Abstract
The mammalian gastro-intestinal (GI) tract is colonized by trillions of beneficial commensal bacteria that are essential for promoting normal intestinal physiology. While the majority of commensal bacteria are found in the intestinal lumen, many species have also adapted to colonize different anatomical locations in the intestine, including the surface of intestinal epithelial cells (IECs) and the interior of gut-associated lymphoid tissues. These distinct tissue localization patterns permit unique interactions with the mammalian immune system and collectively influence intestinal immune cell homeostasis. Conversely, dysregulated localization of commensal bacteria can lead to inappropriate activation of the immune system and is associated with numerous chronic infectious, inflammatory and metabolic diseases. Therefore, regulatory mechanisms that control proper anatomical containment of commensal bacteria are essential to maintain tissue homeostasis and limit pathology. In this review, we propose that commensal bacteria associated with the mammalian GI tract can be anatomically defined as (i) luminal, (ii) epithelial-associated or (iii) lymphoid tissue-resident, and we will discuss the role and regulation of these microbial populations in health and disease.
Introduction
The mammalian GI tract is colonized by beneficial microbes such as commensal bacteria, which collectively influence host intestinal physiology. Levels of commensal bacteria in the GI tract have been reported to be as high as 1014 organisms, with over 1000 different bacterial species represented (1, 2). Some of the most well characterized roles of commensal bacteria include promoting efficient host nutrient absorption and protection from pathogen colonization (reviewed in (3–8)). In addition, an expanding body of literature has identified critical roles for commensal bacteria in the development of the host immune system and maintenance of immune cell homeostasis (reviewed in (9, 10)).
In contrast to the ability of commensal bacteria to confer beneficial properties, dysregulated interactions between commensal bacteria and the host are also associated with many chronic inflammatory diseases such as inflammatory bowel disease (IBD), chronic viral infection, obesity, cancer and cardiovascular disease (9, 11). This has been proposed to occur either by changes in the composition of the microbiota known as dysbiosis and/or bacterial translocation (9, 11–18). Although many studies have identified and characterized dybiosis during disease, much less is known about the role of commensal bacteria localization in disease development and pathogenesis.
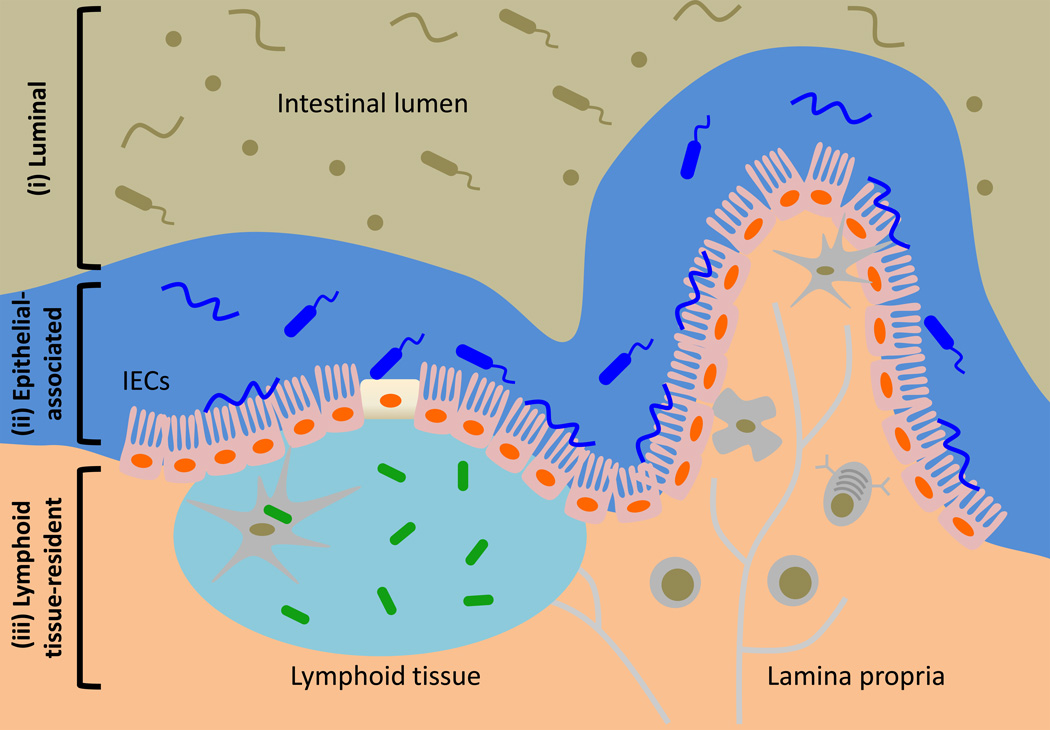
Much of our current knowledge on the role of the microbiota in health and disease is derived from studies on luminal commensal bacteria. However, emerging studies suggest that, in the steady state, specific commensal populations have been shown colonize distinct compartments of the intestine and alter immune cell homeostasis to provide host protection from disease. For example, the Firmicutes phylum member Clostridium spp. was demonstrated to colonize the lumen of the colon while the Bacteroidetes phylum member Bacteroides fragilis was shown to colonize both the lumen and crypts of the colon (19–23). In contrast, the gram-positive segmented filamentous bacteria (SFB) colonized the intestine by adhering tightly to epithelial cells of the terminal ileum in mice (24, 25). These studies highlight that the anatomical localization of commensal bacteria in the intestine can be categorized into at least two groups: (i) luminal and (ii) epithelial-associated. More recently, two studies have described gut-associated lymphoid tissues as a novel and unexpected site for commensal bacterial colonization in healthy mammals. These commensal species, herein referred to as lymphoid tissue-resident commensal bacteria, were shown to colonize the interior of Peyer’s patches (PPs) of healthy mice, primates and humans (26, 27). Despite our growing understanding of commensal-immune system relationships, how these interactions are influenced by commensal bacteria colonization in different compartments of the intestine is not well understood. In this review, we will discuss how anatomically distinct commensal populations, including (i) luminal, (ii) epithelial-associated or (iii) lymphoid tissue-resident (Figure 1), are recognized by the immune system, influence immune cell function and are anatomically restricted via host and bacterial intrinsic mechanisms. Moreover, we will highlight current literature involving both human and mouse studies on how dysregulated commensal bacteria localization may contribute to a variety of chronic inflammatory diseases.
Figure 1.
Commensal bacteria in the mammalian GI tract can be classified by their anatomical localization as (i) luminal, (ii) epithelial-associated or (iii) lymphoid tissue-resident. Commensal bacteria are important for promoting normal host physiology. In the healthy mammals, most commensal bacteria occupy the lumen of the intestine while some are found associated with the intestinal epithelium. Recent studies have identified that commensal bacteria can also inhabit gut-associated lymphoid tissues in the steady state.
Luminal commensal bacteria
Analyses of commensal bacteria composition in the mammalian GI tract have mainly focused on 16S rDNA sequencing of luminal contents from the intestine. In healthy mice, luminal commensal bacteria are largely represented by three phyla: Bacteroidetes, Firmicutes and Proteobacteria. Within these groups, approximately 70% are Bacteroidetes, 20% are Firmicutes and less than 2% are Proteobacteria (28). Consistent with the intestine being a hypoxic environment, most luminal commensal bacteria, especially Bacteroidetes and Firmicutes members, are obligate anaerobes (29, 30). Furthermore, many of these bacteria are not culturable using current techniques and have only been identified through 16S rDNA sequencing (31, 32). Using this technique to interrogate interactions between the host and luminal commensal bacteria, an increasing body of data has revealed that members of both Bacteroidetes and Firmicutes have profound roles in shaping the host immune system.
The Bacteroidetes phylum is comprised predominantly of gram-negative, rod-shaped, anaerobic bacteria. Within this phylum, the genus Bacteroides is highly represented along the mammalian GI tract and has been reported to reach as high as 1010 – 1011 organisms per gram of human feces (33). Many members of the Bacteroides genus including B. thetaiotaomicron and B. fragilis are well adapted to thrive in the GI tract (33) and have been described to aid in host metabolism by processing complex polysaccharides into smaller compounds for host absorption (34).
