Abstract
Purpose
Cytochrome b5 (encoded by CYB5A) and NADH cytochrome b5 reductase (encoded by CYB5R3) detoxify aromatic and heterocyclic amine mammary carcinogens found in cigarette smoke. We hypothesized that CYB5A and CYB5R3 polymorphisms would be associated with breast cancer risk in women.
Methods
We characterized the prevalence of 18 CYB5A and CYB5R3 variants in genomic DNA from African American (AfrAm) and Caucasian (Cauc) women from the Carolina Breast Cancer Study population (1946 cases and 1747 controls), and determined their associations with breast cancer risk, with effect modification by smoking.
Results
A CYB5R3 variant, I1M+6T (rs8190370) was significantly more common in breast cancer cases (MAF 0.0238) compared to controls (0.0169, P =0.039); this was attributable to a higher MAF in AfrAm cases (0.0611) compared to AfrAm controls (0.0441, P=0.046; adjusted OR 1.41, CI 0.98-2.04; P=0.062). When smoking was considered, I1M+6T was more strongly associated with breast cancer risk in AfrAm smokers (adjusted OR 2.10, 1.08-4.07; P=0.028) compared to never-smokers (OR=1.21; 0.77-1.88; P for interaction=0.176). I1M+6T and three additional CYB5R3 variants, -251T, I8-1676C, and *392C, as well as two CYB5A variants, 13G and I2-992T, were significantly more common in AfrAms compared to Caucs.
Conclusions
CYB5R3 I1M+6 C>T should be considered in future molecular epidemiologic studies of breast cancer risk in AfrAms. Further, variants in CYB5A and CYB5R3 should be considered in the evaluation of other tumors in AfrAms that are associated with aromatic and heterocyclic amine exposures, to include prostate, bladder, and colon cancers.
Keywords: Heterocyclic amines, aromatic amines, PhIP, 4-aminobiphenyl, smoking
Introduction
Breast cancer has a high incidence in industrialized countries, and women who move from low-risk to high-risk countries acquire a breast cancer risk of the host country in as little as two generations [1,2]. Environmental factors including diet, smoking, and pollutants appear to play an etiologic role. The aromatic amine 4-aminobiphenyl (4-ABP), is a mammary carcinogen in rodents that is found in cigarette smoke [3,4], and the heterocyclic amine 2-amino-1-methyl-6-phenylimidazo[4,5-b] pyridine (PhIP), is another mammary carcinogen found in cigarette smoke and well done meats [5-7]. Both 4-ABP and PhIP lead to DNA adducts that have been found in the breast tissue and milk of women [8,9]. These adducts have also been correlated with smoking exposures [10,11]. Further, some studies have found higher levels of DNA adducts in breast cancer patients versus controls [12,13], although an established relationship between these DNA adducts and breast cancer remains to be proven. Epidemiologic studies of smoking and breast cancer risk have been mixed, with some studies finding modest positive associations [14-20], and other studies yielding no association [21-28]. These inconsistent results could be due, in part, to individual differences in the disposition of tobacco carcinogens after similar smoking exposures.
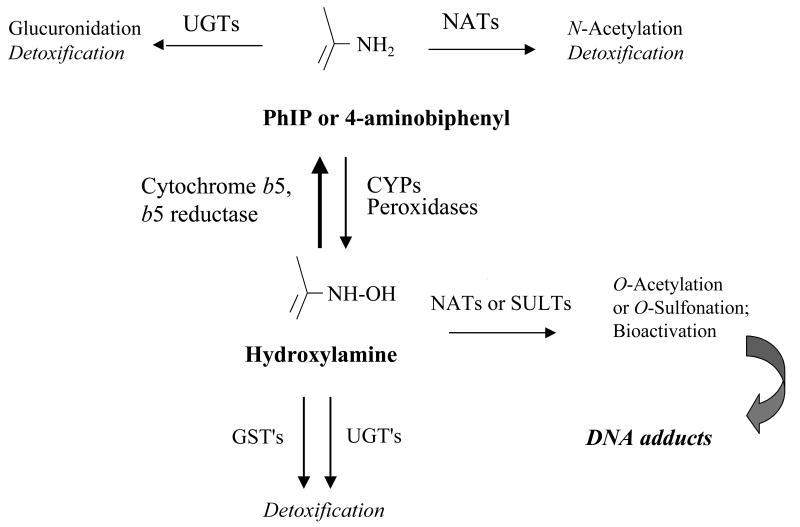
Both PhIP and 4-ABP are bioactivated to arylhydroxylamine metabolites [29-31], which ultimately form mutagenic DNA adducts (Figure 1). The disposition of these arylhydroxylamines appears to vary among individuals, in that exposure of individual primary human mammary epithelial cell lines to standard concentrations of PhIP hydroxylamine results in more than 75-fold variability in DNA adduct formation [32]. The hydroxylamines of PhIP and 4-ABP are reduced back to their parent compounds, which are not directly mutagenic, by cytochrome b5 (b5) and NADH cytochrome b5 reductase (b5R) [33]. Reduction of arylhydroxylamines by this pathway is substantially more efficient than generation of these metabolites by oxidation of the parent compound [34], which suggests that variability in the b5/b5R pathway could modulate the amount of arylhydroxylamine available for DNA adduct formation.
Figure 1.
Metabolism and bioactivation of carcinogenic aromatic and heterocyclic amines. Aromatic and heterocyclic amines may initially be detoxified by either glucuronidation or N-acetylation (by NAT1 or NAT2). Alternatively, the parent amines can be bioactivated to their hydroxylamine metabolites via cytochrome P450′s (CYP1A2, 1A1, and 1B1), myeloperoxidases, or lactoperoxidases. Hydroxylamine metabolites are reduced back to their parent compound by NADH cytochrome b5 reductase (b5R) and cytochrome b5 (b5) [33]. Reverse reduction by the b5/b5R pathway is up to 55 times more efficient than forward oxidation by P450′s [34]. Hydroxylamine metabolites may also be detoxified by glutathione S-transferases (GST′s) or UDP-glucuronosyltransferases (UGT's), or can be further bioactivated by O-acetylation or O-sulfonation. This final bioactivation step can lead to DNA adducts via arylnitrenium ion formation.
We have found individual differences in cytochrome b5 and NADH cytochrome b5 reductase protein expression and arylhydroxylamine detoxification activities in both human liver and breast samples, with more than 75-fold variability in activities in 70 breast samples from women undergoing reduction mammoplasty or lumpectomy [35,36]. We further found polymorphisms in the genes CYB5A and CYB5R3, which encode b5 and b5R, in tissues with outlier low protein expression and activities [35,36]. Many of these variants were found only or predominantly in African American samples.
