Abstract
The human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) is a heterodimer comprised of two structurally distinct subunits (p51 and p66). Since p51 and p66 are derived from the same coding region, subunit-specific structure-function studies of RT have been conducted exclusively by in vitro biochemical approaches. To study RT subunit function in the context of infectious virus, we constructed an LTR-vpr-p51-IRES-p66 expression cassette in which the HIV-1 vpr gene was fused in frame with p51, followed by an internal ribosome entry site (IRES) sequence and the p66 coding region. By coexpression with RT-deficient proviral DNA, we demonstrated that the p66 subunit is specifically and selectively packaged into virions as a Vpr-p51/p66 complex. Our analysis showed that cleavage by the viral protease liberates Vpr and generates functional heterodimeric RT (p51/p66) that supports HIV-1 reverse transcription and virus infection. By exploiting this novel trans-complementation approach, we demonstrated, for the first time with infectious virions, that the YMDD aspartates of p66 are both required and sufficient for RT polymerase function. Mutational analyses of the p51 YMDD aspartates indicated that they play an important structural role in p51 folding and subunit interactions that are required for the formation of an active RT heterodimer within infected cells. Understanding the role of the individual RT subunits in RNA- and DNA-dependent DNA synthesis is integral to our understanding of RT function. Our findings will lead to important new insights into the role of the p51 and p66 subunits in HIV-1 reverse transcription.
The biologically relevant, catalytically active form of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) is a heterodimer consisting of a 51-kDa and a 66-kDa subunit. RT has both DNA polymerase and RNase H activities that are required to convert the single-stranded viral RNA genome into double-stranded DNA upon entry into host cells. RT is translated and assembled into virions as part of a precursor Gag-Pol polyprotein (Pr160Gag-Pol). Proteolytic processing of Pr160Gag-Pol by the pol-encoded protease (PR) generates the heterodimeric RT enzyme (p51/p66) (24, 44). The N-terminal 440 amino acids of p51 and p66 are colinear (8, 31). The p66 subunit contains the DNA polymerase and RNase H domains, while the p51 subunit lacks the RNase H domain (16, 38). The polymerase domain can be divided into the fingers, palm, thumb, and connection subdomains (26). However, the relative arrangement of these subdomains differs markedly between the p51 and p66 subunits. Therefore, the effects of a mutation in the RT coding region on subunit structure and function are not equivalent (2, 26). Thus, the heterodimeric nature of the HIV-1 RT has impeded detailed molecular genetic analyses of p51 and p66 subunit function in vivo.
The YXDD motif of retroviral RTs is highly conserved and has been described in the active site of many viral and cellular polymerases (23, 45). The HIV-1 YMDD motif is situated in the palm subdomain (26, 40). The Y183 and M184 amino acid residues contribute to the deoxynucleoside triphosphate (dNTP) binding pocket of p66 (19). While some mutations of these residues are tolerated, most impair polymerase function (14, 46). The most conserved feature of the YMDD motif is the aspartates (D185 and D186), which together with a third aspartate (D110) form the polymerase catalytic triad. Mutation of the aspartates abolishes RT catalytic function and virus infectivity (6, 7, 29, 32). The role of the catalytic aspartates in each RT subunit has been studied by expressing p51 and p66 separately in Escherichia coli (17, 30). Recombinant heterodimers containing polymerase active site mutations exclusive to p51 retain near-wild-type levels of polymerase activity, whereas heterodimers containing the same mutation(s) in p66 are defective. These results imply that the aspartate residues of the p66 subunit are solely responsible for catalysis, while those of p51 are not important for RT function.
Our laboratory previously described an approach to incorporate viral and foreign proteins into HIV virions in trans by expression as Vpr fusion proteins. Incorporation is mediated by Vpr and its interaction with the p6 domain of the cognate Gag precursor polyprotein during virus assembly (36, 48). Using this approach, our group has shown that HIV-1 RT and IN functions can be provided when expressed independently of Pr160Gag-Pol (35, 49, 50). In this report, we describe a novel trans-complementation approach that enables the function of each RT subunit to be studied in the context of an infectious virus. By cotransfecting cells with RT-defective proviral DNA and an LTR-vpr-p51-IRES-p66 expression cassette, we demonstrate that Vpr-p51 interacts with p66 and mediates virion incorporation of a Vpr-p51/p66 heterodimeric complex. Cleavage by the viral PR liberates Vpr, generating functional heterodimeric RT (p51/p66) and infectious virions. By mutating the YMDD aspartates of p66, we demonstrate, for the first time in vivo, that these aspartates are required and sufficient for polymerase function of the RT heterodimer. Our analyses of the p51 aspartates (D185 and D186) indicate that these residues participate in multiple interdomain interactions which are important for the formation of an active RT.
MATERIALS AND METHODS
Cells and antibodies.
The 293T, JC53 (37), and TZM-bl (47) cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), penicillin (100 U/ml), and streptomycin (0.1 mg/ml). Antibodies used included monoclonal antibodies (MAbs) to HIV-1 capsid (183-H12-5C; contributed by Bruce Chesebro and Hardy Chen) and to HIV-1 RT (8C4 and 7E5; contributed by Dag E. Helland), obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health. 8C4 is reactive to the polymerase domain and 7E5 is reactive to the RNase H domain of RT.
Construction of heterodimeric RT expression plasmid.
For independent expression of the RT subunits in trans, the pLR2P-vpr-p51-IRES-p66 (vpr-p51/p66) plasmid was constructed (abbreviations of plasmids are provided in Table 1). Briefly, the sense (5′-TAGATCAGATCTGTTGACTCAGATTGGTTGCA-3′) and antisense (5′-ATCTACACGCGTTTAGAAGGTTTCTGCGCCTT-3′) primers containing the BglII and MluI restriction sites (underlined), respectively, were used to PCR amplify a p51-containing DNA fragment from pSG3wt (SG3) (13). The internal ribosome entry site (IRES) was PCR amplified from the encephalomyocarditis virus (10) (GenBank accession no. NC_001479) using sense (5′-TTATTAACGCGTCCGCCCCTCTCCCTCCCCCC-3′) and antisense (5′-CCATCCCGGGCTTTAATTTTACTGGTACAGTTTCAATAGGACTAATGGGTCCCATGGTATTATCGTCTT-3′) primers containing MluI and XmaI sites (underlined), respectively. The p51 and IRES DNA fragments were digested with corresponding endonucleases and ligated into BglII-XmaI-cut pLR2P-vprRT (50). This construction (Fig. 1A) placed vpr and RT in frame, while preserving the N-terminal protease cleavage site of RT by including 33 bp of PR sequence 5′ of RT. The antisense primer introduced a translational stop codon (TAA) to terminate RT expression at amino acid 440, which is the full-length p51 subunit. The vpr-p51 reading frame was followed by the IRES and then p66. To enable efficient expression of p66, an artificial Kozak sequence was included 5′ of p66 (27), adding a Met-Gly onto the p66 N terminus. The pLR2P-vpr-Δp51-IRES-p66 (vpr-Δp51/p66) control plasmid was constructed to contain a translational stop codon at the first amino acid position of p51 by amplification of a BglII-MluI DNA fragment from the pSG3S-RT (S-RT) clone (50). Other derivatives of vpr-p51/p66 were constructed using PCR-based site-directed mutagenesis and cloning into the BglII-MluI or XmaI-XhoI sites for p51 or p66, respectively. All clones were confirmed by sequencing. The pLR2P-vprIN (vpr-IN) expression vector has been described previously (50).
