Abstract
Assembly of the β-barrel outer membrane proteins (OMPs) is an essential cellular process in Gram negative bacteria and in the mitochondria and chloroplasts of eukaryotes—two organelles of bacterial origin. Central to this process is the conserved β-barrel OMP that belongs to the Omp85 superfamily. In Escherichia coli, BamA is the core β-barrel OMP, and together with four outer membrane lipoproteins, BamBCDE, constitute the β-barrel assembly machine (BAM). In this paper, we investigated the roles of BamD, an essential lipoprotein, and BamB in BamA biogenesis. Depletion of BamD caused impairment in BamA biogenesis and cessation of cell growth. These defects of BamD depletion were partly reversed by single amino acid substitutions mapping within the β-barrel domain of BamA. However, in the absence of BamB, the positive effects of the β-barrel substitutions on BamA biogenesis under BamD depletion conditions were nullified. By employing a BamA protein bearing one such substitution, F494L, it was demonstrated that the mutant BamA protein could not only assemble without BamD, but it could also facilitate the assembly of wild-type BamA expressed in trans. Based on these data, we propose a model in which the Bam lipoproteins, which are localized to the outer membrane by the BAM-independent Lol pathway, aid in the creation of new BAM complexes by serving as outer membrane receptors and folding factors for nascent BamA molecules. The newly assembled BAM holocomplex then catalyzes the assembly of substrate OMPs and BamA. These in vivo findings are corroborated by recently published in vitro data.
Keywords: β-barrel outer membrane protein assembly, BAM complex, genetics
Introduction
The two major classes of Gram negative outer membrane proteins (OMPs) are lipoproteins and β-barrels [1]. The localization of lipoprotein (Lol) pathway targets mature, N-terminal lipid-modified, proteins from the periplasmic side of the inner membrane to the periplasmic side of the outer membrane [2]. The Lol pathway is essential because it is involved in the biogenesis of cellular machineries that catalyze the assembly of β-barrel OMPs and lipopolysaccharide (LPS) in the outer membrane [36].
The Escherichia coli β-barrel assembly machine (BAM) contains four outer membrane lipoproteins, BamBCDE, and a β-barrel OMP, BamA (for recent reviews, see [5-7]; BamD and BamA are the two indispensable components of the BAM complex [8-11]. BamA belongs to the Omp85 superfamily of proteins that mediate protein transport/assembly functions [12-14]. In contrast, the BamBCDE homologs are found only in Gram negative bacteria [15,16].
Both in vivo and in vitro studies have shown that the BAM holocomplex can exist as two major subcomplexes, BamAB and BamACDE [11,17,18]. These two subcomplexes carry out partially overlapping functions [15,19]. Of the three non-essential lipoproteins, the absence of BamB produces the most significant OMP biogenesis defect [17,20-22]. The defect of ΔbamB is further exacerbated when BamC or BamE is also simultaneously absent [17], with the ΔbamB ΔbamE mutant displaying the most drastic growth and OMP biogenesis defects [15].
The high-resolution structures of all Bam proteins have been solved and they have begun to provide mechanistic insights into the BAM activity. All five proteins contain motifs known to facilitate protein-protein interactions. The N-terminal soluble domain of BamA is exposed to the periplasm, while its C-terminal domain is embedded in the outer membrane [12-14]. The soluble N-terminal end of BamA folds into five polypeptide-transport-associated (POTRA) domains [23] whose structure revealed possible interactions with substrate polypeptides via β-augmentation [24-26]. Mutational and pull-down assays have also revealed that the POTRA domains of BamA are critical for interactions with the Bam lipoproteins [24] and a periplasmic chaperone, SurA [27,28]. Thus, the POTRA domains appear to serve as the initial staging ground for substrate interaction and folding. The C-terminal β-barrel domain of BamA is thought to catalyze the final steps of OMP assembly [29]. An important feature of this domain is a large loop, L6, which contains the ‘VRGF/Y’ motif conserved in all members of the Omp85 superfamily [13,14,30]. The functional importance of the BamA L6 region and its conserved VRGF motif was first determined through the genetic analysis [15] and subsequently by site-directed mutagenesis [29,31]. Exactly how the L6 region participates in the β-barrel OMP assembly remains to be elucidated.
Three-dimensional structures of BamB [32-34], BamD [35-37], BamCD [38], and BamE [35,39] have been solved. However, the roles of these lipoproteins in OMP assembly remained largely speculative. It has been hypothesizes that BamB may channel substrate OMPs to BamA [32], but this has not been supported experimentally [34]. The structural and mutational data suggest that the N-terminus of BamD might interact with substrate OMPs [11,35]. Interestingly, the same region of BamD is also shown to interact with BamC [38], thus prompting a suggestion that BamC may play a role in regulating BamD activity. BamE has been proposed to interact with the cell envelope components to stabilize and/or regulate the activity of the BAM complex. A recent study suggested a role for BamE in modulating BamA's conformation during its functional cycle [40].
In a recent in vitro study, Hagan et al. [19] showed that BamB and BamD can bind to unfolded BamA and facilitate its assembly and insertion into proteoliposomes. Their study also showed that BamCDE-mediated assembly of unfolded BamA does not require preassembled BamA in proteoliposomes, although the presence of BamA in the BamCDE complex greatly enhanced assembly of the unfolded BamA substrate. These results reinforce a previous suggestion that BamA aids in its own assembly [24,27,31]. It is unclear whether the nascent OMPs enter the BamA channel [41] or stay outside the BamA barrel at the protein-lipid interface. An attractive model outlined by Kim et al. [42] and Noinaj et al. [29] envisions that the genesis of a substrate β-barrel begins with its β-strands aligning with the first and last β-strands of the transiently open BamA barrel, and culminating in the substrate β-sheet budding off into the lipid phase of the membrane to form an enclosed β-barrel.
In this paper, we investigated the in vivo roles of the BamD and BamB lipoproteins in BamA biogenesis, and determined the effects of BamA β-barrel substitutions that were previously shown to improve BamA biogenesis in a ΔbamB ΔbamE double mutant background [15]. Because BamA itself has a β-barrel domain, it is logical to consider that the pre-assembled BAM complex would also catalyze the assembly of nascent BamA molecules. Consequently, a defective BAM complex should affect the assembly of all β-barrel OMPs, including BamA [27,31,43]. We show here that BamA misfolds when BamD is depleted from the cell and this defect is reversed by the β-barrel substitutions in BamA, but only in the presence of BamB. Moreover, the mutant BamA, which assembles without BamD, in turn facilitates the assembly of wild-ype BamA expressed in trans under acute BamD depletion conditions. These data reveal that BamD and BamB play crucial roles in BamA folding and biogenesis in vivo and give physiological credence to the recent in vitro study by Hagan et al. [19].
Results
bamA suppressor alleles allow growth under BamD depletion conditions
We have previously reported the isolation of six novel bamA suppressor alleles from a strain simultaneously lacking the two non-essential lipoproteins of the BAM complex, BamB and BamE, [15]. The double mutant, expressing wild-type BamA (BamAWT), failed to grow on rich medium, grew poorly on minimal medium, and displayed a severe OMP biogenesis defect [15]. Moreover, the data revealed that without BamB and BamE, BamA could not fold correctly and was present in reduced levels presumably due to proteolysis of misfolded BamA. These data suggested a role for the Bam lipoproteins in BamA biogenesis. The presence of the suppressor mutations in bamA in a ΔbamB ΔbamE background improved bacterial growth, OMP biogenesis, and restored BamA levels and folding to various degrees [15]. Since all six suppressor alterations map in the BamA β-barrel domain, it stands to reason that BamB and BamE influence folding of the BamA β-barrel domain, and thus the OMP biogenesis defect stems from defects caused by misfolded BamA and the loss of BamB and BamE activities.
