of stromal-epithelial interactions in prostate cancer identifies PTPCAAX2 as a potential oncogene. Cancer Letters. 2002;175:63–69. doi: 10.1016/s0304-3835(01)00703-0. [DOI] [PubMed] [Google Scholar]
- 53.Werner SR, Lee PA, DeCamp MW, et al. Enhanced cell cycle progression and down regulation of p21(Cip1/Waf1) by PRL tyrosine phosphatases. Cancer Letters. 2003;202:201–211. doi: 10.1016/s0304-3835(03)00517-2. [DOI] [PubMed] [Google Scholar]
- 54. Liang F, Liang J, Wang WQ, et al. PRL3 promotes cell invasion and proliferation by down-regulation of Csk leading to Src activation. Journal of Biological Chemistry. 2007;282:5413–5419. doi: 10.1074/jbc.M608940200. One of several papers connecting PRL to Src pathway. Evidence is shown that PRL-3 regulates Src pathway through Csk.
- 55. Bardelli A, Saha S, Sager JA, et al. PRL-3 expression in metastatic cancers. Clinical Cancer Research. 2003;9:5607–5615. Study of PRL-3 in metastatic cancers
- 56.Peng L, Ning J, Meng L, Shou C. The association of the expression level of protein tyrosine phosphatase PRL-3 protein with liver metastasis and prognosis of patients with colorectal cancer. Journal of Cancer Research and Clinical Oncology. 2004;130:521–526. doi: 10.1007/s00432-004-0563-x. [DOI] [PubMed] [Google Scholar]
- 57.Song R, Qian F, Li YP, et al. Phosphatase of regenerating liver-3 localizes to cyto-membrane and is required for B16F1 melanoma cell metastases in vitro and in vivo. PLoS ONE. 2009;4(2):e4450. doi: 10.1371/journal.pone.0004450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu X, Zeng H, Zhang X, et al. Phosphatase of regenerating liver-3 promotes motility and metastasis of mouse melanoma cells. American Journal of Pathology. 2004;164:2039–2054. doi: 10.1016/S0002-9440(10)63763-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qian F, Li YP, Sheng X, et al. PRL-3 siRNA inhibits the metastasis of B16-BL6 mouse melanoma cells in vitro and in vivo. Molecular Medicine. 2007;13:151–159. doi: 10.2119/2006-00076.Qian. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guo K, Li J, Tang JP, Koh V, Gan BQ, Zeng Q. Catalytic domain of PRL-3 plays an essential role in tumor metastasis: Formation of PRL-3 tumors inside the blood vessels. Cancer Biology & Therapy. 2004;3:945–951. doi: 10.4161/cbt.3.10.1111. [DOI] [PubMed] [Google Scholar]
- 61.Basak S, Jacobs SBR, Krieg AJ, et al. The Metastasis-Associated Gene Prl-3 Is a p53 Target Involved in Cell-Cycle Regulation. Molecular cell. 2008;30.3:303–314. doi: 10.1016/j.molcel.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Min SH, Kim DM, Heo YS, et al. Downregulation of p53 by phosphatase of regenerating liver 3 is mediated by MDM2 and PIRH2. Life Sciences. 2010;86(1–2):66–72. doi: 10.1016/j.lfs.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 63.Min SH, Kim DM, Heo YS, et al. New p53 target, phosphatase of regenerating liver 1(PRL-1) downregulates p53. Oncogene. 2009;28(4):545–554. doi: 10.1038/onc.2008.409. [DOI] [PubMed] [Google Scholar]
- 64.Guo K, Li J, Wang H, et al. PRL-3 initiates tumor angiogenesis by recruiting endothelial cells in vitro and in vivo. Cancer Research. 2006;66:9625–9635. doi: 10.1158/0008-5472.CAN-06-0726. [DOI] [PubMed] [Google Scholar]
- 65.Rouleau C, Roy A, St Martin T, et al. Protein tyrosine phosphatase PRL-3 in malignant cells and endothelial cells: expression and function. Molecular Cancer Therapeutics. 2006;5:219–229. doi: 10.1158/1535-7163.MCT-05-0289. [DOI] [PubMed] [Google Scholar]
- 66.Xu J, Cao S, Wang L, Xu R, Chen G, Xu Q. VEGF promotes the transcription of the human PRL-3 gene in HUVEC through transcription factor MEF2C. PLoS ONE. 2011;6(11):e27165. doi: 10.1371/journal.pone.0027165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Liang F, Luo Y, Dong Y, et al. Translational Control of C-terminal Src kinase (Csk) expression by PRL3 phosphatase. Journal of Biological Chemistry. 2008;283(16):10339–10346. doi: 10.1074/jbc.M708285200. Study connecting PRL to Src.
