Abstract
Complete envelope genes were amplified from autopsy brain tissue of five individuals who had died of AIDS and had neurological complications. Lymph node samples were included for two of the patients. Nineteen different envelope clones from the five patients had distinct V1V2 sequences. Thirteen of the envelopes were functional and conferred fusigenicity and infectivity for CD4+ CCR5+ cells. Infectivity and cell-cell fusion assays showed that most envelopes used both CCR5 and CCR3. One brain-derived envelope used a broad range of coreceptors, while three other brain envelopes from one individual were restricted to CCR5. However, there was no correlation between tissue of origin and coreceptor use. Envelopes showed two very distinct phenotypes depending on their capacity to infect macrophages and to exploit low levels of CD4 and/or CCR5 for infection. Envelopes that were highly fusigenic and tropic for macrophages were identified in brain tissue from four of the five patients. The enhanced macrophage tropism correlated with reduced sensitivity to inhibition by Q4120, a CD4-specific antibody, but not with sensitivity to the CCR5 inhibitor, TAK779. The highly macrophage-tropic envelopes were able to infect cells expressing low levels of CD4 and/or CCR5. Comparison with several well-characterized macrophage-tropic envelopes showed that the four identified patient envelopes were at the top limit of macrophage tropism. In contrast, all four lymph node-derived envelopes exhibited a non-macrophage-tropic phenotype and required high levels of CD4 for infection. Our data support the presence of envelopes that are highly fusigenic and tropic for macrophages in the brains of patients with neurological complications. These envelopes are able to infect cells that express low levels of CD4 and/or CCR5 and may have adapted for replication in brain macrophages and microglia, which are known to express limited amounts of CD4.
Human immunodeficiency virus type 1 (HIV-1) replication in the brain results in the development of severe neurological disorders known as AIDS dementia complex in about 30% of AIDS patients (29). The mechanisms that cause the loss of up to 40% of neurons (10) as well as result in dementia are unclear. In the rhesus macaque simian immunodeficiency virus (SIV) model, both neurotropic and neurovirulent SIVMAC variants have been described (11, 22, 38). It is therefore suspected that specific neurotropic and neurovirulent HIV-1 variants exist and are associated with dementia.
CCR5-using HIV-1 strains are predominant in the brain (13), and CCR5+ perivascular macrophages and microglia are the cell types most frequently infected (19, 35, 37, 43, 49, 50). The role of other cell types resident in the brain is less clear. There is evidence that CD4− astrocytes become infected, particularly in pediatric AIDS cases (32, 36, 37, 43). Astrocyte infection is relatively unproductive with early HIV mRNAs (e.g., for rev and nef) detectable, but no late mRNAs encoding the structural gag and env proteins needed to produce progeny virus particles (36, 37). Nevertheless, abundant expression of nef and rev in brain astrocytes has been associated with dementia (32). Infection of microvascular endothelial cells of the blood brain barrier could represent a direct route for HIV entry into the brain. However, few studies support more than sporadic infection (27, 46). Neurons have not been considered a significant target for virus replication in vivo, although occasional infection has been reported (25, 43). Two recent studies using laser capture microscopy detected HIV-1 sequences in isolated neurons (44, 45). These reports are intriguing and could indicate latent infection but are currently difficult to reconcile with the consensus from earlier studies indicating only rare infection.
If neurotropic HIV-1 strains do exist, then they would be expected to infect macrophages and microglia efficiently. Several observations are consistent with this view. Shieh et al. described a variant of the HIV-1 R5 strain, BORI, that replicated more efficiently in microglial cultures following selection in vitro (39). This increased microglial tropism was conferred by mutations in the V1 loop, an envelope region that influences cell tropism and coreceptor use (5, 34). Gorry et al. (16) reported a brain isolate (UK1-br) that carried an enhanced fusigenicity and tropism for macrophages. Both Gorry's brain isolate and Shieh's microglia-passaged BORI variant had increased capacity to infect cells expressing low levels of CD4 and CCR5 (16, 23, 39).
Analysis of the phenotypes of envelopes derived from brain tissue has been limited mainly to viruses isolated into blood-derived lymphocytes (15, 16, 42, 51) or envelope fragments cloned into chimeric viruses (3). A recent report by Ohagen et al. (28) described envelope genes that were amplified by PCR from brain tissue without culture. Here, we describe genotypic and biological characterization of full-length envelope genes also amplified from brain tissue of AIDS patients taken at autopsy. PCR-amplified envelopes have not been through isolation procedures that potentially select for virus strains able to replicate fastest in donor peripheral blood mononuclear cell (PBMC) cultures and may leave behind viruses adapted for replication in cell types resident in the brain. We describe several brain-derived envelopes that have enhanced fusigenicity and tropism for macrophages as well as the capacity to infect cells that express low levels of CD4 and/or CCR5. We believe that these envelopes have adapted for replication in macrophages and microglia in the brain that express low levels of CD4.
MATERIALS AND METHODS
Patient tissue samples.
Tissue samples dissected at autopsy were frozen at −80°C. Genomic DNA was extracted as described previously (40). Samples from frontal lobe tissue of five late-term AIDS patients (NA118, NA420, NA20, NA176, and NA353) and from the lymph node (LN) tissue of two of these patients (NA118 and NA420) were used in this study. Information regarding each patient is shown in Table 1.
TABLE 1.
Patient details
| Patient no. | Statusa | Characteristics | Sample(s) |
|---|---|---|---|
| NA118 | IVDU | Dementia, florid giant-cell encephalitis | Frontal lobe, LN |
| NA420 | Heterosexual | No cognitive impairment, sparse giant-cell encephalitis | Frontal lobe, LN |
| NA20 | Hemophiliac | Mild cognitive impairment, sparse giant-cell encephalitis, cerebral toxoplasmosis | Frontal lobe |
| NA176 | IVDU | Dementia, focal giant-cell encephalitis | Frontal lobe |
| NA353 | IVDU | Mild cognitive impairment, florid giant-cell encephalitis | Frontal lobe |
IVDU, intravenous drug user.
PCR amplification of complete envelope genes.
The gp160 PCRs were performed using the Expand high fidelity DNA polymerase system (Roche Inc.) according to the manufacturer's instructions. This kit contains a mixture of a polymerase with proofreading capability and Taq polymerase. NA118, NA420, NA20, NA176, and NA353 envelope sequences were amplified by nested PCR as described by Gao et al. (14). Outer primers were Env A (GGC TTA GGC ATC TCC TAT GGC AGG AAG AA) and Env N (CTG CCA ATC AGG GAA GTA GCC TTG TGT), and inner primers were Env B (AGA AAG AGC AGA AGA CAG TGG CAA TGA) and Env M (TAG CCC TTC CAG TCC CCC CTT TTC TTT TA). Conditions for both rounds of PCR were as follows: 94°C for 45 s, 50°C for 45 s, and 72°C for 4 min (20 cycles); 94°C for 45 s, 50°C for 45 s, and 72°C for 4 min 15 s (15 cycles). Between 0.01 and 1 μg of patient DNA per reaction was included. The PCR products were fractionated by agarose gel electrophoresis, and 3-kb bands were excised and recovered using a gel extraction kit (QIAGEN Inc.). PCRs were carried out using a 5′ chemically phosphorylated PCR primer (Env B) in the second round of the nested PCR. The 3-kb fragments thus contained a phosphate at the 5′ end. These were then ligated into the pCR3.1-Uni vector (Invitrogen Inc.) (see Fig. S1A in the supplemental material). pCR3.1-Uni was provided in a linear form with overhanging Ts at each end. The 5′ overhang was also predephosphorylated, enabling the 5′-phosphorylated PCR fragments to ligate only in the correct orientation for expression from an upstream cytomegalovirus promoter. Portions (5 to 10 μl) of the ligation reaction were used to transform competent TOP10F bacteria. Minipreps were prepared from resulting colonies, and the presence of inserts was screened by digestion using appropriate restriction enzymes. Variable V1V2 loop sequences were obtained for all patient envelopes amplified to confirm the presence of novel sequences and the lack of contamination with standard HIV-1 envelopes.
Molecular constructs required for pseudotype virus production.