BamB and BamE form two BAM subcomplexes, BamAB and BamACDE [11,17,18]. Whereas BamB interacts with BamA directly [8,22], BamE does so indirectly via BamD [17]. Since BamD is the essential lipoprotein and directly interacts with BamA [11], we reasoned that it is likely to play a more critical role in BamA biogenesis than the non-essential BamB or BamE protein individually. Therefore, we investigated BamA biogenesis under BamD depletion conditions and tested the effects of the six bamA suppressor alleles on bacterial growth, BamA levels and folding, and general OMP biogenesis.
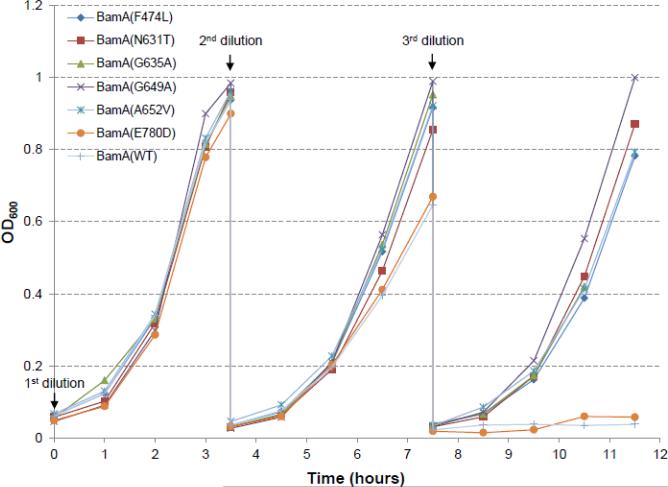
The parental E. coli strain used in this study had its chromosomal bamD deleted and expressed bamD from an arabinose-inducible allele inserted at the lambda attachment site [11]. Consequently, growth of the parental strain is dependent on arabinose but is inhibited by glucose, which lowers the level of cAMP required for the activity of the araBAD promoter [11]. However, despite this arabinose dependence, the parental strain can grow normally for several generations in a minimal medium containing glycerol, where BamD levels are depleted at a much slower rate than that in the minimal medium supplemented with glucose. Therefore, bacterial cultures were first grown overnight in glycerol minimal medium at 37°C to initiate depletion of BamD, followed by growth next day in glucose medium to completely deplete BamD. The cell densities of all overnight grown cultures were nearly identical. Next day, cultures were diluted 1:100 in glucose minimal medium and grown at 37°C. After 3.5 hours, when OD600 reached around 1.0 (Fig. 1), cultures were diluted 1:50 in glucose minimal medium and incubated at 37°C for 4 hours. At the end of this incubation period, cultures were diluted 1:50 one final time in glucose minimal medium and grown for 4 hours. After the third (final) dilution, the two cultures that failed to grow expressed either BamAWT or BamAE780D (Fig. 1). In contrast, the remaining cultures expressing five different BamA suppressor variants grew at rates very similar to that seen after the second dilution. Interestingly, BamAE780D was also the weakest among the six BamA suppressors in terms of improving the growth of the ΔbamB ΔbamE mutant [15]. Growth after the fourth dilution was sporadic with some cultures occasionally growing weakly (data not shown). It is worth mentioning that BamD levels under these growth conditions became undetectable at the end of the first dilution in glucose minimal medium (e.g. see Fig. 3). These data show that the expression of the BamA suppressor variants, except that bearing an E780D substitution, afforded bacterial growth even under acute BamD depletion conditions. Note that BamA residue numbers are relative to the mature BamA sequence.
Fig 1.
Effects of BamD depletion on cell growth. Overnight grown cultures in glycerol minimal medium were diluted 1:100 in glucose minimal medium and grown for 3.5 hours. At the end of this period, cultures were diluted 1:50 in the same medium and grown for 4 hours, and then diluted 1:50 one last time in fresh glucose minimal medium and grown for another 4 hours. BamD expression is repressed in glucose minimal medium. Cell densities were measured in a spectrophotometer at 600 nm. All cultures were grown at 37°C in baffled flasks in a water bath incubator shaker set at 180 rpm. Labeled are cells expressing different BamA proteins. Residue numbers are relative to the mature BamA protein sequence.
Fig. 3.
Analyses of OMP levels and BamA folding from BamD-depleted cells. Envelopes were purified from cultures grown in glucose- or arabinose-supplemented glycerol minimal medium to repress or induce bamD expression, respectively. (a) BamA folding status was determined from envelopes obtained from cultures after first and second dilutions. Prior to SDS-PAGE analysis, SDS-sample buffer-solubilized samples were either left at room temperature (25°C) or heated (100°C). Detection of BamA was carried out by Western blot analysis using BamA-specific antibodies. Electrophoresis was carried out at room temperature. BamAU and BamAF refer to unfolded and folded forms of BamA, respectively. (b) SDS(urea)-PAGE analysis of purified envelopes. Protein bands were visualized after Coomassie blue staining. Positions of the three major OMPs are shown. (c) Western blot analysis to detect BamD. The Coomassie blue-stained gel in (b) was electro-blotted and the membrane blot was probed with BamD-specific antibodies. (d) Western blot analysis to detect OMPs from BamD-depleted envelope samples.
Suppressors stabilize BamA under BamD depletion conditions
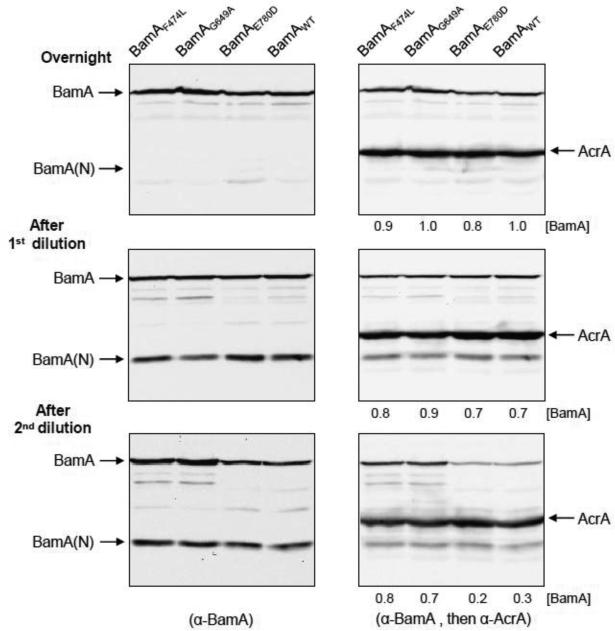
BamD has been known to stabilize the BAM complex [11], but its effect on BamA biogenesis is not known. BamA levels were examined by Western blot analysis from cultures grown overnight in glycerol minimal medium and after dilutions in glucose minimal medium. Figure 2 shows data from BamAWT and three BamA mutants; the data from the entire set of BamA mutants are presented in Fig. S1. As mentioned earlier, BamD was only partially depleted when cultures were grown overnight in glycerol minimal medium, but depleted rapidly during growth in glucose minimal medium (Figs. 3 and 5). BamA levels were very similar in all cultures grown overnight and after the first dilution (Fig. 2), which is consistent with their similar overnight cell densities and first dilution growth rates. However, after the second dilution, BamA levels in cultures expressing BamAWT and BamAE780D dropped more than two fold compared to those in cultures expressing BamAF474L and BamAG649A (Figs. 2 and S1). The drop in BamA levels in these two strains coincided with their growth decline during the same period (Fig. 1). It is important to mention that the decrease in BamAWT and BamAE780D levels (Figs. 2 and S1) is an indication of significant defects in BamA assembly since envelope stress should increase under BamD depletion conditions, elevating bamA expression by the σE-regulated bamA promoter [27,44,45].
Fig. 2.