TABLE 1.
Abbreviations for plasmids
| Plasmid | Abbreviation |
|---|---|
| pSG3wt | SG3 |
| pSG3M7 | M7 |
| pLR2P-vpr-p66 | vpr-p66 |
| pLR2P-vpr-Δp51-IRES-p66 | vpr-Δp51/p66 |
| pLR2P-vpr-p51-IRES-p66 | vpr-p51/p66 |
| pLR2P-vpr-p51-IRES-p66YMNN | vpr-p51/p66NN |
| pLR2P-vpr-p51YMNN-IRES-p66 | vpr-p51NN/p66 |
| pLR2P-vpr-p51YMAA-IRES-p66 | vpr-p51AA/p66 |
| pLR2P-vpr-p51YMEE-IRES-p66 | vpr-p51EE/p66 |
| pLR2P-vpr-p51YMKK-IRES-p66 | vpr-p51KK/p66 |
| pLR2P-vpr-p51D185A-IRES-p66 | vpr-p51D185A/p66 |
| pLR2P-vpr-p51D186A-IRES-p66 | vpr-p51D186A/p66 |
| pLR2P-vpr-p51T409A-IRES-p66 | vpr-p51T409A/p66 |
| pLR2P-vpr-p51W410A-IRES-p66 | vpr-p51W410A/p66 |
| pLR2P-vpr-p51T409A,W410A-IRES-p66 | vpr-p51T409A,W410A/p66 |
| pLR2P-vpr-p51D185A,T409A,W410A- IRES-p66 | vpr-p51D185A,T409A,W410A/p66 |
| pLR2P-vpr-p51D186A,T409A,W410A- IRES-p66 | vpr-p51D186A,T409A,W410A/p66 |
| pLR2P-vpr-IN | vpr-IN |
FIG. 1.
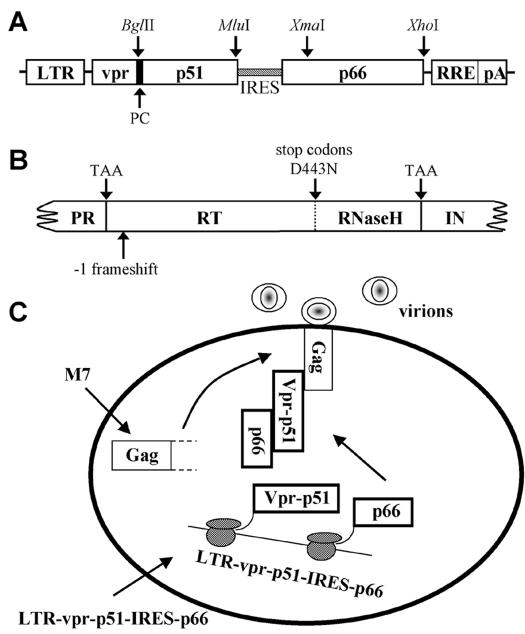
Analysis of RT subunit function by trans-complementation. (A) Illustration of the vpr-p51/p66 expression plasmid. The vpr-p51/p66 expression plasmid was constructed to allow independent expression and subunit-specific analysis of p51 and p66. The vpr-p51/p66 expression plasmid was used to construct various p51 and p66 mutants. Unless otherwise indicated, this was accomplished by inserting p51 or p66 DNA fragments at the BglII-MluI or XmaI-XhoI sites, respectively. (B) The M7 proviral clone. M7 contains a TAA stop codon at the first amino acid positions of RT and IN, three stop codons, 441 (TAA), 444 (TGA), and 447 (TAG), in the RNase H domain, a −1 frameshift at amino acid position 14 of RT, and the RNase H catalytic site mutation, D443N. (C) Model for trans expression and packaging of heterodimeric RT. Cells were cotransfected with the M7 and vpr-p51/p66 expression plasmids. Vpr-p51 incorporates p66 through interaction and stable association of the two subunits (Vpr-p51 and p66) within the cellular cytoplasm. Specific interaction between Vpr and Pr55Gag leads to the incorporation of the Vpr-p51/p66 complex into progeny virions. Subsequent cleavage by the viral PR generates wild-type RT heterodimer (p51/p66).
HIV-1 proviral clones.
The HIV-1 SG3 proviral clone (GenBank accession no. L02317) was used to produce wild-type virus and to construct RT-deficient proviral clones and all recombinant RT and IN expression plasmids. The pSG3M7 (M7) proviral construct (Fig. 1B) was created from S-RT. Briefly, the sense (5′-AAGCCCGGGATGGATGGCCCAAAAGT-3′) and antisense (5′-TCCTAAACGCGTCTCCCTCTAAGCTGCTCAATTTACTTAGAAAGT-3′)primers containing XmaI and MluI sites, respectively, and the sense (5′-ACTTTCTAAGTAAATTGAGCAGCTTAGAGGGAGACGCGTTTAGGA-3′) and antisense (5′-TATGTCGACACCCAATTATGAAAAG-3′) primers containing MluI and SalI sites, respectively, were used to amplify two DNA fragments from the S-RT constructs (nucleotides 2132 to 3455 and 3410 to 5340). Each one was digested with corresponding restriction endonucleases, purified, and ligated together into an XmaI-SalI-cut S-RT plasmid. This introduced three additional stop codons at amino acid positions 441 (TAA), 444 (TGA), and 447 (TAG) and a D443N RNase H catalytic mutation in the RNase H reading frame.
Transfections and analysis of virus infectivity.
DNA transfections were performed on monolayer cultures of 293T cells grown in six-well plates using the calcium phosphate DNA precipitation method. Unless otherwise noted, each cell monolayer (well) was transfected with 6 μg of proviral DNA, 3 μg of vpr-p51/p66 construct, and 1 μg of vpr-IN construct. Culture supernatants from the 293T cells were collected 60 h posttransfection, clarified by low-speed centrifugation (1,000 × g, 10 min), and filtered through 0.45-μm-pore-size sterile filters. The clarified supernatants were analyzed for HIV-1 p24 antigen concentration by enzyme-linked immunosorbent assay (Beckman-Coulter Inc.).
Virus infectivity was assessed using the TZM-bl reporter cell line as described earlier (47). Briefly, virus-containing supernatants were normalized for p24 antigen concentration, serially diluted (fivefold dilutions), and used to infect monolayer cultures of TZM-bl cells. At 48 h postinfection, the cells were fixed and stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside reagent as described earlier (25). The blue-stained cells were counted using a light microscope. Wells containing between 30 and 300 blue cells were used to calculate the infectious units of virus per nanogram of p24 antigen.
