Abstract
Epigenetic regulation utilizes different mechanisms to convey heritable traits to progeny cells that are independent of DNA sequence, including DNA silencing, post-translational modifications of histone proteins and the post-transcriptional modulation of RNA transcript levels by non-coding RNAs. Although long non-coding RNAs have recently emerged as important regulators of gene imprinting, but their functions during osteogenesis are as yet unexplored. In contrast, microRNAs (miRNAs) are well characterized for their control of osteogenic and osteoclastic pathways; thus, further defining how gene regulatory networks essential for skeleton functions are coordinated and finely tuned through the activities of miRNAs. Roles of miRNAs are constantly expanding as new studies uncover associations with skeletal disorders. The distinct functions of epigenetic regulators and evidence for integrating their activities to control normal bone gene expression and bone disease will be presented. In addition, potential for using “signature microRNAs” to identify, manage and therapeutically treat osteosarcoma will be discussed in this review.
Keywords: Osteosarcoma miRNAs, Epigenetic marks in bone, Bone osteoblasts, miRNA, Histone modifications, DNA methylation
Introduction
Since discovery of the DNA code, transcriptional control by transmissible DNA regulatory elements in gene promoters was established as the fundamental determinate of gene regulation and tissue-specific commitment and progression of cells to establishment and maintenance of a differentiated phenotype. This dogma of gene regulation changed with the characterization of several reversible, post-translation modifications of chromatin that were able to orchestrate heritable changes in gene expression and provide a new level of understanding for regulation of gene expression that continues to expand. The variations in expression of genes among individuals, referred to as “trait-associated DNA” is important for understanding biological differences not solely transmitted through DNA sequence [1]. In addition to their role in normal biological variance, epigenetic pathways are being recognized for their associations with diseases [2], including diabetes [3], cardiovascular disease [4] and obesity [5]. For skeletal disorders, there are few comprehensive studies, however recent reviews have called attention for the need to better understand the contribution of epigenetic control mechanisms to osteoporosis, osteoarthritis and other skeletal disorders [6–9].
This review will cover the spectrum of epigenetic mechanisms that modulate gene transcription, including DNA methylation, histone post-translational modifications, long non-coding RNA binding, as well as epigenetic mechanisms regulating mRNA translation, such as microRNAs. Recent studies have provided compelling evidence that each of these epigenetic levels are integrated in cooperative pathways and regulatory networks to control biological change [10]. Several excellent reviews have documented characterization of many miRNAs regulating distinct aspects of bone formation [7, 11–15]. In this review, emphasis will be on findings from numerous in vitro and several in vivo studies that have contributed to defining modes of epigenetic control of genes from a systems biology perspective. In addition, potential and ongoing future directions will be discussed that will lead to new modes of diagnosis and treatment of osteosarcoma and other bone-related diseases.
Interrelated Epigenetic Mechanisms: Multifaceted Biological Control of the Genome
DNA Methylation
The earliest recognition of epigenetic control was the silencing of genes by DNA methylation which accounts for much of the inherited transcription of genes, as well as the deregulation, and frequent abrogation of gene silencing that contributes to cancer [16]. Large stretches of cytosine and guanine dinucleotides (CpG islands) flanking genes and gene promoters are primary targets for methylation by a small family of DNA methyltransferases (e.g., DNMT1, DNMT3, DNMT3b, etc.) resulting in repressed gene expression. This mode of epigenetic control contributes to genome stability, development and the transcription of genes. Discovery of Ten Eleven Translocation (TET) enzymes that can actively reverse DNA methylation revealed that functions of DNA methylation can be regulated through demethylation [17]. To date, the most well studied mechanism is by oxidation of 5-methylcytosine to 5-hydroxymethyl cytosine; and there is much yet to learn regarding biological states that are dynamically controlled by the methylation-hydroxymethylation status [17]. Further, recent studies indicate that microRNAs contribute to resulting methylation of DNA. Such findings underscore the integration of epigenetic pathways [18, 19].
Studies have examined the role of DNA methylation in bone, during bone development [20] and directly regulating bone-related genes (e.g., osteocalcin) [21], Dlx and Osx [22]. Most recently, a genome-wide methylation profile during osteoblast differentiation showed that the promoters of the genes highly methylated in non-osseous cells, must be demethylated for their activation of transcription at the onset of osteoblast differentiation [23]. In addition, osteogenic signaling factors such as BMP and hormones regulating bone metabolism can affect DNA methylation mechanisms. Dexamethasone, well known for its negative effects on bone formation, was discovered to influence promoter methylation that favored adipogenesis over osteoblastogenesis [24]. Studies have also indicated that the methylation levels of the two cytochrome P450 enzymes that regulate vitamin D hydroxylation are informative for providing vitamin D responses in subjects [25–27].
DNA methylation is linked to a large number of diseases and has a widely studied role in numerous cancers. DNA methylation patterns in breast cancer subtypes are being examined for the correlation with risk, racial disparity and other parameters of disease outcome [28]. In contrast, examination of DNA methylation in aging bone disorders such as osteoporosis and osteoarthritis has been minimal. However a few notable, studies have highlighted the importance of this mechanism in these diseases [29–32]. Recently, deregulated chondrocyte DNA methylation was observed in arthritis [33]. Additionally, it was demonstrated that Type IX collagen was hypermethylated in OA chondrocytes and exhibited reduced expression, contributing to compromised capability to maintain cartilage tissue in the diseased state [34].
Histone Modifications
Dynamic and stable changes in DNA structure are induced by the post-translational modifications of core histone proteins that comprise the nucleosome unit in addition to coiled DNA. Numerous enzymes chemically modify amino terminal “tails” of histone proteins, thereby altering interaction with DNA and allowing for interactions with chromodomain and bromodomain-containing proteins that recognize lysine acetylation and methylation sites, respectively. These interactions stabilize large chromatin remodeling complexes (e.g., SWI/SNF) or modulate nucleosome positioning. The interactions of chromatin with SWI/SNF complexes allow for displacement of nucleosomes near translational start sites, thereby facilitating transcriptional activation [35]. The fine regulatory roles for these chromatin modifications have been linked with developmental activation of the osteocalcin gene [36, 37].
The H3 and H4 core histones tails are the main target for acetylations (Ac) and methylations (Me), primarily at lysine and arginine residues. Methylation of specific lysine residues have defined roles in regulating gene expression. Actively transcribed or genes poised to be expressed are generally marked by trimethylated H3K4 (H3K4me3) near transcription start sites. In contrast, H3K27me3 modifications are mechanistically associated with repression of genes [38]. Prior to differentiation, genes that are poised but not yet expressed, may contain both H3K4me3 and H3K27me3 marks at gene promoters in a concept referred to as bivalency [39]. Regulation of methylation on lysine residues on histones, specifically, H3K9me3 and H3K27me3, by lysine demethylases (KDMs) have been demonstrated to be important in commitment of mesenchymal cells to osteoblasts [40].
Acetylation of lysine residues on histones is associated with gene activation and these modifications are conferred by a broad spectrum of histone acetylating enzymes (HATs). In general, HAT activity results in increased gene expression, while removal of acetyl groups by histone deacetylases (HDAC) decreases gene transcription. Studies have identified the significance of histone modifying enzymes on skeletal development. For example, HDAC4 is a potent inhibitor of endochondral bone formation (EBF) in vivo [41]. The phenotype of excess bone in HDAC null mice, as well as inhibited EBF by overexpression of HDAC4 in chondrocytes, has been attributed to modifications in Runx2 activity. There is a requirement for a transcriptional complex that includes Runx2 and HDAC4 to regulate genes essential for normal endochondral bone formation. More recently, conditional knock-out of HDAC3 in chondro-osteoprogenitors (Osx-Cre positive osteoprogenitors) was revealed to be a positive regulator of osteoprogenitors [42]. HDAC3 null mice exhibited a marrow filled with massive numbers of adipocytes. The importance specifically of lysine acetylation (e.g., H3K9Ac, H3K27ac) and the actions of HDACs in regulating osteogenesis have been recognized [43–45].