Effects of BamD depletion on BamA. Cell lysates, prepared from samples collected during the growth experiments in Fig. 1, were analyzed by SDS-PAGE. For clarity, data from only four strains are shown here; data from the entire set, including samples collected after the third dilution, are shown in Fig. S1. BamA was detected by Western blot analysis using BamA-specific antibodies (left panels). After developing with BamA antibodies, the same blot was probed with antibodies against AcrA (right panels). AcrA, an inner membrane lipoprotein, is used as a gel loading control. Each lane contains protein lysate from equal number of cells based on OD600. Positions of AcrA, BamA and a major BamA fragment [BamA-(N)] are shown. [BamA] refers to BamA/AcrA ratios. Residue numbers are relative to the mature BamA protein sequence.
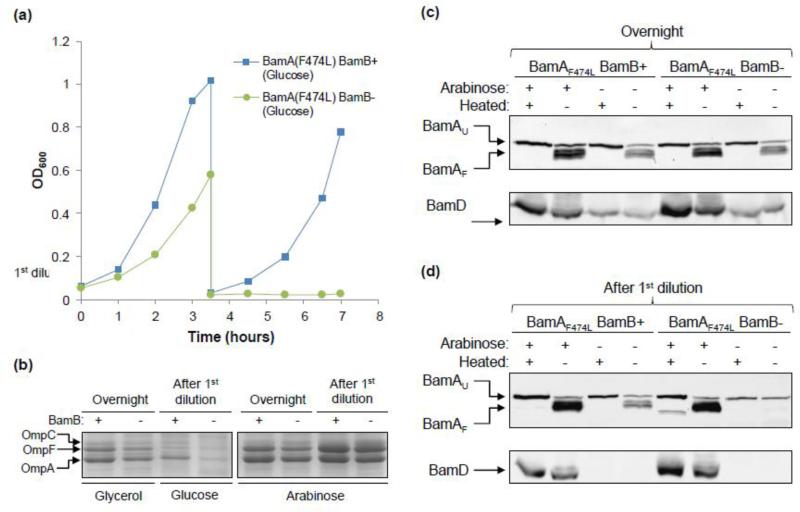
Fig 5.
Effects of the presence or absence of BamB on BamAF474L folding in BamD-depleted cells. (a) Overnight cultures were grown in glycerol minimal medium, supplemented with or without arabinose. Cultures grown without arabinose were diluted 1:100 in glucose minimal medium and grown for 3.5 hours. At the end of this period, cultures were diluted 1:50 in fresh glucose minimal medium and grown for another 3.5 hours. Cell densities were measured at 600 nm. Cell samples were collected from overnight grown cells and at the end of the first dilution for protein analysis. All cultures were grown at 37°C in baffled flasks in a water bath incubator shaker set at 180 rpm. (b) Envelopes obtained from cultures described in Fig. 5a were analyzed by SDS(urea)-PAGE. BamD is expressed in the presence of arabinose in glycerol minimal medium and repressed without arabinose in glucose minimal medium. Protein bands were visualized after Coomassie blue staining. Positions of the three major OMPs are shown. (c) and (d) BamA folding status was determined using envelopes obtained from cultures grown overnight in glycerol minimal medium (c) or after the first dilution in glucose minimal medium (d). Prior to SDS-PAGE analysis, SDS-solubilized samples were either left at room temperature (25°C) or heated (100°C). Detection of BamA was carried out by Western blot analysis using BamA-specific antibodies. The same blot was subsequently probed with BamD antibodies (lower panels). BamD is expressed in the presence of arabinose and repressed in its absence in glucose minimal medium. Electrophoresis was performed at room temperature. BamAU and BamAF refer to unfolded and folded forms of BamA, respectively.
A prominent BamA antibody-reacting protein band, migrating at the 35 kDa position, consistently appeared in our work (Figs. 2 and S1). Although the 35 kDa band appeared in the overnight cultures, its relative intensity increased substantially after the first dilution, when bamD expression was inhibited by glucose, and stayed high after the second and third dilutions during continual BamD depletion (Figs. 2 and S1). Interestingly, a similar 35 kDa BamA fragment was also observed by Ricci et al. [43] in a strain expressing a mutant BamA protein defective in its ability to interact with BamD, but not in a strain expressing BamAWT. In that study and ours the 35 kDa fragment appears to be a proteolysis product of BamA resulting from the loss of BamA-BamD interactions due to a defective BamA protein [43] or BamD depletion (this work). Consistent with Ricci et al. [43], we found that the 35 kDa fragment is derived from an N-terminal POTRA domain of BamA, since it interacts with the antibodies raised against a N-terminal derived mature BamA peptide (A21-Q35; [24]) (data not shown). For this reason, the 35 kDa is labeled as BamA(N) in Figs. 2 and S1.
In summary, these data showed that with the exception of BamAWT and BamAE780D, the remaining five BamA variants assumed conformations that resisted in vivo destabilization under BamD depletion conditions. Since these five BamA variants also improved bacterial growth without BamD, they are likely to restore some OMP biogenesis function in cells. It is important to mention that despite these positive effects of the BamA suppressor variants under BamD depletion conditions, cell growth deteriorated dramatically after the fourth dilution and we were unable to obtain stable ΔbamD transductants in these mutant bamA backgrounds. Therefore, while the BamA suppressor variants lower the dependence on BamD, they do not completely eliminate it.
Suppressor alteration renders BamA folding independent of BamD, but not OMP biogenesis
Next we examined the effects of BamA suppressors on BamA folding and OMP levels under BamD depletion conditions. Folded BamA can be separated from unfolded, heat-denatured, BamA on a SDS-polyacrylamide gel since the folded more compact form of BamA migrates faster than the unfolded more extended form [15,24,31,46]. We have shown previously that with the exception of F474L and E780D, the other four BamA substitutions altered the heat-modifiability property of BamA [15], thus making it difficult to assess the folding status of BamA in these four mutants. Also, as observed in the previous section, the strain expressing BamAE780D behaved very similar to that expressing BamAWT under BamD depletion conditions. Given these two observations, we examined BamA folding and OMP biogenesis status from cultures expressing BamAF474L and BamAWT. Envelopes were extracted from cultures expressing BamD (arabinose added) or depleted of it (glucose added) (Fig. 3).
BamA's folding status was determined from cultures grown after first and second dilutions. In cultures expressing BamD (plus arabinose; Fig. 3), both BamAWT and BamAF474L were present at the same levels and displayed an identical heat-modifiability behavior (Fig. 3a). Likewise, levels and folding behaviors of the two proteins were very similar after the first dilution in glucose medium (minus arabinose; Fig. 3). However, after the second dilution in which BamD was further depleted, nearly all of the BamAWT protein from unheated samples migrated at the unfolded/denatured position (Fig. 3a), indicating severe BamA misfolding. Strikingly, under the identical BamD depletion condition, a significant amount of the BamAF474L protein maintained the normal heat-modifiability property (Fig. 3a), indicating normal folding status. Thus, the F474L alteration in the BamA β-barrel domain renders BamA folding independent of BamD, just as it did in the absence of BamB and BamE [15]. We surmise from these results that BamD directly or indirectly, via the assembled BAM complex, assists folding of the BamA β-barrel domain, and that the growth defects under BamD depletion conditions are due in part to misfolding and thus loss of BamA function.