Semiquantitative detection of viral DNA.
The PCR method used to analyze the synthesis of nascent viral DNA in infected cells was similar to those described earlier (11, 28, 51). Briefly, 500-ng equivalents (p24 antigen) of transfection-derived virions were incubated with DNase I (4 μg/ml; Worthington Inc.) at 37°C for 1 h to minimize plasmid DNA carryover. The treated virus was then used to infect one million JC53 cells for 4 h in DMEM (1% FBS, 10 μg of DEAE-dextran/ml). The cells were washed twice with DMEM, and the medium was replaced with complete DMEM (10% FBS). At 18 h postinfection, the cells were lysed and total DNA was extracted by organic methods, resuspended in 200 μl of distilled water, and treated with the DpnI restriction endonuclease to digest bacterially derived plasmid DNA. Each PCR subjected 250 pg of DNA extract to 30 rounds of amplification with primers designed to detect early (R-U5, sense, nucleotides 79 to 99, AGCTTGCCTTGAGTGCTTCAA; antisense, nucleotides 182 to 157, CTGCTAGAGATTTTCCACACTGACTA) and late (R-gag, sense, nucleotides 43 to 63, GGCTAGCTAGGGAACCCACTG; antisense, nucleotides 355 to 334, ATACTGACGCTCTCGCACCCAT) viral DNA. The PCR products were separated on a 1.0% agarose gel and visualized by ethidium bromide staining. The relative amount of amplified DNA was determined by comparison to known standards (serial dilutions of pSG3 DNA).
Western blot (immunoblot) analysis.
Transfection-derived virions were concentrated by ultracentrifugation through a 20% sucrose cushion (125,000 × g, 2 h, 4°C) using an SW41 rotor (Beckman Inc.). Pellets were solubilized in loading buffer (62.5 mM Tris-HCl [pH 6.8], 0.2% sodium dodecyl sulfate, 5% 2-mercaptoethanol, 10% glycerol) and boiled, and proteins were separated on 12.0% polyacrylamide gels containing sodium dodecyl sulfate. Following electrophoresis, proteins were transferred to nitrocellulose (0.2-μm pore size) by electroblotting and incubated for 1 h at room temperature in blocking buffer (5% nonfat dry milk in phosphate-buffered saline). The blocked blots were exposed to the appropriate primary antibody for 1 h in blocking buffer with constant mixing. After extensive washing, bound antibodies were detected by chemiluminescence using horseradish peroxidase-conjugated species-specific secondary antibodies (Southern Biotechnology Associates, Inc.) as described by the manufacturer (Amersham Biosciences).
RESULTS
Expression and virion incorporation of heterodimeric RT in trans.
Our strategy for analyzing RT subunit function necessitates Vpr-p51-mediated selective incorporation of p66. Therefore, the pSG3M7 (M7) proviral clone (for abbreviations of plasmids, see Table 1) was constructed with multiple mutations in the RT and IN coding regions (Fig. 1B) to minimize the chance of encoding functional RT, including that which conceivably could be generated via intermolecular genetic recombination with the pLR2P-vpr-p51-IRES-p66 (vpr-p51/p66) plasmid. Our overall strategy for studying subunit-specific RT function in vivo is illustrated in Fig. 1C. Effective trans-complementation would require expression of the two subunits (Vpr-p51 and p66), dimerization, and stable association of the p51 (Vpr-p51) and p66 subunits within the cytosol of the cell, specific interaction of Vpr with Pr55Gag, incorporation of the Vpr-p51/p66 heterodimeric complex into virions, proteolytic cleavage to liberate Vpr from p51/p66, and proper interaction of RT with the template-primer.
To determine the feasibility of this strategy, we tested whether the Vpr-p51 fusion protein could selectively incorporate p66 into virions. Virions generated by cotransfection of M7 with vpr-p51/p66 were shown to contain the p51 and p66 proteins, detectable with anti-RT MAb (Fig. 2A, lane 5). Virions generated by cotransfecting 293T cells with M7 and vpr-Δp51/p66 did not contain detectable p66 (lane 4), indicating that very little, if any, non-Vpr-p51-mediated p66 incorporation occurs. Probing blots with p66-specific MAb confirmed selective packaging of p66 (Fig. 2B, lane 5). Probing a replica blot with MAb against CA confirmed that approximately the same amount of each virus was analyzed and that the M7 virus did not have detectable abnormalities in either virion assembly or proteolytic processing (Fig. 2C). These results demonstrate that the Vpr-p51 fusion protein selectively incorporates the p66 RT subunit into HIV-1 virions. Moreover, they indicate that the heterodimeric trans-RT is relatively stable subsequent to virion incorporation. If not, free p66 might be expected to form homodimers that would be processed by the viral PR. Figure 2A suggests that a similar amount of each subunit was present in the M7 virions.
FIG. 2.
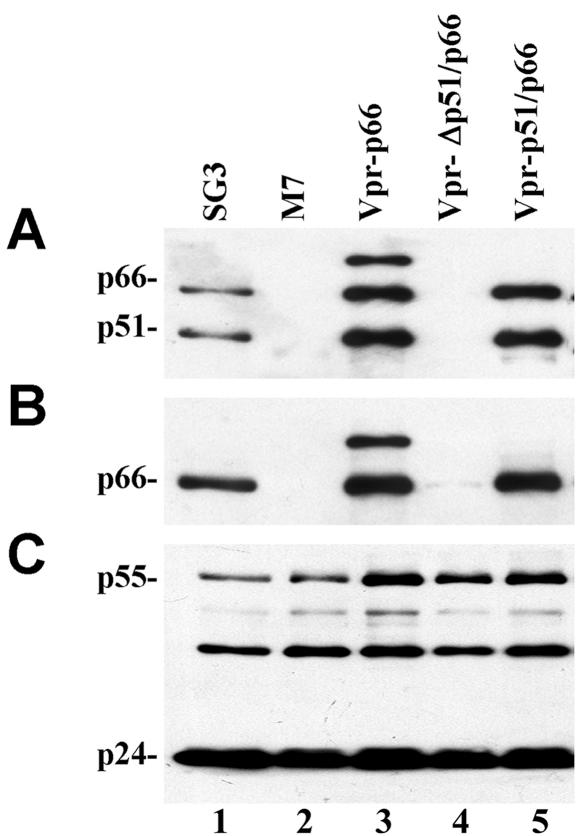
Vpr-p51-mediated p66 virion incorporation and proteolytic processing. M7 proviral DNA was transfected alone or together with the vpr-IN expression plasmid and either the vpr-p66, vpr-p51/p66, or vprΔp51/p66 expression plasmid. Wild-type SG3 was included as a control. Transfection-derived virions were concentrated by ultracentrifugation, lysed, and analyzed by immunoblotting for p51/p66 (A), p66 (B), or p24 (C) as described in Materials and Methods.
Heterodimeric trans-RT rescues the infectivity of RT-deficient virus.