Constructs used to prepare pseudotype viruses are shown in the supplemental material. Figure S1 shows pseudotype viruses carrying each patient envelope that were produced by cotransfection of env+ pSVIIIenv (14) with an env− pNL43 molecular clone into 293T cells using calcium phosphate. Pseudotype viruses prepared using the pSVIIIenv vector for envelope expression contained infectivity that was more than 100-fold higher than that of pCR3.1-Uni pseudotypes (data not shown). All patient-derived envelope genes were therefore cloned into pSVIIIenv via conserved KpnI sites for the production of pseudotype viruses. Cell supernatants carrying progeny pseudotype virions were harvested 48 h after transfection, clarified (1,000 × g for 10 min), aliquoted, and stored at −152°C.
Preparation of PBMCs and macrophages.
Fresh blood (from volunteer donors) was diluted 1:1 in RPMI 1640 medium and carefully layered onto 15 ml of Ficoll-Hypaque (Pharmacia) in a 50-ml Falcon tube and centrifuged at 350 × g for 30 min at room temperature. Erythrocytes sedimented at the bottom of the tube, while the white cells were harvested from the interphase between Ficoll and diluted plasma. These cells were washed twice in RPMI 1640 and resuspended to give 1 × 106 cells/ml in RPMI 1640 containing 10% fetal calf serum (FCS) and 0.5 μg of phytohemagglutinin/ml. The cells were then grown for 2 to 3 days. Cells were then diluted to 3 × 105 to 5 × 105 cells/ml and cultured for a further 2 to 3 days in media supplemented with 10% FCS and human interleukin-2 (from human lymphocytes, 20 U/ml; Boehringer) prior to infection. Macrophages were also prepared from the Ficoll white cell layer as described previously (41). Briefly, 108 cells were plated onto a 14-cm-diameter bacterial petri dish in 15 ml of Dulbecco's modified Eagle medium (DMEM) containing 10% human serum. Cells were incubated for 2 h at 37°C. The adherent cells were then gently washed, first with the media on the plate and then twice with fresh DMEM before 15 ml of DMEM containing 10% human serum was added, and cells were incubated at 37°C overnight. The following day, adherent cells were washed again, and fresh media was added. Macrophages were ready for infection 5 to 7 days later. The day prior to infection, the macrophages were washed three times in EDTA and incubated at 37°C for 10 min to loosen cell attachments. Macrophages were then gently scraped off the dish by using a cell scraper. Cells were resuspended in DMEM containing 10% human serum, counted, and reseeded into 48-well tissue culture trays at 1.25 ×105 cells per well. Alternatively, elutriated monocytes (18) were resuspended in medium containing macrophage colony stimulating factor (R&D Systems) and cultured for 7 days before use for virus infections.
Cell lines expressing CD4 and coreceptors.
Coreceptors used by pseudotype viruses carrying patient-derived envelopes were investigated by testing infection of the CD4+ GHOST (2a) and NP2 (42a) sets of cell lines. Infectivity via CCR2b, CCR3, CCR5, CX3CR1, CXCR6, and GPR15 was tested on GHOST cells. Infection via CCR3, CCR5, CCR8, GPR1, APJ, and CXCR4 was tested on NP2/CD4 cells. CD4-independent infection was assessed on NP2/CCR5 cells. GHOST and NP2/CD4 cell lines and their derivatives were seeded into 48-well trays on the day prior to infection; 0.5 ml of GHOST (4 × 104 cells/ml) or NP2/CD4 (5 × 105 cells/ml) cells were seeded per well. Infections were performed in duplicate and repeated at least once. One hundred microliters of serially diluted, cell-free virus supernatant was incubated with the appropriate cell lines for 3 h at 37°C before addition of 400 μl of growth medium. Cells were fixed and immunostained for virus antigens 3 days postinfection, and the foci of infection per milliliter was calculated (see below).
p24 antigen immunostaining.
Immunostaining was carried out as previously described (6). HIV-infected cells were washed once in phosphate-buffered saline (PBS) and then fixed in cold (−20°C) methanol-acetone at a ratio of 1:1 for 10 min at room temperature. The cells were then washed once with PBS and once with PBS containing 1% FCS before adding anti-HIV-1 p24 monoclonal antibodies (38:96K and EF7; Centralised Facility for AIDS Reagents, Potters Bar, United Kingdom). These monoclonal antibodies were added as hybridoma cell culture fluid at a 1:1 ratio diluted 1 in 40 in PBS-1% FCS and incubated at room temperature for at least an hour. Cells were washed once in PBS-1% FCS before incubating for 1 h at room temperature with anti-mouse F(ab′)2 fragments conjugated to β-galactosidase (Southern Biotechnology Associates, Inc.) at a dilution of 1 in 400 in PBS-1% FCS to detect the first antibody layer. Cells were then washed once with PBS-1% FCS and twice with PBS. Infected cells were stained blue on addition of X-Gal substrate (0.5 mg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside/ml [Fisher Bioreagents Inc.], 3 mM potassium ferrocyanide, 3 mM potassium ferricyanide, 1 mM magnesium chloride). The prepared pseudotype viruses were competent only for single rounds of replication. Thus, individuals or groups of recently divided blue-stained cells were regarded as foci of infection, and virus infectivity was estimated as focus-forming units (FFU) per milliliter. Supplemental Fig. S1D and E show immunostained NP2/CD4/CCR5 cells and primary macrophages following challenge with an R5 envelope-positive pseudotype virus. Foci formation required both CD4 and CCR5 and was blocked by 1 μM 3′-azido-3′-deoxythymidine.
Cell-cell fusion assays.
Cell-cell fusion was evaluated by cocultivating 293T cells transfected with env+ pSVIIIenv and env− pNL43 with appropriate test cells. Thus, coreceptor-positive NP2/CD4 or GHOST cells were seeded at 4 × 104 to 5 × 104 cells per well in 48-well culture dishes, while primary macrophages were prepared as described above and seeded at 2.5 × 105 per well in 48-well culture dishes and incubated overnight. 293T cells were transfected as described above and also incubated overnight. Each transfected 293T cell culture was trypsinized, and 50 μl of cells (5 × 105 cells/ml) was cocultivated with coreceptor-positive NP2/CD4 cells, GHOST cells, or macrophages. After 6 h of incubation, cocultivations with coreceptor-positive NP2/CD4 or GHOST cells were washed in PBS and fixed for 10 min in methanol containing 1% methylene blue and 0.25% basic fuchsin. Fixed cells were destained in PBS and examined by light microscopy for the presence of syncytia. Macrophage cocultivations were observed over 3 days and fixed when syncytia were apparent.
Nucleotide sequence accession numbers.
GenBank accession numbers for the V1V2 and V3 loops presented here are as follows: V1V2 loops, AY569981 to AY569999; V3 loops, AY570000 to AY570018.
RESULTS
Sequence analysis of envelope genes amplified from patient brain and LN tissue samples.
We first sequenced the variable V1V2 loops of each envelope clone obtained to identify individual genotypes and to omit replicate clones from further study. A total of 19 distinct V1V2 genotypes were identified from the five patients (Table 2). V1V2 sequences were distinct for each patient. NA118 and NA420 sequences are also closely related with previously published envelope sequences from these individuals (17). For NA420, brain and LN V1V2 sequences were clearly distinct genotypes consistent with tissue-specific evolution. For NA118, brain and LN V1V2 sequences revealed distinct genotypes; however, specific compartmentalization was less clear, as previously reported for this individual (17).
TABLE 2.
