Abstract
Vitamin A, retinol, circulates in blood bound to retinol-binding protein (RBP). At some tissues, RBP is recognized by STRA6, a plasma membrane protein that serves a dual role: it transports retinol from extracellular RBP into cells and it transduces a signaling cascade mediated by the Janus kinase JAK2 and the transcription factors STAT3 and STAT5. We show here that expression of RBP and STRA6 is markedly upregulated in human breast and colon tumors, that holo-RBP/STRA6 signaling promotes oncogenic properties, and that STRA6 expression is critical for tumor formation by colon carcinoma cells in vivo. The holo-RBP/STRA6 pathway also efficiently induces fibroblasts to undergo oncogenic transformation rendering them highly tumorigenic. The data establish that holo-RBP and its receptor STRA6 are potent oncogenes and suggest that the pathway is a novel target for therapy of some human cancers.
Keywords: retinol, JAK/STAT signaling, colon cancer, breast cancer
Introduction
Vitamin A, retinol (ROH) is essential for multiple biological functions both during embryonic development and in the adult. Many of the biological activities of retinol are exerted by its active metabolite all-trans-retinoic acid (RA), which regulates transcription by activating the nuclear receptors retinoic acid receptors (RARs) and peroxisome proliferator activated receptor β/δ (PPARβ/δ) (1, 2). Vitamin A is stored in various tissues, including adipose tissue, lung, and retinal pigment epithelium in the eye, but its major storage site is in the liver. ROH is secreted from storage pools into the circulation bound to serum retinol-binding protein (RBP). In turn, ROH-bound RBP (holo-RBP) associates in blood with another plasma protein, transthyretin (TTR), to form a holo-RBP-TTR complex which circulates at approximately 1:1 molar stoichiometry under normal physiological conditions (3, 4).
ROH enters target cells from circulating holo-RBP by two distinct mechanisms. Due to its lipophilic nature, it readily diffuses through the plasma membrane, a process that, in most tissues, accounts for the major fraction of vitamin A uptake by cells (4–7). ROH can also be transported into cells by an integral plasma membrane protein termed Stimulated by RA 6 (STRA6) which binds extracellular holo-RBP and transports ROH into cells (8). While STRA6-mediated ROH uptake is not necessary for maintaining adequate ROH supply in most tissues, it’s contribution to ROH supply becomes significant in cells that express a very high level of the receptor, primarily the retinal pigment epithelium (RPE) in the eye (7, 9).
Our recent studies surprisingly revealed that, in addition to serving as a vitamin A transporter, STRA6 is a cytokine receptor. We thus found that binding of holo-RBP triggers phosphorylation of a tyrosine residue in the cytosolic domain of STRA6, resulting in recruitment and activation of the Janus kinase JAK2 and, in a cell-dependent manner, the transcription factors STAT3 or STAT5 (10, 11). Holo-RBP thus activates STRA6-mediated signaling that culminates in upregulation of STAT target genes. Interestingly, the two functions of STRA6 were found to be interdependent, i.e. STRA6-mediated ROH transport is required for activation of STRA6 signaling, and vice versus, phosphorylation of STRA6 is essential for retinol transport mediated by the receptor (11). The observations further demonstrated that STRA6 directly transfers ROH from extracellular RBP to cellular retinol-binding protein 1 (CRBP1), which directly binds to STRA6, accepts ROH, and dissociates from the receptor upon ligation (11). STRA6-mediated ROH uptake also critically requires the presence of lecithin: retinol acyl transferase (LRAT), an enzyme that catalyzes the conversion of retinol to its storage species retinyl esters. Transfer of ROH from CRBP1 to LRAT regenerates apo-CRBP1, enabling the binding protein to re-associate with STRA6 and further promote ROH uptake and STRA6 signaling (11). ROH metabolism by LRAT also maintains an inward directed ROH concentration gradient, necessary for continuing influx. Hence, STRA6 orchestrates a multi-component “machinery” that couples vitamin A homeostasis and metabolism to activation of a signaling cascade.
Importantly, the binding partner for RBP in blood, TTR, competes with STRA6 for holo-RBP and thus prevents holo-RBP from interacting with the receptor. Consequently, STRA6 is only active either when holo-RBP levels in serum exceed that of TTR, e.g. in obese animals (5, 12, 13), or in cells that express a very high level of the receptor, e.g. the RPE. Other physiological circumstances that enable STRA6 signaling remain to be identified but it has been reported that STRA6 is upregulated in several human cancers, including Wilm’s kidney tumors, melanomas, and colorectal, ovarian, and endometrium cancers (14). The functional significance of the increased expression of STRA6 in carcinoma cells is unknown but the discovery that STRA6 signaling triggered by holo-RBP activates a JAK2/STAT3/5 cascade may provide a clue. These STATs are associated with inflammation, cellular transformation, survival, proliferation, invasion, and angiogenesis and they are considered to be oncogenes (15–18). Tumor-promoting STAT target genes include the cell cycle regulators cyclin D1 and cyclin D3, the oncogene c-Myc, the growth factor VEGF, genes involved in migration and invasion such as MMP-9, and anti-apoptotic genes including survivin, Mcl-1, and Bcl-XL (16, 19). Hence, an intriguing possibility is that STRA6 and its associated machinery may be involved in oncogenic activities. Work described in this manuscript aimed to examine this possibility.
Materials and Methods
Reagents
Antibodies were obtained from: STAT3, pSTAT3, PCNA: Cell Signaling; actin, histidine: Santa Cruz; STRA6: Novus Biologicals; STRA6 polyclonal antibody: see (7). Expression vector for his-STRA6 was from Genecopia. Mutants were generated using Quik Change mutagenesis kit (Stratagene). Breast (BCRT101) and colon (HC105) cancer arrays were purchased from OriGene. RBP4 shRNAs were from Open Biosystems (TRCN0000060040) or Sigma (TRCN0000060038 and TRCN0000060042). STRA6 shRNAs (TRCN0000128799 and TRCN0000129158) were obtained from Open Biosystems. AG490 and ROH were from Calbiochem and Sigma Chemical Co., respectively. Transfections were carried out using PolyFact (Qiagen)
RBP was expressed in E. coli and purified as described in (20). The preparation typically yields holo-RBP at an ROH:RBP mole ratio of 0.8–1:0.
Cells
HCT116, SW480, and SW620 cells were purchased from ATCC and cultured in McCoy’s 5A media supplemented with 10% fetal bovine serum (FBS). NIH3T3 fibroblasts were purchased from ATCC and cultured in Dulbecco’s modified Eagle’s medium supplemented with FBS. Generation of NIH3T3 fibroblasts stably overexpressing LRAT was previously described (21). Generation of HCT116 cells expressing the Y705F dominant negative mutant of STAT3 was previously described (22). HCT116 cells stably overexpressing STRA6 were generated by transfection of vector encoding his-STRA6 and selection using G418 (10 mg/ml). Colonies were pooled. SW480 stable lines with reduced expression of STRA6 or RBP were generated using lentiviral vectors pLKO.1-puro encoding respective shRNA targeting GFP, human STRA6 (Open Biosystems, AL, USA), or human RBP (Sigma). Viruses were packaged in HEK293T cells and used as per manufacturer’s protocols. Cells were selected using 10 μg/ml puromycin. NIH3T3-L1 cells were differentiated as previously described (23).
BrdU proliferation assay kit was purchased from EMD Millipore.
Quantitative real-time PCR (Q-PCR)
RNA was extracted using TriZol. cDNA was generated using GeneAmp RNA PCR (Applied Biosystems). Q-PCR was carried out using TaqMan chemistry and Assays-on-Demand probes (Applied Biosystems): MMP9 (Hs00957562_m1), MYC (Hs99999003_m1), STRA6 (Hs00980261_g1), VEGFA (Hs00900055_m1), C-FOS (Hs00170630_m1), CCND1 (Hs00765553_m1). 18s (4352930) rRNA.
