Abstract
The prevalence of life-threatening anaphylactic responses to food is rising at an alarming rate. The emerging role of the gut microbiota in regulating food allergen sensitization may help explain this trend. The mechanisms by which commensal bacteria influence sensitization to dietary antigens are only beginning to be explored. We have found that a population of mucosa-associated commensal anaerobes prevents food allergen sensitization by promoting an IL-22-dependent barrier protective immune response that limits the access of food allergens to the systemic circulation. This early response is followed by an adaptive immune response mediated in part by an expansion of Foxp3+ Tregs that fortifies the tolerogenic milieu needed to maintain non-responsiveness to food. Bacterial metabolites, such as short-chain fatty acids, may contribute to the process through their ability to promote Foxp3+ Treg differentiation. This work suggests that environmentally induced alterations of the gut microbiota offset the regulatory signals conferred by protective bacterial species to promote aberrant responses to food. Our research presents exciting new possibilities for preventing and treating food allergies based on interventions that modulate the composition of the gut microbiota.
Keywords: intestinal microbiota, short chain fatty acids, food allergy, oral tolerance, Foxp3+ Tregs, IL-22
Introduction
Differentiating innocuous environmental antigens from serious threats is a particular challenge in the intestinal mucosa, which is constantly exposed to antigens derived from both food and the commensal microbiota. Oral tolerance typically refers to the process by which the immune system limits the response to dietary antigens. Experimentally, administration of antigen by the oral route induces mucosal and systemic non-responsiveness to subsequent peripheral challenge [1]. Rodent models have provided insight into the physiological processes required to maintain tolerance to food and suggest that sensitization to dietary allergens increases when these processes fail.
Since nutrients are absorbed in the small intestine studies on the mechanisms regulating oral tolerance have focused primarily on antigen-specific immune responses in the gut associated lymphoid tissues (GALT) that drain this site. Emerging evidence, however, points to a central role for commensal bacteria in preventing food allergen sensitization. Dietary antigens share the intestinal lumen with trillions of astoundingly diverse bacteria comprising approximately one thousand different species [2]. These bacteria colonize the gut at increasing densities from stomach to rectum. In the small intestine, bacterial load is kept low due to the high motility of intestinal contents and the bactericidal activity of bile salts, with densities of 104–105 bacteria per milliliter of effluent in the proximal small intestine and 108 bacteria per milliliter of effluent in the ileum [3]. Bacterial load is by far greatest in the colon, where densities can reach 1011 organisms per gram of luminal content [3]. The high microbial content of the ileum and colon exerts a formidable influence on the framework of the mucosal immune system in the gut, not only to promote local homeostatic interactions, but also to modulate immune responses to antigens at peripheral sites beyond the mucosa. We have found that particular populations of bacteria are required to prevent sensitization to dietary antigens; when protective bacteria-derived signals are lacking, tolerance to dietary antigen is not induced. Our data support the idea that an environmentally induced alteration of the commensal microbiota is driving the rapidly increasing prevalence of allergic responses to food in Western societies [4].
In this review, we will first discuss what is currently known about the regulation of tolerance to dietary antigen before discussing how commensal bacteria influence this process.
Mechanisms of orally induced non-responsiveness
The mechanisms governing non-responsiveness to dietary antigens remain poorly understood; the ways in which dietary antigens cross the intestinal epithelial barrier, are presented to the immune system, and elicit a response are still being defined. It is clear, however, that the maintenance of tolerance involves both cellular and humoral processes that are induced in the intestinal lamina propria (LP) and the mesenteric lymph node (MLN). In order to be recognized by the immune system dietary antigen must first cross the epithelium of the small intestine. Antigen can be transcytosed by specialized epithelial cells called microfold (M) cells, which reside in the follicle-associated epithelium of Peyer’s patches (PP) [5]. M cells lack the brush border glycocalyx present on other enterocytes and instead contain large vesicles in their cytoplasm that enable transepithelial transport of luminal antigens [6, 7]. The basolateral surface of M cells is in close contact with the underlying GALT where antigen presenting cells (APCs) can take up and process these luminal antigens for presentation to naïve T cells [8].
M cells were initially thought to be the major route by which dietary antigens cross the epithelial barrier. More recent evidence, however, suggests that other types of epithelial cells may assist in this process. Goblet cells, best known for their ability to produce the thick mucus glycocalyx overlaying the epithelium, may play a role in antigen transport. Fluorescent imaging techniques indicate that goblet cell-associated passages (GAPs) allow labeled, orally delivered antigen to cross from the lumen into the LP [9]. This process is constitutively active in the small intestine, suggesting that GAPs may be an important, underappreciated route for antigen uptake. In addition to active transport through enterocytes, antigen can pass between epithelial cells. Intestinal epithelial cells (IECs) are joined by tight junctions, adherens junctions, and desmosomes that together form a “molecular gasket” to seal the paracellular space [10]. Under homeostatic conditions, solutes, proteins, and certain microbial components can infiltrate this complex and pass between cells. Paracellular transport is regulated closely by the cytokine milieu, which can dramatically alter the expression of tight junction proteins under conditions of inflammation [10]. Some evidence also suggests that extensions from a subset of dendritic cells (DC) in the small intestine reach through epithelial tight junctions to sample luminal antigens [11–13].
Once antigen has passed from the intestinal lumen into the GALT it is taken up by APCs for presentation to naïve T cells. Several functionally distinct subsets of APCs in the GALT can be defined by their surface marker expression. Two major populations have been implicated in oral tolerance: a DC subset identified by the expression of CD11c and CD103 and a macrophage subset defined by the expression of CD11b and CX3CR1 [1]. CD11c+ CD103+ DCs express high levels of MHC class II and the homing receptor CCR7 that enables migration to the MLN [14, 15]. CD11c+ CD103+ CCR7+ DCs are the major cell population responsible for picking up and processing antigens crossing the epithelial barrier [16, 17]. Once antigen has been processed, these DCs traffic through the lymph to the MLN where they encounter and stimulate naïve CD4+ T cells. The fate of an antigen-specific CD4+ T cell following stimulation depends upon the cytokine milieu in which the encounter occurs. Under homeostatic conditions, high concentrations of TGF-β and retinoic acid (RA) in the MLN promote a tolerogenic environment that favors the differentiation of regulatory T cells (Tregs) [18, 19]. The TGF-β required for this process is produced both by CD103+ DCs themselves as well as by IECs [20, 21]. Interestingly, IEC-derived TGF-β has been shown to be important for promoting TGF-β production by CD103+ DCs and enhancing their ability to drive Treg differentiation [22]. RA is derived from dietary vitamin A and is metabolized primarily by CD11c+CD103+ DCs, which express high levels of retinaldehyde dehydrogenase (RALDH) and aldehyde dehydrogenase (ALDH) enzymes [18, 19]. RA enhances the TGF-β dependent upregulation of Foxp3, the transcription factor that controls Treg differentiation [18]. In the absence of dietary vitamin A, the frequency of Tregs in the MLN and intestinal LP is significantly reduced [23, 24].
Of the two major APC subsets in the GALT, the role of CX3CR1+ macrophages has been controversial. Using an in vitro transwell system, Rescigno et al first visualized the ability of a subset of DCs to extend their dendrites between epithelial tight junctions [12]. This finding was corroborated using confocal microscopy, which found the dendrites to extend primarily in the villi of the terminal ileum. The DC subset involved was later characterized as CD11c+ CD11b+ CX3CR1+ DCs that derived from myeloid precursors [11]. However, some question remained as to the role of these cells in oral tolerance, as they were shown to be poor APCs for the stimulation of T cell proliferation in vitro [25]. Moreover, CX3CR1+ DCs do not migrate under homeostatic conditions and have been observed in the MLN only after infection with an intestinal pathogen or following antibiotic treatment [26]. Given these characteristics, CX3CR1+ cells were thought to be more representative of a macrophage rather than a DC subset. Yet, their importance in establishing oral tolerance is clear; in the absence of CX3CR1 expression, the uptake of fed antigen and the expansion of cognate T cells is reduced, resulting in increased delayed-type hypersensitivity (DTH) reactions in response to antigen challenge [27]. Recent work suggests that CX3CR1+ macrophages are the first to acquire luminal antigen, which they then pass via cell-to-cell contact and gap junctions to CD103+ DCs that migrate and interact with naïve T cells [27]. Other work suggests that the major function of these cells is to produce IL-10, which supports the proliferation and expansion of antigen-specific Foxp3+ Tregs in the LP [28].