We also analyzed OMP levels from cultures expressing BamD or depleted of it (Fig. 3b-d). When arabinose was added to the growth medium, all cultures showed identical protein profiles from purified envelopes (Fig. 3b) and expressed readily detectable amounts of BamD (Fig. 3c). In contrast, significantly reduced levels of OMPs were present in envelopes isolated from cultures grown in glucose minimal medium after the first dilution (Fig. 3b and d), while BamD could not be detected from purified envelopes (Fig. 3c). Interestingly, cultures expressing either BamAWT or BamAF474L displayed very similar OMP profiles after the first dilution (Fig. 3b and d). This is consistent with the observation that all cultures exhibited almost identical growth rates after the first dilution in glucose minimal medium, thus presumably similar OMP biogenesis status (Fig. 1). However, there was an appreciable difference in the OMP profile from cultures isolated after the second dilution in glucose minimal medium (Fig. 3b and d) where cells expressing BamAF474L grew better than those expressing BamAWT (Fig. 1). The most noticeable difference was in the levels of OmpA and OmpC, which were not significantly affected in a culture expressing BamAF474L compared to that expressing BamAWT (Fig. 3b and d). In contrast, OmpF levels were severely reduced in both cultures (Fig. 3b and d).
These data indicated that even though the F474L alteration can render BamA folding and biogenesis independent of BamD, these changes do not make the activity of the BAM complex fully independent of BamD. In other words, in addition to assisting BamA folding, BamD must perform some indispensible function in OMP biogenesis.
BamAF474L assists assembly of BamAWT under BamD depletion conditions
BamD-independent assembly of BamAF474L allowed us to ask a crucial question of whether the assembled BamAF474L molecules are functionally active by examining their abilities to assemble BamAWT. To answer this, we built two partially diploid strains expressing either BamAWT or BamAF474L from the chromosome and a His-tagged copy of wild-type BamA (BamAWT-His) from a low copy number plasmid, pZS21 [24,27]. A null recA allele was introduced to prevent recombination between the wild-type and mutant alleles of bamA. Use of His-Probe allowed for the detection of only the plasmid-expressed BamAWT-His and not the chromosomally expressed BamA.
Under BamD depletion conditions, the strain expressing only the wild-type versions of tagged and untagged BamA proteins are expected to stop growing after the second dilution, as observed when using the haploid strain (Fig. 1). Similarly, BamAWT folding is expected to become acutely defective after the second dilution, as observed in Fig. 3. In contrast, the strain expressing BamAF474L from the chromosome and BamAWT-His from the plasmid may grow past the second dilution and correctly fold BamAWT-His, if folded BamAF474L is functionally active. The data from these experiments are presented in Fig. 4.
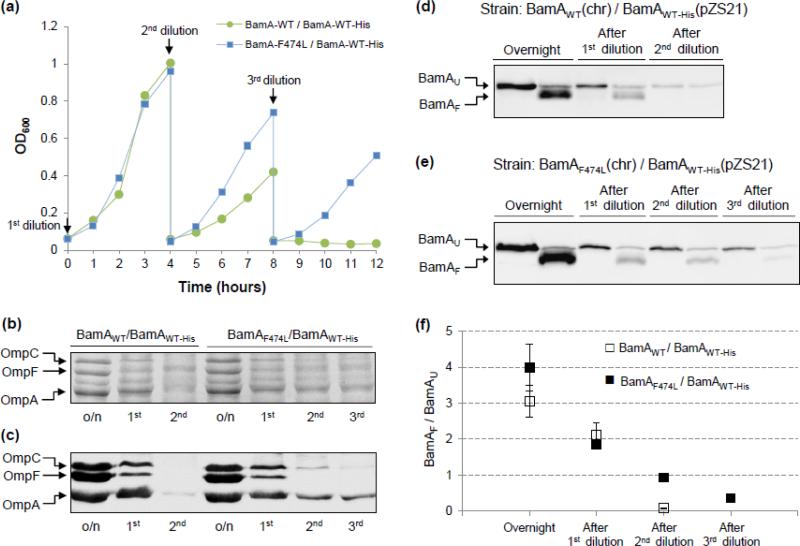
Fig. 4.
Effects of BamD depletion on bacterial growth, OMP levels, and BamA folding in strains partially diploid for bamA. (a) Overnight grown cultures in glycerol minimal medium were diluted three times and grown in a 37°C water bath as described in Fig. 1. BamD expression is repressed in glucose minimal medium. Cell densities were measured in a spectrophotometer at 600 nm. The untagged BamA proteins were expressed from the chromosome, while BamAWT-His was expressed from pZS21 (Kmr). Medium was supplemented with kanamycin to retain the pZS21 plasmid and with tetracycline to induce the expression of bamA from the plasmid replicon. Without tetracycline, the TetR repressor protein from a chromosomally localized zae::Tn10 inhibits the expression of bamA from the pZS21 plasmid. (b) and (c) SDS(urea)-PAGE and Western blot analyses of envelopes purified from glycerol-grown overnight (o/n) cultures and after three dilutions (1st, 2nd, and 3rd) in glucose minimal medium (Fig. 4a). Protein bands were visualized either after Coomassie blue staining (b) or Western blot analysis using OmpF/C/A antibodies (c). BamA compositions of the partially diploid strains are labeled on top. (d) and (e) BamA folding status was determined from envelopes obtained from cultures grown overnight in glycerol minimal medium and after dilutions in glucose minimal medium (Fig. 4a). Prior to SDS-PAGE analysis, SDS-solubilized samples were either left at room temperature (25°C) or heated (100°C). Detection of BamAWT-His was carried out by Western blot analysis using His-Probe. BamAU and BamAF refer to unfolded and folded forms of BamA, respectively. (f) Quantification of BamA folding. The graph shows the ratios of folded (BamAF) to unfolded (BamAU) species. Data were obtained from two independent experiments.
As expected, the partially diploid strain expressing BamAF474L from the chromosome grew significantly better than that expressing BamAWT after the second and third dilutions in glucose minimal medium to deplete BamD (Fig. 4a). However, growth rates after these dilutions were noticeably lower than those observed with the haploid strain (compare Figs. 1 and 4a), presumably due to the extra burden imposed by the presence of a plasmid and a null recA allele in the partial diploid strains. The strain expressing tagged and untagged versions of wild-type BamA failed to grow after the third dilution (Fig. 4a).
We analyzed the levels of major OMPs (Fig. 4b and c) from samples collected during the growth experiments shown in Fig. 4a. The OMP profiles of the two partially diploid strains from cultures obtained after overnight growth and after the first dilution were almost identical. However, after the second dilution, although the major OMP levels deteriorated further, their levels were higher in the strain expressing BamAF474L than that expressing BamAWT (Fig. 4b and c). We then determined BamAWT-His folding from purified envelopes (Fig. 4d-f). In the strain expressing the chromosomally-encoded BamAWT protein, folding of the plasmid-born BamAWT-His deteriorated rapidly after the second dilution, as no folded species was detected (Fig. 4d and f). However, in the strain expressing the chromosomally-encoded BamAF474L protein, almost 50% of the BamAWT-His protein folded correctly after the second dilution (Fig. 4e and f). Even after the third dilution, between 30 and 40% of the BamAWT-His protein folded correctly (Fig. 4e and f). Therefore, while the chromosomally-encoded BamAWT failed to fold and function correctly in the absence of BamD (Figs. 3 and S2), the BamAF474L protein not only folded correctly without BamD (Figs. 3 and S2), it maintained a significant functional activity, as demonstrated by its ability to catalyze folding of the substrate BamAWT-His protein, whose folding is normally dependent on BamD (Fig. 4d and f). These data showed good correlation between the presence of correctly folded BamA species (Fig. 4d-f) and the major OMP levels (Fig. 4b and c). Therefore, BamAF474L could catalyze the assembly of BamA and major OMPs under acute BamD depletion conditions, albeit not as efficiently as in the presence of BamD.