Soft Agar assays
0.8% agar was placed in a 6 well plate, allowed to solidify and topped with 2500 cells/ml in 0.3% agar and growth media. 0.3% agar media was replaced every 48 h. for 21 days and colonies were visualized using 0.5% crystal violet.
Migration assays were performed using 8 μm pore size Transwell migration plates (Corning) plated with 104 cells/cm2 of HCT116 and NIH3T3 fibroblasts, and 105 cells/cm2 of SW480 cells. Cells were placed on the bottom of the transwell plate at 100% confluence. Cells in the top chamber were allowed to migrate for 12 h, the top portion of the insert was wiped with a cotton swab, inserts were fixed in 4% formalin (30 min.) and washed twice with PBS. Cells were stained with 0.5% crystal violet (10–15 min.), washed twice with PBS, visualized and counted.
Wound healing assays
Cells were grown in growth media to 100% confluence and scratched using a 200 μl pipette tip. Cells were washed extensively with PBS to remove floating cells and images were taken at initial, 12 h and 24 h after scratching. Similar results were obtained using Ibidi™ wound healing chambers.
Invasion assays were performed using BD bioscience invasion assay following manufacture’s protocol.
Secondary focus formation assays
10,000 NIH3T3 fibroblasts and 1,000 HCT116 or SW480 cells were plated. Media was replaced every 48 h. for 14 days. Media was removed and cells washed with PBS, fixed in 10% formalin and stained for 30 min. in 0.5% crystal violet.
Immunoblots
Cell protein was extracted using RIPA buffer (25 mM Tris-HCl, 150 mM NaCl, 1% NP40, 1% sodium deoxycholate, 0.1% SDS). Proteins were resolved by SDS-PAGE, transferred onto nitrocellulose membrane and immunoblotted. Band intensities were quantified using Image J 1.40g software (WayenRasband, NIH, USA).
Immunohistochemistry
Excised tumors were placed in 10% formalin, embedded in paraffin and sectioned and stained with hematoxylin and eosin (H&E). Slides were rehydrated in xylene and ethanol and boiled in 1 mM citrate. Slides were blocked and stained with Dako Rabbit immunohistochemistry kit.
Mice
NCr athymic mice were housed in accordance with ARC protocol and IACUC regulations. Six week old male athymic mice were subcutaneously injected with 2×106 HCT116, 5×106 SW480, or 5×106 NIH3T3 cells. Tumors were measured using calipers and tumor volume calculated by (length × width2/2).
Database analysis
Normalized data from Oncomine Cancer Microarray database were plotted as a log median change and standard deviation determined for each independent array.
Statistics
Statistical significance of treatments was analyzed using an independent sample T-test.
Results
Expression of STRA6 and RBP is upregulated in breast and colon cancers
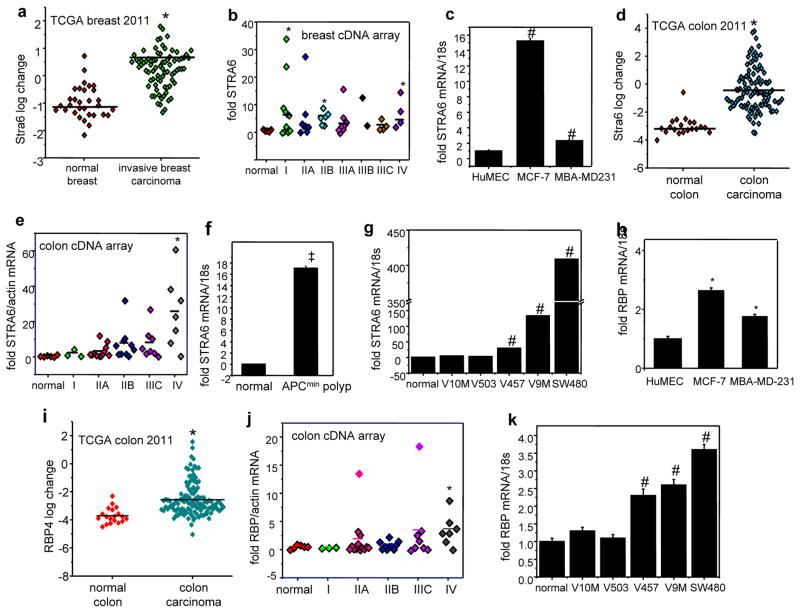
Expression array profiles deposited in the database Oncomine™ (Compedia Bioscience, Ann Arbor, Michigan) indicate that 3 independent studies documented that the level of STRA6 mRNA is elevated in human breast tumors vs. normal breast tissue (Fig. 1a, Fig. S1a, S1b). Accordingly, analysis of a human breast cancer cDNA array (OriGene) showed that STRA6 expression is significantly higher in breast tumors as compared with normal mammary tissue (Fig. 1b). The data showed marked variability and, likely due to the small number of samples on the array, the increase was not statistically significant in all stages. However, a clear trend indicating that STRA6 was upregulated early during tumor development was seen, and it reached statistical significance in stages I, IIB, and IV (Fig. 1b). STRA6 expression was higher in mammary carcinoma MCF-7 and MBA-MD-231 cells than in normal breast epithelium (HuMEC) cells (Fig. 1c). Similarly, STRA6 was undetectable in normal colon and significantly elevated in colon carcinoma (Fig. 1d, S1c, S1d). Analysis of a human colon cancer cDNA array further suggested that the level of STRA6 increases progressively along with tumor development (Fig. 1e). STRA6 was also found to be upregulated during polyp formation in APCmin mouse model of colon adenoma (Fig. 1f), and correlated with the tumorigenic potential of human colon cancer cell lines (Fig. 1g). The level of RBP, the STRA6 ligand, was also elevated in MCF-7 and MBA-MD-231 mammary carcinoma cells (Fig. 1h), and was associated with the metastatic potential of the 67 NR series of isogenic breast cancer cell lines (24)(Fig. S1e). RBP expression was higher in human colon tumors vs. normal colon (Fig. 1i, S1f, S1g), and, similarly to STRA6, analysis of a cDNA array suggested that RBP levels increase progressively with colon cancer development (Fig. 1j). Also similarly to STRA6, RBP expression was found to be associated with the aggressiveness of human colon cancer cell lines (Fig. 1k).
Figure 1. Expression of STRA6 and RBP in tumors.
a) Levels of STRA6 mRNA in samples of normal human breast and invasive breast carcinoma (36). Data were obtained from Oncomine™ (Compedia Bioscience, Ann Arbor, Michigan). *p<0.05. b) STRA6 expression in a TissueScan™ tissue qPCR array consisting of cDNA derived from samples of normal breast and denoted stages of breast tumors (OriGene). *p<0.05. c) Relative STRA6 mRNA levels in denoted cell lines and in normal human mammary epithelial cells (HuMEC) measured by Q-PCR. #p<0.05 vs. HuMEC. d) Levels of STRA6 mRNA in samples of normal human colon and colon carcinoma (36). *p<0.05 vs. normal colon. e) Analysis of TissueScan™ tissue qPCR array consisting of cDNA derived from samples of normal colon and denoted stages of colon tumors (OriGene). *p<0.05 vs. normal colon. f) Levels of STRA6 mRNA in normal colon tissue and in colon polyps in 60 day old APCmin mice, measured by Q-PCR. Mean±S.E.M. (n=3), ‡p<0.0001 vs. normal mouse colon. g) Relative STRA6 mRNA levels in denoted human colon carcinoma cell lines and in normal human colon cells (normal), measured by Q-PCR. Data are mean±S.E.M (n=3). #p<0.05 vs. normal epithelium cells. h) Relative RBP mRNA levels in denoted cell lines and in normal human mammary epithelial cells (HuMEC) measured by Q-PCR.*p<0.05 vs. HuMEC. i) Levels of STRA6 mRNA in samples of normal human colon cells and colon carcinoma (36)). *p<0.05 vs. normal colon. j) RBP expression in a TissueScan™ tissue qPCR array consisting of cDNA derived from samples of normal colon and denoted stages of colon tumors (OriGene).*p<0.05 vs. normal colon. k) Relative RBP mRNA levels in denoted colon carcinoma cell lines and in normal human colon cells (normal), measured by Q-PCR. Data are mean±S.E.M (n=3). #p<0.05 vs. normal epithelium cells. All data are mean±S.E.M. (n=3).