Based on these findings, we hypothesized that polymorphisms in CYB5A or CYB5R3 contribute to breast cancer risk, particularly in women who smoke. We further hypothesized that such polymorphisms would be more common in African American compared to Caucasian women. The purpose of this study, therefore, was to screen for the prevalence of 18 CYB5A and CYB5R3 variants in African American and Caucasian women with breast cancer (invasive or carcinoma in situ (CIS)), compared to age-matched unaffected controls, using the Carolina Breast Cancer Study population [37], and to determine whether CYB5A or CYB5R3 polymorphisms were associated with breast cancer risk in relationship to smoking.
Materials and Methods
Carolina Breast Cancer Study population
The Carolina Breast Cancer Study (CBCS) is a case-control study population of women with breast cancer from 24 counties of central and eastern North Carolina [37]. Women who were diagnosed with invasive breast cancer or CIS between the ages of 20-74 were identified through the North Carolina Central Cancer Registry; population-based controls were frequency matched to cases by age (within 5 years) and race [37]. Race was self-reported, and 144 ancestry informative markers (AIMs) were used to estimate West African genetic ancestry and control for population stratification [38]. The majority of subjects in the CBCS were Caucasian (Cauc) or African American (AfrAm) [39], with 1.5% of the population from Native American, Asian, Hispanic, or multi-racial groups. Only Cauc and AfrAm subjects were analyzed in the present study, to include DNA samples from 1946 breast cancer cases (742 AfrAm and 1204 Cauc) and 1747 controls (658 AfrAm and 1089 Cauc). Smoking history was recorded for all subjects, to include active status, packs per day, duration, and time since cessation [27]. Characteristics of CBCS participants included in the study are listed in Table 1.
Table 1.
Characteristics of Carolina Breast Cancer Study (CBCS) patients with invasive carcinomas and carcinoma in situ, and unaffected controls included in CYB5A and CYB5R3 genotyping.
| Controls | Cases | |||
|---|---|---|---|---|
| N | % | N | % | |
| African American | 658 | 100 | 742 | 100 |
| Median age in years (range) | 50 (26–74) | 51 (23-74) | ||
| Mean proportion of African ancestry | 0.774 | 0.778 | ||
| Age (years) | ||||
| <50 | 314 | 47.7 | 355 | 47.8 |
| >=50 | 344 | 52.3 | 387 | 52.2 |
| Menopausal status | ||||
| Premenopausal | 290 | 44.1 | 324 | 43.7 |
| Postmenopausal | 368 | 55.9 | 418 | 56.3 |
| Education | ||||
| <High school (HS) | 198 | 30.1 | 215 | 29.0 |
| HS & Post HS | 348 | 53.0 | 406 | 54.8 |
| >=College | 111 | 16.9 | 120 | 16.2 |
| Smoking | ||||
| Ever | 262 | 39.8 | 322 | 43.4 |
| Never | 396 | 60.2 | 420 | 56.6 |
| Caucasian | 1089 | 100 | 1204 | 100 |
| Median age in years (range) | 51 (21–74) | 50 (24-74) | ||
| Mean proportion of African ancestry | 0.066 | 0.064 | ||
| Age (years) | ||||
| <50 | 491 | 45.1 | 592 | 49.2 |
| >=50 | 598 | 54.9 | 612 | 50.8 |
| Menopausal status | ||||
| Premenopausal | 456 | 41.9 | 540 | 44.9 |
| Postmenopausal | 633 | 58.1 | 664 | 55.2 |
| Education | ||||
| <High school | 115 | 10.6 | 103 | 8.6 |
| HS & Post HS | 613 | 56.3 | 671 | 55.7 |
| >=College | 361 | 33.1 | 430 | 35.7 |
| Smoking | ||||
| Ever | 546 | 50.1 | 587 | 48.8 |
| Never | 543 | 49.9 | 617 | 51.3 |
Genotyping
Genomic DNA samples from all women were genotyped for 18 single nucleotide polymorphisms (SNPs), 8 in CYB5A (Table 2) and 10 in CYB5R3 (Table 3). SNPs were selected based on the following criteria: 1) a variant previously observed in breast or liver samples with low protein expression or arylhydroxylamine reduction activities [35,36]; 2) a reported non-synonymous cSNP (NCBI SNP database; www.ncbi.nlm.nih.gov/); or 3) a reported SNP in a region predicted to affect protein function, splicing, or transcription factor or miRNA binding, even if apparently rare. For the latter predictions, the possible effects of coding polymorphisms on protein function were evaluated using PMut (mmb.pcb.ub.es/PMut/), SIFT (sift.jcvi.org/), and PolyPhen-2 (genetics.bwh.harvard.edu/pph2/ index.shtml) software. The effects of intronic variants on splicing were modeled with the Human Splicing Finder suite of software (HSF, MaxEnt, and ESE Finder; www.umd.be/HSF/). The influence of variants on transcription factor binding was predicted with the MATCH program (www.bioinfo.de/isb/gcb01/poster/goessling.html) and the ENCODE data base [40] (http://genome.cse.ucsc.edu/encode/), and the effects of 3′UTR (untranslated region) SNPs on miRNA binding were predicted with UTRscan (itbtools.ba.itb.cnr.it/utrscan).
Table 2.
Single nucleotide polymorphisms in CYB5A selected for genotyping in the Carolina Breast Cancer Study population, along with rationale for selection.
| SNP | SNP ID | Rationale for selection |
|---|---|---|
| -206G>- | rs36082929 | Reported in the NCBI SNP database.Within binding sites of several transcription factors (YY1, NFκ-B, NF-κB, NF- κA, and c-Fos).a |
| 13T>GSer5Ala | rs75160992 | Found in 2 of 111 liver samples (both heterozygous African Americans) in association with outlier low protein expression and activity [35]. |
| 65A>GHis22Arg | rs74339771 | Found in 1 of 70 breast samples (heterozygous African American woman) in association with low protein expression and activity [36]. Predicted to be deleterious to protein function. b |
| I2-992C>T | rs1790894 | Reported in the NCBI SNP database. Within binding sites for NF κ-B. a |
| 155G>A Arg52Lys | rs1803366 | Reported in the NCBI SNP database Predicted to be deleterious to protein function. c |
| 178A>G Thr60Ala | rs78009726 | Found in 1 of 63 leukocyte cDNA samples in a heterozygous African American subject [59]. Variant protein undergoes accelerated proteasomal degradation [59]. |
| 390C>ATyr130stop | rs1803364 | Reported in the NCBI SNP database.Predicted to be deleterious due to stop codon. d |
| *246T>C | rs76241580 | Found in 1 of 69 liver samples (Caucasian heterozygote)in association with outlier low b5 protein expression andactivity; e Located in internal ribosomal entry site [60]; may affect translation initiatio [61]. |
ENCODE database and ChIP experimental data (www.genome.ucsc.edu);
PMut;
SIFT;
Polyphen-2;
Unpublished data from Sacco et al. 2010.
