Abstract
A pool of murine cytomegalovirus (MCMV) mutants was previously generated by using a Tn3-based transposon mutagenesis approach (X. Zhan, M. Lee, J. Xiao, and F. Liu, J. Virol. 74:7411-7421, 2000). In this study, one of the MCMV mutants, Rvm155, which contained the transposon insertion in open reading frame m155, was characterized in vitro for its replication in tissue culture and in vivo for its growth and virulence in immunodeficient SCID mice. Compared to the wild-type strain and a rescued virus that restored the m155 region, the mutant is significantly deficient in growth in many organs of the infected animals. At 21 days postinfection the titers of Rvm155 in the salivary glands, lungs, spleens, livers, and kidneys of the intraperitoneally infected SCID mice were lower than the titers of the wild-type virus and the rescued virus by 50-, 1,000-, 500-, 100-, and 500-fold, respectively. Moreover, the viral mutant was attenuated in killing the SCID mice, as none of the SCID mice that were intraperitoneally infected with Rvm155 died until 38 days postinfection while all the animals infected with the wild-type and rescued viruses died at 27 days postinfection. Our results provide the first direct evidence that a disruption of m155 expression leads to attenuation of viral virulence and growth in animals. Moreover, these results suggest that m155 is a viral determinant for optimal MCMV growth and virulence in vivo.
Human cytomegalovirus (HCMV) is a ubiquitous herpesvirus that causes mild or subclinical diseases in immunocompetent adults but may lead to severe complications in neonates and immunocompromised individuals (18, 23). Disseminated HCMV infection, common in AIDS patients, is usually associated with gastroenteritis, pneumonia, and retinitis (8, 22). HCMV infection continues to be a major cause of morbidity and mortality in bone marrow and solid-organ transplant recipients (18, 23). Studies on the functions of viral genes in HCMV replication in vivo are essential for understanding viral pathogenesis and developing new strategies to combat the viral infection. However, there are presently no suitable animal models for HCMV infection. HCMV only propagates in human cells and grows slowly due to a long lytic replication cycle (18, 23). These properties of HCMV have hampered the studies of HCMV pathogenesis and gene function.
Infection of the mouse with murine cytomegalovirus (MCMV) provides a valuable in vivo model for studying the biology of CMV infection. This is because infection of mice by MCMV resembles in many ways its human counterpart with respect to pathogenesis (11, 12, 18, 23). For example, tropism for the spleen and liver is believed to be important in the infection of both HCMV and MCMV, as the spleen and liver are the sites for primary acute infections as well as persistent and latent infections (18, 23). The MCMV genome is 230 kb long and is predicted to encode more than 170 open reading frames, 78 of which have extensive homology to those of HCMV (4, 25). A clear understanding of the biology of MCMV and the function of its genes will provide insight into the pathogenesis of HCMV.
Numerous studies have demonstrated the power of mutagenesis in studying the function(s) of herpesvirus genes (for reviews see references 18 and 28). By specifically mutagenizing a gene and then analyzing the phenotype of the mutated virus both in tissue culture and in animal models, it has been possible to map on the herpesvirus genome specific viral functions, such as those involved in replication, tropism, virulence, and other biological phenomena, such as viral immune evasion. The construction of herpesvirus mutants has been possible by using site-directed homologous recombination and transposon-mediated insertional mutagenesis (5, 13, 19, 27, 32, 33, 36). Herpesvirus genomes have also been cloned into a bacterial artificial chromosome (BAC), and viral mutants have been successfully generated from the BAC-based viral genome by using a bacterial mutagenesis procedure (3, 7, 17, 29, 30, 35). These studies have facilitated the identification of the functions of viral genes in tissue culture and in animals.
Many of the CMV genes have been found to be dispensable for growth in cultured cells. Their presence in the viral genome indicates that they are likely needed to perform functions involved in modulating the interactions between the virus and its respective human or animal hosts. Some of these genes from a particular CMV (e.g., MCMV) might have coevolved with its respective host and interacted with specific components of the host and, therefore, are unique and may not share significant sequence homologies with CMVs from other species (e.g., HCMV). Studies on the function of these species-specific viral genes will provide insight into the general mechanism of how CMV interacts with its respective host and establishes infection. For example, HCMV US11, which is dispensable for viral replication, functions to downregulate the expression and presentation of the major histocompatibility complex (MHC) class I molecules (37). Meanwhile, MCMV open reading frame m157, a nonessential protein, modulates natural killer (NK) cell functions during MCMV infection (1). Thus, studies of viral mutants carrying mutations in genes that are found to be dispensable in tissue culture, including those species-specific open reading frames, are invaluable to the understanding of gene function in viral pathogenesis and virus-host interactions, including tissue tropism and virulence.
We have recently applied a Tn3 transposon-mediated shuttle mutagenesis system to generate a pool of MCMV mutants with specific open reading frames disrupted by transpositional insertion (39). Our further characterization of one of the viral mutants (i.e., Rvm09) in NIH 3T3 cells and in immunodeficient SCID mice indicated that the presence of the transposon sequence per se in the viral genome does not significantly affect viral replication in vitro and viral growth and virulence in vivo (40). In the present study, we report both in vitro and in vivo characterization of a MCMV mutant, Rvm155, which contains a transposon insertion in open reading frame m155. Little is presently known about the function of m155 in viral replication and pathogenesis. Indeed, this open reading frame has not been extensively characterized transcriptionally or translationally. We provide the first direct evidence to suggest that a disruption of the m155 open reading frame leads to attenuation of viral virulence and deficient growth in vivo. When the mutant virus was used to infect immunodeficient SCID mice intraperitoneally, the viral titers in the salivary glands, lungs, spleens, livers, and kidneys were significantly lower than those in mice inoculated with the wild type virus and a revertant virus that rescued the mutation and restored the m155 open reading frame. In particular, the titers of the mutant found in the spleens and livers of the infected animals at 21 days postinfection were less than 8 × 101 PFU/ml of tissue homogenate, while those of the wild-type Smith strain were at least 8 × 103 PFU/ml of tissue homogenate. Moreover, the viral mutant was attenuated in killing SCID mice. These results suggest that m155 is a viral determinant of MCMV pathogenecity and is required for optimal viral virulence and growth in vivo.
MATERIALS AND METHODS
Cells and viruses.
Mouse NIH 3T3 cells and STO cells (American Type Culture Collection [ATCC], Rockville, Md.) were maintained in complete growth medium consisting of Dulbecco's modified Eagle medium (DMEM) supplemented with 10% NuSerum (Becton Dickson, Bedford, Mass.), amino acids, and antibiotics (Life Technologies Inc., Gibco BRL, Grand Island, N.Y.). The wild-type Smith strain (ATCC), mutant Rvm155, and the rescued virus Rqm155 were propagated in NIH 3T3 cells as described previously (40).