Other post-translational modifications of the histone proteins can include phosphorylation, methylation, sumoylation and ubiquination at distinct amino acid residues. A combination of specific histone marks provide a signature or “histone code” for a cell phenotype or a disease state. Generally, such histone marks are found on regulating regions of lineage-specific genes that together indicate cell commitment or activation of a differentiation program and can be informative as to which genes are expressed in normal or altered in a disease state. In addition, several other histone modifying proteins including WDR5 [46], NO66 [47] and others [48] have been demonstrated to play a role in osteogenesis and/or bone formation. These studies highlight the importance of understanding the epigenetic contribution of histone modifications to normal bone biology and pathologic disorders. Such knowledge has the potential to provide a basis for therapy to reverse skeletal disorders by targeting the enzymes responsible to for the deregulated histone modification.
Not to be overlooked is the combinatorial role of transcription factors in mediating epigenetic modifications. Transcription factors can both activate or repress target genes in pluripotent cells and following commitment to a phenotype. Thus, the functional coupling of transcriptional regulators binding to sequence-specific DNA regulatory elements with co-regulatory factors that are histone modifiers, underscores the importance of transcriptional regulation through chromatin modification in developmental cell fate decisions and in disease pathogenesis. Runx2, an essential transcription factor for osteoblast differentiation, has demonstrated properties in forming complexes with SWI/SNF proteins, e.g., Brg1 [36, 49–51], regulating nucleosome sliding for transcription factor accessibility [36], forming complexes with both HATs and HDACs [45], and directly regulating expression of other chromatin remodeling factors, as Ezh2 [52••]. Epigenetic control of gene expression by histone modifications coupled with transcription factors is the major contributor to dynamic changes required for gene expression during cellular differentiation programs or in response to physiological signals.
Non-coding RNAs (ncRNA)
The mammalian transcriptome is highly complex and involves a large number of non-coding RNAs. Recent large scale transcriptome studies have determined that less than 5% of the entire human genome is transcribed from DNA into messenger RNA [53]. More than 75% of the cellular transcriptome is comprised of ncRNA. This abundant class of RNA molecules includes many different types which are classified either by their size (e.g., long non-coding (lnc), microRNA (miRNA)) or cellular location and activity (e.g., snoRNA, piRNA) [54, 55]. Of these groups, the most well-characterized are the miRNAs [56].
The central importance of miRNAs in epigenetic control of cellular properties is their functional ability to bind to many target mRNAs due to their small seed sequence, providing sprawling regulatory control of diverse biological processes. Targeting of miRNAs occurs primarily at the 3′ UTR but also in some 5′ UTR domains). The complementary binding of the miRNA to a targeted mRNA transcript results in degradation of the mRNA (perfect complementary sequence match) or stable binding (imperfect match) resulting in blocked translation of protein. In this manner, miRNAs regulate signaling pathways by targeting multiple components of a pathway and can be viewed as masterminds of cellular processes, regulatory pathways and network connections. The canonical pathway of microRNAs biogenesis has been well studied with respect to their processing by enzymes to pre-miRNAs in the nucleus by Drosha and to mature functional miRNAs in the cytoplasm by Dicer. The argonaute complex that allows interaction of the miRNA with its target messenger RNA. However, recent studies have revealed non-canonical pathways and even more complex mechanisms involving modifications of miRNA, expanding their functional activities [57]. Functional roles of microRNAs in regulating osteogenesis are presented below in an expanded section.
Like miRNAs, lncRNAs function in lineage commitment but through an entirely different set of mechanisms. lncRNA are more recently discovered and have multiple and complex regulatory mechanisms [58]. They can function much like transcriptional activators, binding to gene promoters or enhancers to activate gene expression [59]. In the case of the XIST gene, this lncRNA functions in female cells to inactivate the second copy of the X-chromosome [60]. Studies are now emerging that indicate substantial cross regulation between miRNA and lncRNA, with the large lncRNA acting as “sponges” to sequester miRNAs, thereby inhibiting their activities [61]. To date, very few papers have examined lncRNAs function in with bone, which is surprising given the number of excellent studies on lncRNA functions in muscle [62–65].
Mitotic Bookmarking
Phenotype stability is maintained during cell division through a process known as “mitotic bookmarking”. During mitosis the majority of phenotypic genes are not transcribed; rather genes are poised to resume transcription immediately after completion of cell division. This suggests that the cell has a “memory” of the transcriptional program and the chromatin state prior to cell division (hence “bookmarked”). This assures that genes required to maintain a lineage specific phenotype or induce a differentiation program are rapidly transcribed in post-mitotic cells.
The mechanism of maintaining phenotype stability during mitosis by the retention of tissue-specific transcription factors at target gene loci on mitotic chromosomes arose from the initial discovery that Runx2 remained bound to ribosomal genes that are transcribed by RNA Pol I acrocentric chromosomes, and equally important, Runx2 is concentrated on RNA Pol II-transcribed genes on mitotic chromosomes [66]. This observation was seminal to understanding how phenotype stability is maintained during cell division. Association of Runx2 with ribosomal genes in the nucleolus imaging cells following cell division indicates coordinate control of protein synthesis and phenotype. Since this discovery, other tissue-specific transcription factors have been identified to function in a similar manner. For example, C/EBPα (an archetypical tissue-specific transcription factor regulating adipogenesis) and not PPARγ, was observed to be associated with mitotic chromosomes for pre-adipocyte stability during proliferation. This level of control underscores the importance for architectural organization of tissue-specific regulatory machinery reflected by the strategic location of transcription factors at multiple sites on target genes in focal microenvironments in the interphase nuclease. Several reviews have described the importance of compartmentalization of transcriptional machinery in unique subnuclear domains for specific biological processes [67–69].
microRNAs: An Epigenetic Mechanism by Translational Control of Proteins
The discovery of microRNAs (miRNA) led to seminal findings in human biology and disease states. The importance of miRNAs in organ development in mouse models was first established by tissue-specific conditional deletion of the Dicer enzyme essential for the biogenesis of functional miRNAs. The advent of miRNA profiling studies revealed miRNAs functions in maintenance of cell stemness, commitment and differentiation of cell phenotypes and in the cancer, the characterization of “oncomiRs” associated with disease progression paved the road to miRNAs as therapeutic possibilities [70, 71]. The hundreds of studies examining differentiation of mesenchymal lineages, including adipocytes, myoblasts, chondro- and osteo-progenitors regulated by miRNAs added a new dimension for understanding cellular plasticity and the regulation of lineage commitment. From this knowledge base, studies exploring miRNAs linked to degenerative disease that includes osteoporosis and osteoarthritis [6–9] and bone cancers, particularly metastatic bone disease [72–74] osteosarcomas [75] and multiple myeloma [76].
MicroRNAs have powerful properties in regulating commitment to the osteoblast phenotype. They drive progression of differentiation and, at the same, time support the timing of expressed genes and stability of subpopulations of bone cells. Understanding their complex control of osteogenic signaling pathways in relation to other epigenetic control mechanisms is crucial to developing miRNAs for therapeutic intervention of skeletal disorders. As a result of hundreds of studies characterizing miRNA functions in relation to a specific target(s) during osteoblast or osteoclast differentiation [5–7, 11–15], new concepts developed expanding our knowledge of miRNA control of bone formation. The following experimental findings point to mechanisms and osteogenic activities controlled by miRNAs.