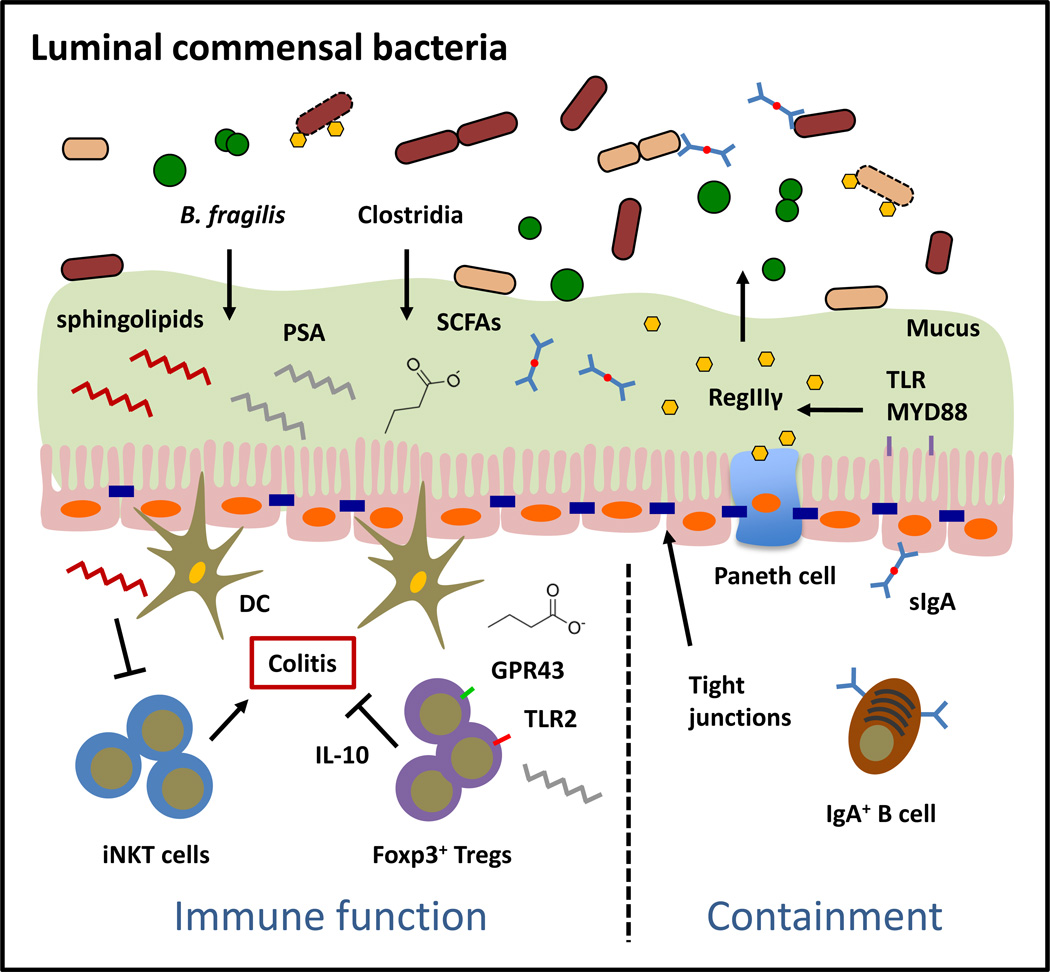
Studies interrogating the interactions between Bacteroides and the host immune system have focused primarily on one species, B. fragilis (Figure 2). An identifying feature of this bacterium is the presence of a capsular polysaccharide complex composed of eight capsular polysaccharides, of which polysaccharide A (PSA) is the most highly expressed (35). While PSA can elicit pro-inflammatory responses such as production of interleukin (IL)-12, tumor necrosis factor alpha (TNFα) and interferon gamma (IFNγ) through recognition of PSA by dendritic cells (DCs) (35), PSA also has potent anti-inflammatory properties and can stimulate interleukin (IL)-10-producing CD4+ Foxp3+ Tregs in the colon (21). Surprisingly, this response was independent of antigen presenting cells (APCs) but dependent on T cell recognition of PSA by Toll-like receptor (TLR) 2 (22). In mice, this immunomodulatory response was required for intestinal colonization by B. fragilis and was associated with protection from experimental colitis (21). B. fragilis could also produce glycosphingolipids that inhibit invariant natural killer T (iNKT) cell proliferation, likely by interfering with lipid loading on CD1d molecules (36). Functionally, modulation of iNKT cell homeostasis in the colon by B. fragilis glycosphingolipids protected mice from iNKT cell-mediated intestinal inflammation. In a separate report, B. fragilis can produce immune-stimulatory glycolipids that bind CD1d and activate iNKT cells in mice and humans (37), suggesting a dual role for Bacteroides glycolipids in regulating immune cell homeostasis.
Figure 2.
Luminal commensal bacteria comprise of the majority of commensal bacteria in the mammalian intestine and are physically separated from immune cells. Colonization of the GI tract by luminal commensal bacteria promotes tissue protective and inhibits pathological immune responses. Anatomical containment of luminal commensal populations is mediated by physical and biochemical barriers such as IECs, tight junctions, mucus, antimicrobial peptides (AMPs) and secretory immunoglobulin A.
Patients with autism spectrum disorder (ASD) often present with GI symptoms (38–41). In recent findings, B. fragilis was demonstrated to correct behavioral abnormalities and restore increased intestinal permeability and altered luminal commensal bacteria composition in a mouse model of ASD (42). Notably, oral treatment with PSA-deficient B. fragilis or wild-type B. thetaiotaomicron but not Enterococcus faecalis also improved anxiety-like, repetitive and communicative behavior of mice with ASD. Furthermore, it was demonstrated that oral treatment of ASD mice with B. fragilis could limit elevated levels of the neurotoxic, commensal bacteria-derived serum metabolite 4-ethylphenylsulfate. These findings suggest that Bacteroides may play important roles in host physiology beyond the intestine by modulating the metabolic activity of other commensal bacteria.
Within the Firmicutes phylum, which is comprised of gram-positive, spore forming, obligate anaerobes, the most studied group of commensal bacteria in regard to modulation of immune cell function is the class Clostridia (Figure 2). Similar, to B. fragilis, Clostridium spp. isolated from mouse and human feces can promote the differentiation and function of CD4+ Foxp3+ Tregs in the mouse colon and are sufficient to protect mice from experimental colitis (19, 20). Colonization of germ free mice, which have enlarged cecums, by chloroform-resistant Clostridium spp. resulted in significantly reduced cecum size due to Clostridium-dependent fermentation of dietary fiber and production of short-chain fatty acids (SCFAs) as metabolic by-products (43). Consistent with the hypothesis that gut metabolism and immune cell homeostasis are two related processes, optimal induction of colonic Treg responses by Clostridium spp. was dependent on metabolism of dietary fiber and subsequent production of SCFAs (20, 43–45). The cellular and molecular mechanisms by which SCFAs influence immune cell function have begun to be elucidated. SCFAs could directly promote Treg immune-suppressive function through Treg-intrinsic expression of the SCFA receptor, GPR43 (45). Butyrate also promoted acetylation at the Foxp3 locus and the Foxp3 protein (43, 44), which have been previously shown to enhance Foxp3 stability and function (46, 47). Alternatively, butyrate and propionate could function independently of Tregs by suppressing DC production of pro-inflammatory cytokines and augmenting their ability to induce peripheral Treg differentiation (44). Furthermore, the SCFA butyrate was shown to promote Tregs at extra-intestinal tissues such as the spleen and peripheral lymph nodes (44). Collectively, Clostridium spp. are prominent members of the luminal microbiota that induce tissue protective, immunoregulatory responses through production of microbial-derived metabolites.
Anatomical restriction of B. fragilis and Clostridium spp. to the lumen of the intestine is dependent on the presence of physical and biochemical barriers that limit interactions of commensal bacteria with IECs and potential translocation to the underlying connective tissues of the intestinal lamina propria (Figure 2). Mucus, which is secreted by specialized epithelial cells known as goblet cells, forms a dense gel-like layer on the apical surface of the intestinal epithelium and is composed of mucin glycoproteins including the highly glycosylated mucin MUC2 (48). In the colon, mucus is organized into an inner mucus layer, which is approximately 50 µm thick in mice and is firmly attached to the surface of epithelial cells, and an outer non-epithelial attached layer, which is approximately 100 µm thick in mice (48). Luminal commensal bacteria in the colon are restricted to either the lumen or the outer mucus layer whereas the inner mucus is mostly devoid of commensal bacteria with the exception of some epithelial-associated species (48), which will be discussed in the following section. Although B. fragilis could colonize the lumen, it could bind intestinal mucins and was found within the colon mucous layer of gnotobiotic mice dual-colonized with E. coli and B. fragilis (49). In the small intestine, the mucus layer, although much less defined than in the colon, is about 50 µm thick in mice and also promotes physical separation between most luminal commensal bacteria and the epithelium (48). In mice deficient in the mucin glycoprotein MUC2, physical separation between commensal bacteria and the intestinal epithelium was lost resulting in the development of spontaneous colitis (50, 51). More recently, it was described that MUC2 in the mucus layer of the small intestine also actively participated in promoting immune tolerance to luminal antigens by conditioning intestinal DCs (51). Recognition and uptake of MUC2 by DCs enhanced their expression of the immunoregulatory cytokines Il10, Tgfb1 and limited their ability to induce T cell proliferation and production of the proinflammatory cytokine IFNγ (51). These data indicate that mucus secretion is an important mechanism that restricts dissemination of luminal commensal bacteria and supports immunological tolerance to luminal commensal antigens.
In addition to mucus, antimicrobial peptides (AMPs) are small molecular weight proteins secreted into the lumen of the intestine that limit colonization of commensal bacteria and promote physical separation between commensal bacteria and the intestinal epithelium. AMPs are produced by virtually all IECs but the expression of a subset of AMPs such as the C-type lectin regenerating islet-derived protein 3γ (REGIIIγ) is enriched in specialized IECs such as Paneth cells in the small intestine (9). AMPs are concentrated to the epithelial surface to limit apical attachment of commensal bacteria normally restricted to the lumen (52). These studies also demonstrated that production of REGIIIγ could be induced by TLR signaling, as IEC-intrinsic deletion of Myd88 resulted in loss of commensal-epithelium segregation (52). Collectively, these data suggest that recognition of commensal bacteria by TLRs initiates regulatory pathways that promote anatomical restriction. Recently, it was demonstrated that the REGIII family proteins recognize and target commensal bacteria by binding to surface carbohydrates and forming an oligomeric membrane-permeabilizing pore (53). The gram-negative bacteria-derived component lipopolysaccharide (LPS) inhibited REGIIIα pore-forming activity, providing a mechanism by which REGIII family proteins are bactericidal to gram-positive but not gram-negative bacteria. Other AMPs such as lipocalin 2 and calprotectin (S100A8/9), which limit microbial access to micronutrients such as zinc, manganese and iron, are likely candidates that control the gram-negative commensal populations in the intestinal lumen (54–57).
A third mechanism by which the host limits commensal attachment to the epithelium is through secretory IgA. IgA is the most abundant immunoglobulin produced in mammals and play a critical role in immune defense at the intestinal barrier surface. Induction of IgA in the intestine occurs predominantly in the PPs and isolated lymphoid follicles (ILFs) of the small intestine and, similar to mucins and AMPs, is highly dependent on the microbiota (58–60). IgA secreted by plasma cells in these tissues is covalently bound to the polymeric immunoglobulin receptor (pIgR) and transported through IECs via intracellular vesicles. Once released into the lumen, IgA restricts commensal bacteria from associating with the epithelium by binding to the surface of bacteria and neutralizing their ability to trigger inflammation (58, 60). Mice-deficient in pIgR displayed increased penetration of commensal bacteria to the mesenteric lymph node (mLN) and systemic antibody responses to commensal bacteria. IgA can be induced by both T-dependent and T-independent mechanisms. While high affinity IgA from T-dependent pathways is important for protection from mucosal pathogen colonization, low-affinity, polyreactive IgA from T-independent pathways is generally necessary for confining commensal bacteria to the intestinal lumen (58, 59). Taken together, mucus, AMPs and secretory IgA are three of the main host mechanisms that cooperatively serve as important regulators that restrict commensal bacteria to the lumen of the GI tract.