To analyze the functionality of the trans-RT, the M7 proviral construct was cotransfected into 293T cells with vpr-p51/p66 and vpr-IN. The vpr-IN expression plasmid was included, since the M7 clone does not express the IN protein and integration of the nascent viral cDNA is required to detect infection using the TZM-bl reporter cell line. Moreover, IN is also required for efficient initiation of reverse transcription (49). In three independent experiments, virus infectivity was rescued to about 15% of that of wild-type virus (Fig. 3A, lane 5). Virus derived by cotransfecting 293T cells with M7, vpr-p66, and vpr-IN exhibited a similar level of infectivity (lane 3), consistent with earlier reports (50). The infectivity of M7 virus derived by cotransfection with vpr-Δp51/p66 and vpr-IN was less than 0.05% of that of wild-type virus (lane 4), or 0.2% compared with M7 virus complemented with Vpr-p51/p66. These results demonstrated that the heterodimeric trans-RT is functional and, with the M7 proviral clone, minimal complementation of virus infectivity was due to non-Vpr-p51-mediated packaging of p66. Neither virus infectivity (lane 6) nor DNA synthesis (data not shown) was efficiently complemented by Vpr-p51/p66 without the IN protein.
FIG. 3.
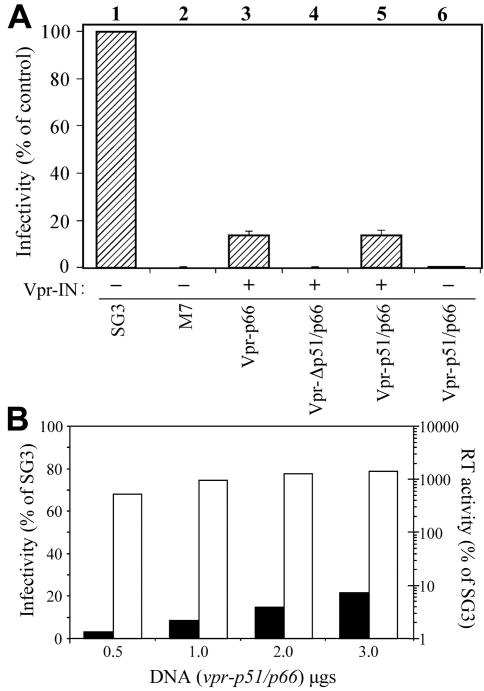
trans-complementation analysis of RT-IN-deficient virus. (A) Analysis of infectivity. Transfection-derived viruses were analyzed for infectivity using the TZM-bl reporter cell line as described in Materials and Methods. Infectivity is expressed as a percentage of the wild-type virus control. Viruses were derived from 293T cells with (+) or without (−) cotransfection of vpr-IN. The results of three independent experiments are shown. (B) Analysis of heterodimeric trans-RT activity. Increasing DNA concentrations of vpr-p51/p66 (ranging from 0.5 to 3.0 μg) were transfected into 293T cells together with a constant amount of M7 and vpr-IN. Culture supernatants were collected and treated as described in Materials and Methods. The transfection-derived virions were analyzed for infectivity using the TZM-bl reporter cell line (black bars) and for RT activity using the chemiluminescent RT assay (Roche) (white bars). Results are expressed as a percentage relative to an equal amount of wild-type SG3 virus.
Increasing DNA concentrations of vpr-p51/p66 (ranging from 0.5 to 3.0 μg) were transfected into 293T cells along with constant amounts of M7 and vpr-IN (Fig. 3B). This correlated with transfection-derived virions containing increasing concentrations of the RT heterodimer (p51/p66) when analyzed by immunoblotting (data not shown). The virions were analyzed for infectivity using the TZM-bl reporter cell line and RT activity using the chemiluminescent RT assay (Roche). Results are expressed as a percentage relative to an equal amount of wild-type SG3 virus. Infectivity increased with increasing amounts of the trans-RT heterodimer plasmid, reaching a maximum of approximately 20% of that of wild-type virus. The RT activity exceeded that of wild-type by ∼14-fold at the highest DNA concentration of vpr-p51/p66 used in the assay.
Subunit-specific analysis of the YMDD motif.
There exists a preponderance of evidence from biochemical and structural studies that suggests HIV-1 reverse transcription is catalyzed by the p66 subunit of RT. To study the function of the D185 and D186 aspartate residues in each subunit, either the p66 or the p51 coding region of the vpr-p51/p66 plasmid was mutated in both aspartates of the YMDD motif. Transfection-derived virus was analyzed for infectivity and cDNA synthesis following acute infection of JC53 cells. Virions containing the p51/p66NN mutant RT (D185N and D186N mutations in p66) were severely defective in infectivity (Fig. 4A, lane 3). Analysis of infected cells for viral DNA revealed a severe defect in reverse transcription (Fig. 4B and C, lanes 5). The severity of this defect suggested that the p51 subunit of the heterodimer does not catalyze viral DNA synthesis in vivo. Moreover, when the equivalent catalytic site mutation was analyzed in p51 (p51NN/p66), virus infectivity was only modestly reduced (Fig. 4A, lane 4). Consistent with this result, only a small reduction in viral DNA synthesis was observed (Fig. 4B and C, lanes 6). The severe defect caused by mutating the p66 YMDD aspartates shows that the trans-incorporated p51/p66 heterodimer mimics a well-established enzymatic property of the endogenous HIV-1 RT.
FIG. 4.
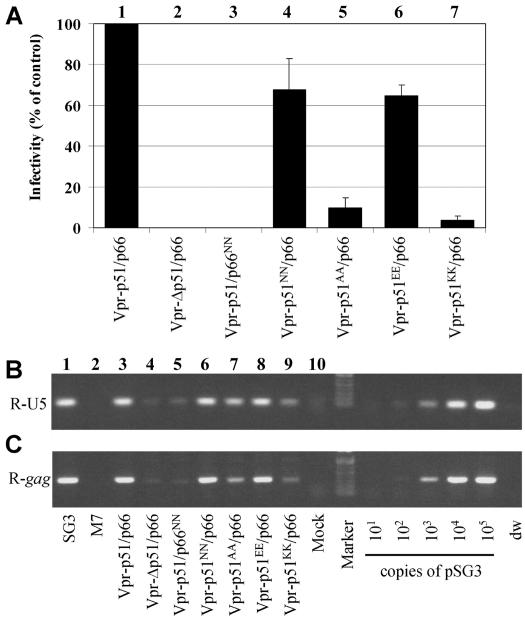
Subunit-specific analysis of the YMDD aspartates. The wild-type, control, and mutated vpr-p51 expression plasmids were cotransfected into 293T cells with the M7 and vpr-IN DNAs to generate virions. (A) Analysis of infectivity. Infectivity, expressed as a percentage of wild-type virus, was analyzed from three independent experiments. (B and C) Analysis of viral DNA synthesis. DNA products of reverse transcription were analyzed as described in Materials and Methods. Early (R-U5) (B) and late (R-gag) (C) DNA products were amplified by PCR, resolved on 1.0% agarose gels, and stained with ethidium bromide. To approximate the relative amount of each of the amplified DNA products, 10-fold serial dilutions of pSG3 DNA (ranging from 101 to 105 copies) were prepared and analyzed in parallel. Distilled water (dw) was included as a negative control.