Subsets of miRNAs are Required for Formation of Bone during Development and for Regulating Bone Mass in the Adult Skeleton
Conditional ablation of the Dicer gene in early chondro-osteoprogenitors by a Col1a-Cre driver and in osteoblasts using Osteocalcin (OC)-Cre mice established the requirement for functional miRNAs in bone formation [77]. DicerΔcol/Δcol mice resulted in embryonic lethality at E 12.5, and neither calcified cartilage nor bone was evident compared to WT mice, suggesting a subset of miRNAs are critical for induced osteogenesis. In contrast conditional knock-out (CKO) of Dicer in mature osteoblasts increased bone mass nearly two fold continuously during post-natal growth up to eight months in DicerΔcol/Δcol. The phenotype was attributed to robust synthesis of bone matrix proteins. Bone resorption was not impaired, but kept up the pace for bone remodeling. Several miRNAs characterized for their roles in osteoblasts, explain the high bone mass phenotype: miR-29a,b inhibits collagens, osteonectin and Wnt inhibitors, miR-218 [78] and miR-335-5p [79] target different inhibitors of Wnt and BMP/TGFβ, and miR-338-39 represses FGF2 signaling [80]. In addition miRNAs present in osteoblasts inhibits bone essential transcription factors including 11 Runx2 miRNAs [81], 5 Osterix miRNAs [82–85], ATF4 by miR-214 [86••], DLX5 by miR-141, miR-200a [87] SATB2 targeted by miR-31 [87] and the miR-23a~27a~24-2 cluster [88]. Accelerated bone formation in Dicer CKO osteoblasts indicates that the deficiency in functional miRNAs relieved repression of essential osteogenic factors, thereby increasing osteoblast activity and bone volume. An important conclusion from the above studies is that a cohort of microRNAs regulate the pace of bone formation and limits bone mass in the adult skeleton. Such knowledge is opportune for development of translational approaches for skeletal complications.
microRNAs Function as Regulators of Osteogenic Pathways through Different Mechanisms
During osteogenesis, the same miRNA can target different genes, dependent on the stage of osteoblast differentiation. This ability supports the temporal expression of genes. The specificity for a miRNA to inhibit protein translation at one stage and allow for transcription of the gene at a different stage is not clear, but for many targets in osteoblasts and osteoclasts a reciprocal expression pattern between the miRNA and the target is observed [89, 90]. This notion is clearly evident from the diversity of miR-29 a,b targets [91] and the miR cluster miR-23a~27a~24-2 targeting three essential transcriptional regulators [88].
MicroRNAs are a mechanism to epigenetically support pluripotency of a cell, as well as control lineage commitment to a specific phenotype. For example, specific miRNAs expressed at high levels in the mesenchymal stem cell (MSCs) target tissue-specific regulators to prevent phenotype differentiation. These miRNAs become downregulated in response to a signaling cue for a differentiation program. For osteogenesis, it is well documented that BMP2 downregulates many miRNAs that target Runx2, Osterix, Satb2 and Smad receptors and others [92]. BMPs operate similarly in other differentiation programs, e.g., in muscle by upregulating and downregulating miRNAs in progenitors [93]. Importantly, phenotype stability of the differentiated cell is maintained through a mechanism by which a single miRNA targets transcription factors essential for inhibition of a competing phenotype. For example, miR-133a promotes myogenesis, but this miRNA also sharply downregulates Runx2 to block osteogenesis [92]. This complementary regulation is also observed by miRNAs that maintain a balance between adipocytes and osteoblasts. With aging, fatty marrow develops. Investigations have focused on identifying molecular mechanism that switch bone marrow mesenchymal cells (BMSCs) from adipogenic to osteogenic differentiation [94, 95]. This decision process and maintaining a physiological balance between fat and bone is controlled in part by multiple miRNAs including miR-204, miR-30a, miR-17-5p, miR-106a, miR-22, miR-705; miR-3077-5p, miR-637. These miRNAs either decrease BMP2 signaling or inhibit the bone essential Runx2 and Osterix/Sp7 regulators, resulting in cells that default to adipocyte differentiation and/or the miRNAs may actively promote adipogenesis [24, 81, 92, 94, 96–99].
MiRNA Regulatory Circuits within Osteoblasts
MiRNAs are involved in intricate feedforward and feedback pathways in regulation of biological processes. One example is the miR cluster 23a~27a~24-2 [88] that is transiently upregulated prior to osteoblast commitment, at which time Runx2 inhibits expression of the cluster at Runx2 regulatory elements in the proximal promoter of the cluster. Each miRNA in the cluster directly represses Satb2, an important co-activator with Runx2 for bone formation. Thus, Runx2 downregulation of the cluster promotes differentiation by increasing Satb2. However, the cluster begins to increase in expression when reaching the mineralization stage to downregulate Runx2 directly via miR-23a, as well as Satb2 and Hoxa 10 by miR-27a. In this manner, these osteogenic factors are decreased and maintained at physiological levels when differentiation is completed.
Runx2 is involved is several complex circuits; e.g., miR-218 upregulates Wnt signaling by downregulating three inhibitors of Wnt signaling (Sost, SFRP2, DKK2) and in turn, Wnt signaling increases the level of Runx2 creating a positive feedback loop to recruit more progenitors cells into the osteoblast lineage. MiR-31 controls a loop which directly decreases expression of Osx and Satb2, but not Runx2. In fact, Runx2 represses miR-31, accounting for the decreased expression of miR-31 during BMSC differentiation [87]. Osterix/Sp7 and miR-93 also function in an autoregulatory loop where miR-93 directly targets Osx/Sp7, downregulating expression of this transcription factor [100]. miRNA activities can also regulate cellular protein levels by a mechanism in which miRNAs protect bone-essential transcription factors from proteosomal degradation; miR-322 protects Osx and miR-15b protects Runx2 from Smurf mediated protein degradation [99, 101].
Regulating the miRNAs
The osteogenic signaling pathways TGFβ/BMP and Wnt have demonstrated consequences in down and upregulating miRNAs [78, 92]. Hormonal and cytokine signals impact on miRNAs levels. As well, drugs that affect the skeleton, are expected to change homeostatic miRNAs levels. The precise molecular mechanisms for regulating miRNA cellular levels require further study. MicroRNAs can regulate their biogenesis by targeting the Dicer and Drosha enzymes and components of the RISC complex. As nearly 70% of miRNAs are located in intronic sequences, cellular levels of many miRNAs can be controlled by transcription of the host gene that occurs under physiological control and/or in pathological conditions [102, 103]. Conversely, a miRNA could negatively regulate the host gene. Elucidation of the relationship between intronic microRNAs and their host genes is important for understanding the functional activity of a miRNA, but is often overlooked in studies where emphasis is place on a target mRNA of the miRNA. Other regulatory mechanisms include lncRNAs that promote skeletal muscle differentiation by transcribing miRNAs as shown for the miR675-3p and miR-675-5p [63]. MicroRNA regulation of DNA and vice versa DNA methylation of host genes will control cellular levels of miRNAs, and this mechanism is reported in a number of cancers [104].
Clinical Relevance of miRNAs in the Skeleton
miRNA Contributing to Osteoporosis
Few genetic links of miRNAs to osteoporosis have been reported: a homozygous mutation in miR-2861 [105] and polymorphisms at target sites [106]. Both deficiencies in bone formation by osteoblasts as well as increased activity of bone resorbing osteoclasts, are reflected by changes in miRNAs that are potential targets for intervention. Recently reported studies have identified miR-705 and miR-3077-5p as two upregulated miRNAs in BMSCs from osteoporotic (OP) bone [96] that target Runx2 and Hoxa10 [107, 108]. An important study identified that miR-214 levels are elevated in bone specimens from aged patients with fractures and are correlated with a lower degree of bone formation [86••]. This miRNA targets the ATF-4 transcription factor that promotes bone formation. While transgenic mice expressing miR-214 had reduced bone mass, targeted delivery of the antagomiR of miR-214, to osteoblast provided protection from bone loss in the OVX mouse model [86••].