After dietary antigen-specific T cells recognize their cognate antigen and differentiate into Foxp3+ Tregs, they upregulate the homing molecules CCR9 and α4β7 that direct migration back to the small intestinal LP [28, 29]. Once there, Foxp3+ Tregs expand and suppress aberrant responses to dietary antigens through the production of IL-10, TGF-β, and IL-35 [30]. In the absence of induced Tregs, the cytokine milieu of the MLN is highly Th2-skewed with increases in CD4+ T cells producing IL-4, IL-13 and IL-5 [31]. Food allergen sensitization occurs when naïve CD4+ T cells differentiate into Th2 cells in the presence of IL-4 [32]. Th2 cells then help to promote an allergic response by inducing B cell class-switching to the IgE isotype. IgE subsequently binds to its high affinity Fc receptor, FcεRI, which is expressed predominantly on mast cells. Re-exposure to dietary antigen crosslinks bound IgE, inducing mast cell degranulation and the release of mediators that precipitate an allergic, and potentially anaphylactic, reaction [33].
The cytokine environment may not be the only factor regulating tolerance to food. Recent work suggests that mucus is more than just a physical barrier between IECs and the intestinal lumen and actively promotes tolerance by repressing the expression of inflammatory cytokines by DCs [34]. In the presence of the mucin protein MUC2, DCs produce more IL-10 and express higher levels of RALDH and ALDH enzymes. Mice deficient in mucus production (Muc2−/− mice) have a decreased proportion of Foxp3+ Tregs in the LP and exhibit increased DTH responses after antigen feeding. This effect of MUC2 is mediated by its binding to a galectin-3-dectin-1-FcγRIIB complex on DCs, activating β-catenin signaling to induce a tolerogenic phenotype that contributes to mucosal homeostasis [34].
Antibody secreting B cells also contribute to the induction of oral tolerance. IgA is the most abundantly produced immunoglobulin isotype and can be found in two forms: a monomeric form in the systemic circulation and a dimeric form at mucosal sites [35]. IgA is uniquely suited for protection of mucosal sites like the GALT because it is neutralizing but not inflammatory and binds luminal antigens to prevent them from contacting the epithelial barrier or the underlying immune system. In this way, IgA may reinforce oral tolerance by excluding bound dietary components from the LP [36]. Its necessity is debated, however, because there is no impairment of tolerance when IgA is eliminated [36].
Commensal bacteria and sensitization to food allergens
What, then, is the role of the commensal bacteria in regulating sensitization to dietary antigens? The mechanisms regulating orally induced systemic non-responsiveness described above focus solely on interactions between immune cells and dietary antigen in the local environment of the small intestine and ignore the abundance of bacteria colonizing the length of the intestinal tract. The high microbial content of the intestine (and of the colon in particular) undoubtedly influences the underlying immune system in the GALT. Indeed the GALT exhibits several specialized adaptations, such as the secretion of antimicrobial proteins by IECs and the production of secretory IgA, that act in concert to maintain tolerance to the gut microbiota [37–39]. Whether commensal bacteria also interact with the GALT to maintain non-responsiveness to the other major luminal constituent – food – is only beginning to be explored.
We have found that administration of broad-spectrum antibiotics to neonatal mice induces sensitization to orally administered peanut allergens, as characterized by increased levels of circulating peanut-specific IgE and IgG1 [40, 41]. 16S rRNA sequencing demonstrates that antibiotic treatment markedly alters the composition of the colonic and ileal microbiota, depleting most members of the Bacteroidetes and Firmicutes phyla usually found to be prevalent in the gut. Notably, reconstituting the gut microbiota by transfer of feces from an untreated mouse rescues antibiotic-treated mice from food allergen sensitization. Moreover, colonizing antibiotic-treated mice with a microbiota enriched in Clostridia, an indigenous class of anaerobic spore-forming Firmicutes that resides in close proximity to the colonic epithelium, confers the same protection [41]. These results indicate that certain signals from the microbiota can prevent food allergen sensitization. This concept is supported by work from other laboratories. Food allergy-prone Il4raF709 mice with a gain-of-function mutation in the IL-4 receptor attain a unique microbial signature upon OVA-sensitization that is non-overlapping with that of sensitized wild-type (WT) mice [42]. Transplantation of germ-free (GF) mice with the microbiota of OVA-sensitized Il4raF709 mice, but not of OVA-sensitized WT mice, results in a more severe anaphylactic response upon challenge [42]. In another study, GF mice colonized with the microbiota of a healthy human infant and sensitized with whey protein exhibited milder allergic symptoms following challenge with β-lactoglobulin than did their GF counterparts [43].
As mentioned above, the prevalence of allergic responses to food has been increasing in Western societies at an unprecedented rate, rising by as much as 20% in a recent ten year period [44–47]. The rapidity of this trend makes it unlikely that genetic drift alone is responsible. Adherence to a Westernized lifestyle introduces several environmental risk factors that can disturb the homeostatic balance of the gut microbiota. These include excessive antibiotic use, a shift towards formula-feeding and Caesarean births, and consumption of a Western high fat diet. In particular, antibiotic use during infancy has often been cited as a major factor contributing to the rising prevalence of allergic disease and is considered among the most potent stimuli for perturbing bacterial communities throughout the gastrointestinal tract [48, 49]. Indeed, increased urinary levels of the antimicrobial agent triclosan are correlated with a heightened risk of sensitization to food [50], and maternal use of antibiotics before and during pregnancy is positively associated with an increased risk of cow’s milk allergy among newborns [51]. Aside from antibiotic use, a greater risk of developing IgE-mediated sensitization to food allergens has also been noted among children delivered by Caesarean section [52]. Several studies have already reported significant differences in the composition of the microbiota of allergic versus non-allergic infants [53–56]. Collectively, these data provide clinical and epidemiological findings in support of a role for commensal dysbiosis in driving the increasing prevalence of food allergy.
Few reports have built on this foundation, however, to examine how microbiota-derived signals regulate sensitization to dietary antigen. Our studies in gnotobiotic and antibiotic-treated murine models have provided novel mechanistic insight into this question. We have found that commensal bacteria prevent sensitization to food allergens by inducing the differentiation of Foxp3+ Tregs and the secretion of IgA and by regulating allergen uptake into the systemic circulation through bacteria-induced IL-22 production.
Bacteria-induced Foxp3+ Tregs
In our mouse model, the protection against allergic sensitization to food conferred by a Clostridia-containing microbiota is associated with an increase in the proportion of Foxp3+ Tregs among CD4+ T cells in the colonic LP and an increase in the concentration of IgA in feces [41]. Both of these responses are indicative of a protective adaptive immune response initiated by Clostridia colonization. In particular, the Clostridia-induced expansion of Foxp3+ Tregs may promote oral tolerance by quenching aberrant dietary antigen-specific Th2 responses in the small intestine. Other work has also reported that commensal bacteria direct naïve T cell differentiation to the peripheral Foxp3+ Treg compartment. Colonic Tregs possess a TCR repertoire distinct from that used by Tregs in other locations, with some TCRs displaying reactivity to bacterial antigens [57]. Furthermore, Clostridium clusters IV, XIVa and XVIII [58, 59] as well as Bacteroides fragilis [60] are potent inducers of Foxp3+ Treg differentiation. The mechanisms by which bacteria promote Foxp3+ Treg differentiation are starting to be elucidated. Some evidence suggests that, as part of the indigenous mucosa-associated microbiota, Clostridia induce a TGF-β rich environment within the colon to enhance Foxp3+ Treg differentiation [59]. Bacteria-specific Foxp3+ Tregs may then direct anti-inflammatory responses in the gut through the secretion of TGF-β, IL-10, and IL-35, behaving similarly to their dietary antigen-specific counterparts [30].
Bacteria-induced IL-22 production
Our work suggests that Clostridia also play a novel role in preventing food allergen sensitization by inducing the production of the barrier protective cytokine IL-22. IL-22 is unusual in that it is produced exclusively by hematopoietic cells while the expression of its receptor is limited to non-hematopoietic cells and particularly to epithelial cells of the mucosa [61, 62]. IL-22 therefore mediates a critical line of communication between the immune system and the intestinal epithelium and is important for maintaining epithelial barrier integrity during homeostasis and in response to infection-mediated damage. Both CD4+ T cells and RORγt+ innate lymphoid cells (ILCs) produce IL-22. At steady state, mice lacking RORγt+ ILCs are more susceptible to microbial translocation from the gut lumen to peripheral organs [63, 64], an effect that can be reversed by the administration of IL-22 [63]. During infection with Citrobacter rodentium, mice lacking IL-22 exhibit defects in the repair of epithelial damage and suffer greater systemic bacterial burdens [65, 66]. In both scenarios, the barrier protective effect of IL-22 has been attributed to its ability to increase the production of antimicrobial peptides including RegIIIβ, RegIIIγ, S100A8, and S100A9, thereby fortifying the intestinal barrier by keeping pathogens and commensals at bay [63, 65].