V1V2 loop amino acid sequences of patient envelopes
| Patient | Source | Clone | V1V2 sequencea |
|---|---|---|---|
| NA118 | LN | LN9 | CTDARNTTTN....TTGD.....SWKRMEKGELKNCSFNITTSMRDKMQKEYALFYKLDVVPIDDN.SSNNTSNYSSYRLISC |
| LN27 | ----T---GGSGGN--G.NTTSG---M----------------I-----------------S--K-N.-G...---------- | ||
| LN33 | --------R-...N--DG.....--QK--Q------------------------------------.---------------- | ||
| Brain | B12 | CTDATNNTSGSSKNTTG.NTTSGSWKMMEKGELKNCSFNITTSIRDKMQKEYALFYKLDVVQIDNNNSSHDNSNYSSYRLVSC | |
| B18 | ---WN-TATSDR--D-..----AG--------I----------------------------P--D-S-.NNT----D---I-- | ||
| B44 | ------T-G--GG----.-------------------------------------------P---D---.--...-----I-- | ||
| B45 | ------T-G--GG---SD--------I-----I-----------K-------------------KD---K-.--------I-- | ||
| NA420 | LN | LN40 | CTDLRNATNTNSSSERK.MEEGEIKNCSFYVTPTLRDKKQKEYATFYKLDVMPIDKDNTSYRLISC |
| LN85 | --------------GRIM-EG----------S-N---R---------------------------- | ||
| Brain | B3 | CTDLKNATNTNSSSGR.MMEGGAIKNCSFNI..SIRGKVQKEYAFFYKLDVIPIENDTTSYRLISC | |
| B13 | ----R-----------.--------------..--------------------------------- | ||
| B33 | ---FR-----------.-----E--------..---D----------------------------- | ||
| B42 | ---F------------.V----E--------..--------------------------------- | ||
| NA20 | Brain | B59 | CTDWRKNATAITTATPSSSEGTLEGGEMKNCSFNITTSIRDKVKKEYALFYRLDVVPIENDNTSYRLISC |
| B76 | ----------T-------RG------------------------------------------------N- | ||
| NA176 | Brain | B72 | CTDWKSNANTTNTTSSTNTTNSSGETMEGGDMKNCSFNITSNIRDKMQKEYALFYKLDVVPIGNSNTSYRIINC |
| B93 | ---------I---------------I------------------------------------------------ | ||
| NA353 | Brain | B13 | CTNANVTS------NTTSNSWETMEKGEIKNCSFNITTNIGGKMQKEYALFYKLDVIPIDNDTISYRLINC |
| B27 | --D--A--SSGNTT--------------------------S------------------------------ |
Dashes indicate homology, dots indicate spaces added to maintain alignment.
V3 loop charge.
The overall positive charge of the V3 loop has been used as an important indicator of HIV-1 tropism as either syncytium inducing or non-syncytium inducing (12). Syncytium-inducing and non-syncytium-inducing phenotypes are now known to represent viruses that use CXCR4 or are predominantly CCR5 using, respectively (2). All V3 loop sequences carried an overall charge of 3 or 4, consistent with a CCR5-using phenotype (Table 3).
TABLE 3.
V3 loop amino acid sequences of patient envelopes
| Sample | Clone | V3 loop sequenceb | Charge |
|---|---|---|---|
| Consensusa | CTRPNNNTRKSIHIGPGRAFYTATGEIIGDIRQAHC | 3 | |
| NA118 | LN9 | CTRPNNNTRKSIHIGPGRAFYATGDIIGDIRQAHC | 3 |
| LN27 | ----------------------------------- | 3 | |
| LN33 | ----------------------------------- | 3 | |
| B12 | CTRPNNNTRKSIHIGPGRAFYATGDIIGDIRQAHC | 3 | |
| B18 | ----------------------------------- | 3 | |
| B44 | ----------------------------------- | 3 | |
| B45 | ------------P-----------E---------- | 3 | |
| NA420 | LN40 | CTRPNNNTRKSIHLGPGRAFYTTGEITGDIRQAHC | 3 |
| LN85 | ----------------------------------- | 3 | |
| B3 | CTRPNNNTRKSINLGPGRALYTTGEITGDIRQAHC | 3 | |
| B13 | -------------------F------I-------- | 3 | |
| B33 | ----------------------------------- | 3 | |
| B42 | --------------------------I-------- | 3 | |
| NA20 | B59 | CTRPNNNTRKSIHLGPGSAFYTTGQILGDIRQAHC | 3 |
| B76 | -------------M---RV--A--A-T-------- | 4 | |
| NA176 | B72 | CTRPNNNTRKGIHIGPGRAFYTTGEIIGNIRQAHC | 4 |
| B93 | ----------------------------------- | 4 | |
| NA353 | B13 | CTRPNNNTRKGIHIGPGRAFYATGEIIGDIRQAHC | 3 |
| B27 | ----------------------------------- | 3 |
Consensus V3 sequence for R5 envelopes (4).
Dashes indicate homology.
Infectivity and coreceptor use of pseudotype viruses carrying LN- and brain-derived envelopes.
Pseudotype viruses were prepared by cotransfecting 293T cells with env+ pSVIIIenv together with env− pNL43. Pseudotype viruses were harvested after 48 h, and their infectivity was analyzed for the GHOST and NP2/CD4 series of coreceptor-expressing cell lines. Infectivity data for envelope-positive pseudotype viruses are shown in Table 4. Thirteen of the 19 distinct envelope genotypes yielded pseudotypes with infectivity for both GHOST/CCR5 and NP2/CD4/CCR5 cells. Infectivity of these pseudotypes was comparable to pseudotypes carrying well-characterized HIV-1 envelopes, e.g., SF162. Some envelopes (NA20 B76, NA353 B13, NA118 B18, NA118 B44, NA118 LN9, and NA420 B3) failed to yield infectious pseudotype viruses, while others consistently yielded pseudotypes with lower levels of infectivity (NA118 B45, NA176 B72, NA420 LN85, and NA420 LN40). Three distinct phenotypes of coreceptor use were observed. First, some envelopes, e.g., NA420 brain envelopes B33, B42, and B13, were mainly restricted for CCR5 use and unable to infect cells expressing other coreceptors. Second, the majority of functional envelopes consistently used CCR3 in addition to CCR5 but did not use other coreceptors. Third, a single envelope genotype from NA20 (B59) showed a broader use of coreceptors, including CCR3, CCR8, GPR1, GPR15, CXCR6, and CCR5 (Table 4).
TABLE 4.
Summary of macrophage syncytia induction by patient and control envelopes
| Patient | Envelopea | Infectivity (FFU/ml) for:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NP2/CD4
|
GHOST
|
|||||||||||
| CCR3 | CCR5 | CCR8 | CXCR4 | GPR1 | APJ | CCR2b | CCR5 | GPR15 | CXCR6 | CX3CR1 | ||
| NA118 | LN27 | 41,000 | 8,900 | —b | — | — | — | — | 23,500 | — | — | — |
| LN33 | 6,650 | 24,000 | — | — | — | — | — | 12,200 | — | — | — | |
| B12 | 32,100 | 40,100 | — | — | — | — | — | 179,000 | — | — | — | |
| B45 | — | 210 | — | — | — | — | — | 565 | — | — | — | |
| NA420 | LN40 | 120 | 1,105 | — | — | — | — | — | 590 | — | — | — |
| LN85 | 20 | 120 | — | — | — | — | — | 510 | — | — | — | |
| B13 | 230 | 37,850 | — | — | — | — | — | 44,000 | — | — | — | |
| B33 | — | 72,500 | — | — | — | — | — | 68,000 | — | — | — | |
| B42 | — | 12,400 | — | — | — | — | — | 14,500 | — | — | — | |
| NA20 | B59 | 40,500 | 56,500 | 3,750 | — | 420 | — | — | 38,000 | 710 | 215 | — |
| B76c | 6,000 | 16,300 | — | — | — | — | — | 15,650 | — | — | — | |
| NA176 | B72 | 139 | 7,650 | — | — | — | — | — | 8,100 | — | — | — |
| B93 | 13,480 | 43,600 | — | — | — | — | — | 32,000 | — | — | — | |
| NA353 | B27 | 19,100 | 114,500 | — | — | — | — | — | 187,000 | — | — | — |
Only results for functional envelopes are shown. Other envelopes (NA118 LN09, NA118 B18, NA118 B44, NA420 B03, and NA353 B13) failed to confer infection of the coreceptor-positive cells shown.
—, <10 FFU/ml.
Repaired envelope (see text on the next page).
Cell-cell fusion induced by patient envelopes.