Table 3.
Single nucleotide polymorphisms in CYB5R3 selected for genotyping in the Carolina Breast Cancer Study population, along with rationale for selection.
| SNP | SNP ID | Rationale for selection |
|---|---|---|
| -251G>T | rs73888347 | Found in 1 of 69 livers in association with low (outside the 95% confidence interval of the mean) b5R expression and reduction activity; heterozygous African American subject. a Predicted deletion of VDR/CAR/PXR binding sites. b |
| -231C>A | rs75133903 | Found in 1 of 69 livers in association with low b5R expression; heterozygous Caucasian subject.a |
| I1M+6C>T | rs8190370 | Found in 1 of 69 livers, a and 2 of 70 breast samples [36], in association with low b5R expression; all heterozygous African American.Predicted to create a cryptic splice site.c |
| I1M+6072C>T | rs8190414 | Reported in the NCBI SNP database. Within binding sites of NFκ-B, Max. d |
| 176G>AArg59His | rs111154229 | Observed in a heterozygous Caucasian liver with low b5R expression [35]. Predicted to be deleterious to protein function.e,f |
| I8-1676T>C | rs751153 | Reported in the NCBI SNP database. Within binding sites of Rad21.d |
| 890G>AArg297His | rs76458556 | Observed in heterozygous Caucasian liver with low b5R expression and activity [35]. Predicted to be deleterious to protein function. g |
| *138G>A | ss159816065 | Found in 1 of 69 livers with outlier low b5R expression and activity; heterozygous African American subject. a |
| *392G>C | rs7284807 | Found in 5 of 69 livers with predominantly low b5R activity; all African American heterozygotes.a |
| *863T>C | ss159830807 | Found in 1 of 69 livers with low b5R expression and activity; heterozygous Caucasian subject.a |
Unpublished data from Sacco et al. 2010;
MATCH;
Human Splicing Finder;
ENCODE database and ChIP experimental data (www.genome.ucsc.edu);
PMut;
SIFT;
PolyPhen-2.
The PCR-based Taqman Genotyping Assay (Applied Biosytems, Foster City, CA) was utilized for most polymorphisms, and was performed at the University of North Carolina at Chapel Hill. SNPs that failed the Taqman assay were evaluated by pyrosequencing, using the PSQTM96MA System (Biotage AB, Uppsala, Sweden), at the University of Wisconsin Carbone Comprehensive Cancer Center. Both SNP screening techniques were validated by running positive and negative genomic DNA controls from liver or breast samples, where available, in which the allele of interest had been previously established by direct sequencing [35,36]. A total of 144 ancestry informative markers (AIMs) were also genotyped to estimate African and European ancestry [41]. These AIMs were selected from a panel that has been used by others to estimate ancestry in African Americans [42,43].
Data analyses
Prior to association analysis, SNPs were checked for genotyping efficiency and tested for deviation from genotype proportions expected within control groups, stratified by race under Hardy-Weinberg equilibrium (HWE) conditions using Chi Square testing. Minor allele frequencies (MAFs) and genotypes for each SNP were then compared between AfrAm and Cauc women in the control groups, and between women with breast cancer and controls (both across all subjects and within the two racial groups), using Chi Square testing with P < 0.05. Breast cancer risk was calculated using unconditional logistic regression (SAS software program) in order to quantify the association with CYB5A and CYB5R3 genotypes, with adjustment for age and African ancestry. An offset term was included in models to account for randomized recruitment sampling. The sample size provided 80% power to detect odds ratios of 1.2 or greater for alleles with a frequency of 5% or greater in the population overall. We also estimated odds ratios stratified by smoking status (ever, never). As an exploratory analysis, we modeled multiplicative interactions between genotype and smoking using a likelihood ratio test; however, power for testing interactions between genotype and smoking, stratified by race, was fairly limited.
Results
Allele detection
Five previously reported CYB5A variants, rs36082929 (-206G>-), rs74339771 (65A>G), rs1803366 (155G>A), rs78009726 (178A>G, Thr60Ala), and rs76241580 (*246T>C) were not detected in the CBCS population. These rare variants have not been found in population screens reported in either the NCBI or the 1000 Genomes databases (www.1000genomes.org/). Two CYB5R3 variants, rs111154229 (176G>A), and rs76458556 (890G>A), previously found by direct resequencing of liver cDNA samples [35], were also not detected in the CBCS population. Both appear to be rare; 176G>A has not been reported in other SNP databases (either NCBI or 1000 Genomes), while 890G>A was reported with an MAF of only 0.002 overall (browser.1000genomes.org). All detected alleles were in HWE in the AfrAm population. Two variants, rs7284807 (*392 G>C) and rs1790894 (I2 -992 C>T) were not in HWE in Cauc controls, and were not analyzed further.
Allele frequencies by cancer outcome
Allele frequencies for the remaining 11 variants were compared between breast cancer cases and controls, both overall and within racial groups. CYB5R3 I1M+6C>T was significantly more common in breast cancer cases compared to controls (MAF 0.0238 versus 0.0169, P=0.039), with an odds ratio of 1.42 (0.99-2.04), P=0.057. This difference was attributable to a significantly higher prevalence in AfrAm cases compared to AfrAm controls (MAF 0.0611 versus 0.0441, P=0.046; OR 1.41 (0.98-2.04); P=0.062, Tables 4 and 5). An elevated, but very imprecise odds ratio was found between I1M+6C>T and breast cancer in Cauc women (OR 1.57 (0.13-19.71), P=0.726)), due to very low allele frequencies. No other variant was significantly different between cases and controls, either in the population overall or as stratified by race.
Table 4.