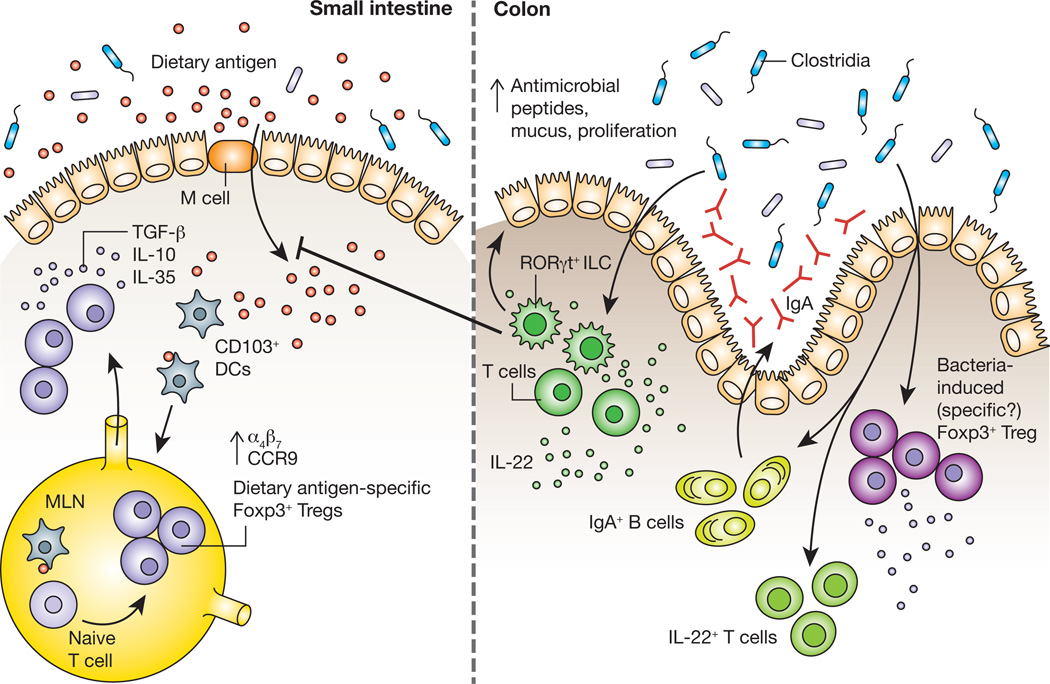
Although implicated in aberrant allergic responses to food, the factors regulating epithelial barrier function remain unclear [67]. To gain insight into the mechanisms by which commensal bacteria influence epithelial permeability to dietary antigens we isolated IECs from mice colonized with Clostridia, which conferred maximal protection against peanut allergen sensitization in our model, and from mice colonized with a representative member of the Bacteroidetes, B. uniformis, which provided no protection against sensitization [41]. Because our microarray analysis showed that Clostridia colonization differentially upregulates the expression of RegIIIβ and RegIIIγ in isolated IECs, we examined whether it also regulated IL-22 production in the colonic LP. We found that Clostridia colonization induces the production of IL-22 by both RORγt+ ILCs and CD4+ T cells in the colonic LP. Since antigen uptake from the intestinal lumen is the first step in sensitization to a food allergen, we reasoned that Clostridia-induced IL-22 production in the intestinal LP reinforces the epithelial barrier to reduce permeability to dietary proteins. Ara h 2 and Ara h 6, the immunodominant allergens of peanut (Arachis hypogaea), are resistant to proteolytic degradation in vitro; their passage into the bloodstream with secondary protein structure intact may potentiate their allergenicity [68]. We therefore utilized sensitive capture ELISAs to detect the transient presence of these allergens in the serum within hours of oral gavage. We found that both Clostridia colonization and treatment with an IL-22 Fc fusion protein decreases intestinal permeability to Ara h 2/6. Treatment of Clostridia colonized mice with a neutralizing antibody to IL-22 demonstrated that the effects of Clostridia on epithelial permeability to Ara h 2/6 are attributable to the actions of this cytokine. IL-22 from RORγt+ ILCs is primarily responsible for this effect, since Rag1−/− mice colonized with Clostridia maintain barrier function while Rag1−/− mice treated with an anti-CD90.2 monoclonal antibody to deplete ILCs exhibit increased permeability to Ara h 2/6 [41]. In addition to increasing the expression of antimicrobial peptides, IL-22 is known to promote IEC proliferation [69, 70] and mucus secretion by goblet cells [71]. Given these general effects on barrier integrity, it is likely that IL-22 blocks allergen uptake in an antigen non-specific manner. Our data therefore suggest that Clostridia protect against food allergen sensitization, in part, by regulating the access of dietary antigen to the systemic circulation. In our working model (Figure 1), colonization with Clostridia induces an early, innate, barrier protective response mediated by IL-22 producing RORγt+ ILCs, followed by an adaptive immune response involving IL-22+CD4+ T cells, Foxp3+CD4+ T cells and IgA secreting B cells that reinforces the immunoregulatory environment needed to mediate tolerance to dietary antigens.
Figure 1.
The process of maintaining non-responsiveness to food begins when dietary antigen crosses the intestinal epithelium. CD103+ DCs that have acquired antigen subsequently migrate to the mesenteric lymph node (MLN), where they encounter naïve T cells. Within the tolerogenic environment of the MLN, T cells differentiate into dietary antigen-specific Foxp3+ Tregs and upregulate expression of the gut-homing receptors α4β7 and CCR9, which drive their migration back to the intestinal lamina propria. Dietary antigen-specific Foxp3+ Tregs can then suppress aberrant responses to dietary antigens through the production of the cytokines TGF-β, IL-10 and IL-35. Work from our laboratory demonstrates that signals derived from Clostridia, a class of anaerobic spore-forming Firmicutes that resides in close proximity to the colonic epithelium are also required to prevent sensitization to food allergens. Studies in gnotobiotic mice showed that Clostridia colonization induces an early barrier protective response mediated by a surge in IL-22 production from RORγt+ innate lymphoid cells (ILCs). IL-22 acts on intestinal epithelial cells to promote the production of antimicrobial peptides and mucus and reduce uptake of dietary antigen into the systemic circulation. This initial barrier protective response is then reinforced by a bacteria-induced adaptive immune response that includes IL-22+ CD4+ T cells, an expansion of Foxp3+ Tregs in the colonic lamina propria and increased secretion of IgA into the intestinal lumen.
A potential role for short-chain fatty acids
Microbial metabolites are increasingly appreciated as potent regulators of immunity. Indeed, the gut is not only exposed to the trillions of bacteria colonizing its surface but also to a myriad of bacteria-derived compounds produced through the fermentation of dietary substrates [72]. In particular, short-chain fatty acids (SCFAs) have received much attention for their immunoregulatory capacity. SCFAs are a subset of fatty acids produced during the bacterial fermentation of indigestible dietary fiber. SCFA concentrations are highest in the proximal colon, where they can be used locally as an energy source for enterocytes or be absorbed across the epithelium and into the bloodstream [73]. Acetate, propionate and butyrate (acetic, propionic and butyric acids, respectively) are the major SCFAs produced in the mammalian gut [73]. Recent work has revealed an important role for SCFAs in promoting the differentiation of colonic Foxp3+ Tregs. Administration of acetic, propionic and butyric acids in the drinking water, either in combination or individually, increases the frequency of Foxp3+ Tregs among CD4+ T cells in the colonic LP, but not the spleen, MLN or thymus of GF mice [74]. More recently, two groups have found butyrate to be the most potent inducer of colonic Foxp3+ Tregs. Furusawa et al reported that a diet rich in butyrylated high-amylose maize starches increases the proportion of IL-10-producing colonic Foxp3+ Tregs in specific pathogen-free (SPF) mice to a greater extent than diets containing propionylated and acetylated starches [75]. In line with these results, Arpaia et al showed that administration of butyrate by diet or by enema leads to an increase in colonic Foxp3+ Tregs in SPF mice [76]. This increase was dependent on CNS1, an intronic enhancer required for the extrathymic conversion of Tregs, demonstrating that butyrate specifically promotes Treg development by the extrathymic induced pathway [76]. In light of these findings, we are exploring the possibility that the ability of a Clostridia-containing microbiota to protect against food allergen sensitization is mediated, at least in part, through SCFA activity. Of note, Clostridium species are major producers of SCFAs [73] and GF mice colonized with a mixed population of Clostridia have higher levels of cecal acetate and butyrate than GF controls [75] (unpublished data).
