Abstract
PURPOSE
Loss of function of PTEN is a frequent event in melanoma, particularly in tumors with BRAFV600 mutations. The prevalence, pathological features, and clinical outcomes associated with PTEN loss in patients with stage IIIB/C melanoma were interrogated to improve our understanding of the clinical significance of this molecular event.
EXPERIMENTAL DESIGN
Archival tissue from lymphadenectomy specimens among patients (n=136) with stage IIIB or IIIC melanoma were assessed by DNA sequencing for activating BRAF and NRAS mutations, and by immunohistochemistry (IHC) for the expression of PTEN protein. Associations of these molecular aberrations with demographics, tumor characteristics, and clinical outcomes were determined.
RESULTS
The prevalence of BRAFV600 mutations (40% overall), NRAS mutations (10%), and PTEN loss (25%) did not vary by pathological substage. BRAF/NRAS mutation status did not correlate with distant disease-free survival (DDFS) or overall survival (OS). Complete loss of PTEN expression correlated with shorter OS but not DDFS. When stratified by specific sites of distant metastasis, PTEN loss was associated with significantly shorter time to melanoma brain metastasis (MBM), but not to liver, lung, or bone. Analysis of PTEN in mutationally-defined subsets showed that PTEN loss was significantly associated with OS and time to MBM in patients with BRAFV600 mutations.
CONCLUSIONS
Loss of PTEN protein expression correlates significantly with decreased OS and time to MBM in stage IIIB/C melanoma patients with BRAFV600 mutations. The findings add to evidence supporting a significant role for PTEN loss and the PI3K-AKT pathway in melanoma.
Keywords: melanoma, BRAF, NRAS, PTEN, brain metastasis
Introduction
The clinical management of melanoma is evolving rapidly due to improved understanding of the molecular drivers of this disease. Substitutions affecting the V600 residue of the BRAF serine-threonine kinase are the most common activating mutation detected in cutaneous melanoma patients (40–45%).(1, 2) BRAFV600 mutations increase the kinase activity of BRAF and result in constitutive activation of the RAS-RAF-MAPK pathway. Recurrent point mutations affecting residues G12, G13, and Q61 of NRAS are the second most common activating event in melanoma (~20%).(2) Previous studies have demonstrated that NRAS mutations are essentially mutually exclusive with BRAFV600 mutations, likely due to the fact that they also activate signaling through RAS-RAF-MAPK.(3, 4) In addition to the high prevalence of oncogenic mutations, the importance of this pathway in melanoma is supported by the clinical results observed with inhibitors targeting it. Mutation-selective BRAF inhibitors (vemurafenib, dabrafenib) and MEK inhibitors (trametinib) have been approved by the FDA for the treatment of metastatic melanoma patients with BRAFV600 mutations.(5–7) MEK inhibitors have also demonstrated clinical activity in patients with activating NRAS mutations, and clinical trials of MEK inhibitors alone and in combination with other agents are planned and/or ongoing.(8)
Mutant RAS proteins utilize multiple signaling pathways to mediate their oncogenic effects. One such pathway is the PI3K-AKT pathway. In addition to NRAS mutations, this pathway can be activated in melanoma by loss of function of the phosphatase PTEN.(9) Genetic analyses of melanoma cell lines and clinical specimens have demonstrated that inactivating mutations and deletions of the PTEN gene are relatively rare (~10%), and are largely mutually exclusive with NRAS mutations in melanoma, but frequently overlap with BRAFV600 mutations.(3, 4) Loss of PTEN mRNA and protein expression has been detected more frequently (20–30%), potentially due to epigenetic mechanisms.(10–12) Our previous quantitative proteomic analyses of cell lines and frozen tumor samples in multiple tumor types have shown that loss of PTEN protein expression correlates with greater activation of AKT and other PI3K pathway effectors than a number of other oncogenic events, including PIK3CA and NRAS mutations.(9, 13) Multiple inhibitors against targets in the PI3K-AKT pathway are in various phases of clinical testing in melanoma and other malignancies.(14)
In addition to serving as therapeutic targets, molecular aberrations can impact patient management in cancer by improving risk models of disease progression and mortality. Several previous studies have examined the clinical associations and prognostic significance of BRAFV600 and NRAS mutations in primary melanomas.(15–19) We and others have also reported their prevalence and prognostic significance in patients with stage IV melanoma.(20, 21) However, currently there is very little information about the clinical significance of BRAF, NRAS, or PTEN in stage III melanoma patients. Patients with stage III melanoma have highly variable outcomes, and currently there are no validated molecular markers that add to existing prognostic models utilizing clinical and pathological features.(22) The identification of markers that could improve risk stratification in stage III melanoma patients is important, as the only currently approved adjuvant therapies for these patients (interferon-α-2B and pegylated-interferon-α-2b) are given for prolonged periods of time and have significant associated toxicities. Further, evaluating the prognostic significance of such markers in stage III melanoma patients will also strengthen the design and interpretation of new clinical trials that have been designed for this population based on observed activity in patients with stage IV disease.
In order to test the hypothesis that molecular markers can improve existing prognostic models, and to improve our understanding of their potential role in the pathogenesis of this disease, we determined the mutational status of BRAF and NRAS, and the protein expression of PTEN, in lymphadenectomy specimens from patients who underwent standard-of-care surgical treatment for stage IIIB/C melanoma. This study includes analysis of PTEN expression (by IHC) in a cohort of tumors that previously underwent quantitative proteomic analysis. This analysis demonstrates that only complete loss of PTEN protein expression correlates with increased expression of activation-specific markers in the PI3K-AKT pathway, thus providing a molecular rationale to guide the evaluation of PTEN expression in melanoma clinical samples. Our subsequent analysis of BRAF, NRAS, and PTEN status in a cohort of clinically annotated lymphadenectomy specimens from patients with stage IIIB/C melanoma identifies a significant association for loss of expression of PTEN with shorter overall survival and shorter time to melanoma brain metastasis.