Minor allele frequencies (MAF) for CYB5A and CYB5R3 variants in African American (AfrAm) and Caucasian (Cauc) women from the Carolina Breast Cancer Study [37]. Subjects are cases (with a diagnosis of invasive breast cancer or carcinoma in situ) or controls (without a breast cancer or CIS diagnosis and frequency-matched by age and race).
| CYB5A | Minor allele frequencies Cases | Minor allele frequencies Controls | ||
|---|---|---|---|---|
| AfrAm | Cauc | AfrAm | Cauc | |
| 13 T>G(Ser5Ala) | 0.0088 | 0.0008 | 0.0069 a | 0.0000 a |
| I2-992 C>T | 0.1577 | 0.0096 c | 0.1689 | 0.0069 c |
| 390 C>A | 0.0000 | 0.0004 | 0.0000 | 0.0000 |
| CYB5R3 | Minor allele frequencies Cases | Minor allele frequencies Controls | ||
| AfrAm | Cauc | AfrAm | Cauc | |
| -251 G>T | 0.0674 | 0.0017 | 0.0727 a | 0.0005 a |
| -231 C>A | 0.0000 | 0.0000 | 0.0000 | 0.0009 |
| I1M+6 C>T | 0.0611 b | 0.0008 | 0.0441a, b | 0.0005 a |
| I1M+6072 C>T | 0.0007 | 0.0000 | 0.0000 | 0.0000 |
| I8-1676 T>C | 0.1493 | 0.0025 | 0.1531a | 0.0014 a |
| *138 G>A | 0.0027 | 0.0025 | 0.0000 | 0.0028 |
| *392 G>C | 0.1154 | 0.0033 c | 0.1353 | 0.0042 c |
| *863 T>C | 0.0000 | 0.0000 | 0.0000 | 0.0005 |
Significantly different between AfrAm and Cauc controls (P ≤ 0.0001).
Significantly different between cases and controls (P < 0.05).
Not in HWE in Caucasians.
Table 5.
Odds ratios (OR) with 95% confidence intervals (CI) for breast cancer risk among African American (AfrAm) and Caucasian (Cauc) women from the Carolina Breast Cancer Study population, in relation to CYB5R3 I1M+6 genotype and smoking status.
| I1M+6 C>T genotype (rs8190370) | OR (95% CI)* | P value | ||||
|---|---|---|---|---|---|---|
| Cases | Controls | |||||
| All subjects | CC CT + TT |
1847 89 |
CC CT + TT |
1685 57 |
Referent 1.42 (0.99-2.04) |
0.0565 |
| Ever smokers | CC CT + TT |
868 34 |
CC CT + TT |
792 15 |
Referent 1.97 (1.03-3.77) |
0.0400 |
| Never smokers | CC CT + TT |
979 55 |
CC CT + TT |
893 42 |
Referent1.25 (0.81-1.95) | 0.3155 |
| Caucasians | CC CT + TT |
1198 2 |
CC CT + TT |
1084 1 |
Referent 1.57 (0.13-19.71) |
0.7257 |
| Ever mokers | CC CT + TT |
583 0 |
CC CT + TT |
544 1 |
Referent Not calculated |
|
| Never smokers | CC CT + TT |
615 2 |
CC CT + TT |
540 0 |
Referent Not calculated |
|
| African American | CC CT + TT |
649 87 |
CC CT + TT |
601 56 |
Referent 1.41 (0.98-2.04) |
0.0623 |
| Ever smokers | CC CT + TT |
285 34 |
CC CT + TT |
248 14 |
Referent 2.10 (1.08 – 4.07) |
0.0284 |
| Never smokers | CC CT + TT |
364 53 |
CC CT + TT |
353 42 |
Referent 1.21 (0.77-1.88) |
0.4096 |
Adjusted for age, race, and African ancestry. An offset term is included in the model to account for randomized recruitment probabilities.
Breast cancer risk was further evaluated for I1M+6C>T in relationship to smoking status (Table 5). Because of relatively low numbers of carriers for most alleles, smoking status was collapsed by necessity into two separate categories, ever smokers and never smokers (defined as less than 100 lifetime cigarettes). The CYB5R3 I1M+6T allele was significantly associated with breast cancer risk in ever smokers (OR 1.97, 95% CI 1.03-3.77), and in particular in AfrAm ever smokers (OR 2.10, 95% CI 1.08-4.07). This relationship was not as strong among AfrAm never smokers (OR 1.21, 95% CI 0.77-1.88; P = 0.4096). In Cauc women, allele frequencies for I1M+6T were too low to determine risk when stratified by smoking status (Table 5). Interactions between the I1M+6 genotype and smoking for breast cancer risk were also modeled. The P value for interactions was 0.162 for all subjects and 0.176 for the AfrAm population.
In silico analysis of the 1M+6 locus
In silico analyses were performed on the IM+6 polymorphic locus. The reference C allele was conserved among primates, rodents, and rabbits (UCSC Genome Browser; genome.ucsc.org). The T variant was predicted by Human Splicing Finder to create a cryptic splice site 4 nucleotides downstream from the natural donor site at the junction between alternative exon 1M and intron 1. The resulting mRNA was predicted to be 4 bp longer than wild type (NCBI Ref Seq ID: NM_000398), and to create a frame-shift leading to a premature stop codon. Further, this transcript was predicted to undergo nonsense-mediated decay, since the predicted stop codon was located more than 50 nucleotides upstream of the exon 2/3 splice junction [44].
Allele frequencies by race
Minor allele frequencies for CYB5A and CYB5R3 SNPs by race are shown in Table 4. I1M+6C>T was almost 90-fold more common in AfrAm compared to Cauc controls (MAF 0.0441 versus 0.0005, P < 0.0001). Three other CYB5R3 variants were also over-represented in AfrAm versus Cauc controls. The promoter variant -251G>T was ∼140-fold more prevalent in AfrAm women (MAF 0.0727; P < 0.0001), and was predicted to delete binding sites for several transcription factors (Table 3). The intronic SNP I8-1676T>C was more than 100-fold more prevalent in AfrAm women (MAF 0.1531, P < 0.0001), and was within an experimentally demonstrated binding site for the DNA repair protein RAD21. The 3′UTR variant *392G>C was found with an MAF of 0.1353 in AfrAm women, but is of unknown functional significance.
As for CYB5A, two variants were more prevalent in AfrAm women (Table 4). The intronic SNP I2-992T was found with a MAF of 0.1689 in AfrAm controls, and was predicted to be within a binding site for NF-κB. The non-synonymous cSNP 13T>G (Ser5Ala) was found only in AfrAm subjects, but with a very low frequency (MAF 0.0069).