- 68. Luo Y, Liang F, Zhang ZY. PRL 1 promotes cell migration and invasion by increasing MMP2 and MMP9 expression through Src and ERK1/2 pathways. Biochemistry. 2009;48(8):1838–1846. doi: 10.1021/bi8020789. First evidence of PRL connection to Src pathway and MMPs
- 69.Nakashima M, Lazo JS. Phosphatase of regenerating liver-1 promotes cell migration and invasion and regulates filamentous actin dynamics. Journal of Pharmacology and Experimental Therapeutics. 2010;334(2):627–633. doi: 10.1124/jpet.110.167809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wang H, Quah SY, Dong JM, et al. PRL-3 down-regulates PTEN expression and signals through PI3K to promote epithelial-mesenchymal transition. Cancer Research. 2007;67:2922–2926. doi: 10.1158/0008-5472.CAN-06-3598. First evidence of PRL interaction with PI3K pathway through PTEN
- 71.Liu H, Al-aidaroos AQ, Wang H, et al. PRL-3 suppresses c-Fos and integrin alpha2 expression in ovarian cancer cells. BMC Cancer. 2013;13:80. doi: 10.1186/1471-2407-13-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marchio S, Soster M, Cardaci S, et al. A complex of [alpha]6 integrin and E-cadherin drives liver metastasis of colorectal cancer cells through hepatic angiopoietin-like 6. Embo Molecular Medicine. 2012;4(11):1156–1175. doi: 10.1002/emmm.201101164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Peng L, Jin G, Wang L, Guo J, Meng L, Shou C. Identification of integrin alpha1 as an interacting protein of protein tyrosine phosphatase PRL-3. Biochemical and Biophysical Research Communications. 2006;342:179–183. doi: 10.1016/j.bbrc.2006.01.102. [DOI] [PubMed] [Google Scholar]
- 74.Tian W, Qu L, Meng L, Liu C, Wu J, Shou C. Phosphatase of regenerating liver-3 directly interacts with intergrin 1 and regulates its phosphorylation at tyrosine 783. BMC Biochemistry. 2012;13.1:22. doi: 10.1186/1471-2091-13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Walls CD, Iliuk A, Bai Y, Wang M, Tao WA, Zhang ZY. Phosphatase of Regenerating Liver 3 (PRL3) Provokes a Tyrosine Phosphoproteome to Drive Prometastatic Signal Transduction. Molecular and Cellular Proteomics. 2013;12(12):3759–3777. doi: 10.1074/mcp.M113.028886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Dong Y, Zhang L, Zhang S, et al. Phosphatase of regenerating liver 2 (PRL2) is essential for placental development by down-regulating PTEN (Phosphatase and Tensin Homologue Deleted on Chromosome 10) and activating Akt protein. Journal of Biological Chemistry. 2012;287(38):32172–32179. doi: 10.1074/jbc.M112.393462. PRL-2 null mice exhibit placental and growth defect and evidence of interaction with PTEN in vivo.
- 77. Dong Y, Zhang L, Bai Y, et al. Phosphatase of regenerating liver 2 (PRL2) deficiency impairs Kit signaling and spermatogenesis. J. Biol. Chem. 2014;289 doi: 10.1074/jbc.M113.512079. in press. PRL-2 null male mice exhibit testicular and reproductive defect.
- 78. Al-Aidaroos AQ, Yuen HF, Guo K, et al. Metastasis-associatedPRL-3 induces EGFR activation and addiction in cancer cells. Journal of Clinical Investigation. 2013;123(8):3459–3471. doi: 10.1172/JCI66824. PRL-3 found to interact with EGFR.