Methods
Patients
Under an Institution Review Board-approved protocol, patients were identified who underwent standard-of-care lymphadenectomy for stage IIIB/C melanoma, for whom archival material was available, and that had clinical follow-up of at least 5 years for surviving patients. The patients underwent lymphadenectomy surgery between 1988 to 2009. Patient data and primary tumor characteristics were collected for all patients. Patient records were reviewed for clinical events including development of local recurrence, development and pattern(s) of distant metastasis(es), and overall survival.
DNA Mutation Detection
DNA was extracted from unstained formalin-fixed paraffin embedded (FFPE) slides that had undergone H&E-guided macrodissection to isolate portions with at least 70% viable tumor cells. DNA was extracted by the MD Anderson Biospecimen Extraction Core Facility. Samples were screened for hotspot mutations in BRAF and NRAS using the Sequenom MassArray Platform by the MD Anderson Characterized Cell Line Core facility, as previously described.(9)
PTEN Immunohistochemistry
PTEN protein expression in FFPE samples was assessed by immunohistochemistry (IHC) (6H2.1 antibody, Cascade Bioscience) as previously described.(23) Tumor samples were assessed as part of a tissue microarray (TMA), which included 3 cores for each tumor. Complete absence of PTEN was defined as <10% of tumor cells with any immunoreactivity in tumors with staining observed in internal positive controls (i.e. endothelial cells).
Statistical Analysis
Patient and tumor characteristics were compared to molecular features using Fisher’s exact, Wilcoxon rank sum or Kruskal-Wallis tests. Time to any (local or distant) relapse (RFS), time to first distant metastasis (DDFS), and overall survival (OS) from diagnosis of stage III disease were estimated using Kaplan-Meier method. For organ-site specific analyses, patients without metastases were censored at the date of their last imaging by CT and/or MRI scan. Log-rank testing was used to assess differences between groups. The association between PTEN status and OS and time to brain metastasis was further assessed using multivariable Cox proportional hazards regression models that included age, clinical stage, and gender.
Results
Evaluation of PTEN expression by immunohistochemistry
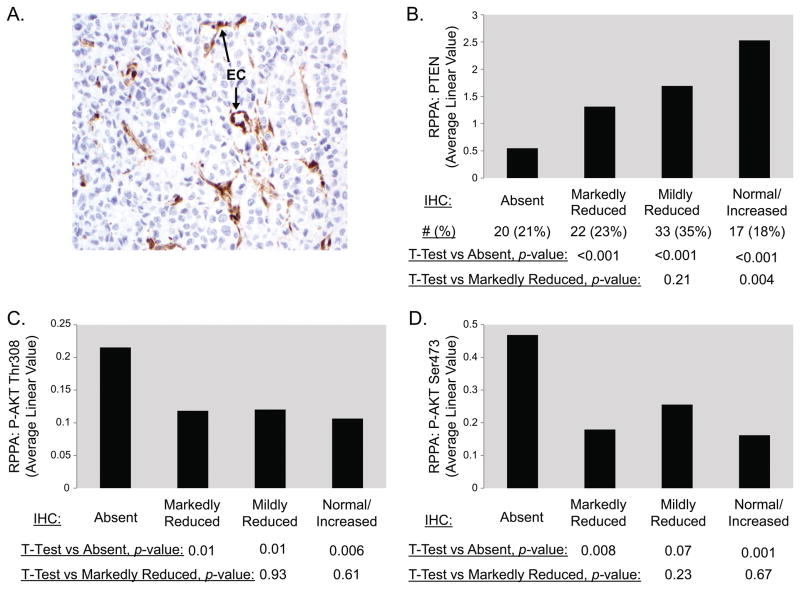
Previously we used reverse phase protein arrays (RPPA) to quantitatively assess protein expression levels of PTEN and PI3K-AKT pathway activation-specific markers (i.e., Phospho-AKTSer473, Phospho-AKTThr308) in a cohort (n=96) of frozen melanoma regional and distant metastases.(9) To determine the expression and molecular correlations of PTEN expression as assessed by IHC, we performed staining for PTEN protein expression on available FFPE slides from the same surgical accessions (n=94) used in the RPPA analysis. Five patterns of PTEN protein expression were observed. Complete absence of PTEN protein expression in the tumor cells (Figure 1A) was observed in 20 samples (21%). PTEN expression was markedly reduced compared to internal positive controls (i.e., endothelial cells) in 22 tumors (23%), mildly reduced in 33 tumors (35%), and normal or increased in 17 tumors (18%) (representative images are shown in Figure S1). The staining in virtually all samples was homogenous; however, 2 tumors (2.1%) demonstrated distinct areas of PTEN absence and PTEN presence (termed ‘clonal-like’ due to its appearance) (Figure S1). Levels of PTEN measured by RPPA showed a good correlation with IHC-based PTEN measurement, and quantitative levels of PTEN (RPPA) demonstrated statistically significant differences among the four prevalent IHC staining patterns (Figure 1B). In contrast, expression levels of P-AKTSer473 and P-AKTThr308 (RPPA) were significantly elevated only in the tumors with complete absence of PTEN expression (Figure 1C/D). The levels of both P-AKT proteins did not differ significantly between the other 3 groups. Based on these findings, PTEN expression in subsequent studies was categorized as Absent or Not Absent; tumors with ‘clonal-like’ expression were also identified but were excluded from analyses due to their rarity and uncertain molecular category.
Figure 1.
Evaluation of PTEN expression in melanoma by immunohistochemistry (IHC). (A) Representative image of a melanoma with complete loss of PTEN protein expression in the tumor cells. Endothelial cells (labeled “EC”) serve as an internal positive control and comparator. (B) Comparison of PTEN protein expression measurement by RPPA (on frozen tumor tissue) and by IHC (FFPE from the same surgical accessions). PTEN expression in the melanomas was categorized (x-axis) based on the observed results of IHC analysis. The average PTEN protein expression for each group of tumors as measured by RPPA is shown on the y-axis (arbitrary units). The RPPA results for each group were compared by un-paired t-tests, p-values are shown below the bar graph. Similar comparison of PTEN IHC results to levels of AKTThr308 and P-AKTSer473, each measured by RPPA, are show in panels (C) and (D), respectively. Melanomas with Clonal PTEN expression (by IHC) were not analyzed due to insufficient number of samples (n=2) to make meaningful conclusions.
