Abstract
Severe acute respiratory syndrome (SARS) is a life-threatening disease caused by a newly identified coronavirus (CoV), SARS-CoV. The spike (S) glycoprotein of CoV is the major structural protein responsible for induction of host immune response and virus neutralization by antibodies. Hence, knowledge of neutralization determinants on the S protein is helpful for designing protective vaccines. To analyze the antigenic structure of the SARS-CoV S2 domain, the carboxyl-terminal half of the S protein, we first used sera from convalescent SARS patients to test the antigenicity of 12 overlapping fragments spanning the entire S2 and identified two antigenic determinants (Leu 803 to Ala 828 and Pro 1061 to Ser 1093). To determine whether neutralizing antibodies can be elicited by these two determinants, we immunized animals and found that both of them could induce the S2-specific antisera. In some animals, however, only one determinant (Leu 803 to Ala 828) was able to induce the antisera with the binding ability to the native S protein and the neutralizing activity to the SARS-CoV pseudovirus. This determinant is highly conserved across different SARS-CoV isolates. Identification of a conserved antigenic determinant on the S2 domain of the SARS-CoV S protein, which has the potential for inducing neutralizing antibodies, has implications in the development of effective vaccines against SARS-CoV.
Severe acute respiratory syndrome (SARS) is a life-threatening form of atypical pneumonia (27, 43) that was first reported in Guangdong Province of China in November 2002 (19). According to the records of the World Health Organization, this epidemic had resulted globally in 8,439 cases, of which 812 were fatal, by 3 July 2003. Although the first outbreak of SARS is over, the inadequate research laboratory safety procedures, such as those that caused the recent two SARS cases in Singapore and Taiwan (11, 26), make a new outbreak possible. So far, no available vaccine against SARS has been developed. Hence, there is an urgent need for effective vaccines for the prevention and control of this disease.
There is now clear evidence that SARS is caused by a newly identified coronavirus (CoV), SARS-CoV, which belongs to the Coronaviridae family of enveloped, positive-stranded RNA viruses (10, 17, 19, 24, 33, 43). CoVs cause many diseases of the respiratory, hepatic, gastrointestinal, and neurological systems in mammalian and avian species, exhibiting a broad host range (4, 35). The genome of SARS-CoV is 29,727 nucleotides in length and has 11 open reading frames, including the open reading frames that encode four typical structural proteins of CoVs: the spike (S) glycoprotein, the small membrane (M) protein, the envelope (E) protein, and the nucleocapsid (N) protein (8, 33, 34). The S glycoprotein is an important determinant of CoV virulence and tissue tropism and is crucial for viral attachment and entry into host cells (4, 28, 34). In many CoVs, proteolytic cleavage of the S glycoprotein yields the amino-terminal S1 and the carboxyl-terminal S2 subunits (3, 23, 25, 36, 38). The S1 subunit binds to host cell receptors, while the S2 subunit is responsible for membrane fusion (1, 12, 13, 39). Amino acid sequence analysis has shown that the S2 subunit is more conserved than the S1 subunit among the CoVs (33). Although there is no clear evidence so far that the S protein of SARS-CoV is cleaved into two subunits, the S1 and S2 domains of the SARS-CoV S protein can be identified through their homology with the S1 and S2 subunits of other CoVs (21, 37), such as murine hepatitis virus (MHV) and bovine CoV, which belong to the group 2 CoVs (8, 31).
The S protein of CoV is known to be the primary protein responsible for inducing host immune response and virus neutralization by antibodies (5, 16, 36); hence, knowledge of its antigenic structure, especially the location of conserved neutralization epitopes, is helpful for designing vaccines (30). Epitope-based vaccines can avoid any possibility of reversion to virulence and may be able to avoid the vaccine-induced enhancement of disease (8). Much attention has been focused on identifying neutralization epitopes on the S glycoprotein, including conformational and linear epitopes (7, 18, 25, 40, 42, 44, 45). In previous studies of MHV (7), bovine CoV (44), and avian infectious bronchitis CoV (IBV) (20), a linear immunodominant region was identified near the amino terminus of the S2 subunit by the method of expressing fragments in a prokaryotic system and analyzing their antigenicity with monoclonal antibodies (MAbs) or polyclonal antisera. One epitope contained in that immunodominant region of MHV can be recognized by several neutralizing MAbs (7, 22, 41, 42) and is able to induce an in vivo protective immune response in immunized animals (6, 15, 16, 47).
In this study, we used sera from convalescent SARS patients to identify two linear antigenic determinants (Leu 803 to Ala 828 and Pro 1061 to Ser 1093) through antigenicity analysis of 12 overlapping fragments covering the S2 domain. In addition, we immunized animals with the protein containing these two determinants and found that the determinant (Leu 803 to Ala 828) was capable of inducing neutralizing antibodies in some animals. These findings will have implications for developing an effective SARS-CoV vaccine.
MATERIALS AND METHODS
Expression and purification of fragments of the SARS-CoV S protein.
The full-length cDNA of the SARS-CoV S gene (strain BJ01; GenBank accession no. AY278488) was a gift provided by Shengli Bi at the Institute of Virology, Chinese Center for Disease Control and Prevention. It was used as a template to amplify different fragments, including the EcoRI (5′) and XhoI (3′) restriction sites appended to the primers. The PCR products were first cloned into pGEM-T vector (Promega) and sequenced. Then the desired fragments were generated from digestion of the recombinant T vector and subcloned into pET-32a(+) vector (Novagen) fused with the 3′ terminal of the thioredoxin (Trx) gene. After screening for positive-testing clones, the recombinant plasmids were transfected into Escherichia coli JM109(DE3)-competent cells. All the Trx fusion proteins were induced to express by isopropyl-β-d-thiogalactopyranoside (IPTG) and were purified according to the manufacturer's protocols for Ni-nitrilotriacetic acid (Ni-NTA) resin (QIAGEN). The fragments used for immunization experiments were also cloned into pGEX-4T-1 vector (Amersham Biosciences) and were induced to express as glutathione-S-transferase (GST) fusion proteins, which were purified by glutathione Sepharose 4B (Pharmacia Biotech).