- 79.Lai W, Liu L, Zrng Y, et al. KCNN4 channels participate in EMT induced by PRL-3 in colorectal cancer. Medical Oncology. 2013;30(2):566. doi: 10.1007/s12032-013-0566-z. [DOI] [PubMed] [Google Scholar]
- 80.McParland V, Varsano G, Li X, et al. The metastasis-promoting phosphatase PRL-3 shows activity toward phosphoinositides. Biochemistry. 2011;50(35):7579–7590. doi: 10.1021/bi201095z. [DOI] [PubMed] [Google Scholar]
- 81.Zhang J, Xiao Z, Lai D, et al. miR-21, miR-17 and miR-19a induced by phosphatase of regenerating liver-3 promote the proliferation and metastasis of colon cancer. British journal of cancer. 2012;107.2:352–359. doi: 10.1038/bjc.2012.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li Z, Cao Y, Jie Z, et al. miR-495 and miR551a inhibit the migration and invasion of human gastric cancer cells directly interacting with PRL-3. Cancer Letters. 2012;323(1):41–47. doi: 10.1016/j.canlet.2012.03.029. [DOI] [PubMed] [Google Scholar]
- 83.Amann RP. A critical review of methods for evaluation of spermatogenesis from seminal characteristics. Journal of Andrology. 1981;2.1:37–58. [Google Scholar]
- 84.Anton GJ, Siep M, Baarends WM. Molecular and cellular mechanisms in spermatogenesis. Best Practice & Research Clinical Endocrinology & Metabolism. 2000;14.3:331–343. doi: 10.1053/beem.2000.0083. [DOI] [PubMed] [Google Scholar]
- 85. Kobayashi M, Bai Y, Dong Y. PRL2/PTP4A2 phosphatase is important for hematopoietic stem cell self-renewal. Stem Cells. 2014;32 doi: 10.1002/stem.1672. in press. First evidence of hematopoietic phenotype in PRL-2 null mice. Reduction in Stem Cell maintenance.
- 86. Daouti S, Li WH, Qian H, Huang KS, Holmgren J, Levin W, Reik L, McGady, Gillespie DL, Perrotta P, Bian A, et al. A selective phosphatase of regenerating liver phosphatase inhibitor suppresses tumor cell anchorage-independent growth by a novel mechanism involving p130Cas cleavage. Cancer Res. 2008;68:1162–1169. doi: 10.1158/0008-5472.CAN-07-2349. Discovery of a promising PRL small-molecule inhibitor.
- 87.Min G, Lee SK, Kim HN, et al. Rhodanine-based PRL-3 inhibitors blocked the migration and invasion of metastatic cancer cells. Bioorganic and Medicinal Chemistry Letters. 2013;23(13):3769–3774. doi: 10.1016/j.bmcl.2013.04.092. [DOI] [PubMed] [Google Scholar]
- 88.Han YM, Lee SK, Jeong DG, et al. Emodin inhibits migration and invasion of DLD-1 (PRL-3) cells via inhibition of PRL-3 phosphatase activity. Bioorg Med Chem Lett. 2012;22:323–326. doi: 10.1016/j.bmcl.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 89.Choi SK, Oh HM, Lee SK, et al. Biflavonoids inhibited phosphatase of regenerating liver-3 (PRL-3) Natural Product Research. 2006;20(4):341–346. doi: 10.1080/14786410500463312. [DOI] [PubMed] [Google Scholar]
- 90.Park H, Jung SK, Jeong DG, Ryu SE, Kim SJ. Discovery of novel PRL-3 inhibitors based on the structure-based virtual screening. Bioorg Med Chem Lett. 2008;18:2250–2255. doi: 10.1016/j.bmcl.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 91. Jeong DG, Kim SJ, Kim JH, et al. Trimeric structure of PRL-1phosphatase reveals an active enzyme conformation and regulation mechanisms. Journal of Molecular Biology. 2005;345:401–413. doi: 10.1016/j.jmb.2004.10.061. Evidence of trimer structure and structural analysis
- 92.Guo K, Li J, Tang JP, et al. Targeting intracellular oncoproteins with antibody therapy or vaccination. Science Translational Medicine. 2011;3(99):99ra85. doi: 10.1126/scitranslmed.3002296. [DOI] [PubMed] [Google Scholar]
- 93. Lv J, Liu C, Huang H, et al. Suppression of breast tumor growth by DNA vaccination against phosphatase of regenerating liver 3. Gene Therapy. 2013;20(8):834–845. doi: 10.1038/gt.2013.5. Proof-of-concept treatment of PRL-driving metastasis with antibodies against PRL
- 94. Guo K, Tang JP, Tan CPB, Wang H, Zeng Q. Monoclonal antibodies target intracellular PRL phosphatases to inhibit cancer metastases in mice. Cancer Biology and Therapy. 2008;7(5):750–757. doi: 10.4161/cbt.7.5.5764. Proof-of-concept treatment of PRL-driving metastasis with antibodies against PRL
- 95.Blanchetot C, Chagnon M, Dube, et al. Substrate-trapping techniques in the identification of cellular PTP targets. Methods. 2005;35.1:44–53. doi: 10.1016/j.ymeth.2004.07.007. [DOI] [PubMed] [Google Scholar]