Abstract
The Ribosomal Protein (RP)-HDM2-p53 pathway has been shown to play key roles in oncogene-induced apoptosis and senescence, but the mechanism regulating this pathway remains elusive. The Proline-Rich Akt Substrate of 40 kDA (PRAS40) has recently been identified as a binding partner and inhibitor of the mechanistic Target of Rapamycin Complex 1 (mTORC1). Although other inhibitors of mTORC1 are known tumor suppressors, PRAS40 promotes cell survival and tumorigenesis. Here we demonstrate that Akt- and mTORC1-mediated phosphorylation of PRAS40 at T246 and S221, respectively, promotes nuclear-specific association of PRAS40 with Ribosomal Protein L11 (RPL11). Importantly, silencing of PRAS40 induces upregulation of p53 in a manner dependent upon RPL11. This effect is rescued by wild type PRAS40, but not by the RPL11 binding-null PRAS40 T246A mutant. We find that PRAS40 negatively regulates the RPL11-HDM2-p53 nucleolar stress response pathway and suppresses induction of p53-mediated cellular senescence. This work identifies nuclear PRAS40 as a dual-input signaling checkpoint that links cell growth and proliferation to inhibition of cellular senescence. These findings may help to explain the pro-tumorigenic effect of PRAS40 and identify the PRAS40-RPL11 complex as a promising target for p53-restorative anti-cancer drug discovery.
Keywords: PRAS40, RPL11, mTOR, Akt, senescence, nucleolar stress
Introduction
In order to maintain homeostasis cells must respond efficiently to environmental conditions such as stress, growth factors, and nutrients. The tumor suppressor p53 orchestrates the cellular response to a wide variety of stressors, including DNA damage, oncogene expression, and disruption of ribosome biogenesis – a condition known as nucleolar stress.1, 2 Recently it has been shown that ribosomal protein L11 (RPL11) plays a key role in coordinating the p53 response to nucleolar stress. Specifically, when ribosome production is disturbed, RPL11 translocates from the nucleoli to the nucleoplasm where it binds and inhibits the p53-directed E3 ubiquitin ligase HDM2, resulting in increased p53 protein stability and transcriptional activity.3–6 This enables activation of a transcription program promoting cell cycle arrest, senescence, or apoptosis. Although the RP-HDM2-p53 pathway has been shown to play key roles in oncogene-induced apoptosis and senescence,6, 7 the molecular mechanisms responsible for its regulation remain largely unknown.8
In response to growth signals, the Ser/Thr protein kinase Akt phosphorylates a wide array of substrates including Bad, TSC2, and FOXO1 to inhibit apoptosis and promote survival.9 The main intracellular pathway responsible for nutrient sensing is centered on the mechanistic (formerly referred to as mammalian) Target of Rapamycin (mTOR). mTOR is a Ser/Thr protein kinase that serves as the catalytic subunit of two multi-protein complexes with distinct functions – mTOR Complexes 1 and 2 (mTORC1/2). mTORC1 is a pro-growth/survival kinase that is activated only when nutrients, such as glucose and amino acids, and growth factors are available in a cell’s microenvironment. Once activated, mTORC1 phosphorylates p70S6K and 4EBP1 to promote cap-dependent translation and cell growth. Growth factor-stimulated Akt promotes mTORC1 activation through two pathways. In the first, Akt phosphorylates and inhibits the GTPase-activating protein TSC2, thereby allowing the small GTPase Rheb to remain GTP-bound and activate mTORC1 (ref. 10). The second depends upon the Proline-Rich Akt Substrate of 40kDa (PRAS40). When in its non-phosphorylated form, PRAS40 binds the mTORC1 component Raptor and is believed to inhibit mTORC1 kinase activity by competing for substrate binding. In response to growth factors, PRAS40 binds the scaffolding protein 14-3-3 in a phosphorylation-dependent manner and dissociates from Raptor, thereby allowing mTORC1 access to its downstream effector substrates. Interestingly, both Akt- and mTORC1-mediated phosphorylation of PRAS40 are required for 14-3-3 binding in response to growth factors.11–19 The fate or function, if any, of mTORC1-dissociated, phospho-PRAS40 remains unknown.
PRAS40 is a major target of both Akt and mTORC1 (refs. 18–21) and, as such, is used extensively as a biomarker for development of PI3K-Akt and mTORC1 pathway inhibitors.22–24 Owing to its reported role as a negative regulator of mTORC1, PRAS40 may be expected to suppress cell growth and proliferation. Indeed, other inhibitors of mTORC1, such as LKB1 and TSC2, are bona fide tumor suppressors.25–28 However, in nearly all studies of PRAS40 in disease models, PRAS40 upregulation has been found to promote cell survival, tumorigenesis, or tumor progression.29–31 Therefore, we hypothesize that phosphorylated, mTORC1-dissociated PRAS40 has its own pro-survival function, independent of mTORC1 inhibition. To explore this possibility we interrogated the subcellular localization and interactome of PRAS40. Our findings indicate that dual phosphorylation of PRAS40 by Akt and mTORC1 promotes formation of a nuclear-specific PRAS40- and RPL11-containing complex distinct from mTORC1 that inhibits the RPL11-HDM2-p53 pathway and thereby suppresses induction of senescence.
Results
PRAS40 dynamically shuttles between the cytoplasm and the nucleus
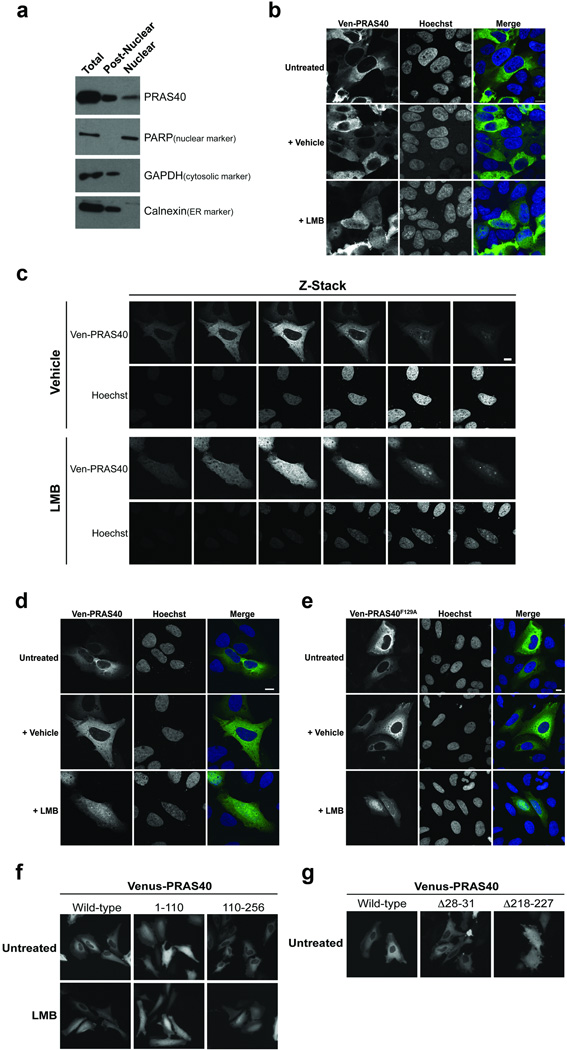
Although mTORC1 is thought to function mainly in the cytoplasm, it has been reported that PRAS40 and other mTORC1 components are also found in the nucleus.32–41 To rigorously test the subcellular localization of PRAS40, we purified nuclei from HeLa cells by centrifuging through a sucrose cushion. The resultant nuclear fraction is void of cytosolic and endoplasmic reticulum contamination as evidenced by lack of the marker proteins GAPDH and calnexin, respectively. As expected, PRAS40 is abundant in the post-nuclear fraction. Importantly, we also observe a distinct population of PRAS40 in the nuclear fraction, albeit in lower abundance (Fig. 1A). Similarly, exogenously expressed Venus- and Flag-tagged-PRAS40 are detected in both the cytoplasm and nuclei of U2OS and HeLa cells via confocal microscopy (Fig. 1B–E and Fig. 2D). To test if an active nuclear shuttling process is responsible for the nucleocytoplasmic concentration differential of PRAS40, we treated HeLa and U2OS cells expressing Venus-PRAS40 with the nuclear export inhibitor Leptomycin B (LMB). PRAS40 accumulates in the nuclei of cells treated with LMB, but not vehicle control, suggesting that PRAS40 subcellular localization is controlled at least in part by active, dynamic nucleocytoplasmic shuttling (Fig. 1B–D). In all LMB experiments a fraction of Ven-PRAS40 remains in the cytoplasm regardless of treatment time, suggesting that only a sub-population of PRAS40 is involved in shuttling. Importantly, no difference is observed between wild-type (WT) Venus-PRAS40 and Raptor-binding null Venus-PRAS40F129A in response to LMB treatment, suggesting that PRAS40 nucleocytoplasmic shuttling does not require interaction with mTORC1 (Fig. 1E).
Figure 1. PRAS40 is a cytoplasmic and nuclear protein that undergoes active, dynamic nucleocytoplasmic shuttling independent of mTORC1.
A) HeLa cells were fractionated via swelling in hypotonic buffer, Dounce homogenization, and centrifugation through a sucrose gradient. HeLa (B) or U2OS (C–E) cells were transfected and treated with 100 nM Leptomycin B (LMB) or vehicle control (EtOH) for 6 h as indicated. Images were obtained via confocal microscopy. Any slight differences between analogous images presented in D and E are not representative of a specific trend. Scale bars = 10 µm. F,G) HeLa cells were transfected and treated with 20 nM LMB for 1 h (F) or not treated (G) as indicated. Images in F and G were acquired via epifluorescence microscopy.
