Abstract
Oral squamous cell carcinoma (OSCC), a subset of head and neck squamous cell carcinoma (HNSCC), is the eighth most common cancer in the U.S.. Amplification of chromosomal band 11q13 and its association with poor prognosis has been well established in OSCC. The first step in the breakage-fusion-bridge (BFB) cycle leading to 11q13 amplification involves breakage and loss of distal 11q. Distal 11q loss marked by copy number loss of the ATM gene is observed in 25% of all Cancer Genome Atlas (TCGA) tumors, including 48% of HNSCC. We showed previously that copy number loss of distal 11q is associated with decreased sensitivity (increased resistance) to ionizing radiation (IR) in OSCC cell lines. We hypothesized that this radioresistance phenotype associated with ATM copy number loss results from upregulation of the compensatory ATR-CHEK1 pathway, and that knocking down the ATR-CHEK1 pathway increases the sensitivity to IR of OSCC cells with distal 11q loss. Clonogenic survival assays confirmed the association between reduced sensitivity to IR in OSCC cell lines and distal 11q loss. Gene and protein expression studies revealed upregulation of the ATR-CHEK1 pathway and flow cytometry showed G2-M checkpoint arrest after IR treatment of cell lines with distal 11q loss. Targeted knockdown of the ATR-CHEK1 pathway using CHEK1 or ATR siRNA or a CHEK1 small molecule inhibitor (SMI, PF-00477736) resulted in increased sensitivity of the tumor cells to IR. Our results suggest that distal 11q loss is a useful biomarker in OSCC for radioresistance that can be reversed by ATR-CHEK1 pathway inhibition.
INTRODUCTION
Cancer is a critical public health problem accounting for one in four deaths in the U.S. and more worldwide despite advances in diagnosis and treatment (Heron et al., 2009; Siegel et al., 2013). Oral (oropharyngeal) squamous cell carcinoma (OSCC) is the eighth most common cancer in the U.S., accounting for an estimated 41,380 new cases (2.5%) in 2013 and 7,890 deaths (1.4%) (Siegel et al., 2013). Worldwide, HNSCC (worldwide statistics include lip, oral cavity, larynx, and pharynx) was diagnosed in 550,319 new patients (4.4%) in 2008 and resulted in 305,096 deaths (4.0%) (Ferlay et al., 2010; Bray et al., 2013). Over the past four decades, the 5-year relative survival rates have improved substantially for oropharyngeal SCC in both Caucasians (from 54 to 67%) and in African Americans (from 36 to 45%), although disturbing disparities remain (Desantis et al., 2013).
Carcinomas are frequently characterized by chromosomal instability, resulting in loss of tumor suppressor genes and gain or amplification of oncogenes (Ha and Califano, 2002). One of the most frequent chromosomal abnormalities in OSCC and other carcinomas is amplification of chromosomal band 11q13, which includes the cyclin D1 gene (CCND1) (Schraml et al., 1999; Gollin, 2001; Huang et al., 2002; Albertson, 2006; Jin et al., 2006; Gibcus et al., 2007). 11q13 amplification results from breakage-fusion-bridge (BFB) cycles (Reshmi et al., 2007) and/or chromosome breakage and rearrangement resulting from palindromic segmental duplications flanking 11q13 (Gibcus et al., 2007). The first step in the BFB cycle is a chromosome break distal to the amplified region, possibly at the FRA11F chromosomal fragile site or as a result of rearrangement involving segmental duplications resulting in loss of some or all of the distal segment of chromosome 11q (Shuster et al., 2000; Reshmi et al., 2007).
Loss of distal 11q and amplification of chromosomal band 11q13 are associated with poor prognosis in OSCC (Michalides et al., 1995; Akervall et al., 1997; Jin et al., 1998; Jin et al., 2006). Although numerous investigators identified copy number loss of distal 11q, from 11q14→11qter, centered primarily on 11q22→q23 in a variety of primary tumors and cell lines (George et al., 2007; Parikh et al., 2007; Ambatipudi et al., 2011; Swarts et al., 2011; Edelmann et al., 2012), we showed that distal 11q (ATM gene) loss is associated with reduced sensitivity (resistance) to ionizing radiation (IR) in OSCC cell lines (Parikh et al., 2007; Henson et al., 2009). In silico copy number analysis of the ATM gene shows that distal 11q loss is present a focal peak of deletion containing 60–80 genes in 25% of The Cancer Genome Atlas (TCGA) more than 7,200 tumors, including 47–54% of melanomas, HNSCC, esophageal SCC, breast and cervical carcinomas, 30–36% of lung SCC, ovarian, prostate, and bladder carcinomas, and 18–28% of lung, stomach, colorectal, hepatocellular, and rectal carcinomas (Beroukhim et al., 2010; Mermel et al., 2011). Thus, based on the American Cancer Society statistics (Siegel et al., 2013) and the TCGA frequencies, at least 330,000 of the 1,660,290 new cancer cases expected in the U.S. in 2013 may have distal 11q loss.
Distal 11q contains a block of critical DNA damage response (DDR) genes, including ATM (11q22.3), MRE11A (11q21), H2AFX (11q23.3) and CHEK1 (11q24.2). The cornerstone of the DDR to IR is the ATM gene, which is mutated in the rare, pleiotropic autosomal recessive disorder, ataxia telangiectasia (AT) (Harnden, 1994; Savitsky et al., 1995; Baskaran et al., 1997; Lavin and Shiloh, 1997; Pandita et al., 1999). ATM encodes a 370 kDa protein that is a member of the family of lipid/protein kinases related to phosphatidylinositol 3-kinase (PI3K), known as the PI3K-related kinases (Keith and Schreiber, 1995; Shiloh, 2003; Abraham, 2004). In IR-induced double strand breaks (DSB), the MRE11A-RAD50-NBS1 (MRN) complex plays the role of a ‘sensor’ (Stracker et al., 2004). The primary ‘transducer’ of the DSB signal is ATM (Shiloh, 2003). In response to DNA DSB induced by IR, ATM is rapidly phosphorylated at the serine 1981 (ser1981) residue, which facilitates the phosphorylation of various other proteins involved in the regulation and repair of DNA damage (Bakkenist and Kastan, 2003). These ‘effectors’ of ATM include ABL1, BRCA1, TP53, and CHEK2 (Baskaran et al., 1997; Khanna and Jackson, 2001). An important substrate of ATM is the histone H2AX, a member of the histone H2A subfamily that maps to distal 11q (Fernandez-Capetillo et al., 2004). ATM activates H2AX at the DSB site converting it into the phosphorylated form, γH2AX, which then anchors DNA damage response proteins to the sites of damage (Stucki et al., 2005). The signaling cascade activated by ATM culminates in cell cycle arrest, apoptosis and DNA repair. Ataxia Telangiectasia and Rad3-related (ATR) is another PI3K family protein involved primarily in signaling the presence of stalled replication forks and maintenance of genomic integrity during S phase, along with its partners, ATR interacting protein (ATRIP) and replication protein A (RPA) (Cortez et al., 2001; Zou and Elledge, 2003; Byun et al., 2005). Although ATR plays a primary role in responding to ultraviolet light (UV) and chemotherapy-induced genomic insults, it also appears to play a role in responding to IR-induced DNA damage (Adams et al., 2006; Myers and Cortez, 2006). The presence of ATR in nuclear foci after IR treatment suggests recruitment of this kinase to sites of DNA damage; ATM is known to regulate the loading of ATR to sites of DNA DSB (Zou and Elledge, 2003; Cuadrado et al., 2006). Unlike the rapid phosphorylation of ATM post-treatment, ATR recruitment and activation is comparatively delayed, but also requires a functional MRN complex (Adams et al., 2006). Even though the ATM and ATR pathways are thought to respond primarily to different types of DNA damage, the two pathways appear to be intimately intertwined.
Based on our observation that distal 11q loss leads to radioresistance in OSCC cell lines, this study tests the hypotheses that 1) distal 11q loss, including loss of the ATM gene is associated with an upregulated ATR-CHEK1 pathway after IR, 2) upregulation of the ATR-CHEK1 pathway results in G2-M checkpoint arrest after IR treatment and radioresistance, and 3) knockdown of the ATR-CHEK1 pathway by RNA interference or a targeted small molecule inhibitor resensitizes cancer cells with distal 11q loss to IR.
MATERIALS AND METHODS
Cell Culture
The UPCI:SCC OSCC cell lines developed by us (White et al., 2007) were cultured in M10 medium comprised of Minimal Essential Medium (MEM) supplemented with 1% non-essential amino acids (NEAA), 1% L-glutamine, 0.05 mg/ml gentamicin and 10% fetal bovine serum (FBS) (all from GIBCO Invitrogen, Grand Island, NY). Normal human oral keratinocytes grown in our laboratory (NHOK) and TERT-transfected human keratinocytes (OKF6/TERT-1 cells, a gift of James Rheinwald, Brigham and Women‧s Hospital, Harvard Institutes of Medicine (Dickson et al., 2000)) cell lines were used as controls. NHOK were isolated from normal oral epithelial tissue obtained and deidentified by the University of Pittsburgh Head & Neck SPORE Tissue Bank from consenting non-cancer patients under our unified Tissue Bank Institutional Review Board approval. NHOK were cultured in serum-free keratinocyte growth medium-2 (KGM-2) medium (Lonza, Walkersville, MD) supplemented with bovine pituitary extract (BPE), human epidermal growth factor (hEGF), insulin (bovine), hydrocortisone, GA-1000 (Gentamicin, Amphotericin B), epinephrine and transferrin supplied in the KGM-2 BulletKit™ (Lonza) as per the manufacturer’s instructions. OKF6/TERT-1 cells were cultured in Keratinocyte-SFM supplemented with 25 µg/ml bovine pituitary extract, 0.2 ng/ml EGF, 0.3 mM CaCl2, and penicillin-streptomycin (GIBCO Invitrogen).
Fluorescence In Situ Hybridization (FISH)
Molecular cytogenetic analysis was carried out to determine the relative copy numbers of ATM, MRE11A, H2AFX and CHEK1 in OSCC cell lines compared to the chromosome 11 centromere (CEP11/D11Z1, Abbott Molecular Inc., Des Plaines, IL). Molecular cytogenetic analyses by FISH were carried out and analyzed in the University of Pittsburgh Cell Culture and Cytogenetics Facility as described previously (Parikh et al., 2007). The FISH probes were prepared from the following BAC clones purchased from Children’s Hospital of Oakland Research Institute (CHORI, Oakland, CA; http://www.chori.org/bacpac): RP11-241D13 (ATM), RP11-685N10 (MRE11A), RP11-892K21 (H2AFX), and RP11-712D22 (CHEK1). BAC DNA was labeled with Spectrum Orange™ using a nick translation kit from Abbott Molecular Inc.. The percentage of cells with relative loss, gain, or amplification was determined after counting signals in at least 200 nuclei per slide. Copy number gain or loss of genes was determined by the relative ratio of the BAC probe to centromere 11. A ratio of <1.0 was considered to have relative gene copy loss, a ratio of 1 = equal, a ratio between 1 and 2.5 = gain, and a ratio ≥ 2.5 = amplified with respect to CEP11.
Clonogenic Survival Assay
Clonogenic (also called colony) survival assays were carried out to determine cell survival in response to treatment as described earlier (Parikh et al., 2007). Cell lines that formed clearcut colonies were selected for this study. Untreated cells, seeded at the same density as the treated cells, served as controls to determine relative plating efficiency. All experiments were done in triplicate. Results were reported as a ‘Surviving Fraction,’ which is the ratio of the number of colonies observed at a particular dose to that observed in the untreated control, represented as a percentage calculated using the formulae below.