To further analyze the role of these p51 aspartates, they were mutated to alanines, glutamates, or lysines. Virus stocks containing each mutant RT were prepared by cotransfection of 293T cells and analyzed for infectivity and DNA synthesis. Similar to the asparagine mutations, the glutamic acid mutations (p51EE/p66) decreased virus infectivity and DNA synthesis only slightly (Fig. 4). More dramatic decreases in both DNA synthesis and virus infectivity were observed for viruses containing either the alanine (p51AA/p66) or the lysine (p51KK/p66) p51 mutations. Viral DNA synthesis was analyzed using primer pairs to detect either early (R-U5) or late (R-gag) products of reverse transcription, and the results indicated that the defect is at or prior to initiation.
The p51 YMDD aspartates are important for RT subunit interaction.
To determine whether mutations at the D185 and D186 residues of p51 affected interaction with p66, virions were analyzed for p66 content. Virions generated with the more conservative mutations, p51NN/p66 and p51EE/p66 (Fig. 5A, lanes 3 and 5), contained a similar amount of p66 compared to the wild-type p51/p66 (lane 1). Notably, the more-disruptive p51AA/p66 and p51KK/p66 mutations (lanes 4 and 6) showed reduced levels of p66 incorporation. The negative control (lane 2) had no detectable p66. The amount of p51 in these virions incorporated via Vpr was approximately the same (Fig. 5B). Analysis with a MAb against CA confirmed that a similar amount of each virus was analyzed (Fig. 5C). Immunoblot analysis of the transfected cells indicated that the expression of p66 from the vpr-p51AA/p66 and vpr-p51KK/p66 plasmids was not impaired (data not shown).
FIG. 5.
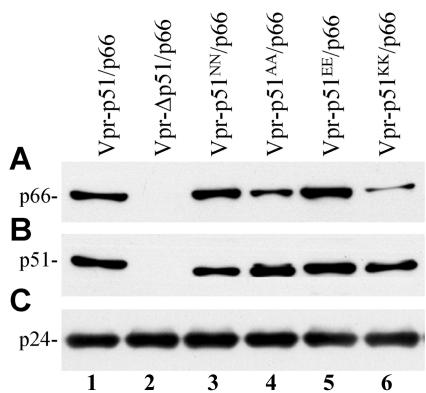
Analysis of p66 virion incorporation. M7 proviral DNA was cotransfected into 293T cells with the vpr-p51/p66 plasmids containing various mutations in the YMDD aspartates. The transfection-derived virions were concentrated by ultracentrifugation, lysed, and examined for p66 (A), p51 (B), or p24 (C) by immunoblot analysis as described in Materials and Methods.
Putative interactions of the p51 YMDD aspartates.
In the p51 subunit, the connection subdomain folds into the expanded cleft between its fingers and thumb subdomains, resulting in its closed conformation (26). Since the connection subdomain of p51 makes multiple contacts with three other subdomains of p51 (fingers, palm, and thumb), we predicted that the more-disruptive p51 mutations (YMAA and YMKK) destabilize these interactions and consequently have global effects on RT folding (Fig. 6). D185 has a possible backbone interaction with Q151 and side chain interaction with R72, while the D186 residue has possible side chain interactions with T409 and W410. Mutation of these two aspartates individually (p51D185A/p66 and p51D186A/p66) did not have a significant effect on viral infectivity (Fig. 7, lanes 3 and 4). The predicted interaction of D186 with the T409 and W410 residues was further analyzed by complementation analysis using mutants p51T409A/p66, p51W410A/p66, and p51T409A,W410A/p66. Our results suggested that the loss of these interactions either individually (lanes 5 and 6) or together (lane 7) does not reduce viral infectivity to the extent exhibited by p51AA/p66. Analysis of the p51D185A,T409A,W410A/p66 mutant, which should disrupt most if not all the putative interactions associated with aspartates D185 and D186 of p51, showed a more substantial decrease in viral infectivity (lane 8). Notably, the p51D186A,T409A,W410A/p66 mutant also caused a similar decrease in virus infectivity (lane 9), which could be due to an ability of the nonmutated aspartate to partially compensate for the loss of these interactions.
FIG. 6.
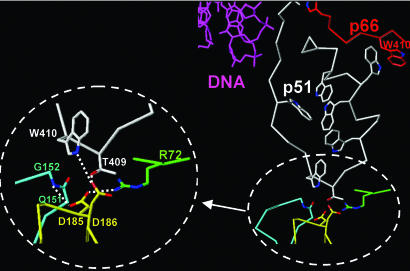
Putative interactions of the p51 D185/D186 aspartates, including interactions of the p51 D185/D186 (yellow) of HIV-1 RT at the junction of the p51 palm (cyan and yellow), p51 connection (white), and p51 fingers (green) subdomains in the structure of the RT/DNA/dNTP complex (PDB code 1RTD). The Trp-rich region is shown at the interface of the p51 (white) and p66 (red) subunits and proximal to the DNA binding cleft (red).
FIG. 7.
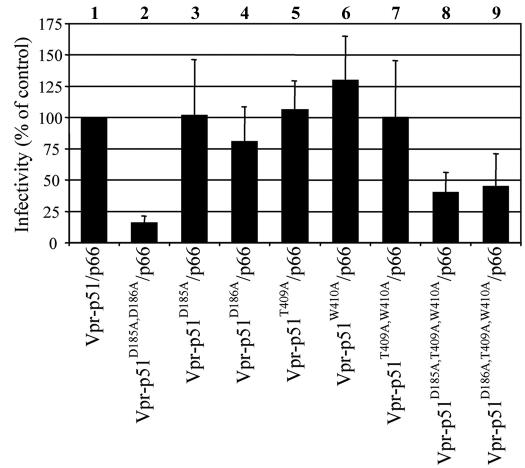
Analysis of putative interactions of p51 D185/D186. Virions containing p51 mutations at amino acid residues within interacting distance of its YMDD aspartates were analyzed for infectivity on TZM-bl reporter cells as described in Materials and Methods. Infectivity is expressed as a percentage of the wild-type virus control. The results of five independent experiments are shown.