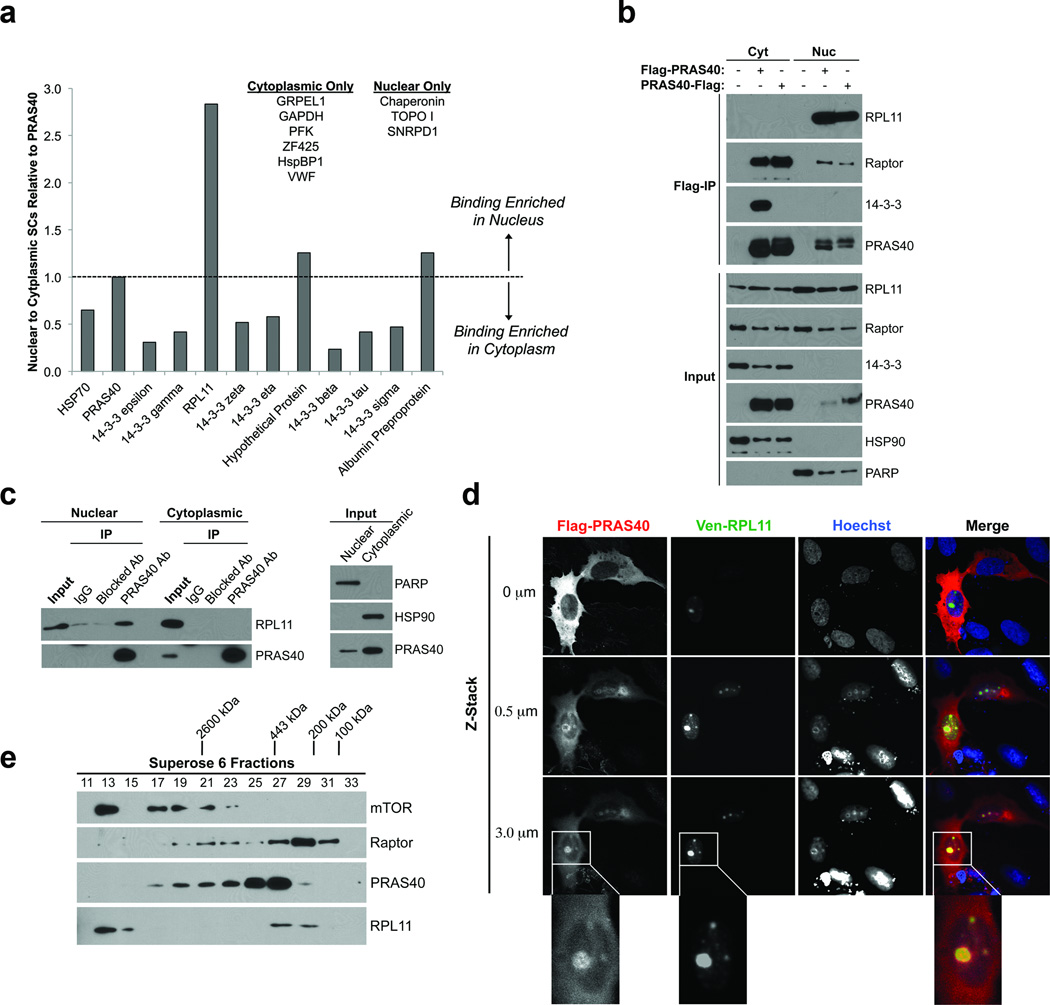
Figure 2. PRAS40 associates with RPL11 in a nuclear-specific, non-mTORC1 complex.
A) Putative PRAS40 binding partners identified via Flag-PRAS40 IP and mass spectrometry analysis. Background-corrected average spectral counts of putative binding partners (“prey”) in each subcellular fraction were normalized to those of PRAS40 (“bait”) from the same fraction and used to determine normalized nuclear to cytoplasmic binding ratios. B) HeLa cells were transfected and fractionated as indicated. Input and Flag-immunocomplexes were resolved by SDS-PAGE and analyzed by Western Blotting (WB). C) Endogenous PRAS40 was IPed from nuclear and cytoplasmic HeLa extract using a PRAS40-specific monocolonal antibody. Non-specific, species-matched IgG or PRAS40 antibody pre-blocked with a PRAS40 peptide were used as negative controls. D) U2OS cells growing on poly-D-Lys-coated cover slips were transfected as indicated, fixed, processed for immunofluorescent detection of Flag-PRAS40, and mounted on glass slides. Images were obtained via confocal microscopy. E) HeLa nuclear extract was resolved via gel filtration chromatography using a Superose® 6 column (GE Healthcare). 0.5 mL fractions were collected.
PRAS40 residues 218–227 serve as a Nuclear Export Signal (NES) Sequence
We next sought to identify Nuclear Localization or Nuclear Export Signal (NLS or NES) sequences responsible for PRAS40’s dynamic nucleocytoplasmic shuttling. Deletion of the C-terminus of PRAS40 abolishes its subcellular concentration differential and renders it unresponsive to LMB treatment (Fig. 1F). Conversely, deletion of the N-terminus of PRAS40 has no such effects, suggesting that sequence motifs critical to subcellular localization lie in the C-terminus of PRAS40. Sequence analysis identifies at least two putative NES sequences in PRAS40 – 28LVLL31 in the N-terminus and, as noted by others,42–44 218IAASMRALVL227 in the C-terminus, but no classical NLS sequence. Deletion of residues 218–227, but not 28–31, results in diffuse cytoplasmic and nuclear localization of PRAS40, suggesting that residues 218–227 serve as a functional NES sequence in PRAS40 (Fig. 1G).
PRAS40 forms a nuclear-specific complex with RPL11
It is unknown what function PRAS40 may serve in the nucleus. The existence of Pro-rich domains in its N-terminus and 14-3-3-binding phospho-sites in its C-terminus suggests that PRAS40 may participate in various protein-protein interactions. Therefore, we probed the PRAS40 interactome of the cytoplasm and nucleus. To achieve this, we immunoprecipitated (IPed) N-terminally or C-terminally tagged Flag-PRAS40 from post-nuclear and nuclear fractions of HeLa cells. The eluates were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS). Eluates from post-nuclear and nuclear fractions of untransfected HeLa cells were used as negative controls. As expected, the known PRAS40-binding partner 14-3-3 was identified in eluates from both the cytoplasm and nucleus, supporting the validity of our approach. Table 1 lists all the potential PRAS40-associated proteins identified and their spectral counts averaged between N- and C-terminally tagged PRAS40 IPs in the nuclear and cytoplasmic fractions. To identify binding partners enriched in the nucleus we normalized spectral counts of putative binding partners (“prey”) in each subcellular fraction to those of PRAS40 (“bait”) from the same fraction. Any putative binding partner with a normalized nuclear to cytoplasmic ratio greater than 1 (or with zero cytoplasmic spectral counts) was identified as a potential nuclear-enriched PRAS40 binding partner (Fig. 2A). Of the proteins identified by this analysis, we chose to focus on Ribosomal Protein L11 (RPL11) because its average spectral count value is similar to that of some 14-3-3 isoforms, which are known PRAS40-binding proteins. Notably, despite their high cellular concentrations, no RPs other than RPL11 were identified in our eluates. As predicted from the IP-MS results, endogenous RPL11 robustly co-precipitates with N- and C-terminally-tagged Flag-PRAS40 from nuclear, but not cytoplasmic extract (Fig. 2B). It is important to note that PRAS40 and RPL11 are present in each subcellular fraction, supporting the notion that the PRAS40-RPL11 association is nuclear-specific. Furthermore, endogenous RPL11 co-precipitates with endogenous PRAS40 from nuclear, but not cytoplasmic HeLa extract (Fig. 2C). Neither non-specific IgG nor PRAS40-specific antibody pre-blocked with a PRAS40 peptide is capable of precipitating RPL11, supporting the specificity of the endogenous PRAS40-L11 association (Fig. 2C). In further support of these findings, Flag-PRAS40 co-localizes with Venus-L11 in the nucleoli of U2OS cells (Fig. 2D).
Table 1.
Putative PRAS40-interacting proteins identified by IP-MS.
| Protein | Accession Number | Average Nuclear SCs1 |
Average Cytoplasmic SCs1 |
|
|---|---|---|---|---|
| Nuclear and Cytoplasmic | HSP70 | NP_006588.1 | 153 | 297 |
| PRAS40 | NP_001092102.1 | 87 | 110 | |
| 14-3-3 ε | NP_006752.1 | 22 | 88 | |
| 14-3-3 γ | NP_036611.2 | 6 | 18 | |
| RPL11 | NP_000966.2 | 5 | 2 | |
| 14-3-3 ζ | NP_003397.1 | 4 | 9 | |
| 14-3-3 η | NP_003396.1 | 3 | 7 | |
| Hypothetical Protein | XP_001716117.1 | 3 | 3 | |
| 14-3-3 β | NP_003395.1 | 2 | 8 | |
| 14-3-3 τ | NP_006817.1 | 2 | 5 | |
| 14-3-3 σ | NP_006133.1 | 2 | 4 | |
| Albumin Preproprotein | NP_000468.1 | 1 | 1 | |
| Nuclear Only | Chaperonin | NP_955472.1 | 2 | 0 |
| TOPO1 | NP_003277.1 | 2 | 0 | |
| SNRPD1 | NP_008869.1 | 1 | 0 | |
| Cytoplasmic Only | GRPEL1 | NP_079472.1 | 0 | 4 |
| GAPDH | NP_002037.2 | 0 | 3 | |
| PFK | NP_002618.1 | 0 | 2 | |
| ZF425 | NP_001001661.1 | 0 | 2 | |
| HspBP1 | NP_036399.3 | 0 | 1 | |
| VWF | NP_000543.2 | 0 | 1 | |
Spectral Counts
The nuclear PRAS40- and RPL11-containing complex is distinct from mTORC1
To test whether nuclear PRAS40-RPL11 association represents a variant of mTORC1 or a novel complex, we processed HeLa cell nuclear extract via gel filtration chromatography using a Superose 6 column. At least two separate peaks of PRAS40 were observed, representing different high MW complexes (Fig. 2E). The larger is approximately 2,600 kDa and co-migrates with the mTORC1 components mTOR and Raptor, but not RPL11. The smaller PRAS40 complex is approximately 450–700 kDa and co-migrates with RPL11, but not mTOR. A second Raptor peak was observed around 100–200 kDa (with slight overlap of the lower MW PRAS40 peak), most likely representing monomeric Raptor (predicted MW = 150 kDa). These results suggest that the nuclear-specific PRAS40- and RPL11-containing complex is distinct from mTORC1 and may contain other, as-of-yet unidentified components.