Clinical associations of BRAFV600 and NRAS mutations in stage IIIB/C melanoma
An independent cohort of patients (n=136) with archival specimens available from standard of care lymphadenectomy surgery for clinically apparent regional lympadenopathy were identified for molecular analysis (Figure S2). Approximately two-thirds of the patients were male, and the median age at the time of surgery was 55.4 years (Table 1). Thirty five patients previously presented with stage I or II melanoma, and three patients were previously treated for stage IIIA disease, before presenting with clinically apparent regional lymphadenopathy. For the purpose of this study, the patients were categorized as stage IIIB or IIIC based on AJCC criteria at the time of surgery for their clinically apparent lymphadenopathy, along with the 98 patients who were stage IIIB/C at presentation. Thirty eight patients were classified as stage IIIB disease, and ninety eight as stage IIIC. Consistent with validated prognostic models, compared to patients with stage IIIB disease, patients with stage IIIC disease had shorter OS (median 1.9 versus 5.0 years, p=0.005) and DDFS (median 0.8 versus 1.6 years, p=0.057). Thirty two patients received adjuvant interferon, which did not correlate significantly with OS.
Table 1.
Demographics, Tumor Characteristics, and Clinical Outcomes in Stage IIIB and IIIC Patients.
| Measure | All Patients (N=136) | Stage IIIB (N=38) | Stage IIIC (N=98) | p-value |
|---|---|---|---|---|
| Gender, n (%) | ||||
| Male | 94 (69) | 25 (66) | 69 (70) | 0.680a |
| Female | 42 (31) | 13 (34) | 29 (30) | |
| Age at Stage III diagnosis (years), median (range) | 55.4 (24.9, 85.4) | 54.3 (26.1, 85.4) | 55.7 (24.9, 84.0) | 0.684b |
| Mutation, n (%) | ||||
| BRAF | 55 (40) | 15 (39) | 40 (41) | 1.000a |
| NRAS | 13 (10) | 4 (11) | 9 (9) | |
| Wild-type | 68 (50) | 19 (50) | 49 (50) | |
| PTEN, n (%) | ||||
| Absent | 31 (25) | 8 (24) | 23 (25) | 1.000a |
| Not Absent | 93 (75) | 25 (76) | 68 (75) | |
| Missing | 12 | 5 | 7 | |
| Time to any distant recurrence (years), median (95% CI) | 1.1 (0.8, 1.4) | 1.6 (1.0, 3.3) | 0.8 (0.6, 1.3) | 0.057c |
| Overall survival (years), median (95% CI) | 2.2 (1.9, 3.2) | 5.0 (2.3, 6.5) | 1.9 (1.5, 2.4) | 0.005c |
Fisher’s exact test.
Wilcoxon rank sum test.
Log-rank test.
In the full cohort, 40% of the patients had BRAFV600 mutations, 10% had NRAS mutations, and 50% had no hotspot mutation in either gene (“WT”). No patients had mutations in both BRAFV600 and NRAS. Over 90% (51/55) of the detected BRAF mutations resulted in BRAFV600E substitution, which is the most common change observed in melanoma. Mutation status was significantly associated with primary melanoma subtype (p=0.026). Relatively low rates of BRAFV600 mutations were detected in acral and mucosal melanomas compared to cutaneous melanomas (Table S1). The rate of BRAF mutations was very similar in the melanomas from known cutaneous primaries (n=94, 47.9%) and those with no known primary tumor (n=20, 40%). Mutation status was not significantly associated with the primary tumor Breslow thickness (p=0.42) or ulceration (p=0.26). Mutation status was also not significantly associated with pathological substage (IIIB/C) (p=1.00), DDFS (p=0.34), or OS (p=0.89) (Table S1).
Clinical associations of PTEN expression
PTEN IHC was performed on a tissue microarray (TMA) comprised of lymph node metastases from the 136 patients (Table 1) with stage IIIB or IIIC metastatic melanoma. All samples had tissue cores from 3 different regions of the tumor. Nine samples were not evaluable for PTEN expression due to a lack of viable tumor tissue in the TMA samples (7%) (Figure S2). Three samples (2%) demonstrated ‘clonal-like’ PTEN expression and were excluded from subsequent analyses due to their rarity and uncertain molecular category. Absent PTEN expression was observed in 31 (25%) of the 124 evaluable tumors. All evaluable cores for these 31 tumors demonstrated concordant results. The prevalence of PTEN Absent was similar in the stage IIIB (24.2%) and IIIC (25.0%) patients (Table 1).
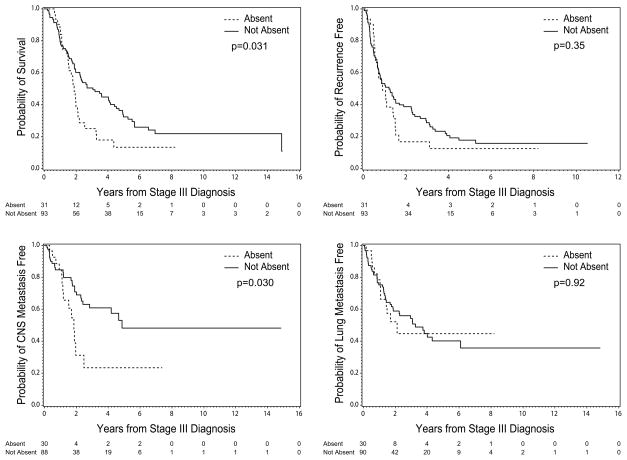
PTEN status did not correlate significantly with age, gender, primary tumor type, or primary tumor characteristics (Table 2). Patients with Absent PTEN survived a median of 1.9 years from stage IIIB/C diagnosis, which was significantly shorter than patients with PTEN Not Absent (median 3.1 years, p=0.03) (Figure 2A). However, PTEN status did not correlate with DDFS (PTEN Absent vs PTEN Not Absent, 0.9 years vs. 1.2 years, p=0.35) (Figure 2B), nor RFS (median 0.7 years for both, p=0.91).