Growth analysis of the viruses in animals.
We intraperitoneally infected 4- to 6-week-old CB17 SCID mice (National Cancer Institute, Bethesda, Md.) with 104 PFU of each virus and sacrificed the animals at 1, 3, 7, 10, 14, and 21 days postinoculation. For each time point, we used at least three animals as a group and infected them with the same virus. The salivary glands, lungs, spleens, livers, and kidneys were harvested and sonicated as a 10% (wt/vol) suspension in a 1:1 mixture of DMEM and skim milk. The sonicates were stored at −80°C until plaque assays were performed.
Organs harvested from the mice were titered for viral growth on NIH 3T3 cells in 6-well tissue culture plates (Corning Inc., Corning, N.Y.). We first inoculated the cells with 1 ml of the sonicated organs in 10-fold serial dilutions. After 1 h of incubation at 37°C with 5% CO2, we washed the plates with complete medium and then overlaid the cells with fresh complete medium containing 1% agarose and cultured them for 4 to 6 days before the plaques were counted under an inverted microscope. We titered each sample in triplicate and recorded viral titers as PFU per milliliter of organ homogenates. The limit of virus detection in the organ homogenates was 10 PFU/ml of the sonicated mixture. Those samples that were negative at a 10−1 dilution were designated a titer value of 10 (101) PFU/ml.
Virulence assays.
We studied the virulence of the viruses by determining the mortality of the animals infected with the Smith strain, Rvm155, or Rqm155. The CB17 SCID mice (5 animals per group) were infected intraperitoneally with 104 PFU of each virus. We observed the animals twice daily and monitored the mortality of the infected animals for at least 52 days postinfection and determined the survival rates.
Growth analysis of the viruses in vitro.
The growth kinetics of the viruses was determined as described previously (40). We grew NIH 3T3 cells to 50 to 60% confluence and then inoculated the cells with the viruses at a multiplicity of infection (MOI) of either 0.5 or 5 PFU per cell. At 0, 1, 2, 4, and 7 days postinfection we harvested the infected cells together with medium and mixed the cell lysates with an equal volume of 10% skim milk before sonication. We subsequently determined virus titers by plaque assays in NIH 3T3 cells. The values of the viral titers were the average of triplicate experiments.
Transposon shuttle mutagenesis of MCMV and generation of MCMV recombinant mutants.
Following the procedures described by Zhan et al. (39), we constructed a MCMV genomic subclone pool and carried out transposon-based shuttle mutagenesis to generate a pool of viral DNA fragments containing a Tn3-based transposon (Fig. 1A). To generate a library of MCMV mutants that contained the transposon sequence, we cotransfected intact MCMV genomic DNAs and plasmid DNAs containing the Tn3-gpt-MCMV fragments into NIH 3T3 cells by using a calcium phosphate precipitation protocol (Gibco BRL). We then selected the recombinant MCMV mutants in the presence of mycophenolic acid (MPA) (25 μg/ml; Gibco BRL) and xanthine (50 μg/ml; Sigma, St. Louis, Mo.). The viral mutants were plaque purified six times in the presence of MPA and xanthine, as described previously (39). To confirm the integration of the transposon in the viral genome and to identify the genes that were disrupted by the transposon insertion, we purified the viral DNA and directly sequenced the DNA using the primer FL110PRIM (5′-GCAGGATCCTATCCATATGAC-3′) with the fmol cycle-sequencing kit (Promega, Inc., Madison, Wis.).
FIG. 1.
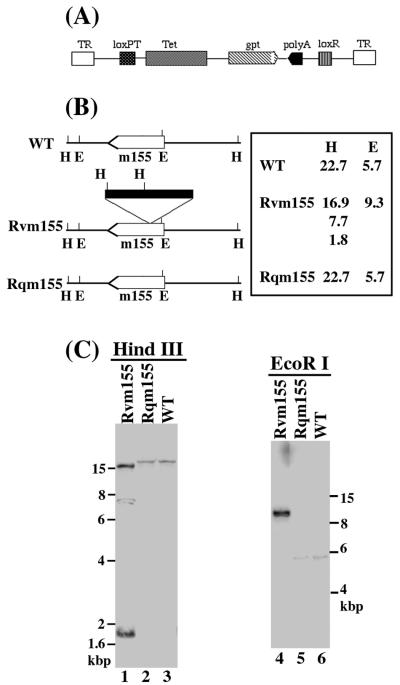
(A) Structure of the transposon construct used for mutagenesis. TR, terminal repeat; Tet, tetracycline resistance gene; gpt, gene that encodes guanine phosphoribosyltransferase (gpt); poly(A), transcription termination signal. (B) Transposon insertion in the recombinant virus. The open arrow shows the coding sequence of open reading frame m155; the filled bar represents the transposon sequence. The orientation of the arrow represents the direction of translation and transcription as predicted from the nucleotide sequence (25). The numbers show the sizes of the DNA fragments of the viruses that were generated by digestion with HindIII (H) or EcoRI (E). (C) Southern analyses of the DNAs of the recombinant viruses. The DNA fractions were isolated from cells infected with the wild-type (WT) virus, Rvm155 (Rvm155), or Rqm155 (Rqm155). The DNA samples (20 μg) were digested with either HindIII (H) or EcoRI (E), separated on 0.8% agarose gels, transferred to a Zeta probe membrane, and hybridized to a 32P-radiolabeled DNA probe that contained the MCMV DNA fragment that was inserted with the transposon sequence.
To construct the rescued virus Rqm155, we isolated the full-length genomic DNA of Rvm155 from infected cells as described previously (39). Using a calcium phosphate precipitation protocol (Gibco BRL), we cotransfected NIH 3T3 cells with the full-length intact Rvm155 viral genomic DNA (8 to 12 μg) and PCR products that contained the m155 coding sequence (1 to 3 μg). The recombinant virus was selected in STO cells in the presence of 25 μg of 6-thioguanine (Sigma)/ml and was purified by six rounds of amplification and plaque purification, following the protocol described previously (9, 38). We prepared viral stocks by growing the viruses in NIH 3T3 cells.
Southern and Northern analyses of recombinant viruses.
Cells that exhibited 100% cytopathic effect were washed with phosphate-buffered saline (PBS) and were subjected to proteolysis with a solution containing sodium dodecyl sulfate (SDS) and proteinase K. We then purified the genomic DNA by extraction with phenol-chloroform followed by precipitation with 2-propanol (34, 39). The DNA was then digested with HindIII or EcoRI, separated on agarose gels (0.8%), transferred to Zeta-Probe nylon membranes (Bio-Rad, Hercules, Calif.), and hybridized with 32P-labeled DNA probes specific for both the transposon and the MCMV sequences. We analyzed the results with a STORM840 PhosphorImager.