The nuclear-specific PRAS40-RPL11 association requires PRAS40 residues S221 and T246 and is phosphorylation-dependent
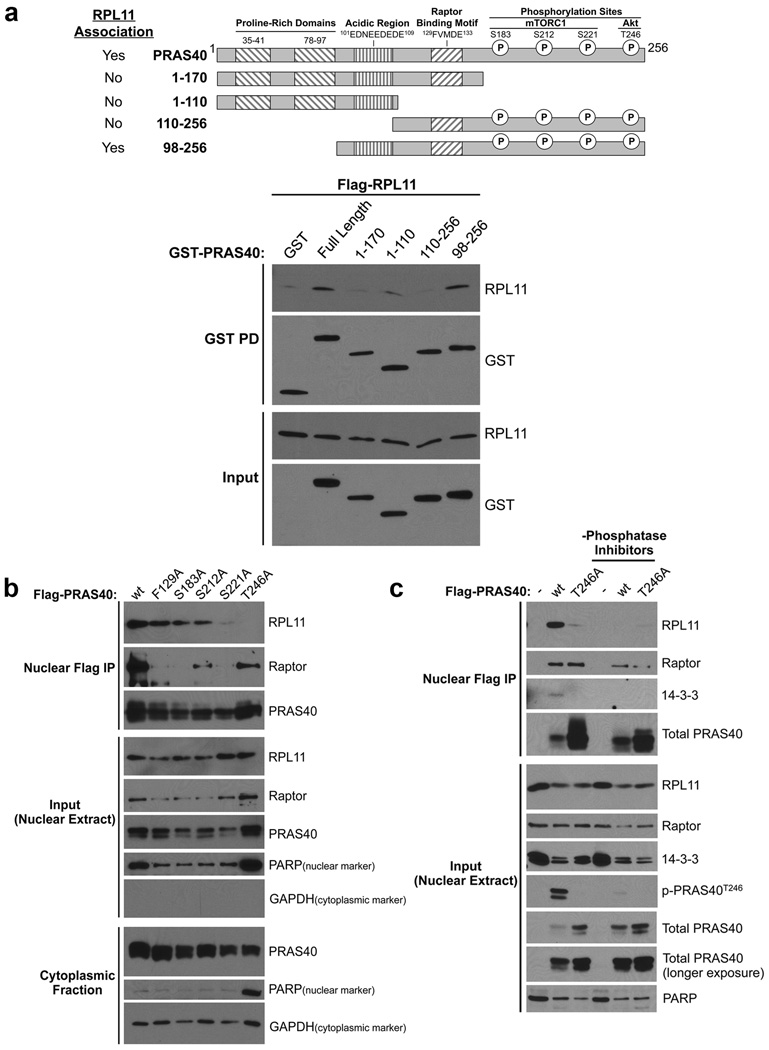
We next sought to identify elements of PRAS40 important for nuclear RPL11 association. Immunoblot analysis of GST-PRAS40 truncation complexes identifies critical regions in the Glu/Asp-rich domain (98–109) (Fig. 3A, compare Lanes 5 and 6), suggesting the importance of electrostatic forces, and in the C-terminus (171–256) (Fig. 3A, compare Lanes 2 and 3). Interaction of PRAS40 with its established binding partners, Raptor and 14-3-3, is controlled by phosphorylation within this C-terminal region (Fig. 3A).42, 43 Therefore, we hypothesized that phosphorylation may also control the novel PRAS40- and RPL11-containing complex. To test this, we generated Flag-PRAS40 plasmids harboring single, non-phosphorylatable Ser or Thr to Ala point mutations of all the major identified PRAS40 phosphorylation sites, including mTORC1-targeted S183, S212, and S221, as well as Akt-targeted T246 (refs. 12, 15, 16, 19, 45). We also generated an F129A mutation in the Tor Signaling motif of PRAS40. This mutation is known to drastically attenuate the interaction of PRAS40 with Raptor/mTORC1 (refs. 12, 15, 16). We assessed the effect of each of these mutations on PRAS40-RPL11 and PRAS40-Raptor association by co-IP. Mutation of PRAS40 F129, S183, and S212 has little or no effect on RPL11 co-precipitation. However, mutation of the mTORC1-targeted residue S221 or the Akt-targeted residue T246 abolishes RPL11 binding, suggesting that these residues are each necessary for PRAS40-RPL11 association (Fig. 3B). Notably, the T246A mutation disrupts RPL11 co-precipitation while maintaining moderate interaction with Raptor, whereas the F129A mutation abolishes Raptor binding but leaves RPL11 association intact, providing further evidence that PRAS40-RPL11 association does not require Raptor/mTORC1 binding (Fig. 3B). Although PRAS40 residues S221 and T246 are clearly important, this finding does not directly implicate phosphorylation in regulation of PRAS40-RPL11 association. To address this, we performed IPs of WT or T246A Flag-PRAS40 from HeLa nuclear extracts using buffers either containing or lacking phosphatase inhibitors (PIs). We find that the PRAS40WT-RPL11 co-precipitation, while robust in the presence of PIs, is abolished in the absence of PIs (Fig. 3C, Lanes 2 and 5). As anticipated from our previous results, Flag-PRAS40T246A fails to co-precipitate RPL11 regardless of PI status (Fig. 3C, Lanes 3 and 6). Importantly, use of a phospho-specific antibody confirms that absence of PIs abolishes PRAS40 phosphorylation at T246 (Fig. 3C, compare Input Lanes 2 and 5). Notably, although PRAS40T246A was expressed at much greater levels than WT, this mutant still fails to precipitate RPL11, underscoring the importance of the T246 site in PRAS40-RPL11 association. Taken together, these results indicate that the nuclear PRAS40- and RPL11-containing complex not only requires PRAS40 residues S221 and T246, but is also phosphorylation-dependent.
Figure 3. Association of nuclear PRAS40 with RPL11 requires PRAS40 residues S221 and T246 and is phosphorylation-dependent.
A) HeLa cells were transfected as indicated. GST-tagged PRAS40 truncations were precipitated from whole-cell lysates (WCL) using glutathione-conjugated beads. Inputs and eluates were resolved via SDS-PAGE and analyzed by WB. B) HeLa cells were transfected and fractionated as indicated. Inputs and Flag-immunocomplexes from nuclear extracts were resolved by SDS-PAGE and analyzed by WB. C) HeLa cells were transfected as indicated. Nuclear extracts were isolated and analyzed as in B using buffers either containing or lacking phosphatase inhibitors.