DISCUSSION
Understanding the role of the individual RT subunits in RNA- and DNA-dependent DNA synthesis is integral to our understanding of RT and has been the focus of several studies. Most of these used in vitro biochemical methods to analyze the enzymatic activity of recombinant HIV-1 RT heterodimers, wherein either the p51 or p66 subunit was selectively mutated (5, 17, 30). However, due to inherent limitations in applying a subunit-specific mutagenesis approach to infectious virus, our understanding of RT structure-function is incomplete. Therefore, we developed a novel Vpr fusion protein targeting strategy to study the RT heterodimer in a higher-ordered, biologic context. Our findings indicate that Vpr-p51 and p66 form an intracellular dimer (Vpr-p51/p66) that is specifically incorporated into virions, processed by the viral PR to liberate p51/p66, and rescues the infectivity of RT-deficient HIV-1. By analyzing mutations in the aspartates D185 and D186 of p51 and p66, respectively, we were able to analyze the function of these residues in the context of an infectious virus. The absence of minus-strand strong-stop DNA synthesis in cells infected with virus, in which the YMDD aspartates of p66 were mutated, corroborated findings from previous in vitro studies and demonstrated that as a heterodimer p66 is solely responsible for the catalytic or polymerase function of RT in vivo. Our analysis of the p51 subunit indicated that mutation of D185 and D186 impaired virus infection and DNA synthesis due to an effect on RT structure rather than catalytic function. Analysis of putative interactions with surrounding amino acids further suggested that these aspartates play an important role in p51 folding and its interaction with the p66 subunit, including the formation of the enzymatically active heterodimer.
It is noteworthy that the trans-heterodimeric RT rescued infectivity to a level approximately 15% of that of wild-type virus, although the virion-associated RT activity was greater than that of wild-type SG3. This is consistent with earlier reports on the complementation of DNA synthesis and infectivity of RT-IN-deficient virus. When this virus was complemented with Vpr-RT and Vpr-IN, infectivity peaked at approximately 15% while complementation with Vpr-RT-IN peaked at about 85%, although RT activity in both cases was greater than wild type (50). Shehu-Xhilaga et al. found that the expression of RT-IN may facilitate normal virion genomic RNA conformation and core morphogenesis (41). We also considered the possibility that virus infectivity was reduced by a dominant-negative effect exerted by the Vpr-p51/p66 and/or the Vpr-IN fusion protein. By cotransfecting different concentrations of the vpr-p51/p66 and the vpr-IN expression plasmids with wild-type virus, we did not detect a significant negative effect, even at concentrations higher than those used in this study (data not shown). Based on the findings of Wu et al. and Shehu-Xhilaga et al., we are considering other explanations that may limit the efficiency of complementation with the trans-heterodimeric RT. Nevertheless, with a complementation efficiency (for infectivity) of approximately 15% and a background of about 0.05% of that of wild-type virus, this approach appears to provide sufficient sensitivity for subunit-specific studies of RT function.
Mutational analysis of the aspartates D185 and D186 of p51 included conservative and nonconservative amino acid substitutions. The D185N/D186N mutant was relatively conservative, since Asn is almost isosteric to, but not negatively charged like, Asp. The D185E/D186E mutant increased side chain length by one methylene group without changing the negative charge. Similar to aspartate, asparagine and glutamine can participate in hydrogen bond interactions through their side chains. The D185N/D186N and D185E/D186E p51 mutants caused a slight reduction in infectivity and viral DNA synthesis. Substitution of the p51 aspartates with either alanines (D185A/D186A) or lysines (D185K/D186K) drastically reduced infectivity and DNA synthesis. The alanines have a short hydrophobic side chain that cannot hydrogen bond with neighboring polar residues. The lysines present an opposite polarity and have a lengthened side chain. Our analysis of the p51 aspartates demonstrated that small changes in charge and/or length of the side chain may be tolerated, while a charge shift and/or substantial changes in side chain length are not. These findings indicate that aspartates D185 and D186 of p51 are important for RT activity.
The D185 and D186 of p51 lie in the expanded cleft between the fingers and thumb subdomains. Folding of the p51 connection subdomain produces the closed conformation of p51 in the RT heterodimer. Based on the coordinates of the crystal structure of HIV-1 RT in complex with DNA and dNTP (PDB code 1RTD), we predicted putative interactions of the p51 aspartates (D185 and D186) with residues that lie within interacting distance (approximately 3 Å). These include interdomain interactions with “tryptophan motif” (Trp-motif) residues T409 and W410 of the p51 connection subdomain (Fig. 6). It is plausible that mutation at the p51 D185/D186 could affect multiple interactions involving the Trp-motif and the heterodimer interface (4, 9, 43). These residues are also in the proximity of the floor of the DNA binding cleft-RNase H primer grip, which includes K390, K395, and E396 of p51, which interact directly with the template-primer (19, 40). Additionally, the p51 D185/D186 residues have putative interactions with R72 of the p51 fingers subdomain and Q151-G152 at the tip of α-helix E in the palm subdomain. Decreased p66 virion incorporation observed with certain p51 D185/D186 mutations (AA and KK) indicates that these p51 aspartates may mediate interactions that are required for heterodimer formation and/or stability. Our mutational analyses support the model (Fig. 6) that aspartates D185 and D186 of p51 are important for local structural integrity and contribute to multiple interdomain interactions. Mutations predicted to affect interactions between only a single aspartate (either D185 or D186) had only limited effect on virus infectivity. The significant decrease in viral infectivity observed with the p51D185A, T409A, W410A/p66 mutant supports our model, since all four predicted interactions of p51 D185/D186 are expected to be disrupted. Notably, the effect of these mutations is not as great as that with p51YMAA/p66. This could be due to an ability of the nonmutated aspartate to partially compensate for these interactions through minor rearrangement of its side chain and other local structural elements.
Our finding that subunit-specific RT function can be studied using infectious virus has broad implications. While the p51 subunit was believed to function primarily as a scaffold to maintain the active structure of p66 (20, 44), other functions have been suggested, including involvement in tRNA primer binding (3, 21), loading of p66 onto the template-primer (15), and enhancement of strand displacement (1, 18). The dimer interface between p51 and p66 is critical for reverse transcription, and it has been proposed as an ideal target for therapeutic intervention (9, 34, 39). This notion was supported by several studies demonstrating that mutation of amino acid residues involved in subunit interactions alter the arrangement of the RT subdomains and disrupt RT function (12, 43). Mutations in p51 have also been implicated in resistance to nonnucleoside reverse transcriptase inhibitors and inhibitors of RNase H activity. The E138K mutation, which confers resistance to 2′,5′-bis-O-(tert-butyldimethylsilyl)-3′-spiro-5"-(4"-amino-1",2"-oxathiole-2",2"-dioxide), has been mapped to the p51 subunit (22, 42). The C280S mutation in RT causes resistance to the RNase H inhibitor N-ethylmaleimide (33). Interestingly, both the p51 and p66 subunits were found to contribute to the resistance to N-ethylmaleimide in vitro. The inherent ability of HIV-1 RT to mutate under selective pressure is widely recognized; however, the effects of these mutations on viral fitness, and specifically RT function, are not completely understood. We expect that the trans-heterodimeric RT complementation approach described here will provide new opportunities to probe RT function in vivo and lead to new insights into our understanding of HIV-1 reverse transcription. Finally, since the YMDD aspartates are invariant and contribute to interactions required for an active RT heterodimer, our results may suggest a new target for inhibiting RT function.
Acknowledgments
We thank Kenneth Zammit, Lakeasha McTear, and Kristen Boydston for technical assistance.