RBP and STRA6 facilitate carcinoma cell proliferation
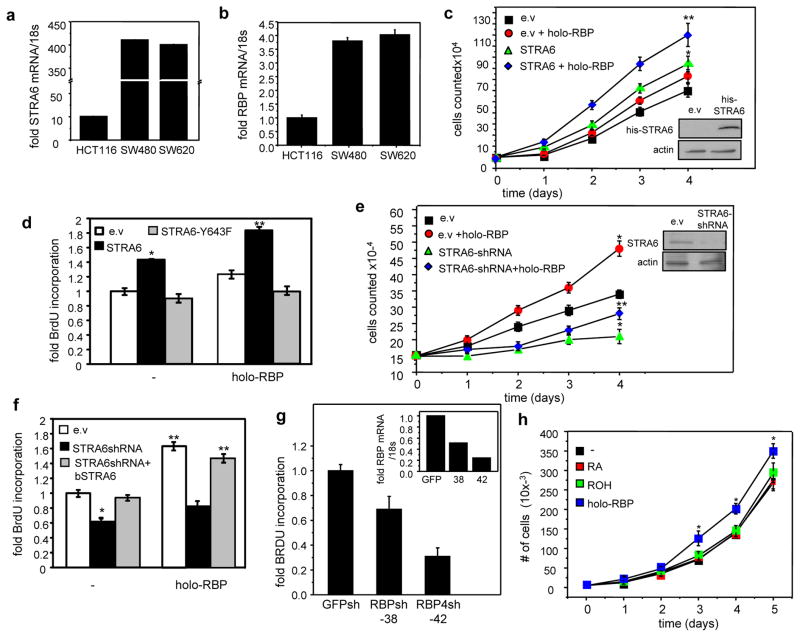
HCT116 colon carcinoma cells which express low levels of STRA6 and RBP, and SW480 and the highly metastatic SW620 colon carcinoma cell lines, which highly express both proteins (Fig. 2a, 2b), were used to begin to examine the role of RBP and STRA6 in tumor development. An HCT116 cell line that stably over-expresses STRA6 was generated (Fig. 2c). Treatment of HCT116 cells with holo-RBP or over-expression of STRA6 modestly facilitated their growth but combining the two factors by treating STRA6-overexpressing cells with holo-RBP markedly enhanced proliferation (Fig. 2c). In accordance, treatment of STRA6-overexpressing cells with holo-RBP enhanced DNA synthesis, reflected by BrdU incorporation, by almost 2 fold (Fig. 2d). In contrast, expression of the signaling defective mutant STRA6-Y643F abolished the ability of holo-RBP to induce DNA synthesis (Fig. 2d). We also generated SW480 cell lines in which STRA6 expression was stably decreased by two independent lentiviral vectors encoding STRA6 shRNAs and which displayed 40% and 80% reduction in receptor expression, respectively (Fig. S2a, inset). The decrease in STRA6 expression suppressed DNA synthesis in a dose-dependent manner (Fig. S2a). A third SW480 cell line which stably expresses both constructs was then generated. Treatment of these cells with holo-RBP markedly facilitated their proliferation, and reducing STRA6 expression inhibited growth and hampered the pro-proliferative activity of holo-RBP (Fig. 2e). Accordingly, reducing the expression of STRA6 suppressed DNA synthesis and abolished the ability of holo-RBP to enhance BrdU incorporation (Fig. 2f). Ectopic expression of bSTRA6 rescued holo-RBP-induced DNA synthesis in cells with reduced expression of the endogenous hSTRA6 (Fig. 2f). STRA6 and holo-RBP also cooperated in promoting DNA synthesis and proliferation in MCF-7 mammary carcinoma cells (Fig. S2b, S2c) and promoted BrdU incorporation in SW620 colon carcinoma cells (Fig. S2e). Similarly, reducing the expression of RBP in SW480 cells suppressed DNA synthesis and did so in a dose dependent manner (Fig. 2g). While holo-RBP induced proliferation of SW480 cells, neither ROH alone nor RA exerted such an activity (Fig. 2h), demonstrating that holo-RBP exerts pro-proliferative activities independently from generation of ROH-derived RA.
Figure 2. RBP and STRA6 promote colon carcinoma cell proliferation.
a, b) Levels of mRNA for STRA6 (a) and RBP (b) in HCT116, SW480 and SW620 colon cancer cells, measured by Q-PCR. c) Growth of HCT116 cells transfected with empty vector (e.v) or a vector encoding STRA6 and treated with 1 μM holo-RBP every 24 h. *p<0.05 vs. e.v.-expressing cells, **p<0.05 vs. corresponding untreated cells. Inset: immunoblot demonstrating over-expression of STRA6. d) BrdU incorporation in HCT116 cells transfected with e.v, or vectors encoding STRA6 or STRA6-Y643F. Cells were cultured in the absence or presence of 1 μM holo-RBP. *p<0.05 vs. e.v.-expressing cells. **p<0.05 vs. corresponding untreated cells. e) Growth of SW480 cells stably expressing e.v or vectors harboring STRA6 shRNA. Cells were treated with 1 μM holo-RBP every 24 h. *p<0.05 vs. e.v.-expressing cells, **p<0.05 vs. corresponding untreated cells. Inset: Immunoblot demonstrating reduced expression of STRA6. f) BrdU incorporation in SW480 cells transfected with e.v, a vector encoding STRA6 shRNA or co-transfected with a vector encoding STRA6 shRNA and an expression vector for bSTRA6. Cells were cultured in the absence or presence of 1 μM holo-RBP.*p<0.05 vs. e.v.-expressing cells, **p<0.05 vs. corresponding untreated cells. g) BrdU incorporation in SW480 cells stably expressing GFP shRNA or vectors encoding RBP shRNA. Inset: expression of STRA6 in cells expressing GFP shRNA or two RBP shRNA constructs: TRCN0000060038 or TRCN0000060042 (Sigma). h) Growth of SW480 cells treated with vehicle or 1 μM RA, ROH or holo-RBP. Ligands were replenished every 24 h. All data are mean±S.E.M. (n=3). *p<0.05 vs. untreated cells. All data are mean±S.E.M. (n=3).