PRAS40-RPL11 association is controlled by amino acids and serum factors through the kinase activities of mTORC1 and Akt
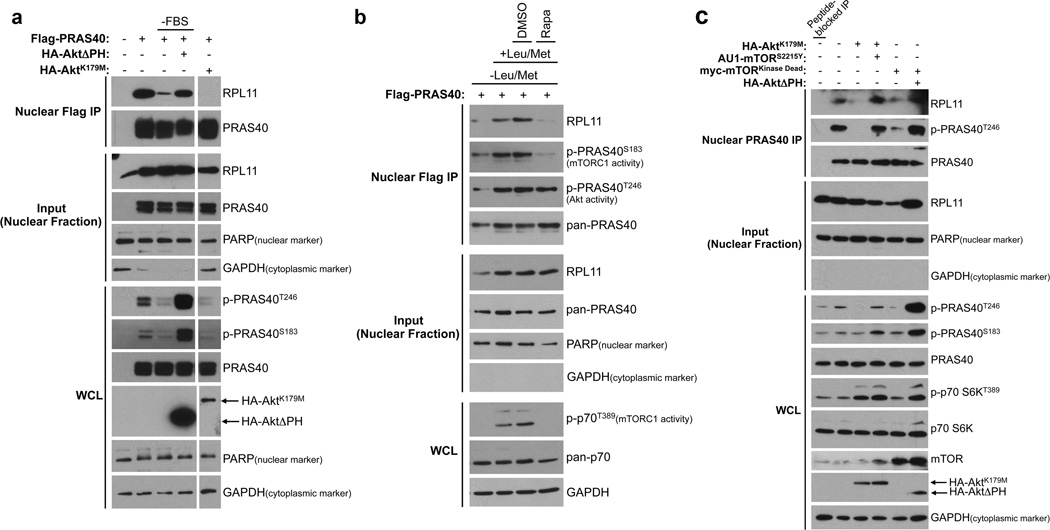
Because PRAS40 residues S221 and T246 are known targets of mTORC1 (ref. 45) and Akt,19 respectively, we wondered if these kinases and their upstream activating signals can regulate the nuclear PRAS40- and RPL11-containing complex. PI3K-Akt signaling is potently activated by growth factors found in serum. As demonstrated by Flag-PRAS40 coIP from HeLa nuclear extracts, serum withdrawal reduces PRAS40-RPL11 co-precipitation to background levels. This effect is negated by expression of constitutively active Akt (HA-AktΔPH) during serum withdrawal, whereas expression of dominant negative Akt (HA-AktK179M) abolishes PRAS40-RPL11 co-precipitation in the presence of serum (Fig. 4A). mTORC1 kinase activity requires both growth factors and an ample supply of extracellular nutrients such as amino acids and glucose. To manipulate mTORC1 activity without inhibiting Akt, HeLa cells were incubated in media lacking Leu and Met but replete with serum. Nuclear PRAS40-RPL11 co-precipitation is enhanced when amino acid-starved cells are returned to Leu/Met-containing media (Fig. 4B, Lanes 1 and 2). This effect is robustly inhibited by treatment with the potent and specific mTORC1 inhibitor rapamycin, but not vehicle (DMSO) control (Fig. 4B). Importantly, upon rapamycin treatment (Fig. 4B, Lane 4) mTORC1 activity is suppressed as evidenced by loss of p70S6K and PRAS40S183 phosphorylation, while Akt remains active as demonstrated by maintenance of PRAS40T246 phosphorylation. This finding suggests that inhibition of mTORC1 activity alone is sufficient to disrupt the nuclear PRAS40- and RPL11-containing complex. Because mTORC1 is activated downstream of Akt, experimental scenarios in which Akt is inhibited while mTOR remains active are more limited. Indeed, we observe a decrease in mTORC1 activity (measured by PRAS40S183 phosphorylation) with expression of dominant negative Akt (Fig. 4A). To attempt to circumvent this issue and also to test whether endogenous PRAS40 behaves similarly to exogenous Flag-PRAS40, endogenous PRAS40 was IPed from nuclear extract of HeLa cells expressing dominant negative Akt or mTOR with or without constitutively active mTOR or Akt, respectively (Fig. 4C). We find that while dominant negative Akt or mTOR alone disrupts the endogenous PRAS40-RPL11 association as expected (Fig. 4C, Lanes 3 and 5), co-expression of constitutively active mTOR or Akt, respectively, can rescue these effects (Fig. 4C, Lanes 4 and 6). Notably, both constitutively active Akt and mTOR were able to promote phosphorylation of Akt- and mTORC1-activity indicator sites on PRAS40 (T246 and S183, respectively). Although somewhat unexpected, this result is consistent with our point mutation and phosphatase inhibitor studies that indicate a critical role for phosphorylation of Akt- and mTORC1-targeted PRAS40 residues in PRAS40-RPL11 association. Taken together, these findings indicate that formation of the nuclear PRAS40- and RPL11-containing complex is regulated by the amino acid- and serum-stimulated kinase activities of mTORC1 and Akt.
Figure 4. Formation of the nuclear-specific PRAS40- and RPL11-containing complex requires the kinase activities of Akt and mTORC1.
A) HeLa cells were either untransfected or transfected with plasmids encoding Flag-PRAS40 and constitutively active (ΔPH) or dominant negative (K179M) HA-Akt as indicated. Indicated samples (-FBS) were serum-starved for 24 h. Flag-immunocomplexes from nuclear extracts were resolved by SDS-PAGE and analyzed by WB. All bands shown are from the same gel. B) HeLa cells expressing Flag-PRAS40 were grown in media lacking Leu and Met for 6.5 h. Cells were either left in amino acid limiting media or returned to Leu- and Met-replete media for 2 h in the presence of either no additive, vehicle (DMSO), or 20 nM rapamycin (Rapa) as indicated. Nuclear extracts were analyzed as in A. C) HeLa cells were transfected with plasmids encoding dominant negative Akt (K179M) or mTOR (Kinase Dead) with or without constitutively active mTOR (S2115Y) or Akt (ΔPH). Endogenous PRAS40 immunocomplexes from nuclear extracts were resolved by SDS-PAGE and analyzed by WB. (WCL = Whole Cell Lysate)
PRAS40 depletion induces p53 protein stability in an RPL11-dependent manner
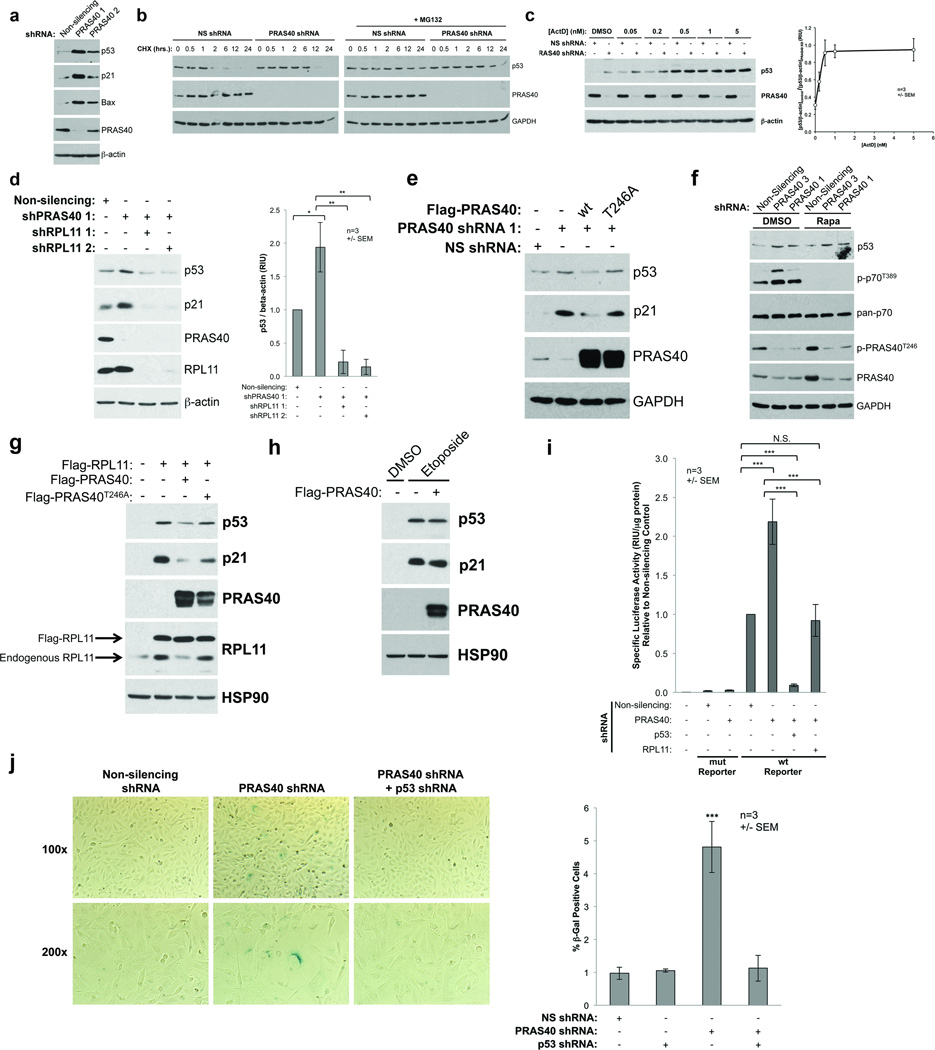
Next, we sought to determine what effect PRAS40 might have on nuclear RPL11 function. In addition to its role in protein synthesis as a component of the ribosome, RPL11 is a key signaling molecule in communicating aberrant ribosomal biogenesis to the cellular stress response machinery. When ribosome assembly is disrupted (a condition referred to as nucleolar stress),2 RPL11 translocates from nucleoli to the nucleoplasm where it binds and inhibits the E3 ubiquitin ligase HDM2. This triggers protein stabilization of the tumor suppressor p53, leading to cell cycle arrest, senescence, or apoptosis mediated by p53’s transcriptional targets.3–6, 8, 46, 47 Because PRAS40 displays pro-survival30, 34, 35 and pro-tumorigenic29, 31 activity through an unknown mechanism, we hypothesized that PRAS40 negatively regulates p53 through association with RPL11 and inhibition of the RPL11-HDM2-p53 pathway. To explore this possibility, we depleted U2OS human osteosarcoma cells of PRAS40 using shRNAs targeted to various regions of the PRAS40 transcript. U2OS cells express wildtype p53 and possess a functional RPL11-HDM2-p53 pathway, but do not express p14ARF 3–5. Interestingly, PRAS40 KD induces an increase in p53 protein levels compared to control cells harboring non-silencing (NS) shRNA (Fig. 5A). This p53 upregulation is accompanied by an increase in protein levels of the p53 transcriptional targets p21 and Bax. These effects are proportional to the efficiency of PRAS40 KD by two different shRNAs, suggesting that the observed effects are directly related to PRAS40 KD (Fig. 5A). Because p53 is a critical mediator of cell fate, it is subject to multiple layers of regulation, including transcriptional, translational, and post-translational mechanisms. The RPL11-HDM2 pathway acts specifically at the post-translational level by inhibiting the ubiquitin-ligase activity of HDM2, thereby increasing p53 protein stability. Considering this, we sought to test whether PRAS40 also controls p53 through regulation of protein stability. We treated cells harboring either NS or PRAS40-targeted shRNAs with the translation inhibitor cycloheximide for various times. PRAS40 KD increases p53 protein half-life approximately 6-fold compared to the NS control (Fig. 5B). This effect, and p53 degradation in general, is abolished by co-treatment with the proteasome inhibitor MG132, indicating that PRAS40 negatively regulates p53 protein levels in a proteasome-dependent manner (Fig. 5B).
Figure 5. PRAS40 negatively regulates p53 through association with RPL11.