This research was supported by National Institutes of Health grants CA73470 and AI47714 and facilities of the Central AIDS Virus, Genetic Sequencing, and Protein Expression Cores of the Birmingham Center for AIDS Research (P30-AI-27767) and the UAB Arthritis and Musculoskeletal Diseases Center. This research was also supported by a Merit Review Award funded by the Office of Research and Development, Medical Research Services, Department of Veterans Affairs.
REFERENCES
- 1.Amacker, M., M. Hottiger, and U. Hubscher. 1995. Feline immunodeficiency virus reverse transcriptase: expression, functional characterization, and reconstitution of the 66- and 51-kilodalton subunits. J. Virol. 69:6273-6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnold, E., A. Jacobo-Molina, R. G. Nanni, R. L. Williams, X. Lu, J. Ding, A. D. Clark, Jr., A. Zhang, A. L. Ferris, P. Clark, et al. 1992. Structure of HIV-1 reverse transcriptase/DNA complex at 7 Å resolution showing active site locations. Nature 357:85-89. [DOI] [PubMed] [Google Scholar]
- 3.Arts, E. J., X. Li, Z. Gu, L. Kleiman, M. A. Parniak, and M. A. Wainberg. 1994. Comparison of deoxyoligonucleotide and tRNA(Lys-3) as primers in an endogenous human immunodeficiency virus-1 in vitro reverse transcription/template-switching reaction. J. Biol. Chem. 269:14672-14680. [PubMed] [Google Scholar]
- 4.Baillon, J. G., N. T. Nashed, A. Kumar, S. H. Wilson, and D. M. Jerina. 1991. A leucine zipper-like motif may mediate HIV reverse transcriptase subunit binding. New Biol. 3:1015-1019. [PubMed] [Google Scholar]
- 5.Boyer, P. L., J. Ding, E. Arnold, and S. H. Hughes. 1994. Subunit specificity of mutations that confer resistance to nonnucleoside inhibitors in human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 38:1909-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyer, P. L., A. L. Ferris, and S. H. Hughes. 1992. Cassette mutagenesis of the reverse transcriptase of human immunodeficiency virus type 1. J. Virol. 66:1031-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chao, S. F., V. L. Chan, P. Juranka, A. H. Kaplan, R. Swanstrom, and C. A. Hutchison III. 1995. Mutational sensitivity patterns define critical residues in the palm subdomain of the reverse transcriptase of human immunodeficiency virus type 1. Nucleic Acids Res. 23:803-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.di Marzo Veronese, F., T. D. Copeland, A. L. DeVico, R. Rahman, S. Oroszlan, R. C. Gallo, and M. G. Sarngadharan. 1986. Characterization of highly immunogenic p66/p51 as the reverse transcriptase of HTLV-III/LAV. Science 231:1289-1291. [DOI] [PubMed] [Google Scholar]
- 9.Divita, G., T. Restle, R. S. Goody, J. C. Chermann, and J. G. Baillon. 1994. Inhibition of human immunodeficiency virus type 1 reverse transcriptase dimerization using synthetic peptides derived from the connection domain. J. Biol. Chem. 269:13080-13083. [PubMed] [Google Scholar]
- 10.Duke, G. M., M. A. Hoffman, and A. C. Palmenberg. 1992. Sequence and structural elements that contribute to efficient encephalomyocarditis virus RNA translation. J. Virol. 66:1602-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fassati, A., and S. P. Goff. 2001. Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J. Virol. 75:3626-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghosh, M., P. S. Jacques, D. W. Rodgers, M. Ottman, J. L. Darlix, and S. F. Le Grice. 1996. Alterations to the primer grip of p66 HIV-1 reverse transcriptase and their consequences for template-primer utilization. Biochemistry 35:8553-8562. [DOI] [PubMed] [Google Scholar]
- 13.Ghosh, S. K., P. N. Fultz, E. Keddie, M. S. Saag, P. M. Sharp, B. H. Hahn, and G. M. Shaw. 1993. A molecular clone of HIV-1 tropic and cytopathic for human and chimpanzee lymphocytes. Virology 194:858-864. [DOI] [PubMed] [Google Scholar]
- 14.Gotte, M., D. Arion, M. A. Parniak, and M. A. Wainberg. 2000. The M184V mutation in the reverse transcriptase of human immunodeficiency virus type 1 impairs rescue of chain-terminated DNA synthesis. J. Virol. 74:3579-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris, D., P. N. Yadav, and V. N. Pandey. 1998. Loss of polymerase activity due to Tyr to Phe substitution in the YMDD motif of human immunodeficiency virus type-1 reverse transcriptase is compensated by Met to Val substitution within the same motif. Biochemistry 37:9630-9640. [DOI] [PubMed] [Google Scholar]
- 16.Hizi, A., C. McGill, and S. H. Hughes. 1988. Expression of soluble, enzymatically active, human immunodeficiency virus reverse transcriptase in Escherichia coli and analysis of mutants. Proc. Natl. Acad. Sci. USA 85:1218-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hostomsky, Z., Z. Hostomska, T. B. Fu, and J. Taylor. 1992. Reverse transcriptase of human immunodeficiency virus type 1: functionality of subunits of the heterodimer in DNA synthesis. J. Virol. 66:3179-3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hottiger, M., V. N. Podust, R. L. Thimmig, C. McHenry, and U. Hubscher. 1994. Strand displacement activity of the human immunodeficiency virus type 1 reverse transcriptase heterodimer and its individual subunits. J. Biol. Chem. 269:986-991. [PubMed] [Google Scholar]
- 19.Huang, H., R. Chopra, G. L. Verdine, and S. C. Harrison. 1998. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 282:1669-1675. [DOI] [PubMed] [Google Scholar]
- 20.Hughes, S. H. 2001. Molecular matchmaking: NNRTIs can enhance the dimerization of HIV type 1 reverse transcriptase. Proc. Natl. Acad. Sci. USA 98:6991-6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacques, P. S., B. M. Wohrl, K. J. Howard, and S. F. Le Grice. 1994. Modulation of HIV-1 reverse transcriptase function in “selectively deleted” p66/p51 heterodimers. J. Biol. Chem. 269:1388-1393. [PubMed] [Google Scholar]
- 22.Jonckheere, H., J. M. Taymans, J. Balzarini, S. Velazquez, M. J. Camarasa, J. Desmyter, E. De Clercq, and J. Anne. 1994. Resistance of HIV-1 reverse transcriptase against [2′,5′-bis-O-(tert-butyldimethylsilyl)-3′-spiro-5"-(4"-amino-1",2"-oxathiole-2",2"-dioxide)] (TSAO) derivatives is determined by the mutation Glu138→Lys on the p51 subunit. J. Biol. Chem. 269:25255-25258. [PubMed] [Google Scholar]
- 23.Kamer, G., and P. Argos. 1984. Primary structural comparison of RNA-dependent polymerases from plant, animal and bacterial viruses. Nucleic Acids Res. 12:7269-7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katz, R. A., and A. M. Skalka. 1994. The retroviral enzymes. Annu. Rev. Biochem. 63:133-173. [DOI] [PubMed] [Google Scholar]