RBP and STRA6 promote oncogenic properties
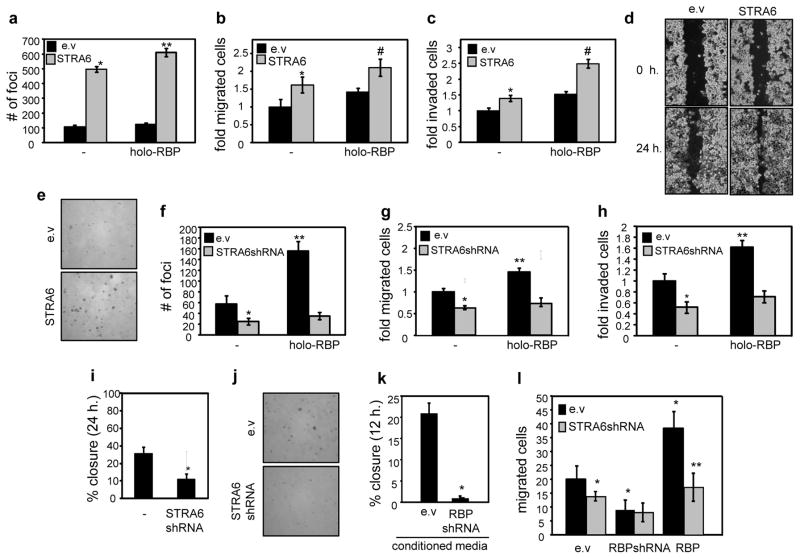
HCT116 and SW480 cells were also used to investigate the effects of RBP and STRA6 on various hallmarks of oncogenic transformation. Parental HCT116 cells, which express a low level of STRA6, showed minimal activity in secondary focus formation assays (Fig. 3a). Over-expression of STRA6 markedly enhanced the ability of these cells to form foci and holo-RBP further potentiated the activity (Fig. 3a). Over-expression of STRA6 and holo-RBP also facilitated the rate of migration of HCT116 cells (Fig. 3b), and their invasion through matrix gel (Fig. 3c). In accordance, STRA6-overexpressing HCT116 cells displayed facilitated wound closure behavior in scratch assays (Fig. 3d, Fig. S2g). Over-expression of STRA6 markedly increased both the number and the size of colonies formed by HCT116 cells in soft agar (Fig. 3e, Fig. S2h). In SW480 cells, holo-RBP promoted foci formation (Fig. 3f) and facilitated cell migration (Fig. 3g) and invasion (Fig. 3h). Strikingly, decreasing the expression of STRA6 in SW480 cells completely abolished these responses (Fig. 3f–3h). The reduced expression of STRA6 also hampered the wound closure behavior of the cells (Fig. 3i) and the number and diameter of colonies formed in soft agar (Fig. 3j, Fig. S2i). STRA6 and holo-RBP also promoted oncogenic activities in MCF-7 mammary carcinoma cells (Fig. S2d), and in SW620 colon cancer cells (Fig. S2e, S2f).
Figure 3. STRA6 and extracellular RBP promote oncogenic properties in colon carcinoma cells.
a) Secondary focus formation by HCT116 cells stably expressing an empty vector (e.v.) or a vector encoding STRA6. Assays were carried out and foci counted as described under Experimental Procedures. Cells were treated with vehicle or 1 μM holo-RBP every 48 h. for 14 days. *p<0.05 vs. e.v.-expressing cells, **p<0.05 vs. corresponding untreated cells. b) Migration of HCT116 cells stably expressing an e.v. or a vector encoding STRA6. Cells were treated with vehicle or 1 μM holo-RBP for 24 h. prior to analyses.*p<0.05 vs. e.v.-expressing cells, #p<0.05 vs. e.v.-expressing, holo-RBP-treated cells. c) Invasion assays using HCT116 cells stably expressing an e.v. or a STRA6-encoding vector. Cells were treated with vehicle or 1 μM holo-RBP for 24 h. prior to analyses. *p<0.05 vs. e.v.-expressing cells, #p<0.05 vs. e.v.-expressing, holo-RBP-treated cells. d) Scratch assays using HCT116 stably expressing an e. v. or a vector encoding STRA6. Images were taken immediately following and 24 h after scratching. See Fig. S2g for quantification. e) Colony formation in soft agar by HCT116 cells stably expressing e.v. or vector encoding STRA6. See Fog. S1h for quantification. f) Secondary focus formation by SW480 cells stably expressing an e.v. or a vector harboring STRA6 shRNA. Cells were treated with vehicle or 1 μM holo-RBP every 48 h. for 14 days. *p<0.05 vs. e.v.-expressing cells, **p<0.05 vs. corresponding untreated cells. g) Rate of migration of SW480 cells stably expressing an e.v. or a vector harboring STRA6 shRNA. Cells were treated with vehicle or 1 μM holo-RBP for 24 h. prior to analyses.*p<0.05 vs. e.v.-expressing cells, **p<0.05 vs. corresponding untreated cells. h) Invasion assays using SW480 cells stably expressing an e.v. or a vector harboring STRA6 shRNA. Cells were treated with vehicle or 1 μM holo-RBP for 24 h. prior to analyses. *p<0.05 vs. e.v.-expressing cells, **p<0.05 vs. corresponding untreated cells. i) Quantification of wound healing assays conducted with SW480 cells stably expressing an e.v or a vector harboring STRA6 shRNA. Images were taken immediately following and 24 h after scratching. Wound closure from 3 independent experiments quantified using image J software.*p<0.05 vs. e.v.-expressing cells. j) Colony formation in soft agar by SW480 cells stably expressing e.v. or STRA6 shRNA. See Fig. S1i for quantification. k) Scratch assays using parental SW480 cells treated with conditioned media from SW480 cells expressing empty lentivirus (e.v.) or lentivirus encoding RBP shRNA. Wound closure in 3 independent experiments was assessed at 12 h. *p<0.05 vs. e.v.-expressing cells. l) SW480 cells transfected with e.v, a vector harboring RBP shRNA, or a vector encoding RBP were plated at the bottom of a trans-well plate, and SW480 cells that express an empty vector or a vector encoding STRA6 shRNA were plated at the top of the trans-well plate. Migration of the cells from the top across the membrane was assessed 12 h. later. Mean±S.E.M. (n=3). *p<0.05 vs. e.v.-expressing cells. **p<0.05 vs. RBP-expressing cells. All data are mean±S.E.M. (n=3).
The observations that, in addition to STRA6, SW480 cells also express a high level of RBP (Fig. 2b) raise the question of whether RBP functions by activating STRA6 following its secretion from the cells or whether it has intracellular activities. SW480 cells and counterparts expressing reduced level of RBP (Fig. S3a) were cultured in serum-free medium for 24 h., and conditioned media were collected. Media were placed on parental SW480 cells and scratch assays were carried out. Conditioned media from parental SW480 cells efficiently promoted wound healing while media from cells with reduced expression of RBP did not (Fig. 3k). Similarly, SW480 cells cultured in conditioned media from cultured adipocytes displayed a more efficient wound closure behavior as compared with cells treated with media from adipocytes with reduced RBP expression (Fig. S3b, S3c). The ability of secreted RBP to promote the motility of neighboring cells was then examined by co-culture experiments. SW480 cells that overexpress RBP (Fig. S3d) or express RBP shRNA were plated at the bottom of a trans-well plate. SW480 cells that express an empty vector or a vector encoding STRA6 shRNA were plated at the top of the trans-well plate and their migration towards the bottom assessed 12 h. later. Over-expression of RBP in cells in the lower chamber facilitated, and down-regulation of RBP inhibited the movement of cells from the top chamber. This activity of RBP was found to critically require expression of STRA6 (Fig. 3l). Notably, as STRA6 is activated by holo- and not apo-RBP (5, 10), these observations indicate that RBP was secreted from cells in complex with retinol. The observations thus demonstrate that holo-RBP can signal in a paracrine fashion.