The capacity of the different patient-derived envelopes to induce cell-cell fusion was evaluated. 293T cells were cotransfected with the pNL43 env− plasmid together with each patient env+ pSVIIIenv construct. After overnight incubation, the transfected 293T cells were trypsinized and cocultivated with different CD4+, coreceptor-positive cell lines (the GHOST and NP2/CD4 series of coreceptor-expressing cell lines). After incubations between 4 h and overnight, cocultivations were fixed and stained as described in Materials and Methods. Typical spreading syncytia were observed in cocultivations (Fig. 1). Syncytium induction assays confirmed the coreceptor use detected by infectivity assays described above. For example, envelopes that conferred infection of NP2/CD4/CCR3 also induced cell-cell fusion with this cell line (data not shown). The broad coreceptor use by NA20 envelope B59 was also confirmed by induction of syncytia via CCR5, CCR3, CCR8, GPR1, and very weakly via CXCR4 (but not via APJ) on NP2/CD4 cells and via GPR15 and CXCR6 on GHOST cells (Fig. 1). Cell-cell fusion was not detected for any other patient envelope via CXCR4 expressed either on NP2/CD4 or on GHOST cells. However, these CXCR4+ cells supported extensive syncytium formation when cocultivated with 293T cells expressing the X4 envelope of NL43 (data not shown).
FIG. 1.

Coreceptor use of NA20 B59 envelope evaluated by cell-cell fusion assays. NA20 B59-induced syncytia in GHOST and NP2/CD4 cells expressing a range of different coreceptors but not in the parental cell lines lacking coreceptors.
Patient envelopes that failed to yield infectious pseudotypes also failed to induce cell-cell fusion. This result suggests that these envelopes carry a defect(s) that eliminates their capacity to function in these assays, as reported for other envelopes PCR amplified from patient tissue (28). This conclusion is supported by the NA20 B76 envelope, where a single premature stop codon, identified in the extracellular domain of gp41 (data not shown), precluded the production of a full-length functional envelope. Repair of this stop codon restored full function to the NA20 B76 envelope. The repaired NA20 B76 was shown to be an R3R5 envelope similar to the majority of patient envelopes described here but distinct from the broadly tropic B59 envelope also from NA20 (Table 4).
Two patient envelopes, NA420 LN40 and NA420 LN85, consistently yielded pseudotypes that carried only low-level infectivity for GHOST/CCR5 and NP2/CD4/CCR5 cells. Interestingly, these envelopes were fully functional for cell-cell fusion and induced syncytia formation on both GHOST/CCR5 and NP2/CD4/CCR5 cells. Moreover, both low-infectivity envelopes conferred cell-cell fusion in NP2/CD4/CCR3 cells, confirming an R5R3 phenotype. Both NA420 LN40 and NA420 LN85 envelopes contained mutations in the gp41 cytoplasmic domain, mutations that severely reduced their incorporation onto budding virions (data not shown). These envelopes were also restored to full infectivity by preparing chimeric constructs that contained gp120 sequences from either NA420 LN40 or NA420 LN85 and gp41 sequence from NA420 B33.
Infection and cell-cell fusion of primary macrophage cultures.
Both Gorry's UK1-br brain isolate (16) and Shieh's microglia-passaged BORI variant (39) were reported to be more infectious and fusigenic for microglial cells and macrophages. We therefore compared the capacity of the patient-derived envelopes to confer infectivity and syncytium induction in primary macrophage cultures. Patient env+ pseudotypes with high infectivity titers on CD4+ CCR5+ cell lines also infected primary macrophages (Fig. 2A). Ratios of patient env+ pseudotype infectivity for macrophages compared to those for GHOST/CCR5 are shown in Fig. 2B. These results indicate that four brain envelopes infected primary macrophages efficiently, while other patient envelopes conferred less-efficient or no infection. We also tested envelopes of several well-characterized HIV-1 strains including macrophage-tropic (SF162 and AD8) and brain- or cerebrospinal fluid-derived (YU2, JRFL, and JRCSF) viruses. These envelopes were cloned from infectious molecular clones into pSVIIIenv. Four of these (YU2, JRFL, SF162, and AD8) conferred infection of primary macrophage cultures. However, at least two of the brain-derived patient envelopes (NA20 B59 and NA353 B27) conferred even higher macrophage-GHOST/CCR5 infectivity ratios, indicating that these envelopes carry an enhanced macrophage tropism. Both R5 and R5R3 patient envelopes segregated with macrophage-tropic and non-macrophage-tropic envelopes, and thus no obvious association was observed between CCR3 use and macrophage infection.
FIG. 2.

Infection of primary macrophages by patient LN and brain envelopes. (A) Infectivity titers for patient envelope-positive pseudotype viruses titrated on primary macrophages from different donors and on GHOST/CCR5 cells. Arrows indicate that infection is below the level of detection. Data are derived from duplicate infections on two donor macrophage batches. GHOST/CCR5 titers were derived from infectivity titrations carried out with the same batch of virus as in macrophage infections. (B) Macrophage-GHOST/CCR5 infectivity ratios indicate that patient envelopes NA20 B59, NA353 B27, NA420 B33, and NA176 B93 are highly macrophage tropic.
NA118 B12, LN27, and LN33 envelopes failed to confer macrophage infection, even though they conferred high infectivity for GHOST/CCR5. The three NA420 brain envelopes conferred macrophage infection with NA420 B33 infecting most efficiently of the three. In contrast, both repaired NA420 LN envelopes failed to confer any macrophage infection. Taken together, these results show that all LN-derived envelopes were non-macrophage tropic, while most brain-derived envelopes (all except NA118 B12) conferred macrophage infection (Fig. 2A and B).
We next evaluated the capacity of patient envelopes to induce cell-cell fusion in primary macrophage cultures. These fusion assays were carried out by cocultivation of primary macrophage cultures with 293T cells cotransfected with patient env+ pSVIIIenv and env− pNL43 vectors as described in Materials and Methods. A minority of envelopes induced very large and spreading macrophage syncytia containing numerous nuclei (Fig. 3). The same four patient envelopes that efficiently infected macrophages consistently induced large spreading syncytia over 2 to 3 days of coculture. The extent of macrophage syncytia induction was donor dependent. Of the well-characterized control envelopes (YU2, JRCSF, JRFL, SF162, and AD8), only AD8 conferred levels of macrophage syncytia similar to the patient brain envelopes described above. Macrophage syncytium induction is summarized in Table 5.
FIG. 3.
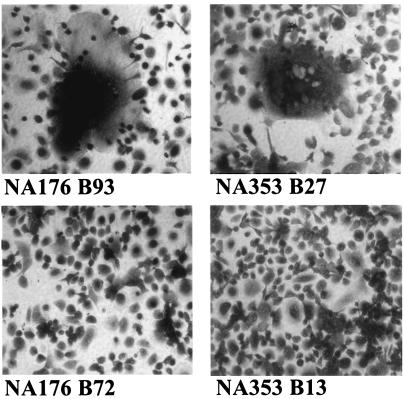
Syncytium induction in primary macrophages by patient brain-derived envelopes. Large spreading syncytia induced by patient brain envelopes NA176 B93 and NA353 B27 are shown. Note that the syncytia shown contain hundreds of nuclei (top panels). No syncytia were induced by NA176 B72 or the nonfunctional NA353 B13 (bottom panels).
TABLE 5.
Summary of macrophage syncytia induction by patient and control envelopes
| Patient envelopes | Phenotypea | Cell-cell fusion in macrophagesb |
|---|---|---|
| NA118 LN27 | R5R3 | − |
| NA118 LN33 | R5R3 | − |
| NA118 B12 | R5R3 | − |
| NA420 LN40 | R5R3 | − |
| NA420 LN85 | R5R3 | − |
| NA420 B13 | R5 | − |
| NA420 B33 | R5 | + |
| NA420 B42 | R5 | − |
| NA20 B59 | R5R3*** | ++ |
| NA20 B76 | R5R3 | − |
| NA176 B72 | R5R3 | − |
| NA176 B93 | R5R3 | +++ |
| NA353 B27 | R5R3 | +++ |
| SF162 | R5* | + |
| JRCSF | R5 | − |
| JRFL | R5R3 | + |
| AD8 | R5R3* | +++ |
| YU2 | R5R3* | + |
Coreceptor use of control envelopes (SF162, JRCSF, JRFL, AD8, and YU2) was taken from the HIV sequence database (http://www.hiv.lanl.gov/). *, use of additional coreceptors; ***, use of several additional coreceptors.