For Northern blot analysis, we infected cells with viruses at an MOI of 5 and harvested them at different time points postinfection. In experiments to assay the expression of immediate-early (IE) transcripts, we pretreated cells with 100 μg of cycloheximide (Sigma Co.)/ml and then infected them with viruses and harvested these cells at 6 h postinfection. Total cytoplasmic RNA was isolated from NIH 3T3 cells infected with the viruses as described previously (15). Viral RNAs were separated in 1% agarose gels that contained formaldehyde, were transferred to nitrocellulose membranes, hybridized with the 32P-radiolabeled DNA probes that contained specific MCMV sequences, and finally were analyzed with a STORM840 PhosphorImager. Using PCR with viral DNA as the template, we generated the DNA probes that were used for Northern analyses. The 5′ PCR primers used in the construction of DNA probes for the Northern analysis of M25 and m155 transcripts were M25/1 (5′-AATCCATCTCCGCATCCGAACCCTG-3′) and m155-1 (5′-CGAGTATGTGCTCTCCTGCTCTTGAT-3′), respectively. The 3′ PCR primers used were M25/2 (5′-CCTCAGACGGGATGCTCAATGGCTT-3′) and m155-2 (5′-TACAGACGACAGGTGTCAGACCAAA-3′), respectively.
RESULTS
Generation of MCMV mutant Rvm155 containing a transposon insertion at open reading frame m155 and its rescued virus Rqm155.
We constructed a pool of MCMV mutants with an Escherichia coli Tn3-based transposon mutagenesis system (39). In our mutagenesis procedure, a library of MCMV genomic fragments was cloned in E. coli. A shuttle mutagenesis procedure was carried out on this MCMV library in E. coli, as described previously, to generate a pool of MCMV DNA constructs that contain a randomly inserted transposon in each viral DNA fragment (39). The transposon contained the expression cassette encoding guanine phosphoribosyltransferase (gpt) to allow selection of the transposon-containing MCMV mutants in mammalian cells (Fig. 1A). We cotransfected NIH 3T3 cells with the full-length genomic DNA of the wild-type virus (Smith strain) and the pool of MCMV genomic fragments that contain the inserted Tn3-gpt sequence, allowing homologous recombination to occur. The cells that harbored the progeny viruses and expressed the gpt gene were selected for growth in the presence of mycophenolic acid and xanthine (9, 20, 34). After multiple rounds of selection and plaque purification, we were able to isolate individual recombinant viruses and determined the location of the inserted transposon by directly sequencing the genomic DNA of the recombinants.
Figure 1A shows the structure of the transposon used to generate MCMV mutants. In addition to the gpt expression cassette, the transposon also contains an additional transcription termination site that leads to transcription truncation of the disrupted gene (39). In the constructed transposon, the gpt expression cassette and the additional poly(A) signal in the transposon are in opposite orientation. Thus, the expression of nearby genes that may share a common poly(A) signal with the transposon-targeted gene would not be disrupted (Fig. 1A). One of the recombinant viruses, which contained the transposon sequence inserted within open reading frame m155, was designated Rvm155 and was further characterized (Fig. 1B). Sequencing analyses of the junction between the transposon and the viral sequence revealed that the location of the transposon in Rvm155 is at nucleotide position 215228 in reference to the genome sequence of the wild-type Smith strain (25) (Fig. 1B and data not shown).
Spontaneous mutations within the viral genome, including deletion and rearrangement, have been observed in previous studies on construction of viral mutants using a homologous recombination approach (16 and G. Abenes, X. Zhan, M. Lee, and F. Liu, unpublished data). Thus, it is possible that the phenotype observed with Rvm155 might be due to some other adventitious mutations in the genome of the viral mutant rather than due to the disruption of the m155 open reading frame. In order to exclude this possibility, we constructed a rescued virus, Rqm155, which was derived from Rvm155 by restoration of the wild-type m155 sequence in Rvm155 (Fig. 1B). Construction of the rescued virus was carried out by using a procedure similar to that used for generating the viral mutant. We cotransfected the full-length Rvm155 genomic DNA and a DNA fragment that contained the m155 coding region into NIH 3T3 cells, allowing for homologous recombination to occur. We then grew the progeny viruses in STO cells in the presence of 6-thioguanine, which selects against gpt expression (9, 20). The rescued virus, designated Rqm155, which did not express the gpt protein and no longer contained the transposon, was isolated after multiple rounds of selection and plaque purification.
Characterization of mutant virus Rvm155 and rescued virus Rqm155 by Southern analysis.
We examined the genomic structure of Rvm155 and the location of the transposon insertion in the viral genome by using a DNA probe containing both the transposon and the viral sequence (Fig. 1B and C). When the viral DNA samples were digested with HindIII and subjected to Southern analyses, a small fragment of 1.8 kb representing the gpt gene was detected, indicating the presence of the transposon sequence within the viral genome (Fig. 1C, lane 1). This finding was further supported by the results of Southern analyses of the Rvm155 DNA samples digested with EcoRI (Fig. 1C, lanes 4). The EcoRI-digested genomic fragment containing the transposon was found to be larger than that of the wild-type virus by 3.6 kb, which is the size of the transposon (Fig. 1B and C).
The Southern blots also showed that the stocks of the mutant virus were pure and free of the wild-type strain. This was evident as the hybridizing DNA fragments from the mutant did not comigrate with those of the wild-type Smith strain (Fig. 1C, lanes 1, 3 to 4, and 6). For example, the hybridization patterns of the Rvm155 and Smith strain DNAs digested with HindIII gave rise to three (16.9, 7.7, and 1.8 kb) and one DNA band (22.7 kb), respectively (Fig. 1C, lanes 1 and 3). We also observed that the hybridized species (9.3 kb) of the EcoRI-digested Rvm155 DNA migrated differently from that (5.7 kb) of the wild-type viral DNA digested with the same enzyme (Fig. 1C, lanes 4 and 6). The sizes of the hybridized DNA fragments (Fig. 1C) were consistent with the predicted digestion patterns of the recombinant virus based on the MCMV genomic sequence (25) and the location of the transposon insertion in the viral genome as determined by sequence analysis (Fig. 1B). By ethidium bromide staining of the digested DNAs, we observed that the restriction enzyme digestion patterns of the regions of the mutant Rvm155 genomic DNA other than the transposon insertional site appeared to be identical to those of the parental Smith strain (data not shown). Southern analyses using probes covering the adjacent ORFs suggest that there is no disruption of the adjacent gene sequences in Rvm155. These observations imply that regions of the viral genome other than that containing the transposon insertion remained intact in Rvm155.