Plating Efficiency, PE = colonies counted in untreated / cells seeded
Surviving Fraction, SF = colonies counted in treated / (cells seeded * PE/100)
Correlation of Distal 11q Loss with Patient Outcome
The effect of distal 11q loss was dichotomized into 80% or greater vs. less than 80% loss. Disease-specific survival was defined as time from the date of tissue procurement (surgery or biopsy) until death from cancer. Patients who were alive at last follow-up or had died from causes unrelated to their disease were censored. Disease-specific survival by 11q loss status was estimated by the Kaplan-Meier method and tested for a difference by a two-tailed log rank test. Four covariates were available for Pittsburgh patients and tested for influence on disease – free survival: age, T stage, N stage, or exposure to chemo- or radiotherapy. The effect of 11q loss upon disease-specific survival was assessed with proportional hazards regression by adjusting for these covariates as needed to determine if 11q loss could be considered independently associated with cancer mortality.
Quantitative Real Time PCR (QRT-PCR)
QRT-PCR was carried out to assess the relative ATR and CHEK1 expression in untreated cell lines and those treated with IR or transfected with CHEK1 or ATR siRNA. RNA extraction for real-time PCR was performed using TRIzol reagent (Gibco Invitrogen) according to the manufacturer’s instructions. The extracted RNA was purified using the RNeasy Mini kit (QIAGEN, Germantown, MD) and resuspended in 100 µl RNase-free water. The RNA samples were purified of contaminating DNA using a DNA–free DNase kit (Ambion, Austin, TX) according to the manufacturer’s instructions. RNA concentrations were assessed using a SmartSpec 3000 (Bio-Rad Laboratories, Hercules, CA) and normalized to 40 ng/µl. Reverse transcription was carried out as described earlier (Huang et al., 2002).
QRT-PCR of cDNA obtained after reverse transcription was carried out on a 7300 Real-Time PCR System (Applied Biosystems), and analysis was done using the relative quantitation method as in Huang et al. (2002). The final concentrations of the QRT-PCR reaction components were as follows: 1X Taqman Gene Expression Master Mix and 1X Taqman Gene Expression Assays (Probe/Primer mix for ATR, CHEK1 and 18s RNA) (Applied Biosystems). Thermocycler conditions for QRT-PCR were as follows: 95°C for 10 min, 40 cycles of 95°C for 15 s and 60°C for 60 s using the (Applied Biosystems). The RNA expression levels were quantified relative to Universal Reference cDNA obtained from Clontech (Mountain View, CA).
Immunoblotting
Immunoblotting was carried out as described previously (Parikh et al., 2007) to evaluate expression of ATR-CHEK1 pathway proteins including ATR, CHEK1 and pCHEK1, and to study the function of TP53 in our cell lines by analyzing TP53 and p21 protein expression after Adriamycin treatment (0.4 µg/ml in culture medium for 4–5 h at 37°C). The primary antibodies utilized were: ATR (Affinity Bioreagents, Golden, CO, 1:1000 dilution), CHEK1 (Cell Signaling, Danvers, MA, 1:1000 dilution), pCHEK1 ser345 (Cell Signaling, 1:1000 dilution), pCHEK1 ser317 (Cell Signaling, 1:1000 dilution), TP53 (Santa Cruz Biotechnology, Inc., 1:750 dilution), and p21 (Santa Cruz Biotechnology, Inc., 1:750 dilution). Appropriate species-specific secondary antibodies (Santa Cruz Biotechnology, Inc., 1:5000 dilution) were applied and target proteins were visualized using the Western Lightning™ Chemiluminescence Reagent Plus kit (PerkinElmer Life Sciences, Boston, MA) according to the manufacturer‧s instructions. To verify equal protein loading in the gels, membranes were stripped and re-probed with antibodies against β-actin (Sigma Immunochemicals, St. Louis, MO, 1:1000 dilution) or α-Actinin (Santa Cruz Biotechnology, Inc., 1:1000 dilution).
Densitometric Analysis of Protein Bands
Image J software from NIH was used for analyzing densities of protein bands in western blots. Relative optical densities were calculated for the protein being evaluated and the loading control protein for all lanes using one of the lanes as a reference. Adjusted densities were then calculated for each sample by normalizing the relative density of the protein of interest to the loading control for the same.
Cell Cycle Analysis by Flow Cytometry
For cell cycle analysis, cells were seeded in 35 mm or 60 mm dishes and allowed to attach overnight. Following the relevant treatments, mock or IR, floating and adherent cells were collected at the end of 24 h, washed with phosphate-buffered saline (PBS), and fixed with 70% ethanol. The cells were then treated with 80 µg/ml RNase A and 50 µg/ml propidium iodide (Invitrogen-Molecular Probes, Carlsbad, CA) for 45 min at 37°C. The stained cells were analyzed using a Coulter Epics XL Flow Cytometer in the UPCI Flow Cytometry Facility.
Assessment of Mitotic Segregation Defects
To assess genotoxic stress and chromosomal instability, we analyzed mitotic segregation defects in OSCC cell lines with or without distal 11q loss that were grown on coverslips. Cells were either untreated or treated with IR and then grown for 18 or 36 h. At the specified timepoint, the cells on coverslips were fixed with 100% methanol, dried, stained with DAPI, and mounted onto slides using antifade. The slides were coded and 1000 cells were analyzed from each cell line. The frequencies of anaphase and interphase bridges and micronuclei, markers of chromosomal segregation defects resulting from chromosomal aberrations, were recorded.
CHEK1 Knockdown by RNA Interference
RNA interference to CHEK1 and ATR was performed using Smartpool duplexes for each of the specific genes, obtained from Dharmacon (Lafayette, CO). Nonspecific (scrambled) control duplexes (Dharmacon) were used as controls. The duplexes were reconstituted in DNA–free RNA resuspension buffer provided by Dharmacon according to the manufacturer’s instructions. For transfection, the carcinoma cell lines were seeded in 60 mm dishes or T25 flasks and transfected with siRNA duplexes using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. The final working siRNA concentration ranged from 90–100 nM. We examined cells treated without vector (untreated controls), cells transfected with the nonspecific (scrambled) control siRNA (mock transfected controls), and cells transfected with smartpool siRNA duplexes for either CHEK1 (siCHEK1 transfected) or ATR (siATR transfected) for all of our siRNA experiments. Peak siRNA transfection was seen at the end of 24–96 h as assessed by RT-PCR and immunoblotting. At 72 h post-transfection, cells were seeded for clonogenic survival assay.
CHEK1 Knockdown by Small Molecule Inhibitor
PF-00477736, a potent, specific CHEK1 small molecule inhibitor (SMI) was a gift from Pfizer, Inc. (Groton, CT). PF-00477736 selectively inhibits enzymatic activity of CHEK1, abrogates DNA damage-induced cell cycle arrest, and increases the cytotoxic effect of DNA damaging agents in TP53-defective cell lines (Blasina et al., 2008). Dose-response curves for the SMI, assessment of the optimal time for addition of the SMI in relation to IR treatment, and the effects of monotherapy with the drug or combined treatment with IR were determined using clonogenic survival assays.
RESULTS
Copy Number Loss of Genes on Distal 11q in OSCC Cell Lines is Associated with Decreased Sensitivity to IR
Dual-color FISH with BAC probes to the ATM, MRE11A, H2AFX, and CHEK1 genes was carried out in a selected series of our OSCC cell lines based on their utility in clonogenic survival assays to determine the relative gene copy number compared to the chromosome 11 centromere enumeration probe (CEP11). Table 1 summarizes these FISH results, which suggest that copy number loss of genes on distal 11q is observed in a substantial fraction of OSCC cell lines.
Table 1.
Summary of the interphase FISH results in the selected OSCC cell lines. Interphase FISH was used to assess copy number of ATM, MRE11A, H2AFX and CHEK1 genes relative to the chromosome 11 centromere.
| Percentage of Cells with Copy Number Loss of * | ||||
|---|---|---|---|---|
| Cell Line |
MRE11A 11q21 (94150466 - 94227040) |
ATM 11q22-q23 (108093559 - 108239826) |
H2AFX 11q23.3 (118964584 - 118966177) |
CHEK1 11q24.2 (125495031- 125546150) |
| UPCI:SCC029B+ | 90 | 92 | 97 | 99 |
| UPCI:SCC040+ | 100 | 99 | 99 | 99 |
| UPCI:SCC084+ | 95 | 93 | 98 | 97 |
| UPCI:SCC131+ | 98 | 92 | 96 | 98 |
| UPCI:SCC136+ | 99 | 92 | 91 | 96 |
| UPCI:SCC104 | 1 | 98 | 96 | 1 |
| UPCI:SCC081+ | 8 | 22 | 97 | 4 |
| UPCI:SCC066 | 20 | 3 | 13 | 15 |
| UPCI:SCC116 | 14 | 15 | 16 | 10 |
Lower ratio of test gene relative to the copy number of the chromosome 11 centromere.
These cell lines express CCND1 amplification.
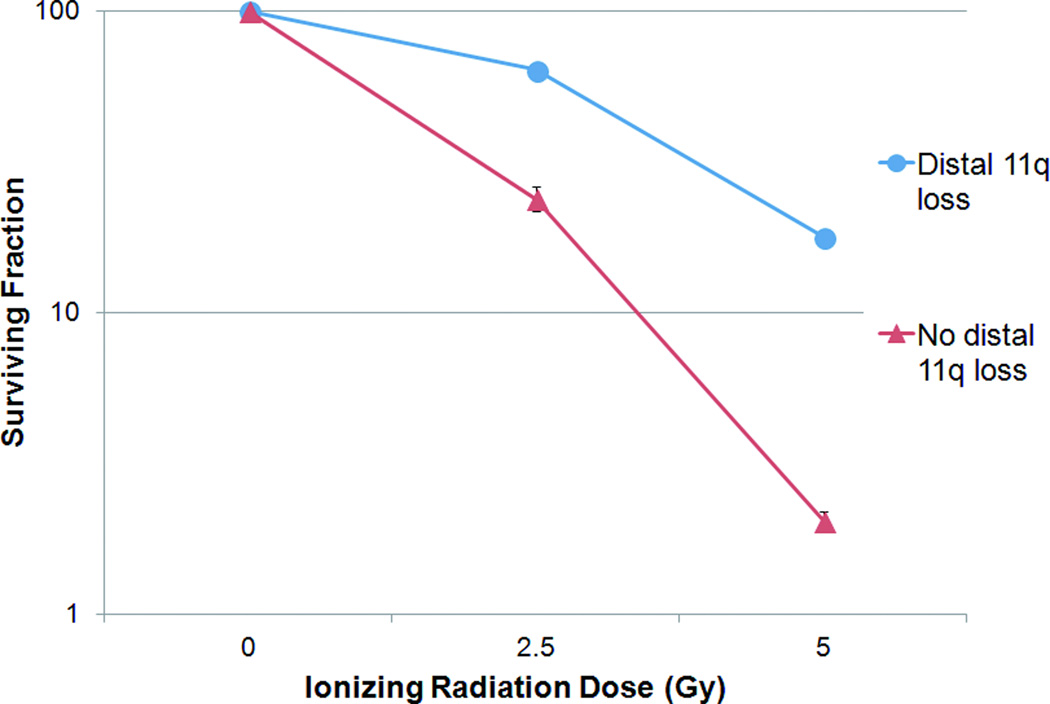
The sensitivity of OSCC cells to IR was assessed by clonogenic survival assay. The cell lines were divided into two groups: ‘distal 11q loss’ and ‘no distal 11q loss.’ The IR doses used for our experiments were 2.5 Gy and 5 Gy. Results are reported as plots of ‘Surviving Fraction’ vs. radiation dose on a semilog scale. OSCC cell lines with distal 11q loss showed 63.8±1.9% and 17.8±0.9% survival at 2.5 and 5 Gy of IR, respectively, whereas the cell lines without distal 11q loss showed 23.9±2.4% and 2.1±0.1% survival, respectively at the same two IR doses (Fig. 2). These results translated to three-fold higher survival at 2.5 Gy and eight-fold higher survival at 5 Gy IR in the OSCC cell lines with distal 11q loss compared to the OSCC cell lines without distal 11q loss. Similar to our previous results (Parikh et al., 2007), these results showed clear evidence of radioresistance (or increased survival) in OSCC cell lines with distal 11q loss compared to OSCC cell lines without distal 11q loss at both radiation doses tested. Hence, radioresistance appears to be associated with distal 11q loss marked by the ATM gene.