Cell-cell fusion data are representative of at least three independent experiments. Cell-cell fusion in GHOST/CCR5 cells and primary macrophages was assessed by the extent of syncytium formation following overnight cocultivation with 293T cells transfected with env+ pSVIIIenv and env mutant pNL43. For GHOST/CCR5 cells, all envelopes showed extensive syncytium formation involving over 50% of plated cells. Syncytium formation for primary macrophages is represented as follows: −, no syncytia formed; +, 0 to 10 large syncytia observed per well of a 48-well culture dish; ++, 10 to 25 syncytia per well; +++, >25 syncytia per well.
Brain-derived HIV-1 envelopes are not CD4 independent.
Several studies have correlated SIV macrophage tropism with the capacity to infect CCR5+ cell lines in vitro in the absence of CD4 (24, 31). We therefore tested whether pseudotype viruses carrying patient envelopes could infect CD4− NP2/CCR5 cells. We also tested a primary HIV-2 strain (ALI) that was previously shown to infect CCR5+ cells in the absence of CD4 (33). In contrast to HIV-2 ALI, none of the brain or LN envelopes conferred any infection of CD4− NP2/CCR5 cells (data not shown), even though many envelopes conferred high levels of infection on NP2/CD4/CCR5 cells (Table 4). It was previously shown that pretreatment of HIV-2 with soluble CD4 induced infection of CD4− cells (6). However, soluble CD4 (1 μg/ml) had no effect on the capacity of patient envelopes to confer infection or cell-cell fusion of CD4− NP2/CCR5 cells (data not shown). Together, these observations show that all the patient envelopes investigated here, whether from LN or brain, were completely dependent on cell surface CD4 for infection and cell-cell fusion.
Some brain envelopes can infect cells expressing low levels of CD4 and/or CCR5.
Although, none of the patient envelopes studied here were CD4 independent, it was possible that brain envelopes required less cell surface CD4 to trigger infection as described by Gorry et al. (16) and Martin et al. (23). We therefore tested the capacity of pseudotype viruses carrying brain and LN envelopes to infect HeLa cells expressing either high or low levels of CD4 and three different levels of CCR5 (30). Platt et al. reported that the high CD4 clones express approximately 4 × 105 molecules/cell and the low CD4 clones express approximately 104 receptors/cell. Low, medium, and high CCR5 clones were reported to express approximately 2 × 103, 104, and 105 molecules/cell, respectively. Immunostaining and fluorescence-activated cell sorter analysis confirmed that these different clones expressed low or high CD4 and low, medium, or high CCR5 amounts on their cell surfaces.
All envelopes tested were capable of infecting HeLa cells expressing high levels of CD4 and different levels of CCR5 (Fig. 4). Infection of HeLa/HiCD4 cells with the lowest CCR5 was within 1 log of maximum infectivity recorded in the presence of the highest CCR5 levels. In contrast, infectivity recorded on HeLa cells with low levels of CD4 (HeLa/LowCD4) showed that only particular envelopes were able to infect these cells efficiently (Fig. 4). These included the four brain-derived envelopes that scored highest for macrophage fusion and infection. Infection by patient NA118 envelopes (B12, LN27, and LN33) was severely affected by CD4 expression. NA420 brain and LN envelopes clearly segregated into separate phenotypes with NA420 B33, B13, and B42 envelopes conferring infection of HeLa/LowCD4 relatively efficiently, while infection by the repaired NA420 LN envelopes was below the level of detection on cells expressing low levels of CD4. JRCSF showed a CD4 requirement similar to those of the NA118 and NA420 LN envelopes. Several of the control macrophage-tropic HIV-1 envelopes (AD8 as well as brain tissue-derived YU2 and JRFL) also infected in the presence of low CD4 and low CCR5. These envelopes were as efficient on the HeLa/LowCD4/CCR5 cultures as were patient brain envelopes NA20 B59 and NA353 B27.
FIG. 4.

Infection of cells expressing low levels of CD4 and/or CCR5 by patient envelopes. Infection of HeLa/HiCD4 cells expressing high levels of CD4 and three different amounts of CCR5 (top panels). Infection of HeLa/LowCD4 expressing low levels of CD4 and three different amounts of CCR5 (bottom panels). Infectivity was assessed as FFU per milliliter by immunostaining for p24 as described in Materials and Methods. Values plotted are the average FFU counts for duplicate wells in a single experiment. Similar data were obtained from at least one repeat experiment. Results for different envelopes are grouped into separate graphs to avoid congestion.
Dejucq et al. previously reported that particular R5 HIV-1 isolates were able to infect the CD4+ T-cell line MOLT 4. Although these cells express high levels of CD4, CCR5 expression was undetectable by flow cytometry and only weakly detectable by reverse transcription-PCR (7). Nevertheless, MOLT 4 infection by R5 isolates was completely inhibited by CCR5-specific ligands (7). Here, we tested whether the patient envelopes could infect or induce cell-cell fusion of MOLT 4 cells. The four macrophage-tropic brain-derived envelopes conferred infection of MOLT 4 cells (data not shown) and were able to induce the formation of large syncytia (Fig. 5). Syncytium formation by NA20 B59 and by NA176 B93 was completely blocked by TAK779 and unaffected by a range of ligands for other potential coreceptors, including AMD3100, the ligand for CXCR4 (data not shown). Several other patient envelopes induced low levels of small syncytia. Of the control envelopes, only AD8 and YU2 consistently induced low levels of MOLT 4 syncytia.
FIG. 5.

Cell-to-cell fusion induced by patient envelopes in MOLT 4 cells that express very low levels of CCR5. 293T cells expressing different patient envelopes were cocultivated with MOLT 4 cells. The presence of syncytia was assessed by light microscopy after overnight incubation. Arrows mark examples of syncytia. The control panel shows cocultivation of MOLT 4 with 293T cells transfected with a pSVIIIenv carrying a defective envelope.
Together these results show that four brain-derived envelopes were capable of exploiting low levels of CD4 and/or CCR5 for infection and cell-cell fusion of macrophages and T cells.
Sensitivity of patient envelopes to inhibition by ligands to CD4 and CCR5.
We next tested whether the distinct phenotypes of the patient envelopes impacted on their sensitivity to inhibition by reagents that blocked either CD4 or CCR5. We first tested the blockade of CCR5 by TAK779. GHOST/CCR5 cells were treated with twofold dilutions of TAK779 for 1 h before challenging with 100 to 200 FFU of patient envelope-positive pseudotype viruses as described in Materials and Methods. Inhibition was assessed as 90% inhibitory concentrations (IC90s) (Fig. 6). A range of IC90s was evident and no clear association of TAK779 sensitivity and envelope phenotype could be seen (Fig. 6A). IC90s were also obtained for envelope-positive pseudotype inhibition by Q4120 (Fig. 6B). Q4120 is a monoclonal antibody that recognizes the N-terminal domain of CD4 and blocks gp120 binding and infectivity. In contrast to TAK779, Q4120 IC90s showed a strong correlation with patient envelope phenotype. The four brain-derived envelopes that showed enhanced fusigenicity and tropism for macrophages and MOLT 4 cells required the highest doses of Q4120 for inhibition. These results are consistent with an increase in envelope affinity for CD4, conferring an adaptation to infect cells expressing low levels of CD4. Of note, all four LN envelopes clustered along with JRCSF as the most sensitive to Q4120 inhibition. These results are consistent with the hypothesis that envelopes that evolve a higher affinity for CD4 will have an increased tropism for cells expressing low levels of CD4 and/or CCR5, including both macrophages and T cells.
FIG. 6.

Sensitivity of patient envelopes to CCR5 and CD4 ligands. (A) TAK779 (CCR5 inhibitor) IC090 0 inhibitory doses. (B) Q4120 (anti-CD4 domain 1) IC090 0 inhibitory doses. Black symbols represent macrophage-tropic patient envelopes. Gray symbols are other patient envelopes. Striped symbols are control, well-characterized HIV-1 envelopes. Data shown are derived from a single experiment and are representative of data from several experiments.
DISCUSSION
We have characterized the biological properties of complete envelope genes amplified by PCR from brain tissue of five AIDS patients with neurological conditions. For two of the patients, envelope genes were also successfully recovered from LN tissue. We have identified highly macrophage-tropic and fusigenic envelopes in brain tissue of four of the five patients. Two very distinct envelope phenotypes were observed. A group of four brain envelopes were highly macrophage tropic and infected cells expressing low amounts of CD4 or CCR5. The LN envelopes formed a second group that failed to confer macrophage infection and required substantially higher amounts of CD4 for infection.