Using Southern analysis, we also examined the genomic structure of the rescued virus Rqm155. Analysis of the Rqm155 DNA samples digested with HindIII and EcoRI showed that the sizes of the hybridized DNA fragments for Rqm155 were identical to those of the hybridized fragments for the wild-type Smith strain and were different from those for Rvm155 (Fig. 1C), indicating that Rqm155 did not contain the transposon sequence and that the m155 region was restored. Using ethidium bromide staining of the digested DNAs, we also observed that the restriction enzyme digestion patterns of the regions of the rescued Rqm155 genomic DNA samples other than the m155 region appeared to be the same as those of the parental Rvm155 (data not shown). These results suggest that the regions of the Rqm155 genome other than the m155 region are identical to those of Rvm155 and that Rqm155 represents a rescued virus for Rvm155.
Examination of the viral transcripts expressed from the m155 region of Rvm155 and Rqm155.
Because of the presence of the two transcription termination signals within the transposon (Fig. 1A), we anticipated that the transcription from the m155 region of Rvm155 is disrupted and that the region of the m155 open reading frame downstream from the transposon insertion site will not be expressed. To determine whether this is the case, we isolated cytoplasmic RNAs from cells infected with the mutant virus at different time points (e.g., 4, 12, and 24 h) postinfection and carried out Northern analysis to examine the expression of the transcripts from the m155 open reading frame downstream from the transposon insertion site (Fig. 2). The probe (the 3′ probe) used in this analysis contained the DNA sequence complementary to the 3′ m155 coding region that is within 300 nucleotides downstream of the site of the transposon insertion. We detected a ∼2-kb RNA species in cells that were infected with the wild-type Smith strain (Fig. 2, lane 8). This ∼2-kb RNA species was also readily detected in cells infected with the Smith strain by using a DNA probe (the 5′ probe) complementary to the 5′ terminal sequence of the m155 open reading frame that is within 150 nucleotides downstream of the m155 translational initiation site (data not shown). These results suggest that this ∼2-kb RNA species represents the transcript expressed from the m155 open reading frame. However, using the 3′ probe, which is complementary to the m155 coding region downstream from the site of the transposon insertion, we failed to detect this transcript in the RNA fractions isolated from cells infected with Rvm155 (Fig. 2, lane 6). Thus, transcription from the m155 region downstream from the transposon insertion site appeared to be disrupted in Rvm155. Meanwhile, we detected the expression of the m155 transcript in the RNA fractions from cells infected with the rescued virus Rqm155 (Fig. 2, lane 7). In these experiments we used the level of MCMV M25 transcript (6, 39) as the internal control for the expression of the m155 transcript. As shown in Fig. 2, we found that the levels of the M25 transcript detected in cells that were infected with Rvm155 and Rqm155 were similar to that of the m155 transcript in cells infected with the Smith strain (Fig. 2, lanes 2 to 4). These observations suggest that the transposon insertion in Rvm155 disrupted the transcript expressed from the m155 open reading frame, whereas the wild-type expression of the transcript was restored in Rqm155. The detection of a single transcript from the m155 region using both the 5′ and 3′ probes suggests that only the m155 transcript is expressed from the region and that there is no overlapping transcript. During a 36-h time course study, the m155 transcript was detected from cells after 10 h postinfection but not from cells that were treated with cycloheximide, implying that m155 is expressed at early and late but not IE times.
FIG. 2.
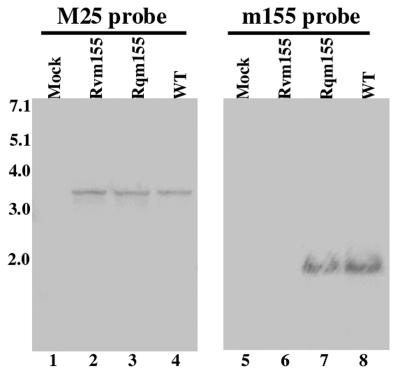
Northern analyses of the expression of the m155 transcript. At 24 h postinfection, RNA fractions were isolated from 106 NIH 3T3 cells that were mock infected (lanes 1 and 5) or infected (MOI, 5) with the wild-type virus (WT) (lanes 4 and 8), Rvm155 (lanes 2 and 6), and Rqm155 (lanes 3 and 7). Equal amounts of RNA samples (25 μg) were separated on agarose gels that contained formaldehyde, transferred to a nitrocellulose membrane, and hybridized to a 32P-radiolabeled probe that contained the sequence of M25 (M25 probe) (lanes 1 to 4) or m155 (m155 probe) (lanes 5 to 8).
Replication of Rvm155 and Rqm155 in vitro in tissue culture.
In order to determine whether these viruses had any growth defects in vitro, we studied the growth rates of the recombinant viruses in NIH 3T3 cells. Cells were infected with the viruses at both low and high MOI, and their growth rates were assayed in triplicate experiments. We found no significant difference in growth rates among Rvm155, Rqm155, and the Smith strain. For example, the peak titers of Rvm155 were similar to those of the Smith strain and Rqm155 under both high- and low-MOI infections (Fig. 3). Previous studies have shown that viral mutants with large deletions of 9 to 15 kb of DNA sequence that may encompass the m155 region replicated in NIH 3T3 cells as efficiently as the Smith stain (2, 31). Our results are consistent with these previous observations that m155 is probably not essential for viral growth in vitro.
FIG. 3.
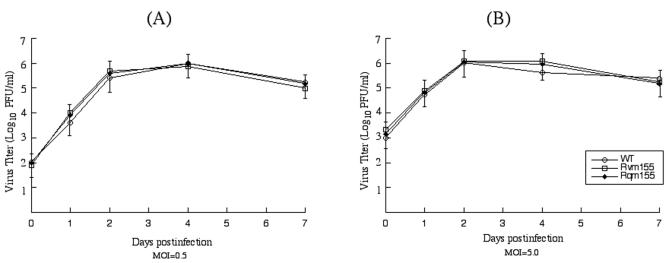
Growth of MCMV mutants in cultured NIH 3T3 cells. NIH 3T3 cells were infected with each virus at a MOI of either 0.5 PFU (A) or 5 PFU per cell (B). At 0, 1, 2, 4, and 7 days postinfection, we harvested cells and culture media and determined the viral titers by plaque assays on NIH 3T3 cells. The error bars indicate the standard deviations. The values of the viral titer represent the average obtained from triplicate experiments.
Virulence of recombinant viruses Rvm155 and Rqm155 in immunodeficient animals.