Figure 2.
Survival of OSCC cell lines after treatment with IR. The surviving fraction of cells at specific IR doses is plotted with error bars (+SEM) on a logarithmic scale. Carcinoma cells in the “distal 11q loss” group (UPCI:SCC029B, 040, and 131) showed three times as much survival as cells in the “no distal 11q loss” group (UPCI:SCC066, 081, and 116).
Distal 11q (ATM) Loss is Associated with Shorter Disease-Specific Survival in Patients with OSCC
To examine the effect on disease-specific survival of distal 11q loss as measured by FISH with the ATM/CEP11 probe set, we carried out Kaplan-Meier survival estimates in a retrospective series of 42 OSCC patients, 35 from our institution and seven from the University of Michigan. All patients had surgical resection of their SCCHN tumors between 1992 and 1996. Thirty-nine patients are deceased, 30 subsequent to head and neck cancer and nine from other causes; three patients were alive without evidence of disease at last follow-up.
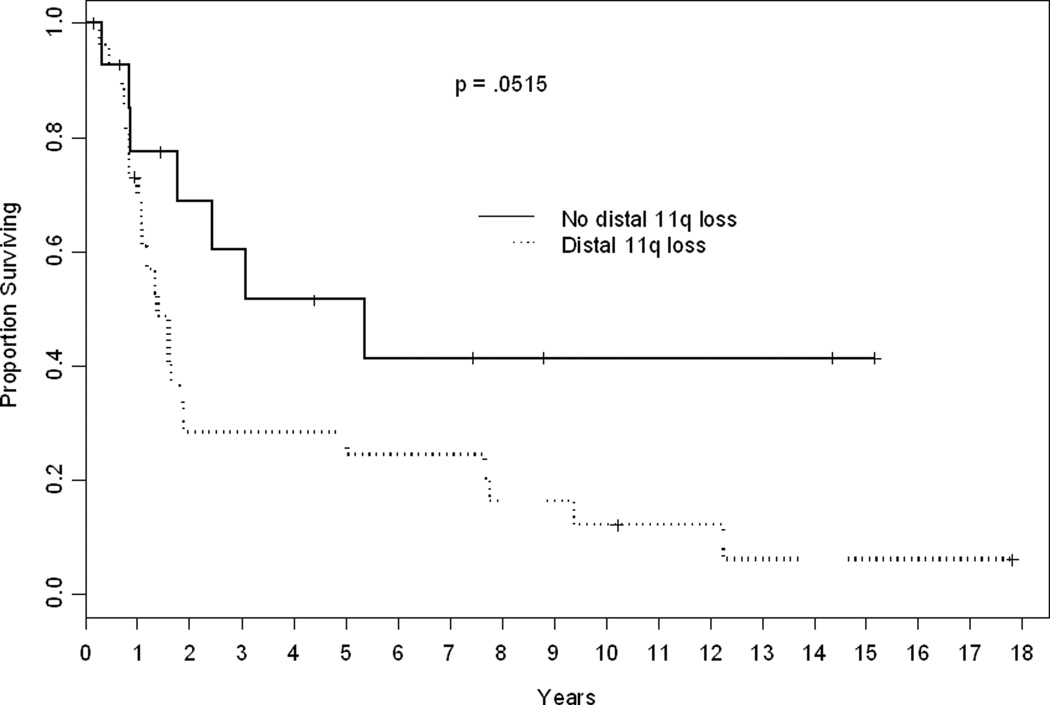
The results show that those 27 OSCC patients with distal 11q loss in at least 80% of cells have a median disease-specific survival of 18 months compared to 65.4 months for the 15 patients with distal 11q loss in less than 80% of cells (log rank p = .0501) (Fig. 3). We investigated whether this difference was influenced by treatment with radiation or chemotherapy. Thirty-one patients underwent chemo- or radiotherapy either in the adjuvant setting or for disease recurrence. Applying Cox proportional hazards regression, we found no interaction between distal 11q loss and exposure to chemo- or radiotherapy (p = 0.183). The hazard ratio for distal 11q loss alone was 2.27 (95% CI = 0.97 to 5.33). The effect of distal 11q loss among the 31 treated patients was similar or slightly more pronounced (p = .022) with a hazard ratio of 3.07 (95% CI = 1.12 – 8.38). We also investigated whether the association between distal 11q loss and disease-specific survival was due to the confounding influence of other clinical variables. Age and T stage were modestly associated with survival, but neither covariate was correlated with distal 11q loss. Nonetheless, we estimated the hazard ratio for distal 11q loss alone and conditional upon age and T stage. The hazard ratio was unchanged (2.0 alone vs. 2.04 adjusting for age and T stage). We conclude that distal 11q loss is associated with increased risk of cancer mortality independently of T and N stage and may have predictive utility for response to chemotherapy or radiotherapy.
Figure 3.
Kaplan-Meier plot of disease specific survival. Forty-two patients were classified by distal 11q loss (+80%). The median followup for 12 censored patients was 5.9 years (range 2 months–18 years). P 5 0.0515 by a log rank test.
Increased Mitotic Segregation Defects after IR Treatment of Cells with Distal 11q Loss
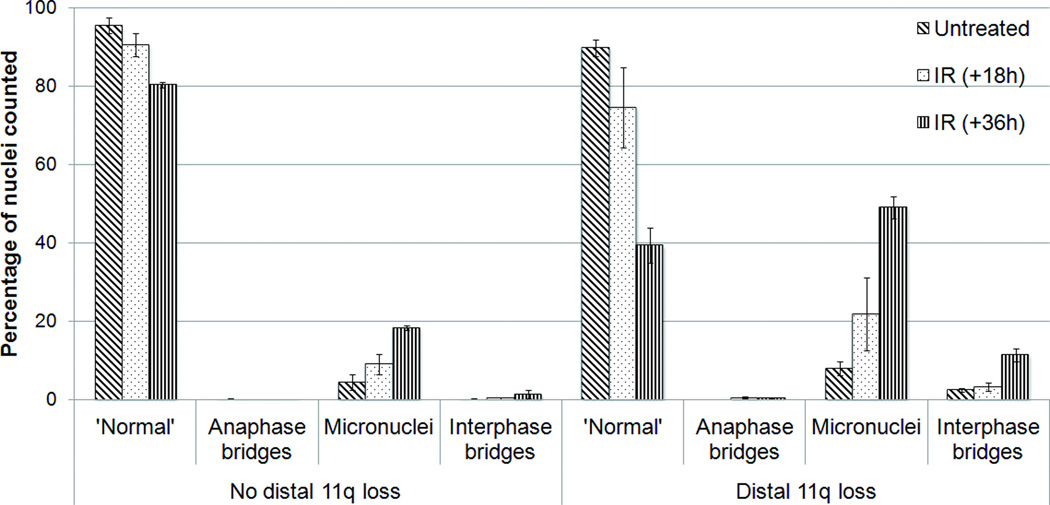
To assess mitotic defects in OSCC cell lines after treatment with IR, we measured the frequencies of micronuclei and anaphase and interphase chromosome bridges, which represent the result of misrepaired DNA and chromosomes and/or defective chromosomal segregation (Fenech et al., 2011). The frequency of each aberration type with or without IR treatment is shown in Figure 4. Cells with distal 11q loss have a greater than two-fold increase in micronuclei and a 10-fold increase in interphase bridges 18 and 36 h post-irradiation compared to cells without distal 11q loss. No significant accumulation of anaphase bridges was observed in either group of cell lines. This is not surprising, since most cells would be expected to continue to cycle, resolving anaphase bridges into aberrant nuclear chromosomes, micronuclei, and/or interphase bridges. This result provides additional support to the proposition that cells with distal 11q loss have a diminished DNA damage response to IR-induced DNA damage compared to cells without distal 11q loss.
Figure 4.
Chromosomal segregation defects after irradiation of tumor cells with distal 11q loss compared with cells without distal 11q loss.
The ATR-CHEK1 Pathway is Upregulated in OSCC Cell Lines with Distal 11q Loss
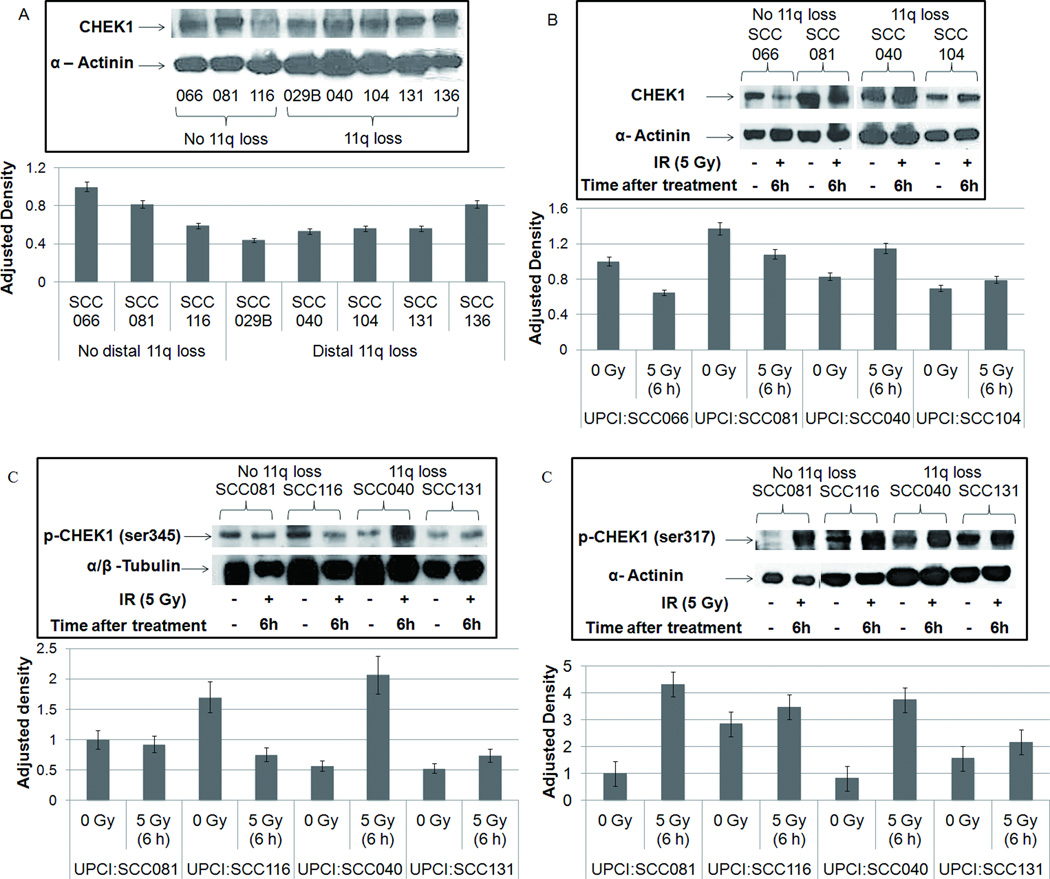
Copy number loss of genes on distal 11q resulted in decreased expression of MRE11A, ATM, and H2AFX and their proteins (Parikh et al., 2007). This observation combined with the radioresistance phenotype suggests that a decrease in the ATM pathway results in increased activity of a compensatory pathway in the cells with distal 11q loss. We evaluated the activity of the related ATR-CHEK1 pathway by examining the protein expression of some members of this pathway, specifically, ATR, and CHEK1 and its phosphorylated form. Immunoblotting for CHEK1 protein revealed that in untreated OSCC cells, there was no strong correlation observed between CHEK1 protein expression and distal 11q loss as was seen between CHEK1 gene expression and distal 11q loss (Parikh et al., 2007). Four of five cell lines with distal 11q loss (UPCI:SCC029B, 040, 104, 131) showed lower CHEK1 protein expression compared to the cell lines without 11q loss (UPCI:SCC066, 081, 116) (Fig. 5A). UPCI:SCC136 (distal 11q loss) had high CHEK1 expression. OSCC cell lines with distal 11q loss that were treated with IR showed an increase in CHEK1 expression 6 h after radiation compared to cell lines without 11q loss (Fig. 5B). This suggests upregulation of the ATR-CHEK1 pathway after irradiation of cells with distal 11q loss.