Immunization experiments.
Groups of two New Zealand White rabbits and four female 6-week-old BALB/c mice were injected with 0.5 to 1 mg (for rabbits) and 50 to 100 μg (for mice) of purified Trx fusion protein. For the first immunization, the immunogen was emulsified in an equal volume of complete Freund's adjuvant (Sigma). Animals were injected at 2-week intervals with the same immunogen mixed with incomplete Freund's adjuvant (Sigma). Antisera of immunized animals were collected 1 week after each booster immunization and examined for the presence of fragment-specific antibodies by enzyme-linked immunosorbent assay (ELISA). Before immunization, the preimmune sera were collected for use as a control.
ELISAs.
The reactivity of the convalescent SARS patients' sera (provided by Beijing Ditan Hospital) and the immunized animals' antisera to the S protein fragments was evaluated by ELISA. Flat-bottomed plates (Maxisorp; Nunc) (96 well) were precoated with 1 μg of purified protein/well in 50 mM carbonate buffer (pH 9.6) for 2 h at 37°C and blocked with 3% bovine serum albumin (BSA) in carbonate buffer. Diluted human sera or immunized animal sera in phosphate-buffered saline (PBS)-0.05% Tween (PBS-T) (pH 7.4) were added to the wells and incubated for 2 h at 37°C. The secondary antibodies conjugated with horseradish peroxidase were diluted 1:2,000 to 1:10,000 in PBS-T-1% BSA. Optical densities were determined at a wavelength of 450 nm in an ELISA plate reader (Model 550; Bio-Rad).
Western blot analysis.
Both the GST fusion proteins and the control GST protein were run on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12% acrylamide) followed by transfer onto a nitrocellulose membrane (Amersham Pharmacia). After blocking, the nitrocellulose membrane was cut into strips and incubated with the immunized animals' sera diluted 1:200 (rabbits) or 1:100 (mice) in PBS-T. The alkaline phosphatase-labeled goat antirabbit or horse antimouse immunoglobulin G (IgG) (Santa Cruz Biotechnology) (1:1,000 dilution) was used as the secondary antibody in the detection.
Preparation of SARS-CoV S protein pseudotyped virus.
The full-length cDNA of the SARS-CoV S gene was optimized according to human codon usage and cloned into the pCDNA3.1(+) vector (Invitrogen). The resulting “humanized” S sequence was identical with that of strain BJ01 at the amino acid level. To promote the expression of the S glycoprotein on the cell membrane surface, a tissue plasminogen activator signal sequence was introduced to replace the original 13-residue signal peptide of the S protein at the 5′-terminal end (24) and the synthetic 42-residue cytoplasmic tail of Sendai virus (14) was appended to the S transmembrane domain (24) at the 3′-terminal end by overlapping PCR. The SARS-CoV S protein pseudotyped virus with luciferase as a reporter was produced by a similar method described previously (9; Y. Nie et al., submitted for publication). Briefly, pHIV-luc (a plasmid containing the human immunodeficiency virus [HIV] genome in which a partial envelope gene was replaced by the luciferase reporter gene; a kind gift from Dan Littman, New York University Medical Center, New York) (12 μg) and the-S expressing plasmid (6 μg) were cotransfected into 2 × 106 293T cells. Supernatants were harvested 48 h later, and the purified pseudoparticles were pelleted by ultracentrifugation through a 20% sucrose cushion at 50,000 ×g for 90 min. The pseudotype virus was normalized by a p24 ELISA kit (Biomerieux bv, Boxtel, The Netherlands).
Neutralization assays.
The SARS-CoV pseudoviruses were used in our neutralization assay to detect the neutralizing activity of various samples as described previously (Y. Nie et al., submitted). Briefly, the human hepatoma cell line Huh-7 (a gift from Stanley M. Lemon, University of Texas Medical Branch, Galveston) was used as the target cell and preseeded in a 96-well plate at a density of 8 × 103 cells/well. The next day, 50 ml of Dulbecco's modified Eagle's medium containing the equivalent of 0.5 ng (p24) of pseudotype virus was incubated with 50 μl of serial dilutions of sera for 30 min at 37°C. After incubation for 48 h at 37°C, the cells were washed with PBS and lysed with 40 μl of cell lysis buffer (Promega, Madison, Wis.)/well. The lysate was measured for luciferase activity in a luminometer (Wallac Multilabel 1450 counter; Perkin-Elmer, Singapore). The infectivity of the pseudotype virus was calculated as a percentage in the average amount of luciferase activity from the test culture relative to that of untreated controls (29, 32), and it could reflect the neutralizing activity of serum samples.
Establishment of the S-expressing SP2/0 cell line.
The “humanized” full-length S cDNA in pCDNA3.1(+) vector was digested with BamHI and EcoRI and cloned into the retroviral vector pBABE-puro. The positive-testing construct (pBabe/S) was cotransfected with pVSV/G into 293T cells by using phosphate calcium to package the retrovirus containing the S gene. After 48 h of transfection, the supernatant was collected and used to infect SP2/0 cells. After incubation for 12 h, the supernatant was replaced with Dulbecco's modified Eagle's medium (10% fetal calf serum). After 32 h, the cells were selected with puromycin at a concentration of 5 μg/ml for 1 week.