Table 2.
Demographics, Tumor Characteristics, and Clinical Outcomes by PTEN Status.
| Measure | PTEN Absent (N=31) | PTEN Not Absent (N=93) | p-value |
|---|---|---|---|
| Gender, n (%) | |||
| Male | 22 (71) | 63 (68) | 0.826a |
| Female | 9 (29) | 30 (32) | |
| Age at Stage III diagnosis (years), median (range) | 48 (26.3, 82.4) | 58.1 (24.9, 85.4) | 0.073b |
| Clinical/Pathological Type, n (%) | |||
| Acral | 4 (13) | 12 (13) | 0.878a |
| Cutaneous | 20 (65) | 65 (70) | |
| Mucosal | 1 (3) | 3 (3) | |
| Unknown Primary | 6 (19) | 13 (14) | |
| Breslow thickness (mm), median (range) | 3.2 (0.4, 25.0) | 3.2 (0.3, 33.0) | 0.462b |
| Ulceration, n (%) | |||
| No | 6 (30) | 10 (15) | 0.188a |
| Yes | 14 (70) | 56 (85) | |
| Missing | 11 | 27 | |
| Time to any recurrence (years), median (95% CI) | 0.7 (0.5, 1.1) | 0.7 (0.5, 0.9) | 0.913c |
| Time to any distant recurrence (years), median (95% CI) | 0.9 (0.6, 1.4) | 1.2 (0.7, 1.7) | 0.352c |
| Overall survival (years), median (95% CI) | 1.9 (1.5, 2.1) | 3.1 (2.0, 4.1) | 0.031c |
| Time to CNS metastasis (years), median (95% CI) | 1.8 (1.2, 2.5) | 4.9 (2.4, NE) | 0.030c |
| Time to lung metastasis (years), median (95% CI) | 2.1 (1.1, NE) | 3.3 (1.9, 6.1) | 0.920c |
| Time to liver metastasis (years), median (95% CI) | NE (1.4, NE) | 6.3 (3.0, NE) | 0.522c |
| Time to bone metastasis (years), median (95% CI) | NE (1.4, NE) | NE (6.3, NE) | 0.102c |
Fisher’s exact test.
Wilcoxon rank sum test.
Log-rank test.
Abbreviations: CI = confidence interval; NE = not estimated.
Figure 2.
Clinical outcomes by PTEN status in stage IIIB/C melanoma patients (N=124). Kaplan-Meier analyses are shown for (A) OS, (B) DDFS, (C) CNS metastasis-free survival, and (D) lung metastasis-free survival.
The anatomic site(s) of distant metastasis are significantly associated with overall survival in stage IV melanoma patients and have been incorporated into AJCC melanoma staging system and prognostic models.(22) Our previous RPPA analysis identified decreased PTEN expression in melanoma brain metastases compared to lung and liver metastases(9). Since brain metastases are associated with poor outcomes, we determined the time to brain, lung, liver, and bone metastasis for all patients. Patients with PTEN Absent developed melanoma brain metastases (MBM) at a median of 1.8 years after stage IIIB/C diagnosis, which was significantly shorter than for patients with PTEN Not Absent (median 4.9 years, p=0.03) (Figure 2C). In contrast, there was no significant difference in the median time to lung (p=0.92) (Figure 2D), liver (p=0.52), or bone (p=0.10) metastasis (Table 2).
Multivariate analysis of OS incorporating AJCC substage (IIIB/C), PTEN (Absent/Not Absent), age, gender, and ipilimumab (Received/Not Received) was performed on all patients (Table S2). Both increased stage (HR 1.8, p=0.016) and PTEN Absent (HR1.6, p=0.046) were significant predictors of OS. No factors were significantly (p<0.05) associated with time to brain metastasis on multivariate analysis (Table S2).
Clinical associations and significance of PTEN in BRAFV600-mutant and BRAF/NRAS-Wild Type melanomas
PTEN mutations are generally mutually exclusive with the presence of hotspot NRAS mutations; however, PTEN loss is observed BRAFV600-mutant and BRAF/NRAS-WT melanomas.(3) In this study, complete loss of PTEN expression was observed in 8% (1 of 13) of NRAS, 31% (16 of 52) of BRAFV600, and 24% (14 of 59) of WT tumors.
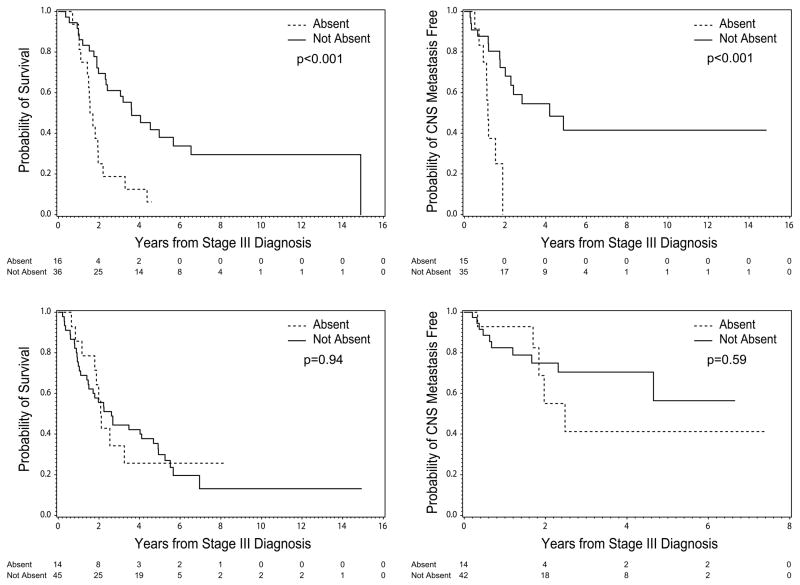
Among the patients with BRAFV600 mutations, PTEN status was not significantly associated with stage (p=0.90), or with treatment (at any time) with either ipilimumab (p=0.98) or a selective BRAF inhibitor (p=0.29). There was also no significant difference between PTEN Absent and PTEN Not Absent patients in RFS (median 0.7 years in both groups, p=0.91) or DDFS (median 0.7 vs, 0.9 years, p=0.10) (Table S3). However, patients with BRAFV600 mutations with Absent PTEN had significantly shorter OS than the patients with PTEN Not Absent (median 1.6 versus 3.6 years, p<0.001) (Figure 3A). BRAFV600 melanoma patients with PTEN Absent also had significantly shorter time to melanoma brain metastasis (1.2 vs. 4.2 years, p<0.001) (Figure 3B) and liver metastasis (1.4 years vs, not estimable p=0.046), and trends for shorter time to lung (1.4 vs. 4.0 years, p= 0.08), and bone (3.0 years vs. not estimable p=0.14) metastasis. In contrast, in patients without activating mutations in BRAFV600 or NRAS, PTEN was not significantly associated with OS (Figure 3C), DDFS, or time to brain metastasis (Figure 3D) (Table S4). The differences in OS and and time to MBM by PTEN status in the patients with BRAFV600 mutations remained significant after adjusting for multiple comparisons (adjusted p=0.02 for both).(24)
Figure 3.