Oncogenic activities of RBP and STRA6 are mediated by STAT3
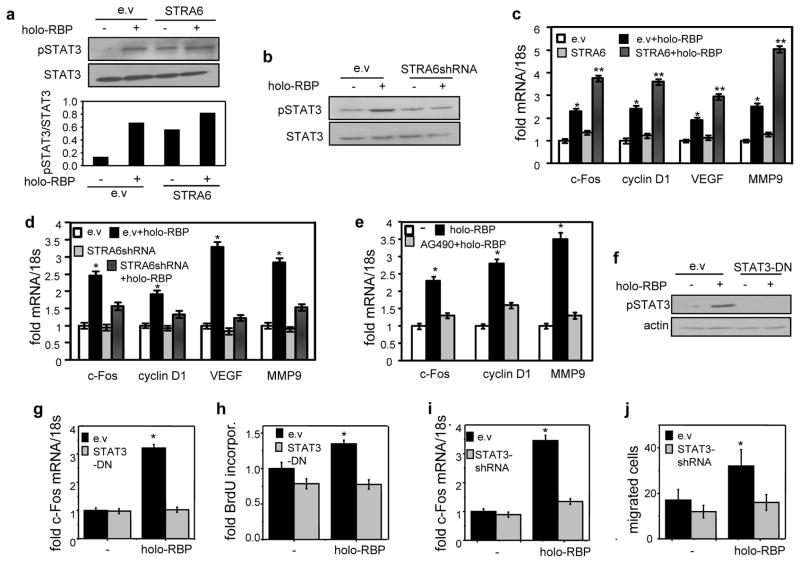
Treatment with holo-RBP increased the phosphorylation level of STAT3 both in HCT116 colon cancer cells (Fig. 4a) and in MCF-7 mammary carcinoma cells (Fig. S3e). Holo-RBP also induced STAT3 phosphorylation in SW480 cells and the effect was lost upon downregulation of STRA6 (Fig. 4b). Attesting to activation of STAT, holo-RBP induced the expression of several pro-oncogenic STAT target genes, including c-Fos, cyclin D1, VEGF-A, and MMP9 in HCT116 (Fig. 4c) and in SW480 cells (Fig. 4d). This activity was augmented upon over-expression of STRA6 in HCT116 cells (Fig. 4c) and suppressed upon decreasing STRA6 expression or ectopic expression of a signaling-defective STRA6 mutant in SW480 cells (Fig. 4d, Fig. S4a). Expression of these genes decreased in a dose-dependent fashion upon reducing the expression of either STRA6 (Fig. S4b) or RBP (Fig. S4c). Induction of these genes by holo-RBP was not inhibited by pre-treatment with the protein synthesis inhibitor cycloheximide, demonstrating that it was exerted by direct transcriptional control (Fig. S4d). Treatment of SW480 cells with the JAK inhibitor AG490 suppressed holo-RBP-induced upregulation of the genes (Fig. 4e). In HCT116 cells, stable over-expression of a dominant-negative construct of STAT3 (STAT3-Y705F, STAT3-DN, (25)) prevented holo-RBP-induced STAT3 phosphorylation (Fig. 4f), and it abolished the ability of holo-RBP to upregulate the expression of c-Fos (Fig. 4g), and to enhance BrdU incorporation (Fig. 4h). Decreasing the expression of STAT3 (Fig. S4e) abolished the ability of holo-RBP to induce c-Fos (Fig. 4i), and to facilitate of cell migration (Fig. 4j). Hence, holo-RBP and STRA6 promote proliferation and migration of these cells by activating STAT3.
Figure 4. Oncogenic activities of RBP and STRA6 are mediated by STAT3.
a) Top: Representative immunoblots of phosphorylated STAT3 (pSTAT3) and total STAT3 in HCT116 stably expressing STRA6 and treated with vehicle (−) or holo-RBP (1 μM, 15 min.). Bottom: quantitation of immunoblots. b) Immunoblots of pSTAT3 and STAT3 in SW480 cells stably expressing e.v. or STRA6 shRNA treated with vehicle (−) or holo-RBP (1 μM, 15 min.). c) Levels of mRNAs of denoted genes in HCT116 cells stably expressing e.v. or STRA6 treated with 1 μM holo-RBP for 4 h. *p<0.05 vs. e.v.-expressing untreated cells, **p<0.05 vs. e.v.-expressing holo-RBP-treated cells. d) Levels of mRNAs of denoted genes in SW480 cells stably expressing e.v. or STRA6 shRNA treated with 1 μM holo-RBP for 4 h. *p<0.05 vs. e.v.-expressing untreated cells. e) Levels of mRNAs of denoted genes in SW480 cells pre-treated with 10 μM AG490 for 24 h. and then treated with 1 μM holo-RBP for 4 h. *p<0.05 vs. e.v.-expressing untreated cells. f) Immunoblots of pSTAT3 and STAT3 in HCT116 stably expressing e.v. or a vector encoding STAT3-DN treated with 1 μM holo-RBP for 15 min. *p<0.05 vs. e.v.-expressing untreated cells. g, i) Levels of c-Fos mRNA in HCT116 cells stably expressing e.v. or STAT3-DN (g) or STAT3 shRNA (i). Cells were treated with 1 μM holo-RBP for 4 h. *p<0.05 vs. e.v.-expressing untreated cells. h) 24 h. BrdU incorporation in HCT116 cells stably expressing e.v. or vector encoding STAT3-DN. Cells were treated with vehicle or 1 μM holo-RBP. *p<0.05 vs. e.v.-expressing untreated cells. j) Rates of migration of HCT116 cells expressing e.v. or a vector encoding STAT3 shRNA and treated with 1 μM holo-RBP. *p<0.05 vs. e.v.-expressing untreated cells. All data are mean±S.E.M. (n=3).
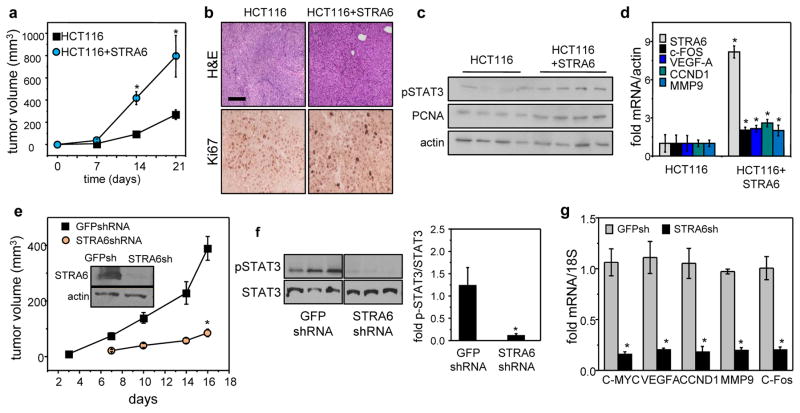
STRA6 is critical for colon tumor development in a xenograft mouse model
NCr athymic mice were used to explore the involvement of STRA6 in colon tumor formation in vivo. To minimize variability between animals, each mouse was injected with the parental HCT116 cells into one flank, and HCT116 cells that stably over-express STRA6 into the opposite flank. Sites injected with cells that over-express STRA6 developed markedly larger tumors than sites injected with parental cells (Fig. 5a). Immunostaining showed that STRA6-overexpressing neoplasm expressed higher levels of the proliferation marker Ki67 (Fig. 5b) and the DNA synthesis marker PCNA (Fig. 5c). The phosphorylation status of STAT3 was higher in STRA6-overexpressing tumors (Fig. 5c) and, accordingly, these tumors expressed higher levels of c-Fos, VEGF-A, cyclin D1 and MMP9 (Fig. 5d).