Figure 5.
(A) CHEK1 protein expression in OSCC cells shows that cells without distal 11q loss tend to have higher CHEK1 expression compared with cells with 11q loss; (B) After IR treatment, cells with distal 11q loss showed an increase in CHEK1 expression compared with cells without distal 11q loss; (C) Phosphorylation status of CHEK1 ser345 and ser317 showing increased ATR-dependent ser345 phosphorylation in cells with distal 11q loss, but no difference in ser317 phosphorylation between cells with or without distal 11q loss.
We then examined the phosphorylation status of CHEK1 using antibodies against the phosphorylation sites ser345 and ser317 to confirm upregulation of the ATR-CHEK1 pathway. Ser317 phosphorylation is required for subsequent ser345 phosphorylation in response to DNA damage (Wang et al., 2012). Ser345 phosphorylation is necessary for CHEK1 activation and a proper checkpoint response after DNA damage and promotes nuclear retention of CHEK1 (Jiang et al., 2003; Niida et al., 2007). Early phosphorylation of ser345 is thought to occur in an ATR-dependent manner, whereas phosphorylation of ser317 in response to IR is reported to be dependent on ATM and NBS1 (Gatei et al., 2003; Wang et al., 2012). As predicted, cells with distal 11q loss showed an increase in CHEK1 ser345 phosphorylation after IR, which was absent in the cells without distal 11q loss (Fig 5C), which expressed increased phosphorylation of CHEK1 ser317, suggesting that ATM is primarily responsible for the IR-induced DDR. There was also an observable increase in ser317 phosphorylation in cells with distal 11q loss, since they retain some ATM expression (Fig 5C). These results confirm increased activity of the ATR-CHEK1 pathway in the cells with distal 11q loss compared to those without distal 11q loss.
Distal 11q Loss is Associated with Increased S and G2M Checkpoint Arrest after IR
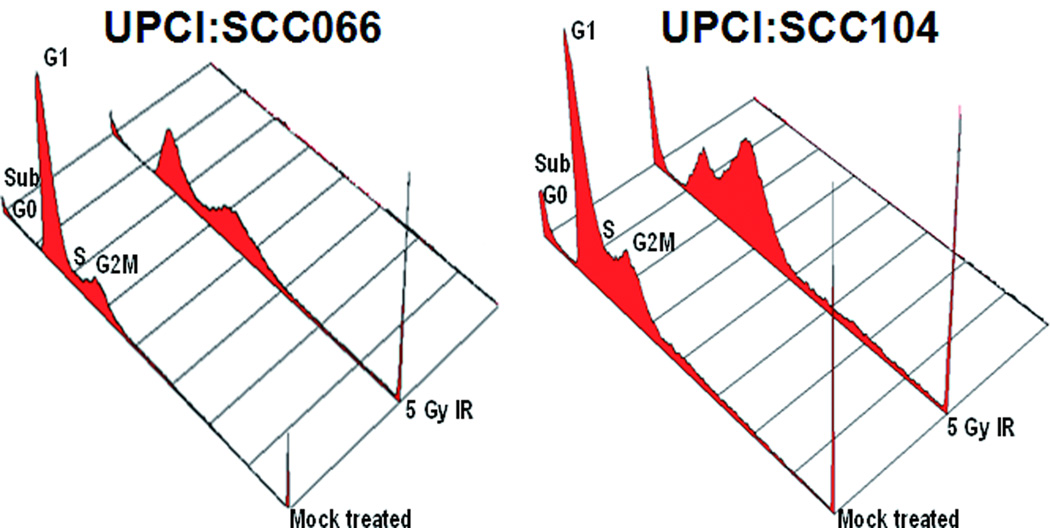
To study the cell cycle profiles of OSCC cells in response to DNA damage, flow cytometry was carried out on cell lines treated with 5 Gy IR (Table 2). We observed loss of the G1 checkpoint and increased G2M accumulation after treatment with IR in the three (of five) cell lines with distal 11q loss. Both cell lines without 11q loss, UPCI:SCC066 and UPCI:SCC116, had an intact G1 checkpoint and when compared to the NHOK control cells, these cell lines showed increased accumulation of cells in the G2M phase. Untreated UPCI:SCC104 (distal 11q loss) cells had a higher percentage of G2M phase cells compared to untreated UPCI:SCC066 (no 11q loss) cells (Fig. 6). Following 5 Gy IR, UPCI:SCC066 showed an accumulation of cells in both the G1 and G2M phases, while UPCI:SCC104 cells showed predominant accumulation in the G2M phase.
Table 2.
Results of cell cycle analysis in OSCC cell lines in response to ionizing radiation.*
| Distal 11q loss status |
Cell Lines | Untreated | 5 Gy IR (24 h) | ||||
|---|---|---|---|---|---|---|---|
| G1 | S | G2M | G1 | S | G2M | ||
| No loss | UPCI:SCC066 | 70% | 13% | 16% | 50% | 16% | 32% |
| UPCI:SCC116 | 62% | 15% | 22% | 49% | 15% | 32% | |
| Loss | UPCI:SCC084 | 60% | 10% | 29% | 32% | 12% | 54% |
| UPCI:SCC104 | 53% | 16% | 30% | 22% | 18% | 57% | |
| UPCI:SCC131 | 67% | 16% | 15% | 32% | 11% | 55% | |
OSCC tumor cells were treated with 5 Gy of IR and grown for 24 hours, after which they were fixed and stained for cell cycle analysis.
Figure 6.
Comparison of cell cycle profiles between UPCI:SCC066 (without distal 11q loss) and UPCI:SCC104 (with distal 11q loss) in response to IR. UPCI:SCC066 and 104 were either mock-treated or treated with 5 Gy IR and allowed to recover for 24 h. At the end of 24 h, flow cytometric analyses show loss of the G1 cell cycle checkpoint in UPCI:SCC104, with most cells accumulating in G2M, and accumulation of UPCI:SCC066 cells in both the G1 and G2M cell cycle phases.
CHEK1 or ATR siRNA Resensitizes OSCC Cells to IR-Induced DNA Damage
Our previous studies suggested that the ATR-CHEK1 pathway plays a role in the radioresistant phenotype observed in the cell lines with distal 11q loss. To confirm the role of the ATR-CHEK1 pathway in radioresistance in cells with distal 11q loss, we used siRNA specific to CHEK1 or ATR to knock down its expression in cell lines and assess the effect of this knockdown on resistance. CHEK1 expression was inhibited to a high level (~ 90%) after knockdown using siRNA specific to CHEK1, while a scrambled non– specific siRNA did not inhibit CHEK1 expression after 72 h, as assessed by QRT-PCR and immunoblotting (data not shown). Similarly, ATR expression was also inhibited to a high level (> 80%) after siRNA-based knockdown. The effect of CHEK1 and ATR knockdown on sensitivity to IR was assessed by clonogenic survival assays. The cells in the clonogenic survival assay were divided into three groups – ‘Untreated,’ ‘Mock transfected,’ and ‘siCHEK1 transfected.’ The ‘Mock transfected’ group represents the average survival values for the two controls, one a control treated with the transfection reagent, Lipofectamine 2000, and the other control treated with a pool of non-specific siRNA. To maximize the effect of CHEK1 or ATR knockdown, cells were harvested and plated for clonogenic survival assay 48–72 hours post-transfection and treated with IR 24 h later.
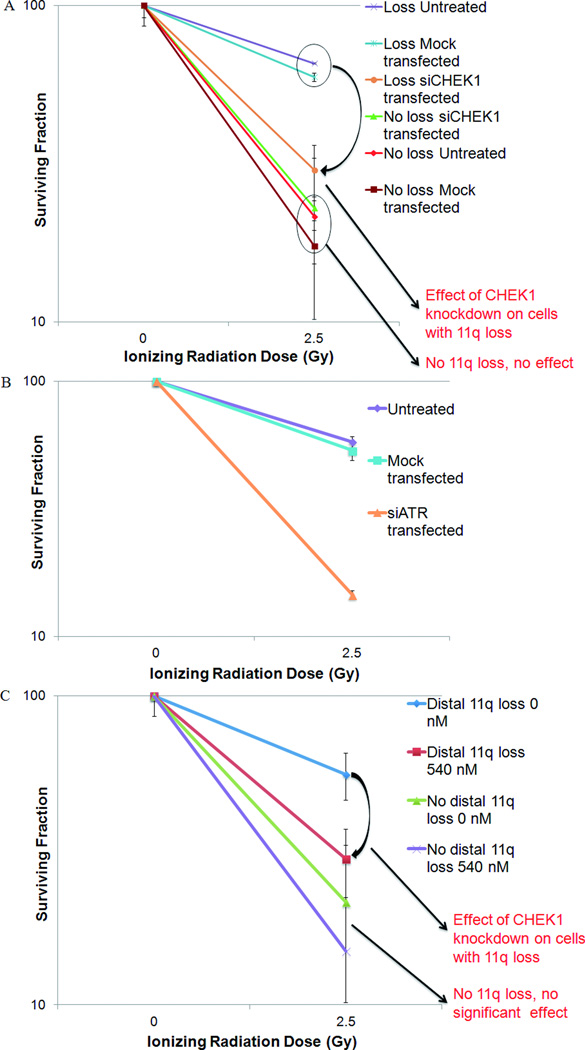
The survival of untransfected OSCC cells with distal 11q loss (UPCI:SCC040, UPCI:SCC029B and UPCI:SCC131) was 65.6±0.23% and 18.8±0.1% at 2.5 and 5 Gy of IR, respectively (Fig. 7A). Mock-transfected cells showed a slight, but not statistically significant decrease in survival compared to the untreated cells. The survival of siCHEK1 transfected cells was 30.2±6.1% and 6.4±2.4% at 2.5 and 5 Gy of IR, respectively. The results show that half as many OSCC cells with distal 11q loss survived an IR dose of 2.5 Gy after transfection with CHEK1 siRNA compared to cells without 11q loss. Radiation treatment of 5 Gy resulted in three-fold lowered survival in CHEK1 siRNA-transfected cells compared to untreated and mock-transfected cells. No significant survival difference was observed between the three treatment conditions in tumor cells without distal 11q loss.