Determining levels of miRNAs in circulating progenitor cells of osteoclast from OP patients has identified a number of deregulated miRNA. One study demonstrated that miR-503 is lacking in CD14 + PBMCs and when the antagomiR was tested in OVX mice, bone mass improved [109]. MiR-133a was identified in circulating monocytes in 20 postmenopausal women and found to be upregulated in 10 of them with low BMD compared to 10 patients with high BMD [110]. Although it is not clear yet how this miRNA relates to disease progression, it would be of interest to know if miR-133 was secreted from monocytes, and therefore represents a circulating serum biomarker, as it is known to inhibit Runx2 expression and promote myogenesis, thus affecting other cellular systems. In another study, free circulating miRNAs were reported to be associated with OP fractures; however these could reflect consequence of the fracture and also may not serve as a potential early biomarker for osteoporosis or for response to intervention of bone loss [111].
MicroRNAs regulating osteoclast differentiation, as well as osteoclastic miRNAs being responsive to estrogen are described [90, 112]. Recently intravenous injection of four miRNAs were tested in vivo: miR-133a, miR-141, miR-190 and miR-219 significantly reduced osteoclast activity in mice [113••]. In this study, two miRNAs, miR-16 and miR-378, which are elevated during normal osteoclast differentiation, were found to be increased in serum and correlated with tumor burden in mouse models. Systemic delivery of these two miRNAs inhibited osteoclast activity and reduced osteolytic bone disease in tumor bearing mice. In another study, miR-34a was identified as a potent osteoclast inhibitor by targeting Tgif2 and was demonstrated to block osteoporosis and bone metastasis in mouse models [114]. These are encouraging studies for considering a miRNA for therapeutic intervention, notwithstanding challenges of delivery, and specificity of the desired effect.
MiRNA Biomarkers for Osteosarcomas
Osteosarcoma tumors are of unknown etiology and classified into different histological subtypes based on the population of cells (osteoblasts, fibroblasts, chondrocytes) forming the tumor. Most osteosarcomas (OS) are high grade and mechanistic when diagnosed, most frequently in lungs. Even with limb amputation recurrence of OS is remains an issue, and further, chemotherapy regimens remain challenging as tumors become chemoresistant during treatments. [115, 116]. Although p53 hereditary mutations have been identified in osteosarcoma and decades of research have demonstrated deregulated expressed genes; however, there is still not a clear understanding of the genomic defects leading to the tumor cell, nor the cell of origin. [117]. Thus, there is a compelling need for new countermeasures for assessing risk, intervention at early stages, and biomarkers for informing treatments strategies.
Deregulated molecular signaling in OS is known, including developmental pathways such as Notch, Wnt, TGFβ and tumor associated CXCR4 signaling [118, 119]. Runx2, essential for osteogenic commitment and differentiation, has a prominent role in supporting osteosarcoma tumor growth [120]. High levels of Runx2 are correlated when poorly differentiated tumors and poor response to chemotherapy [121]. Also, Runx2 was reported to be associated with poor prognosis. Further, Runx2, P53 and pRB status were indicated as diagnostic markers for deregulation of osteoblast differentiation in OS cells [122–124]. These clinical correlations suggest Runx2 no longer functions as an anti-proliferative differentiation factor and the studies point to deregulation of Runx2 activities. One likely mechanism contributing to Runx2 tumor-related properties in OS is the loss of the tumor suppressor WWOX which is lacking in many cancers, including osteosarcomas [124–126]. In mouse models of WWOX deficiency, osteosarcomas occur several months prior to detection of tumors in lung [127].
Numerous profiling studies have revealed miRNAs that are characteristic of human OS tissue or OS cell lines when compared to normal bone marrow derived human stromal cells or osteoblast cell lines [43, 128, 129]. While many studies focus on a single microRNA, other studies showed miRNAs correlated with deregulated cellular pathways, disease subtypes and treatment outcomes [126, 129, 130]. A miRNA global scale profiling study which examined 32 osteosarcoma samples and 12 controls, identified an osteosarcoma signature that were correlated with poor differentiation and distinguished chemoresistant versus chemosensitive groups and associated metastasis [130]. MicroRNAs that were significantly decreased from controls included miR-15,16, a growth suppressor, miR-29b and miR-223 that are pro-differentiation factors and seven other miRNAs targeting tumor suppressors. The upregulated miRNAs that are highly correlated with aggressive tumor growth included miR-27a associated with growth and invasion, and the miR-181, miR-10b, miR-214 and miR-190 that were previously classified as “oncomiRs” in other tumors. Many of these miRNAs commonly appear in other OS profiling studies as highly changed miRNAs from control samples. Further, in a study examining 27 sarcomas compared to seven normal controls in an array containing 723 human miRNAs, 23 closely linked miRNAs in cytoband 14q32 were identified. Based on predicted targets identified from gene expression profiles of osteosarcomas, these miRNAs were found associated with pathways regulating osteoblast functions; specifically, Notch, MAPK, WNT and Jun/Fos signaling which are activated in OS [131].
Future studies will need to ascertain if deregulated miRNAs are a consequence and/or a contributing cause of tumor aggressiveness. MicroRNAs secreted into the circulation could represent potential biomarkers of deregulated genes or pathways and maybe serve to better guide treatment strategies. However such studies are very limited, but one study reported results of miRNAs increased and decreased in plasma that are consistent with expression of miRNAs in osteosarcoma tissue [132]. However aberrant expression of circulating miRNAs could arise in the tumor cell or in other cells in the bone microenvironment in response to the tumor. Either the micro RNA or its targeted pathway could be a therapeutic target for intervention.
Closing Remark
The authors acknowledge in this review the unprecedented discoveries of epigenetic regulation of the genome that had to be presented in a limited fashion. Although there is so much more to be learned, it is hoped that this review will encourage investigators to expand current areas of research aiming to develop a deeper understanding of epigenetic controls in normal bone biology and in skeletal disorders.
Fig. 1.
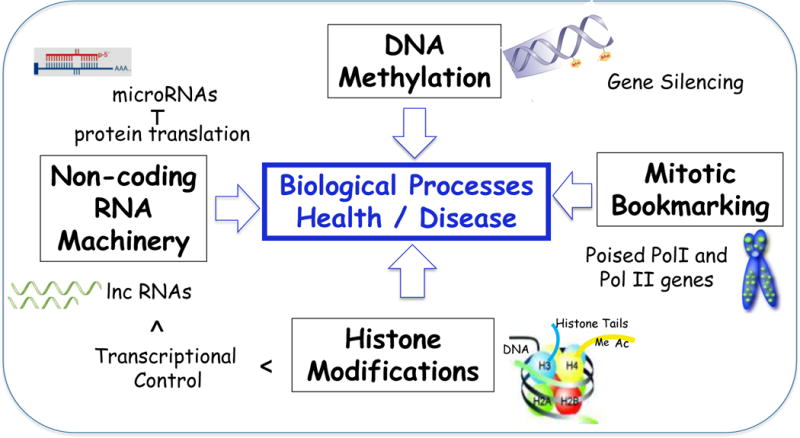
Epigenetic mechanisms regulating biological systems. The four types of epigenetic control of gene expression (black boxes) are illustrated by their effect of DNA structure (schematic) and their functional activity. DNA methylation is the major contributor to heritable traits. Mitotic bookmarking contributes to cell phenotype stability during cell division. Histone modifications on amino acids in tails of histone proteins (e.g., acetylation, methylation, phosphorylation) alter DNA structure to facilitate accessibility of transcription factor binding. RNA machinery involves several classes of small and long non-coding RNAs; the miRNAs are extensively discussed in this review, as they have greatly impacted on bone biology and represent promise of therapeutic applications for bone diseases
Acknowledgments
We thank Jennifer Díaz for manuscript preparation and formatting. The authors acknowledge financial support from the National Institutes of Health (National Cancer Institute P01 CA082834, National Institute of Dental and Craniofacial Research R37 DE012528, National Institute of Arthritis and Musculoskeletal and Skin Diseases R01 AR039588 and P01 AR048818) and the US-Israel Binational Science Foundation (2011300), and FONDAP 15090007.