Figure 5. Colon tumor development in vivo critically depends on expression of STRA6.
a) NCr athymic male mice were injected with 2 x106 HCT116 cells stably transfected with an e.v. into the right flank and HCT116 cells stably expressing STRA6 into the left flank. Tumor growth at both injection sites was monitored by measuring the length and width with calipers and tumor volume calculated as length x width2/2. Data are mean±S.E.M. (n=5)*p<0.05 vs. e.v.-expressing tumors. b) H&E staining and Ki67 immunostaining of tumors that arose from HCT116 cells expressing e.v or STRA6 (bar: 100 μm). c) Immunoblots of pSTAT3 and the proliferation marker PCNA in tumors that arose from HCT116 cells expressing e.v or STRA6. d) Levels of denoted mRNAs in tumors that arose from HCT116 cells expressing e.v or HCT116 cells expressing STRA6. Data are mean±S.E.M. (n=3)*p<0.05 vs. e.v.-expressing tumors. e) NCr male mice were injected with 5×106 SW480 cells stably expressing either GFP shRNA (shGFP, left flank) or STRA6 shRNA (shS6, right flank). Tumor growth at both injection sites was monitored. Data are mean±S.E.M. (n=9). *p<0.01 vs. tumors expressing GFP shRNA. Inset: immunoblots of STRA6 in SW480 lines stably expressing GFP shRNA (shGFP) or STRA6 shRNA (shS6). f) Left: immunoblots of pSTAT3 in tumors that arose from SW480 cells stably expressing GFP shRNA or STRA6 shRNA. Right: quantification of immunoblots. Mean±S.E.M. (n=3). * p<0.05 vs. GFP shRNA-expressing tumors. g) Levels of mRNA for STAT3 target genes in tumors that arose from SW480 cells expressing GFP shRNA or STRA6 shRNA. Data are mean±SD (n=3). * p<0.01 vs. GFP shRNA-expressing tumors. All data are mean±S.E.M. (n=3).
SW480 cells were used to further examine whether STRA6 is critical for tumor development. NCr athymic mice were injected with SW480 cells into the right flank, and SW480 cells in which STRA6 expression is stably down-regulated into the left flank. Tumor development was markedly suppressed by reducing the expression of STRA6 (Fig. 5e). Phosphorylation of STAT3 (Fig. 5f) and expression of pro-oncogenic STAT target genes (Fig. 5g) were both markedly lower in tumors that arose from cells with reduced expression of STRA6.
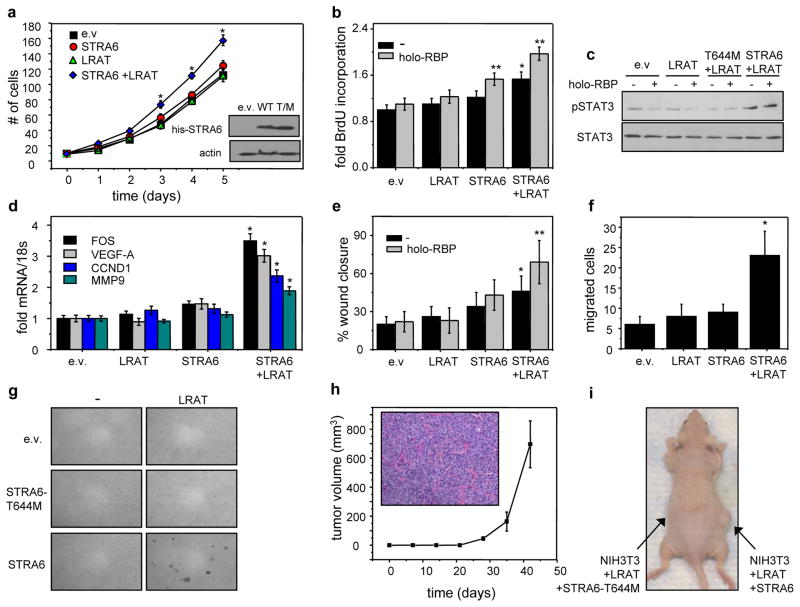
STRA6 and its associated machinery induce oncogenic transformation in NIH3T3 fibroblasts
The ability of STRA6 signaling to transform NIH3T3 fibroblasts, a classical approach for examining the oncogenic potential of proteins, was then examined. STRA6 signaling is coupled to cellular uptake of retinol, and both activities require the expression of the intracellular retinol acceptor CRBP1, and the retinol-metabolizing enzyme LRAT (11). NIH3T3 cells express CRBP1 but not STRA6 or LRAT (5, 11). To examine whether the entire STRA6-associated machinery is necessary for oncogenic activities, NIH3T3 cells that stably express STRA6 or stably co-express STRA6 and LRAT were generated. As a control, an additional cell line that stably expresses co-express LRAT and the signaling-defective STRA6-T644 M mutant was also generated (Fig 6a, inset).
Figure 6. STRA6 and its associated machinery induce oncogenic transformation.
a) Growth curve of NIH3T3 fibroblasts stably overexpressing e.v, STRA6, LRAT, or co-expressing STRA6 and LRAT. Inset: Immunoblots demonstrating expression of his-STRA6 and his-STRA6-T644M in NIH3T3 cells. Data are mean±S.E.M. (n=3)*p<0.05 vs. e.v.-expressing cells. b) BrdU incorporation in NIH3T3 fibroblasts overexpressing e.v, STRA6, LRAT, or both and treated with vehicle or 1 μM holo-RBP. Mean±S.E.M. (n=3). *p<0.05 vs. e.v.-expressing cells, **p<0.05 vs. corresponding untreated cells. c) Immunoblots of pSTAT3 and STAT3 in NIH3T3 cells expressing denoted constructs and treated with vehicle or 1 μM holo-RBP for 15 min. d) Levels of mRNAs of STAT target genes in NIH3T3 cells expressing e.v. or vectors encoding STRA6, LRAT, or both. Cells were treated with 1 μM holo-RBP for 4 h.*p<0.05 vs. e.v.-expressing cells. e) Scratch assay conducted with NIH3T3 fibroblasts expressing denoted constructs and treated with vehicle or 1 μM holo-RBP for 24 h. *p<0.05 vs. e.v.-expressing cells, **p<0.05 vs. corresponding untreated cells. f) Migration assays using NIH3T3 fibroblast sectopically expressing denoted proteins. *p<0.05 vs. e.v.-expressing cells.*p<0.05 vs. e.v.-expressing cells. g) Soft agar colony formation assays using NIH3T3 cells expressing denoted constructs. Colonies were allowed to form for 28 days. Data are mean±S.E.M. (n=3). h) NCr athymic male mice were injected with 5×106 NIH3T3 fibroblast co-expressing LRAT and STRA6 into the right flank and LRAT and STRA6-T644M into the left flank. Tumor growth was monitored for 40 days. No tumors arose in sites injected with cells co-expressing LRAT and the STRA6-T644M mutant. mean±S.E.M. (n=5). Inset: H&E staining of fibrosarcoma that developed from NIH3T3 cells co-expressing STRA6 and LRAT. i) Image of a representative mouse 40 days following injection of denoted cells. Data are Mean±S.E.M. (n=3).
Over-expression of either LRAT or STRA6 alone did not affect NIH3T3 cell growth but co-expression of the receptor and the enzyme significantly facilitated proliferation (Fig. 6a) and potentiated the ability of holo-RBP to promote DNA synthesis (Fig. 6b). Co-expression of STRA6 and LRAT also increased the phosphorylation status of STAT3 (Fig. 6c), upregulated the expression of oncogenic STAT target genes (Fig. 6d), facilitated closure in scratch assays (Fig. 6e), and enhanced cell migration (Fig. 6f). LRAT- and STRA6-expressing cells, but not cells expressing STRA6 alone or LRAT in conjunction of the signaling defective STRA6-T644M mutant, formed colonies in soft agar (Fig. 6g). Finally, when injected into athymic mice, NIH3T3 cells that co-express LRAT and STRA6 efficiently developed tumors resembling fibrosarcoma (Fig. 6h). While tumors developed in 5 out of 10 animals injected with cells co-expressing LRAT and STRA6, no tumors arose in any site injected with cells expressing LRAT in conjunction with STRA6-T644M (Fig. 6i). Hence, STRA6 and its associated machinery potently promote oncogenic transformation.