Figure 7.
Survival after CHEK1 or ATR knockdown using siRNA or the SMI (PF-00477736) in OSCC cell lines. The surviving fraction of treated or untreated cells at doses of 0 and 2.5 Gy is plotted on a logarithmic scale with error bars (6SEM). (A) Cells transfected with siCHEK1 in the “distal 11q loss” group (UPCI:SCC029B, 040, and 131) showed 50% decreased survival compared with the untreated and mock-transfected cells. Survival in the “no distal 11q loss” group was not altered significantly by siCHEK1 transfection. (B) HNSCC cells transfected with siATR in the “distal 11q loss” group (UPCI:SCC029B, 040, 131) showed fourfold decrease in survival at 2.5 Gy and 10-fold decrease at 5 Gy compared with the untreated and mock transfected cells. (C) Combined IR and SMI treatment was twice as effective in killing cells with distal 11q loss (UPCI:SCC040 and UPCI:SCC131) compared with IR alone. UPCI:SCC116 (without distal 11q loss) did not show a significant difference in survival when treated with combined IR and SMI compared with IR alone.
Similarly, ATR knockdown resulted in resensitization exclusively of tumor cells with distal 11q loss to radiation treatment. ATR inhibition in OSCC cells with loss of distal 11q (UPCI:SCC029B, 040 and 131) showed a four-fold decrease in survival at 2.5 Gy and a 10-fold decrease at 5 Gy (Fig. 7B). OSCC cells without loss of distal 11q (UPCI:SCC116) did not show a significant difference in survival after ATR inhibition. These results suggest that the upregulation of the ATR/CHEK1 pathway plays a direct role in the radioresistance of tumor cells with distal 11q loss. Knockdown of CHEK1 or ATR expression by siRNA transfection showed that inhibiting the activity of the ATR-CHEK1 pathway can partially resensitize the cells with distal 11q loss to radiation therapy.
Targeted Inhibition of CHEK1 Radiosensitizes OSCC Cells
To advance these studies towards the clinic, we used a potent and specific targeted CHEK1 small molecule inhibitor (SMI), PF-00477736, to inhibit the enzymatic activity of CHEK1, and then assessed the effect of this inhibition on survival after radiation treatment. CHEK1 inhibitors are reported to selectively increase the efficacy of IR and other DNA damaging agents in cells with defective G1 checkpoints (e.g., as a result of defective TP53 signaling) while producing minimal damage in cells without G1 checkpoint defects (Blasina et al., 2008). Prior to SMI treatment, we tested our cell lines for TP53 functionality by treating the cells with the chemotherapeutic agent, Adriamycin and then assessing the protein expression of TP53 and its downstream target, p21 by immunoblotting. The results of these studies identified OSCC cell lines with distal 11q loss and defective TP53 signaling for the SMI experiments.
PF-00477736 dose response curves generated in OSCC cell lines in our lab and by other investigators (Blasina et al., 2008) showed that the optimal dose range required to achieve maximum effect is between 360 and 540 nM (data not shown). To determine the best time to add the SMI, clonogenic survival assays were carried out after adding the SMI at various timepoints around the radiation therapy. Addition of the SMI 8 h prior to irradiation was determined to have the maximum effect on cell killing (data not shown). OSCC cell lines UPCI:SCC040 and UPCI:SCC131 (both with distal 11q loss and defective TP53 signaling), when treated with 540 nM SMI in combination with IR resulted in a two-fold reduction in survival at 2.5 Gy and >four-fold reduction in survival at 5 Gy, compared to cells treated with IR alone (Fig. 7C). UPCI:SCC116 (no distal 11q loss, defective TP53 signaling) when treated with the same dose of SMI combined with IR showed no difference in survival between the untreated and SMI+IR-treated cells at 2.5 or 5 Gy. The SMI alone did not have a significant deleterious effect on these cells, but was lethal at 360 nM in UPCI:SCC066 cells (no distal 11q loss, defective TP53 signaling) and hTERT-transfected human OKF6 cells (data not shown). Our results indicate that the CHEK1 SMI, PF-00477736 mirrored the effect of the siRNA-based knockdown of the ATR-CHEK1 pathway in OSCC cell lines.
DISCUSSION
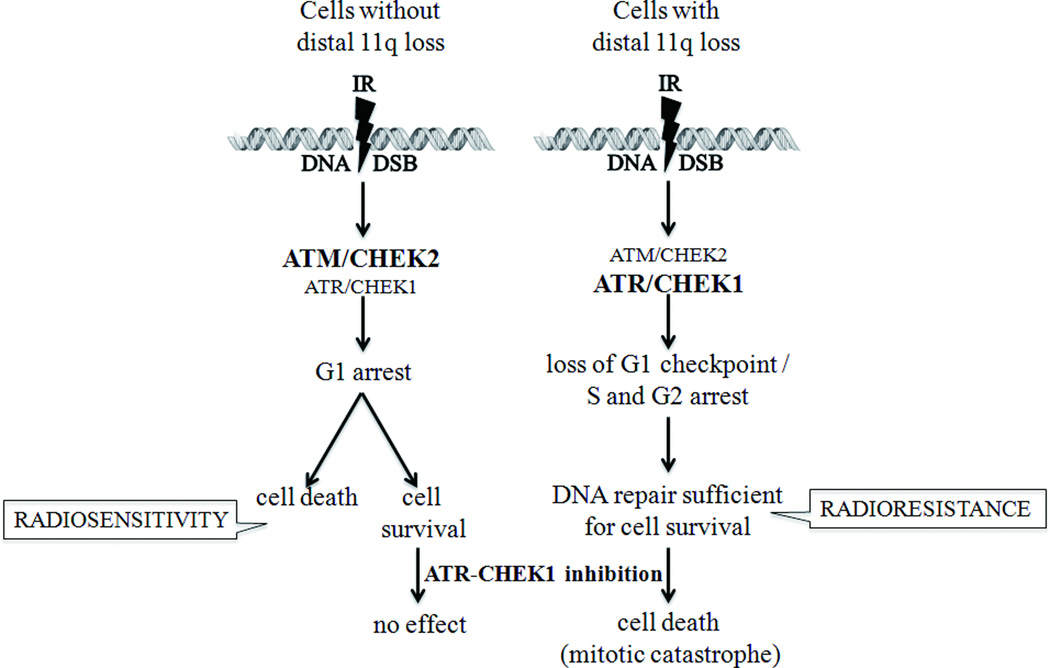
Although distal 11q copy number loss, from band 11q14 to 11qter has been reported in many types of tumors, its significance had not been investigated until our previous study (Parikh et al., 2007). Copy number loss appears to be centered around 11q22 to 11q23, in the vicinity of the ATM gene. We showed copy number loss of the region from 11q21 to 11q24 in about 25% of OSCC, breast and ovarian primary tumors and a high frequency of loss in OSCC cell lines (Parikh et al., 2007). The Broad Institute TCGA and Tumorscape websites support our observations, showing ATM copy number loss in about 20–25% of all cancers and frequencies of loss as high as 54% in cutaneous melanomas and 48% in invasive breast adenocarcinomas and HNSCC. We showed previously that distal 11q loss is associated with resistance to radiation therapy and our current results confirm and extend this finding (Parikh et al., 2007). AT cells, lacking a functional ATM protein, are radiosensitive because the presence of ATM protein 1 h post-irradiation is essential for the survival of cells (Choi et al., 2010). In contrast, tumor cells with distal 11q loss retain some ATM expression that based on our results, is sufficient to provide the cells the ability to ‘repair’ their DSB and survive IR treatment. AT cells that lack ATM protein upregulate the ATR-CHEK1 pathway resulting in a prolonged G2M arrest (Wang et al., 2003). Similarily we observed upregulation of the ATR-CHEK1 pathway and cell cycle arrest in the S and G2M phases after IR treatment of tumor cells with distal 11q loss. S/G2M arrest and subsequent repair prevents the entry into mitosis of cells with DNA damage, which would lead to mitotic catastrophe. Instead, these cells seem to ‘repair’ the damaged DNA, enabling survival and resulting in decreased sensitivity to IR. Knockdown of the ATR-CHEK1 pathway using siRNA or the Pfizer CHEK1 inhibitor resulted in resensitization of the cells with distal 11q loss to IR, which led to cell death by mitotic catastrophe (Fig. 8). Since CHEK1 inhibitors are being developed by several companies, our results suggest that distal 11q loss (with particular emphasis on ATM copy number loss) may be a useful companion diagnostic biomarker to select the subgroup of OSCC patients expected to respond poorly to radiation therapy, but better to combined radiation therapy and CHEK1 or ATR inhibition.
Figure 8.
11q13 amplification correlates with an aggressive tumor phenotype and poor clinical outcome (Jares et al., 1994; Akervall et al., 1995; Michalides et al., 1995; Fracchiolla et al., 1997; Michalides et al., 1997; Mineta et al., 2000; Miyamoto et al., 2003; Wreesmann et al., 2004; Wreesmann et al., 2004; Clark et al., 2009). In a series of 11 OSCC cell lines examined by chromosomal comparative genomic hybridization (cCGH), four of five patients whose tumors had 11q13 amplification/distal 11q loss died, whereas all six patients whose tumors lacked 11q13 amplification/distal 11q loss survived (Martin et al., 2008). Therefore, we examined 42 OSCC patients by Kaplan-Meier disease-specific survival analysis and found that patients whose tumors have distal 11q loss have a significantly worse outcome than patients whose tumors do not have 11q loss. Based on these results, we conclude that distal 11q loss is a frequent finding in carcinomas and correlates with a worse clinical outcome, most likely due to therapeutic resistance.
Copy number loss of ATM, MRE11A, and H2AFX results in decreased expression of these genes, reduced γH2AX focus formation in response to radiation-induced DSBs and chromosomal instability (Parikh et al., 2007). The intrinsic level of chromosomal instability observed in OSCC cell lines with distal 11q loss is higher than in normal keratinocytes or OSCC cells without 11q loss (Parikh et al., 2007). We observed a similar increase in mitotic segregation defects in OSCC cells with distal 11q loss after treatment with IR compared to OSCC cells without 11q loss. Increased chromosomal instability is consistent with a diminished DDR to IR-induced DNA damage in cells with distal 11q loss. The homologous recombination repair (HRR) mechanism is considered to be responsible for repairing IR-induced damage in the S and G2M phases of the cell cycle. Further, the ATR-CHEK1 pathway repairs IR-induced DNA damage by a mechanism independent of non-homologous end joining (NHEJ), but dependent on HRR (Wang et al., 2004; Wang et al., 2005). Although cells with distal 11q loss usually arrest in G2M phase after IR, the observed increased accumulation of mitotic segregation defects shown by our studies suggests repair by an error-prone mechanism that enables the cells to progress through mitosis without undergoing mitotic catastrophe. A third DNA repair pathway gaining significant attention is microhomology-mediated end joining (MMEJ) which might be involved in the repair of DSBs in our model. Further studies are warranted to clarify which of these three DNA repair pathways is responsible for radioresistance of tumor cells with distal 11q loss.