SCFAs mediate their effects through two known mechanisms: (1) direct inhibition of histone deacetylases (HDACs) to influence gene expression and (2) signaling through the G-protein coupled receptors (GPCR) GPR41, GPR43, and GPR109A [73]. To date, studies on the mechanism by which SCFAs induce the differentiation of Foxp3+ Tregs have focused on the former function, with both Furusawa et al and Arpaia et al demonstrating that butyrate treatment enhances the acetylation of histone H3 at the Foxp3 promoter [75, 76]. Whether or not GPCR signaling is also required is less clear. Smith et al reported that GPR43 signaling is needed for propionate induced expansion of Foxp3+ Tregs [74]. With regard to butyrate, one study has implicated a role for signaling via GPR109A, as treatment of WT but not Gpr109a−/− CD11b+ macrophages and CD11c+ DCs with butyrate or niacin (another ligand of GPR109A) enhances their ability to promote the differentiation of Tregs [77]. Notably, the authors found that butyrate and niacin increased the expression of Aldh1a1 in cultured APCs in a Gpr109a-dependent manner, implying that butyrate signaling via GPR109A induces Treg differentiation by promoting RA production from APCs [77]. However, results from Arpaia et al disagree with the requirement for GPR109A, as they demonstrate that pre-treatment of WT and Gpr109a−/− DCs with butyrate induces Tregs to a similar extent [76]. How signaling via these three identified GPCRs cooperate to mediate SCFA function in various contexts will need to be further addressed.
An interesting question arising from these studies is whether the colonic Foxp3+ Tregs induced by SCFAs are dietary antigen-specific and/or bacteria-specific, and how their expansion may influence oral tolerance in the small intestine. It is important to highlight that the studies above have focused on the expansion of Foxp3+ Tregs specifically in the colon. Interestingly, Furusawa et al have demonstrated that a butyrate-containing diet promotes the differentiation of colonic OVA-specific OT-II Foxp3+ T cells following the adoptive transfer of naïve OTII CD4+ T cells and oral administration of OVA [75]. The same was shown by Singh et al with niacin via GPR109A signaling, where administration of niacin to antibiotic-treated WT mice led to a greater increase in the percentage of OT-II Foxp3+ T cells in the colon than seen in antibiotic-treated Gpr109a−/− mice [77]. These results raise the possibility that SCFAs promote oral tolerance by facilitating the conversion of dietary antigen-specific Foxp3+ Tregs. Indeed, the finding that butyrate increases Aldh1a1 expression in gut APCs suggests that this is the case, as RA is a known prerequisite for the differentiation of dietary antigen-specific Foxp3+ Tregs. Whether or not a similar expansion occurs in the small intestine, or how it is that these colonic dietary antigen-specific Tregs influence immune responses to antigen in the small intestine, warrants exploration and is further discussed below. Additionally, it is likely that SCFAs also promote the differentiation of bacteria-specific Foxp3+ Tregs. In support of this, a butyrate-containing diet failed to increase colonic Treg frequencies in GF mice [75], implying that Tregs may be induced by SCFAs following the recognition of bacterial antigens.
Connecting bacteria-derived signals in the colon to non-responsiveness to dietary antigen in the small intestine
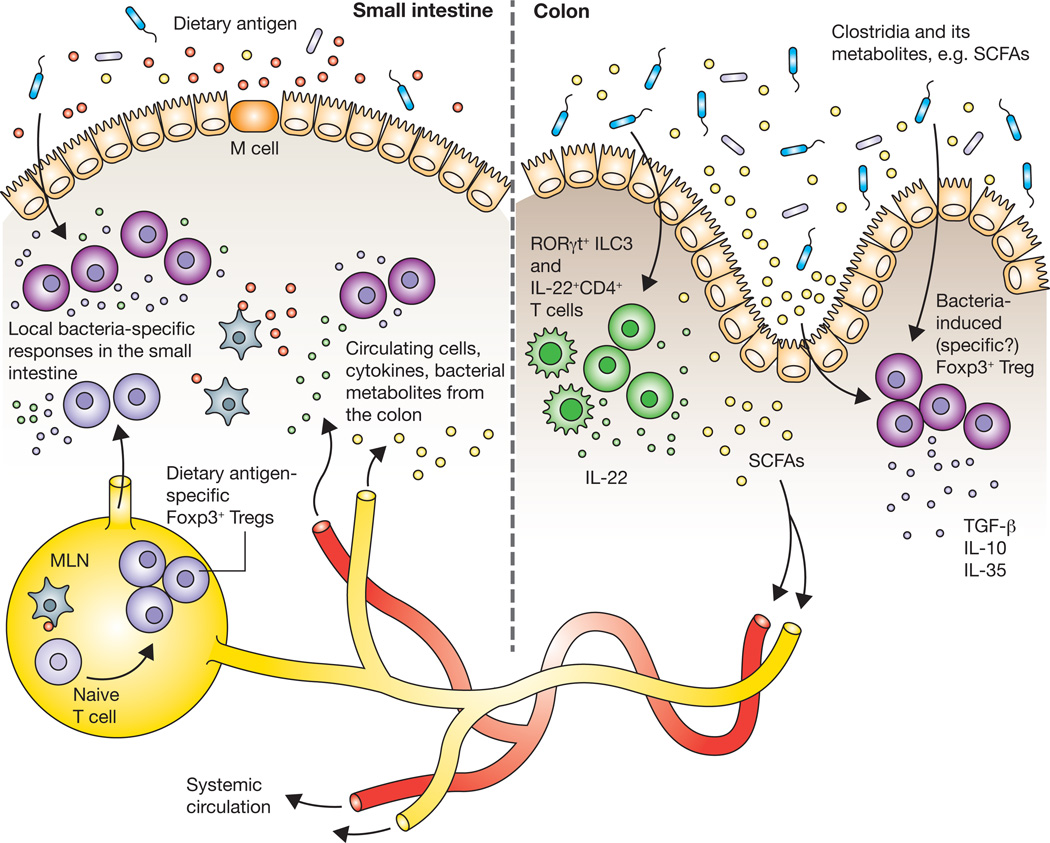
How signals derived from commensal bacteria in the colon shape immune responses to dietary antigens in the small intestine remains an open question (Figure 2). The ability of commensal bacteria to influence immune responses at sites far beyond the colon has been demonstrated in several disease models including diabetes [78], arthritis [79], experimental autoimmune encephalomyelitis [80, 81], and allergic airway inflammation [82]. One mechanism by which commensal bacteria may exert far-reaching effects is through lymphocyte migration. As mentioned earlier, it is accepted that naïve CD4+ T cells, upon activation in the MLN and PP, are imprinted with homing receptors that drive them towards the small intestinal or colonic LP. While homing to the small intestine depends on the cell surface molecules α4β7 and CCR9 [83], homing to the colon relies on α4β7 and signaling via GPR15 [84]. These distinct homing requirements imply that immune responses are highly compartmentalized within the GALT. However, this compartmentalization may not be completely rigid. Lymphocytes from the colon could transiently migrate to the small intestine through the connected network of lymphatics in the GALT [85] or through the systemic circulation [86]. In support of the latter possibility, Wu et al found that α4β7+Th17 cells induced by segmented filamentous bacteria in the small intestine were present in the spleens of arthritic K/BxN mice and contributed to exacerbated arthritis [79]. How it is that effector T cells already imprinted to home to a specific location can then populate other sites of the body was not addressed.
Figure 2.
How do immune responses to bacteria in the colon influence the induction of tolerance to dietary antigens absorbed in the small intestine? Immune cells, such as Foxp3+ Tregs, induced by commensal Clostridia and/or their metabolites, may reach the small intestine by migrating via the connected network of lymphatics in the GALT or through the systemic circulation. Cytokines produced by bacteria-induced immune cells in the colon could similarly circulate and reach the small intestine. Short chain fatty acids (SCFAs) can enter the bloodstream and modulate immune responses at distal sites. Aside from the migration of cells and molecules, Clostridia present in the small intestine itself may induce local immune responses that contribute to non-responsiveness to dietary antigen.