Discussion
4-ABP and PhIP are mammary carcinogens that are found in tobacco smoke. These chemicals are bioactivated to their arylhydroxylamine metabolites, which lead to DNA adducts that are thought to initiate cancer [29-31]. Cytochrome b5 and its reductase oppose this bioactivation step; this pathway, found in both liver and breast tissues, is a potential source of individual variability in response to these carcinogens [33,34]. We hypothesized that variants in the CYB5A and CYB5R3 genes encoding this pathway would be associated with breast cancer risk in women who smoke. We further hypothesized that several CYB5A and CYB5R3 variants that we previously found only in African American liver and breast tissues (Table 2) would be over-represented in AfrAm women in this larger population.
Of the 18 variants screened, an intronic variant in CYB5R3, I1M+6T (rs8190370) was significantly more common in breast cancer cases compared to controls, and this was attributable to a significantly higher allele frequency in AfrAm cases compared to AfrAm controls. When stratified by smoking, I1M+6T was more strongly associated with breast cancer, particularly in AfrAm women where the allele was most prevalent. When we further analyzed for an interaction between the I1M+6T allele and smoking, the P values were not significant for an interaction on the multiplicative scale. However, given the relatively low allele frequencies overall, the study as underpowered to detect such an interaction.
In the control subjects in this CBCS survey, I1M+6C>T was observed with an MAF that was almost 90-fold higher in AfrAm versus Cauc women. This variant was previously observed in the heterozygous state in 1 of 69 livers (unpublished data from Sacco et al. 2010), and 2 of 70 breast samples [36] all samples were from AfrAm subjects, with b5R immunoreactive protein expression below the 95% confidence interval for each tissue. The reference C allele is conserved among mammals, and the T variant is predicted to change the splice site at the first intron-exon splice junction of CYB5R3, leading to a truncated transcript. The CYB5R3 I1M+6C>T variant may therefore lead to impaired b5R expression, and this polymorphism merits further functional characterization.
The risk of breast cancer in African American versus Caucasian women has been the subject of a number of studies, most of which have focused on differences in clinical presentation, tumor behavior, and dietary and hormonal exposures [50,51]. As for interactions between smoking and race, a 1992 study found an association between smoking and breast cancer in white women, but not black women [52]. In the CBCS population used in the present study, smoking was previously found to be a more significant risk factor for breast cancer in AfrAm than Cauc women, based on both duration of active smoking and time since smoking cessation [53,54]. This relationship was found to be stronger when considering multiple polymorphisms in nucleotide excision repair (NER) genes [54], which mediate repair of smoking-induced DNA adducts. These studies, together with our data, suggest that the higher risk associated with smoking among AfrAm women may be partly due to genetic variation in pathways that mediate both the detoxification of arylamine and heterocyclic amine tobacco carcinogens and the repair of resulting DNA adducts.
Overall, CYB5A and CYB5R3 polymorphisms were fairly uncommon, with MAF values ≤ 0.15 for most variants. This may reflect evolutionary pressure to conserve the function of this pathway, which has an important endogenous role in maintaining hemoglobin in its functional, reduced state [45-47]. However, several variants were more prevalent in AfrAm women compared to Cauc women. In addition to I1M+6T, the CYB5R3 promoter SNP -251G>T was more than 140-fold more prevalent in AfrAm women, with an MAF (∼0.070) that was similar to that reported for native African populations (browser.1000genomes.org). We previously observed this variant in a heterozygous liver with outlier low b5R expression and hydroxylamine reduction activity (Table 3). This SNP is predicted to eliminate VDR/CAR/PXR binding sites, and functional analysis of this SNP using a luciferase reported assay showed a 58% decrease in expression compared to the wild type promoter (data not shown).
A third CYB5R3 variant, I8-1676T>C, which was found with > 100-fold higher allele frequency in AfrAm women, is within a binding site for RAD21, which could affect DNA double-strand-break repair. A final CYB5R3 variant, *392G>C, was found with >30-fold higher frequency in AfrAm women. This variant was previously observed in haplotype configuration in 8 breast samples, all from AfrAm women. [36] Although it was not predicted by UTRscan to affect miRNA binding, this 3′UTR variant merits functional characterization because of its relatively high prevalence.
Two CYB5A variants also had significantly higher allele frequencies in AfrAm women. One coding SNP, 13T>G (Ser5Ala) was exclusively found in AfrAm subjects, albeit at a very low frequency. This variant was also reported with low frequency in native Africans (MAF 0.020) and was absent in European subjects in the 1000 Genomes Project (browser.1000genomes.org). Another CYB5A variant, I2-992C>T, was observed with >20-fold higher allele frequency in AfrAm women (MAF 0.169); this frequency is intermediate between those reported for African (0.201) and European (0.007) populations (browser.1000genomes.org). I2-992C>T was predicted to be within a binding site for NF-κB, which could affect gene expression [48,49]. There are several limitations to this study. Because of low allele frequencies, some of the estimates were imprecise and required that the smoking data be categorized as ever or never smokers. This did not allow consideration of age at smoking initiation, cigarette dosage and duration, or exposure to passive smoke [54,55]. Our findings should be confirmed in a larger AfrAm population to allow inclusion of more detailed smoking categories. In addition, samples were not fully re-sequenced, so other, potentially novel polymorphisms or haplotype associations within CYB5A and CYB5R3 were not evaluated. Finally, we have previously found substantial individual variability in b5 and b5R protein expression, as well as 4-ABP hydroxylamine reduction activities in human breast, which could not be attributed to genetic polymorphisms alone [36]. This suggests that there may be tissue-specific, environmentally induced, or epigenetic regulation of this pathway that may also influence breast detoxification of carcinogenic arylhydroxylamines. This possibility deserves further study in breast tissues from women with and without breast cancer.
In summary, we found that the CYB5R3 variant I1M+6 C>T was significantly over-represented in women with breast cancer, in particular AfrAm women. Further, there was a suggestion of an interaction of this variant and smoking among AfrAm women. This intronic variant is predicted to cause aberrant splicing of the CYB5R3 transcript, which may lead to decreased b5R expression and impaired detoxification of carcinogenic arylhydroxylamine metabolites. The CYB5R3 I1M+6 C>T variant should be considered in the molecular epidemiology studies of breast cancer risk in AfrAms. In addition, this and other variants in CYB5A and CYB5R3 that are over-represented in AfrAm subjects should be considered in etiologic studies of prostate, bladder, and colon cancers, which are also associated with exposure to arylamine and heterocyclic amine carcinogens [56-58].