Discussion
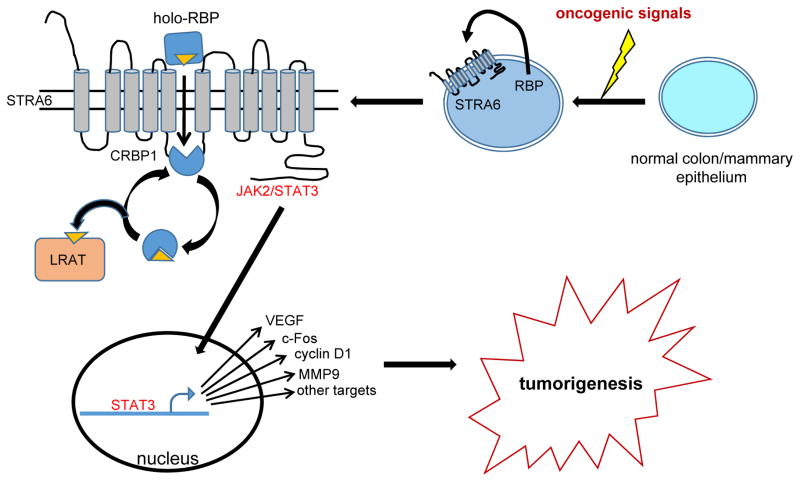
The observations demonstrate that RBP and its plasma membrane receptor STRA6 are undetectable in normal colon and mammary epithelium and are upregulated in some breast and colon cancers. The signals that induce the expression of these genes remain to be investigated but the data suggest that STRA6 is upregulated early in breast cancer development and that expression of both the receptor and its ligand RBP progressively increase throughout colon cancer progression. As shown in Fig. 7, the data show that RBP is secreted from carcinoma cells and activates STRA6 which, in turn, triggers JAK2/STAT3 signaling resulting in induction of multiple pro-oncogenic STAT target genes. Hyper-activation of STRA6 may thus underlie the well-documented observations that STAT3 is constitutively active in multiple cancers (26). In accordance with the observations that STRA6 signaling is coupled to retinol metabolism (11), oncogenic activities of STRA6 require the expression of the retinol-esterifying enzyme LRAT. STRA6 thus links retinol uptake and intracellular metabolism to activation of a JAK/STAT cascade and promotion of oncogenic transformation. In contrast with these conclusions, it was recently reported that STRA6 contributes to p53-induced apoptosis in response to DNA damage (27). The basis for the discrepancy and the mechanism through which STRA6 may exert a proapoptic activity remains to be clarified.
Figure 7. A model describing the oncogenic activities of holo-RBP and STRA6.
Expression of STRA6 and RBP is undetectable in normal mammary and colon epithelium. Oncogenic signals, the nature of which remains to be clarified, induce expression of both proteins. Holo-RBP is secreted from the cells and binds to the extracellular domain of STRA6. Subsequently, ROH transfers from RBP to the intracellular acceptor apo-CRBP1 thereby activating STRA6. In turn, STRA6 transduces JAK2/STAT3 signaling, leading to induction STAT target genes that promote oncogenic transformation. LRAT unloads ROH from holo-CRBP1 thereby regenerating apo-CRBP1 which can re-associate with STRA6 allowing ROH uptake and STRA6 signaling to proceed.
Considering that holo-RBP circulates in blood at 1–2 μM concentrations, mechanisms that protect tissues from hyper-activation of STRA6 must exist. One such mechanism is the association of holo-RBP with its serum binding partner TTR which efficiently competes with STRA6 for holo-RBP and thud inhibits STRA6-initiated cell signaling (5). It could thus be expected that animals would be protected from the oncogenic activities of the pathway when serum holo-RBP level does not exceed that of TTR. Moreover, due to its low molecular weight, free holo-RBP is rapidly excreted by glomeruli filtration. The signaling activities of this cytokine are thus constrained both by its association with TTR and by its short half-life in the circulation. The complete spectrum of physiological situations in which plasma RBP concentration exceeds that of TTR remains to be clarified but it has been reported that blood level of RBP is elevated in obese mice and humans (12). Under these circumstances, the elevated RBP/TTR ratio in plasma results in marked activation of STRA6 and STAT3/5 and, consequently, in insulin resistance (5, 10). Notably, a large body of epidemiological studies indicates that obesity is a risk factor for multiple types of cancers including colon and breast cancers (28, 29). The association is particularly strong with colorectal cancer where convincing evidence identified obesity to be a cause for the disease (30). Breast and colorectal cancer are also associated with insulin resistance. For example, high levels of fasting insulin have been associated with a 2–3-fold increase in risk of mortality from breast cancer (31). The molecular mechanisms that underlie the association between excess body weight and insulin resistance and cancer are incompletely understood but accumulated evidence suggests that the adipose tissue is an important mediator of this link (32). Among adipocyte-derived factors that have been implicated in involvement in cancer development, the most extensively studied is leptin whose expression is upregulated in obese animals and which displays distinct pro-tumorigenic activities (28, 33–35). The findings of the present work suggest that holo-RBP/STRA6 signaling may comprise another important molecular mechanism through which obesity promotes cancer development.
Supplementary Material
Acknowledgments
We thank Joseph Nadeau and Stephanie Doener (Case Western Reserve University) for mRNA from APCmin polyps and matching normal colon tissue, Sanford Markowitz (Case Western Reserve University) for sharing colon cancer cell lines, Hui Jin (Case Western Reserve University) for SW480 cells stably expressing STRA6 shRNA, Krzyszt of Palczewski and Marcin Golczak (Case Western Reserve University) for NIH3T3 fibroblasts stably overexpressing LRAT, Zhenghe Wang (Case Western Reserve University) for HCT116 cells stably expressing STAT3-Y705F, Hui Sun (UCLA) for a bSTRA6 vector, and Silke Vogel (Columbia University) for RBP bacterial expression vector. This work was supported by NIH grants PHS RO1 DK088969 and DK060684.