Role of BamB in BamA biogenesis
In the absence of BamB, BamA can fold efficiently, but the absence of both BamB and BamE results in BamA misfolding and a conditional lethal phenotype [15]. BamB interacts with BamA directly and the stability of this subcomplex remains unaffected under BamD depletion conditions [11]. Even though BamAF474L folds independently of BamD (Fig. 3a), it may still require BamB for correct folding, since BamB and BamD subcomplexes are known to play an overlapping role in BamAWT folding [15]. To test this possibility, we introduced a ΔbamB::Kmr allele in bamD depletion strains expressing either wild-type or the various suppressor bamA alleles. The bamB null allele was introduced by P1 transduction in the presence of arabinose to induce bamD expression from the chromosome. All strains yielded stable ΔbamB::Kmr transductants when bamD expression was induced by arabinose. However, none of the transductants were able to form single colonies on a medium lacking arabinose at 30°C or 37°C. These growth defects were much more severe than those observed in a bamB+ background. A representative growth pattern of strains expressing BamAWT and three BamA mutants is shown in Fig. S3.
We then carried out growth experiments in liquid cultures to assess the optimum time for sample collection and protein analysis. As described earlier, cultures were grown overnight in glycerol minimal medium. Next day, overnight cultures were diluted 1:100 in glucose minimal medium to further deplete BamD levels. However, bamB had a dramatic impact on glucose-grown cells, with the ΔbamB culture reaching only half the cell density compared to the bamB+ culture after 3.5 hours of growth (Fig. 5a). At the end of this growth period, cultures were diluted 1:50 in fresh media and grown for another 3.5 hours. Growth of the ΔbamB strain in glucose minimal medium ceased at the onset of the second dilution, whilst others grew somewhat slower than during the first dilution period (Fig. 5a). Cultures grown in the glycerol medium supplemented with arabinose displayed similar growth rates, regardless of the presence or absence of bamB, but these rates were lower than those observed after the first dilution in medium containing high energy-yielding glucose (data not shown).
The presence of BamB is critical for BamAF474L folding and OMP biogenesis under BamD depletion conditions
The functional consequence of ΔbamB under BamD depletion conditions was determined by analyzing the levels of OmpA, OmpC and OmpF, all of which are BAM substrates [27]. Envelopes isolated from cultures grown above (Fig. 5a) were analyzed on SDS(urea)-polyacrylamide gels and protein bands were visualized after Coomassie blue staining (Fig. 5b). When bamD expression was fully induced by arabinose, the effect of ΔbamB on OMPs were small albeit noticeable (Fig. 5b). However, the effects of ΔbamB on OMP levels were salient in cultures grown overnight in glycerol minimal medium without arabinose, and became more pronounced when BamD levels were further depleted after the first dilution in glucose minimal medium (Fig. 5b).
BamAF474L folding status was assessed from envelopes purified from cultures grown overnight and at the end of the first dilution (Fig. 5c and d). When bamD expression was induced by arabinose, BamAF474L displayed the typical heat-modifiability pattern, regardless of the presence or absence of BamB or the age of the culture (Fig. 5c and d). However, under BamD-depletion conditions, BamAF474L folding differed depending on the presence or absence of BamB and the culture age. Note, however, that BamD was readily detected in cultures grown overnight in glycerol minimal medium, though its level was significantly lower than that in cultures supplemented with arabinose (Fig. 5c). BamAF474L in ΔbamB and bamB+ backgrounds from glycerol-grown overnight cultures folded similarly to those in arabinose-supplemented cultures (Fig. 5c). After further depletion of BamD in glucose minimal medium, the effect of ΔbamB on BamAF474L folding became more apparent, as virtually no heat-modifiable form of BamAF474L could be detected (Fig. 5d). Moreover, BamAF474L was present at a significantly reduced level compared to the bamB+ background under identical growth conditions (Fig. 5d). In contrast, BamAF474L folded correctly in the bamB+ background despite BamD levels being undetectable after the first dilution in glucose minimal medium (Fig. 5d). These data showed that BamAF474L stability and folding were severely compromised in the absence of BamB under BamD depletion conditions. Thus, while BamAF474L folding may become largely independent of BamD (Figs. 3 and 4), it is still dependent on BamB in the absence of BamD.
Collectively, these data showed a good correlation between BamAF474L stability/folding and the OMP biogenesis defect, and revealed a critical role of BamB in the biogenesis of BamA and other OMPs when BamD is depleted in the cell.
Discussion
The in vivo roles of the BamBCDE lipoproteins are currently unknown. Initial clues of their importance came from the genetic studies showing that bamD could not be deleted from the cell [10,11] and that the simultaneous deletion of any two non-essential BamBCE lipoproteins conferred a synthetic and/or conditional lethal phenotype [8,15, 17]. The conditional lethal phenotype of the ΔbamB ΔbamE double mutant was shown to be, in part, due to a severe impairment in BamA biogenesis, causing it to misfold and degrade [15]. Because BamE interacts with BamA indirectly via BamD, it was anticipated that when BamD is depleted, this would severely impair BamA biogenesis. Indeed, we found in this work that when BamD is depleted from the cell, the BamAWT protein failed to fold correctly. The reduced BamAWT level under BamD depletion conditions is most likely due to degradation of misfolded nascent and/or pre-inserted BamA and not its reduced synthesis, since the elevated envelope stress created by these conditions should increase bamA expression by activating the σE regulon [27,44,45].
If the lethal effects of BamD depletion are mainly due to misfolding of BamA, then certain alterations in BamA that make BamA's folding and/or assembly less dependent on BamD should allow for improved bacterial growth and OMP biogenesis under BamD depletion conditions. To test this hypothesis, we introduced six previously characterized BamA alterations that were obtained in a ΔbamB ΔbamE background that improved bacterial growth and stabilized BamA by influencing its folding in the absence of the two Bam lipoproteins [15]. Five of the six BamA alterations significantly improved bacterial growth under BamD depletion conditions (Fig. 1) and stabilized BamA (Figs. 2 and S1). The sole BamA variant that did not improve growth or stabilize BamA carried an E780D alteration and was also the weakest suppressor in the ΔbamB ΔbamE background [15]. Notably, while the BamA suppressor alterations improved bacterial growth and/or OMP biogenesis in ΔbamB, ΔbamB ΔbamE and BamD depletion backgrounds, they produced no negative side effects when the BAM complex contained all four wild-type Bam lipoproteins [15]. Thus, the suppressor alterations’ effects on BamA folding and activity were not specific to defective or partially functional BAM complexes.
Because the F474L alteration does not affect the heat-modifiability property of BamA [15], BamAF474L and BamAWT were chosen to test the effect of BamD depletion on BamA folding. Not only was the bacterial growth significantly improved by the F474L alteration, it allowed the mutant BamA protein to maintain the normal folding state, which the BamAWT protein lost when BamD levels dropped below the detection level of Western blot analysis (Fig. 3). Using a partially diploid strain expressing both BamAF474L and BamAWT, we showed the folding and assembly of BamAWT was significantly improved in the presence of BamAF474L under BamD depletion conditions (Fig. 4). These data indicated that the folded BamAF474L molecules without BamD are functionally active and reinforced the idea that BamA assembles BamA [8,15,27]. Curiously, despite allowing normal BamA folding and improving bacterial growth, the positive effect of BamAF474L on OMPs other than BamAWT biogenesis was modest when BamD was depleted (Figs. 3 and 4). One explanation for this observation could be that the F474L substitution specifically influences folding and assembly of the BamA β-barrel to make it less dependent on BamD, but the overall activity of the BAM complex is still BamD dependent. Therefore, a modest improvement in substrate OMP assembly is likely a consequence of improved mutant BamA assembly and activity, which compensated for a compromised BamD-depleted BAM complex. There could be any number of indispensible roles BamD may play in OMP assembly, including serving as an OMP receptor and directly or indirectly assisting in OMP β-barrel formation and insertion into the outer membrane via BamA. It is worth mentioning that unlike in E. coli, BamD in Salmonella was shown to be dispensable for growth, although the bamD null mutant grew poorly [47]. This could be due to the presence of permissive substitutions in the BamA β-barrel sequence of Salmonella. Interestingly, both bacteria contain identical residues at the six BamA suppressor sites identified in E. coli.