Flow cytometric analysis.
The S-expressing SP2/0 cells (SP2/0-S) and the untransduced normal SP2/0 cells were collected and washed with PBS (0.1% BSA). These two groups of cells were then incubated for 1 h with the serum samples. After three washes with PBS (0.1% BSA), the cells were incubated with the fluorescein isothiocyanate-conjugated goat antihuman or antirabbit IgG (Santa Cruz) (1:100) or antimouse IgG (Sigma) (1:100) for 30 min. Finally, the cells were analyzed by a flow cytometer (MoFlo high-performance cell sorter; Dako Cytomation).
RESULTS
Identification of antigenic determinants on the S2 domain of the SARS-CoV S glycoprotein.
According to our unpublished data, few SARS patients' sera could react with the S1 domain expressed by E. coli, which suggested that most antigenic determinants on the S1 domain are conformation dependent. To identify linear antigenic determinants on the S2 domain of the S protein, we first tested the reactivity of 64 sera from convalescent SARS patients to the prokaryote-expressed S2 domain (Asn 733 to Gln 1190 of S protein) with ELISA and found that there were 15 sera exhibiting high-level reactivity (data not shown). Then we designed 11 partially overlapping fragments (F1 to F11) at a length of 48 to 65 amino acid (aa) covering the S2 (Fig. 1A). After expressing these fragments as Trx fusion types in pET32a(+) vector, we purified them by Ni-NTA chromatography and performed SDS-PAGE to check their size and purity (Fig. 1B). Next, we carried out ELISA for antigenicity analysis using the 15 sera that reacted to the S2 domain, with two healthy human sera as controls. The results of the ELISA are shown in Fig. 2. As can be seen, only three fragments, F2 (Ala 765 to Gly 814), F3 (Arg 797 to Pro 844), and F9 (Leu 1045 to Asp 1109), could be recognized by the patients' sera, demonstrating that they contained the main linear antibody binding sites of the SARS-CoV S2 domain.
FIG. 1.
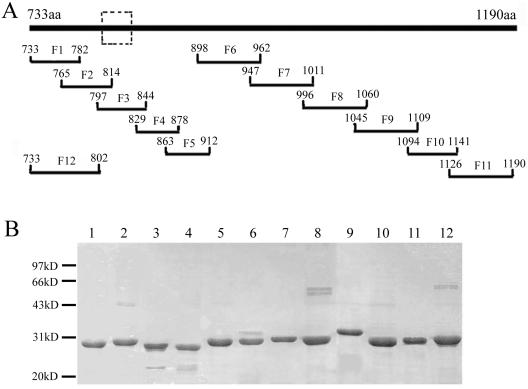
Location and expression of the partially overlapping fragments covering the S2 domain. (A) Schematic representation of the S2 domain (represented by the thick top line) and the twelve fragments F1 to F12 and their amino acid locations on the S protein. The dashed box defines the major antigenic determinant (Leu 803 to Ala 828) described in this paper. (B) SDS-PAGE of the E. coli-expressed and Ni-NTA-purified Trx fusion fragments F1 to F12 (lanes 1 to 12).
FIG. 2.
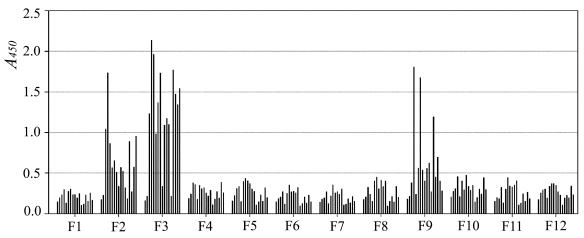
Identification of the antigenic determinants on the S2 domain. A total of 15 patients' sera and 2 healthy human sera were used in ELISA to analyze the antigenicity of F1 to F12. In each group, 17 bars represent the 2 healthy human sera and the 15 different patients' sera, respectively, from left to right. The serum samples were diluted 1:800.
Since F2 and F3 share a common 18-aa sequence (Fig. 1A), we did the following experiments to test whether the antigenicity of F2 was due to the overlapping part with F3 or to a unique antigenic determinant on F2 itself. We generated the 12th fragment (F12) covering F1 and a part of F2 (Asn 733 to Asp 802; Fig. 1A) and tested the reactivity of the sera described above. None of the sera reacted to F12 (Fig. 2), indicating that no unique antigenic determinant was contained in F2 and that its antigenicity was due to the overlapping part with F3. Therefore, F3 and F9 contained two different linear antigenic determinants, Leu 803 to Ala 828 (described as determinant I) and Pro 1061 to Ser 1093 (described as determinant II), on the S2 domain of the SARS-CoV S glycoprotein. Moreover, in comparison to the results seen with F9, the reactivity of patients' sera to F3 was more frequent and stronger. As Fig. 2 shows, 13 of 15 patients' sera reacted strongly to F3, suggesting that determinant I (Leu 803 to Ala 828 of S) is the major antigenic determinant of the S2 domain.
Immunogenicity of the two antigenic determinants.
To analyze the immunogenicity of these two antigenic determinants, we immunized mice and rabbits with prokaryote-expressed Trx-F3 or Trx-F9 proteins. To eliminate the elicited antibodies against the Trx part, we expressed and purified GST-F3 and GST-F9 and then used them to test the reactivity of the antisera collected from the immunized animals in ELISA. According to the results, both Trx-F3 and Trx-F9 proteins could elicit the antibodies specific to F3 or F9 fragment (data not shown).