The potential impact of PCR error on envelope phenotype.
Investigation of HIV-1 cell tropism has generally been carried out using viruses isolated into PBMCs. This approach is likely to be a highly selective process that will amplify virus strains that replicate fastest in CD4+ T cells and may overlook virus strains adapted for replication in specialized cells resident in particular tissues or organs. This possibility is potentially important for HIV in the brain, where perivascular macrophages and microglial cells that express low levels of CD4 (1, 21, 26) are the predominant infected cell types. In this study, we have used PCR techniques to amplify HIV-1 envelope sequences directly from brain tissue samples, thus avoiding possible selection in culture. PCR amplification of viral sequences also has limitations and may result in the introduction of mutations into the DNA products. The proofreading enzymes used here help to minimize this possibility, and the majority of PCR-amplified envelopes described here were fully functional.
We were still concerned that envelopes carrying PCR-introduced errors may appear functional yet carry less-obvious defects. For example, NA118 and NA420 LN envelopes all failed to confer macrophage infection, were most sensitive to Q4120 inhibition, and required high levels of CD4 for infection. The reduced efficiency of these envelopes could potentially be due to PCR-introduced errors that affect the capacity of an envelope to mediate infection. Thus, PCR-introduced sequence errors would be expected to render such envelopes generally more sensitive to all inhibitors of infection. However, these envelopes were not sensitive to all inhibitors and varied considerably in their sensitivity to TAK779 inhibition with the NA118 LN33 envelope, the most resistant (Fig. 6). Alternatively, PCR-introduced errors may impact on the amount of envelope processed into gp120 and gp41 and assembled onto virions. The patient envelopes showed some variation in the amount of envelope processed and on virions, but this did not correlate with other envelope properties reported here. For instance, NA118 envelopes were processed and incorporated into virions as efficiently as the most macrophage-tropic envelope, NA353 B27 (data not shown). Finally, the JRCSF envelope was cloned from an early PBMC culture of isolated virus (20) and was not derived by PCR, yet it showed a similar requirement for high CD4 for infection and failed to confer macrophage infection. Our results show a wide variation in envelope requirement for CD4 to induce infection that is not related to PCR amplification procedures. We therefore believe that these procedures are a viable alternative to virus isolation for the study of HIV envelope phenotype.
Compartmentalized envelope evolution and sampling issues.
Study subjects NA118 and NA420 were previously reported to carry distinct V3 loop envelope sequences in brain and LN consistent with compartmentalized evolution (9). V1V2 and V3 sequences of the NA420 envelopes studied here were also distinct for brain versus LN origins (Tables 2 and 3). Phenotype analyses showed that these sequence differences translated into biological differences. Specifically, the three brain envelopes were specific for infection via CCR5 and could confer infection of cells expressing low levels of CD4, including macrophages. In contrast, NA420 LN envelopes were able to fuse with cells expressing either CCR3 or CCR5 but required high levels of CD4 for infection and failed to confer infection of macrophages. These observations are consistent with compartmentalized phenotype evolution occurring in parallel with genotype evolution. All envelopes described here were amplified from brain or LN tissue samples. Since tissues contain a mixture of cell types and brain tissue also contains blood capillaries and blood cells, it is not possible to know definitively their cellular origin.
Coreceptor use.
There was no correlation between coreceptor use and tissue of origin for the envelopes investigated. Moreover, no correlation was apparent between coreceptor use and other envelope properties, e.g., macrophage infectivity. Only the NA420 brain envelopes were restricted to infection via CCR5. All other functional envelopes conferred infection of CD4+ NP2 cells via CCR3 and CCR5. Intriguingly, infectivity titers recorded on NP2/CD4/CCR3 were nearly as high as those for NP2/CD4/CCR5 cells (Table 4). The significance of CCR3 use by the majority of envelopes amplified is unclear, since there is no current evidence to indicate that CCR3 is used in vivo or is more than a minor coreceptor. NA20 B59 carries a V3 loop that has a low overall charge, consistent with an R5 phenotype. This envelope used a broad range of different coreceptors, including CXCR4, albeit weakly. Ohagen's study (28) also detected several low-V3-charged envelopes that conferred broad coreceptor use. This type of envelope may therefore represent a new category of HIV-1 envelope that needs to be considered in the light of new therapies that target CCR5.
Brain envelopes and macrophage tropism.
We identified envelopes that were highly fusigenic and tropic for primary macrophages in brain tissue from four of the five patients. These envelopes were also able to confer infection of cells expressing low levels of CD4 and CCR5. It was possible that these envelopes represented a distinct category for CCR5-using viruses. Alternatively, they might simply be macrophage tropic, a phenotype expected for envelopes from the brain, where the main cell types infected are of monocyte/macrophage lineage. We therefore included in our study several well-characterized macrophage-tropic HIV-1 envelopes to help evaluate whether the enhanced fusigenicity and macrophage tropism of the four patient envelopes was a distinct outlying phenotype. Our results show that the four brain envelopes are at the upper limit of a range of macrophage fusigenicity and tropism. Of the five control envelopes, the AD8 envelope was the most fusigenic for primary macrophages and showed a capacity for infection of cells with low levels of CD4 similar to those of the four highly macrophage-tropic, brain-derived envelopes.
Broadening of tropism for both macrophages and T cells.
The heightened macrophage fusigenicity and tropism correlated with increased resistance to inhibition by Q4120, an antibody that binds domain 1 of CD4 and blocks infection. However, sensitivity to inhibition by TAK779 was apparently not closely associated with macrophage tropism. These results are consistent with an increased affinity of the brain envelopes for CD4 that confers the capacity to infect cells expressing low levels of CD4. As reported by others (16, 23, 39), this adaptation may confer more-efficient replication in brain microglia and macrophages, which express relatively low levels of CD4. Whether the four brain envelopes have evolved in response to selective pressures in the brain or whether envelopes with similar properties are present in other tissues (17, 48) is unclear from the present study. Indeed, the most macrophage-tropic envelopes were also able to infect and fuse with the T-cell line, MOLT 4, that expresses very low levels of CCR5 (7). These results indicate that such envelopes are broadly tropic for both macrophage and T-cell populations.
In summary, we have amplified complete HIV-1 envelope genes from brain tissue of AIDS patients with neurological conditions. Envelopes that were highly fusigenic and tropic for macrophages were obtained from four of the five patients. These envelopes also conferred infection and fusion of cells expressing low levels of CD4 and/or CCR5. As suggested by others (16, 23), we believe these envelopes are neurotropic and adapted for replication in brain macrophages and microglial cells that express low levels of CD4 (8, 21, 47).
Supplementary Material
Acknowledgments
We thank Bruce Blais for help with flow cytometry, Richard Hudson for supplying white blood cells, Mario Stevenson for primary macrophage cultures, and Ellen Kittler and Maria Zapp for valuable assistance in DNA sequencing. These services were provided by the University of Massachusetts Center for AIDS Research. We thank Navid Madani (Dana-Farber Cancer Institute, Boston, Mass.) and David Kabat (Oregon Health & Science University) for providing the HeLa cell clones. We also thank Hiroo Hoshino (University of Gunma, Gunma, Japan) for kindly providing NP2 cells. Brain and LN autopsy samples were provided by the Edinburgh brain bank. We are highly appreciative of the NIH AIDS Research and Reference Reagent Program and the U.K. Centralised Facility for AIDS Reagents, which provided many reagents for the work presented.
Early work in this study was supported by a program grant funded by the U.K. Medical Research Council. Later (and most) work was supported by NIH grant R01MH64408-01 and amFAR grant 02802-30-RG. S.H. was funded by a U.K. Medical Research Council studentship. P.R.C. is an Elizabeth Glaser Pediatric AIDS Foundation scientist.