Epithelial-associated commensal bacteria
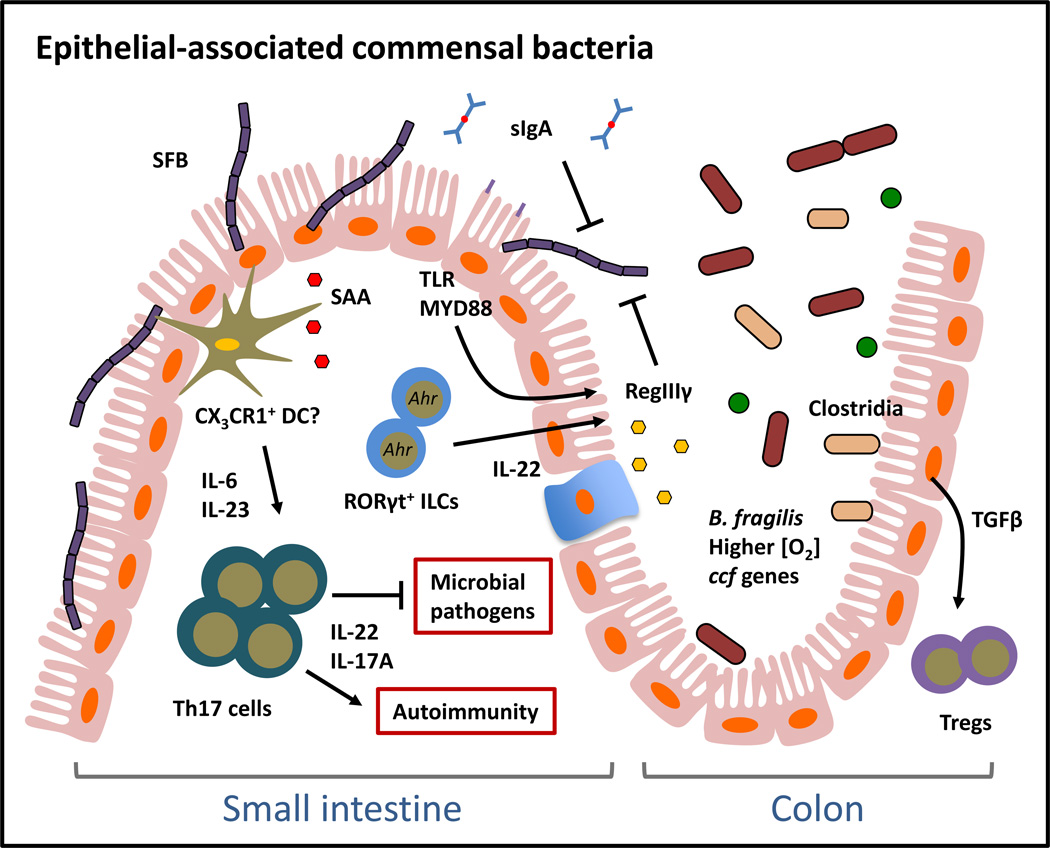
Although most commensal bacteria are restricted to the intestinal lumen, a small fraction can penetrate the inner mucus layer and colonize the surface of IECs. Sequencing of 16S rDNA on the surface of the intestinal epithelium has revealed the presence of epithelial-associated commensal bacteria that include members of the Bacteroidetes and Firmicutes phyla at levels of about 2–3 logs less than those observed for luminal bacteria (28, 52). The most notable example in mice is SFB, a member of the Firmicutes phylum, which colonizes and adheres tightly to IECs of the terminal ileum (24, 25). Colonization of mice with SFB results in elevated Th17 cell responses in the small intestine, which are associated with both intestinal tissue protection from mucosal pathogen infection and susceptibility to mouse models of autoimmune arthritis and experimental autoimmune encephalopathy (61, 62). Studies addressing the mechanism(s) by which SFB induces Th17 cells demonstrated that SFB-colonized mice exhibited increased intestinal expression of the acute-phase response protein serum amyloid A (SAA), which conditions lamina propria DCs to produce Th17 cell-inducing cytokines IL-6 and IL-23 (25). Therefore, SFB is an epithelial-associated commensal bacterium that can elicit inflammatory immune responses that are, depending on context, tissue-protective or pathologic.
Direct recognition of SFB is likely mediated by lamina propria DCs that actively sample bacteria at the apical surface of epithelial cells (Figure 3). Subsets of lamina propria DCs that express the fractalkine receptor CX3CR1 have been shown to sample antigens by extending transepithelial dendrites across the intestinal epithelium (63). Although these DCs are believed to be non-migratory and only stimulate local mucosal immune responses, one report suggests that they could carry commensal bacteria antigens to the mLN to stimulate both T cell and IgA responses (64). Interestingly, migration of CX3CR1-expressing DCs to the mLN and subsequent mucosal immune responses were regulated by recognition of commensal bacteria through MYD88 (64). In contrast, it was recently demonstrated that CX3CR1+ macrophages in the small intestine can acquire and transfer soluble antigens via the gap junction protein connexin 43 to CD103+ DCs, which then migrate to draining lymph nodes to induce antigen-specific T cell responses (65).
Figure 3.
Colonization of the mammalian GI tract by epithelial-associated commensal bacteria promotes pro-inflammatory immune responses and is regulated by distinct host mechanisms. SFB is a model epithelial-associated commensal bacterium that colonizes the small intestine by adhering to IECs. SFB can promote pro-inflammatory immune responses that are associated with both pathogen immunity and autoimmunity. Some members of the luminal commensal flora such as B. fragilis and Clostridia can also colonize the epithelial surface of the colon and may interact with IECs.
Similar to their functions in regulating luminal commensal bacteria, host mechanisms such as AMPs and IgA are critical regulators of epithelial-associated commensal bacteria. Consistent with the specificity of REGIIIγ for gram-positive bacteria, REGIIIγ downstream of MYD88 signaling was demonstrated to restrict expansion of SFB on the epithelial surface as Reg3g-deficient mice had increased epithelial colonization of SFB and another epithelial-associated bacterium Eubacterium rectale in the small intestine (52). Increased commensal colonization at intestinal epithelial surfaces due to loss of REGIIIγ resulted in increased frequencies of lamina propria T helper 1 (Th1) but not Th17 cells. Since total levels of SFB were not altered in Reg3g-deficient mice, these data suggest that dysregulated epithelial colonization by SFB may skew towards a Th1 response. A role for AMPs in restricting colonization of epithelial-associated commensal bacteria was also demonstrated in mice ectopically expressing the human α-defensin gene Defa5 (66). Analyses of the microbiota composition in the distal small intestine of these mice demonstrated that while the amount of total bacteria was similar to littermate controls, Defa5-transgenic (Defa5-Tg) mice had significantly reduced colonization of SFB. In support of a role for AMPs in regulating epithelial-associated bacterial species, cytokine responses that promote AMP expression, such as IL-22, were recently demonstrated to suppress SFB colonization (67, 68). In addition, pathways and molecules associated with IL-22 production in the intestine, such as the aryl hydrocarbon receptor (Ahr) and RORγt+ ILC responses also direct colonization of the intestinal epithelial surface (67). Ahr-deficient mice, which have reduced RORγt+ ILCs, demonstrated expansion of SFB and increased small intestine Th17 cells (67). Finally, IgA is also involved in restricting colonization by SFB as demonstrated by expansion of this commensal bacterium in the feces of Iga-deficient mice (69). Collectively, these data highlight a critical role for these biochemical and cellular mechanisms in regulating colonization of SFB on the intestinal epithelial surface. Further investigation may reveal whether other epithelial-associated commensal bacteria are similarly restricted by AMPs and IgA.
Intriguingly, commensal bacteria that are found frequently in the lumen of the intestine can also be epithelial-associated. Electron microscopy (EM) and immunofluorescence revealed that B. fragilis could penetrate the inner mucus layer and colonize the crypts of the large intestine. This was demonstrated to be dependent on both host-derived and bacterial-intrinsic factors. Colonic crypt colonization by B. fragilis was dependent on induction of Tregs by PSA (22). In a more recent study, it was shown that expression of commensal colonization factor (ccf) genes by B. fragilis could promote colonization of the bacteria at colonic crypts. It was hypothesized that host-derived glycan structures could induce the expression of ccf genes, consistent with the dependence of Bacteroides speces on host-derived carbohydrates for intestinal colonization and survival (33). It has also been reported that B. fragilis, although an anaerobe, can tolerate and benefit from nanomolar concentrations of oxygen, which is higher at the epithelial surface relative to the lumen of the intestine (70). Additionally, although Clostridium spp. typically reside in the intestinal lumen, EM analysis of the colon of Clostridium spp. colonized mice also demonstrated their presence on the epithelial surface (20). In support of a role for Clostridium spp. in modulating epithelial cell responses, cecal extracts from Clostridium-colonized mice could induce primary IECs to produce the Treg-inducing cytokine TGF-β (20). Taken together, these data suggest that commensal bacteria-elicited immune responses and commensal bacteria-intrinsic factors coordinate to permit stable intestinal colonization by epithelial-associated commensal bacteria. Further analyses are required to delineate whether the immune cell responses promoted by Bacteroides and Clostridia members are associated with their localization in either the lumen or the epithelial surface.