Further, we used the GST-S2, GST-F3/F9, and GST proteins in Western blot analysis to test the reactivity of the antisera to them. As shown in Fig. 3, the antisera induced by Trx-F3 could recognize GST-F3 and the antisera induced by Trx-F9 could recognize GST-F9 whereas none of them could react with the control GST protein. Besides, the antisera induced by either Trx-F3 or Trx-F9 could recognize the GST-S2 protein (Fig. 3) and none of the preimmune sera reacted with any protein we tested in Western blot analysis (data not shown). All of these results suggested that both fragment F3 and fragment F9 were immunogenic and could elicit the antibodies specific to the S2 domain.
FIG. 3.
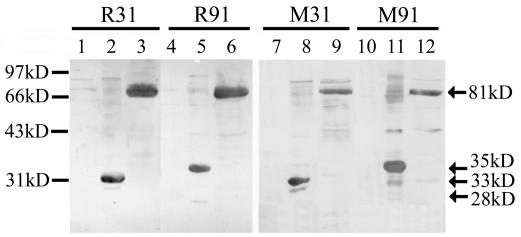
Western blot analysis of polyclonal antisera from animals immunized with the Trx-fusion proteins. R31 and M31 represent the antisera of rabbit 1 and mouse 1 immunized by Trx-F3; R91 and M91 represent the antisera of rabbit 1 and mouse 1 immunized by Trx-F9. 28kD, 33kD, 35kD, and 81kD represent the molecular masses of GST (lanes 1, 4, 7, and 10), GST-F3 (lanes 2 and 8), GST-F9 (lanes 5 and 11), and GST-S2 (lanes 3, 6, 9, and 12) protein, respectively. Molecular mass standards are shown on the left. The antisera from other immunized animals showed results similar to those obtained with antisera R31 and M31 or antisera R91 and M91.
The ability of the antisera to bind with the cell surface-expressed S glycoprotein.
To confirm the correct expression of the S protein on the surface of the SP2/0-S cells, which were transfected with S cDNA-contained retrovirus and selected with drugs, we first submitted the SP2/0-S cell lysates to Western blot analysis using the anti-S protein rabbit sera (antisera from the rabbits immunized with the S14-670, N-terminal domain of the S protein) for detection. Two major protein bands reacted with the anti-S sera: the 180-kDa mature S glycoprotein and the 130-kDa nonglycosylated precursors of the S protein (our unpublished data). We further stained the SP2/0-S cells and the untransfected normal SP2/0 cells with healthy human sera and sera of SARS patients and submitted the cells to flow cytometric analysis. As shown in Fig. 4A, the SP2/0-S cells could be specifically recognized only by the serum of the patients, suggesting that the S glycoprotein on the cell surface has a conformation similar to that of the native S protein on SARS-CoV.
FIG. 4.
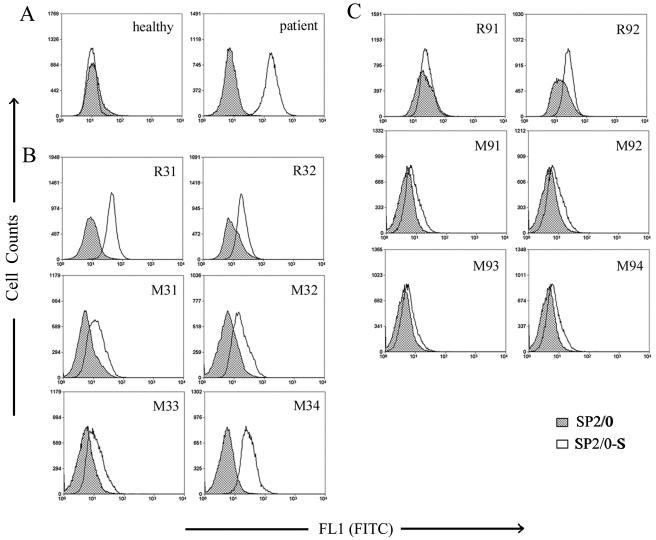
Ability of the antisera to bind to the cell surface-expressed S protein. The S-expressing SP2/0 cells (SP2/0-S) and the normal control cells (SP2/0) submitted to flow cytometric analysis were stained with the healthy human serum (healthy) and the patient serum (patient) (A); the antisera from two rabbits (R31 and R32) and four mice (M31 to M34) immunized with Trx-F3 (B); or the antisera from two rabbits (R91 and R92) and four mice (M91 to M94) immunized with Trx-F9 (C). Data of flow cytometry were analyzed with Summit version 3.1 software (Dako Cytomation).
To study whether the polyclonal antisera from the immunized animals were able to recognize the above-described S protein expressed by mammalian cells, we stained both the SP2/0-S cells and control SP2/0 cells with these antisera and the corresponding preimmune sera and performed flow cytometric analysis. The results showed that the antisera from some animals immunized with Trx-F3 (rabbit 1 and mouse 4) could specifically bind to the SP2/0-S compared with the results seen with SP2/0 (Fig. 4B) and that all the antisera from animals immunized with Trx-F9 bound poorly to SP2/0-S (Fig. 4C). As we expected, no preimmune serum could bind to the SP2/0-S or SP2/0 cells (data not shown). The above-described results indicate that the antisera from some animals immunized with Trx-F3 could specifically recognize the native S glycoprotein expressed on the surface of SP2/0-S cells.
Neutralizing activity of the polyclonal antisera.