Clinical outcomes by PTEN status in stage IIIB/C melanoma patients with BRAFV600 mutations (A, B) and in patients without BRAFV600 or NRAS mutations (C, D). Kaplan-Meier analysis are shown for (A, C) OS and (B, D) CNS metastasis-free survival.
Multivariate analysis of patients with BRAFV600 mutations incorporating stage (IIIB/C), PTEN (Absent/Not Absent), age, gender, subsequent ipilimumab (Received/Not Received), and subsequent BRAF inhibitor treatment identified PTEN Absent (HR 2.7, p=0.008) as the only factor significantly associated with OS (Table 3). Multivariate analysis using the same factors also identified PTEN Absent (HR 4.2, p=0.008) as the only significant predictor of time to brain metastasis (Table 3). After adjusting for multiple comparisons the p-value for both OS and time to MBM by PTEN status was 0.07.
Table 3.
Multivariable analysis of Overall Survival and Time to CNS metastasis in Patients with BRAFV600 Mutations
| Overall Survival | Time to CNS Metastasis | |||
|---|---|---|---|---|
| Measure | Hazard Ratios (95% CI) | p-value | Hazard Ratios (95% CI) | p-value |
| Stage III status (IIIC v IIIB) | 2 (0.9, 4.2) | 0.09 | 1.4 (0.5, 3.7) | 0.503 |
| PTEN (Absent v Not Absent) | 2.7 (1.3, 5.5) | 0.008 | 4.2 (1.5, 12.0) | 0.008 |
| BRAF Inhibitor use (No v Yes) | 3 (0.4, 23.3) | 0.285 | 3 (0.4, 23.8) | 0.299 |
| Ipilimumab use (No v Yes) | 1 (0.3, 3.0) | 0.976 | 1.2 (0.4, 4.2) | 0.741 |
| Age | 1 (1.0, 1.0) | 0.549 | 1 (1.0, 1.0) | 0.98 |
| Gender (Male v Female) | 1.5 (0.7, 3.8) | 0.293 | 2.1 (0.7, 6.2) | 0.179 |
Discussion
Activating BRAFV600 and NRAS mutations, and loss of expression of the tumor suppressor PTEN, are three of the most common oncogenic events in melanoma. Multiple clinical trials are planned or ongoing to evaluate the benefit of targeting these molecules and their associated pathways. In addition to providing benefit as therapeutic targets, molecular markers can enhance patient management by improving prognostic models. This study represents the first integrated analysis of BRAF, NRAS and PTEN with clinical outcomes in stage III melanoma patients. The results of this analysis add to existing data from preclinical models and clinical trials implicating loss of PTEN as a clinically significant event in this disease, specifically in tumors with BRAFV600 mutations.
PTEN was discovered in the minimal region of loss of chromosome 10 that occurs frequently in glioblastoma multiforme (GBM).(25, 26) Subsequent studies identified loss of function deletions and mutations of the PTEN gene in multiple tumor types, including melanoma.(27) PTEN is a lipid phosphatase that dephosphorylates the 3′-position of the inositol ring of phosphatidylinositols. Thus, PTEN is a negative regulator of PI3K, and loss of PTEN increases signaling through the PI3K-AKT signaling pathway, which is commonly assessed by measuring levels of phosphorylated (activated) AKT.
Our previous RPPA-based proteomic analysis of frozen metastatic melanoma tissue samples demonstrated that PTEN protein levels were significantly associated and inversely correlated with levels of multiple activation-specific markers in the PI3K-AKT pathway.(9) PTEN loss was detected in BRAFV600-mutant and in BRAFV600/NRAS Wild-Type tumors. While RPPA is a powerful technology, its use is predominantly limited to frozen tumor samples. In contrast, in the current study we used an IHC assay performed in a Clinical Laboratory Improvement Amendments (CLIA)-certified lab to measure PTEN expression in FFPE tissues, which are widely available. One challenge with IHC assays is the determination of rational and reproducible scoring criteria. Our comparison of IHC and RPPA results demonstrates a strong correlation between PTEN measurements by these two methods, supporting their technical validity. In addition, the finding that only the PTEN Absent IHC pattern correlated with AKT activation provides a molecular rationale for a dichotomous scoring system. Assessment of the interobserver agreement for PTEN IHC scoring of the pilot set of FFPE samples (n=94) by two dermatopathologists using the kappa statistic identified a kappa coefficient of 0.73. Thus the dichotomous scoring system also has the benefit of good reproducibility compared to more complex criteria. Previous studies in other tumor types also support the rationale to perform protein-based measurement of PTEN, as PTEN expression can regulated epigenetically.(11, 12, 28) It will be important in future studies to integrate and compare PTEN protein, mRNA, and genetic status with each other and with clinical outcomes.