It has been shown that immunodeficient animals are extremely susceptible to MCMV infection (10, 21, 24, 26). For example, the CB17 SCID mice, which lack functional T and B lymphocytes, are extremely sensitive to viral infection and can succumb to as few as 10 PFU of MCMV (21, 24). In contrast, immunocompetent animals such as BALB/c mice, when infected with wild-type MCMV, are usually asymptomatic unless given a very high viral inoculation dose (e.g., >5 × 106 PFU). Thus, analysis of viral replication in the immunodeficient SCID mice serves as an excellent model for determining the virulence of different MCMV strains and mutants and for studying the mechanism of how they cause pathogenesis and opportunistic infections in animals. To determine whether the m155 open reading frame plays a significant role in CMV virulence, we examined the ability of the mutant virus Rvm155 to kill SCID mice by comparing the survival rates of animals infected with Rvm155, Rqm155, and the wild-type Smith strain. For each virus, we intraperitoneally injected 5 SCID mice with 104 PFU of Rvm155, Rqm155, or the Smith strain. All the mice that were infected with either Smith strain or Rqm155 died within 26 to 27 days postinfection (Fig. 4). In contrast, none of the animals infected with Rvm155 died until 38 days postinfection (Fig. 4). This observation indicated that Rvm155 was attenuated in viral virulence in killing SCID mice. Previous studies in our laboratory have shown that the virulence of a viral mutant (i.e., Rvm09) carrying the transposon sequence was similar to that of the wild-type Smith strain in killing SCID mice, suggesting that the presence of the transposon sequence per se within the viral genome does not significantly affect MCMV virulence (40). Thus, our results suggest that disruption of the m155 open reading frame attenuates viral virulence and that m155 is important to MCMV virulence in SCID mice.
FIG. 4.
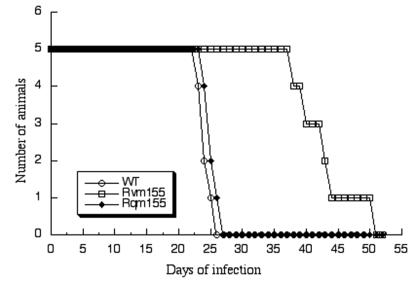
Survival rate of the SCID mice infected with the Smith strain, Rvm155, and Rqm155. CB17 SCID mice (5 animals per group) were infected intraperitoneally with 104 PFU of each virus. Survival of mice was monitored for at least 53 days postinfection, and survival rates were determined.
Growth of Rvm155 and Rqm155 in immunodeficient animals.
To determine whether the disruption of m155 significantly affects viral replication in animals and to investigate the function of m155 in vivo, we studied the pathogenesis of the mutant virus as well as the wild-type Smith strain and rescued mutant Rqm155 in SCID mice. We examined the replication of Rvm155 in different organs of the animals during a 21-day infection period before the mortality of the infected animals became apparent. In these experiments we intraperitoneally injected SCID mice with 104 PFU of each virus (Rvm155, Rqm155, or the wild-type Smith strain). At 1, 3, 7, 10, 14, and 21 days postinfection, 3 mice from each virus group were sacrificed and the salivary glands, lungs, spleens, livers, and kidneys were harvested. By assaying the viral titers in the organs, we determined the levels of viral growth in these five organs. The titers of Rqm155 in each of the organs examined were similar to those of the Smith strain (Fig. 5). However, the titers of the mutant virus Rvm155 were consistently lower than those of the wild-type virus at every time point examined. At 21 days postinfection, the titers of Rvm155 in the salivary glands, lungs, spleens, livers, and kidneys of the infected animals were lower than the titers of the wild-type virus by 50-, 1,000-, 500-, 100-, and 500-fold, respectively (Fig. 5). Our results suggest that Rvm155 is deficient in growth in the organs of the immunodeficient animals. Previous studies have shown that the presence of the transposon sequence per se within the viral genome does not significantly affect viral growth in SCID mice in vivo (40). Thus, these results suggest that the attenuated growth of Rvm155 in these organs is probably due to the disruption of m155 and that open reading frame m155 may be required for optimal growth of MCMV in these organs in immunodeficient hosts.
FIG. 5.
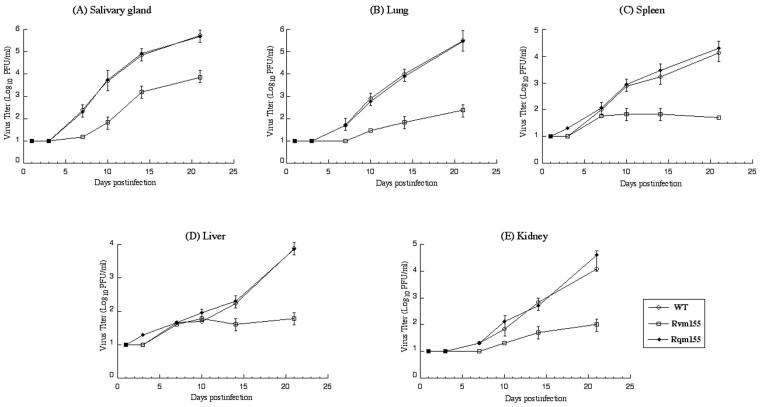
Titers of MCMV mutants in the salivary glands (A), lungs (B), spleens (C), livers (D), and kidneys (E) of the CB17 SCID mice that were infected intraperitoneally with 104 PFU of each virus. At 1, 3, 7, 10, 14, and 21 days postinfection, we sacrificed the animals (3 mice per group); collected the salivary glands, lungs, spleens, livers, and kidneys; and sonicated these organs. We subsequently determined the viral titers in the tissue homogenates by standard plaque assays in NIH 3T3 cells. The limit of detection was 10 PFU/ml of the tissue homogenate. The viral titers represent the average obtained from triplicate experiments. The error bars indicate the standard deviation, and those that are not evident indicate that the standard deviation was less than or equal to the height of the symbols.
The stability of the genome and transposon insertion of Rvm155 during viral replication in SCID mice.