To study the specificity of our neutralization assay, we first preincubated the SARS-CoV S protein pseudotyped particles (HIV/SARS) with the sera from convalescent SARS patients or the healthy human sera before incubating them with Huh-7 cells for infection. Huh-7 cells have been found to express a high level of the SARS-CoV receptor ACE2 and could be effectively infected by HIV/SARS (46). We have also demonstrated that Huh-7 cells could be infected by the pseudotype particles bearing the vesicular stomatitis virus G glycoprotein (HIV/VSV-G) (our unpublished data). As shown in Fig. 5A, the sera of convalescent SARS patients had an obvious neutralizing activity on HIV/SARS but failed to neutralize HIV/VSV-G, whereas the healthy human sera could not neutralize either of these two pseudoviruses. This suggested that our neutralization assay could be used to detect SARS-CoV-specific neutralizing antibodies.
FIG. 5.
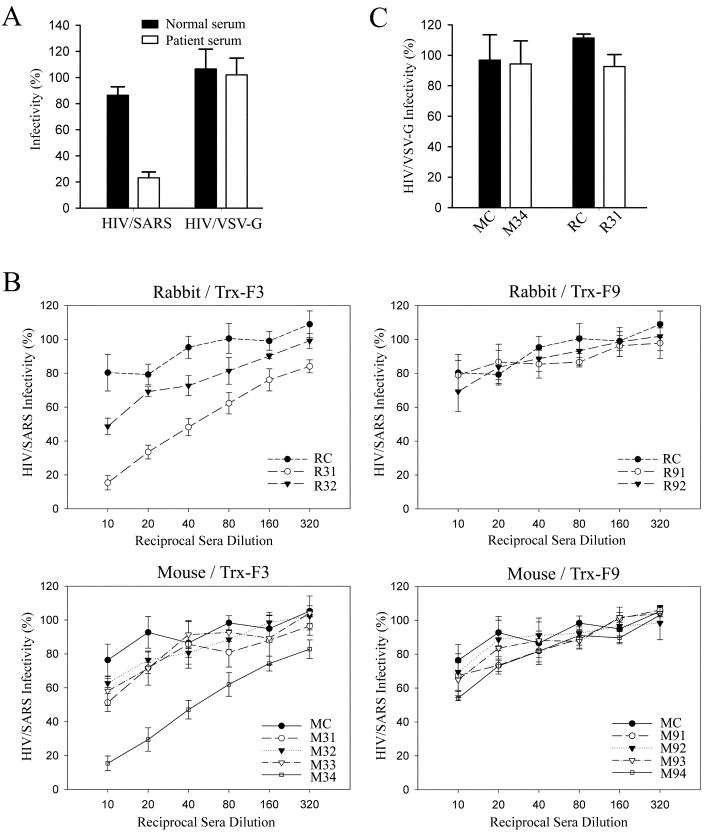
Neutralization assay of pseudotype viruses. (A) The neutralizing activity of SARS patient serum and normal human serum (diluted 1:100) for HIV/SARS or HIV/VSV-G pseudovirus. (B) The neutralizing activity of the animal sera (1:10 to 1:320 diluted) for HIV/SARS pseudovirus. R31 and R32, antisera from the two rabbits immunized with Trx-F3; M31 to M34, antisera from the four mice immunized with Trx-F3; R91 and R92, antisera from the two rabbits immunized with Trx-F9; M91 to M94, antisera from the four mice immunized with Trx-F9; RC and MC, the preimmune sera from rabbits and mice. (C) The neutralizing activity of the two neutralizing antisera (M34 and R31) and their corresponding preimmune sera (MC and RC) (diluted 1:40) for HIV/VSV-G pseudovirus. All results of neutralization assays represent four repeated experiments.
To determine whether immunization with Trx-F3 and Trx-F9 could induce neutralizing antibodies against SARS-CoV, we analyzed the neutralizing activities of the animal antisera with the neutralization assay. We observed that none of the antisera from the animals immunized with Trx-F9 could inhibit the HIV/SARS infection and that the antisera from two animals immunized with Trx-F3 (rabbit 1 and mouse 4) showed neutralizing activity and could inhibit about 50% of the HIV/SARS pseudotype virus infectivity at the 1:50 dilution (Fig. 5B).
To further prove that the neutralizing activity of these two antisera was specific to the SARS-CoV S protein, we used the HIV/VSV-G pseudovirus instead of HIV/SARS in the same neutralization assay and found that these antisera could not neutralize the HIV/VSV-G, having no obvious differences with the control preimmunized sera (Fig. 5C). The fact that only these two antisera were able to bind to the mammalian cell-expressed S protein (Fig. 4B) also supported the idea of their neutralizing activity. It was therefore concluded that Trx-F3 protein, which contains determinant I between Leu 803 and Ala 828 of the S protein, could induce neutralizing antibodies against SARS-CoV at least in some animals.
DISCUSSION
We have localized two linear antigenic determinants (determinants I [Leu 803 to Ala 828] and II [Pro 1061 to Ser 1093]) on the S2 domain of the SARS-CoV S glycoprotein (Fig. 2). The proteins containing either determinant I or II can elicit S2-specific antibodies from immunized animals (Fig. 3). However, only immunization with the protein containing determinant I, the major antigenic determinant on the S2 domain, was able to induce polyclonal antisera that could recognize the native S glycoprotein and showed in vitro neutralizing activity in some animals (Fig. 4 and 5).