- 25.Kimpton, J., and M. Emerman. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J. Virol. 66:2232-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohlstaedt, L. A., J. Wang, J. M. Friedman, P. A. Rice, and T. A. Steitz. 1992. Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science 256:1783-1790. [DOI] [PubMed] [Google Scholar]
- 27.Kozak, M. 1987. At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J. Mol. Biol. 196:947-950. [DOI] [PubMed] [Google Scholar]
- 28.Lai, L., H. Liu, X. Wu, and J. C. Kappes. 2001. Moloney murine leukemia virus integrase protein augments viral DNA synthesis in infected cells. J. Virol. 75:11365-11372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larder, B. A., D. J. Purifoy, K. L. Powell, and G. Darby. 1987. Site-specific mutagenesis of AIDS virus reverse transcriptase. Nature 327:716-717. [DOI] [PubMed] [Google Scholar]
- 30.Le Grice, S. F., T. Naas, B. Wohlgensinger, and O. Schatz. 1991. Subunit-selective mutagenesis indicates minimal polymerase activity in heterodimer-associated p51 HIV-1 reverse transcriptase. EMBO J. 10:3905-3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lightfoote, M. M., J. E. Coligan, T. M. Folks, A. S. Fauci, M. A. Martin, and S. Venkatesan. 1986. Structural characterization of reverse transcriptase and endonuclease polypeptides of the acquired immunodeficiency syndrome retrovirus. J. Virol. 60:771-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lowe, D. M., V. Parmar, S. D. Kemp, and B. A. Larder. 1991. Mutational analysis of two conserved sequence motifs in HIV-1 reverse transcriptase. FEBS Lett. 282:231-234. [DOI] [PubMed] [Google Scholar]
- 33.Loya, S., H. Q. Gao, O. Avidan, P. L. Boyer, S. H. Hughes, and A. Hizi. 1997. Subunit-specific mutagenesis of the cysteine 280 residue of the reverse transcriptase of human immunodeficiency virus type 1: effects on sensitivity to a specific inhibitor of the RNase H activity. J. Virol. 71:5668-5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morris, M. C., V. Robert-Hebmann, L. Chaloin, J. Mery, F. Heitz, C. Devaux, R. S. Goody, and G. Divita. 1999. A new potent HIV-1 reverse transcriptase inhibitor. A synthetic peptide derived from the interface subunit domains. J. Biol. Chem. 274:24941-24946. [DOI] [PubMed] [Google Scholar]
- 35.Padow, M., L. Lai, C. Deivanayagam, L. J. DeLucas, R. B. Weiss, D. M. Dunn, X. Wu, and J. C. Kappes. 2003. Replication of chimeric human immunodeficiency virus type 1 (HIV-1) containing HIV-2 integrase (IN): naturally selected mutations in IN augment DNA synthesis. J. Virol. 77:11050-11059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paxton, W., R. I. Connor, and N. R. Landau. 1993. Incorporation of Vpr into human immunodeficiency virus type 1 virions: requirement for the p6 region of gag and mutational analysis. J. Virol. 67:7229-7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Platt, E. J., K. Wehrly, S. E. Kuhmann, B. Chesebro, and D. Kabat. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophage-tropic isolates of human immunodeficiency virus type 1. J. Virol. 72:2855-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prasad, V. R., and S. P. Goff. 1989. Linker insertion mutagenesis of the human immunodeficiency virus reverse transcriptase expressed in bacteria: definition of the minimal polymerase domain. Proc. Natl. Acad. Sci. USA 86:3104-3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Restle, T., B. Muller, and R. S. Goody. 1990. Dimerization of human immunodeficiency virus type 1 reverse transcriptase. A target for chemotherapeutic intervention. J. Biol. Chem. 265:8986-8988. [PubMed] [Google Scholar]
- 40.Sarafianos, S. G., K. Das, C. Tantillo, A. D. Clark, Jr., J. Ding, J. M. Whitcomb, P. L. Boyer, S. H. Hughes, and E. Arnold. 2001. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J. 20:1449-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shehu-Xhilaga, M., M. Hill, J. A. Marshall, J. Kappes, S. M. Crowe, and J. Mak. 2002. The conformation of the mature dimeric human immunodeficiency virus type 1 RNA genome requires packaging of Pol protein. J. Virol. 76:4331-4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sluis-Cremer, N., G. I. Dmitrienko, J. Balzarini, M. J. Camarasa, and M. A. Parniak. 2000. Human immunodeficiency virus type 1 reverse transcriptase dimer destabilization by 1-[spiro[4"-amino-2",2"-dioxo-1",2"-oxathiole-5",3′-[2′,5′-bis-O-(tert-butyldimethylsilyl)-beta-D-ribofuranosyl]]]-3-ethylthy-mine. Biochemistry 39:1427-1433. [DOI] [PubMed] [Google Scholar]
- 43.Tachedjian, G., H. E. Aronson, M. de los Santos, J. Seehra, J. M. McCoy, and S. P. Goff. 2003. Role of residues in the tryptophan repeat motif for HIV-1 reverse transcriptase dimerization. J. Mol. Biol. 326:381-396. [DOI] [PubMed] [Google Scholar]
- 44.Telesnitsky, A., and S. P. Goff. 1997. Reverse transcriptase and the generation of retroviral DNA, p. 121-160. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 45.Toh, H., H. Hayashida, and T. Miyata. 1983. Sequence homology between retroviral reverse transcriptase and putative polymerases of hepatitis B virus and cauliflower mosaic virus. Nature 305:827-829. [DOI] [PubMed] [Google Scholar]
- 46.Wakefield, J. K., S. A. Jablonski, and C. D. Morrow. 1992. In vitro enzymatic activity of human immunodeficiency virus type 1 reverse transcriptase mutants in the highly conserved YMDD amino acid motif correlates with the infectious potential of the proviral genome. J. Virol. 66:6806-6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu, X., J. A. Conway, J. Kim, and J. C. Kappes. 1994. Localization of the Vpx packaging signal within the C terminus of the human immunodeficiency virus type 2 Gag precursor protein. J. Virol. 68:6161-6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu, X., H. Liu, H. Xiao, J. A. Conway, E. Hehl, G. V. Kalpana, V. Prasad, and J. C. Kappes. 1999. Human immunodeficiency virus type 1 integrase protein promotes reverse transcription through specific interactions with the nucleoprotein reverse transcription complex. J. Virol. 73:2126-2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu, X., H. Liu, H. Xiao, J. A. Conway, E. Hunter, and J. C. Kappes. 1997. Functional RT and IN incorporated into HIV-1 particles independently of the Gag/Pol precursor protein. EMBO J. 16:5113-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zack, J. A., S. J. Arrigo, S. R. Weitsman, A. S. Go, A. Haislip, and I. S. Chen. 1990. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell 61:213-222. [DOI] [PubMed] [Google Scholar]