Footnotes
The authors declare that they do not have any conflict of interest with the work described in this manuscript
References
- 1.Germain P, Chambon P, Eichele G, Evans RM, Lazar MA, Leid M, et al. International Union of Pharmacology. LX. Retinoic acid receptors. Pharmacol Rev. 2006;58:712–25. doi: 10.1124/pr.58.4.4. [DOI] [PubMed] [Google Scholar]
- 2.Schug TT, Berry DC, Shaw NS, Travis SN, Noy N. Opposing effects of retinoic acid on cell growth result from alternate activation of two different nuclear receptors. Cell. 2007;129:723–33. doi: 10.1016/j.cell.2007.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raghu P, Sivakumar B. Interactions amongst plasma retinol-binding protein, transthyretin and their ligands: implications in vitamin A homeostasis and transthyretin amyloidosis. Biochimica et biophysica acta. 2004;1703:1–9. doi: 10.1016/j.bbapap.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 4.Noy N. Retinoid-binding proteins: mediators of retinoid action. The Biochemical journal. 2000;348(Pt 3):481–95. [PMC free article] [PubMed] [Google Scholar]
- 5.Berry DC, Croniger CM, Ghyselinck NB, Noy N. Transthyretin blocks retinol uptake and cell signalling by the holo-retinol-binding protein receptor STRA6. Molecular and cellular biology. 2012;32:3851–9. doi: 10.1128/MCB.00775-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Terra R, Wang X, Hu Y, Charpentier T, Lamarre A, Zhong M, et al. To Investigate the Necessity of STRA6 Upregulation in T Cells during T Cell Immune Responses. PloS one. 2013;8:e82808. doi: 10.1371/journal.pone.0082808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berry DC, Jacobs H, Marwarha G, Gely-Pernot A, O’Byrne SM, DeSantis D, et al. The STRA6 receptor is essential for retinol-binding protein-induced insulin resistance but not for maintaining vitamin A homeostasis in tissues other than the eye. The Journal of biological chemistry. 2013;288:24528–39. doi: 10.1074/jbc.M113.484014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawaguchi R, Yu J, Honda J, Hu J, Whitelegge J, Ping P, et al. A membrane receptor for retinol binding protein mediates cellular uptake of vitamin A. Science. 2007;315:820–5. doi: 10.1126/science.1136244. [DOI] [PubMed] [Google Scholar]
- 9.Ruiz A, Mark M, Jacobs H, Klopfenstein M, Hu J, Lloyd M, et al. Retinoid content, visual responses and ocular morphology are compromised in the retinas of mice lacking the retinol-binding protein receptor, STRA6. Investigative ophthalmology & visual science. 2012 doi: 10.1167/iovs.11-8476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berry DC, Jin H, Majumdar A, Noy N. Signaling by vitamin A and retinol-binding protein regulates gene expression to inhibit insulin responses. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:4340–5. doi: 10.1073/pnas.1011115108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berry DC, O’Byrne SM, Vreeland AC, Blaner WS, Noy N. Cross Talk between Signaling and Vitamin A Transport by the Retinol-Binding Protein Receptor STRA6. Molecular and cellular biology. 2012;32:3164–75. doi: 10.1128/MCB.00505-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Q, Graham TE, Mody N, Preitner F, Peroni OD, Zabolotny JM, et al. Serum retinol binding protein 4 contributes to insulin resistance in obesity and type 2 diabetes. Nature. 2005;436:356–62. doi: 10.1038/nature03711. [DOI] [PubMed] [Google Scholar]
- 13.Mody N, Graham TE, Tsuji Y, Yang Q, Kahn BB. Decreased clearance of serum retinol-binding protein and elevated levels of transthyretin in insulin-resistant ob/ob mice. Am J Physiol Endocrinol Metab. 2008;294:E785–93. doi: 10.1152/ajpendo.00521.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szeto W, Jiang W, Tice DA, Rubinfeld B, Hollingshead PG, Fong SE, et al. Overexpression of the retinoic acid-responsive gene Stra6 in human cancers and its synergistic induction by Wnt-1 and retinoic acid. Cancer research. 2001;61:4197–205. [PubMed] [Google Scholar]
- 15.Koptyra M, Gupta S, Talati P, Nevalainen MT. Signal transducer and activator of transcription 5a/b: biomarker and therapeutic target in prostate and breast cancer. Int J Biochem Cell Biol. 2011;43:1417–21. doi: 10.1016/j.biocel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aggarwal BB, Kunnumakkara AB, Harikumar KB, Gupta SR, Tharakan ST, Koca C, et al. Signal transducer and activator of transcription-3, inflammation, and cancer: how intimate is the relationship? Annals of the New York Academy of Sciences. 2009;1171:59–76. doi: 10.1111/j.1749-6632.2009.04911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferbeyre G, Moriggl R. The role of Stat5 transcription factors as tumor suppressors or oncogenes. Biochimica et biophysica acta. 2011;1815:104–14. doi: 10.1016/j.bbcan.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 18.Quesnelle KM, Boehm AL, Grandis JR. STAT-mediated EGFR signaling in cancer. Journal of cellular biochemistry. 2007;102:311–9. doi: 10.1002/jcb.21475. [DOI] [PubMed] [Google Scholar]
- 19.Leeman RJ, Lui VW, Grandis JR. STAT3 as a therapeutic target in head and neck cancer. Expert Opin Biol Ther. 2006;6:231–41. doi: 10.1517/14712598.6.3.231. [DOI] [PubMed] [Google Scholar]
- 20.Xie Y, Lashuel HA, Miroy GJ, Dikler S, Kelly JW. Recombinant human retinol-binding protein refolding, native disulfide formation, and characterization. Protein Expr Purif. 1998;14:31–7. doi: 10.1006/prep.1998.0944. [DOI] [PubMed] [Google Scholar]
- 21.Golczak M, Maeda A, Bereta G, Maeda T, Kiser PD, Hunzelmann S, et al. Metabolic basis of visual cycle inhibition by retinoid and nonretinoid compounds in the vertebrate retina. The Journal of biological chemistry. 2008;283:9543–54. doi: 10.1074/jbc.M708982200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao Y, Zhang X, Guda K, Lawrence E, Sun Q, Watanabe T, et al. Identification and functional characterization of paxillin as a target of protein tyrosine phosphatase receptor T. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:2592–7. doi: 10.1073/pnas.0914884107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berry DC, DeSantis D, Soltanian H, Croniger CM, Noy N. Retinoic acid upregulates preadipocyte genes to block adipogenesis and suppress diet-induced obesity. Diabetes. 2012;61:1112–21. doi: 10.2337/db11-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aslakson CJ, Miller FR. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer research. 1992;52:1399–405. [PubMed] [Google Scholar]
- 25.Kaptein A, Paillard V, Saunders M. Dominant negative stat3 mutant inhibits interleukin-6-induced Jak-STAT signal transduction. The Journal of biological chemistry. 1996;271:5961–4. doi: 10.1074/jbc.271.11.5961. [DOI] [PubMed] [Google Scholar]
- 26.Bromberg J, Darnell JE., Jr The role of STATs in transcriptional control and their impact on cellular function. Oncogene. 2000;19:2468–73. doi: 10.1038/sj.onc.1203476. [DOI] [PubMed] [Google Scholar]
- 27.Carrera S, Cuadrado-Castano S, Samuel J, Jones GD, Villar E, Lee SW, et al. Stra6, a retinoic acid-responsive gene, participates in p53-induced apoptosis after DNA damage. Cell death and differentiation. 2013;20:910–9. doi: 10.1038/cdd.2013.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park J, Euhus DM, Scherer PE. Paracrine and Endocrine Effects of Adipose Tissue on Cancer Development and Progression. Endocrine reviews. 2011 doi: 10.1210/er.2010-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Demark-Wahnefried W, Platz EA, Ligibel JA, Blair CK, Courneya KS, Meyerhardt J, et al. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2012. The Role of Obesity in Cancer Survival and Recurrence. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vainio H, Bianchini F, editors. Weight Control and Physical Activity. Lyon, France: IARC Press; 2002. [Google Scholar]
- 31.Goodwin PJ, Ennis M, Pritchard KI, Trudeau ME, Koo J, Madarnas Y, et al. Fasting insulin and outcome in early-stage breast cancer: results of a prospective cohort study. J Clin Oncol. 2002;20:42–51. doi: 10.1200/JCO.2002.20.1.42. [DOI] [PubMed] [Google Scholar]
- 32.Donohoe CL, Doyle SL, Reynolds JV. Visceral adiposity, insulin resistance and cancer risk. Diabetol Metab Syndr. 2011;3:12. doi: 10.1186/1758-5996-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Renehan AG, Roberts DL, Dive C. Obesity and cancer: pathophysiological and biological mechanisms. Arch Physiol Biochem. 2008;114:71–83. doi: 10.1080/13813450801954303. [DOI] [PubMed] [Google Scholar]
- 34.Zheng Q, Dunlap SM, Zhu J, Downs-Kelly E, Rich J, Hursting SD, et al. Leptin deficiency suppresses MMTV-Wnt-1 mammary tumor growth in obese mice and abrogates tumor initiating cell survival. Endocrine-related cancer. 2011;18:491–503. doi: 10.1530/ERC-11-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoda MR, Theil G, Mohammed N, Fischer K, Fornara P. The adipocyte-derived hormone leptin has proliferative actions on androgen-resistant prostate cancer cells linking obesity to advanced stages of prostate cancer. J Oncol. 2012;2012:280386. doi: 10.1155/2012/280386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.TCGA. The Cancer Genome Atlas. 2011 http://tcga-data.nci.nih.gov/tcga/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.