When BamD is depleted from the cell, the whole BamACDE subcomplex should be effectively disabled because BamCE interact with BamA via BamD [11,17]. However, the effects of BamD depletion on cell viability and OMP biogenesis are by far more catastrophic, as shown in this work, than that caused by the simultaneous absence of BamCE [40]. Thus, while the whole BamACDE subcomplex is rendered inoperative without BamD, it is the absence of the essential BamD protein alone that inflicts the most devastating effects on BamA and OMP biogenesis. As mentioned earlier, the BamA suppressor alterations were isolated for their abilities to allow a ΔbamB ΔbamE mutant to grow on a rich medium [15]. In this background, the BamA suppressors also significantly improved BamA and OMP biogenesis. The fact that the absence of BamE and BamB individually does not affect BamA folding but their simultaneous absence does [15] suggests that both BamB and BamCDE subcomplexes play overlapping roles in BamA biogenesis, with the latter playing a more critical role than the former. This same conclusion was reached in this study while examining BamAF474L folding under BamD depletion conditions in a ΔbamB background. Several observations made in our in vivo work here are consistent with a recent in vitro analysis carried out by Hagan et al. [19] which showed that BamB and BamCDE subcomplexes are individually sufficient to facilitate BamA assembly into proteoliposomes. Their analysis also showed that the presence of pre-assembled BamA in the BamCDE subcomplex allowed more efficient assembly of a FLAG-tagged substrate BamA protein than the BamCDE subcomplex alone, again indicating a role for BamA in its own assembly.
Both BamB and BamD make direct contacts with BamA [8,11,17]. BamA-BamB interaction sites, which were identified by mutagenesis and pull-down assays [22], are clustered closely in the folded structure [32-34]. It remains to be determined whether or not these sites of BamB participate in BamA assembly. The essential POTRA 5 domain of BamA is shown to be critical for BamA-BamD interactions [8,43]. Interestingly, the BamCDE subcomplex was shown in vitro to efficiently assemble a truncated BamA substrate lacking the POTRA 5 domain, indicating that the site of BamD-BamA interactions during BamA assembly may be different than those that occur in the fully assembled BAM holocomplex. A similar possibility was previously raised based on the structural data for BamD [35,36,38]. Of the five tetratricopeptide repeats (TPR; [48]) of BamD, the first three N-terminal TPR domains have been implicated in the formation of a structural scaffold for substrate peptide binding [35,36], while a mutational analysis revealed the involvement of the C-terminus of BamD in BamA binding [11]. The last β-strand of BamA contains residues that are highly conserved in almost all bacterial β-barrel OMPs [49]. These conserved sequences of OMPs are suggested to form a common motif recognized by the N-terminus substrate binding scaffold of BamD [35,36]. Thus, the last β-strand of nascent BamA molecules may transiently interact with the N-terminal TPR domains of BamD during assembly.
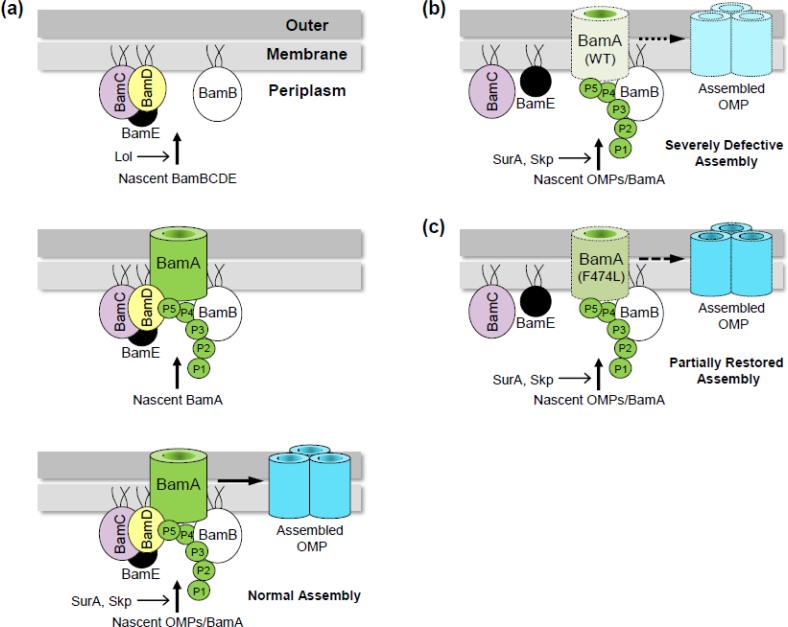
While the folding and insertion of BamAWT are dependent on the Bam lipoproteins, the reverse is not true [24,43]. This is because lipoproteins rely on a well-defined, Lol-dependent sorting and targeting pathway that does not involve any β-barrel proteins [2]. Consequently, the outer membrane lipoproteins, including all four lipoproteins of the BAM complex, will be stably localized and anchored to the outer membrane by the Lol system, even in the absence of the BamA protein (Fig. 6a, top panel). Once localized in the outer membrane by the Lol system, the BamB/CDE lipoproteins in the outer membrane could then potentially act as receptors for the nascent BamA molecules in the periplasm to guide their folding and insertion into the outer membrane (Fig. 6a, middle panel). In turn, the newly assembled BAM holocomplex can catalyze the assembly of all β-barrel OMPs, including BamA (Fig. 6a, bottom panel). When BamD is depleted from the cell, the remaining BamAB subcomplex is not adequate to support BamA and OMP assembly (Fig. 6b), except in the presence of suppressor alterations within the BamA β-barrel domain (Fig. 6c). However, even in the presence of BamA β-barrel suppressor alterations, growth and OMP assembly are not restored to wild-type levels, reflecting an indispensable role of BamD in E. coli BamA and OMP assembly. Perhaps distinct and/or multiple BamA alterations might be required to render BamD dispensable in E. coli, like in Salmonella [47].
Fig. 6.
A hypothetical model for the in vivo assembly of an active BAM complex and β-barrel OMPs. (a) The outer membrane lipoproteins, including the four lipoproteins of the BAM complex, BamDCDE, are localized to the outer membrane via the localization of lipoprotein (Lol) system [2] (top panel). The BamB and BamDCE subcomplexes facilitate the assembly of the nascent BamA molecules (middle papel). The assembled BAM holocomplex then catalyzes the assembly of all β-barrel OMPs, including BamA, which is more efficiently assembled in the presence of the BAM holocomplex (bottom panel) than in the presence of the Bam lipoproteins alone. (b) When BamD is depleted, the assembly of BamAWT and β-barrel OMPs is severely compromised. (c) Certain alterations within BamA β-barrel, such F474L, significantly improve assembly of BamA when BamD is depleted (bottom panel). In contrast to BamAF474L, a relatively modest improvement in the assembly of other β-barrel OMPs under BamD depletion conditions may reflect their continued dependence on BamD.
The SAM complex, which catalyzes β-barrel OMPs assembly in yeast mitochondria, also contains two essential proteins, Sam50 and Sam35, which are analogous to BamA and BamD in bacteria ([6]; and references therein). Both BamA and Sam50 belong to the Omp85 superfamily of proteins [14,50]. Just as BamD is required for proper BamA assembly, Sam50 assembly has been reported to be dependent on Sam35 [51]. Interestingly, however, unlike BamA and BamD, Sam50 and Sam35 are interdependent for their assembly and stability [51]. This is because Sam35 is a peripheral mitochondrial OMP [52,53] whose outer membrane localization appears to be catalyzed primarily by direct protein-protein interactions with Sam50. Therefore, when Sam50 is depleted, Sam35 cannot be stabilized in the outer membrane from the cytoplasmic side. Conversely, when Sam35 is depleted, the resulting SAM complex cannot assemble nascent Sam50 molecules. Although the bulk of the Sam35 protein is apparently exposed to the cytosolic side of the mitochondrial outer membrane, data suggest that Sam35 acts as a receptor for nascent β-barrel OMPs, including Sam50, by recognizing their β-signal in the intermembrane space [54]. BamD, like Sam35, may also serve as a general receptor for all nascent β-barrel OMPs, including BamA. Additionally, our data and those of Hagan et al. [19] suggest that BamD serves to fulfill a special folding/assembly requirement of BamA.