Lymphoid tissue-resident commensal bacteria
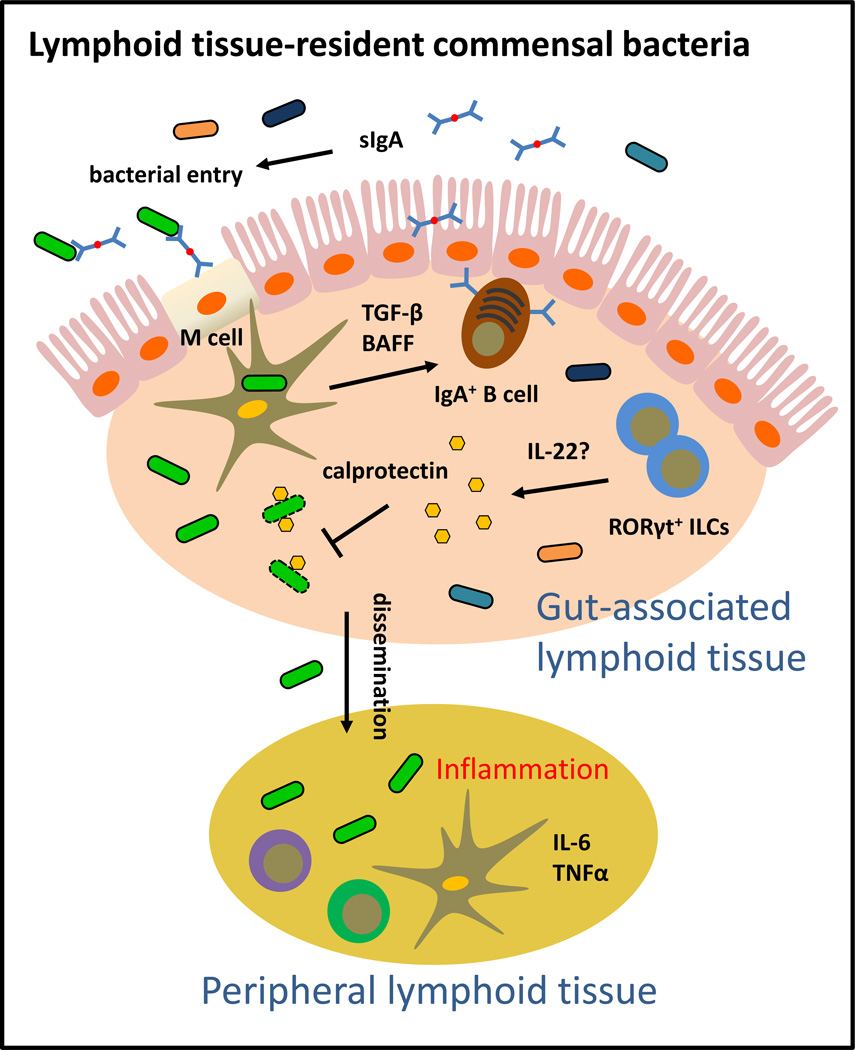
Tissues beyond the intestinal epithelium are not thought of as typical sites for commensal bacteria colonization. Although pathogens can penetrate the intestinal epithelium, evade macrophage killing and in some cases thrive in intestinal tissues, commensal bacteria that breach the intestinal barrier are typically phagocytosed and cleared by lamina propria macrophages (71–73). However, recent data provided evidence that some commensal species could stably colonize the interior of gut-associated lymphoid tissues such as PPs and mLN of healthy mice and humans (26, 27) (Figure 4). In a seminal study by Kiyono and colleagues, sequencing of 16S rDNA in the interior of intestinal lymphoid tissues revealed the presence of specific groups of commensal bacteria including Alcaligenes spp., Ochrobactrum spp., Serratia spp. and Burkholderia spp. Alcaligenes spp. represented the most abundant lymphoid tissue-resident commensal in the PPs. It was observed that 16S rDNA of some of these bacteria were detected in the PP DCs of healthy mice, suggesting the possibility of intracellular survival in DCs. These data support a hypothesis in which lymphoid tissue-resident commensal bacteria can preferentially access host lymphoid tissues and potentially evade clearance by host immune cells. Indeed, Alcaligenes spp. can be transcytosed by specialized IECs known as M cells from the lumen of the small intestine to PPs (74).
Figure 4.
Lymphoid-tissue resident commensal bacteria can colonize gut-associated lymphoid tissues such as Peyer’s patches, isolated lymphoid follicles and mesenteric lymph nodes. Lymphoid tissue-resident commensal bacteria share many properties: they are typically aerobic, environmental microbes that are broadly resistant to antibiotics. One member of this group, Alcaligenes spp., is a potent inducer of intestinal IgA responses. Anatomical containment of Alcaligenes spp. is mediated by IL-22-producing RORγt+ innate lymphoid cells. Loss of this pathway results in dissemination of Alcaligenes spp., systemic inflammation and tissue pathology.
It is interesting to note that all of the lymphoid tissue-resident species identified so far are also aerobic, environmental microbes, resistant to a broad spectrum of antibiotics and associated with opportunistic nosocomial infections, particularly in the respiratory tract (75–78). Some of these species, such as Burkholderia spp. and Serratia spp. are well adapted to survive on inanimate surfaces and also in host cells (79, 80). Whether these features are related to their unique colonization pattern in the intestine is not well understood and requires further investigation. However, analyses in Alcaligenes-colonized mice demonstrated that Alcaligenes spp. is sufficient to promote robust Alcaligenes-specific and non-Alcaligenes-specific IgA (26). Furthermore, B cell-deficient CBA/N xid and Iga-deficient mice had reduced numbers of Alcaligenes spp. in the PP. These data suggest that in contrast to luminal and epithelial-associated commensal bacteria, which are restricted by IgA, optimal colonization by Alcaligenes spp. is dependent on IgA-producing B cells (26). Consistent with the notion that mucosal DCs are restricted from migrating beyond the mLN to systemic lymphoid tissues (81), Alcaligenes-specific IgG responses were not detected in the periphery of Alcaligenes-colonized mice (26).
Recently, anatomical containment of the lymphoid tissue-resident-commensal bacteria Alcaligenes xylosoxidans (also referred to as Achromobacter xylosoxidans) was demonstrated to be dependent on IL-22 produced by RORγt+ ILCs (27) (Figure 4). Rag1-deficient mice depleted of RORγt+ ILCs experienced low-grade systemic inflammation that was associated with dissemination of Alcaligenes spp. to the liver, spleen and blood (27). Administration of IL-22 or the AMP calprotectin to ILC-depleted Rag1-deficient mice partially restored anatomical containment of Alcaligenes xyloxidans, suggesting a role for IL-22 induced factors such as AMPs in restricting Alcaligenes to intestinal lymphoid tissues (27). More recently, it was revealed that secretory IgA from breast milk could restrict translocation of Ochrobactrum anthropi from the neonatal gut to the draining lymph node, suggesting a role for IgA in anatomical containment of other lymphoid tissue-resident commensal bacteria (82). These data provide potential mechanisms for anatomical restriction of Alcaligenes spp. and also highlight that dysregulated localization of normally beneficial commensal bacteria can promote pathological inflammation, a condition often associated with human chronic inflammatory diseases. This idea will be discussed further in the following section. However, despite these advances, lymphoid tissue-residexnt commensal bacteria are still the least understood class of bacteria that colonize the intestine. Since most studies on the composition of the mammalian microbiota have focused on fecal specimens, future studies would benefit from 16S rDNA sequencing of not only intestinal luminal contents but also intestinal lymphoid tissues. Further investigation will be necessary to define the precise host and microbial pathways that promote tissue colonization patterns of lymphoid-resident commensal species as well as determine the functional significance of lymphoid tissue-resident commensal colonization in the context of health and disease.
Anatomical localization of commensal bacteria in disease
Analyses of the human and mouse microbiota have established that in some contexts, commensal bacteria can promote chronic inflammatory disease through dysbiosis and/or bacterial translocation (9, 11). Most studies thus far have focused on how changes in the composition of the luminal microbiota can influence the development of disease. For example, IBD and obesity were associated with a dysbiotic microbiota that was sufficient to transfer disease to healthy hosts following cohousing or fecal transfer (83, 84). The role of dysbiosis in other chronic diseases such as AIDS and cancer has been extensively reviewed elsewhere (10, 14, 85–89). Alternatively, prebiotic and probiotic administration of beneficial microbes have been reported to promote intestinal health and limit pathologic inflammation (90, 91). However, an emerging theme of the studies performed in the last decade is that dysregulated localization, not just dysbiosis, of normally beneficial commensal bacteria can be detrimental to the host and is often a major contributing factor to the development and pathogenesis of chronic inflammatory diseases. Chronic viral infections, IBD, metabolic syndromes and cancer have all been associated with dysregulated localization of commensal bacteria. In the following sections, we will highlight disease settings where anatomical restriction of luminal, epithelial-associated and lymphoid tissue-resident commensal bacteria is disrupted and may contribute to chronic inflammatory disease.
Chronic viral infections
Chronic viral infections are strongly associated with the loss of intestinal barrier function, commensal bacteria translocation to peripheral tissues and induction of systemic inflammation in humans (15, 92–95). A seminal finding by Douek and colleagues demonstrated that patients chronically infected with HIV, and those that have progressed to AIDS, exhibit elevated plasma LPS levels, suggesting that translocation of gram-negative bacteria components or whole gram-negative bacteria occurs in these diseases (15, 95). Consistent with elevated plasma LPS, the peripheral blood of AIDS patients exhibited signs of systemic immune activation such as the presence of elevated serum IFNα, activated monocytes and activated circulating CD8+ T cells, hallmarks that contributes to viral replication, dissemination and disease progression (15, 95, 96). Interestingly, SIV-infected sooty mangabeys, which are natural hosts of SIV that do not develop AIDS, did not exhibit loss of intestinal barrier function, bacterial translocation or systemic inflammation (97). Bacterial translocation has been observed in other chronic viral infections such as HCV and HBV. Patients infected with HCV and HBV had elevated plasma LPS, which was associated with parameters of systemic immune activation such as elevated serum levels of the TLR4-co-receptor CD14 and IL-6 (98). Taken together, these data suggest that loss of anatomical restriction of gram-negative commensal bacteria-derived products contribute to the pathogenesis of chronic viral infections.