It is also possible that cytokines produced by bacteria-induced lymphocyte populations circulate to other locations where they may direct immune responses. We found that Clostridia promote the production of IL-22 by CD4+ T cells and RORγt+ ILCs in the colon and regulate allergen uptake in the small intestine. Much lower numbers of Clostridia colonize the ileum of both mice [41] and humans [3], although in our hands, induction of IL-22 is undetectable in this site. It may be that IL-22 circulates through the bloodstream or lymph to reach the small intestine, enriching the concentration of this cytokine to strengthen the epithelial barrier against allergen uptake. Colonic Foxp3+ Tregs may also exert their effects in a similar fashion, releasing anti-inflammatory cytokines that reach the small intestine to promote a tolerogenic environment (Figure 2).
Another possibility is that bacteria-derived metabolites travel outside the colon via the bloodstream and exert effects at distal sites. Of note, SCFAs are known to circulate in the blood and have been detected in the portal, hepatic and peripheral veins [87]. Recently, circulating SCFAs have been shown to protect against allergic airway inflammation. Mice fed a high-fiber diet had increased levels of SCFAs in both the cecum and the blood and were protected against inflammation following intranasal administration of house dust mite extract [88]. Thus, to promote tolerance to dietary antigens, SCFAs produced by bacteria in the colon could feedback through the bloodstream to induce Foxp3+ Treg differentiation in the small intestine. Of note, Trompette et al found that SCFA-mediated protection against allergic airway inflammation was not due to Foxp3+ Tregs, as populations of Foxp3+ Tregs did not expand in the lung draining lymph nodes [88]. Instead, they propose that circulating SCFAs influence DC hematopoiesis, populating the lungs with DCs that have an altered activation state and an impaired ability to promote Th2 cells. While the effects of SCFAs on hematopoiesis may also influence non-responsiveness to dietary antigen, it is likely that circulating SCFAs shape immune responses in the lung and gut environments via different mechanisms.
Beyond the movement of cells, cytokines, and metabolites from the colon, it is important not to overlook the immunoregulatory potential of commensal bacteria in the small intestine itself. Though present at much lower microbial densities, bacteria in the small intestine may regulate tolerogenic immune responses that influence oral tolerance.
Implications for the development of novel immunotherapeutics to prevent or treat food allergy
The rising prevalence of food allergies in Western nations has become a public health crisis that requires immediate attention. The urgency of the problem is exacerbated by the fact that no effective treatment for food allergies is currently available. 90% of food allergies are attributable to only eight major types of food: milk, eggs, shellfish (particularly crustaceans), fish, peanuts, soybeans, tree nuts and wheat [44]. Clinically, allergic disease follows a characteristic natural history, often referred to as the “allergic march”, which appears first as atopic dermatitis in infancy and is then followed by food allergy, allergic rhinitis and asthma in later years [89]. Atopic children typically exhibit transient allergic responses to milk, soybeans, eggs and wheat. Allergic responses to peanuts, tree nuts and shellfish, however, are likely to result in life-long anaphylactic hypersensitivity. Most of the immunodominant allergens in each of these foods have been characterized but their biochemistry has not yet yielded clues that explain their potent allergenicity [44].
Our rapidly growing understanding of the role of our gut microbiota in allergic disease opens up an exciting avenue of research into therapeutic interventions that can harness the immunoregulatory potential of host-commensal interactions. There is already an extensive literature documenting the efficacy of live, health promoting microorganisms (probiotics) as a treatment for allergic disease [90]. Much of this work has focused on readily culturable Lactobacillus and Bifidobacterium species that predominate in the infant gut [91]. Treatment seems to be most efficacious when administered to pregnant mothers both pre- and post-natally during their children’s infancy [92–98]. Indeed, infancy marks a period of rapid development for the gut microbiota, when its composition is inherently unstable and likely to be receptive to therapeutic manipulation [99].
Lactobacillus rhamnosus GG (LGG) is among the probiotic formulations most often associated with clinical efficacy [94, 95, 98]. Berni Canani et al have recently demonstrated that dietary management with an extensively hydrolyzed casein formula (EHCF) containing LGG results in a higher rate of tolerance acquisition in infants with cow’s milk allergy (CMA) than treatment with EHCF alone or with other non-milk based formulas [100]. We hypothesized that this effect is attributable, in part, to an influence of this dietary intervention on the composition of the gut microbiota and collaborated with this group to test this hypothesis [101]. We found that, before treatment, the gut microbiota of allergic infants exhibited an accelerated ecological succession to a community structure more typical of adults. When we examined the influence of LGG treatment on the composition of the gut microbiota we found that it does not result in an increased abundance of lactobacilli detectable in the feces of the treated infants. Instead, treatment with EHCF plus LGG, but not a rice protein hydrolyzed formula (RHF), is associated with changes in microbial community structure that include the expansion of butyrate-producing Clostridia. CMA infants treated with EHCF plus LGG, but not RHF, had significantly higher levels of butyrate detectable in their feces and an accelerated acquisition of tolerance to cow’s milk [101].
These findings are particularly relevant in light of the recent advances in our understanding, discussed above, of how specific bacterial species and their metabolites influence food allergen sensitization. The discovery that butyrate-producing Clostridia become more prevalent in this cohort of treated infants corroborates the findings in our mouse model that a Clostridia-containing microbiota confers protection against food allergen sensitization. Oral desensitization protocols are already showing some efficacy for the treatment of food allergy in clinical trials [102]. Our data suggest that strategies that pair Clostridia enrichment of the gut microbiota with these tolerance-inducing protocols may potentiate antigen-specific tolerance to prevent or treat food allergy.
In this regard, our mouse model provides a valuable pre-clinical foundation for elucidating more effective strategies to restore or enhance the immunoregulatory capacity of commensal bacteria. For instance, further research into how Clostridia initiate protective immune responses, be it through metabolites such as SCFAs or through direct signaling to receptors on host cells, will accelerate the development of novel forms of probiotic intervention. The relevance of SCFAs in intestinal health may inform strategies to modulate fiber intake as a means of promoting tolerance to dietary antigens. Indeed, butyrate-producing Clostridia species are known to be particularly sensitive to dietary manipulation, increasing in abundance with increased fiber intake [103]. In addition, the development of agonists that mimic Clostridia’s interaction with its host could help to maintain immunoregulatory signals when the composition of the microbiota is disturbed. Overall, a deeper understanding of the role of the gut microbiota in regulating responses to food holds much promise for new immunotherapeutics that can combat the precipitous rise in the prevalence of food allergies.