Whereas Sam35 is analogous to BamD, the third core component of the SAM complex, Sam37, does not appear to be the counterpart of BamB. Sam37 is thought to act downstream of Sam35 to assist in the release of substrates bound to Sam35/Sam50 [55]. In contrast, BamB has been suggested to act at an early step of OMP biogenesis [21,22]. The absence of Sam37 destabilizes the SAM complex, as the levels of both Sam35 and Sam50 are reduced [51], while the absence of BamB has no effect on the BamACDE subcomplex [8] or BamA folding or levels [15]. However, BamA does become unstable when BamBE are simultaneously missing [15] or BamD is depleted (this work). One characteristic Sam37 and BamB do share is that they both are nonessential, indicating either their activities are redundant or regulatory in nature.
Materials and methods
Strains, culture conditions and chemicals
Bacterial strains used in the study are listed in Table 1. M63-salt based minimal medium was prepared as described by Silhavy et al. [56]. The medium was supplemented with thiamine (0.1 mg ml−1), MgSO4 (1 mM), and casamino acids (0.1%). Glucose or glycerol (both at 0.2%) was used as a carbon source. When necessary, L-arabinose was added to final concentration of 0.2%. All media ingredients were purchased from Difco. All other chemicals were of analytical grade. All cultures were grown at 37°C unless specified.
Table 1.
Bacterial strains used in this study.
| Strains | Relevant characteristics | Reference |
|---|---|---|
| MC4100 | F- araD139 △(argF-lac)U139 rspL150 relAl flbB5301 ptsF25 deoCl thi-1 rbsR | [57] |
| RAM1821 | JCM290: MC4100 ardr/- △bamD △(λatt-lom)::bla PBAD bamD araC | [11] |
| RAM1823 | RAM1821 degP::Kmr | This study |
| RAM2253 | RAM1821 zae::Tn10 (4.75’) BamAF474L degP+ | This study |
| RAM2254 | RAM1821 zae::Tn10 (4.75’) BamAN631T degP | This study |
| RAM2255 | RAM1821 zae::Tn10 (4.75’) BamAG635A degP+ | This study |
| RAM2256 | RAM1821 zae::Tn10 (4.75’) BamAG649A degP+ | This study |
| RAM2257 | RAM1821 zae::Tn10 (4.75’) BamAA652V degP+ | This study |
| RAM2258 | RAM1821 zae::Tn10 (4.75’) BamAE780D degP+ | This study |
| RAM2259 | RAM1821 zae::Tn10 (4.75’) BamAWT degP+ | This study |
| RAM2268 | RAM2253 △bamB::Kmr | This study |
| RAM2269 | RAM2254 △bamB::Kmr | This study |
| RAM2270 | RAM2255 △bamB::Kmr | This study |
| RAM2271 | RAM2256 △bamB::Kmr | This study |
| RAM2272 | RAM2257 △bamB::Kmr | This study |
| RAM2273 | RAM2258 △bamB::Kmr | This study |
| RAM2274 | RAM2259 △bamB::Kmr | This study |
| RAM2308 | RAM2259 recA::Cmr | This study |
| RAM2309 | RAM2253 recA::Cmr | This study |
| RAM2310 | RAM2308 (pZS21-BamAWT-His) | This study |
| RAM2311 | RAM2309 (pZS21-BamAWT-His) | This study |
Protein methods
Envelopes were isolated by the French press method as described previously (Bennion et al., 2010). Detergent solubilized envelopes were analyzed by mini sodium dodecyl sulfate (SDS)-polyacrylamide (11%) gel electrophoresis (PAGE). For a better separation of OmpC and OmpF, 4M urea was added to the running gel. Protein bands were visualized by staining gels with Coomassie blue.
For Western blot analysis, samples from whole cell extracts or purified envelopes were analyzed by SDS-PAGE and transferred onto Immobilon-P polyvinylidene difluoride membranes (Millipore). Membranes were blocked overnight in 5% (w/v) non-dairy cream. After overnight blocking, membranes were incubated with primary antibodies for 1.5 hours. Primary antibodies used were raised in rabbits against purified AcrA (1:16,000), BamA (1:5,000), and BamD (1:2,000), OmpF/C/A (1:25,000). BamA and BamD antibodies were gifts from Tom Silhavy, Princeton University. After incubation with primary antibodies, membranes were washed twice for 15 minutes each and incubated for 1 hour in secondary antibodies (goat anti-rabbit horseradish peroxidase-conjugated IgG). Protein bands were visualized with Bio-Rad Molecular Imager ChemiDoc XRS System and quantified using Quantity One Software (Bio-Rad).
Genetic methods
Bacteriophage P1-mediated transductions were carried out using a protocol described by Silhavy et al. [56].The bamA alleles were transduced into a ΔbamD Δ(λatt-lom)::bla PBAD bamD araC degP::Kmr strain (RAM1823) using a bamA-linked zae::Tn10 (Tcr; 4.75’). Tcr transductants were screened for kanamycin sensitivity to ensure the replacement of degP::Kmr (3.90’), which is located upstream of bamA (4.27’) by the degP+ and bamA alleles. DNA sequence analysis of the bamA gene confirmed that all Tcr Kms transductants had replaced the recipient bamA allele with that linked to the zae::Tn10 from the donor P1 lysates. Strains partially diploid for bamA were constructed in two steps. First, a recA::Cmr allele was introduced via P1 transduction so as to prevent homologous recombination between the two alleles of bamA. These strains were then transformed with pZS21, which expresses a His-tagged BamA protein.
Supplementary Material
Highlights.
Without essential BamD, the core BamA protein of the BAM complex becomes misfolded.
Compensatory alterations within BamA β-barrel restore BamA folding without BamD.
Folded mutant BamA without BamD then facilitates OMP and wild type BamA assembly.