Although it remains unclear whether whole commensal bacteria are found in the periphery of these patients or which commensal species promote disease progression, recent studies suggest that HIV and HCV-infected patients have elevated serum IgG responses to the lymphoid tissue-resident commensal bacteria Alcaligenes (27, 99). In support of a role for these bacteria in disease pathogenesis and systemic inflammation, levels of Alcaligenes/Achromobacter-specific IgG were associated with hallmarks of AIDS such as reduced CD4+ T cell counts and increased proportion of activated CD4+ T cells, and parameters of HCV progression including increased serum bilirubin and decreased serum albumin and platelets. Potential translocation of Achromobacter/Alcaligenes members in HIV-infected patients agrees with previous reports that Burkholderiales 16S rDNA, which includes the genus Achromobacter and Alcaligenes, was present in the peripheral blood of some patients (100). Other groups of bacteria such as members of the orders Enterobacteriales, Lactobacillales and Pseudomonadales were also identified in the serum of these patients.
Recent findings suggest that loss of cellular pathways that promote intestinal epithelial barrier function may contribute to bacterial translocation observed in chronic HIV and HCV infection. RORγt+ ILCs, which are potent sources of IL-17 and IL-22, and the CD103+ immunoregulatory DC subset were found to be dramatically reduced in SIV infection (101–103). These data are consistent with the role of RORγt+ ILCs in regulating the anatomical containment of SFB and Alcaligenes spp. (27, 67). Given that antiretroviral therapy is inadequate to completely control bacterial translocation (100), further work is required to define the pathways that regulate intestinal barrier integrity and systemic inflammation in chronic viral infections. It is also important to highlight that patients with chronic viral infections often exhibit intestinal, metabolic and cardiovascular symptoms, which have all been associated with dysregulated localization of commensal bacteria (104–107). Therefore, it is possible that bacterial translocation observed in HIV and HCV patients may be related to the onset of these other diseases and thus warrants additional investigation.
Inflammatory bowel disease
IBD, which includes Crohn’s disease (CD) and ulcerative colitis (UC), is also strongly associated with dysregulated localization of commensal bacteria in humans. It is well documented that dysregulated host-commensal bacteria interactions, as indicated by parameters such as elevated serum LPS and bacterial DNA, plays a major role in the pathogenesis of IBD by promoting inappropriate immune activation and chronic inflammation (17, 108–111). Changes in localization of commensal bacteria in IBD have been observed in the form of both bacterial translocation and increased bacterial colonization on the surface of the intestinal epithelium. Studies have implicated a role for increased colonization of the ileum by an adherent-invasive strain of Escherichia coli (AIEC). The predominance of AIEC epithelial colonization was demonstrated to contribute to chronic inflammation and immunopathology in human IBD (61, 112, 113). To analyze the composition of disseminated commensal bacteria in gut-associated lymphoid tissues, one recent report identified that Escherichia and the mucosal pathogen Shigella were enriched in the mLN of CD patients (113, 114). Similar findings were reported by a study that sequenced 16S rDNA from microdissected intestinal biopsies. Facultative anaerobes such as Enterobacteriaceae were enriched in microdissected ulcer biopsies of CD patients (115). Collectively, these data suggest support the hypothesis that increased epithelial and lymphoid tissue colonization by commensal bacteria are hallmarks of and may contribute to the pathogenesis of IBD. To further investigate bacterial translocation to extra-intestinal sites in IBD, one study examined and identified the presence of Enterococcus faecalis, a common member of the luminal flora, to the visceral fat of UC and CD patients (116). In addition, it was recently demonstrated that pediatric CD patients exhibit increased serum IgG specific for Alcaligenes spp. (27), suggesting that impaired localization and systemic dissemination of lymphoid tissue-resident commensal bacteria is also associated with disease pathogenesis.
While it is unclear whether translocation of commensal bacteria is the cause or effect of IBD, several genetic polymorphisms in genes involved in host-microbial interactions are strongly associated with IBD (117). Among these, the most prevalent are polymorphisms in Atg16l1, Il23r, Nod2 and Il10r (118, 119). ATG16L1, a component of the autophagy machinery, is involved in intracellular degradation of bacteria (120). IL-23R is expressed on innate and adaptive immune cells that promote immunity to intestinal pathogens and maintain intestinal barrier function (121). NOD2 is a pattern recognition receptor (PRR) that recognizes peptidoglycan from bacteria and triggers anti-bacterial immune responses (122). IL-10 is a potent anti-inflammatory cytokine produced by innate and adaptive immune cells that maintain tissue homeostasis by suppressing pro-inflammatory responses (123, 124). Therefore, loss of function mutations in these genes may contribute to enhanced bacterial translocation and intestinal inflammation. More studies will be required to delineate the functional role of these genetic polymorphisms and disseminated species in disrupting the intestinal immune microenvironment and promoting chronic intestinal inflammation.
Metabolic diseases
Diabetes and obesity were two of the first extra-intestinal diseases to be associated with commensal bacteria-derived chronic inflammation (1, 125). In a mouse model of diet-induced obesity, mice fed a high-fat diet (HFD) for four weeks had increased concentrations of bacterial DNA and LPS in the blood, mesenteric adipose tissue and mLN as well as increased adherence of commensal bacteria to the intestinal epithelial (126, 127). Analysis of translocated bacteria species in the mesenteric adipose tissue identified members of the family Enterobacteriaceae and Enterococcus spp. and Lactobacillus spp. (127), which are commensal bacteria commonly found in the lumen of the intestine. Translocation of commensal bacteria was associated with a loss of intestinal barrier integrity (127), increased levels of IL-1, IL-6 and TNFα in adipose depots and the onset of diabetes. Many of these parameters were markedly reduced in HFD-fed mice lacking the intracellular PRR Nod1 (127, 128), TLR4 (128) or the TLR4-co-receptor CD14 (126, 127), suggesting that recognition of translocated commensal bacteria can drive tissue inflammation and disease pathology. In support of these findings, repeated subcutaneous injections of LPS to mice fed a normal chow (NC) was sufficient to recapitulate many of the diabetic symptoms observed in mice fed a HFD (126). Altogether, these findings indicate that impaired anatomical localization is a hallmark and significant contributor of diet-induced obesity.
Obesity is associated with significant changes in the composition of the luminal microbiota. For example, mice deficient in TLR5 are highly susceptible to diet-induced obesity, which was associated with a dysbiotic microbiota that could confer disease to wild-type germ free mice following fecal transfer (84). In contrast, mice deficient in lymphotoxin β signaling are resistant to diet-induced obesity, and cecal contents from HFD-fed lymphotoxin β receptor (Ltbr)-deficient mice were less efficient in promoting weight gain in wild-type germ free recipients compared to cecal contents from HFD-fed wild-type mice (68). Interestingly, normal chow diet (NCD)-fed Ltbr-deficient mice, which lacked IL-23 and IL-22, demonstrated increased colonization by SFB. Whereas SFB levels were reduced in HFD-fed littermates, SFB levels in HFD-fed Ltbr-deficient mice remained high relative to littermates. Furthermore, administration of IL-22 or IL-23-Ig restored HFD-induced changes in SFB and obesity in Ltbr-deficient mice. Although bacterial translocation was not analyzed in these studies, it is interesting to speculate that specific groups of commensal bacteria that promote or limit disease are more or less likely, respectively, to disseminate to adipose depots and subsequently trigger chronic inflammation. Nevertheless, these data support the hypothesis that specific immune pathways and/or commensal communities can promote (e.g. LTβ signaling, Enterobacteriaceae) or limit (e.g. TLR5, SFB) the development of obesity.
Other metabolic diseases are also associated with signs of dysbiosis and loss of normal localization of commensal bacteria in the intestine. For example, plasma LPS levels are significantly elevated in patients with chronic hepatitis, alcoholic and non-alcoholic liver disease and have been associated with severe liver dysfunction (129–131). In a mouse model of non-alcoholic fatty liver disease (NAFLD), it was demonstrated that a dysbiotic microbiota in inflammasome-deficient mice could promote hepatic steatosis and Tnf expression due to translocation of TLR4 and TLR9 agonists through the portal circulation to the liver (18). Although translocation of intact bacteria was not detected in the portal vein or liver, these data suggest that translocation of commensal-derived products alone to extra-intestinal sites can promote immune activation and tissue pathology in this mouse model of liver disease.
Cardiovascular disease
An increasing body of literature has revealed that the inflammatory and metabolic effects of the gut microbiota contribute to the development and pathogenesis of cardiovascular disease (132, 133). Several studies have identified changes in the composition of luminal commensal bacteria and investigated the mechanisms by which commensal-dependent metabolic pathways can impact disease. In one study, fecal microbiota analyses of healthy control and atherosclerosis patients demonstrated a selective enrichment of the genus Collinsella and reduction in the genus Eubacterium and Roseburia in patients (134). By measuring the metabolic activity of the luminal microbiota, it was determined that the microbiota of healthy controls was enriched for pathways involved in the synthesis of anti-inflammatory and anti-oxidant compounds whereas the microbiota of atherosclerosis patients was enriched for genes in the peptidoglycan biosynthesis pathway. These data suggest that changes in composition of luminal commensal bacteria can alter the inflammatory and metabolic status of the host to promote atherosclerosis. In addition, a separate study demonstrated that increased levels of a commensal-dependent dietary metabolite, trimethylamine-N-oxide (TMAO), were associated with an increased risk of adverse cardiovascular events (135). This is supported by studies in which metabolism of the dietary metabolite L-carnitine, which is abundant in red meat, produces TMAO and accelerates onset of atherosclerosis (136). Taken together, these findings establish a link between luminal commensal bacteria, metabolism and atherosclerosis. Despite these findings, much less is known about the role of anatomical localization of commensal bacteria in cardiovascular disease. However, one study sequenced 16S rDNA from atherosclerotic plaques of human patients and demonstrated the presence of bacteria that are normal inhabits of the intestinal lumen including members of the Bacteroidaceae, Enterobacteriaceae and Lachnospiraceae families (137). These data suggest that dissemination of commensal bacteria or commensal bacteria-derived products to the circulation may be associated with the development and pathogenesis of atherosclerosis. However, additional studies are necessary to interrogate the origin, cause and contribute of commensal bacteria in atherosclerotic plaques.