U2OS cells were transfected with plasmids encoding shRNAs as indicated and selected with puromycin (A–D). A) WCLs were resolved by SDS-PAGE and analyzed by WB. B) Cells were treated with 20 µg/mL cycloheximide (CHX) in the absence or presence of 20 µM MG132 and lysed at various time points as indicated. C and D) U2OS cells were treated with Actinomycin D (ActD) as indicated (C). WB bands were quantified by densitometry using Image J software. E and F) U2OS cells stably expressing inducible non-silencing (NS) or PRAS40-targeted shRNA were induced with Doxycycline (Dox) for 48 h, during which time the cells were either transfected with the indicated Flag-PRAS40 plasmids for 24h (E) or treated with 0.01% DMSO or 20 nM Rapamycin (F). G) U2OS cells were transfected with the indicated plasmids. H) U2OS cells were transfected with either empty vector or Flag-PRAS40 plasmids and treated with either 0.1% DMSO or 20 µM Etoposide for 24 h. I) U2OS cells were transfected with plasmids encoding shRNAs as indicated and selected with puromycin. p53 transcriptional activity was assessed using luciferase reporter plasmids containing wild-type (WT) or mutated (mut) repeats of the p53 consensus binding element. WCLs were prepared and normalized to equal total protein concentration. Luciferase activity was measured using the Envision® Multilabel plate reader (PerkinElmer). J) U2OS cells were transfected with plasmids encoding shRNAs as indicated and selected with puromycin. Cells were fixed and stained for senescence-associated β-galactosidase X-gal cleavage activity at pH 6.0. Cells were counted and scored by two blinded, impartial investigators. (*p < 0.5, **p < 0.01, ***p < 0.001)
To further explore the possibility that PRAS40 regulates p53 through inhibition of the RPL11-HDM2 pathway, we employed Actinomycin D (ActD), a polypeptide antibiotic known to induce nucleolar stress and upregulate p53 exclusively through the RPL11-HDM2 pathway when used at concentrations < 10 nM.6, 8, 46 While PRAS40 KD and ActD have an additive effect at low ActD concentrations, there is no combinatorial effect at higher ActD concentrations (Fig. 5C), suggesting that PRAS40 KD and ActD activate p53 through a similar mechanism, namely the RPL11-HDM2 nucleolar stress response pathway. Importantly, we also find the effect of PRAS40 KD on p53 protein level is abolished by co-KD of RPL11 using two different shRNAs (Fig. 5D). Taken together, these results indicate that PRAS40 KD induces p53 protein stabilization in an RPL11-dependent manner and imply that PRAS40 may act as an endogenous regulator of the RPL11-HDM2-p53 pathway.
PRAS40 suppresses p53 accumulation and activity in a manner dependent upon PRAS40-RPL11 association
To decrease experimental variation and limit the possibilities of transfection- or selection-related artifacts, we generated U2OS cells stably expressing Doxycyline (Dox)-inducible NS or PRAS40-targeted shRNAs. In agreement with our transient transfection studies, Dox-induced PRAS40 KD results in p53 and p21 upregulation compared to NS control cells (Figs. 5E,F). Importantly, re-introduction of WT PRAS40 completely rescues this effect, whereas RPL11 binding-null PRAS40T246A fails to do so (Fig. 5E), indicating that the ability of PRAS40 to suppress p53 is dependent upon the capacity of PRAS40 to associate with RPL11. Because PRAS40 is an established negative regulator of mTORC1, we sought to test whether PRAS40 KD-induced mTORC1 activation contributes to p53 upregulation. Although rapamycin treatment robustly blocks mTORC1 activity as indicated by loss of p70S6K T389 phosphorylation, it has no effect on PRAS40 KD-induced upregulation of p53 (Fig. 5F). Together, these results reveal that PRAS40-mediated suppression of p53 is dependent upon PRAS40’s ability to associate with RPL11, but independent of its ability to inhibit mTORC1. This provides further support for the hypothesis that PRAS40 negatively regulates p53 through inhibition of the RPL11-HDM2 pathway. To more directly test this, we activated the RPL11-HDM2 pathway by overexpression of RPL11 as previously reported.3–5 RPL11-inducd p53 upregulation is attenuated by co-expression of WT PRAS40, but not RPL11 binding-null PRAS40T246A (Fig. 6G). Interestingly, PRAS40 overexpression has no effect on p53 induced by the DNA damage-causing agent etoposide (Fig. 6F), indicating that PRAS40’s regulatory effect on p53 is at least partially specific to the RPL11-HDM2 pathway.
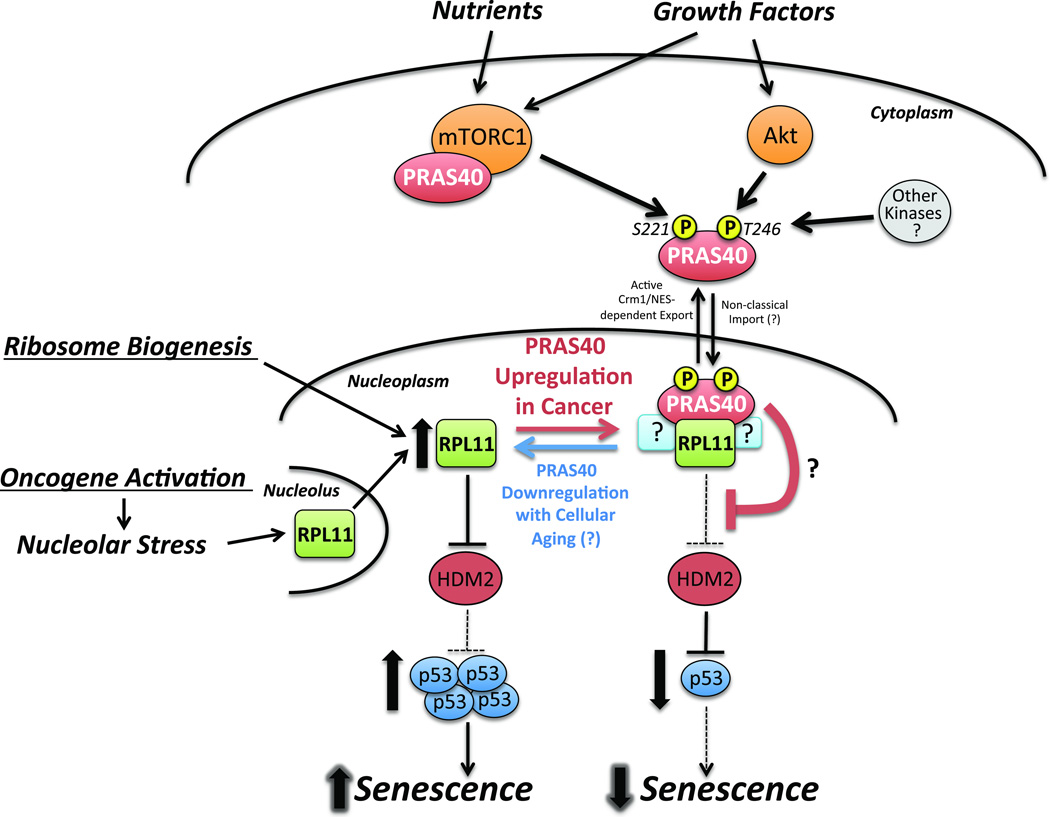
Figure 6. A working model for the role of PRAS40 in regulating the RPL11-HDM2-p53 pathway.
In response to growth factors and nutrients, Akt and mTORC1 phosphorylate PRAS40 leading to the formation of a nuclear-specific PRAS40- and RPL11-containing complex. PRAS40 suppresses the RPL11-HDM2-p53 pathway and inhibits the induction of cellular senescence under growth conditions. In healthy cells, this may be one mechanism by which mTORC1 and Akt prevent aberrant activation of the RPL11-HDM2-p53 pathway during routine ribosome biogenesis. Cancer cells that overexpress PRAS40 may exploit this mechanism to overcome oncogene-induced senescence.
PRAS40 negatively regulates p53-dependent transcriptional activity and senescence
To test if the observed PRAS40 KD-induced p53 and p21 upregulation (Figs. 5A,D,E) corresponds to increased p53 transcriptional activity we used a p53 reporter plasmid in which luciferase transcription is driven by the consensus p53-binding promoter sequence. PRAS40 KD significantly increases p53 transcriptional activity compared to the NS control. Importantly, this effect is abolished by co-KD of either p53 or RPL11 (Fig. 5I). Finally, we tested whether PRAS40 KD-induced p53 upregulation has any effect on cell survival or growth. In order to suppress tumorigenesis, p53 transcriptional targets are known to induce cell death, temporary cell cycle arrest, or a permanent loss of replicative potential known as senescence. While we are not able to detect significant induction of cell death in U2OS cells, PRAS40 depletion significantly increases senescence-associated β-galactosidase activity compared to NS control cells. Importantly, this effect is abrogated by co-KD of p53 (Fig. 5J). Taken together, our results suggest that PRAS40 negatively regulates p53 stability and activity by inhibiting the RPL11-HDM2 pathway in a manner dependent upon PRAS40-RPL11 association, thereby helping to suppress p53-mediated induction of senescence in unstressed cells.