Previous studies have shown that MCMV mutants with an insertional sequence may not be stable and can generate spontaneous mutations during replication in vitro and in vivo (16 and G. Abenes, X. Zhan, M. Lee, and F. Liu, unpublished data). Indeed, a viral mutant with a spontaneous deletion at the viral 3′-terminal region encompassing m155 has been reported after extensive passage of the virus in tissue-cultured cells (2). It is possible that the transposon sequence in Rvm155 is not stable during viral replication in SCID mice, and the introduction of an adventitious mutation may be responsible for the observed phenotypes of the virus in the immunodeficient animals. To exclude this possibility, we harvested the salivary glands and the lungs from the Rvm155-infected SCID mice at 21 days postinfection. We subsequently infected NIH 3T3 cells with the sonicated tissue homogenates. Viral DNAs were purified from the infected cells, and their restriction profiles were analyzed in agarose gels. Moreover, we also performed Southern analysis of the Rvm155 viral DNAs with a DNA probe that contained the transposon and the m155 open reading frame sequence. We found no change in the hybridization patterns of Rvm155 after viral growth in animals for 21 days (Fig. 6). Moreover, ethidium bromide staining of the digested viral DNAs suggests that the overall HindIII digestion patterns of Rvm155 DNA isolated from either infected cultured cells or animals were identical to those of the original recombinant virus Rvm155 (data not shown). These observations imply that the transposon insertion in Rvm155 is stable and the genome of Rvm155 remains intact during replication in the infected SCID mice.
FIG. 6.
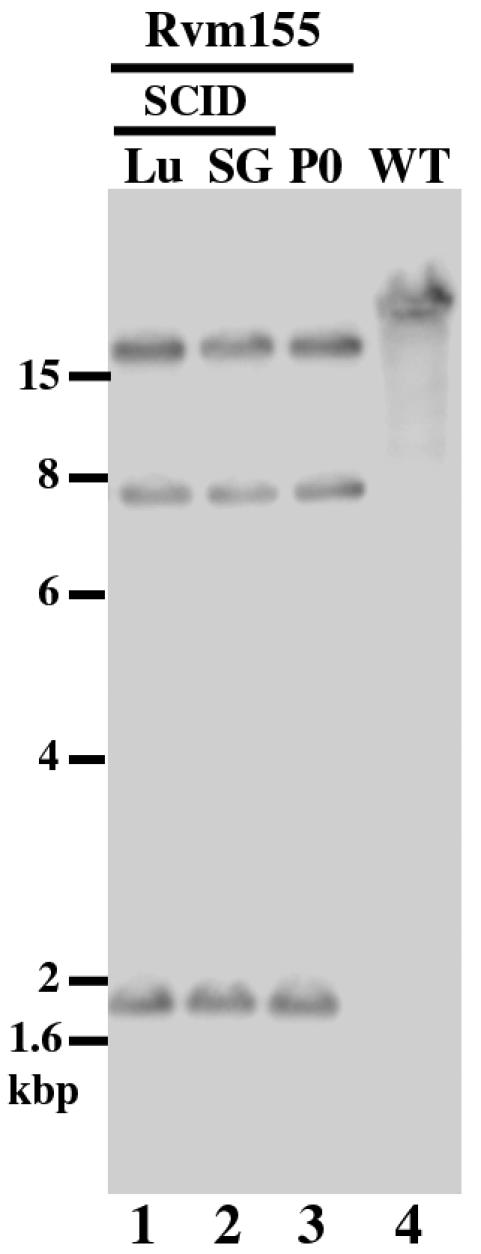
The stability of the transposon insertional mutation of Rvm155 in CB17 SCID mice. We isolated viral DNAs from cells that were infected with Rvm155 (MOI < 0.01) and allowed to grow in culture for 5 days (P0) (lane 3) or from cells that were infected with the virus collected from the lungs (LU, lane 1) and salivary glands (SG, lane 2) of the infected SCID mice 21 days after intraperitoneal inoculation with 104 PFU of Rvm155. Southern analyses of the viral DNA fractions digested with HindIII are shown. The 32P-radiolabeled probe was derived from the same plasmid that was used for Southern analyses of Rvm155 shown in Fig. 1 and contained the transposon and the m155 sequence. The DNA of the wild-type virus (WT) is shown in lane 4.
DISCUSSION
In this study, we have characterized the virulence and growth of a recombinant virus in vitro in cultured NIH 3T3 fibroblasts and in vivo in SCID mice. This viral mutant, Rvm155, contained an insertional mutation in open reading frame m155. Our results provide the first direct evidence to suggest that disruption of the m155 open reading frame results in attenuated virulence and reduced growth of the virus in SCID mice. Moreover, the results presented in this study suggest that m155 functions in supporting efficient viral replication in vivo and is required for optimal viral virulence in immunodeficient hosts.
Our results indicate that the transposon sequence is present in the Rvm155 genome and disrupts the coding sequence of open reading frame m155 (Fig. 1B). It is possible that the functional m155 protein product might still be synthesized from the transposon-disrupted region. However, several lines of evidence strongly suggest that this is not the case. First, the transposon sequence was inserted into the coding sequence of the m155 open reading frame (Fig. 1B). Second, transcription from the region downstream from the transposon insertion site was not detected in cells infected with the mutant virus (Fig. 2). These results indicate that the region of the target open reading frame downstream from the transposon insertion site, which includes more than 70% of the m155 coding sequence, was not expressed. Thus, it is likely that no functional m155 protein was expressed from the viral mutant. The growth rate of Rvm155 in NIH 3T3 cells was similar to that of the Smith strain (Fig. 3). These results provide evidence that the carboxyl 264-amino-acid sequence of m155 is not essential for viral replication in vitro. Moreover, these results are consistent with previous observations (2, 31) that m155 is probably dispensable for MCMV growth in vitro in NIH 3T3 cells.
The function of m155 is presently unknown. Our results showed that Rvm155 was deficient in replication in the salivary glands, lungs, spleens, livers, and kidneys of the SCID mice that were intraperitoneally infected. For example, at 21 days postinfection, the titers of Rvm155 in the salivary glands, lungs, spleens, livers, and kidneys of the infected SCID mice were lower than the titers of the wild-type virus by 50-, 1,000-, 500-, 100-, and 500-fold, respectively (Fig. 5). Moreover, no death occurred among the SCID mice infected with Rvm155 up to 38 days postinfection, while all mice infected with the Smith strain or Rqm155 died within 27 days postinfection (Fig. 4). These results strongly suggest that m155 is a viral determinant for MCMV growth in vivo and for viral virulence in killing SCID mice.