Cancer
As previously discussed, translocation of commensal bacteria is commonly observed in chronic liver diseases and can contribute to liver fibrosis and cirrhosis (129–131). Hepatocellular carcinoma (HCC) is a leading cause of death in patients that develop fibrotic and cirrhotic livers due to chronic inflammation. In support of the hypothesis that bacterial translocation promotes liver tumor development, Tlr4-deficient mice, germ free mice and mice treated with oral antibiotics were protected from hepatocarcinogenesis in a mouse model of chronic liver injury (138). This is consistent with a previous report that LPS-induced liver fibrosis was reduced in Myd88-deficient mice and that hepatic satellite cells are the dominant target by which TLR4 ligands promote liver fibrosis (139). Another cancer associated with dysregulated anatomical location of commensal bacteria is colorectal cancer (CRC). Bacterial translocation has been observed in the mLN and serum of CRC patients (140–142). These patients also exhibited cachexia and elevated serum pro-inflammatory cytokine levels and had a poor prognosis. However, studies on specific groups of commensal bacteria that disseminate in human colon cancer are limited. Fusobacterium nucleatum, a rare member of the human intestinal microbiota, was demonstrated to be strongly associated with human colorectal carcinoma and was found to be enriched in human colonic adenomas (143). In contrast to commensal bacteria such as the enterotoxigenic B. fragilis and AIEC which promote colitis-associated cancer (144, 145), F. nucleatum did not promote colitis in mice, but rather increased tumor multiplicity, recruited tumor-promoting myeloid cells and generated a pro-inflammatory environment that favored tumor development. Since F. nucleatum was also enriched in the fecal microbiota of CRC patients, it is likely that increased luminal burden of F. nucleatum contributes to the accumulation of this bacterium in colonic adenomas. Further studies are required to investigate whether other commensal bacteria can colonize cancerous tissues in the intestine and how their colonization influence cancer development.
Concluding remarks
Mammals have evolved to coexist with over 100 trillion commensal bacteria in the GI tract. It is clear that interactions between commensal bacteria and the host are tightly coordinated to promote numerous biological processes important for mammalian health ranging from metabolism, pathogens immunity to immune cell homeostasis. However, dysregulated host-commensal bacteria interactions are detrimental to host health and are associated with an expanding list of chronic inflammatory diseases. While current studies on the role of the microbiota in disease have focused on transient or permanent shifts in the luminal composition of the commensal flora and microbiota-derived metabolites, altered anatomical localization of commensal bacteria is often associated with and may contribute to the development of chronic inflammatory diseases. However, it remains unclear how these bacteria with altered anatomical localization patterns in the context of inflammatory diseases become dysregulated. It is possible that these species disseminated from intestinal compartments, were acquired from the environment, or were present at very low levels but can expand under inflammatory conditions. To improve our understanding of host-commensal relationships during health and disease, further investigation is necessary to 1) profile not only luminal but also epithelial-associated and lymphoid tissue-resident commensal bacteria during the steady state and disease, 2) profile and determine the source of bacteria that are present at extra-intestinal tissues during disease and 3) examine whether settings of impaired commensal bacteria localization is associated with loss of immune pathways that maintain intestinal barrier function. Studying the immunological mechanisms that promote anatomical containment of commensal bacteria in health and disease may identify new therapeutic targets for the treatment of commensal bacteria-associated chronic inflammation.
Acknowledgements
We thank members of the Sonnenberg and Artis laboratories for discussions and critical reading of the manuscript. TCF is supported by the American Heart Association Predoctoral Fellowship and the Cancer Research Institute Student Training and Research in Tumor Immunology grant. Research in the Artis lab is supported by the National Institutes of Health (R01AI095466 and R01AI102942). Research in the Sonnenberg laboratory is supported by the National Institutes of Health (DP5OD012116), the NIAID Mucosal Immunology Studies Team (MIST) Scholar Award in Mucosal Immunity, a pilot grant from the NIDDK P30 center for Molecular Studies in Digestive and Liver Diseases (P30DK50306) and the Institute for Translational Medicine and Therapeutics Transdisciplinary Program in Translational Medicine and Therapeutics (UL1-RR024134 from the US National Center for Research Resources).
Abbreviations used
- GI
gastrointestinal
- IBD
inflammatory bowel disease
- SFB
segmented filamentous bacteria
- PP
Peyer’s patch
- mLN
mesenteric lymph node
- PSA
polysaccharide A
- IL
interleukin
- TNFα
tumor necrosis factor alpha
- IFNγ
interferon gamma
- DC
dendritic cell
- APC
antigen presenting cell
- TLR
Toll-like receptor
- iNKT cell
invariant natural killer T cell
- ASD
autism spectrum disorder
- AMP
antimicrobial peptide
- REGIIIγ
regenerating islet-derived protein 3 gamma
- ILF
isolated lymphoid follicle
- ILC
innate lymphoid cell
- SAA
serum amyloid A
- Ahr
aryl hydrocarbon receptor
- EM
electron microscopy
- ccf
commensal colonization factor
- HIV
human immunodeficiency virus
- AIDS
acquired immune deficiency syndrome
- HCV
hepatitis C virus
- HBV
hepatitis B virus
- CD
Crohn’s disease
- UC
ulcerative colitis
- AIEC
adherent-invasive Escherichia coli
- HFD
high-fat diet
- NCD
normal chow diet
- TMAO
trimethylamine-N-oxide
- LPS
lipopolysaccharide
- CRC
colorectal cancer
- IEC
intestinal epithelial cell
- PRR
pattern recognition receptor
References
- 1.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 2.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nature reviews Immunology. 2013;13:790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamada N, Chen GY, Inohara N, Nunez G. Control of pathogens and pathobionts by the gut microbiota. Nature immunology. 2013;14:685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Musso G, Gambino R, Cassader M. Interactions between gut microbiota and host metabolism predisposing to obesity and diabetes. Annual review of medicine. 2011;62:361–380. doi: 10.1146/annurev-med-012510-175505. [DOI] [PubMed] [Google Scholar]
- 6.Flint HJ, Scott KP, Louis P, Duncan SH. The role of the gut microbiota in nutrition and health. Nature reviews Gastroenterology & hepatology. 2012;9:577–589. doi: 10.1038/nrgastro.2012.156. [DOI] [PubMed] [Google Scholar]
- 7.Brestoff JR, Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nature immunology. 2013;14:676–684. doi: 10.1038/ni.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cani PD. Metabolism in 2013: The gut microbiota manages host metabolism. Nature reviews Endocrinology. 2014;10:74–76. doi: 10.1038/nrendo.2013.240. [DOI] [PubMed] [Google Scholar]
- 9.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ivanov II, Honda K. Intestinal commensal microbes as immune modulators. Cell host & microbe. 2012;12:496–508. doi: 10.1016/j.chom.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamada N, Seo SU, Chen GY, Nunez G. Role of the gut microbiota in immunity and inflammatory disease. Nature reviews Immunology. 2013;13:321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- 12.Blumberg R, Powrie F. Microbiota, disease, and back to health: a metastable journey. Science translational medicine. 2012;4:137rv137. doi: 10.1126/scitranslmed.3004184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loh G, Blaut M. Role of commensal gut bacteria in inflammatory bowel diseases. Gut microbes. 2012;3:544–555. doi: 10.4161/gmic.22156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nature reviews Gastroenterology & hepatology. 2012;9:599–608. doi: 10.1038/nrgastro.2012.152. [DOI] [PubMed] [Google Scholar]
- 15.Brenchley JM, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nature medicine. 2006;12:1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 16.Wang Z, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brenchley JM, Douek DC. Microbial translocation across the GI tract. Annual review of immunology. 2012;30:149–173. doi: 10.1146/annurev-immunol-020711-075001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henao-Mejia J, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–185. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Atarashi K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 21.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Round JL, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, Mazmanian SK. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501:426–429. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaboriau-Routhiau V, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 25.Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Obata T, et al. Indigenous opportunistic bacteria inhabit mammalian gut-associated lymphoid tissues and share a mucosal antibody-mediated symbiosis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:7419–7424. doi: 10.1073/pnas.1001061107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sonnenberg GF, et al. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science. 2012;336:1321–1325. doi: 10.1126/science.1222551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hill DA, et al. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal immunology. 2010;3:148–158. doi: 10.1038/mi.2009.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colgan SP, Taylor CT. Hypoxia: an alarm signal during intestinal inflammation. Nature reviews Gastroenterology & hepatology. 2010;7:281–287. doi: 10.1038/nrgastro.2010.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glover LE, Colgan SP. Hypoxia and metabolic factors that influence inflammatory bowel disease pathogenesis. Gastroenterology. 2011;140:1748–1755. doi: 10.1053/j.gastro.2011.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salzman NH, de Jong H, Paterson Y, Harmsen HJ, Welling GW, Bos NA. Analysis of 16S libraries of mouse gastrointestinal microflora reveals a large new group of mouse intestinal bacteria. Microbiology (Reading, England) 2002;148:3651–3660. doi: 10.1099/00221287-148-11-3651. [DOI] [PubMed] [Google Scholar]