Acknowledgments
Funding for this work was provided by Food Allergy Research and Education (FARE) and by the NIH (NIAID). We thank Andrew Stefka for critical review of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pabst O, Mowat AM. Oral tolerance to food protein. Mucosal Immunol. 2012;5:232–239. doi: 10.1038/mi.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu J, Gordon JI. Honor thy symbionts. Proc Natl Acad Sci U S A. 2003;100:10452–10459. doi: 10.1073/pnas.1734063100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walter J, Ley R. The human gut microbiome: ecology and recent evolutionary changes. Annu Rev Microbiol. 2011;65:411–429. doi: 10.1146/annurev-micro-090110-102830. [DOI] [PubMed] [Google Scholar]
- 4.Feehley T, Stefka AT, Cao S, Nagler CR. Microbial regulation of allergic responses to food. Semin Immunopathol. 2012;34:671–688. doi: 10.1007/s00281-012-0337-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mabbott NA, Donaldson DS, Ohno H, Williams IR, Mahajan A. Microfold (M) cells: important immunosurveillance posts in the intestinal epithelium. Mucosal Immunol. 2013;6:666–677. doi: 10.1038/mi.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neutra MR, Mantis NJ, Frey A, Giannasca PJ. The composition and function of M cell apical membranes: implications for microbial pathogenesis. Semin Immunol. 1999;11:171–181. doi: 10.1006/smim.1999.0173. [DOI] [PubMed] [Google Scholar]
- 7.Mowat AM. Anatomical basis of tolerance and immunity to intestinal antigens. Nat Rev Immunol. 2003;3:331–341. doi: 10.1038/nri1057. [DOI] [PubMed] [Google Scholar]
- 8.Lelouard H, Fallet M, de Bovis B, Meresse S, Gorvel JP. Peyer's patch dendritic cells sample antigens by extending dendrites through M cell-specific transcellular pores. Gastroenterology. 2012;142:592–601. doi: 10.1053/j.gastro.2011.11.039. [DOI] [PubMed] [Google Scholar]
- 9.McDole JR, Wheeler LW, McDonald KG, Wang B, Konjufca V, Knoop KA, Newberry RD, Miller MJ. Goblet cells deliver luminal antigen to CD103+ dendritic cells in the small intestine. Nature. 2012;483:345–349. doi: 10.1038/nature10863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. 2009;9:799–809. doi: 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- 11.Niess JH, Brand S, Gu X, Landsman L, Jung S, McCormick BA, Vyas JM, Boes M, Ploegh HL, Fox JG, Littman DR, Reinecker HC. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science. 2005;307:254–258. doi: 10.1126/science.1102901. [DOI] [PubMed] [Google Scholar]
- 12.Rescigno M, Urbano M, Valzasina B, Francolini M, Rotta G, Bonasio R, Granucci F, Kraehenbuhl JP, Ricciardi-Castagnoli P. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol. 2001;2:361–367. doi: 10.1038/86373. [DOI] [PubMed] [Google Scholar]
- 13.Chieppa M, Rescigno M, Huang AY, Germain RN. Dynamic imaging of dendritic cell extension into the small bowel lumen in response to epithelial cell TLR engagement. J Exp Med. 2006;203:2841–2852. doi: 10.1084/jem.20061884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bogunovic M, Ginhoux F, Helft J, Shang L, Hashimoto D, Greter M, Liu K, Jakubzick C, Ingersoll MA, Leboeuf M, Stanley ER, Nussenzweig M, Lira SA, Randolph GJ, Merad M. Origin of the Lamina Propria Dendritic Cell Network. Immunity. 2009;31:513–525. doi: 10.1016/j.immuni.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edelson BT, Kc W, Juang R, Kohyama M, Benoit LA, Klekotka PA, Moon C, Albring JC, Ise W, Michael DG, Bhattacharya D, Stappenbeck TS, Holtzman MJ, Sung SS, Murphy TL, Hildner K, Murphy KM. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8α+ conventional dendritic cells. J Exp Med. 2010;207:823–836. doi: 10.1084/jem.20091627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Worbs T, Bode U, Yan S, Hoffmann MW, Hintzen G, Bernhardt G, Forster R, Pabst O. Oral tolerance originates in the intestinal immune system and relies on antigen carriage by dendritic cells. J Exp Med. 2006;203:519–527. doi: 10.1084/jem.20052016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jang MH, Sougawa N, Tanaka T, Hirata T, Hiroi T, Tohya K, Guo Z, Umemoto E, Ebisuno Y, Yang BG, Seoh JY, Lipp M, Kiyono H, Miyasaka M. CCR7 is critically important for migration of dendritic cells in intestinal lamina propria to mesenteric lymph nodes. J Immunol. 2006;176:803–810. doi: 10.4049/jimmunol.176.2.803. [DOI] [PubMed] [Google Scholar]
- 18.Coombes JL, Siddiqui KR, Arancibia-Carcamo CV, Hall J, Sun CM, Belkaid Y, Powrie F. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-β and retinoic acid-dependent mechanism. J Exp Med. 2007;204:1757–1764. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun CM, Hall JA, Blank RB, Bouladoux N, Oukka M, Mora JR, Belkaid Y. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J Exp Med. 2007;204:1775–1785. doi: 10.1084/jem.20070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Worthington JJ, Czajkowska BI, Melton AC, Travis MA. Intestinal dendritic cells specialize to activate transforming growth factor-b and induce Foxp3+ regulatory T cells via integrin αvβ8. Gastroenterology. 2011;141:1802–1812. doi: 10.1053/j.gastro.2011.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paidassi H, Acharya M, Zhang A, Mukhopadhyay S, Kwon M, Chow C, Stuart LM, Savill J, Lacy-Hulbert A. Preferential expression of integrin αvβ8 promotes generation of regulatory T cells by mouse CD103+ dendritic cells. Gastroenterology. 2011;141:1813–1820. doi: 10.1053/j.gastro.2011.06.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iliev ID, Mileti E, Matteoli G, Chieppa M, Rescigno M. Intestinal epithelial cells promote colitis-protective regulatory T-cell differentiation through dendritic cell conditioning. Mucosal Immunol. 2009;2:340–350. doi: 10.1038/mi.2009.13. [DOI] [PubMed] [Google Scholar]
- 23.Hall JA, Cannons JL, Grainger JR, Dos Santos LM, Hand TW, Naik S, Wohlfert EA, Chou DB, Oldenhove G, Robinson M, Grigg ME, Kastenmayer R, Schwartzberg PL, Belkaid Y. Essential role for retinoic acid in the promotion of CD4(+) T cell effector responses via retinoic acid receptor alpha. Immunity. 2011;34:435–447. doi: 10.1016/j.immuni.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hill JA, Hall JA, Sun CM, Cai Q, Ghyselinck N, Chambon P, Belkaid Y, Mathis D, Benoist C. Retinoic acid enhances Foxp3 induction indirectly by relieving inhibition from CD4+CD44hi Cells. Immunity. 2008;29:758–770. doi: 10.1016/j.immuni.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schulz O, Jaensson E, Persson EK, Liu X, Worbs T, Agace WW, Pabst O. Intestinal CD103+, but not CX3CR1+, antigen sampling cells migrate in lymph and serve classical dendritic cell functions. J Exp Med. 2009;206:3101–3114. doi: 10.1084/jem.20091925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diehl GE, Longman RS, Zhang JX, Breart B, Galan C, Cuesta A, Schwab SR, Littman DR. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX3CR1hi cells. Nature. 2013;494:116–120. doi: 10.1038/nature11809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mazzini E, Massimiliano L, Penna G, Rescigno M. Oral Tolerance Can Be Established via Gap Junction Transfer of Fed Antigens from CX3CR1(+) Macrophages to CD103(+) Dendritic Cells. Immunity. 2014;40:248–261. doi: 10.1016/j.immuni.2013.12.012. [DOI] [PubMed] [Google Scholar]
- 28.Hadis U, Wahl B, Schulz O, Hardtke-Wolenski M, Schippers A, Wagner N, Muller W, Sparwasser T, Forster R, Pabst O. Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity. 2011;34:237–246. doi: 10.1016/j.immuni.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 29.Cassani B, Villablanca EJ, Quintana FJ, Love PE, Lacy-Hulbert A, Blaner WS, Sparwasser T, Snapper SB, Weiner HL, Mora JR. Gut-Tropic T Cells That Express Integrin α4β7 and CCR9 Are Required for Induction of Oral Immune Tolerance in Mice. Gastroenterology. 2011;141:2109–2118. doi: 10.1053/j.gastro.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bollrath J, Powrie FM. Controlling the frontier: regulatory T-cells and intestinal homeostasis. Semin Immunol. 2013;25:352–357. doi: 10.1016/j.smim.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 31.Josefowicz SZ, Niec RE, Kim HY, Treuting P, Chinen T, Zheng Y, Umetsu DT, Rudensky AY. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature. 2012;482:395–399. doi: 10.1038/nature10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wills-Karp M, Finkelman FD. Untangling the complex web of IL-4- and IL-13-mediated signaling pathways. Science signaling. 2008;1:pe55. doi: 10.1126/scisignal.1.51.pe55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gould HJ, Sutton BJ. IgE in allergy and asthma today. Nat Rev Immunol. 2008;8:205–217. doi: 10.1038/nri2273. [DOI] [PubMed] [Google Scholar]