In this study, complete loss of PTEN expression correlated with OS in stage IIIB/C patients with BRAFV600 mutations, but not in patients with wild-type BRAFV600 and NRAS. Notably, the prognostic significance of PTEN appears to due to shorter survival after the diagnosis of stage IV disease, and PTEN status did not correlate significantly with time to first distant metastasis. The interaction between PTEN and BRAF in melanoma is also supported by functional preclinical studies. In genetically engineered mice the expression of the BRAFV600 mutation in melanocytes resulted in hyperplasia only. However, concurrent loss of PTEN in the melanocytes with BRAFV600 mutations resulted in 100% penetrance of invasive, metastatic melanomas.(29) Loss of PTEN in murine melanocytes without BRAFV600 mutations caused no melanocytic phenotype in that study. Similarly, loss of PTEN was not significantly associated with OS in the stage IIIB/C patients with BRAFV600/NRAS Wild-Type melanomas in our cohort. We and other groups have also observed that human melanoma cell lines with BRAFV600 mutations and loss of PTEN expression are resistant to apoptosis following BRAF inhibition compared to cell lines with normal PTEN.(30–32) Analysis of patients enrolled in early phase clinical trials with the FDA-approved BRAF inhibitor dabrafenib found that patients with genetic alterations in the PTEN gene had a strong trend for shorter PFS compared to patients with a normal PTEN.(33) Lower PTEN expression was also observed in non-responders compared to responders in the phase II clinical trial of vemurafenib.(34) Interestingly, recent data also suggests that PTEN loss can also promote resistance to the anti-tumor immune response.(35) Together, these findings support the rationale to evaluate PTEN status in ongoing and planned clinical trials of targeted and immune therapies in patients with stage III melanoma.
Brain metastases are one of the most common and devastating complications of melanoma. In this study, we observed that complete loss of PTEN expression correlated with markedly shorter time to brain metastasis among patients with BRAFV600 mutations and stage IIIB/C melanoma. As melanoma patients with CNS involvement have historically had very poor outcomes,(36, 37) this association is likely primarily responsible for the observed shorter survival in patients with PTEN loss in this study. This finding also adds to a growing body of literature implicating the PI3K-AKT pathway in brain metastasis in melanoma. Previously, in a pilot RPPA analysis of a small number of frozen distant metastases, we found significantly lower levels of PTEN, and higher levels of P-AKT, in melanoma brain metastases (n=10) compared to lung (n=5) or liver (n=5) metastases.(9) An independent IHC analysis of the expression of P-AKT and P-MAPK protein levels in tumors from patients who underwent synchronous resection of brain and extracranial metastases reported higher P-AKT in the brain metastases of 8 of 9 patients.(38) We have recently also reported the RPPA analysis of an independent set of cohort of patients with resected brain and extracranial metastases.(39) In this study, P-AKT had the highest ratio of expression in brain metastases compared to extracranial metastases (p=0.008 by paired t-test). Preliminary studies have also shown that activated forms of AKT can promote melanoma brain metastasis formation in preclinical models.(40) Together these finding suggest that the PI3K-AKT pathway may be a rational, actionable therapeutic target for melanoma brain metastases.
In this cohort of patients with stage IIIB/C melanoma we did not detect a significant association between BRAF/NRAS mutation status and time to first distant metastasis or overall survival. The low prevalence of NRAS mutations limited our power to detect significant clinical associations with this molecular event. The rate of BRAFV600K mutations (n=4) was also too low to make meaningful comparisons to BRAFV600E mutations, which have demonstrated significant differences in larger cohorts of stage IV patients.(41, 42) Notably, retrospective analyses of melanoma patients with regional disease have reported varying conclusions about the prognostic significance of BRAF and NRAS mutations.(43–45) The varying results support the need for multicenter prospective collection and integrated analysis of clinical, histological, and molecular data to further refine risk models for, and ultimately to improve the management of, melanoma patients with regionally advanced disease. Based on the results of our study, we believe that such studies should not be restricted to mutational analysis alone.
In summary, this study represents the first integrated analysis of BRAFV600 mutations, NRAS mutations, and PTEN loss in patients with stage IIIB/C melanoma. Our findings implicate PTEN loss, which was detected by a routine IHC assay, as a significant predictor of overall survival and time to brain metastasis in patients with BRAFV600 mutations. These findings support the rationale for evaluation of PTEN protein expression in additional cohorts of patients, and the significance of the PI3K-AKT pathway in this disease.
Supplementary Material
Statement of Translational Relevance.
Multiple clinical trials with new agents are now underway or planned for patients with regional metastases from melanoma. The appropriate design and interpretation of these and future investigations will be strengthened developing an improved understanding of the clinical characteristics and outcomes associated with molecular aberrations in this disease. This integrated analysis of BRAF and NRAS mutations, PTEN loss, and clinical outcomes in patients with stage IIIB/C disease adds to growing evidence that melanomas with loss of PTEN represent a clinically significant subpopulation. PTEN loss was associated with shorter time to brain metastasis and overall survival specifically in melanomas with concurrent BRAFV600mutations, supporting functional interactions observed preclinically between these oncogenic events. The novel association of PTEN loss with shorter time to brain metastasis also supports the rationale to further examine the role of the PI3K-AKT pathway in this common and devastating complication of advanced melanoma.