Acknowledgments
The authors thank Jessica Tse, at the Mammalian Genotyping Core at the University of North Carolina at Chapel Hill, for genotype analyses; and Dr. Elim Lau at the University of Wisconsin Carbone Comprehensive Cancer Center (UWCCC) for performing pyrosequencing. The authors also acknowledge the kind assistance of Dr. Richard Weinshilboum, whose laboratory performed initial screening of selected allele frequencies in Coriell DNA samples from African American subjects. Finally, the PI thanks Dr. Andrew Olshan, who was instrumental in the completion of this manuscript following the unexpected death of Dr. Robert Millikan.
Financial support: This work was supported by the Prevent Cancer Foundation and R01 GM61753 from the National Institutes of Health. The Carolina Breast Cancer Study was funded by the Specialized Program of Research Excellence (SPORE) in Breast Cancer at UNC (NIH/NCI P50-CA58223) and the Lineberger Comprehensive Cancer Center Core Grant (P30-CA16086).
Grant support: This study was supported by a grant from the Prevent Cancer Foundation, and in part by R01 GM61753 from the National Institutes of Health. Kristina Blanke was supported by an NIH/NIEHS training grant in Molecular and Environmental Toxicology (T32 ES007015). The Carolina Breast Cancer Study is supported by a Program of Research Excellence (SPORE) in Breast Cancer (NIH/NCI P50-CA58223). The University of Wisconsin Carbone Comprehensive Cancer Center (UWCCC) facilities are supported by NIH/NCI P30 CA014520. Dr. Richard Weinshilboum's contributions to preliminary data were supported by NIH/NIGMS grant U19 GM061388.
Footnotes
Conflicts of Interest: The authors assert that they have no relationships that could be construed as resulting in an actual, potential, or perceived conflict of interest relative to the work in this manuscript.
References
- 1.Kelsey JL, Horn-Ross PL. Breast cancer: magnitude of the problem and descriptive epidemiology. Epidemiol Rev. 1993;15:7–16. doi: 10.1093/oxfordjournals.epirev.a036118. [DOI] [PubMed] [Google Scholar]
- 2.McPherson K, Steel CM, Dixon JM. ABC of breast diseases. Breast cancer-epidemiology, risk factors, and genetics. BMJ. 2000;321:624–8. doi: 10.1136/bmj.321.7261.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hecht SS. Tobacco smoke carcinogens and breast cancer. Environ Mol Mutagen. 2002;39:119–26. doi: 10.1002/em.10071. [DOI] [PubMed] [Google Scholar]
- 4.Shan L, Yu M, Snyderwine EG. Gene expression profiling of chemically induced rat mammary gland cancer. Carcinogenesis. 2005;26:503–9. doi: 10.1093/carcin/bgh330. [DOI] [PubMed] [Google Scholar]
- 5.Ghoshal A, Preisegger KH, Takayama S, Thorgeirsson SS, Snyderwine EG. Induction of mammary tumors in female Sprague-Dawley rats by the food-derived carcinogen 2-amino-1-methyl-6-phenylimidazo[ 4,5-b]pyridine and effect of dietary fat. Carcinogenesis. 1994;15:2429–33. doi: 10.1093/carcin/15.11.2429. [DOI] [PubMed] [Google Scholar]
- 6.Ito N, Hasegawa R, Sano M, Tamano S, Esumi H, Takayama S, et al. A new colon and mammary carcinogen in cooked food, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) Carcinogenesis. 1991;12:1503–6. doi: 10.1093/carcin/12.8.1503. [DOI] [PubMed] [Google Scholar]
- 7.Snyderwine EG, Thorgeirsson UP, Venugopal M, Roberts-Thomson SJ. Mammary gland carcinogenicity of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine in Sprague-Dawley rats on high- and low-fat diets. Nutr Cancer. 1998;31:160–7. doi: 10.1080/01635589809514698. [DOI] [PubMed] [Google Scholar]
- 8.Lightfoot TJ, Coxhead JM, Cupid BC, Nicholson S, Garner RC. Analysis of DNA adducts by accelerator mass spectrometry in human breast tissue after administration of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine and benzo[a]pyrene. Mutat Res. 2000;472:119–27. doi: 10.1016/s1383-5718(00)00134-0. [DOI] [PubMed] [Google Scholar]
- 9.Pfau W, Stone EM, Brockstedt U, Carmichael PL, Marquardt H, Phillips DH. DNA adducts in human breast tissue: association with N-acetyltransferase-2 (NAT2) and NAT1 genotypes. Cancer Epidemiol Biomarkers Prev. 1998;7:1019–25. [PubMed] [Google Scholar]
- 10.Faraglia B, Chen S, Gammon M, Zhang Y, Teitelbaum S, Neugat A, et al. Evaluation of 4-aminobiphenyl-DNA adducts in human breast cancer: the influence of tobacco smoke. Carcinogenesis. 2003;24:719–25. doi: 10.1093/carcin/bgg013. [DOI] [PubMed] [Google Scholar]
- 11.Firozi PF, Bondy ML, Sahin AA, Chang P, Lukmanji F, Singletary E, et al. Aromatic DNA adducts and polymorphisms of CYP1A1, NAT2, and GSTM1 in breast cancer. Carcinogenesis. 2002;23:301–6. doi: 10.1093/carcin/23.2.301. [DOI] [PubMed] [Google Scholar]
- 12.Li D, Wang M, Dhingra K, Hittelman WN. Aromatic DNA adducts in adjacent tissues of breast cancer patients: clues to breast cancer etiology. Cancer Res. 1996;56:287–93. [PubMed] [Google Scholar]
- 13.Zhu J, Chang P, Bondy ML, Sahin AA, Singletary SE, Takahashi S, et al. Detection of 2-amino-1-methyl-6-phenylimidazo[4,5-b]-pyridine-DNA adducts in normal breast tissues and risk of breast cancer. Cancer Epidemiol Biomarkers Prev. 2003;12:830–7. [PubMed] [Google Scholar]
- 14.Bennicke K, Conrad C, Sabroe S, Sorensen HT. Cigarette smoking and breast cancer. BMJ. (Clinical research) 1995;310:1431–3. doi: 10.1136/bmj.310.6992.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brownson RC, Blackwell CW, Pearson DK, Reynolds RD, Richens JW, Jr, Papermaster BW. Risk of breast cancer in relation to cigarette smoking. Arch Intern Med. 1988;148:140–4. [PubMed] [Google Scholar]
- 16.Calle EE, Miracle-McMahill HL, Thun MJ, Heath CW., Jr Cigarette smoking and risk of fatal breast cancer. American Journal of Epidemiology. 1994;139:1001–7. doi: 10.1093/oxfordjournals.aje.a116939. [DOI] [PubMed] [Google Scholar]