REFERENCES
- 1.Bannert, N., D. Schenten, S. Craig, and J. Sodroski. 2000. The level of CD4 expression limits infection of primary rhesus monkey macrophages by a T-tropic simian immunodeficiency virus and macrophagetropic human immunodeficiency viruses. J. Virol. 74:10984-10993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger, E. A., R. W. Doms, E.-M. Fenyo, B. T. M. Korber, D. R. Littman, J. P. Moore, Q. J. Sattentau, H. Schuitemaker, J. Sodroski, and R. A. Weiss. 1998. A new classification for HIV-1. Nature 391:240. [DOI] [PubMed] [Google Scholar]
- 2a.Cecilia, D., V. N. KewalRamani, J. O’Leary, B. Volsky, P. Nyambi, S. Burda, S. Xu, D. R. Littman, and S. Zolla-Pazner. 1998. Neutralization profiles of primary human immunodeficiency virus type 1 isolates in the context of coreceptor usage. J. Virol. 72:6988-6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan, S. Y., R. F. Speck, C. Power, S. L. Gaffen, B. Chesebro, and M. A. Goldsmith. 1999. V3 recombinants indicate a central role for CCR5 as a coreceptor in tissue infection by human immunodeficiency virus type 1. J. Virol. 73:2350-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chesebro, B., K. Wehrly, J. Nishio, and S. Perryman. 1992. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J. Virol. 66:6547-6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho, M. W., M. K. Lee, M. C. Carney, J. F. Berson, R. W. Doms, and M. A. Martin. 1998. Identification of determinants on a dualtropic human immunodeficiency virus type 1 envelope glycoprotein that confer usage of CXCR4. J. Virol. 72:2509-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clapham, P. R., A. McKnight, and R. A. Weiss. 1992. Human immunodeficiency virus type 2 infection and fusion of CD4-negative human cell lines: induction and enhancement by soluble CD4. J. Virol. 66:3531-3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dejucq, N., G. Simmons, and P. R. Clapham. 1999. Expanded tropism of primary human immunodeficiency virus type 1 R5 strains to CD4+ T-cell lines determined by the capacity to exploit low concentrations of CCR5. J. Virol. 73:7842-7847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dick, A. D., M. Pell, B. J. Brew, E. Foulcher, and J. D. Sedgwick. 1997. Direct ex vivo flow cytometric analysis of human microglial cell CD4 expression: examination of central nervous system biopsy specimens from HIV-seropositive patients and patients with other neurological disease. AIDS 11:1699-1708. [DOI] [PubMed] [Google Scholar]
- 9.Donaldson, Y. K., J. E. Bell, E. C. Holmes, E. S. Hughes, H. K. Brown, and P. Simmonds. 1994. In vivo distribution and cytopathology of variants of human immunodeficiency virus type 1 showing restricted sequence variability in the V3 loop. J. Virol. 68:5991-6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Everall, I. P., P. J. Luthert, and P. L. Lantos. 1991. Neuronal loss in the frontal cortex in HIV infection. Lancet 337:1119-1121. [DOI] [PubMed] [Google Scholar]
- 11.Flaherty, M. T., D. A. Hauer, J. L. Mankowski, M. C. Zink, and J. E. Clements. 1997. Molecular and biological characterization of a neurovirulent molecular clone of simian immunodeficiency virus. J. Virol. 71:5790-5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fouchier, R. A., M. Groenink, N. A. Kootstra, M. Tersmette, H. G. Huisman, F. Miedema, and H. Schuitemaker. 1992. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J. Virol. 66:3183-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gabuzda, D., and J. Wang. 2000. Chemokine receptors and mechanisms of cell death in HIV neuropathogenesis. J. Neurovirol. 6(Suppl. 1):S24-S32. [PubMed] [Google Scholar]
- 14.Gao, F., S. G. Morrison, D. L. Robertson, C. L. Thornton, S. Craig, G. Karlsson, J. Sodroski, M. Morgado, B. Galvao-Castro, H. von Briesen, S. Beddows, J. Weber, P. M. Sharp, G. M. Shaw, B. H. Hahn, and the WHO and NIAID networks for HIV isolation and characterization. 1996. Molecular cloning and analysis of functional envelope genes from human immunodeficiency virus type 1 sequence subtypes A through G. J. Virol. 70:1651-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorry, P. R., G. Bristol, J. A. Zack, K. Ritola, R. Swanstrom, C. J. Birch, J. E. Bell, N. Bannert, K. Crawford, H. Wang, D. Schols, E. De Clercq, K. Kunstman, S. M. Wolinsky, and D. Gabuzda. 2001. Macrophage tropism of human immunodeficiency virus type 1 isolates from brain and lymphoid tissues predicts neurotropism independent of coreceptor specificity. J. Virol. 75:10073-10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorry, P. R., J. Taylor, G. H. Holm, A. Mehle, T. Morgan, M. Cayabyab, M. Farzan, H. Wang, J. E. Bell, K. Kunstman, J. P. Moore, S. M. Wolinsky, and D. Gabuzda. 2002. Increased CCR5 affinity and reduced CCR5/CD4 dependence of a neurovirulent primary human immunodeficiency virus type 1 isolate. J. Virol. 76:6277-6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hughes, E. S., J. E. Bell, and P. Simmonds. 1997. Investigation of the dynamics of the spread of human immunodeficiency virus to brain and other tissues by evolutionary analysis of sequences from the p17gag and env genes. J. Virol. 71:1272-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalter, D. C., M. Nakamura, J. A. Turpin, L. M. Baca, D. L. Hoover, C. Dieffenbach, P. Ralph, H. E. Gendelman, and M. S. Meltzer. 1991. Enhanced HIV replication in macrophage colony-stimulating factor-treated monocytes. J. Immunol. 146:298-306. [PubMed] [Google Scholar]
- 19.Koenig, S., H. E. Gendelman, J. M. Orenstein, M. C. Dal Canto, G. H. Pezeshkpour, M. Yungbluth, F. Janotta, A. Aksamit, M. A. Martin, and A. S. Fauci. 1986. Detection of AIDS virus in macrophages in brain tissue from AIDS patients with encephalopathy. Science 233:1089-1093. [DOI] [PubMed] [Google Scholar]
- 20.Koyanagi, Y., S. Miles, R. T. Mitsuyasu, J. E. Merrill, H. V. Vinters, and I. S. Chen. 1987. Dual infection of the central nervous system by AIDS viruses with distinct cellular tropisms. Science 236:819-822. [DOI] [PubMed] [Google Scholar]
- 21.Lee, B., M. Sharron, L. J. Montaner, D. Weissman, and R. W. Doms. 1999. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc. Natl. Acad. Sci. USA 96:5215-5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mankowski, J. L., M. T. Flaherty, J. P. Spelman, D. A. Hauer, P. J. Didier, A. M. Amedee, M. Murphey-Corb, L. M. Kirstein, A. Munoz, J. E. Clements, and M. C. Zink. 1997. Pathogenesis of simian immunodeficiency virus encephalitis: viral determinants of neurovirulence. J. Virol. 71:6055-6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin, J., C. C. LaBranche, and F. Gonzalez-Scarano. 2001. Differential CD4/CCR5 utilization, gp120 conformation, and neutralization sensitivity between envelopes from a microglia-adapted human immunodeficiency virus type 1 and its parental isolate. J. Virol. 75:3568-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Means, R. E., T. Matthews, J. A. Hoxie, M. H. Malim, T. Kodama, and R. C. Desrosiers. 2001. Ability of the V3 loop of simian immunodeficiency virus to serve as a target for antibody-mediated neutralization: correlation of neutralization sensitivity, growth in macrophages, and decreased dependence on CD4. J. Virol. 75:3903-3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michaels, J., L. R. Sharer, and L. G. Epstein. 1988. Human immunodeficiency virus type 1 (HIV-1) infection of the nervous system: a review. Immunodefic. Rev. 1:71-104. [PubMed] [Google Scholar]