Acknowledgments
Financial Support: Supported by NIH/NCI 1R01CA154710-01, Melanoma Research Alliance Young Investigator Award, MD Anderson Goodfellow Scholar Award, P50 CA093459-06, and the MD Anderson Melanoma Moon Shot Program
Footnotes
Disclosures: M.A.D. has served as a consultant for GlaxoSmithKline, Genentech, Novartis, and Sanofi-Aventis, and has received research funding from GlaxoSmithKline, Genentech, AstraZeneca, Oncothyreon, and Myriad Genetics. Data previously presented in part at European Cancer Congress, 2013, and the American Association for Cancer Research Annual Meeting, 2012
References
- 1.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 2.Hocker T, Tsao H. Ultraviolet radiation and melanoma: a systematic review and analysis of reported sequence variants. Hum Mutat. 2007;28:578–88. doi: 10.1002/humu.20481. [DOI] [PubMed] [Google Scholar]
- 3.Goel VK, Lazar AJ, Warneke CL, Redston MS, Haluska FG. Examination of mutations in BRAF, NRAS, and PTEN in primary cutaneous melanoma. J Invest Dermatol. 2006;126:154–60. doi: 10.1038/sj.jid.5700026. [DOI] [PubMed] [Google Scholar]
- 4.Tsao H, Goel V, Wu H, Yang G, Haluska FG. Genetic Interaction Between NRAS and BRAF Mutations and PTEN//MMAC1 Inactivation in Melanoma. J Investig Dermatol. 2004;122:337–41. doi: 10.1046/j.0022-202X.2004.22243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hauschild A, Grob JJ, Demidov LV, Jouary T, Gutzmer R, Millward M, et al. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012 doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- 7.Flaherty KT, Robert C, Hersey P, Nathan P, Garbe C, Milhem M, et al. Improved Survival with MEK Inhibition in BRAF-Mutated Melanoma. New England Journal of Medicine. 2012;367:107–14. doi: 10.1056/NEJMoa1203421. [DOI] [PubMed] [Google Scholar]
- 8.Ascierto PA, Schadendorf D, Berking C, Agarwala SS, van Herpen CML, Queirolo P, et al. MEK162 for patients with advanced melanoma harbouring NRAS or Val600 BRAF mutations: a non-randomised, open-label phase 2 study. The Lancet Oncology. 2013;14:249–56. doi: 10.1016/S1470-2045(13)70024-X. [DOI] [PubMed] [Google Scholar]
- 9.Davies MA, Stemke-Hale K, Lin E, Tellez C, Deng W, Gopal YN, et al. Integrated Molecular and Clinical Analysis of AKT Activation in Metastatic Melanoma. Clin Cancer Res. 2009;15:7538–46. doi: 10.1158/1078-0432.CCR-09-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aguissa-Touré A-H, Li G. Genetic alterations of PTEN in human melanoma. Cellular and Molecular Life Sciences. 2011:1–17. doi: 10.1007/s00018-011-0878-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mirmohammadsadegh A, Marini A, Nambiar S, Hassan M, Tannapfel A, Ruzicka T, et al. Epigenetic silencing of the PTEN gene in melanoma. Cancer Res. 2006;66:6546–52. doi: 10.1158/0008-5472.CAN-06-0384. [DOI] [PubMed] [Google Scholar]
- 12.Zhou X-P, Gimm O, Hampel H, Niemann T, Walker MJ, Eng C. Epigenetic PTEN Silencing in Malignant Melanomas without PTEN Mutation. Am J Pathol. 2000;157:1123–8. doi: 10.1016/S0002-9440(10)64627-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vasudevan KM, Barbie DA, Davies MA, Rabinovsky R, McNear CJ, Kim JJ, et al. AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer Cell. 2009;16:21–32. doi: 10.1016/j.ccr.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwong LN, Davies MA. Navigating the Therapeutic Complexity of PI3K Pathway Inhibition in Melanoma. Clinical Cancer Research. 2013;19:5310–9. doi: 10.1158/1078-0432.CCR-13-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akslen LA, Angelini S, Straume O, Bachmann IM, Molven A, Hemminki K, et al. BRAF and NRAS mutations are frequent in nodular melanoma but are not associated with tumor cell proliferation or patient survival. J Invest Dermatol. 2005;125:312–7. doi: 10.1111/j.0022-202X.2005.23788.x. [DOI] [PubMed] [Google Scholar]
- 16.Edlundh-Rose E, Egyhazi S, Omholt K, Mansson-Brahme E, Platz A, Hansson J, et al. NRAS and BRAF mutations in melanoma tumours in relation to clinical characteristics: a study based on mutation screening by pyrosequencing. Melanoma Res. 2006;16:471–8. doi: 10.1097/01.cmr.0000232300.22032.86. [DOI] [PubMed] [Google Scholar]
- 17.Bauer J, Buttner P, Murali R, Okamoto I, Kolaitis NA, Landi MT, et al. BRAF mutations in cutaneous melanoma are independently associated with age, anatomic site of the primary tumor, and the degree of solar elastosis at the primary tumor site. Pigment Cell Melanoma Res. 2011;24:345–51. doi: 10.1111/j.1755-148X.2011.00837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Devitt B, Liu W, Salemi R, Wolfe R, Kelly J, Tzen C-Y, et al. Clinical outcome and pathological features associated with NRAS mutation in cutaneous melanoma. Pigment Cell Melanoma Res. 2011;24:666–72. doi: 10.1111/j.1755-148X.2011.00873.x. [DOI] [PubMed] [Google Scholar]
- 19.Ellerhorst JA, Greene VR, Ekmekcioglu S, Warneke CL, Johnson MM, Cooke CP, et al. Clinical correlates of NRAS and BRAF mutations in primary human melanoma. Clin Cancer Res. 2011;17:229–35. doi: 10.1158/1078-0432.CCR-10-2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jakob JA, Bassett RL, Ng CS, Curry J, Joseph R, Alvarado G, et al. NRAS mutation status is an independent prognostic factor in metastatic melanoma. Cancer. 2012;118:4014–23. doi: 10.1002/cncr.26724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Long GV, Menzies AM, Nagrial AM, Haydu LE, Hamilton AL, Mann GJ, et al. Prognostic and Clinicopathologic Associations of Oncogenic BRAF in Metastatic Melanoma. Journal of Clinical Oncology. 2011;29:1239–46. doi: 10.1200/JCO.2010.32.4327. [DOI] [PubMed] [Google Scholar]