References
- 1.Kilpinen H, Dermitzakis ET. Genetic and epigenetic contribution to complex traits. Hum Mol Genet. 2012;21:R24–8. doi: 10.1093/hmg/dds383. [DOI] [PubMed] [Google Scholar]
- 2.Butler JS, Koutelou E, Schibler AC, Dent SY. Histone-modifying enzymes: regulators of developmental decisions and drivers of human disease. Epigenomics. 2012;4:163–77. doi: 10.2217/epi.12.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stankov K, Benc D, Draskovic D. Genetic and epigenetic factors in etiology of diabetes mellitus type 1. Pediatrics. 2013;132:1112–22. doi: 10.1542/peds.2013-1652. [DOI] [PubMed] [Google Scholar]
- 4.Aslibekyan S, Claas SA, Arnett DK. Clinical applications of epigenetics in cardiovascular disease: the long road ahead. Transl Res. 2014 Apr 8; doi: 10.1016/j.trsl.2014.04.004. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Dijk SJ, Molloy PL, Varinli H, Morrison JL, Muhlhausler BS, members of Epi S Epigenetics and human obesity. Int J Obes (Lond) 2014 Feb 25; doi: 10.1038/ijo.2014.34. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 6.Vrtacnik P, Marc J, Ostanek B. Epigenetic mechanisms in bone. Clin Chem Lab Med. 2014;52:589–608. doi: 10.1515/cclm-2013-0770. [DOI] [PubMed] [Google Scholar]
- 7.Gamez B, Rodriguez-Carballo E, Ventura F. MicroRNAs and post-transcriptional regulation of skeletal development. J Mol Endocrinol. 2014;52:R179–97. doi: 10.1530/JME-13-0294. [DOI] [PubMed] [Google Scholar]
- 8.Delgado-Calle J, Garmilla P, Riancho JA. Do epigenetic marks govern bone mass and homeostasis? Curr Genomics. 2012;13:252–63. doi: 10.2174/138920212800543129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barter MJ, Young DA. Epigenetic mechanisms and non-coding RNAs in osteoarthritis. Curr Rheumatol Rep. 2013;15:353. doi: 10.1007/s11926-013-0353-z. [DOI] [PubMed] [Google Scholar]
- 10.Murr R. Interplay between different epigenetic modifications and mechanisms. Adv Genet. 2010;70:101–41. doi: 10.1016/B978-0-12-380866-0.60005-8. [DOI] [PubMed] [Google Scholar]
- 11.van der Eerden BC. MicroRNAs in the skeleton: Cell-restricted or potent intercellular communicators? Arch Biochem Biophys. 2014 May 14; doi: 10.1016/j.abb.2014.04.016. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 12.Dong S, Yang B, Guo H, Kang F. MicroRNAs regulate osteogenesis and chondrogenesis. Biochem Biophys Res Commun. 2012;418:587–91. doi: 10.1016/j.bbrc.2012.01.075. [DOI] [PubMed] [Google Scholar]
- 13.Lian JB, Stein GS, van Wijnen AJ, et al. MicroRNA control of bone formation and homeostasis. Nat Rev Endocrinol. 2012;8:212–27. doi: 10.1038/nrendo.2011.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taipaleenmaki H, Bjerre Hokland L, Chen L, Kauppinen S, Kassem M. Mechanisms in endocrinology: micro-RNAs: targets for enhancing osteoblast differentiation and bone formation. Eur J Endocrinol. 2012;166:359–71. doi: 10.1530/EJE-11-0646. [DOI] [PubMed] [Google Scholar]
- 15.van Wijnen AJ, van de Peppel J, van Leeuwen JP, et al. MicroRNA functions in osteogenesis and dysfunctions in osteoporosis. Curr Osteoporos Rep. 2013;11:72–82. doi: 10.1007/s11914-013-0143-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 17.Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472–9. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lehmann U. Aberrant DNA methylation of microRNA genes in human breast cancer – a critical appraisal. Cell Tissue Res. 2014;356:657–64. doi: 10.1007/s00441-014-1793-0. [DOI] [PubMed] [Google Scholar]
- 19.Wu L, Zhou H, Zhang Q, et al. DNA methylation mediated by a microRNA pathway. Mol Cell. 2010;38:465–75. doi: 10.1016/j.molcel.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 20.de Andres MC, Kingham E, Imagawa K, et al. Epigenetic regulation during fetal femur development: DNA methylation matters. PLoS One. 2013;8:e54957. doi: 10.1371/journal.pone.0054957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Villagra A, Gutierrez J, Paredes R, et al. Reduced CpG methylation is associated with transcriptional activation of the bone-specific rat osteocalcin gene in osteoblasts. J Cell Biochem. 2002;85:112–22. [PubMed] [Google Scholar]
- 22.Lee JY, Lee YM, Kim MJ, et al. Methylation of the mouse DIx5 and Osx gene promoters regulates cell type-specific gene expression. Mol Cells. 2006;22:182–8. [PubMed] [Google Scholar]
- 23.Hakelien AM, Bryne JC, Harstad KG, et al. The regulatory landscape of osteogenic differentiation. Stem Cells. 2014 Jun 4; doi: 10.1002/stem.1759. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 24.Li J, Zhang N, Huang X, et al. Dexamethasone shifts bone marrow stromal cells from osteoblasts to adipocytes by C/EBPalpha promoter methylation. Cell Death Dis. 2013;4:e832. doi: 10.1038/cddis.2013.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prosser DE, Jones G. Enzymes involved in the activation and inactivation of vitamin D. Trends Biochem Sci. 2004;29:664–73. doi: 10.1016/j.tibs.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 26.Fetahu IS, Hobaus J, Kallay E. Vitamin D and the epigenome. Front Physiol. 2014;5:164. doi: 10.3389/fphys.2014.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Y, Zhao LJ, Xu X, et al. DNA methylation levels of CYP2R1 and CYP24A1 predict vitamin D response variation. J Steroid Biochem Mol Biol. 2013 Oct 12; doi: 10.1016/j.jsbmb.2013.10.004. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramaniam D, Thombre R, Dhar A, Anant S. DNA methyltransferases: a novel target for prevention and therapy. Front Oncol. 2014;4:80. doi: 10.3389/fonc.2014.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richardson BC, Patel DR. Epigenetics in 2013. DNA methylation and miRNA–key roles in systemic autoimmunity. Nat Rev Rheumatol. 2014;10:72–4. doi: 10.1038/nrrheum.2013.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeffries MA, Donica M, Baker L, et al. Genome-wide DNA methylation study identifies significant epigenomic changes in osteoarthritic cartilage. Arthritis Rheumatol. 2014 Jun 30; doi: 10.1002/art.38762. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 31.Delgado-Calle J, Fernandez AF, Sainz J, et al. Genome-wide profiling of bone reveals differentially methylated regions in osteoporosis and osteoarthritis. Arthritis Rheum. 2013;65:197–205. doi: 10.1002/art.37753. [DOI] [PubMed] [Google Scholar]
- 32.Barter MJ, Bui C, Young DA. Epigenetic mechanisms in cartilage and osteoarthritis: DNA methylation, histone modifications and microRNAs. Osteoarthritis Cartilage. 2012;20:339–49. doi: 10.1016/j.joca.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 33.Hashimoto K, Otero M, Imagawa K, et al. Regulated transcription of human matrix metalloproteinase 13 (MMP13) and interleukin-1beta (IL1B) genes in chondrocytes depends on methylation of specific proximal promoter CpG sites. J Biol Chem. 2013;288:10061–72. doi: 10.1074/jbc.M112.421156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Imagawa K, de Andres MC, Hashimoto K, et al. Reduced type IX collagen gene expression in human osteoarthritic chondrocytes is associated with epigenetic silencing by DNA hypermethylation. Arthritis Rheumatol. 2014 Jul 21; doi: 10.1002/art.38774. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gutierrez J, Paredes R, Cruzat F, et al. Chromatin remodeling by SWI/SNF results in nucleosome mobilization to preferential positions in the rat osteocalcin gene promoter. J Biol Chem. 2007;282:9445–57. doi: 10.1074/jbc.M609847200. [DOI] [PubMed] [Google Scholar]