- 34.Shan M, Gentile M, Yeiser JR, Walland AC, Bornstein VU, Chen K, He B, Cassis L, Bigas A, Cols M, Comerma L, Huang B, Blander JM, Xiong H, Mayer L, Berin C, Augenlicht LH, Velcich A, Cerutti A. Mucus enhances gut homeostasis and oral tolerance by delivering immunoregulatory signals. Science. 2013;342:447–453. doi: 10.1126/science.1237910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagler-Anderson C. Man the barrier! Strategic defenses in the intestinal mucosa. Nature Reviews Immunology. 2001;1:59–67. doi: 10.1038/35095573. [DOI] [PubMed] [Google Scholar]
- 36.Pabst O. New concepts in the generation and functions of IgA. Nat Rev Immunol. 2012;12:821–832. doi: 10.1038/nri3322. [DOI] [PubMed] [Google Scholar]
- 37.Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10:159–169. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- 38.Brown EM, Sadarangani M, Finlay BB. The role of the immune system in governing host-microbe interactions in the intestine. Nat Immunol. 2013;14:660–667. doi: 10.1038/ni.2611. [DOI] [PubMed] [Google Scholar]
- 39.Maynard CL, Elson CO, Hatton RD, Weaver CT. Reciprocal interactions of the intestinal microbiota and immune system. Nature. 2012;489:231–241. doi: 10.1038/nature11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bashir ME, Louie S, Shi HN, Nagler-Anderson C. Toll-like receptor 4 signaling by intestinal microbes influences susceptibility to food allergy. J Immunol. 2004;172:6978–6987. doi: 10.4049/jimmunol.172.11.6978. [DOI] [PubMed] [Google Scholar]
- 41.Stefka AT, Feehley T, Tripathi P, Qiu J, McCoy KD, Mazmanian SK, Seo G-Y, Cao SC, Theriault BR, Antonopoulos D, Zhou L, Chang EB, Fu Y-X, Nagler C. Commensal bacteria induce barrier protective responses to prevent food allergen sensitization. 2014 doi: 10.1073/pnas.1412008111. in revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Noval Rivas M, Burton OT, Wise P, Zhang YQ, Hobson SA, Garcia Lloret M, Chehoud C, Kuczynski J, DeSantis T, Warrington J, Hyde ER, Petrosino JF, Gerber GK, Bry L, Oettgen HC, Mazmanian SK, Chatila TA. A microbiota signature associated with experimental food allergy promotes allergic sensitization and anaphylaxis. J Allergy Clin Immunol. 2013;131:201–212. doi: 10.1016/j.jaci.2012.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodriguez B, Prioult G, Hacini-Rachinel F, Moine D, Bruttin A, Ngom-Bru C, Labellie C, Nicolis I, Berger B, Mercenier A, Butel MJ, Waligora-Dupriet AJ. Infant gut microbiota is protective against cow's milk allergy in mice despite immature ileal T-cell response. FEMS microbiology ecology. 2012;79:192–202. doi: 10.1111/j.1574-6941.2011.01207.x. [DOI] [PubMed] [Google Scholar]
- 44.Wang J, Sampson HA. Food allergy. J Clin Invest. 2011;121:827–835. doi: 10.1172/JCI45434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Branum AM, Lukacs SL. Food allergy among children in the United States. Pediatrics. 2009;124:1549–1555. doi: 10.1542/peds.2009-1210. [DOI] [PubMed] [Google Scholar]
- 46.Gupta R, Sheikh A, Strachan DP, Anderson HR. Time trends in allergic disorders in the UK. Thorax. 2007;62:91–96. doi: 10.1136/thx.2004.038844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Osborne NJ, Koplin JJ, Martin PE, Gurrin LC, Lowe AJ, Matheson MC, Ponsonby AL, Wake M, Tang ML, Dharmage SC, Allen KJ. Prevalence of challenge-proven IgE-mediated food allergy using population-based sampling and predetermined challenge criteria in infants. J Allergy Clin Immunol. 2011;127:668–676. doi: 10.1016/j.jaci.2011.01.039. [DOI] [PubMed] [Google Scholar]
- 48.Marra F, Lynd L, Coombes M, Richardson K, Legal M, Fitzgerald JM, Marra CA. Does antibiotic exposure during infancy lead to development of asthma?: a systematic review and metaanalysis. Chest. 2006;129:610–618. doi: 10.1378/chest.129.3.610. [DOI] [PubMed] [Google Scholar]
- 49.Blaser M. Antibiotic overuse: Stop the killing of beneficial bacteria. Nature. 2011;476:393–394. doi: 10.1038/476393a. [DOI] [PubMed] [Google Scholar]
- 50.Savage JH, Matsui EC, Wood RA, Keet CA. Urinary levels of triclosan and parabens are associated with aeroallergen and food sensitization. J Allergy Clin Immunol. 2012;130:453–460. doi: 10.1016/j.jaci.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Metsala J, Lundqvist A, Virta LJ, Kaila M, Gissler M, Virtanen SM. Mother's and offspring's use of antibiotics and infant allergy to cow's milk. Epidemiology. 2013;24:303–309. doi: 10.1097/EDE.0b013e31827f520f. [DOI] [PubMed] [Google Scholar]
- 52.Koplin J, Allen K, Gurrin L, Osborne N, Tang ML, Dharmage S. Is caesarean delivery associated with sensitization to food allergens and IgE-mediated food allergy: a systematic review. Pediatr Allergy Immunol. 2008;19:682–687. doi: 10.1111/j.1399-3038.2008.00731.x. [DOI] [PubMed] [Google Scholar]
- 53.Bjorksten B, Naaber P, Sepp E, Mikelsaar M. The intestinal microflora in allergic Estonian and Swedish 2-year-old children. Clin Exp Allergy. 1999;29:342–346. doi: 10.1046/j.1365-2222.1999.00560.x. [DOI] [PubMed] [Google Scholar]
- 54.Bjorksten B, Sepp E, Julge K, Voor T, Mikelsaar M. Allergy development and the intestinal microflora during the first year of life. J Allergy Clin Immunol. 2001;108:516–520. doi: 10.1067/mai.2001.118130. [DOI] [PubMed] [Google Scholar]
- 55.Thompson-Chagoyan OC, Fallani M, Maldonado J, Vieites JM, Khanna S, Edwards C, Dore J, Gil A. Faecal microbiota and short-chain fatty acid levels in faeces from infants with cow's milk protein allergy. Int Arch Allergy Immunol. 2011;156:325–332. doi: 10.1159/000323893. [DOI] [PubMed] [Google Scholar]
- 56.Thompson-Chagoyan OC, Vieites JM, Maldonado J, Edwards C, Gil A. Changes in faecal microbiota of infants with cow's milk protein allergy--a Spanish prospective case-control 6-month follow-up study. Pediatr Allergy Immunol. 2010;21:e394–e400. doi: 10.1111/j.1399-3038.2009.00961.x. [DOI] [PubMed] [Google Scholar]
- 57.Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, Santacruz N, Peterson DA, Stappenbeck TS, Hsieh CS. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250–254. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, Kim S, Fritz JV, Wilmes P, Ueha S, Matsushima K, Ohno H, Olle B, Sakaguchi S, Taniguchi T, Morita H, Hattori M, Honda K. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 59.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, Taniguchi T, Takeda K, Hori S, Ivanov II, Umesaki Y, Itoh K, Honda K. Induction of Colonic Regulatory T Cells by Indigenous Clostridium Species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sonnenberg GF, Fouser LA, Artis D. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat Immunol. 2011;12:383–390. doi: 10.1038/ni.2025. [DOI] [PubMed] [Google Scholar]
- 62.Sabat R, Ouyang W, Wolk K. Therapeutic opportunities of the IL-22-IL-22R1 system. Nature reviews Drug discovery. 2014;13:21–38. doi: 10.1038/nrd4176. [DOI] [PubMed] [Google Scholar]
- 63.Sonnenberg GF, Monticelli LA, Alenghat T, Fung TC, Hutnick NA, Kunisawa J, Shibata N, Grunberg S, Sinha R, Zahm AM, Tardif MR, Sathaliyawala T, Kubota M, Farber DL, Collman RG, Shaked A, Fouser LA, Weiner DB, Tessier PA, Friedman JR, Kiyono H, Bushman FD, Chang KM, Artis D. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science. 2012;336:1321–1325. doi: 10.1126/science.1222551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lochner M, Ohnmacht C, Presley L, Bruhns P, Si-Tahar M, Sawa S, Eberl G. Microbiota-induced tertiary lymphoid tissues aggravate inflammatory disease in the absence of RORgamma t and LTi cells. J Exp Med. 2011;208:125–134. doi: 10.1084/jem.20100052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, Abbas AR, Modrusan Z, Ghilardi N, de Sauvage FJ, Ouyang W. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 66.Satoh-Takayama N, Vosshenrich CA, Lesjean-Pottier S, Sawa S, Lochner M, Rattis F, Mention JJ, Thiam K, Cerf-Bensussan N, Mandelboim O, Eberl G, Di Santo JP. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 2008;29:958–970. doi: 10.1016/j.immuni.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 67.Menard S, Cerf-Bensussan N, Heyman M. Multiple facets of intestinal permeability and epithelial handling of dietary antigens. Mucosal Immunol. 2010;3:247–259. doi: 10.1038/mi.2010.5. [DOI] [PubMed] [Google Scholar]