- 22.Balch CM, Gershenwald JE, Soong SJ, Thompson JF, Atkins MB, Byrd DR, et al. Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol. 2009;27:6199–206. doi: 10.1200/JCO.2009.23.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al-Zaid T, Ditelberg JS, Prieto VG, Lev D, Luthra R, Davies MA, et al. Trichilemmomas show loss of PTEN in Cowden syndrome but only rarely in sporadic tumors. J Cutan Pathol. 2012;39:493–9. doi: 10.1111/j.1600-0560.2012.01888.x. [DOI] [PubMed] [Google Scholar]
- 24.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Statistical Society. 1995;57:289–300. [Google Scholar]
- 25.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–7. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 26.Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15:356–62. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 27.Teng DH, Hu R, Lin H, Davis T, Iliev D, Frye C, et al. MMAC1/PTEN mutations in primary tumor specimens and tumor cell lines. Cancer Res. 1997;57:5221–5. [PubMed] [Google Scholar]
- 28.Djordjevic B, Hennessy BT, Li J, Barkoh BA, Luthra R, Mills GB, et al. Clinical assessment of PTEN loss in endometrial carcinoma: immunohistochemistry outperforms gene sequencing. Mod Pathol. 2012;25:699–708. doi: 10.1038/modpathol.2011.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dankort D, Curley DP, Cartlidge RA, Nelson B, Karnezis AN, Damsky WE, Jr, et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet. 2009;41:544–52. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deng W, Yennu-Nanda VG, Scott A, Chen G, Woodman SE, Davies MA. Role and Therapeutic Potential of PI3K-mTOR Signaling in De Novo Resistance to BRAF Inhibition. Pigment Cell & Melanoma Research. 2012;25:248–58. doi: 10.1111/j.1755-148X.2011.00950.x. [DOI] [PubMed] [Google Scholar]
- 31.Paraiso KH, Xiang Y, Rebecca VW, Abel EV, Chen A, Munko AC, et al. PTEN loss confers BRAF inhibitor resistance to melanoma cells through the suppression of BIM expression. Cancer Res. 2011;71:2750–60. doi: 10.1158/0008-5472.CAN-10-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xing F, Persaud Y, Pratilas CA, Taylor BS, Janakiraman M, She QB, et al. Concurrent loss of the PTEN and RB1 tumor suppressors attenuates RAF dependence in melanomas harboring (V600E)BRAF. Oncogene. 2012;31:248–58. doi: 10.1038/onc.2011.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nathanson KL, Martin A-M, Wubbenhorst B, Greshock J, Letrero R, D’Andrea K, et al. Tumor genetic analyses of patients with metastatic melanoma treated with the BRAF inhibitor Dabrafenib (GSK2118436) Clinical Cancer Research. 2013;19:4868–78. doi: 10.1158/1078-0432.CCR-13-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trunzer K, Pavlick AC, Schuchter L, Gonzalez R, McArthur GA, Hutson TE, et al. Pharmacodynamic Effects and Mechanisms of Resistance to Vemurafenib in Patients With Metastatic Melanoma. Journal of Clinical Oncology. 2013;31:1767–74. doi: 10.1200/JCO.2012.44.7888. [DOI] [PubMed] [Google Scholar]
- 35.Dong Y, Richards JA, Gupta R, Aung PP, Emley A, Kluger Y, et al. PTEN functions as a melanoma tumor suppressor by promoting host immune response. Oncogene. 2013 doi: 10.1038/onc.2013.409. [DOI] [PubMed] [Google Scholar]
- 36.Staudt M, Lasithiotakis K, Leiter U, Meier F, Eigentler T, Bamberg M, et al. Determinants of survival in patients with brain metastases from cutaneous melanoma. Br J Cancer. 2010;102:1213–8. doi: 10.1038/sj.bjc.6605622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davies MA, Liu P, McIntyre S, Kim KB, Papadopoulos N, Hwu WJ, et al. Prognostic factors for survival in melanoma patients with brain metastases. Cancer. 2011;117:1687–96. doi: 10.1002/cncr.25634. [DOI] [PubMed] [Google Scholar]
- 38.Niessner H, Forschner A, Klumpp B, Honegger JB, Witte M, Bornemann A, et al. Targeting hyperactivation of the AKT survival pathway to overcome therapy resistance of melanoma brain metastases. Cancer Medicine. 2013;2:76–85. doi: 10.1002/cam4.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen G, Chakravarti N, Aardalen K, Lazar AJ, Tetzlaff M, Wubberhorst B, et al. Molecular Profiling of Patient-Matched Brain and Extracranial Melanoma Metastases Implicates the PI3K Pathway as a Therapeutic Target. Clinical Cancer Research. 2014 doi: 10.1158/1078-0432.CCR-13-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.VanBrocklin MW, Robinson JP, Holmen S. Differential metastatic potential of activated AKT isoforms in a mouse model of melanoma. Pigment Cell & Melanoma Research. 2013;26:960. [Google Scholar]
- 41.Bucheit AD, Syklawer E, Jakob JA, Bassett RL, Jr, Curry JL, Gershenwald JE, et al. Clinical characteristics and outcomes with specific BRAF and NRAS mutations in patients with metastatic melanoma. Cancer. 2013;119:3821–9. doi: 10.1002/cncr.28306. [DOI] [PubMed] [Google Scholar]
- 42.Menzies AM, Haydu LE, Visintin L, Carlino MS, Howle JR, Thompson JF, et al. Distinguishing Clinicopathologic Features of Patients with V600E and V600K BRAF-Mutant Metastatic Melanoma. Clinical Cancer Research. 2012;18:3242–9. doi: 10.1158/1078-0432.CCR-12-0052. [DOI] [PubMed] [Google Scholar]
- 43.Moreau S, Saiag P, Aegerter P, Bosset D, Longvert C, Helias-Rodzewicz Z, et al. Prognostic value of BRAF V600 mutations in melanoma patients after resection of metastatic lymph nodes. Ann Surg Oncol. 2012;19:4314–21. doi: 10.1245/s10434-012-2457-5. [DOI] [PubMed] [Google Scholar]
- 44.Mann GJ, Pupo GM, Campain AE, Carter CD, Schramm SJ, Pianova S, et al. BRAF mutation, NRAS mutation, and the absence of an immune-related expressed gene profile predict poor outcome in patients with stage III melanoma. J Invest Dermatol. 2013;133:509–17. doi: 10.1038/jid.2012.283. [DOI] [PubMed] [Google Scholar]
- 45.Rutkowski P, Jurkowska M, Gos A, Tysarowski A, Michej W, Switaj W, et al. Correlations and molecular alterations in clinical stage III cutaneous melanoma with clinical-pathological features and patient outcomes. J Clin Oncol. 2012;30:8548. doi: 10.3892/ol.2014.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.