- 37.Montecino M, Lian J, Stein G, Stein J. Changes in chromatin structure support constitutive and developmentally regulated transcription of the bone-specific osteocalcin gene in osteoblastic cells. Biochemistry. 1996;35:5093–102. doi: 10.1021/bi952489s. [DOI] [PubMed] [Google Scholar]
- 38.Kimura H. Histone modifications for human epigenome analysis. J Hum Genet. 2013;58:439–45. doi: 10.1038/jhg.2013.66. [DOI] [PubMed] [Google Scholar]
- 39.Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 40.Ye L, Fan Z, Yu B, et al. Histone demethylases KDM4B and KDM6B promotes osteogenic differentiation of human MSCs. Cell Stem Cell. 2012;11:50–61. doi: 10.1016/j.stem.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vega RB, Matsuda K, Oh J, et al. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 2004;119:555–66. doi: 10.1016/j.cell.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 42.Razidlo DF, Whitney TJ, Casper ME, et al. Histone deacetylase 3 depletion in osteo/chondroprogenitor cells decreases bone density and increases marrow fat. PLoS One. 2010;5:e11492. doi: 10.1371/journal.pone.0011492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou S, Geng S, Glowacki J. Histone deacetylation mediates the rejuvenation of osteoblastogenesis by the combination of 25(OH)D3 and parathyroid hormone in MSCs from elders. J Steroid Biochem Mol Biol. 2013;136:156–9. doi: 10.1016/j.jsbmb.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu X, Zhang X, Dai L, et al. Histone deacetylase inhibitor trichostatin A promotes the osteogenic differentiation of rat adipose-derived stem cells by altering the epigenetic modifications on Runx2 promoter in a BMP signaling-dependent manner. Stem Cells Dev. 2013;22:248–55. doi: 10.1089/scd.2012.0105. [DOI] [PubMed] [Google Scholar]
- 45.Bradley EW, McGee-Lawrence ME, Westendorf JJ. Hdac-mediated control of endochondral and intramembranous ossification. Crit Rev Eukaryot Gene Expr. 2011;21:101–13. doi: 10.1615/critreveukargeneexpr.v21.i2.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu ED, Demay MB, Gori F. Wdr5 is essential for osteoblast differentiation. J Biol Chem. 2008;283:7361–7. doi: 10.1074/jbc.M703304200. [DOI] [PubMed] [Google Scholar]
- 47.Sinha KM, Yasuda H, Coombes MM, Dent SY, de Crombrugghe B. Regulation of the osteoblast-specific transcription factor Osterix by NO66, a Jumonji family histone demethylase. EMBO J. 2010;29:68–79. doi: 10.1038/emboj.2009.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hemming S, Cakouros D, Isenmann S, et al. EZH2 and KDM6A act as an epigenetic switch to regulate mesenchymal stem cell lineage specification. Stem Cells. 2014;32:802–15. doi: 10.1002/stem.1573. [DOI] [PubMed] [Google Scholar]
- 49.Young DW, Pratap J, Javed A, et al. SWI/SNF chromatin remodeling complex is obligatory for BMP2-induced, Runx2-dependent skeletal gene expression that controls osteoblast differentiation. J Cell Biochem. 2005;94:720–30. doi: 10.1002/jcb.20332. [DOI] [PubMed] [Google Scholar]
- 50.Cruzat F, Henriquez B, Villagra A, et al. SWI/SNF-independent nuclease hypersensitivity and an increased level of histone acetylation at the P1 promoter accompany active transcription of the bone master gene Runx2. Biochemistry. 2009;48:7287–95. doi: 10.1021/bi9004792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Montecino M, Stein JL, Stein GS, et al. Nucleosome organization and targeting of SWI/SNF chromatin-remodeling complexes: contributions of the DNA sequence. Biochem Cell Biol. 2007;85:419–25. doi: 10.1139/O07-070. [DOI] [PubMed] [Google Scholar]
- 52••.Wu H, Whitfield TW, Gordon JA, et al. Genomic occupancy of Runx2 with global expression profiling identifies a novel dimension to control of osteoblastogenesis. Genome Biol. 2014;15:R52. doi: 10.1186/gb-2014-15-3-r52. Identified Runx2 binding to genomic DNA that discovered hundreds of Runx2 target genes related to epigenetic levels of control during stages of osteoblast differentiation. The complete data setand serves as a valuable resource for investigators. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12:847–65. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 56.Flynn RA, Chang HY. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell. 2014;14:752–61. doi: 10.1016/j.stem.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–24. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 58.Vance KW, Ponting CP. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 2014;30:348–55. doi: 10.1016/j.tig.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Orom UA, Shiekhattar R. Long noncoding RNAs usher in a new era in the biology of enhancers. Cell. 2013;154:1190–3. doi: 10.1016/j.cell.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152:1308–23. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 61.Paci P, Colombo T, Farina L. Computational analysis identifies a sponge interaction network between long non-coding RNAs and messenger RNAs in human breast cancer. BMC Syst Biol. 2014;8:83. doi: 10.1186/1752-0509-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu L, Xu PC. Downregulated LncRNA-ANCR promotes osteoblast differentiation by targeting EZH2 and regulating Runx2 expression. Biochem Biophys Res Commun. 2013;432:612–7. doi: 10.1016/j.bbrc.2013.02.036. [DOI] [PubMed] [Google Scholar]
- 63.Dey BK, Pfeifer K, Dutta A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev. 2014;28:491–501. doi: 10.1101/gad.234419.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li L, Liu B, Wapinski OL, et al. Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell Rep. 2013;5:3–12. doi: 10.1016/j.celrep.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dasen JS. Long noncoding RNAs in development: solidifying the Lncs to Hox gene regulation. Cell Rep. 2013;5:1–2. doi: 10.1016/j.celrep.2013.09.033. [DOI] [PubMed] [Google Scholar]
- 66.Young DW, Hassan MQ, Pratap J, et al. Mitotic occupancy and lineage-specific transcriptional control of rRNA genes by Runx2. Nature. 2007;445:442–6. doi: 10.1038/nature05473. [DOI] [PubMed] [Google Scholar]
- 67.Zaidi SK, Grandy RA, Lopez-Camacho C, et al. Bookmarking target genes in mitosis: a shared epigenetic trait of phenotypic transcription factors and oncogenes? Cancer Res. 2014;74:420–5. doi: 10.1158/0008-5472.CAN-13-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sarge KD, Park-Sarge OK. Gene bookmarking: keeping the pages open. Trends Biochem Sci. 2005;30:605–10. doi: 10.1016/j.tibs.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 69.Zaidi SK, Young DW, Javed A, et al. Nuclear microenvironments in biological control and cancer. Nat Rev Cancer. 2007;7:454–63. doi: 10.1038/nrc2149. [DOI] [PubMed] [Google Scholar]