- 17.Lash TL, Aschengrau A. Active and passive cigarette smoking and the occurrence of breast cancer. Am J Epidemiol. 1999;149:5–12. doi: 10.1093/oxfordjournals.aje.a009727. [DOI] [PubMed] [Google Scholar]
- 18.Marcus PM, Newman B, Millikan RC, Moorman PG, Baird DD, Qaqish B. The associations of adolescent cigarette smoking, alcoholic beverage consumption, environmental tobacco smoke, and ionizing radiation with subsequent breast cancer risk (United States) Cancer Causes Control. 2000;11:271–8. doi: 10.1023/a:1008911902994. [DOI] [PubMed] [Google Scholar]
- 19.Meara J, McPherson K, Roberts M, Jones L, Vessey M. Alcohol, cigarette smoking and breast cancer. Brit J Cancer. 1989;60:70–3. doi: 10.1038/bjc.1989.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Terry PD, Rohan TE. Cigarette smoking and the risk of breast cancer in women: a review of the literature. Cancer Epidemiol Biomarkers Prev. 2002;11:953–71. [PubMed] [Google Scholar]
- 21.Baron JA, Byers T, Greenberg ER, Cummings KM, Swanson M. Cigarette smoking in women with cancers of the breast and reproductive organs. Journal of the National Cancer Institute. 1986;77:677–80. doi: 10.1093/jnci/77.3.677. [DOI] [PubMed] [Google Scholar]
- 22.Braga C, Negri E, La Vecchia C, Filiberti R, Franceschi S. Cigarette smoking and the risk of breast cancer. Eur J Cancer Prev. 1996;5:159–64. doi: 10.1097/00008469-199606000-00003. [DOI] [PubMed] [Google Scholar]
- 23.Chu SY, Stroup NE, Wingo PA, Lee NC, Peterson HB, Gwinn ML. Cigarette smoking and the risk of breast cancer. Am J Epidemiol. 1990;131:244–53. doi: 10.1093/oxfordjournals.aje.a115494. [DOI] [PubMed] [Google Scholar]
- 24.Egan KM, Stampfer MJ, Hunter D, Hankinson S, Rosner BA, Holmes M, et al. Epidemiology. Vol. 13. Cambridge, Mass: 2002. Active and passive smoking in breast cancer: prospective results from the Nurses′ Health Study; pp. 138–45. [DOI] [PubMed] [Google Scholar]
- 25.Field NA, Baptiste MS, Nasca PC, Metzger BB. Cigarette smoking and breast cancer. Int Journal Epidemiol. 1992;21:842–8. doi: 10.1093/ije/21.5.842. [DOI] [PubMed] [Google Scholar]
- 26.Hunter DJ, Hankinson SE, Hough H, Gertig DM, Garcia-Closas M, Spiegelman D, et al. A prospective study of NAT2 acetylation genotype, cigarette smoking, and risk of breast cancer. Carcinogenesis. 1997;18:2127–32. doi: 10.1093/carcin/18.11.2127. [DOI] [PubMed] [Google Scholar]
- 27.Millikan RC, Pittman GS, Newman B, Tse CK, Selmin O, Rockhill B, et al. Cigarette smoking, N-acetyltransferases 1 and 2, and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 1998;7:371–8. [PubMed] [Google Scholar]
- 28.Vatten LJ, Kvinnsland S. Cigarette smoking and risk of breast cancer: a prospective study of 24,329 Norwegian women. Eur J Cancer. 1990;26:830–3. doi: 10.1016/0277-5379(90)90164-o. [DOI] [PubMed] [Google Scholar]
- 29.Bartsch H. Metabolic activation of aromatic amines and azo dyes. Vol. 1. IARC Sci Pub; 1981. pp. 13–30. [PubMed] [Google Scholar]
- 30.Fan L, Schut HA, Snyderwine EG. Cytotoxicity, DNA adduct formation and DNA repair induced by 2-hydroxyamino-3-methylimidazo[4,5-f]quinoline and 2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b]pyridine in cultured human mammary epithelial cells. Carcinogenesis. 1995;16:775–9. doi: 10.1093/carcin/16.4.775. [DOI] [PubMed] [Google Scholar]
- 31.Turesky RJ, Lang NP, Butler MA, Teitel CH, Kadlubar FF. Metabolic activation of carcinogenic heterocyclic aromatic amines by human liver and colon. Carcinogenesis. 1991;12:1839–45. doi: 10.1093/carcin/12.10.1839. [DOI] [PubMed] [Google Scholar]
- 32.Stone EM, Williams JA, Grover PL, Gusterson BA, Phillips DH. Interindividual variation in the metabolic activation of heterocyclic amines and their N-hydroxy derivatives in primary cultures of human mammary epithelial cells. Carcinogenesis. 1998;19:873–9. doi: 10.1093/carcin/19.5.873. [DOI] [PubMed] [Google Scholar]
- 33.Kurian JR, Chin NA, Longlais BJ, Hayes KL, Trepanier LA. Reductive detoxification of arylhydroxylamine carcinogens by human NADH cytochrome b5 reductase and cytochrome b5. Chem Res Toxicol. 2006;19:1366–73. doi: 10.1021/tx060106t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.King R, Teitel C, Shaddock J, Casciano D, Kadlubar F. Detoxification of carcinogenic aromatic and heterocyclic amines by enzymatic reduction of the N-hydroxy derivative. Cancer Letters. 1999;143:167–71. doi: 10.1016/s0304-3835(99)00119-6. [DOI] [PubMed] [Google Scholar]
- 35.Sacco JC, Trepanier LA. Cytochrome b5 and NADH cytochrome b5 reductase: genotype-phenotype correlations for hydroxylamine reduction. Pharmacogenet Genomics. 2010;20:26–37. doi: 10.1097/FPC.0b013e3283343296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rhoads K, Sacco JC, Drescher N, Wong A, Trepanier LA. Individual variability in the detoxification of carcinogenic arylhydroxylamines in human breast. Toxicol Sci. 2011;121:245–56. doi: 10.1093/toxsci/kfr073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newman B, Moorman PG, Millikan R, Qaqish BF, Geradts J, Aldrich T, et al. The Carolina Breast Cancer Study: integrating population-based epidemiology and molecular biology. Breast Cancer Res Treat. 1995;35:51–60. doi: 10.1007/BF00694745. [DOI] [PubMed] [Google Scholar]
- 38.Nyante SJ, Gammon MD, Kaufman JS, Bensen JT, Lin DY, Barnholtz-Sloan JS, et al. Common genetic variation in adiponectin, leptin, and leptin receptor and association with breast cancer subtypes. Breast Cancer Res Treat. 2011;129:593–606. doi: 10.1007/s10549-011-1517-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moorman PG, Newman B, Millikan RC, Tse CK, Sandler DP. Participation rates in a case-control study: the impact of age, race, and race of interviewer. Annal Epidemiol. 1999;9:188–95. doi: 10.1016/s1047-2797(98)00057-x. [DOI] [PubMed] [Google Scholar]