- 32.A framework for human microbiome research. Nature. 2012;486:215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wexler HM. Bacteroides: the good, the bad, and the nitty-gritty. Clinical microbiology reviews. 2007;20:593–621. doi: 10.1128/CMR.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Comstock LE. Importance of glycans to the host-bacteroides mutualism in the mammalian intestine. Cell host & microbe. 2009;5:522–526. doi: 10.1016/j.chom.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 35.Surana NK, Kasper DL. The yin yang of bacterial polysaccharides: lessons learned from B. fragilis PSA. Immunological reviews. 2012;245:13–26. doi: 10.1111/j.1600-065X.2011.01075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.An D, et al. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell. 2014;156:123–133. doi: 10.1016/j.cell.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wieland Brown LC, et al. Production of alpha-galactosylceramide by a prominent member of the human gut microbiota. PLoS biology. 2013;11:e1001610. doi: 10.1371/journal.pbio.1001610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buie T, et al. Evaluation, diagnosis, and treatment of gastrointestinal disorders in individuals with ASDs: a consensus report. Pediatrics. 2010;125(Suppl 1):S1–S18. doi: 10.1542/peds.2009-1878C. [DOI] [PubMed] [Google Scholar]
- 39.Adams JB, Johansen LJ, Powell LD, Quig D, Rubin RA. Gastrointestinal flora and gastrointestinal status in children with autism--comparisons to typical children and correlation with autism severity. BMC gastroenterology. 2011;11:22. doi: 10.1186/1471-230X-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coury DL, et al. Gastrointestinal conditions in children with autism spectrum disorder: developing a research agenda. Pediatrics. 2012;130(Suppl 2):S160–S168. doi: 10.1542/peds.2012-0900N. [DOI] [PubMed] [Google Scholar]
- 41.Kohane IS, et al. The co-morbidity burden of children and young adults with autism spectrum disorders. PloS one. 2012;7:e33224. doi: 10.1371/journal.pone.0033224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hsiao EY, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Furusawa Y, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 44.Arpaia N, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith PM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Loosdregt J, et al. Regulation of Treg functionality by acetylation-mediated Foxp3 protein stabilization. Blood. 2010;115:965–974. doi: 10.1182/blood-2009-02-207118. [DOI] [PubMed] [Google Scholar]
- 47.Zhang H, Xiao Y, Zhu Z, Li B, Greene MI. Immune regulation by histone deacetylases: a focus on the alteration of FOXP3 activity. Immunology and cell biology. 2012;90:95–100. doi: 10.1038/icb.2011.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johansson ME, Larsson JM, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(Suppl 1):4659–4665. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang JY, Lee SM, Mazmanian SK. The human commensal Bacteroides fragilis binds intestinal mucin. Anaerobe. 2011;17:137–141. doi: 10.1016/j.anaerobe.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van der Sluis M, et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology. 2006;131:117–129. doi: 10.1053/j.gastro.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 51.Shan M, et al. Mucus enhances gut homeostasis and oral tolerance by delivering immunoregulatory signals. Science. 2013;342:447–453. doi: 10.1126/science.1237910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vaishnava S, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mukherjee S, et al. Antibacterial membrane attack by a pore-forming intestinal C-type lectin. Nature. 2014;505:103–107. doi: 10.1038/nature12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Corbin BD, et al. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science. 2008;319:962–965. doi: 10.1126/science.1152449. [DOI] [PubMed] [Google Scholar]
- 55.Kehl-Fie TE, et al. Nutrient metal sequestration by calprotectin inhibits bacterial superoxide defense, enhancing neutrophil killing of Staphylococcus aureus. Cell host & microbe. 2011;10:158–164. doi: 10.1016/j.chom.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu JZ, et al. Zinc sequestration by the neutrophil protein calprotectin enhances Salmonella growth in the inflamed gut. Cell host & microbe. 2012;11:227–239. doi: 10.1016/j.chom.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Diaz-Ochoa VE, Jellbauer S, Klaus S, Raffatellu M. Transition metal ions at the crossroads of mucosal immunity and microbial pathogenesis. Frontiers in cellular and infection microbiology. 2014;4:2. doi: 10.3389/fcimb.2014.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cerutti A, Rescigno M. The biology of intestinal immunoglobulin A responses. Immunity. 2008;28:740–750. doi: 10.1016/j.immuni.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Macpherson AJ, McCoy KD, Johansen FE, Brandtzaeg P. The immune geography of IgA induction and function. Mucosal immunology. 2008;1:11–22. doi: 10.1038/mi.2007.6. [DOI] [PubMed] [Google Scholar]
- 60.Macpherson AJ, Geuking MB, McCoy KD. Homeland security: IgA immunity at the frontiers of the body. Trends in immunology. 2012;33:160–167. doi: 10.1016/j.it.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 61.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(Suppl 1):4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu HJ, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Niess JH, et al. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science. 2005;307:254–258. doi: 10.1126/science.1102901. [DOI] [PubMed] [Google Scholar]
- 64.Diehl GE, et al. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX(3)CR1(hi) cells. Nature. 2013;494:116–120. doi: 10.1038/nature11809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mazzini E, Massimiliano L, Penna G, Rescigno M. Oral Tolerance Can Be Established via Gap Junction Transfer of Fed Antigens from CX3CR1(+) Macrophages to CD103(+) Dendritic Cells. Immunity. 2014;40:248–261. doi: 10.1016/j.immuni.2013.12.012. [DOI] [PubMed] [Google Scholar]
- 66.Salzman NH, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nature immunology. 2010;11:76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Qiu J, et al. Group 3 innate lymphoid cells inhibit T-cell-mediated intestinal inflammation through aryl hydrocarbon receptor signaling and regulation of microflora. Immunity. 2013;39:386–399. doi: 10.1016/j.immuni.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Upadhyay V, et al. Lymphotoxin regulates commensal responses to enable diet-induced obesity. Nature immunology. 2012;13:947–953. doi: 10.1038/ni.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Suzuki K, et al. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:1981–1986. doi: 10.1073/pnas.0307317101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Baughn AD, Malamy MH. The strict anaerobe Bacteroides fragilis grows in and benefits from nanomolar concentrations of oxygen. Nature. 2004;427:441–444. doi: 10.1038/nature02285. [DOI] [PubMed] [Google Scholar]
- 71.Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nature reviews Immunology. 2004;4:478–485. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- 72.Varol C, Zigmond E, Jung S. Securing the immune tightrope: mononuclear phagocytes in the intestinal lamina propria. Nature reviews Immunology. 2010;10:415–426. doi: 10.1038/nri2778. [DOI] [PubMed] [Google Scholar]
- 73.Smith PD, Smythies LE, Shen R, Greenwell-Wild T, Gliozzi M, Wahl SM. Intestinal macrophages and response to microbial encroachment. Mucosal immunology. 2011;4:31–42. doi: 10.1038/mi.2010.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sato S, et al. Transcription factor Spi-B-dependent and -independent pathways for the development of Peyer's patch M cells. Mucosal immunology. 2013;6:838–846. doi: 10.1038/mi.2012.122. [DOI] [PubMed] [Google Scholar]
- 75.Davies JC, Rubin BK. Emerging and unusual gram-negative infections in cystic fibrosis. Seminars in respiratory and critical care medicine. 2007;28:312–321. doi: 10.1055/s-2007-981652. [DOI] [PubMed] [Google Scholar]
- 76.Chain PS, et al. Genome of Ochrobactrum anthropi ATCC 49188 T, a versatile opportunistic pathogen and symbiont of several eukaryotic hosts. Journal of bacteriology. 2011;193:4274–4275. doi: 10.1128/JB.05335-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Amoureux L, et al. Detection of Achromobacter xylosoxidans in hospital, domestic, and outdoor environmental samples and comparison with human clinical isolates. Applied and environmental microbiology. 2013;79:7142–7149. doi: 10.1128/AEM.02293-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mahenthiralingam E, Baldwin A, Dowson CG. Burkholderia cepacia complex bacteria: opportunistic pathogens with important natural biology. Journal of applied microbiology. 2008;104:1539–1551. doi: 10.1111/j.1365-2672.2007.03706.x. [DOI] [PubMed] [Google Scholar]
- 79.Valvano MA, Keith KE, Cardona ST. Survival and persistence of opportunistic Burkholderia species in host cells. Current opinion in microbiology. 2005;8:99–105. doi: 10.1016/j.mib.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 80.Fedrigo GV, Campoy EM, Di Venanzio G, Colombo MI, Garcia Vescovi E. Serratia marcescens is able to survive and proliferate in autophagic-like vacuoles inside non-phagocytic cells. PloS one. 2011;6:e24054. doi: 10.1371/journal.pone.0024054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- 82.Rogier EW, et al. Secretory antibodies in breast milk promote long-term intestinal homeostasis by regulating the gut microbiota and host gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:3074–3079. doi: 10.1073/pnas.1315792111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Garrett WS, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vijay-Kumar M, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nature reviews Genetics. 2012;13:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pflughoeft KJ, Versalovic J. Human microbiome in health and disease. Annual review of pathology. 2012;7:99–122. doi: 10.1146/annurev-pathol-011811-132421. [DOI] [PubMed] [Google Scholar]
- 87.Kaur N, Chen CC, Luther J, Kao JY. Intestinal dysbiosis in inflammatory bowel disease. Gut microbes. 2011;2:211–216. doi: 10.4161/gmic.2.4.17863. [DOI] [PubMed] [Google Scholar]
- 88.Schwabe RF, Jobin C. The microbiome and cancer. Nature reviews Cancer. 2013;13:800–812. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Elinav E, Nowarski R, Thaiss CA, Hu B, Jin C, Flavell RA. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nature reviews Cancer. 2013;13:759–771. doi: 10.1038/nrc3611. [DOI] [PubMed] [Google Scholar]
- 90.Lebeer S, Vanderleyden J, De Keersmaecker SC. Host interactions of probiotic bacterial surface molecules: comparison with commensals and pathogens. Nature reviews Microbiology. 2010;8:171–184. doi: 10.1038/nrmicro2297. [DOI] [PubMed] [Google Scholar]
- 91.Bron PA, van Baarlen P, Kleerebezem M. Emerging molecular insights into the interaction between probiotics and the host intestinal mucosa. Nature reviews Microbiology. 2012;10:66–78. doi: 10.1038/nrmicro2690. [DOI] [PubMed] [Google Scholar]