Discussion
Although other negative regulators of mTORC1 such as TSC1/2 and LKB1 function as tumor suppressors, PRAS40 has a pro-tumorigenic function and is positively correlated with proliferative disease progression.29, 31 To clarify the tumorigenic function of PRAS40 we have interrogated its subcellular localization and interactome. Here we experimentally demonstrate that PRAS40 subcellular distribution is controlled at least in part by Crm1-dependent nuclear export directed by a NES sequence - 218IAASMRALVL227. These results, which were obtained in the human cancer cell lines HeLa and U2OS, are consistent with findings in A14 mouse fibroblasts published during the preparation of this manuscript.44 Interestingly, PRAS40 contains neither a classical NLS (cNLS)48 nor a nucleolar localization signal.49, 50 Although the MW of PRAS40 may be low enough to allow nuclear entry via passive diffusion, gel filtration analyses indicate that PRAS40 exits in high MW complexes in cells. Therefore, we speculate that PRAS40 may be actively imported to the nucleus either through interaction with a cNLS-containing protein or in a manner dependent upon a non-classical NLS.51
Using IP-MS coupled with subcellular fractionation we identify RPL11 as a nuclear-specific PRAS40-associated protein. This is confirmed by Flag-PRAS40 and endogenous coIPs. Furthermore, PRAS40 and RPL11 are found to co-localize in nucleoli via confocal microscopy. The molecular mechanism for nuclear specificity remains undetermined at this point; however, we speculate that there may exist cytoplasmic factors that inhibit, or nuclear factors that are required for, the association of PRAS40 and RPL11. Size exclusion chromatography reveals that the nuclear PRAS40- and RPL11-containing complex is distinct from mTORC1, has a high molecular weight (~300–700 kDa), and therefore likely contains other as-of-yet unidentified members. It is unlikely that PRAS40 binds mature ribosomes because 1) the apparent size of the PRAS40-RPL11 complex is far less than that of a ribosome, and 2) no PRAS40-RPL11 co-precipitation occurs in the cytoplasm despite the abundance of mature ribosomes that reside therein. Therefore, it is possible that inclusion in fully formed ribosomes may sequester RPL11 away from PRAS40 in the cytoplasm, thus contributing to the nuclear specificity of the PRAS40-RPL11 association.
The nuclear PRAS40-RPL11 complex is phosphorylation-dependent and requires PRAS40 residues S221 and T246, known mTORC1 and Akt target sites, respectively; however, the precise roles of phosphorylation and residues S221 and T246 remain to be clarified. Importantly, while mutation of S221 and T246 drastically decreases PRAS40-RPL11 association, these mutations do not appear to have a significant effect on PRAS40 subcellular localization (Fig. 3B). This finding is in agreement with results published by Wiza et al.44 Likewise, manipulation of Akt and mTORC1 activity has little effect on PRAS40 subcellular localization (Fig. 4). Together with the fact that purified nuclear extract was used for PRAS40 IPs in this study, these findings indicate that phosphorylation of PRAS40 S221 and T246 does not influence PRAS40-RPL11 association through control of subcellular localization. Although S221 and T246 are both critical to PRAS40-14-3-3 binding,19, 45 it appears unlikely that 14-3-3 is required for the PRAS40-RPL11 association. Serendipitously, we find that a C-terminal Flag epitope tag disrupts PRAS40-14-3-3, but has little effect on PRAS40-RPL11 co-precipitation (Fig. 2B), suggesting that PRAS40-RPL11 association does not require PRAS40-14-3-3 binding. In summary, both phosphorylation and residues S221 and T246 are required for PRAS40-RPL11 association; however, it remains undetermined whether S221/T246 phosphorylation is required for physical association of PRAS40 with RPL11, only for dissociation of PRAS40 from mTORC1, or both.
Our results indicate that the RPL11-PRAS40 complex is responsive to extracellular serum and nutrient conditions and requires the activities of both mTORC1 and Akt. Experiments testing the effect of dominant negative and constitutively active Akt and mTOR on endogenous PRAS40-RPL11 association (Fig. 4C) suggest that in some cases, possibly depending on the strength and nature of the stimulus, activation of either Akt or mTOR alone can be sufficient to promote PRAS40-RPL11 association, likely through activation of redundant signaling pathways that results in dual S221/T246 phosphorylation of PRAS40.
In U2OS cells with a functional RPL11-HDM2 nucleolar stress response pathway and inactive p14ARF, PRAS40 KD leads to an RPL11-dependent increase in p53 protein level and transcriptional activity. Consistent with a model in which Akt- and mTORC1-phosphorylated PRAS40 inhibits the RPL11-HDM2 pathway, PRAS40 KD-induced p53 upregulation is achieved through decreased p53 proteasomal degradation. Together these data imply that PRAS40 is a dual-input signal integrator and effector of mTORC1 and Akt that suppresses the RPL11-HDM2-p53 pathway in the presence of extracellular growth factors and nutrients (Fig. 6). Although PICT1 has been identified as a regulator of the nucleolar stress response pathway,52 the present findings represent the first known regulatory link between extracellular stimuli, the Akt/mTORC1 signaling axis, and the RPL11-HDM2-p53 pathway. The precise mechanism by which PRAS40 regulates the RPL11-HDM2 pathway remains to be determined. One possibility is that PRAS40 may block translocation of RPL11 from nucleoli to the nucleoplasm.
Because ribosome biogenesis requires a massive influx of RPs through the nucleoplasm, it has been suggested that cells possess an unidentified mechanism for suppressing the RPL11-HDM2 pathway during this process.8 Based on the present results, PRAS40 seems to be a potential candidate, as the mTORC1 activity that promotes ribosome biogenesis may simultaneously induce nuclear PRAS40-RPL11 association and suppression of the RPL11-HDM2 pathway.
Oncogene-induced p53 activation is a major tumor suppressive mechanism.53–55 It has been shown that the RP-HDM2-p53 pathway is a critical mediator of oncogene-induced senescence and apoptosis.6, 7 Because our results imply that PRAS40 is a negative regulator of the RPL11-HDM2 pathway that suppresses p53-dependent senescence, it is plausible that upregulation of PRAS40 is one mechanism by which cancer cells overcome the tumor suppressive effects of oncogene-induced p53. Expression of many oncogenes including RAS, B-RAF, E2F3, and SRSF1 can trigger p53-mediated senescence.7, 53–55 For example, progression from benign, BRAFV600E-harboring nevi to malignant melanoma depends on the ability of cells to overcome oncogene-induced senescence.56–58 Activation of the PI3K-Akt pathway is sufficient to reverse oncogenic BRAFV600E-induced senescence and cause progression to malignant melanoma.59 However, the exact mechanism by which PI3K-Akt achieves this is not fully understood. Considering the reported upregulation of PRAS40 in melanoma and Ewing sarcoma cells,29, 31 it will be interesting to determine if PRAS40 plays a role downstream of Akt in overcoming oncogene-induced p53 activation.
In summary, while PRAS40 was previously known to serve as a binding partner and inhibitor of mTORC1, our findings identify PRAS40 as a novel effector of Akt and mTORC1 that negatively regulates the RPL11-HDM2-p53 nucleolar stress response pathway to control cellular senescence. These findings suggest a mechanism for suppression of the RPL11-HDM2 pathway during routine ribosome biogenesis in healthy cells and provide a potential explanation for the pro-tumorigenic effects of upregulated PRAS40 (Fig. 6). As such, the PRAS40-RPL11 complex may serve as a novel target for the discovery of p53-restorative anti-cancer therapeutics.
EXPERIMENTAL PROCEDURES
Plasmids and Stable Cell Lines
All Flag-, GST-, Venus-, and His6-, tagged PRAS40 and RPL11 plasmids for mammalian expression were generated using the Gateway® cloning system (Invitrogen). Human PRAS40 cDNA was obtained by PCR from a human tumor cDNA library. Human RPL11 cDNA was provided by Dr. Yue Xiong (Addgene Plasmid 20936).4 PRAS40 truncations were generated via introduction of a stop codon during PCR amplification. PRAS40 deletions and point mutations were generated via site-directed mutagenesis using the QuikChange Lightning Kit (Stratagene). All plasmids generated were confirmed by sequencing. PRAS40- and RPL11-targeted and non-silencing shRNA plasmids were purchased from Open Biosystems/Thermo Scientific (PRAS40 shRNAs 1,2,3 = V3LHS_340055, V2LHS_138819, and V3LHS_408646; RPL11 shRNAs 1,2 = V2LHS_131577, V3LHS_383206). Plasmids for creating inducible KD cell lines were generated by ligating shRNA sequences into the pTRIPZ plasmid (Thermo Scientific). The resulting plasmids were transfected into U2OS cells using XtremeGene HP (Roche) and were selected in puromycin for 2 months. Akt plasmids were provided by Dr. Michael Greenberg. Constitutively active AU1-mTOR S2215Y plasmid was provided by Dr. Fuyuhiko Tamanoi (Addgene 26037).60 Kinase Dead myc-mTOR was provided by Dr. David Sabatini (Addgene 8482). p53 shRNA was provided by Dr. William Hahn (Addgene 10672).61, 62 p53 luciferase reporters – PG13Py-Luc (wt) and MG15Py-Luc (Mut) – were provided by Dr. Bert Vogelstein.63
Cell Culture
HeLa human cervical adenocarcinoma cells (ATCC CCL-2) and U2OS human osteosarcoma cells (ATCC HTB-96) were maintained in DMEM or McCoy’s (Cellgro – 10013CV and 10050CV, respectively) media, respectively, supplemented with 10% FBS and 1× Penicillin/Streptomycin Solution (Cellgro). Cells were incubated at 37°C in humidified conditions with 5% CO2. For Leu/Met starvation and rescue, cells were rinsed twice in DMEM lacking Leu and Met (Pierce 30030) supplemented with 10% dialyzed FBS (Sigma F0392) and grown in this media for 6.5 h. The media was changed back to complete DMEM containing 10% FBS and either no additive, 0.01% DMSO, or 20 nM Rapamycin (LC Laboratories) for 2 h.
Transfections
FuGene HD (Roche) was used in a ratio of 3 µL to 1 µg DNA for HeLa cells and XtremeGene HP (Roche) was used in a ratio of 2 µL to 1 µg DNA for U2OS cells according to the manufacturers’ instructions.