Our results show that distal 11q loss is associated with decreased expression of MRE11A, ATM, and H2AFX, which are important components of the ATM-CHEK2 DDR pathway (Parikh et al., 2007). Upregulation of the ATR-CHEK1 pathway may be compensating for the decreased functioning of the ATM-CHEK2 pathway. Despite copy number loss of the CHEK1 gene, CHEK1 expression, especially after IR treatment, appears to be increased in cells with distal 11q loss. CHEK1 expression is regulated by E2F transcription factors, with CHEK1 and E2F1 (transcriptional activator) expression showing a strong correlation (Verlinden et al., 2007). It has also been reported that E2F1 levels are higher in OSCC cell lines from highly invasive tumors (Zhang et al., 2000), and that phosphorylation of E2F1 by ATM and CHEK2 result in protein stabilization (Inoue et al., 2007). These findings suggest that transcriptional upregulation of the CHEK1 gene could be responsible for increased CHEK1 expression in tumor cell lines. This would explain the observed increase in CHEK1 expression in spite of copy number loss in OSCC cell lines. Increased ATR-dependent phosphorylation of CHEK1 ser345 in cells with distal 11q loss after IR treatment demonstrates upregulation of ATR at the apex of the ATR-CHEK1 pathway. Activated CHEK1 phosphorylates members of the CDC25 family, resulting in their inhibition. CDC25 phosphatases are known to dephosphorylate cyclin-dependent kinases (CDK), thereby activating them, resulting in cell cycle progression (Uto et al., 2004). Inhibition of CDC25C phosphatase results in cell cycle arrest at the G2M phase. Thus, upregulation of the ATR-CHEK1 pathway is likely responsible for the G2M arrest observed in tumor cells with distal 11q loss. This arrest would enable the tumor cells with distal 11q loss to ‘repair’ their DNA damage and progress through the cell cycle without undergoing mitotic catastrophe. Loss of the G1 checkpoint in OSCC leads to an increased number of cells with unrepaired DNA damage entering the S and the G2M phases of the cell cycle. Thus, OSCC cells with enhanced G2M phase cell cycle checkpoints may be able to avoid TP53-independent cell death by premature chromosome condensation/mitotic catastrophe (Fragkos and Beard, 2011) and may have a survival advantage as demonstrated by our radioresistant OSCC cells.
Our results demonstrate that distal 11q loss is associated with a decreased DDR, radioresistance, an upregulated ATR-CHEK1 pathway, and loss of the G1 checkpoint accompanied by G2M arrest. Since the ATR-CHEK1 pathway appears to function as a compensatory mechanism for a downregulated ATM-CHEK2 pathway, knocking it down with siRNA or a SMI should reverse the observed phenotype. Our results demonstrate that CHEK1 or ATR knockdown using siRNA or a CHEK1 SMI results in radiosensitization of cells with distal 11q loss. Increased cell death in OSCC cells with distal 11q loss is thought to be due to synthetic lethality resulting from loss of the G1 checkpoint and induced loss of the G2M checkpoint by CHEK1 or ATR knockdown or inhibition. Synthetic lethality after loss of cell cycle checkpoints by CHEK1 inhibition and TP53/p21 loss has been described recently (Origanti et al., 2013). Synthetic lethality-inducing drugs in the form of several targeted CHEK1 SMIs with promising effects are in the pipeline or clinical trials (Busby et al., 2000; Graves et al., 2000; Tao et al., 2009; Shaheen et al., 2011; Bennett et al., 2012; Borst et al., 2012; Ferrao et al., 2012; Karp et al., 2012; Schenk et al., 2012; Wu et al., 2012).
In conclusion, our results clearly show that cells with distal 11q loss have a decreased DDR and decreased sensitivity to treatment with IR. Upregulation of the ATR-CHEK1 pathway was also observed in these cells and knockdown of CHEK1 or ATR resulted in radiosensitization. Since inhibition of CHEK1 and ATR using SMIs is being developed as a promising targeted anti-tumor therapy, our results have substantial translational value. Distal 11q loss could be used as a biomarker predictive of a less favorable prognosis and decreased response to conventional radiation therapy. It could also be developed as a companion diagnostic to determine response to CHEK1 or ATR inhibition in combination with radiation or chemotherapy in the treatment of OSCCs and other tumors.
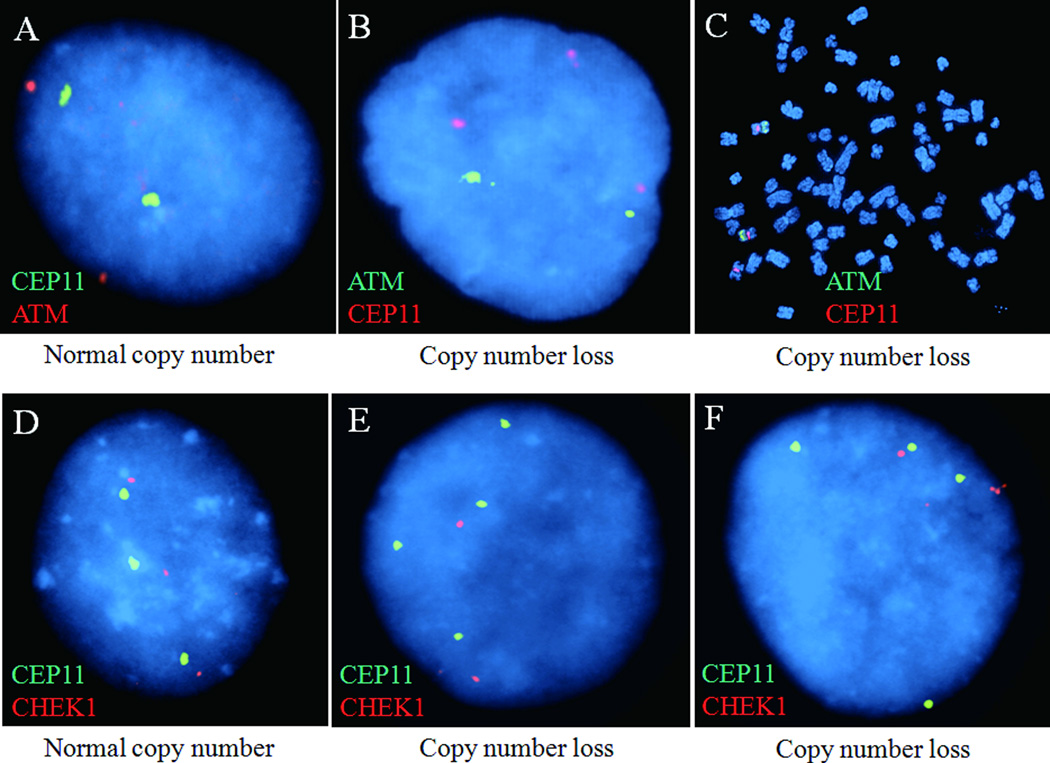
Figure 1.
Representative FISH images (A) showing normal copy number of ATM compared with CEP11; (B) and (C) showing loss of ATM copy number compared with CEP11; (D) showing normal copy number of CHEK1 compared with CEP11; and (E) and (F) showing loss of CHEK1 copy number compared with CEP11.
Table 3.
Frequency of mitotic segregation defects in OSCC cell lines.
| Cell Lines | Nuclear Appearance | Untreated | IR (+18 h) | IR (+36 h) |
|---|---|---|---|---|
| No Distal 11q loss (UPCI:SCC066, UPCI:SCC116) |
‧Normal’ | 95.5 | 90.6 | 80.3 |
| Anaphase bridges | 0.1 | 0.9 | 0.1 | |
| Micronuclei | 4.3 | 9 | 18.3 | |
| Interphase bridges | 0.1 | 0.4 | 1.2 | |
| Distal 11q loss (UPCI:SCC029B, UPCI:SCC040) |
‘Normal’ | 89.7 | 74.5 | 39.3 |
| Anaphase bridges | 0 | 0.5 | 0.3 | |
| Micronuclei | 7.9 | 21.8 | 49 | |
| Interphase bridges | 2.4 | 3.2 | 11.4 |
ACKNOWLEDGMENTS
The authors are grateful to Ms. Jianhua Zhou for assistance with the FISH analysis and other technical support. The authors are also grateful for the gift of PF-00477736 from Pfizer, Inc., helpful discussions from Sreesha Srinivasan, Ph.D., Wendy Levin, M.D., Indrawan McAlpine, Ph.D., Bonnie Dutcher, Ph.D., Matthew Cotter, Ph.D., and Ping Chen, Ph.D., and assistance from Mr. Donnie W. Owens and Ms. Donna Stocker. Molecular cytogenetic studies were carried out in the University of Pittsburgh Cell Culture and Cytogenetics Facility. In addition, we thank Dr. Andrew Remes from the University of Pittsburgh Office of Enterprise Development and Dr. Maria Vanegas of the University of Pittsburgh Office of Technology Management for helpful discussions. Flow cytometry was carried out in the UPCI Flow Cytometry Facility, supported in part by NIH award P30CA047904.
Supported in part by: NIH/NIDCR/NCI Grants R01DE014729, P30CA047904, and P50CA097190; University of Pittsburgh Cancer Institute; an Investigator-Initiated Grant (WS594969) from Pfizer, Inc.; Jennie K. Scaife Ovarian Cancer Center of Excellence at the University of Pittsburgh; Pittsburgh Foundation Learmonth Fund; Joan Gaines Cancer Research Fund at the University of Pittsburgh including donations from Mr. Bill Benter and the Teresa and H. John Heinz III Charitable Fund.