- 92.Balagopal A, et al. Human immunodeficiency virus-related microbial translocation and progression of hepatitis C. Gastroenterology. 2008;135:226–233. doi: 10.1053/j.gastro.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jiang W, et al. Plasma levels of bacterial DNA correlate with immune activation and the magnitude of immune restoration in persons with antiretroviral-treated HIV infection. The Journal of infectious diseases. 2009;199:1177–1185. doi: 10.1086/597476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Redd AD, et al. Microbial translocation, the innate cytokine response, and HIV-1 disease progression in Africa. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:6718–6723. doi: 10.1073/pnas.0901983106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ancuta P, et al. Microbial translocation is associated with increased monocyte activation and dementia in AIDS patients. PloS one. 2008;3:e2516. doi: 10.1371/journal.pone.0002516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Marchetti G, et al. Microbial translocation is associated with sustained failure in CD4+ T-cell reconstitution in HIV-infected patients on long-term highly active antiretroviral therapy. AIDS (London, England) 2008;22:2035–2038. doi: 10.1097/QAD.0b013e3283112d29. [DOI] [PubMed] [Google Scholar]
- 97.Estes JD, et al. Damaged intestinal epithelial integrity linked to microbial translocation in pathogenic simian immunodeficiency virus infections. PLoS pathogens. 2010;6:e1001052. doi: 10.1371/journal.ppat.1001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sandler NG, et al. Host response to translocated microbial products predicts outcomes of patients with HBV or HCV infection. Gastroenterology. 2011;141:1220–1230. doi: 10.1053/j.gastro.2011.06.063. 1230 e1221–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tatro ET, Purnajo I, Richman DD, Smith DM, Gianella S. Antibody response to Achromobacter xylosoxidans during HIV infection is associated with lower CD4 levels and increased lymphocyte activation. Clinical and vaccine immunology : CVI. 2014;21:46–50. doi: 10.1128/CVI.00553-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Merlini E, Bai F, Bellistri GM, Tincati C, d'Arminio Monforte A, Marchetti G. Evidence for polymicrobic flora translocating in peripheral blood of HIV-infected patients with poor immune response to antiretroviral therapy. PloS one. 2011;6:e18580. doi: 10.1371/journal.pone.0018580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Klatt NR, et al. Loss of mucosal CD103+ DCs and IL-17+ and IL-22+ lymphocytes is associated with mucosal damage in SIV infection. Mucosal immunology. 2012;5:646–657. doi: 10.1038/mi.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xu H, Wang X, Liu DX, Moroney-Rasmussen T, Lackner AA, Veazey RS. IL-17-producing innate lymphoid cells are restricted to mucosal tissues and are depleted in SIV-infected macaques. Mucosal immunology. 2012;5:658–669. doi: 10.1038/mi.2012.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li H, Reeves RK. Functional perturbation of classical natural killer and innate lymphoid cells in the oral mucosa during SIV infection. Frontiers in immunology. 2012;3:417. doi: 10.3389/fimmu.2012.00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Keating J, et al. Intestinal absorptive capacity, intestinal permeability and jejunal histology in HIV and their relation to diarrhoea. Gut. 1995;37:623–629. doi: 10.1136/gut.37.5.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Samaras K, Wand H, Law M, Emery S, Cooper D, Carr A. Prevalence of metabolic syndrome in HIV-infected patients receiving highly active antiretroviral therapy using International Diabetes Foundation and Adult Treatment Panel III criteria: associations with insulin resistance, disturbed body fat compartmentalization, elevated C-reactive protein, and [corrected] hypoadiponectinemia. Diabetes care. 2007;30:113–119. doi: 10.2337/dc06-1075. [DOI] [PubMed] [Google Scholar]
- 106.Sheikh MY, Choi J, Qadri I, Friedman JE, Sanyal AJ. Hepatitis C virus infection: molecular pathways to metabolic syndrome. Hepatology (Baltimore, Md) 2008;47:2127–2133. doi: 10.1002/hep.22269. [DOI] [PubMed] [Google Scholar]
- 107.Gandhi RT, Sax PE, Grinspoon SK. Metabolic and cardiovascular complications in HIV-infected patients: new challenges for a new age. The Journal of infectious diseases. 2012;205(Suppl 3):S353–S354. doi: 10.1093/infdis/jis202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gardiner KR, et al. Significance of systemic endotoxaemia in inflammatory bowel disease. Gut. 1995;36:897–901. doi: 10.1136/gut.36.6.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Obermeier F, et al. CpG motifs of bacterial DNA essentially contribute to the perpetuation of chronic intestinal inflammation. Gastroenterology. 2005;129:913–927. doi: 10.1053/j.gastro.2005.06.061. [DOI] [PubMed] [Google Scholar]
- 110.Gutierrez A, et al. Cytokine association with bacterial DNA in serum of patients with inflammatory bowel disease. Inflammatory bowel diseases. 2009;15:508–514. doi: 10.1002/ibd.20806. [DOI] [PubMed] [Google Scholar]
- 111.Lakatos PL, et al. Serum lipopolysaccharide-binding protein and soluble CD14 are markers of disease activity in patients with Crohn's disease. Inflammatory bowel diseases. 2011;17:767–777. doi: 10.1002/ibd.21402. [DOI] [PubMed] [Google Scholar]
- 112.Darfeuille-Michaud A, et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology. 2004;127:412–421. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- 113.Small CL, Reid-Yu SA, McPhee JB, Coombes BK. Persistent infection with Crohn's disease-associated adherent-invasive Escherichia coli leads to chronic inflammation and intestinal fibrosis. Nature communications. 2013;4:1957. doi: 10.1038/ncomms2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.O'Brien CL, Pavli P, Gordon DM, Allison GE. Detection of bacterial DNA in lymph nodes of Crohn's disease patients using high throughput sequencing. Gut. 2014 doi: 10.1136/gutjnl-2013-305320. [DOI] [PubMed] [Google Scholar]
- 115.De Hertogh G, et al. Validation of 16S rDNA sequencing in microdissected bowel biopsies from Crohn's disease patients to assess bacterial flora diversity. The Journal of pathology. 2006;209:532–539. doi: 10.1002/path.2006. [DOI] [PubMed] [Google Scholar]
- 116.Zulian A, et al. Differences in visceral fat and fat bacterial colonization between ulcerative colitis and Crohn's disease. An in vivo and in vitro study. PloS one. 2013;8:e78495. doi: 10.1371/journal.pone.0078495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cho JH. The genetics and immunopathogenesis of inflammatory bowel disease. Nature reviews Immunology. 2008;8:458–466. doi: 10.1038/nri2340. [DOI] [PubMed] [Google Scholar]
- 119.Gutierrez A, et al. Genetic susceptibility to increased bacterial translocation influences the response to biological therapy in patients with Crohn's disease. Gut. 2014;63:272–280. doi: 10.1136/gutjnl-2012-303557. [DOI] [PubMed] [Google Scholar]
- 120.Cadwell K, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Duerr RH, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cho JH, Abraham C. Inflammatory bowel disease genetics: Nod2. Annual review of medicine. 2007;58:401–416. doi: 10.1146/annurev.med.58.061705.145024. [DOI] [PubMed] [Google Scholar]
- 123.Saraiva M, O'Garra A. The regulation of IL-10 production by immune cells. Nature reviews Immunology. 2010;10:170–181. doi: 10.1038/nri2711. [DOI] [PubMed] [Google Scholar]
- 124.Shah N, Kammermeier J, Elawad M, Glocker EO. Interleukin-10 and interleukin-10-receptor defects in inflammatory bowel disease. Current allergy and asthma reports. 2012;12:373–379. doi: 10.1007/s11882-012-0286-z. [DOI] [PubMed] [Google Scholar]
- 125.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cani PD, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56:1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 127.Amar J, et al. Intestinal mucosal adherence and translocation of commensal bacteria at the early onset of type 2 diabetes: molecular mechanisms and probiotic treatment. EMBO molecular medicine. 2011;3:559–572. doi: 10.1002/emmm.201100159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Shi H, Kokoeva MV, Inouye K, Tzameli I, Yin H, Flier JS. TLR4 links innate immunity and fatty acid-induced insulin resistance. The Journal of clinical investigation. 2006;116:3015–3025. doi: 10.1172/JCI28898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wiest R, Lawson M, Geuking M. Pathological bacterial translocation in liver cirrhosis. Journal of hepatology. 2014;60:197–209. doi: 10.1016/j.jhep.2013.07.044. [DOI] [PubMed] [Google Scholar]
- 130.Lin RS, et al. Endotoxemia in patients with chronic liver diseases: relationship to severity of liver diseases, presence of esophageal varices, and hyperdynamic circulation. Journal of hepatology. 1995;22:165–172. doi: 10.1016/0168-8278(95)80424-2. [DOI] [PubMed] [Google Scholar]
- 131.Garcia-Alvarez M, et al. Bacterial DNA translocation and liver disease severity among HIV-infected patients with chronic hepatitis C. Journal of acquired immune deficiency syndromes. 2012;61:552–556. doi: 10.1097/QAI.0b013e31826ea109. (1999). [DOI] [PubMed] [Google Scholar]
- 132.Libby P. Inflammation in atherosclerosis. Nature. 2002;420:868–874. doi: 10.1038/nature01323. [DOI] [PubMed] [Google Scholar]
- 133.Libby P. Inflammation and cardiovascular disease mechanisms. The American journal of clinical nutrition. 2006;83:456s–460s. doi: 10.1093/ajcn/83.2.456S. [DOI] [PubMed] [Google Scholar]
- 134.Karlsson FH, et al. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nature communications. 2012;3:1245. doi: 10.1038/ncomms2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tang WH, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. The New England journal of medicine. 2013;368:1575–1584. doi: 10.1056/NEJMoa1109400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Koeth RA, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nature medicine. 2013;19:576–585. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Koren O, et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(Suppl 1):4592–4598. doi: 10.1073/pnas.1011383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dapito DH, et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer cell. 2012;21:504–516. doi: 10.1016/j.ccr.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Seki E, et al. TLR4 enhances TGF-beta signaling and hepatic fibrosis. Nature medicine. 2007;13:1324–1332. doi: 10.1038/nm1663. [DOI] [PubMed] [Google Scholar]
- 140.Lescut D, et al. Bacterial translocation in colorectal cancers. Gastroenterologie clinique et biologique. 1990;14:811–814. [PubMed] [Google Scholar]
- 141.Chin KF, Kallam R, O'Boyle C, MacFie J. Bacterial translocation may influence the long-term survival in colorectal cancer patients. Diseases of the colon and rectum. 2007;50:323–330. doi: 10.1007/s10350-006-0827-4. [DOI] [PubMed] [Google Scholar]
- 142.Klein GL, Petschow BW, Shaw AL, Weaver E. Gut barrier dysfunction and microbial translocation in cancer cachexia: a new therapeutic target. Current opinion in supportive and palliative care. 2013;7:361–367. doi: 10.1097/SPC.0000000000000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kostic AD, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell host & microbe. 2013;14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wu S, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nature medicine. 2009;15:1016–1022. doi: 10.1038/nm.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Arthur JC, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]