It is possible that other adventitious mutations, which may be generated during the construction and growth of the recombinant virus in cultured cells or in animals, rather than the disruption of the m155 open reading frame are responsible for the observed change in the levels of replication of the mutant. However, several lines of evidence strongly suggest that this is unlikely. First, the wild-type phenotypes for growth and virulence in the infected mice were restored in Rqm155 upon restoration of the wild-type sequence into Rvm155 (Fig. 1, 4, and 5). Furthermore, the restoration of the wild-type phenotypes in Rqm155 occurred together with the restoration of the m155 mRNA expression (Fig. 2). These observations suggest that the transposon insertion rather than an adventitious mutation is responsible for the observed attenuation of Rvm155 replication and virulence in vivo. Second, our previous studies indicated that a virus mutant (i.e., Rvm09) with transposon insertion at the m09 open reading frame replicated as efficiently in SCID mice as the wild-type virus (40). Moreover, mutant Rvm09 exhibited a level of virulence in killing SCID mice similar to that of the wild-type virus. Thus, the transposon sequence per se in the viral genome does not significantly affect viral replication and virulence in the infected animals (40). Third, our results showed that the genome and the transposon insertion in the viral mutant were stable during replication in animals. We found no change in the hybridization patterns of the DNAs from the mutant viruses that were recovered from the salivary glands and lungs of the infected animals after 21 days of infection (Fig. 6 and data not shown). Moreover, the HindIII digestion patterns of the Rvm155 mutant DNAs, other than the transposon insertion region, appeared to be identical to those of the wild-type virus DNA (data not shown). The transposon does not appear to disrupt the expression of adjacent genes, as only the m155 mRNA is expressed from the m155 region and there is no overlapping transcript. Thus, the observed change of the level of Rvm155 replication and virulence in the infected mice is probably due to the disruption of the m155 expression as a result of the transposon insertion.
Our results are consistent with previous observations of viral mutants that contained large deletions at the 3′-terminal region that encompass the m155 open reading frame. These mutants, which contained a deletion of a 9.6- and 15.8-kb sequence at the 3′-terminal region, respectively, exhibited attenuated growth in spleens and salivary glands of the infected BALB/c mice (2, 31). However, whether these mutants are deficient in growth in immunodeficient mice has not been reported. Equally unclear is whether their virulence in killing animals is attenuated. Meanwhile, these mutants contained large sizes of the deletions in their genomes, which resulted in removal of multiple open reading frames. It is not known whether the observed attenuated phenotypes are due to the mutation of multiple genes or a single specific open reading frame (2, 31). Our results provide direct evidence that a single mutation at the m155 open reading frame results in a significant reduction of viral growth and virulence in vivo. Moreover, our study indicates for the first time that m155 is important for viral growth and for viral killing in SCID mice.
Little is presently known about the molecular mechanism of m155 in supporting viral growth and virulence in vivo. Indeed, to our knowledge neither the transcript nor the protein product coded by m155 has been extensively characterized. In our experiments a transcript of about 2,000 nucleotides was found to be expressed from the m155 open reading frame. During a 36-h time course study, the m155 transcript was found to be expressed after 10 h postinfection and was not expressed at IE time. The m155 open reading frame belongs to the MCMV m145 gene family and has been predicted to encode a putative membrane glycoprotein (25). The MCMV m145 gene family, consisting of 11 open reading frames, includes m145, m146, m150, m151, m152, m153, m154, 155, m157, m158, and m17. Previous studies have shown that two members of the MCMV m145 gene family (i.e., m152 and m157) are dispensable for MCMV replication in vitro in tissue culture but play an important role in modulating host immune response (1, 14). Specifically, m152, which encodes a glycoprotein designated gp40, downregulates MHC class I expression at the plasma membrane of the infected cells. Moreover, m152 also modulates NK cell activation by downregulating H-60, the high-affinity ligand for the NK cell receptor NKG2D (14). The m157 open reading frame also encodes a membrane glycoprotein that contains an MHC class I-like domain and can be considered as a nonclassical MHC-like molecule. The m157-encoded product has recently been shown to bind to different NK cell receptors, thereby activating or inhibiting NK cell responses (1). It will be interesting to determine whether m155 may also function in a similar way to modulate immune responses, such as NK cell response, and support optimal viral growth and virulence in vivo. Further studies on in vitro and in vivo replication of Rvm155 and other viral mutants will provide further insight into the functions of viral genes in vivo in MCMV virulence and pathogenesis.
Acknowledgments
We thank Jianqiao Xiao, Tuong Tong, John Kim, and Sean Umamoto for their technical assistance.
K.C. is partially supported by an NIAID predoctoral training grant (T32-AI007620). E.H. acknowledges the support of a fellowship from the Biology Fellow program (University of California—Berkeley). F.L. is a Scholar of the Leukemia and Lymphoma Society and a recipient of the Established Investigator award from the American Heart Association. The research has been supported by grants from the March of Dimes National Birth Defects Foundation, the American Heart Association, and the NIH.
REFERENCES
- 1.Arase, H., E. S. Mocarski, A. E. Campbell, A. B. Hill, and L. L. Lanier. 2002. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science 296:1323-1326. [DOI] [PubMed] [Google Scholar]
- 2.Boname, J. M., and J. K. Chantler. 1992. Characterization of a strain of murine cytomegalovirus which fails to grow in the salivary glands of mice. J. Gen. Virol. 73:2021-2029. [DOI] [PubMed] [Google Scholar]
- 3.Brune, W., C. Menard, U. Hobom, S. Odenbreit, M. Messerle, and U. H. Koszinowski. 1999. Rapid identification of essential and nonessential herpesvirus genes by direct transposon mutagenesis. Nat. Biotechnol. 17:360-364. [DOI] [PubMed] [Google Scholar]
- 4.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison, T. Kouzarides, and J. A. Martignetti. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 5.Cohen, J. I., and K. E. Seidel. 1993. Generation of varicella-zoster virus (VZV) and viral mutants from cosmid DNAs: VZV thymidylate synthetase is not essential for replication in vitro. Proc. Natl. Acad. Sci. USA 90:7376-7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dallas, P. B., P. A. Lyons, J. B. Hudson, A. A. Scalzo, and G. R. Shellam. 1994. Identification and characterization of a murine cytomegalovirus gene with homology to the UL25 open reading frame of human cytomegalovirus. Virology 200:643-650. [DOI] [PubMed] [Google Scholar]
- 7.Delecluse, H. J., T. Hilsendegen, D. Pich, R. Zeidler, and W. Hammerschmidt. 1998. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc. Natl. Acad. Sci. USA 95:8245-8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallant, J. E., R. D. Moore, D. D. Richman, J. Keruly, and R. E. Chaisson. 1992. Incidence and natural history of cytomegalovirus disease in patients with advanced human immunodeficiency virus disease treated with zidovudine. J. Infect. Dis. 166:1223-1227. [DOI] [PubMed] [Google Scholar]
- 9.Greaves, R. F., J. M. Brown, J. Vieira, and E. S. Mocarski. 1995. Selectable insertion and deletion mutagenesis of the human cytomegalovirus genome using the Escherichia coli guanosine phosphoribosyl transferase (gpt) gene. J. Gen. Virol. 76:2151-2160. [DOI] [PubMed] [Google Scholar]
- 10.Grundy, J. E., and C. J. Melief. 1982. Effect of Nu/Nu gene on genetically determined resistance to murine cytomegalovirus. J. Gen. Virol. 61:133-136. [DOI] [PubMed] [Google Scholar]
- 11.Hudson, J. B. 1979. The murine cytomegalovirus as a model for the study of viral pathogenesis and persistent infections. Arch. Virol. 62:1-29. [DOI] [PubMed] [Google Scholar]
- 12.Jordan, M. C. 1983. Latent infection and the elusive cytomegalovirus. Rev. Infect. Dis. 5:205-215. [DOI] [PubMed] [Google Scholar]
- 13.Kemble, G., G. Duke, R. Winter, and R. Spaete. 1996. Defined large-scale alterations of the human cytomegalovirus genome constructed by cotransfection of overlapping cosmids. J. Virol. 70:2044-2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krmpotic, A., D. H. Busch, I. Bubic, F. Gebhardt, H. Hengel, M. Hasan, A. A. Scaizo, U. H. Koszinowski, and S. Jonjic. 2002. MCMV glycoprotein gp40 confers virus resistance to CD8+ cells and NK cells in vivo. Nat. Immunol. 3:529-535. [DOI] [PubMed] [Google Scholar]
- 15.Liu, F., and B. Roizman. 1991. The herpes simplex virus 1 gene encoding a protease also contains within its coding domain the gene encoding the more abundant substrate. J. Virol. 65:5149-5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manning, W. C., C. A. Stoddart, L. A. Lagenaur, G. B. Abenes, and E. S. Mocarski. 1992. Cytomegalovirus determinant of replication in salivary glands. J. Virol. 66:3794-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Messerle, M., I. Crnkovic, W. Hammerschmidt, H. Ziegler, and U. H. Koszinowski. 1997. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 94:14759-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mocarski, E. S., and C. T. Courcelle. 2001. Cytomegaloviruses and their replication, p. 2629-2673. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Williams & Wilkins, Philadelphia, Pa.