Sucrose Cushion Subcellular Fractionation
This procedure was performed essentially as detailed in “Current Protocols in Molecular Biology.64”
Indirect Immunofluorescence and Microscopy
Immunofluorescent labeling of fixed cells was performed as previously described.65 Images were captured using Zeiss LSM 510 and Olympus FluoView 1000 inverted confocal microscopes. Live cells were used for images obtained with the Image ExpressMicro automated epifluorescence microscope (Molecular Devices).
Subcellular Fractionation and IP
These methods were performed essentially as previously described.39
Mass Spectrometry and Peptide Identification
The analysis was essentially carried out using a previously optimized proteomics platform.66 †
Gel Filtration Chromatography
Concentrated Hela nuclear extract was resolved over a 25 mL Superose 6 column (GE) at 0.3 mL/min using the Äkta Purifier FPLC system (GE) in running buffer [10 mM HEPES pH 7.45, 137 mM NaCl, 2.5% glycerol, filtered and degassed]. 0.5 mL fractions were collected.
GST Pull-Down
Cells were lysed in CHAPS Buffer [40 mM HEPES pH 7.5, 120 mM NaCl, 1 mM EDTA, 0.3% CHAPS, 1:500 Sigma Phosphatase Inhibitor Cocktails 2 and 3 and Protease Inhibitor Cocktail] (modified from Sarbassov and Sabatini67) and incubated with glutathione-conjugated beads (GE) for 4 h at 4 °C. Beads were washed three times with CHAPS Buffer and eluted by boiling in SDS-PAGE loading buffer.
p53 Transcriptional Reporter Assay
U2OS cells were lysed in Glo Lysis Buffer (Promega) containing protease and phosphatase inhibitor cocktails (Sigma P8340, P2850, and P5726). Protein concentration was measured by the BCA Assay (Pierce) and concentrations were normalized by addition of Glo Lysis Buffer as necessary. Luciferase activity was measured using the Bright-Glo™ system (Promega). Readings were measured immediately after substrate addition using an Envision® Multilabel plate reader (PerkinElmer) with a 0.1 sec integration time.
Senescence-Associated β-Galactosidase Activity Assay
X-Gal staining was performed using the Senescence β-galactosidase Staining Kit (Cell Signaling 9860). Images were captured using a Zeiss Axiovert inverted microscope. Cells were scored and counted by two blinded, impartial investigators. Nine sites per well were counted in each of three independent experiments.
Western Blotting
The following primary antibodies were used: Rabbit anti-PRAS40, IBL; Rabbit anti-PARP, Cell Signaling, 9542; Mouse anti-GAPDH, Chemicon/Millipore; Rabbit anti-Calnexin, Cell Signaling, 2433; Rabbit anti-RPL11, Abcam, ab79352; Rabbit anti-Raptor, Millipore, 09-217; Rabbit anti-14-3-3 epsilon, Santa Cruz, sc-1020; Rabbit anti-pan-14-3-3, Santa Cruz, sc -629; Mouse anti-HSP90, Santa Cruz, sc-13119; Mouse anti-Flag, Sigma M2; Rabbit anti-mTOR, Cell Signaling, 2972; Rabbit anti-GST, Santa Cruz, sc-459; Rabbit anti-phospho-PRAS40 T246, Invitrogen; Rabbit anti-phospho-PRAS40 S183, IBL; Mouse anti-HA, Santa Cruz, sc-7392; Rabbit anti-phospho-p70, Cell Signaling, 9205; Rabbit anti-p70, Cell Signaling, 2708; Mouse anti-p53, Cell Signaling, 2524; Mouse anti-p21, Cell Signaling, 2946; Rabbit anti-Bax, Cell Signaling 5023; Mouse anti-β-actin, Sigma, A2228.
Statistical Methods
Pooled data are reported as the means of three independent experiments +/− SEM. Multiple comparisons were made using the Bonferroni Test. p values are indicated in figure legends.
Acknowledgements
This research was supported in part by NIH pre-doctoral NRSA F31NS067844 and training grant T32GM008602 to JJH, NIH U01CA168449 and a Georgia Cancer Coalition Award to HF, and the Emory University Integrated Cellular Imaging Microscopy Core of the Emory Neuroscience NINDS Core Facilities grant, P30NS055077. We thank Shannon Elf for assistance with plasmid generation, and Anita Corbett and Maureen Powers for helpful discussions.
Footnotes
Conflict of Interest: The authors declare no conflict of interest.
Contributions: JJH and HF designed experiments and analyzed data. JJH performed all experiments except mass spectrometry. DC and JP performed mass spectrometry and data analysis. ZL assisted with localization experiments. JJH and HF wrote the paper.
REFERENCES
- 1.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 2.Pestov DG, Strezoska Z, Lau LF. Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: effects of nucleolar protein Bop1 on G(1)/S transition. Mol Cell Biol. 2001;21:4246–4255. doi: 10.1128/MCB.21.13.4246-4255.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell. 2003;3:577–587. doi: 10.1016/s1535-6108(03)00134-x. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y, Wolf GW, Bhat K, et al. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol. 2003;23:8902–8912. doi: 10.1128/MCB.23.23.8902-8912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhat KP, Itahana K, Jin A, Zhang Y. Essential role of ribosomal protein L11 in mediating growth inhibition-induced p53 activation. EMBO J. 2004;23:2402–2412. doi: 10.1038/sj.emboj.7600247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Macias E, Jin A, Deisenroth C, et al. An ARF-independent c-MYC-activated tumor suppression pathway mediated by ribosomal protein-Mdm2 Interaction. Cancer Cell. 2010;18:231–243. doi: 10.1016/j.ccr.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fregoso OI, Das S, Akerman M, Krainer AR. Splicing-Factor Oncoprotein SRSF1 Stabilizes p53 via RPL5 and Induces Cellular Senescence. Mol Cell. 2013 doi: 10.1016/j.molcel.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, Lu H. Signaling to p53: ribosomal proteins find their way. Cancer Cell. 2009;16:369–377. doi: 10.1016/j.ccr.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thedieck K, Polak P, Kim ML, et al. PRAS40 and PRR5-like protein are new mTOR interactors that regulate apoptosis. PLoS One. 2007;2:e1217. doi: 10.1371/journal.pone.0001217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L, Harris TE, Roth RA, Lawrence JC., Jr PRAS40 regulates mTORC1 kinase activity by functioning as a direct inhibitor of substrate binding. J Biol Chem. 2007;282:20036–20044. doi: 10.1074/jbc.M702376200. [DOI] [PubMed] [Google Scholar]
- 13.Sancak Y, Thoreen CC, Peterson TR, et al. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell. 2007;25:903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 14.Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol. 2007;9:316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- 15.Fonseca BD, Smith EM, Lee VH, MacKintosh C, Proud CG. PRAS40 is a target for mammalian target of rapamycin complex 1 and is required for signaling downstream of this complex. J Biol Chem. 2007;282:24514–24524. doi: 10.1074/jbc.M704406200. [DOI] [PubMed] [Google Scholar]
- 16.Oshiro N, Takahashi R, Yoshino K, et al. The proline-rich Akt substrate of 40 kDa (PRAS40) is a physiological substrate of mammalian target of rapamycin complex 1. J Biol Chem. 2007;282:20329–20339. doi: 10.1074/jbc.M702636200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu H, Subramanian RR, Masters SC. 14-3-3 proteins: structure, function, and regulation. Annu Rev Pharmacol Toxicol. 2000;40:617–647. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- 18.Harthill JE, Pozuelo Rubio M, Milne FC, MacKintosh C. Regulation of the 14-3-3-binding protein p39 by growth factors and nutrients in rat PC12 pheochromocytoma cells. Biochem J. 2002;368:565–572. doi: 10.1042/BJ20020838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kovacina KS, Park GY, Bae SS, et al. Identification of a proline-rich Akt substrate as a 14-3-3 binding partner. J Biol Chem. 2003;278:10189–10194. doi: 10.1074/jbc.M210837200. [DOI] [PubMed] [Google Scholar]
- 20.Hsu PP, Kang SA, Rameseder J, et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332:1317–1322. doi: 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu Y, Yoon SO, Poulogiannis G, et al. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science. 2011;332:1322–1326. doi: 10.1126/science.1199484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yap TA, Walton MI, Hunter LJ, et al. Preclinical pharmacology, antitumor activity, and development of pharmacodynamic markers for the novel, potent AKT inhibitor CCT128930. Mol Cancer Ther. 2011;10:360–371. doi: 10.1158/1535-7163.MCT-10-0760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cassell A, Freilino ML, Lee J, et al. Targeting TORC1/2 enhances sensitivity to EGFR inhibitors in head and neck cancer preclinical models. Neoplasia. 2012;14:1005–1014. doi: 10.1593/neo.121212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andersen JN, Sathyanarayanan S, Di Bacco A, et al. Pathway-based identification of biomarkers for targeted therapeutics: personalized oncology with PI3K pathway inhibitors. Sci Transl Med. 2010;2:43ra55. doi: 10.1126/scitranslmed.3001065. [DOI] [PubMed] [Google Scholar]
- 25.van Slegtenhorst M, de Hoogt R, Hermans C, et al. Identification of the tuberous sclerosis gene TSC1 on chromosome 9q34. Science. 1997;277:805–808. doi: 10.1126/science.277.5327.805. [DOI] [PubMed] [Google Scholar]