- 70.Esquela-Kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 71.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 72.Ell B, Qiu Q, Wei Y, et al. The MicroRNA-23b/27b/24 Cluster Promotes Breast Cancer Lung Metastasis by Targeting Metastasis-suppressive Gene Prosaposin. J Biol Chem. 2014;289:21888–95. doi: 10.1074/jbc.M114.582866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Browne G, Taipaleenmaki H, Stein GS, Stein JL, Lian JB. MicroRNAs in the control of metastatic bone disease. Trends Endocrinol Metab. 2014;25:320–7. doi: 10.1016/j.tem.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Croset M, Santini D, Iuliani M, et al. MicroRNAs and bone metastasis: a new challenge. Molecules. 2014;19:10115–28. doi: 10.3390/molecules190710115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou G, Shi X, Zhang J, Wu S, Zhao J. MicroRNAs in osteosarcoma: from biological players to clinical contributors, a review. J Int Med Res. 2013;41:1–12. doi: 10.1177/0300060513475959. [DOI] [PubMed] [Google Scholar]
- 76.Bi C, Chng WJ. MicroRNA: important player in the pathobiology of multiple myeloma. Biomed Res Int. 2014;2014:521586. doi: 10.1155/2014/521586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gaur T, Hussain S, Mudhasani R, et al. Dicer inactivation in osteoprogenitor cells compromises fetal survival and bone formation, while excision in differentiated osteoblasts increases bone mass in the adult mouse. Dev Biol. 2010;340:10–21. doi: 10.1016/j.ydbio.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hassan MQ, Maeda Y, Taipaleenmaki H, et al. miR-218 directs a Wnt signaling circuit to promote differentiation of osteoblasts and osteomimicry of metastatic cancer cells. J Biol Chem. 2012;287:42084–92. doi: 10.1074/jbc.M112.377515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang J, Tu Q, Bonewald LF, et al. Effects of miR-335-5p in modulating osteogenic differentiation by specifically downregulating Wnt antagonist DKK1. J Bone Miner Res. 2011;26:1953–63. doi: 10.1002/jbmr.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu H, Sun Q, Wan C, Li L, Zhang L, Chen Z. MicroRNA-338-3p regulates osteogenic differentiation of mouse bone marrow stromal stem cells by targeting Runx2 and Fgfr2. J Cell Physiol. 2014;229:1494–502. doi: 10.1002/jcp.24591. [DOI] [PubMed] [Google Scholar]
- 81.Zhang Y, Xie RL, Croce CM, et al. A program of microRNAs controls osteogenic lineage progression by targeting transcription factor Runx2. Proc Natl Acad Sci U S A. 2011;108:9863–8. doi: 10.1073/pnas.1018493108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Baglio SR, Devescovi V, Granchi D, Baldini N. MicroRNA expression profiling of human bone marrow mesenchymal stem cells during osteogenic differentiation reveals Osterix regulation by miR-31. Gene. 2013;527:321–31. doi: 10.1016/j.gene.2013.06.021. [DOI] [PubMed] [Google Scholar]
- 83.Jia J, Tian Q, Ling S, Liu Y, Yang S, Shao Z. miR-145 suppresses osteogenic differentiation by targeting Sp7. FEBS Lett. 2013;587:3027–31. doi: 10.1016/j.febslet.2013.07.030. [DOI] [PubMed] [Google Scholar]
- 84.Li E, Zhang J, Yuan T, Ma B. MiR-143 suppresses osteogenic differentiation by targeting Osterix. Mol Cell Biochem. 2014;390:69–74. doi: 10.1007/s11010-013-1957-3. [DOI] [PubMed] [Google Scholar]
- 85.Chen Q, Liu W, Sinha KM, Yasuda H, de Crombrugghe B. Identification and characterization of microRNAs controlled by the osteoblast-specific transcription factor Osterix. PLoS One. 2013;8:e58104. doi: 10.1371/journal.pone.0058104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86••.Wang X, Guo B, Li Q, et al. miR-214 targets ATF4 to inhibit bone formation. Nat Med. 2013;19:93–100. doi: 10.1038/nm.3026. Demonstrated the therapeutic effectiveness of antagomiR delivery to osteoblasts in vivo to prevent bone loss in mouse models of osteoporosis. [DOI] [PubMed] [Google Scholar]
- 87.Deng Y, Zhou H, Zou D, et al. The role of miR-31-modified adipose tissue-derived stem cells in repairing rat critical-sized calvarial defects. Biomaterials. 2013;34:6717–28. doi: 10.1016/j.biomaterials.2013.05.042. [DOI] [PubMed] [Google Scholar]
- 88.Hassan MQ, Gordon JA, Beloti MM, et al. A network connecting Runx2, SATB2, and the miR-23a~27a~24-2 cluster regulates the osteoblast differentiation program. Proc Natl Acad Sci U S A. 2010;107:19879–84. doi: 10.1073/pnas.1007698107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li Z, Hassan MQ, Jafferji M, et al. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J Biol Chem. 2009;284:15676–84. doi: 10.1074/jbc.M809787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sugatani T, Vacher J, Hruska KA. A microRNA expression signature of osteoclastogenesis. Blood. 2011;117:3648–57. doi: 10.1182/blood-2010-10-311415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kapinas K, Delany AM. MicroRNA biogenesis and regulation of bone remodeling. Arthritis Res Ther. 2011;13:220. doi: 10.1186/ar3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li Z, Hassan MQ, Volinia S, et al. A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc Natl Acad Sci U S A. 2008;105:13906–11. doi: 10.1073/pnas.0804438105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang J, Greene SB, Bonilla-Claudio M, et al. Bmp signaling regulates myocardial differentiation from cardiac progenitors through a MicroRNA-mediated mechanism. Dev Cell. 2010;19:903–12. doi: 10.1016/j.devcel.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Berendsen AD, Olsen BR. Osteoblast-adipocyte lineage plasticity in tissue development, maintenance and pathology. Cell Mol Life Sci. 2014;71:493–7. doi: 10.1007/s00018-013-1440-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kawai M, de Paula FJ, Rosen CJ. New insights into osteoporosis: the bone-fat connection. J Intern Med. 2012;272:317–29. doi: 10.1111/j.1365-2796.2012.02564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liao L, Yang X, Su X, et al. Redundant miR-3077-5p and miR-705 mediate the shift of mesenchymal stem cell lineage commitment to adipocyte in osteoporosis bone marrow. Cell Death Dis. 2013;4:e600. doi: 10.1038/cddis.2013.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Huang S, Wang S, Bian C, et al. Upregulation of miR-22 promotes osteogenic differentiation and inhibits adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells by repressing HDAC6 protein expression. Stem Cells Dev. 2012;21:2531–40. doi: 10.1089/scd.2012.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Huang J, Zhao L, Xing L, Chen D. MicroRNA-204 regulates Runx2 protein expression and mesenchymal progenitor cell differentiation. Stem Cells. 2010;28:357–64. doi: 10.1002/stem.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gamez B, Rodriguez-Carballo E, Bartrons R, Rosa JL, Ventura F. MicroRNA-322 (miR-322) and its target protein Tob2 modulate Osterix (Osx) mRNA stability. J Biol Chem. 2013;288:14264–75. doi: 10.1074/jbc.M112.432104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yang L, Cheng P, Chen C, et al. miR-93/Sp7 function loop mediates osteoblast mineralization. J Bone Miner Res. 2012;27:1598–606. doi: 10.1002/jbmr.1621. [DOI] [PubMed] [Google Scholar]
- 101.Vimalraj S, Partridge NC, Selvamurugan N. A positive role of microRNA-15b on regulation of osteoblast differentiation. J Cell Physiol. 2014;229:1236–44. doi: 10.1002/jcp.24557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Finnegan EF, Pasquinelli AE. MicroRNA biogenesis: regulating the regulators. Crit Rev Biochem Mol Biol. 2013;48:51–68. doi: 10.3109/10409238.2012.738643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gromak N. Intronic microRNAs: a crossroad in gene regulation. Biochem Soc Trans. 2012;40:759–61. doi: 10.1042/BST20120023. [DOI] [PubMed] [Google Scholar]