- 40.ENCODE Project Consortium. A user's guide to the encyclopedia of DNA elements (ENCODE) PLoS biology. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barnholtz-Sloan JS, Shetty PB, Guan X, Nyante SJ, Luo J, Brennan DJ, et al. FGFR2 and other loci identified in genome-wide association studies are associated with breast cancer in African-American and younger women. Carcinogenesis. 2010;31:1417–23. doi: 10.1093/carcin/bgq128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barnholtz-Sloan JS, McEvoy B, Shriver MD, Rebbeck TR. Ancestry estimation and correction for population stratification in molecular epidemiologic association studies. Cancer Epidemiol Biomarkers Prev. 2008;17:471–7. doi: 10.1158/1055-9965.EPI-07-0491. [DOI] [PubMed] [Google Scholar]
- 43.Tian C, Hinds DA, Shigeta R, Kittles R, Ballinger DG, Seldin MF. A genomewide single-nucleotide-polymorphism panel with high ancestry information for African American admixture mapping. Am J Hum Genet. 2006;79:640–9. doi: 10.1086/507954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schell T, Kulozik AE, Hentze MW. Integration of splicing, transport and translation to achieve mRNA quality control by the nonsense-mediated decay pathway. Genome Biol 3:reviews. 2002;1006:1–6. doi: 10.1186/gb-2002-3-3-reviews1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hultquist D, Passon P. Catalysis of methaemoglobin reduction by erythrocyte cytochrome b5 and cytochrome b5 reductase. Nature New Biol. 1971;229:252–4. doi: 10.1038/newbio229252a0. [DOI] [PubMed] [Google Scholar]
- 46.Kitao T, Sugita Y, Yoneyama Y, Hattori K. Methemoglobin reductase (cytochrome b5 reductase) deficiency in congenital methemoglobinemia. Blood. 1974;44:879–84. [PubMed] [Google Scholar]
- 47.Hegesh E, Hegesh J, Kaftory A. Congenital methemoglobinemia with a deficiency of cytochrome b5. N Eng J Med. 1986;314:757–61. doi: 10.1056/NEJM198603203141206. [DOI] [PubMed] [Google Scholar]
- 48.Xu Y, Kiningham KK, Devalaraja MN, Yeh CC, Majima H, Kasarskis EJ, et al. An intronic NF-kappaB element is essential for induction of the human manganese superoxide dismutase gene by tumor necrosis factor-alpha and interleukin-1beta. DNA Cell Biol. 1999;18:709–22. doi: 10.1089/104454999314999. [DOI] [PubMed] [Google Scholar]
- 49.Bentires-Alj M, Barbu V, Fillet M, Chariot A, Relic B, Jacobs N, et al. NF-kappaB transcription factor induces drug resistance through MDR1 expression in cancer cells. Oncogene. 2003;22:90–7. doi: 10.1038/sj.onc.1206056. [DOI] [PubMed] [Google Scholar]
- 50.Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295:2492–502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 51.Hall IJ, Moorman PG, Millikan RC, Newman B. Comparative analysis of breast cancer risk factors among African-American women and White women. Am J Epidemiol. 2005;161:40–51. doi: 10.1093/aje/kwh331. [DOI] [PubMed] [Google Scholar]
- 52.Mayberry RM, Stoddard-Wright C. Breast cancer risk factors among black women and white women: similarities and differences. Am J Epidemiol. 1992;136:1445–56. doi: 10.1093/oxfordjournals.aje.a116465. [DOI] [PubMed] [Google Scholar]
- 53.Li Y, Millikan RC, Bell DA, Cui L, Tse CK, Newman B, et al. Cigarette smoking, cytochrome P4501A1 polymorphisms, and breast cancer among African-American and white women. Breast Cancer Res. 2004;6:R460–73. doi: 10.1186/bcr814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mechanic L, Millikan RC, Player J, RenedeCotret A, Winkel S, Worley K, et al. Polymorphisms in nucleotide excision repair genes, smoking and breast cancer in African Americans and whites: a population-based case-control study. Carcinogenesis. 2006;27:1377–85. doi: 10.1093/carcin/bgi330. [DOI] [PubMed] [Google Scholar]
- 55.Terry PD, Rohan TE. Cigarette smoking and the risk of breast cancer in women: a review of the literature. Cancer Epidemiol Biomarkers Prev. 2002;11:953–71. [PubMed] [Google Scholar]
- 56.Tang D, Liu JJ, Rundle A, Neslund-Dudas C, Savera AT, Bock C, et al. Grilled meat consumption and PhIP-DNA adducts in prostate carcinogenesis. Cancer Epidemiol Biomarkers Prev. 2007;16:803–8. doi: 10.1158/1055-9965.EPI-06-0973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Letasiova S, Medve'ova A, Sovcikova A, Dusinska M, Volkovova K, Mosoiu C, et al. Bladder cancer, a review of the environmental risk factors. Environ Health. 2012;11(Suppl 1):S11. doi: 10.1186/1476-069X-11-S1-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ferrucci LM, Sinha R, Huang WY, Berndt SI, Katki HA, Schoen R, et al. Meat consumption and the risk of incident distal colon and rectal adenoma. Br J Cancer. 2012;106:608–16. doi: 10.1038/bjc.2011.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kurian J, Bajad S, Miller J, Chin N, Trepanier L. NADH cytochrome b5 reductase and cytochrome b5 catalyze the microsomal reduction of xenobiotic hydroxylamines and amidoximes in humans. J Pharmacol Exp Ther. 2004;311 doi: 10.1124/jpet.104.072389. [DOI] [PubMed] [Google Scholar]
- 60.Grillo G, Turi A, Licciulli F, Mignone F, Liuni S, Banfi S, et al. UTRdb and UTRsite (RELEASE 2010): a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Res. 2010;38:D75–80. doi: 10.1093/nar/gkp902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baird SD, Turcotte M, Korneluk RG, Holcik M. Searching for IRES. RNA. 2006;12:1755–85. doi: 10.1261/rna.157806. [DOI] [PMC free article] [PubMed] [Google Scholar]