- 68.Lehmann K, Schweimer K, Reese G, Randow S, Suhr M, Becker WM, Vieths S, Rosch P. Structure and stability of 2S albumin-type peanut allergens: implications for the severity of peanut allergic reactions. Biochem J. 2006;395:463–472. doi: 10.1042/BJ20051728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kirchberger S, Royston DJ, Boulard O, Thornton E, Franchini F, Szabady RL, Harrison O, Powrie F. Innate lymphoid cells sustain colon cancer through production of interleukin-22 in a mouse model. J Exp Med. 2013;210:917–931. doi: 10.1084/jem.20122308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pickert G, Neufert C, Leppkes M, Zheng Y, Wittkopf N, Warntjen M, Lehr HA, Hirth S, Weigmann B, Wirtz S, Ouyang W, Neurath MF, Becker C. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J Exp Med. 2009;206:1465–1472. doi: 10.1084/jem.20082683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sugimoto K, Ogawa A, Mizoguchi E, Shimomura Y, Andoh A, Bhan AK, Blumberg RS, Xavier RJ, Mizoguchi A. IL-22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis. J Clin Invest. 2008;118:534–544. doi: 10.1172/JCI33194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nyangale EP, Mottram DS, Gibson GR. Gut microbial activity, implications for health and disease: the potential role of metabolite analysis. J Proteome Res. 2012;11:5573–5585. doi: 10.1021/pr300637d. [DOI] [PubMed] [Google Scholar]
- 73.Tan J, McKenzie C, Potamitis M, Thorburn AN, Mackay CR, Macia L. The role of short-chain fatty acids in health and disease. Adv Immunol. 2014;121:91–119. doi: 10.1016/B978-0-12-800100-4.00003-9. [DOI] [PubMed] [Google Scholar]
- 74.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, Takahashi M, Fukuda NN, Murukami S, Miyauchi E, Hino S, Atarashi K, Onawa S, Fujimura Y, Lockett T, Clarke JM, Topping DL, Tomita M, Hori S, Ohara O, Morita T, Koseki H, Kikuchi J, Honda K, Hase K, Ohno H. Commensal microbe-derived butyrate induces differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 76.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, Rudensky AS. Metabolites produced by commensal bacteria promote peripheral regulatory T cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, Thangaraju M, Prasad PD, Manicassamy S, Munn DH, Lee JR, Offermanns S, Ganapathy V. Activation of gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 2014;40:128–139. doi: 10.1016/j.immuni.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, Hu C, Wong FS, Szot GL, Bluestone JA, Gordon JI, Chervonsky AV. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Berer K, Mues M, Koutrolos M, Rasbi ZA, Boziki M, Johner C, Wekerle H, Krishnamoorthy G. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature. 2011;479:538–541. doi: 10.1038/nature10554. [DOI] [PubMed] [Google Scholar]
- 81.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hill DA, Siracusa MC, Abt MC, Kim BS, Kobuley D, Kubo M, Kambayashi T, Larosa DF, Renner ED, Orange JS, Bushman FD, Artis D. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nat Med. 2012;18:538–546. doi: 10.1038/nm.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Agace WW. T-cell recruitment to the intestinal mucosa. Trends Immunol. 2008;29:514–522. doi: 10.1016/j.it.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 84.Kim SV, Xiang WV, Kwak C, Yang Y, Lin XW, Ota M, Sarpel U, Rifkin DB, Xu R, Littman DR. GPR15-mediated homing controls immune homeostasis in the large intestine mucosa. Science. 2013;340:1456–1459. doi: 10.1126/science.1237013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pabst O. Trafficking of regulatory T cells in the intestinal immune system. Int Immunol. 2013;25:139–143. doi: 10.1093/intimm/dxs113. [DOI] [PubMed] [Google Scholar]
- 86.Rothkotter HJ, Pabst R, Bailey M. Lymphocyte migration in the intestinal mucosa: entry, transit and emigration of lymphoid cells and the influence of antigen. Vet Immunol Immunopathol. 1999;72:157–165. doi: 10.1016/s0165-2427(99)00128-2. [DOI] [PubMed] [Google Scholar]
- 87.Cummings JH, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28:1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, Blanchard C, Junt T, Nicod LP, Harris NL, Marsland BJ. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 2014;20:159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- 89.Spergel JM. From atopic dermatitis to asthma: the atopic march. Ann Allergy Asthma Immunol. 2010;105:99–106. doi: 10.1016/j.anai.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 90.Yan F, Polk DB. Probiotics and immune health. Curr Opin Gastroenterol. 2011;27:496–501. doi: 10.1097/MOG.0b013e32834baa4d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wickens K, Black PN, Stanley TV, Mitchell E, Fitzharris P, Tannock GW, Purdie G, Crane J. A differential effect of 2 probiotics in the prevention of eczema and atopy: a double-blind, randomized, placebo-controlled trial. J Allergy Clin Immunol. 2008;122:788–794. doi: 10.1016/j.jaci.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 93.Kalliomaki M, Kirjavainen P, Eerola E, Kero P, Salminen S, Isolauri E. Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. J Allergy Clin Immunol. 2001;107:129–134. doi: 10.1067/mai.2001.111237. [DOI] [PubMed] [Google Scholar]
- 94.Kalliomaki M, Isolauri E. Role of intestinal flora in the development of allergy. Curr Opin Allergy Clin Immunol. 2003;3:15–20. doi: 10.1097/00130832-200302000-00003. [DOI] [PubMed] [Google Scholar]
- 95.Rautava S, Kalliomaki M, Isolauri E. Probiotics during pregnancy and breast-feeding might confer immunomodulatory protection against atopic disease in the infant. J Allergy Clin Immunol. 2002;109:119–121. doi: 10.1067/mai.2002.120273. [DOI] [PubMed] [Google Scholar]
- 96.Niers L, Martin R, Rijkers G, Sengers F, Timmerman H, van Uden N, Smidt H, Kimpen J, Hoekstra M. The effects of selected probiotic strains on the development of eczema (the PandA study) Allergy. 2009;64:1349–1358. doi: 10.1111/j.1398-9995.2009.02021.x. [DOI] [PubMed] [Google Scholar]
- 97.Lee J, Seto D, Bielory L. Meta-analysis of clinical trials of probiotics for prevention and treatment of pediatric atopic dermatitis. J Allergy Clin Immunol. 2008;121:116–121. doi: 10.1016/j.jaci.2007.10.043. [DOI] [PubMed] [Google Scholar]
- 98.Kukkonen K, Savilahti E, Haahtela T, Juntunen-Backman K, Korpela R, Poussa T, Tuure T, Kuitunen M. Probiotics and prebiotic galactooligosaccharides in the prevention of allergic diseases: a randomized, doubleblind, placebo-controlled trial. J Allergy Clin Immunol. 2007;119:192–198. doi: 10.1016/j.jaci.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 99.Dominguez-Bello MG, Blaser MJ, Ley RE, Knight R. Development of the human gastrointestinal microbiota and insights from highthroughput sequencing. Gastroenterology. 2011;140:1713–1719. doi: 10.1053/j.gastro.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Berni Canani R, Nocerino R, Terrin G, Frediani T, Lucarelli S, Cosenza L, Passariello A, Leone L, Granata V, Di Costanzo M, Pezella V, Troncone R. Formula selection for managment of children with cow milk allergy influences the rate of acquisition of tolerance: a prospective multicenter study. The Journal of Pediatrics. 2013;163:771–777. doi: 10.1016/j.jpeds.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 101.Berni Canani R, Stefka AT, Nocerino R, Patton TJ, Aitoro R, Paparo L, Calignano A, Meli R, Mattace Raso G, Simeoli R, DiCostanzo M, Guandalini S, Antonopoulos D, Nagler C. Extensively hydroylyzed casein formula containing Lactobacillus rhamnosus GG expands gut immunoregulatory bacteria in infants with cow's milk allergy. 2014 in review. [Google Scholar]
- 102.Begin P, Dominguez T, Wilson SP, Bacal L, Mehrotra A, Kausch B, Trela A, Tavassoli M, Hoyte E, O'Riordan G, Blakemore A, Seki S, Hamilton RG, Nadeau KC. Phase 1 results of safety and tolerability in a rush oral immunotherapy protocol to multiple foods using Omalizumab. Allergy, asthma, and clinical immunology. 2014;10:7. doi: 10.1186/1710-1492-10-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett. 2009;294:1–8. doi: 10.1111/j.1574-6968.2009.01514.x. [DOI] [PubMed] [Google Scholar]