- 26.Hemminki A, Markie D, Tomlinson I, et al. A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature. 1998;391:184–187. doi: 10.1038/34432. [DOI] [PubMed] [Google Scholar]
- 27.Jenne DE, Reimann H, Nezu J, et al. Peutz-Jeghers syndrome is caused by mutations in a novel serine threonine kinase. Nat Genet. 1998;18:38–43. doi: 10.1038/ng0198-38. [DOI] [PubMed] [Google Scholar]
- 28.Makowski L, Hayes DN. Role of LKB1 in lung cancer development. Br J Cancer. 2008;99:683–688. doi: 10.1038/sj.bjc.6604515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang L, Nakai Y, Kuwahara I, Matsumoto K. PRAS40 is a functionally critical target for EWS repression in Ewing sarcoma. Cancer Res. 2012;72:1260–1269. doi: 10.1158/0008-5472.CAN-11-2254. [DOI] [PubMed] [Google Scholar]
- 30.Yu F, Narasimhan P, Saito A, Liu J, Chan PH. Increased expression of a proline-rich Akt substrate (PRAS40) in human copper/zinc-superoxide dismutase transgenic rats protects motor neurons from death after spinal cord injury. J Cereb Blood Flow Metab. 2008;28:44–52. doi: 10.1038/sj.jcbfm.9600501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madhunapantula SV, Sharma A, Robertson GP. PRAS40 deregulates apoptosis in malignant melanoma. Cancer Res. 2007;67:3626–3636. doi: 10.1158/0008-5472.CAN-06-4234. [DOI] [PubMed] [Google Scholar]
- 32.Beausoleil SA, Jedrychowski M, Schwartz D, et al. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci U S A. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nascimento EB, Fodor M, van der Zon GC, et al. Insulin-mediated phosphorylation of the proline-rich Akt substrate PRAS40 is impaired in insulin target tissues of high-fat diet-fed rats. Diabetes. 2006;55:3221–3228. doi: 10.2337/db05-1390. [DOI] [PubMed] [Google Scholar]
- 34.Saito A, Narasimhan P, Hayashi T, Okuno S, Ferrand-Drake M, Chan PH. Neuroprotective role of a proline-rich Akt substrate in apoptotic neuronal cell death after stroke: relationships with nerve growth factor. J Neurosci. 2004;24:1584–1593. doi: 10.1523/JNEUROSCI.5209-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saito A, Hayashi T, Okuno S, Nishi T, Chan PH. Modulation of proline-rich akt substrate survival signaling pathways by oxidative stress in mouse brains after transient focal cerebral ischemia. Stroke. 2006;37:513–517. doi: 10.1161/01.STR.0000198826.56611.a2. [DOI] [PubMed] [Google Scholar]
- 36.Kim W, Youn H, Seong KM, et al. PIM1-activated PRAS40 regulates radioresistance in non-small cell lung cancer cells through interplay with FOXO3a, 14-3-3 and protein phosphatases. Radiat Res. 2011;176:539–552. doi: 10.1667/rr2609.1. [DOI] [PubMed] [Google Scholar]
- 37.Kim W, Youn H, Kwon T, et al. PIM1 kinase inhibitors induce radiosensitization in non-small cell lung cancer cells. Pharmacol Res. 2013;70:90–101. doi: 10.1016/j.phrs.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 38.Zhang X, Shu L, Hosoi H, Murti KG, Houghton PJ. Predominant nuclear localization of mammalian target of rapamycin in normal and malignant cells in culture. J Biol Chem. 2002;277:28127–28134. doi: 10.1074/jbc.M202625200. [DOI] [PubMed] [Google Scholar]
- 39.Rosner M, Hengstschlager M. Cytoplasmic and nuclear distribution of the protein complexes mTORC1 and mTORC2: rapamycin triggers dephosphorylation and delocalization of the mTORC2 components rictor and sin1. Hum Mol Genet. 2008;17:2934–2948. doi: 10.1093/hmg/ddn192. [DOI] [PubMed] [Google Scholar]
- 40.Li H, Tsang CK, Watkins M, Bertram PG, Zheng XF. Nutrient regulates Tor1 nuclear localization and association with rDNA promoter. Nature. 2006;442:1058–1061. doi: 10.1038/nature05020. [DOI] [PubMed] [Google Scholar]
- 41.Wei Y, Tsang CK, Zheng XF. Mechanisms of regulation of RNA polymerase III-dependent transcription by TORC1. EMBO J. 2009;28:2220–2230. doi: 10.1038/emboj.2009.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nascimento EB, Ouwens DM. PRAS40: target or modulator of mTORC1 signalling and insulin action? Arch Physiol Biochem. 2009;115:163–175. doi: 10.1080/13813450902988580. [DOI] [PubMed] [Google Scholar]
- 43.Wiza C, Nascimento EB, Ouwens DM. Role of PRAS40 in Akt and mTOR signaling in health and disease. Am J Physiol Endocrinol Metab. 2012;302:E1453–E1460. doi: 10.1152/ajpendo.00660.2011. [DOI] [PubMed] [Google Scholar]
- 44.Wiza C, Nascimento EB, Linssen MM, et al. Proline-rich Akt substrate of 40-kDa contains a nuclear export signal. Cell Signal. 2013;25:1762–1768. doi: 10.1016/j.cellsig.2013.05.022. [DOI] [PubMed] [Google Scholar]
- 45.Wang L, Harris TE, Lawrence JC., Jr Regulation of proline-rich Akt substrate of 40 kDa (PRAS40) function by mammalian target of rapamycin complex 1 (mTORC1)-mediated phosphorylation. J Biol Chem. 2008;283:15619–15627. doi: 10.1074/jbc.M800723200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deisenroth C, Zhang Y. Ribosome biogenesis surveillance: probing the ribosomal protein-Mdm2-p53 pathway. Oncogene. 2010;29:4253–4260. doi: 10.1038/onc.2010.189. [DOI] [PubMed] [Google Scholar]
- 47.Miliani de Marval PL, Zhang Y. The RP-Mdm2-p53 pathway and tumorigenesis. Oncotarget. 2011;2:234–238. doi: 10.18632/oncotarget.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J Biol Chem. 2007;282:5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scott MS, Boisvert FM, McDowall MD, Lamond AI, Barton GJ. Characterization and prediction of protein nucleolar localization sequences. Nucleic Acids Res. 2010;38:7388–7399. doi: 10.1093/nar/gkq653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scott MS, Troshin PV, Barton GJ. NoD: a Nucleolar localization sequence detector for eukaryotic and viral proteins. BMC Bioinformatics. 2011;12:317. doi: 10.1186/1471-2105-12-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chook YM, Suel KE. Nuclear import by karyopherin-betas: recognition and inhibition. Biochim Biophys Acta. 2011;1813:1593–1606. doi: 10.1016/j.bbamcr.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sasaki M, Kawahara K, Nishio M, et al. Regulation of the MDM2-P53 pathway and tumor growth by PICT1 via nucleolar RPL11. Nat Med. 2011;17:944–951. doi: 10.1038/nm.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mooi WJ, Peeper DS. Oncogene-induced cell senescence--halting on the road to cancer. N Engl J Med. 2006;355:1037–1046. doi: 10.1056/NEJMra062285. [DOI] [PubMed] [Google Scholar]
- 54.Schmitt CA. Cellular senescence and cancer treatment. Biochim Biophys Acta. 2007;1775:5–20. doi: 10.1016/j.bbcan.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 55.Kuilman T, Michaloglou C, Mooi WJ, Peeper DS. The essence of senescence. Genes Dev. 2010;24:2463–2479. doi: 10.1101/gad.1971610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Michaloglou C, Vredeveld LC, Soengas MS, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436:720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- 57.Patton EE, Widlund HR, Kutok JL, et al. BRAF mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Curr Biol. 2005;15:249–254. doi: 10.1016/j.cub.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 58.Dhomen N, Reis-Filho JS, da Rocha Dias S, et al. Oncogenic Braf induces melanocyte senescence and melanoma in mice. Cancer Cell. 2009;15:294–303. doi: 10.1016/j.ccr.2009.02.022. [DOI] [PubMed] [Google Scholar]
- 59.Vredeveld LC, Possik PA, Smit MA, et al. Abrogation of BRAFV600E-induced senescence by PI3K pathway activation contributes to melanomagenesis. Genes Dev. 2012;26:1055–1069. doi: 10.1101/gad.187252.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sato T, Nakashima A, Guo L, Coffman K, Tamanoi F. Single amino-acid changes that confer constitutive activation of mTOR are discovered in human cancer. Oncogene. 2010;29:2746–2752. doi: 10.1038/onc.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Masutomi K, Yu EY, Khurts S, et al. Telomerase maintains telomere structure in normal human cells. Cell. 2003;114:241–253. doi: 10.1016/s0092-8674(03)00550-6. [DOI] [PubMed] [Google Scholar]
- 62.Stewart SA, Dykxhoorn DM, Palliser D, et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.el-Deiry WS, Tokino T, Velculescu VE, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 64.Ausubel FM. Current protocols in molecular biology. Brooklyn, N.Y. Media, Pa: Greene Pub. Associates; J. Wiley, order fulfillment; 1987. [Google Scholar]
- 65.Havel LS, Wang CE, Wade B, Huang B, Li S, Li XJ. Preferential accumulation of N-terminal mutant huntingtin in the nuclei of striatal neurons is regulated by phosphorylation. Hum Mol Genet. 2011;20:1424–1437. doi: 10.1093/hmg/ddr023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu P, Duong DM, Peng J. Systematical optimization of reverse-phase chromatography for shotgun proteomics. J Proteome Res. 2009;8:3944–3950. doi: 10.1021/pr900251d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sarbassov DD, Sabatini DM. Redox regulation of the nutrient-sensitive raptor-mTOR pathway and complex. J Biol Chem. 2005;280:39505–39509. doi: 10.1074/jbc.M506096200. [DOI] [PubMed] [Google Scholar]