The identified major antigenic determinant (Leu 803 to Ala 828) in this study is located on the S2 domain (Asn 733 to Gln 1190) of the SARS-CoV S protein (Fig. 1A). By comparing the S sequences of 45 SARS-CoV isolates which have been published in GenBank, we have discovered that this determinant is highly conserved and found only one amino acid mutation in a Shanghai LY (AY322207) isolate. In previous studies of the MHV S protein, an immunodominant linear neutralization domain (aa 838 to 905; Fig. 6) was identified on its S2 subunit (aa 770 to 1376) (7, 13) and seven MAbs, including five neutralizing MAbs, were shown to recognize this domain (7). Through amino acid sequence alignment of the SARS-CoV S2 domain and the MHV S2 subunit, we find that our defined determinant, determinant I, has 46% identity in amino acid sequence with the immunodominant neutralization domain on the MHV S2 subunit and that their locations on the S protein are in analogous positions-near the amino terminus of the S2 subunit (Fig. 6). This similarity implies that the major antigenic determinant (determinant I) on the S2 domain of the SARS-CoV S protein may be exposed as an effective target for neutralizing antibodies. Such information may have implications for SARS-CoV vaccine development.
FIG. 6.
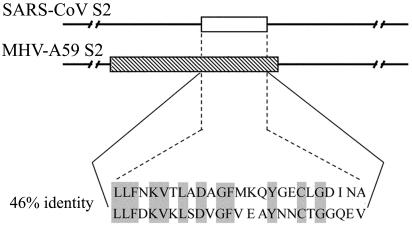
Comparison of the amino acid sequences between antigenic determinant I (26 amino acids; represented by the blank box) on the SARS-CoV S2 domain and the immunodominant domain (68 amino acids; represented by the shaded box) on the MHV-A59 S2 subunit (7).
Through investigation of the antigenicity of a panel of fragments which overlap by 16 to 18 aa and cover the entire 458 residues of the S2 domain (Fig. 1A) and the use of sera of SARS patients for this paper, we have identified a sequence with 26 aa as a major antigenic determinant (Fig. 2). For further mapping of the epitope in this determinant, we have also synthesized a set of consecutive overlapping decapeptides and analyzed the reactivity of the sera of the patients to them in ELISA. Surprisingly, the sera that reacted strongly to this determinant did not recognize any of the decapeptides (data not shown). Similar results have been reported by others in the study of IBV, including results showing that some MAbs reactive with the pEX expression proteins failed to recognize any of the overlapping nonapeptides spanning the antigenic region on the S2 subunit of IBV (20). Whether this is due to the length of synthetic peptides or to fragment-related local conformation (2) remains to be determined.
According to comparisons of the immunogenicity of the proteins Trx-F3 and Trx-F9, only Trx-F3 which contains determinant I has the potential to induce the neutralizing antibodies through immunization (Fig. 5). However, not all the animals immunized with Trx-F3 were able to produce the neutralizing antibodies, possibly because Trx is not a good carrier for immunogens like peptides or small protein fragments. The ability of a neutralizing epitope to induce neutralizing antibodies or protective immune response in immunized animals is influenced greatly by the nature of the protein carrier and the specific conformation of immunogens (6, 7, 16). For example, when conserved neutralizing epitope A of the MHV S protein (22, 41, 42) was expressed by the pET 3x system (7) or coupled to BSA (6), the immunization did not induce a protective virus-specific immune response. However, when epitope A was coupled to keyhole limpet hemocyanin, it showed good immunogenicity and induced protective immunity (6). Moreover, when mice were immunized with epitope A that was carried by the tobacco mosaic virus (15), coupled to the influenza virus T-cell epitope (16), or displayed on the phage surface (47), the mice could be protected against a lethal infection of MHV. On the basis of these studies, better strategies of immunization should be devised for inducing neutralizing antibodies to SARS-CoV with our identified antigenic determinant, determinant I. Besides, our future work will also be designed to identify other conserved neutralizing determinants on the S protein and to use these determinants to develop effective vaccines against SARS-CoV infection.
Acknowledgments
This research was supported by a National Nature Science Foundation of China grant (30340027), a Ministry of Science and Technology grant (2003CB514116), a National Nature Science Foundation of China for Outstanding Young Scientist Award (30125022), a Chinese-German Science Center grant (GZ237[202/10]), a European Commission grant (511063) to H. Deng, and a Ministry of Science and Technology grant (G1999011904) to M. Ding.
We thank Zhaoyi Wang, Eli Sercarz, and Derya Unutmaz for critical reading of the manuscript. We also acknowledge Jianjun Hu, Xiaojing Wang, and Qi Dong for technical assistance, Zai Wang, Fei Yuan, and Aihua Zheng for supplying the HIV/VSV-G pseudotype particles, and Liying Du for providing the technical support of flow cytometric analysis.