- 19.Mocarski, E. S., L. E. Post, and B. Roizman. 1980. Molecular engineering of the herpes simplex virus genome: insertion of a second L-S junction into the genome causes additional genome inversions. Cell 22:243-255. [DOI] [PubMed] [Google Scholar]
- 20.Mulligan, R. C., and P. Berg. 1981. Selection for animal cells that express the Escherichia coli gene coding for xanthine-guanine phosphoribosyltransferase. Proc. Natl. Acad. Sci. USA 78:2072-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okada, M., and Y. Minamishima. 1987. The efficacy of biological response modifiers against murine cytomegalovirus infection in normal and immunodeficient mice. Microbiol. Immunol. 31:45-57. [DOI] [PubMed] [Google Scholar]
- 22.Palella, F. J., Jr., K. M. Delaney, A. C. Moorman, M. O. Loveless, J. Fuhrer, G. A. Satten, D. J. Aschman, and S. D. Holmberg. 1998. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. N. Engl. J. Med. 338:853-860. [DOI] [PubMed] [Google Scholar]
- 23.Pass, R. F. 2001. Cytomegalovirus, p. 2675-2706. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Williams & Wilkins, Philadelphia, Pa.
- 24.Pollock, J. L., and H. W. Virgin. 1995. Latency, without persistence, of murine cytomegalovirus in the spleen and kidney. J. Virol. 69:1762-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rawlinson, W. D., H. E. Farrell, and B. G. Barrell. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833-8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reynolds, R. P., R. J. Rahija, D. I. Schenkman, and C. B. Richter. 1993. Experimental murine cytomegalovirus infection in severe combined immunodeficient mice. Lab. Anim. Sci. 43:291-295. [PubMed] [Google Scholar]
- 27.Roizman, B., and F. J. Jenkins. 1985. Genetic engineering of novel genomes of large DNA viruses. Science 229:1208-1214. [DOI] [PubMed] [Google Scholar]
- 28.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2460. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 29.Smith, G. A., and L. W. Enquist. 1999. Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J. Virol. 73:6405-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stavropoulos, T. A., and C. A. Strathdee. 1998. An enhanced packaging system for helper-dependent herpes simplex virus vectors. J. Virol. 72:7137-7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thale, R., U. Szepan, H. Hengel, G. Geginat, P. Lucin, and U. H. Koszinowski. 1995. Identification of the mouse cytomegalovirus genomic region affecting major histocompatibility complex class I molecule transport. J. Virol. 69:6098-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomkinson, B., E. Robertson, R. Yalamanchili, R. Longnecker, and E. Kieff. 1993. Epstein-Barr virus recombinants from overlapping cosmid fragments. J. Virol. 67:7298-7306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Zijl, M., W. Quint, J. Briaire, T. de Rover, A. Gielkens, and A. Berns. 1988. Regeneration of herpesviruses from molecularly cloned subgenomic fragments. J. Virol. 62:2191-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vieira, J., H. E. Farrell, W. D. Rawlinson, and E. S. Mocarski. 1994. Genes in the HindIII J fragment of the murine cytomegalovirus genome are dispensable for growth in cultured cells: insertion mutagenesis with a lacZ/gpt cassette. J. Virol. 68:4837-4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wagner, M., S. Jonjic, U. H. Koszinowski, and M. Messerle. 1999. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J. Virol. 73:7056-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weber, P. C., M. Levine, and J. C. Glorioso. 1987. Rapid identification of nonessential genes of herpes simplex virus type 1 by Tn5 mutagenesis. Science 236:576-579. [DOI] [PubMed] [Google Scholar]
- 37.Wiertz, E. J., T. R. Jones, L. Sun, M. Bogyo, H. J. Geuze, and H. L. Ploegh. 1996. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell 84:769-779. [DOI] [PubMed] [Google Scholar]
- 38.Xiao, J., T. Tuong, X. Zhan, E. Haghjoo, and F. Liu. 2000. In vitro and in vivo characterization of a murine cytomegalovirus with a transposon insertional mutation at open reading frame M43. J. Virol. 74:9488-9497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhan, X., G. Abenes, M. Lee, I. VonReis, C. Kittinunvorakoon, P. Ross-Macdonald, M. Snyder, and F. Liu. 2000. Mutagenesis of murine cytomegalovirus using a Tn3-based transposon. Virology 266:264-274. [DOI] [PubMed] [Google Scholar]
- 40.Zhan, X., M. Lee, J. Xiao, and F. Liu. 2000. Construction and characterization of murine cytomegaloviruses that contain a transposon insertion at open reading frames m09 and M83. J. Virol. 74:7411-7421. [DOI] [PMC free article] [PubMed] [Google Scholar]