- 104.Baer C, Claus R, Plass C. Genome-wide epigenetic regulation of miRNAs in cancer. Cancer Res. 2013;73:473–7. doi: 10.1158/0008-5472.CAN-12-3731. [DOI] [PubMed] [Google Scholar]
- 105.Li H, Xie H, Liu W, et al. A novel microRNA targeting HDAC5 regulates osteoblast differentiation in mice and contributes to primary osteoporosis in humans. J Clin Invest. 2009;119:3666–77. doi: 10.1172/JCI39832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lei SF, Papasian CJ, Deng HW. Polymorphisms in predicted miRNA binding sites and osteoporosis. J Bone Miner Res. 2011;26:72–8. doi: 10.1002/jbmr.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hassan MQ, Tare R, Lee SH, et al. HOXA10 controls osteoblastogenesis by directly activating bone regulatory and phenotypic genes. Mol Cell Biol. 2007;27:3337–52. doi: 10.1128/MCB.01544-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gordon JA, Hassan MQ, Saini S, et al. Pbx1 represses osteoblastogenesis by blocking Hoxa10-mediated recruitment of chromatin remodeling factors. Mol Cell Biol. 2010;30:3531–41. doi: 10.1128/MCB.00889-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chen C, Cheng P, Xie H, et al. MiR-503 regulates osteoclastogenesis via targeting RANK. J Bone Miner Res. 2014;29:338–47. doi: 10.1002/jbmr.2032. [DOI] [PubMed] [Google Scholar]
- 110.Wang Y, Li L, Moore BT, et al. MiR-133a in human circulating monocytes: a potential biomarker associated with postmenopausal osteoporosis. PLoS One. 2012;7:e34641. doi: 10.1371/journal.pone.0034641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Seeliger C, Karpinski K, Haug AT, et al. Five Freely Circulating miRNAs and Bone Tissue miRNAs Are Associated With Osteoporotic Fractures. J Bone Miner Res. 2014;29:1718–28. doi: 10.1002/jbmr.2175. [DOI] [PubMed] [Google Scholar]
- 112.Sugatani T, Hruska KA. Down-regulation of miR-21 biogenesis by estrogen action contributes to osteoclastic apoptosis. J Cell Biochem. 2013;114:1217–22. doi: 10.1002/jcb.24471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113••.Ell B, Mercatali L, Ibrahim T, et al. Tumor-induced osteoclast miRNA changes as regulators and biomarkers of osteolytic bone metastasis. Cancer Cell. 2013;24:542–56. doi: 10.1016/j.ccr.2013.09.008. First study highlights the translational potential of miRNAs associated with osteoclasts to inhibit metastatic bone disease by systemic delivery in mouse models of tumor growth in bone. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Krzeszinski JY, Wei W, Huynh H, et al. miR-34a blocks osteoporosis and bone metastasis by inhibiting osteoclastogenesis and Tgif2. Nature. 2014 Jun 25; doi: 10.1038/nature13375. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 115.Luetke A, Meyers PA, Lewis I, Juergens H. Osteosarcoma treatment – where do we stand? A state of the art review. Cancer Treat Rev. 2014;40:523–32. doi: 10.1016/j.ctrv.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 116.Zhou W, Hao M, Du X, Chen K, Wang G, Yang J. Advances in targeted therapy for osteosarcoma. Discov Med. 2014;17:301–7. [PubMed] [Google Scholar]
- 117.Mutsaers AJ, Walkley CR. Cells of origin in osteosarcoma: mesenchymal stem cells or osteoblast committed cells? Bone. 2014;62:56–63. doi: 10.1016/j.bone.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 118.Mortus JR, Zhang Y, Hughes DP. Developmental pathways hijacked by osteosarcoma. Adv Exp Med Biol. 2014;804:93–118. doi: 10.1007/978-3-319-04843-7_5. [DOI] [PubMed] [Google Scholar]
- 119.Brennecke P, Arlt MJ, Campanile C, et al. CXCR4 antibody treatment suppresses metastatic spread to the lung of intratibial human osteosarcoma xenografts in mice. Clin Exp Metastasis. 2014;31:339–49. doi: 10.1007/s10585-013-9632-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Martin JW, Zielenska M, Stein GS, van Wijnen AJ, Squire JA. The Role of RUNX2 in Osteosarcoma Oncogenesis. Sarcoma. 2011;2011:282745. doi: 10.1155/2011/282745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sadikovic B, Thorner P, Chilton-Macneill S, et al. Expression analysis of genes associated with human osteosarcoma tumors shows correlation of RUNX2 overexpression with poor response to chemotherapy. BMC Cancer. 2010;10:202. doi: 10.1186/1471-2407-10-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.van der Deen M, Akech J, Lapointe D, et al. Genomic promoter occupancy of runt-related transcription factor RUNX2 in Osteosarcoma cells identifies genes involved in cell adhesion and motility. J Biol Chem. 2012;287:4503–17. doi: 10.1074/jbc.M111.287771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pereira BP, Zhou Y, Gupta A, et al. Runx2, p53, and pRB status as diagnostic parameters for deregulation of osteoblast growth and differentiation in a new pre-chemotherapeutic osteosarcoma cell line (OS1) J Cell Physiol. 2009;221:778–88. doi: 10.1002/jcp.21921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yang J, Zhao L, Tian W, et al. Correlation of WWOX, RUNX2 and VEGFA protein expression in human osteosarcoma. BMC Med Genomics. 2013;6:56. doi: 10.1186/1755-8794-6-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Del Mare S, Kurek KC, Stein GS, Lian JB, Aqeilan RI. Role of the WWOX tumor suppressor gene in bone homeostasis and the pathogenesis of osteosarcoma. Am J Cancer Res. 2011;1:585–94. [PMC free article] [PubMed] [Google Scholar]
- 126.Kurek KC, Del Mare S, Salah Z, et al. Frequent attenuation of the WWOX tumor suppressor in osteosarcoma is associated with increased tumorigenicity and aberrant RUNX2 expression. Cancer Res. 2010;70:5577–86. doi: 10.1158/0008-5472.CAN-09-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Aqeilan RI, Hassan MQ, de Bruin A, et al. The WWOX tumor suppressor is essential for postnatal survival and normal bone metabolism. J Biol Chem. 2008;283:21629–39. doi: 10.1074/jbc.M800855200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Miao J, Wu S, Peng Z, Tania M, Zhang C. MicroRNAs in osteosarcoma: diagnostic and therapeutic aspects. Tumour Biol. 2013;34:2093–8. doi: 10.1007/s13277-013-0940-7. [DOI] [PubMed] [Google Scholar]
- 129.Liang W, Gao B, Fu P, Xu S, Qian Y, Fu Q. The miRNAs in the pathgenesis of osteosarcoma. Front Biosci (Landmark Ed) 2013;18:788–94. doi: 10.2741/4142. [DOI] [PubMed] [Google Scholar]
- 130.Jones KB, Salah Z, Del Mare S, et al. miRNA signatures associate with pathogenesis and progression of osteosarcoma. Cancer Res. 2012;72:1865–77. doi: 10.1158/0008-5472.CAN-11-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Maire G, Martin JW, Yoshimoto M, Chilton-MacNeill S, Zielenska M, Squire JA. Analysis of miRNA-gene expression-genomic profiles reveals complex mechanisms of microRNA deregulation in osteosarcoma. Cancer Genet. 2011;204:138–46. doi: 10.1016/j.cancergen.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 132.Ouyang L, Liu P, Yang S, Ye S, Xu W, Liu X. A three-plasma miRNA signature serves as novel biomarkers for osteosarcoma. Med Oncol. 2013;30:340. doi: 10.1007/s12032-012-0340-7. [DOI] [PubMed] [Google Scholar]