REFERENCES
- 1.Abraham, S., T. E. Kienzle, W. Lapps, and D. A. Brian. 1990. Deduced sequence of the bovine coronavirus spike protein and identification of the internal proteolytic cleavage site. Virology 176:296-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbato, G., E. Bianchi, P. Ingallinella, W. H. Hurni, M. D. Miller, G. Ciliberto, R. Cortese, R. Bazzo, J. W. Shiver, and A. Pessi. 2003. Structural analysis of the epitope of the anti-HIV antibody 2F5 sheds light into its mechanism of neutralization and HIV fusion. J. Mol. Biol. 330:1101-1115. [DOI] [PubMed] [Google Scholar]
- 3.Boireau, P., C. Cruciere, and J. Laporte. 1990. Nucleotide sequence of the glycoprotein S gene of bovine enteric coronavirus and comparison with the S proteins of two mouse hepatitis virus strains. J. Gen. Virol. 71:487-492. [DOI] [PubMed] [Google Scholar]
- 4.Bosch, B. J., R. van der Zee, C. A. M. de Haan, and P. J. M. Rottier. 2003. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 77:8801-8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins, A. R., R. L. Knobler, H. Powell, and M. J. Buchmeier. 1982. Monoclonal antibodies to murine hepatitis virus-4 (strain JHM) define the viral glycoprotein responsible for attachment and cell-cell fusion. Virology 119:358-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daniel, C., M. Lacroix, and P. J. Talbot. 1994. Mapping of linear antigenic sites on the S glycoprotein of a neurotropic murine coronavirus with synthetic peptides: a combination of nine prediction algorithms fails to identify relevant epitopes and peptide immunogenicity is drastically influenced by the nature of the protein carrier. Virology 202:540-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daniel, C., R. Anderson, M. J. Buchmeier, J. O. Fleming, W. J. M. Spaan, H. Wege, and P. J. Talbot. 1993. Identification of an immunodominant linear neutralization domain on the S2 portion of the murine coronavirus spike glycoprotein and evidence that it forms part of complex tridimensional structure. J. Virol. 67:1185-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Groot, A. S. 2003. How the SARS vaccine effort can learn from HIV—speeding towards the future, learning from the past. Vaccine 21:4095-4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng, H., R. Liu, W. Ellmeier, S. Choe, D. Unutmaz, M. Burkhart, P. D. Marzio, S. Marmon, R. E. Sutton, C. M. Hill, C. B. Davis, S. C. Peiper, T. J. Schall, D. R. Littman, and N. R. Landau. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661-666. [DOI] [PubMed] [Google Scholar]
- 10.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. M. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. M. E. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 11.Enserink, M. 2003. Infectious diseases. Singapore lab faulted in SARS case. Science 301:1824. [DOI] [PubMed] [Google Scholar]
- 12.Frana, M. F., J. N. Behnke, L. S. Sturman, and K. V. Holmes. 1985. Proteolytic cleavage of the E2 glycoprotein of murine coronavirus: host-dependent differences in proteolytic cleavage and cell fusion. J. Virol. 56:912-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallagher, T. M., and M. J. Buchmeier. 2001. Coronavirus spike proteins in viral entry and pathogenesis. Virology 279:1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobayashi, M., A. Iida, Y. Ueda, and M. Hasegawa. 2003. Pseudotyped lentivirus vectors derived from simian immunodeficiency virus SIVagm with envelope glycoproteins from paramyxovirus. J. Virol. 77:2607-2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koo, M., M. Bendahmane, G. A. Lettieri, A. D. Paoletti, T. E. Lane, J. H. Fitchen, M. J. Buchmeier, and R. N. Beachy. 1999. Protective immunity against murine hepatitis virus (MHV) induced by intranasal or subcutaneous administration of hybrids of tobacco mosaic virus that carries an MHV epitope. Proc. Natl. Acad. Sci. USA 96:7774-7779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koolen, M. J., M. A. J. Borst, M. C. Horzinek, and W. J. M. Spaan. 1990. Immunogenic peptide comprising a mouse hepatitis virus A59 B-cell epitope and an influenza virus T-cell epitope protects against lethal infection. J. Virol. 64:6270-6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, L. J. Anderson, and SARS Working Group. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 18.Kubo, H., Y. K. Yamada, and F. Taguchi. 1994. Localization of neutralizing epitopes and the receptor-binding site within the amino-terminal 330 amino acids of the murine coronavirus spike protein. J. Virol. 68:5403-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuiken, T., R. A. M. Fouchier, M. Schutten, G. F. Rimmelzwaan, G. van Amerongen, D. van Riel, J. D. Laman, T. de Jong, G. van Doornum, W. Lim, A. E. Ling, P. K. S. Chan, J. S. Tam, M. C. Zambon, R. Gopal, C. Drosten, S. van der Werf, N. Escriou, J. C. Manuguerra, K. Stöhr, J. S. M. Peiris, and A. D. M. E. Osterhaus. 2003. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet 362:263-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lenstra, J. A., J. G. Kusters, G. Koch, and B. A. M. van der Zeijst. 1989. Antigenicity of the peplomer protein of infectious bronchitis virus. Mol. Immunol. 26:7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, W., M. J. Moore, N. Vasilieva, J. Sui, S. K. Wong, M. A. Berne, M. Somasundaran, J. L. Sullivan, K. Luzuriaga, T. C. Greenough, H. Choe, and M. Farzan. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luytjes, W., D. Geerts, W. Posthumus, R. Meloen, and W. Spaan. 1989. Amino acid sequence of a conserved neutralizing epitope of murine coronaviruses. J. Virol. 63:1408-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luytjes, W., L. S. Sturman, P. J. Bredenbeek, J. Charite, B. A. M. van der Zeijst, M. C. Horzinek, and W. J. M. Spaan. 1987. Primary structure of the glycoprotein E2 of coronavirus MHV-A59 and identification of the trypsin cleavage site. Virology 161:479-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marra, M. A., S. J. M. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. N. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 25.Moore, K. M., M. W. Jackwood, and D. A. Hilt. 1997. Identification of amino acids involved in a serotype and neutralization specific epitope within the s1 subunit of avian infectious bronchitis virus. Arch. Virol. 142:2249-2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Normile, D. 2004. Infectious diseases. Second lab accident fuels fears about SARS. Science 303:26. [DOI] [PubMed] [Google Scholar]