- 26.Mori, K., M. Rosenzweig, and R. C. Desrosiers. 2000. Mechanisms for adaptation of simian immunodeficiency virus to replication in alveolar macrophages. J. Virol. 74:10852-10859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moses, A. V., and J. A. Nelson. 1994. HIV infection of human brain capillary endothelial cells—implications for AIDS dementia. Adv. Neuroimmunol. 4:239-247. [DOI] [PubMed] [Google Scholar]
- 28.Ohagen, A., A. Devitt, K. J. Kunstman, P. R. Gorry, P. P. Rose, B. Korber, J. Taylor, R. Levy, R. L. Murphy, S. M. Wolinsky, and D. Gabuzda. 2003. Genetic and functional analysis of full-length human immunodeficiency virus type 1 env genes derived from brain and blood of patients with AIDS. J. Virol. 77:12336-12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petito, C. K., E. S. Cho, W. Lemann, B. A. Navia, and R. W. Price. 1986. Neuropathology of acquired immunodeficiency syndrome (AIDS): an autopsy review. J. Neuropathol. Exp. Neurol. 45:635-646. [DOI] [PubMed] [Google Scholar]
- 30.Platt, E. J., K. Wehrly, S. E. Kuhmann, B. Chesebro, and D. Kabat. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 72:2855-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Puffer, B. A., S. Pohlmann, A. L. Edinger, D. Carlin, M. D. Sanchez, J. Reitter, D. D. Watry, H. S. Fox, R. C. Desrosiers, and R. W. Doms. 2002. CD4 independence of simian immunodeficiency virus Envs is associated with macrophage tropism, neutralization sensitivity, and attenuated pathogenicity. J. Virol. 76:2595-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ranki, A., M. Nyberg, V. Ovod, M. Haltia, I. Elovaara, R. Raininko, H. Haapasalo, and K. Krohn. 1995. Abundant expression of HIV Nef and Rev proteins in brain astrocytes in vivo is associated with dementia. AIDS 9:1001-1008. [DOI] [PubMed] [Google Scholar]
- 33.Reeves, J. D., S. Hibbitts, G. Simmons, A. McKnight, J. M. Azevedo-Pereira, J. Moniz-Pereira, and P. R. Clapham. 1999. Primary human immunodeficiency virus type 2 (HIV-2) isolates infect CD4-negative cells via CCR5 and CXCR4: comparison with HIV-1 and simian immunodeficiency virus and relevance to cell tropism in vivo. J. Virol. 73:7795-7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ross, T. M., and B. R. Cullen. 1998. The ability of HIV type 1 to use CCR-3 as a coreceptor is controlled by envelope V1/V2 sequences acting in conjunction with a CCR-5 tropic V3 loop. Proc. Natl. Acad. Sci. USA 95:7682-7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rostad, S. W., S. M. Sumi, C. M. Shaw, K. Olson, and J. K. McDougall. 1987. Human immunodeficiency virus (HIV) infection in brains with AIDS-related leukoencephalopathy. AIDS Res. Hum. Retroviruses 3:363-373. [DOI] [PubMed] [Google Scholar]
- 36.Saito, Y., L. R. Sharer, L. G. Epstein, J. Michaels, M. Mintz, M. Louder, K. Golding, T. A. Cvetkovich, and B. M. Blumberg. 1994. Overexpression of nef as a marker for restricted HIV-1 infection of astrocytes in postmortem pediatric central nervous tissues. Neurology 44:474-481. [DOI] [PubMed] [Google Scholar]
- 37.Sharer, L. R., Y. Saito, L. G. Epstein, and B. M. Blumberg. 1994. Detection of HIV-1 DNA in pediatric AIDS brain tissue by two-step ISPCR. Adv. Neuroimmunol. 4:283-285. [DOI] [PubMed] [Google Scholar]
- 38.Sharma, D. P., M. C. Zink, M. Anderson, R. Adams, J. E. Clements, S. V. Joag, and O. Narayan. 1992. Derivation of neurotropic simian immunodeficiency virus from exclusively lymphocytetropic parental virus: pathogenesis of infection in macaques. J. Virol. 66:3550-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shieh, J. T., J. Martin, G. Baltuch, M. H. Malim, and F. Gonzalez-Scarano. 2000. Determinants of syncytium formation in microglia by human immunodeficiency virus type 1: role of the V1/V2 domains. J. Virol. 74:693-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simmonds, P., P. Balfe, J. F. Peutherer, C. A. Ludlam, J. O. Bishop, and A. J. Brown. 1990. Human immunodeficiency virus-infected individuals contain provirus in small numbers of peripheral mononuclear cells and at low copy numbers. J. Virol. 64:864-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simmons, G., A. McKnight, Y. Takeuchi, H. Hoshino, and P. R. Clapham. 1995. Cell-to-cell fusion, but not virus entry in macrophages by T-cell line tropic HIV-1 strains: a V3 loop-determined restriction. Virology 209:696-700. [DOI] [PubMed] [Google Scholar]
- 42.Smit, T. K., B. Wang, T. Ng, R. Osborne, B. Brew, and N. K. Saksena. 2001. Varied tropism of HIV-1 isolates derived from different regions of adult brain cortex discriminate between patients with and without AIDS dementia complex (ADC): evidence for neurotropic HIV variants. Virology 279:509-526. [DOI] [PubMed] [Google Scholar]
- 42a.Soda, Y., N. Shimizu, A. Jinno, H. Y. Liu, K. Kanbe, T. Kitamura, and H. Hoshino. 1999. Establishment of a new system for determination of coreceptor usages of HIV based on the human glioma NP-2 cell line. Biochem. Biophys. Res. Commun. 258:313-321. [DOI] [PubMed] [Google Scholar]
- 43.Takahashi, K., S. L. Wesselingh, D. E. Griffin, J. C. McArthur, R. T. Johnson, and J. D. Glass. 1996. Localization of HIV-1 in human brain using polymerase chain reaction/in situ hybridization and immunocytochemistry. Ann. Neurol. 39:705-711. [DOI] [PubMed] [Google Scholar]
- 44.Torres-Munoz, J., P. Stockton, N. Tacoronte, B. Roberts, R. R. Maronpot, and C. K. Petito. 2001. Detection of HIV-1 gene sequences in hippocampal neurons isolated from postmortem AIDS brains by laser capture microdissection. J. Neuropathol. Exp. Neurol. 60:885-892. [DOI] [PubMed] [Google Scholar]
- 45.Trillo-Pazos, G., A. Diamanturos, L. Rislove, T. Menza, W. Chao, P. Belem, S. Sadiq, S. Morgello, L. Sharer, and D. J. Volsky. 2003. Detection of HIV-1 DNA in microglia/macrophages, astrocytes and neurons isolated from brain tissue with HIV-1 encephalitis by laser capture microdissection. Brain Pathol. 13:144-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valle, L. D., S. Croul, S. Morgello, S. Amini, J. Rappaport, and K. Khalili. 2000. Detection of HIV-1 Tat and JCV capsid protein, VP1, in AIDS brain with progressive multifocal leukoencephalopathy. J. Neurovirol. 6:221-228. [DOI] [PubMed] [Google Scholar]
- 47.Wang, J., K. Crawford, M. Yuan, H. Wang, P. R. Gorry, and D. Gabuzda. 2002. Regulation of CC chemokine receptor 5 and CD4 expression and human immunodeficiency virus type 1 replication in human macrophages and microglia by T helper type 2 cytokines. J. Infect. Dis. 185:885-897. [DOI] [PubMed] [Google Scholar]
- 48.Wang, T. H., Y. K. Donaldson, R. P. Brettle, J. E. Bell, and P. Simmonds. 2001. Identification of shared populations of human immunodeficiency virus type 1 infecting microglia and tissue macrophages outside the central nervous system. J. Virol. 75:11686-11699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ward, J. M., T. J. O'Leary, G. B. Baskin, R. Benveniste, C. A. Harris, P. L. Nara, and R. H. Rhodes. 1987. Immunohistochemical localization of human and simian immunodeficiency viral antigens in fixed tissue sections. Am. J. Pathol. 127:199-205. [PMC free article] [PubMed] [Google Scholar]
- 50.Wiley, C. A., R. D. Schrier, J. A. Nelson, P. W. Lampert, and M. B. Oldstone. 1986. Cellular localization of human immunodeficiency virus infection within the brains of acquired immune deficiency syndrome patients. Proc. Natl. Acad. Sci. USA 83:7089-7093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yi, Y., W. Chen, I. Frank, J. Cutilli, A. Singh, L. Starr-Spires, J. Sulcove, D. L. Kolson, and R. G. Collman. 2003. An unusual syncytia-inducing human immunodeficiency virus type 1 primary isolate from the central nervous system that is restricted to CXCR4, replicates efficiently in macrophages, and induces neuronal apoptosis. J. Neurovirol. 9:432-441. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