REFERENCES
- Abraham RT. PI 3-kinase related kinases: ‘big’ players in stress-induced signaling pathways. DNA Repair. 2004;3:883–887. doi: 10.1016/j.dnarep.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Adams KE, Medhurst AL, Dart DA, Lakin ND. Recruitment of ATR to sites of ionising radiation-induced DNA damage requires ATM and components of the MRN protein complex. Oncogene. 2006;25:3894–3904. doi: 10.1038/sj.onc.1209426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akervall JA, Jin Y, Wennerberg JP, Zatterstrom UK, Kjellen E, Mertens F, Willen R, Mandahl N, Heim S, Mitelman F. Chromosomal abnormalities involving 11q13 are associated with poor prognosis in patients with squamous cell carcinoma of the head and neck. Cancer. 1995;76:853–859. doi: 10.1002/1097-0142(19950901)76:5<853::aid-cncr2820760520>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Akervall JA, Michalides RJ, Mineta H, Balm A, Borg A, Dictor MR, Jin Y, Loftus B, Mertens F, Wennerberg JP. Amplification of cyclin D1 in squamous cell carcinoma of the head and neck and the prognostic value of chromosomal abnormalities and cyclin D1 overexpression. Cancer. 1997;79:380–389. [PubMed] [Google Scholar]
- Albertson DG. Gene amplification in cancer. Trends Genet. 2006;22:447–455. doi: 10.1016/j.tig.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Ambatipudi S, Gerstung M, Gowda R, Pai P, Borges AM, Schaffer AA, Beerenwinkel N, Mahimkar MB. Genomic profiling of advanced-stage oral cancers reveals chromosome 11q alterations as markers of poor clinical outcome. PLoS ONE. 2011;6:e17250. doi: 10.1371/journal.pone.0017250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- Baskaran R, Wood LD, Whitaker LL, Canman CE, Morgan SE, Xu Y, Barlow C, Baltimore D, Wynshaw-Boris A, Kastan MB, Wang JY. Ataxia telangiectasia mutant protein activates c-Abl tyrosine kinase in response to ionizing radiation. Nature. 1997;387:516–519. doi: 10.1038/387516a0. [DOI] [PubMed] [Google Scholar]
- Bennett CN, Tomlinson CC, Michalowski AM, Chu IM, Luger D, Mittereder LR, Aprelikova O, Shou J, Piwinica-Worms H, Caplen NJ, Hollingshead MG, Green JE. Cross-species genomic and functional analyses identify a combination therapy using a CHK1 inhibitor and a ribonucleotide reductase inhibitor to treat triple-negative breast cancer. Breast Cancer Res. 2012;14:R109. doi: 10.1186/bcr3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, Mc Henry KT, Pinchback RM, Ligon AH, Cho YJ, Haery L, Greulich H, Reich M, Winckler W, Lawrence MS, Weir BA, Tanaka KE, Chiang DY, Bass AJ, Loo A, Hoffman C, Prensner J, Liefeld T, Gao Q, Yecies D, Signoretti S, Maher E, Kaye FJ, Sasaki H, Tepper JE, Fletcher JA, Tabernero J, Baselga J, Tsao MS, Demichelis F, Rubin MA, Janne PA, Daly MJ, Nucera C, Levine RL, Ebert BL, Gabriel S, Rustgi AK, Antonescu CR, Ladanyi M, Letai A, Garraway LA, Loda M, Beer DG, True LD, Okamoto A, Pomeroy SL, Singer S, Golub TR, Lander ES, Getz G, Sellers WR, Meyerson M. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasina A, Hallin J, Chen E, Arango ME, Kraynov E, Register J, Grant S, Ninkovic S, Chen P, Nichols T, O’Connor P, Anderes K. Breaching the DNA damage checkpoint via PF-00477736, a novel small-molecule inhibitor of checkpoint kinase 1. Mol Cancer Ther. 2008;7:2394–2404. doi: 10.1158/1535-7163.MCT-07-2391. [DOI] [PubMed] [Google Scholar]
- Borst GR, McLaughlin M, Kyula JN, Neijenhuis S, Khan A, Good J, Zaidi S, Powell NG, Meier P, Collins I, Garrett MD, Verheij M, Harrington KJ. Targeted Radiosensitization by the Chk1 Inhibitor SAR-020106. Int J Radiat Oncol Biol Phys. 2012 doi: 10.1016/j.ijrobp.2012.08.006. [DOI] [PubMed] [Google Scholar]
- Bray F, Ren JS, Masuyer E, Ferlay J. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int J Cancer. 2013;132:1133–1145. doi: 10.1002/ijc.27711. [DOI] [PubMed] [Google Scholar]
- Busby EC, Leistritz DF, Abraham RT, Karnitz LM, Sarkaria JN. The radiosensitizing agent 7-hydroxystaurosporine (UCN-01) inhibits the DNA damage checkpoint kinase hChk1. Cancer Res. 2000;60:2108–2112. [PubMed] [Google Scholar]
- Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S, Gamper AM, White JS, Bakkenist CJ. Inhibition of ATM kinase activity does not phenocopy ATM protein disruption: implications for the clinical utility of ATM kinase inhibitors. Cell Cycle. 2010;9:4052–4057. doi: 10.4161/cc.9.20.13471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark ES, Brown B, Whigham AS, Kochaishvili A, Yarbrough WG, Weaver AM. Aggressiveness of HNSCC tumors depends on expression levels of cortactin, a gene in the 11q13 amplicon. Oncogene. 2009;28:431–444. doi: 10.1038/onc.2008.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D, Guntuku S, Qin J, Elledge SJ. ATR and ATRIP: partners in checkpoint signaling. Science. 2001;294:1713–1716. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- Cuadrado M, Martinez-Pastor B, Murga M, Toledo LI, Gutierrez-Martinez P, Lopez E, Fernandez-Capetillo O. ATM regulates ATR chromatin loading in response to DNA double-strand breaks. J Exp Med. 2006;203:297–303. doi: 10.1084/jem.20051923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desantis C, Naishadham D, Jemal A. Cancer statistics for African Americans, 2013. CA Cancer J Clin. 2013;63:151–166. doi: 10.3322/caac.21173. [DOI] [PubMed] [Google Scholar]
- Dickson MA, Hahn WC, Ino Y, Ronfard V, Wu JY, Weinberg RA, Louis DN, Li FP, Rheinwald JG. Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol Cell Biol. 2000;20:1436–1447. doi: 10.1128/mcb.20.4.1436-1447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann J, Holzmann K, Miller F, Winkler D, Buhler A, Zenz T, Bullinger L, Kuhn MW, Gerhardinger A, Bloehdorn J, Radtke I, Su X, Ma J, Pounds S, Hallek M, Lichter P, Korbel J, Busch R, Mertens D, Downing JR, Stilgenbauer S, Dohner H. High-resolution genomic profiling of chronic lymphocytic leukemia reveals new recurrent genomic alterations. Blood. 2012;120:4783–4794. doi: 10.1182/blood-2012-04-423517. [DOI] [PubMed] [Google Scholar]
- Fenech M, Kirsch-Volders M, Natarajan AT, Surralles J, Crott JW, Parry J, Norppa H, Eastmond DA, Tucker JD, Thomas P. Molecular mechanisms of micronucleus, nucleoplasmic bridge and nuclear bud formation in mammalian and human cells. Mutagenesis. 2011;26:125–132. doi: 10.1093/mutage/geq052. [DOI] [PubMed] [Google Scholar]
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM Cancer IAfRo. GLOBOCAN 2008 v1.2, cancer Incidence and Mortality Worldwide: IARC CancerBase No.10. Lyon, France: International Agency for Research on Cancer; 2010. [Google Scholar]
- Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A. H2AX: the histone guardian of the genome. DNA Repair. 2004;3:959–967. doi: 10.1016/j.dnarep.2004.03.024. [DOI] [PubMed] [Google Scholar]
- Ferrao PT, Bukczynska EP, Johnstone RW, McArthur GA. Efficacy of CHK inhibitors as single agents in MYC-driven lymphoma cells. Oncogene. 2012;31:1661–1672. doi: 10.1038/onc.2011.358. [DOI] [PubMed] [Google Scholar]
- Fracchiolla NS, Pruneri G, Pignataro L, Carboni N, Capaccio P, Boletini A, Buffa R, Neri A. Molecular and immunohistochemical analysis of the bcl-1/cyclin D1 gene in laryngeal squamous cell carcinomas: correlation of protein expression with lymph node metastases and advanced clinical stage. Cancer. 1997;79:1114–1121. [PubMed] [Google Scholar]
- Fragkos M, Beard P. Mitotic catastrophe occurs in the absence of apoptosis in p53-null cells with a defective G1 checkpoint. PLoS One. 2011;6:e22946. doi: 10.1371/journal.pone.0022946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatei M, Sloper K, Sorensen C, Syljuasen R, Falck J, Hobson K, Savage K, Lukas J, Zhou BB, Bartek J, Khanna KK. Ataxia-telangiectasia-mutated (ATM) and NBS1-dependent phosphorylation of Chk1 on Ser-317 in response to ionizing radiation. J Biol Chem. 2003;278:14806–14811. doi: 10.1074/jbc.M210862200. [DOI] [PubMed] [Google Scholar]
- George RE, Attiyeh EF, Li S, Moreau LA, Neuberg D, Li C, Fox EA, Meyerson M, Diller L, Fortina P, Look AT, Maris JM. Genome-wide analysis of neuroblastomas using high-density single nucleotide polymorphism arrays. PLoS One. 2007;2:e255. doi: 10.1371/journal.pone.0000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibcus JH, Kok K, Menkema L, Hermsen MA, Mastik M, Kluin PM, van der Wal JE, Schuuring E. High-resolution mapping identifies a commonly amplified 11q13.3 region containing multiple genes flanked by segmental duplications. Hum Genet. 2007;121:187–201. doi: 10.1007/s00439-006-0299-6. [DOI] [PubMed] [Google Scholar]
- Gollin SM. Chromosomal alterations in squamous cell carcinomas of the head and neck: window to the biology of disease. Head Neck. 2001;23:238–253. doi: 10.1002/1097-0347(200103)23:3<238::aid-hed1025>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Graves PR, Yu L, Schwarz JK, Gales J, Sausville EA, O’Connor PM, Piwnica-Worms H. The Chk1 protein kinase and the Cdc25C regulatory pathways are targets of the anticancer agent UCN-01. J Biol Chem. 2000;275:5600–5605. doi: 10.1074/jbc.275.8.5600. [DOI] [PubMed] [Google Scholar]
- Ha PK, Califano JA., 3rd The molecular biology of laryngeal cancer. Otolaryngol Clin North Am. 2002;35:993–1012. doi: 10.1016/s0030-6665(02)00038-5. [DOI] [PubMed] [Google Scholar]
- Harnden DG. The nature of ataxia-telangiectasia: problems and perspectives. Int J Radiat Biol. 1994;66:S13–S19. [PubMed] [Google Scholar]
- Henson BJ, Bhattacharjee S, O’Dee DM, Feingold E, Gollin SM. Decreased expression of miR-125b and miR-100 in oral cancer cells contributes to malignancy. Genes Chromosomes Cancer. 2009;48:569–582. doi: 10.1002/gcc.20666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heron M, Hoyert DL, Murphy SL, Xu J, Kochanek KD, Tejada-Vera B. Deaths: final data for 2006. Natl Vital Stat Rep. 2009;57:1–134. [PubMed] [Google Scholar]
- Huang X, Gollin SM, Raja S, Godfrey TE. High-resolution mapping of the 11q13 amplicon and identification of a gene, TAOS1, that is amplified and overexpressed in oral cancer cells. Proc Natl Acad Sci U S A. 2002;99:11369–11374. doi: 10.1073/pnas.172285799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y, Kitagawa M, Taya Y. Phosphorylation of pRB at Ser612 by Chk1/2 leads to a complex between pRB and E2F-1 after DNA damage. EMBO J. 2007;26:2083–2093. doi: 10.1038/sj.emboj.7601652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jares P, Fernandez PL, Campo E, Nadal A, Bosch F, Aiza G, Nayach I, Traserra J, Cardesa A. PRAD-1/cyclin D1 gene amplification correlates with messenger RNA overexpression and tumor progression in human laryngeal carcinomas. Cancer Res. 1994;54:4813–4817. [PubMed] [Google Scholar]