- 27.Peiris, J. S. M., S. T. Lai, L. L. M. Poon, Y. Guan, L. Y. C. Yam, W. Lim, J. Nicholls, W. K. S. Yee, W. W. Yan, M. T. Cheung, V. C. C. Cheng, K. H. Chan, D. N. C. Tsang, R. W. H. Yung, T. K. Ng, K. Y. Yuen, and SARS Study Group. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phillips, J. J., M. M. Chua, G. F. Rall, and S. R. Weiss. 2002. Murine coronavirus spike glycoprotein mediates degree of viral spread, inflammation, and virus-induced immunopathology in the central nervous system. Virology 301:109-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poignard, P., M. Moulard, E. Golez, V. Vivona, M. Franti, S. Venturini, M. Wang, P. W. H. I. Parren, and D. R. Burton. 2003. Heterogeneity of envelope molecules expressed on primary human immunodeficiency virus type 1 particles as probed by the binding of neutralizing and nonneutralizing antibodies. J. Virol. 77:353-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Posthumus, W. P., J. A. Lenstra, W. M. Schaaper, A. P. van Nieuwstadt, L. Enjuanes, and R. H. Meloen. 1990. Analysis and simulation of a neutralizing epitope of transmissible gastroenteritis virus. J. Virol. 64:3304-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prentice, E., and M. R. Denison. 2001. The cell biology of coronavirus infection. Adv. Exp. Med. Biol. 494:609-614. [DOI] [PubMed] [Google Scholar]
- 32.Richman, D. D., T. Wrin, S. J. Little, and C. J. Petropoulos. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. USA 100:4144-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Peñaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. T. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Günther, A. D. M. E. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 34.Ruan, Y. J., C. L. Wei, A. L. Ee, V. B. Vega, H. Thoreau, S. Y. S. Thoe, J. M. Chia, P. Ng, K. P. Chiu, L. Lim, T. Zhang, K. P. Chan, L. E. O. Oon, M. L. Ng, S. Y. Leo, L. F. P. Ng, E. C. Ren, L. W. Stanton, P. M. Long, and E. T. Liu. 2003. Comparative full-length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection. Lancet 361:1779-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siddell, S., H. Wege, and V. Ter Meulen. 1983. The biology of coronaviruses. J. Gen. Virol. 64:761-776. [DOI] [PubMed] [Google Scholar]
- 36.Spaan, W. J. M., D. Cavanagh, and M. C. Horzinek. 1988. Coronaviruses: structure and genome expression. J. Gen. Virol. 69:2939-2952. [DOI] [PubMed] [Google Scholar]
- 37.Spiga, O., A. Bernini, A. Ciutti, S. Chiellini, N. Menciassi, F. Finetti, V. Causarono, F. Anselmi, F. Prischi, and N. Niccolai. 2003. Molecular modelling of S1 and S2 subunits of SARS coronavirus spike glycoprotein. Biochem. Biophys. Res. Commun. 310:78-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sturman, L. S., C. S. Ricard, and K. V. Holmes. 1985. Proteolytic cleavage of the E2 glycoprotein of murine coronavirus: activation of cell-fusing activity of virions by trypsin and separation of two different 90K cleavage fragments. J. Virol. 56:904-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taguchi, F. 1995. The S2 subunit of the murine coronavirus spike protein is not involved in receptor binding. J. Virol. 69:7260-7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takase-Yoden, S., T. Kikuchi, S. G. Siddell, and F. Taguchi. 1991. Localization of major neutralizing epitopes on the S1 polypeptide of the murine coronavirus peplomer glycoprotein. Virus Res. 18:99-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Talbot, P. J., A. A. Salmi, R. L. Knobler, and M. J. Buchmeier. 1984. Topographical mapping of epitopes on the glycoproteins of murine hepatitis virus-4 (strain JHM): correlation with biological activities. Virology 132:250-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Talbot, P. J., and M. J. Buchmeier. 1985. Antigenic variation among murine coronaviruses: evidence for polymorphism on the peplomer glycoprotein, E2. Virus Res. 2:317-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thiel, V., K. A. Ivanov, A. Putics, T. Hertzig, B. Schelle, S. Bayer, B. Weiβbrich, E. J. Snijder, H. Rabenau, H. W. Doerr, A. E. Gorbalenya, and J. Ziebuhr. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 84:2305-2315. [DOI] [PubMed] [Google Scholar]
- 44.Vautherot, J. F., J. Laporte, and P. Boireau. 1992. Bovine coronavirus spike glycoprotein: localization of an immunodominant region at the amino-terminal end of S2. J. Gen. Virol. 73:3289-3294. [DOI] [PubMed] [Google Scholar]
- 45.Vautherot, J. F., M. F. Madelaine, P. Boireau, and J. Laporte. 1992. Bovine coronavirus peplomer glycoproteins: detailed antigenic analyses of S1, S2 and HE. J. Gen. Virol. 73:1725-1737. [DOI] [PubMed] [Google Scholar]
- 46.Wang, P., J. Chen, A. Zheng, Y. Nie, X. Shi, W. Wang, G. Wang, M. Luo, H. Liu, L. Tan, X. Song, Z. Wang, X. Yin, X. Qu, X. Wang, T. Qing, M. Ding, and H. Deng. 2004. Expression cloning of functional receptor used by SARS coronavirus. Biochem. Biophys. Res. Commun. 315:439-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu, M. W. N., J. K. Scott, A. Fournier, and P. J. Talbot. 2000. Characterization of murine coronavirus neutralization epitopes with phage-displayed peptides. Virology 271:182-196. [DOI] [PMC free article] [PubMed] [Google Scholar]