- Jiang K, Pereira E, Maxfield M, Russell B, Goudelock DM, Sanchez Y. Regulation of Chk1 includes chromatin association and 14-3-3 binding following phosphorylation on Ser-345. J Biol Chem. 2003;278:25207–25217. doi: 10.1074/jbc.M300070200. [DOI] [PubMed] [Google Scholar]
- Jin C, Jin Y, Gisselsson D, Wennerberg J, Wah TS, Stromback B, Kwong YL, Mertens F. Molecular cytogenetic characterization of the 11q13 amplicon in head and neck squamous cell carcinoma. Cytogenet Genome Res. 2006;115:99–106. doi: 10.1159/000095228. [DOI] [PubMed] [Google Scholar]
- Jin Y, Hoglund M, Jin C, Martins C, Wennerberg J, Akervall J, Mandahl N, Mitelman F, Mertens F. FISH characterization of head and neck carcinomas reveals that amplification of band 11q13 is associated with deletion of distal 11q. Genes Chromosomes Cancer. 1998;22:312–320. [PubMed] [Google Scholar]
- Karp JE, Thomas BM, Greer JM, Sorge C, Gore SD, Pratz KW, Smith BD, Flatten KS, Peterson K, Schneider P, Mackey K, Freshwater T, Levis MJ, McDevitt MA, Carraway HE, Gladstone DE, Showel MM, Loechner S, Parry DA, Horowitz JA, Isaacs R, Kaufmann SH. Phase I and pharmacologic trial of cytosine arabinoside with the selective checkpoint 1 inhibitor Sch 900776 in refractory acute leukemias. Clin Cancer Res. 2012;18:6723–6731. doi: 10.1158/1078-0432.CCR-12-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith CT, Schreiber SL. PIK-related kinases: DNA repair, recombination, and cell cycle checkpoints. Science. 1995;270:50–51. doi: 10.1126/science.270.5233.50. [DOI] [PubMed] [Google Scholar]
- Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- Lavin MF, Shiloh Y. The genetic defect in ataxia-telangiectasia. Annu Rev Immunol. 1997;15:177–202. doi: 10.1146/annurev.immunol.15.1.177. [DOI] [PubMed] [Google Scholar]
- Martin C, Reshmi S, Ried T, Gottberg W, Wilson J, Reddy J, Khanna P, Johnson J, Myers E, Gollin S. Chromosomal imbalances in oral squamous cell carcinoma: examination of 31 cell lines and review of the literature. Oral Oncol. 2008;44:369–382. doi: 10.1016/j.oraloncology.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalides R, van Veelen N, Hart A, Loftus B, Wientjens E, Balm A. Overexpression of cyclin D1 correlates with recurrence in a group of forty-seven operable squamous cell carcinomas of the head and neck. Cancer Res. 1995;55:975–978. [PubMed] [Google Scholar]
- Michalides RJ, van Veelen NM, Kristel PM, Hart AA, Loftus BM, Hilgers FJ, Balm AJ. Overexpression of cyclin D1 indicates a poor prognosis in squamous cell carcinoma of the head and neck. Arch Otolaryngol Head Neck Surg. 1997;123:497–502. doi: 10.1001/archotol.1997.01900050045005. [DOI] [PubMed] [Google Scholar]
- Mineta H, Miura K, Takebayashi S, Ueda Y, Misawa K, Harada H, Wennerberg J, Dictor M. Cyclin D1 overexpression correlates with poor prognosis in patients with tongue squamous cell carcinoma. Oral oncology. 2000;36:194–198. doi: 10.1016/s1368-8375(99)00078-0. [DOI] [PubMed] [Google Scholar]
- Miyamoto R, Uzawa N, Nagaoka S, Hirata Y, Amagasa T. Prognostic significance of cyclin D1 amplification and overexpression in oral squamous cell carcinomas. Oral oncology. 2003;39:610–618. doi: 10.1016/s1368-8375(03)00048-4. [DOI] [PubMed] [Google Scholar]
- Myers JS, Cortez D. Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J Biol Chem. 2006;281:9346–9350. doi: 10.1074/jbc.M513265200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niida H, Katsuno Y, Banerjee B, Hande MP, Nakanishi M. Specific role of Chk1 phosphorylations in cell survival and checkpoint activation. Mol Cell Biol. 2007;27:2572–2581. doi: 10.1128/MCB.01611-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Origanti S, Cai SR, Munir AZ, White LS, Piwnica-Worms H. Synthetic lethality of Chk1 inhibition combined with p53 and/or p21 loss during a DNA damage response in normal and tumor cells. Oncogene. 2013;32:577–588. doi: 10.1038/onc.2012.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandita TK, Westphal CH, Anger M, Sawant SG, Geard CR, Pandita RK, Scherthan H. Atm inactivation results in aberrant telomere clustering during meiotic prophase. Mol Cell Biol. 1999;19:5096–5105. doi: 10.1128/mcb.19.7.5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh RA, White JS, Huang X, Schoppy DW, Baysal BE, Baskaran R, Bakkenist CJ, Saunders WS, Hsu LC, Romkes M, Gollin SM. Loss of distal 11q is associated with DNA repair deficiency and reduced sensitivity to ionizing radiation in head and neck squamous cell carcinoma. Genes Chromosomes Cancer. 2007;46:761–775. doi: 10.1002/gcc.20462. [DOI] [PubMed] [Google Scholar]
- Reshmi SC, Roychoudhury S, Yu Z, Feingold E, Potter D, Saunders WS, Gollin SM. Inverted duplication pattern in anaphase bridges confirms the breakage-fusion-bridge (BFB) cycle model for 11q13 amplification. Cytogenet Genome Res. 2007;116:46–52. doi: 10.1159/000097425. [DOI] [PubMed] [Google Scholar]
- Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle DA, Smith S, Uziel T, Sfez S, Ashkenazi M, Pecker I, Frydman M, Harnik R, Patanjali SR, Simmons A, Clines GA, Sartiel A, Gatti RA, Chessa L, Sanal O, Lavin MF, Jaspers NG, Taylor AM, Arlett CF, Miki T, Weissman SM, Lovett M, Collins FS, Shiloh Y. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- Schenk EL, Koh BD, Flatten KS, Peterson KL, Parry D, Hess AD, Smith BD, Karp JE, Karnitz LM, Kaufmann SH. Effects of selective checkpoint kinase 1 inhibition on cytarabine cytotoxicity in acute myelogenous leukemia cells in vitro. Clin Cancer Res. 2012;18:5364–5373. doi: 10.1158/1078-0432.CCR-12-0961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schraml P, Kononen J, Bubendorf L, Moch H, Bissig H, Nocito A, Mihatsch MJ, Kallioniemi OP, Sauter G. Tissue microarrays for gene amplification surveys in many different tumor types. Clin Cancer Res. 1999;5:1966–1975. [PubMed] [Google Scholar]
- Shaheen M, Allen C, Nickoloff JA, Hromas R. Synthetic lethality: exploiting the addiction of cancer to DNA repair. Blood. 2011;117:6074–6082. doi: 10.1182/blood-2011-01-313734. [DOI] [PubMed] [Google Scholar]
- Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- Shuster MI, Han L, Le Beau MM, Davis E, Sawicki M, Lese CM, Park NH, Colicelli J, Gollin SM. A consistent pattern of RIN1 rearrangements in oral squamous cell carcinoma cell lines supports a breakage-fusion-bridge cycle model for 11q13 amplification. Genes Chromosomes Cancer. 2000;28:153–163. [PubMed] [Google Scholar]
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- Stracker TH, Theunissen JW, Morales M, Petrini JH. The Mre11 complex and the metabolism of chromosome breaks: the importance of communicating and holding things together. DNA Repair. 2004;3:845–854. doi: 10.1016/j.dnarep.2004.03.014. [DOI] [PubMed] [Google Scholar]
- Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- Swarts DR, Claessen SM, Jonkers YM, van Suylen RJ, Dingemans AM, de Herder WW, de Krijger RR, Smit EF, Thunnissen FB, Seldenrijk CA, Vink A, Perren A, Ramaekers FC, Speel EJ. Deletions of 11q22.3-q25 are associated with atypical lung carcinoids and poor clinical outcome. Am J Pathol. 2011;179:1129–1137. doi: 10.1016/j.ajpath.2011.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Leteur C, Yang C, Zhang P, Castedo M, Pierre A, Golsteyn RM, Bourhis J, Kroemer G, Deutsch E. Radiosensitization by Chir-124, a selective CHK1 inhibitor: effects of p53 and cell cycle checkpoints. Cell Cycle. 2009;8:1196–1205. doi: 10.4161/cc.8.8.8203. [DOI] [PubMed] [Google Scholar]
- Uto K, Inoue D, Shimuta K, Nakajo N, Sagata N. Chk1, but not Chk2, inhibits Cdc25 phosphatases by a novel common mechanism. EMBO J. 2004;23:3386–3396. doi: 10.1038/sj.emboj.7600328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verlinden L, Vanden Bempt I, Eelen G, Drijkoningen M, Verlinden I, Marchal K, De Wolf-Peeters C, Christiaens MR, Michiels L, Bouillon R, Verstuyf A. The E2F–regulated gene Chk1 is highly expressed in triple-negative estrogen receptor /progesterone receptor /HER-2 breast carcinomas. Cancer Res. 2007;67:6574–6581. doi: 10.1158/0008-5472.CAN-06-3545. [DOI] [PubMed] [Google Scholar]
- Wang H, Hu B, Liu R, Wang Y. CHK1 affecting cell radiosensitivity is independent of non-homologous end joining. Cell Cycle. 2005;4:300–303. [PubMed] [Google Scholar]
- Wang H, Powell SN, Iliakis G, Wang Y. ATR affecting cell radiosensitivity is dependent on homologous recombination repair but independent of nonhomologous end joining. Cancer Res. 2004;64:7139–7143. doi: 10.1158/0008-5472.CAN-04-1289. [DOI] [PubMed] [Google Scholar]
- Wang J, Han X, Zhang Y. Autoregulatory mechanisms of phosphorylation of checkpoint kinase 1. Cancer Res. 2012;72:3786–3794. doi: 10.1158/0008-5472.CAN-12-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Khadpe J, Hu B, Iliakis G, Wang Y. An overactivated ATR/CHK1 pathway is responsible for the prolonged G2 accumulation in irradiated AT cells. J Biol Chem. 2003;278:30869–30874. doi: 10.1074/jbc.M301876200. [DOI] [PubMed] [Google Scholar]
- White JS, Weissfeld JL, Ragin CC, Rossie KM, Martin CL, Shuster M, Ishwad CS, Law JC, Myers EN, Johnson JT, Gollin SM. The influence of clinical and demographic risk factors on the establishment of head and neck squamous cell carcinoma cell lines. Oral Oncol. 2007;43:701–712. doi: 10.1016/j.oraloncology.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wreesmann VB, Shi W, Thaler HT, Poluri A, Kraus DH, Pfister D, Shaha AR, Shah JP, Rao PH, Singh B. Identification of novel prognosticators of outcome in squamous cell carcinoma of the head and neck. J Clin Oncol. 2004;22:3965–3972. doi: 10.1200/JCO.2004.01.094. [DOI] [PubMed] [Google Scholar]
- Wreesmann VB, Wang D, Goberdhan A, Prasad M, Ngai I, Schnaser EA, Sacks PG, Singh B. Genetic abnormalities associated with nodal metastasis in head and neck cancer. Head & Neck. 2004;26:10–15. doi: 10.1002/hed.10344. [DOI] [PubMed] [Google Scholar]
- Wu W, Bi C, Bence AK, Um SL, Yan B, Starling JJ, Marshall MS, Beckmann RP. Antitumor activity of Chk1 inhibitor LY2606368 as a single agent in SW1990 human pancreas orthotopic tumor model. Proceedings of the 103rd Annual Meeting of the American Association for Cancer Research; 2012 Mar 31–Apr 4; Chicago, IL. Philadelphia (PA): AACR. Cancer Res. 2012;72(8 Suppl) 2012. Abstract nr 1776. [Google Scholar]
- Zhang SY, Liu SC, Johnson DG, Klein-Szanto AJ. E2F-1 gene transfer enhances invasiveness of human head and neck carcinoma cell lines. Cancer Res. 2000;60:5972–5976. [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]