Acknowledgements
We are indebted to Phu Vuong for critically reading the manuscript. We would also like to thank Lauren Lynch and Mackenzie Lynch for their assistance in SDS-PAGE analysis, and Rene Tellez Jr. for his involvement in the early phase of the project. RS was supported by a Research Fellowship from the School of Life Sciences Undergraduate Research program. This work was supported by grants from the National Institutes of Health (GM048167) and the School of Life Sciences, Arizona State University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harb Prospect Biol. 2010;2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Okuda S, Tokuda H. Lipoprotein sorting in bacteria. Annu Rev Microbiol. 2011;65:239–259. doi: 10.1146/annurev-micro-090110-102859. [DOI] [PubMed] [Google Scholar]
- 3.Ruiz N, Kahne D, Silhavy TJ. Transport of lipopolysaccharide across the cell envelope, the long road of discovery. Nature Rev Microbiol. 2009;7:677–683. doi: 10.1038/nrmicro2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sperandeo P, Dehò G, Polissi A. The lipopolysaccharide transport system of Gram negative bacteria. Biochim Biophys Acta. 2009;1791:594–602. doi: 10.1016/j.bbalip.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 5.Hagan CL, Silhavy TJ, Kahne D. β-barrel membrane protein assembly by the Bam complex. Annu Rev Biochem. 2011;80:189–210. doi: 10.1146/annurev-biochem-061408-144611. [DOI] [PubMed] [Google Scholar]
- 6.Misra R. Assembly of the β-barrel outer membrane proteins in Gram-negative bacteria, mitochondria, and chloroplasts. ISRN Mol Biol. 2012;2012:1–15. doi: 10.5402/2012/708203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ricci DP, Silhavy TJ. The Bam machine, a molecular cooper. Biochim Biophys Acta. 2012;1818:1067–1084. doi: 10.1016/j.bbamem.2011.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, Kahne D. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 9.Werner J, Misra R. YaeT (Omp85). affects the assembly of lipid-dependent and lipid-independent outer membrane proteins of Escherichia coli. Mol Microbiol. 2005;57:1450–1459. doi: 10.1111/j.1365-2958.2005.04775.x. [DOI] [PubMed] [Google Scholar]
- 10.Onufryk C, Crouch M-L, Fang FC, Gross CA. Characterization of six lipoproteins in the σE regulon. J Bacteriol. 2005;187:4552–4561. doi: 10.1128/JB.187.13.4552-4561.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malinverni JC, Werner J, Kim S, Sklar JG, Kahne D, Misra R, Silhavy TJ. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol Microbiol. 2006;61:151–164. doi: 10.1111/j.1365-2958.2006.05211.x. [DOI] [PubMed] [Google Scholar]
- 12.Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science. 2003;299:262–265. doi: 10.1126/science.1078973. [DOI] [PubMed] [Google Scholar]
- 13.Gentle I, Gabriel K, Beech P, Waller R, Lithgow T. The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J Cell Biol. 2004;164:19–24. doi: 10.1083/jcb.200310092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moslavac S, Mirus O, Bredemeier R, Soll J, Haeseler A von, Schleiff E. Conserved pore-forming regions in polypeptide-transporting proteins. FEBS J. 2005;272:1367–1378. doi: 10.1111/j.1742-4658.2005.04569.x. [DOI] [PubMed] [Google Scholar]
- 15.Tellez R, Jr, Misra R. Substitutions in the BamA β-barrel domain overcome the conditional lethal phenotype of a ΔbamB ΔbamE strain of Escherichia coli. J Bacteriol. 2012;194:317–324. doi: 10.1128/JB.06192-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Webb CT, Heinz E, Lithgow T. Evolution of the β-barrel assembly machinery. Trends Microbiol. 2012;20:612–620. doi: 10.1016/j.tim.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 17.Sklar JG, Wu T, Gronenberg LS, Malinverni JC, Kahne D, Silhavy TJ. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc Natl Acad Sci USA. 2007;104:6400–6405. doi: 10.1073/pnas.0701579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagan CL, Kim S, Khane D. Reconstitution of outer membrane protein assembly from purified components. Science. 2010;328:890–892. doi: 10.1126/science.1188919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagan CL, Westwood DB, Kahne D. Bam lipoproteins assemble BamA in vitro. Biochem. 2013;52:6108–6113. doi: 10.1021/bi400865z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Charlson ES, Werner JN, Misra R. Differential effects of yfgL mutation on the biogenesis of Escherichia coli outer membrane proteins and lipopolysaccharide. J Bacteriol. 2006;188:7186–7194. doi: 10.1128/JB.00571-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ureta AR, Endres RG, Wingreen NS, Silhavy TJ. Kinetic analysis of the assembly of the outer membrane protein LamB in Escherichia coli mutants each lacking a secretion or targeting factor in a different cellular compartment. J Bacteriol. 2007;189:446–454. doi: 10.1128/JB.01103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vuong P, Bennion D, Mantei J, Frost D, Misra R. Analysis of YfgL and YaeT interactions through bioinformatics, mutagenesis, and biochemistry. J Bacteriol. 2008;190:1507–1517. doi: 10.1128/JB.01477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sánchez-Pulido L, Devos D, Genevrois S, Vicente M, Valencia A. POTRA, a conserved domain in the FtsQ family and a class of β-barrel outer membrane proteins. Trends Biochem Sci. 2003;28:523–526. doi: 10.1016/j.tibs.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 24.Kim S, Malinverni JC, Sliz P, Silhavy TJ, Harrison SC, Kahne D. Structure and function of an essential component of the outer membrane protein assembly machine. Science. 2007;317:961–964. doi: 10.1126/science.1143993. [DOI] [PubMed] [Google Scholar]
- 25.Gatzeva-Topalova PZ, Walton TA, Sousa MC. Crystal structure of YaeT, conformational flexibility and substrate recognition. Structure. 2008;16:1873–1881. doi: 10.1016/j.str.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knowles TJ, Jeeves M, Bobat S, Dancea F, McClelland D, Palmer T, et al. Fold and function of polypeptide transport-associated domains responsible for delivering unfolded proteins to membranes. Mol Microbiol. 2008;68:1216–1227. doi: 10.1111/j.1365-2958.2008.06225.x. [DOI] [PubMed] [Google Scholar]
- 27.Bennion D, Charlson ES, Coon E, Misra R. Dissection of β-barrel outer membrane protein assembly pathways through characterizing BamA POTRA 1 mutants of Escherichia coli. Mol Microbiol. 2010;77:1153–1171. doi: 10.1111/j.1365-2958.2010.07280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Workman P, Heide K, Giuliano N, Lee N, Mar J, Vuong P, et al. Genetic, biochemical, and molecular characterization of the polypeptide transport-associated domain of Escherichia coli BamA. J Bacteriol. 2012;194:3512–3521. doi: 10.1128/JB.06740-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noinaj N, Kuszak AJ, Gumbart JC, Lukacik P, Chang H, Easley NC, et al. Structural insight into the biogenesis of β-barrel membrane proteins. Nature. 2013;501:385–392. doi: 10.1038/nature12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacob-Dubuisson F, Villeret V, Clantin B, Delattre A, Saint N. First structural insights into the TpsB/Omp85 superfamily. Biol Chem. 2009;390:675–684. doi: 10.1515/BC.2009.099. [DOI] [PubMed] [Google Scholar]
- 31.Leonard-Rivera M, Misra R. Conserved residues of the putative L6 loop of Escherichia coli BamA play a critical role in the assembly of β-barrel outer membrane proteins, including that of BamA itself. J Bacteriol. 2012;194:4662–4668. doi: 10.1128/JB.00825-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heuck A, Schleiffer A, Clausen T. Augmenting β-augmentation, structural basis of how BamB binds BamA and may support folding of outer membrane proteins. J Mol Biol. 2011;406:659–666. doi: 10.1016/j.jmb.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 33.Kim KH, Paetzel M. Crystal structure of Escherichia coli BamB, a lipoprotein component of the β-barrel assembly machinery complex. J Mol Biol. 2011;406:667–678. doi: 10.1016/j.jmb.2010.12.020. [DOI] [PubMed] [Google Scholar]
- 34.Noinaj N, Fairman JW, Buchanan SK. The crystal structure of BamB suggests interactions with BamA and its role within the BAM complex. J Mol Biol. 2011;407:248–260. doi: 10.1016/j.jmb.2011.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Albrecht R, Zeth K. Structural basis of outer membrane protein biogenesis in bacteria. J Biol Chem. 2011;286:27792–27803. doi: 10.1074/jbc.M111.238931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sandoval CM, Baker SL, Jansen K, Metzner SI, Sousa MC. Crystal structure of BamD, an essential component of the β-barrel assembly machinery of Gram-negative bacteria. J Mol Biol. 2011;409:348–357. doi: 10.1016/j.jmb.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dong C, Hou H-F, Yang X, Shen Y-Q, Dong Y-H. Structure of Escherichia coli BamD and its functional implications in outer membrane protein assembly. Acta Cryst. 2012;68:95–101. doi: 10.1107/S0907444911051031. [DOI] [PubMed] [Google Scholar]
- 38.Kim KH, Aulakh S, Paetzel M. Crystal structure of β-barrel assembly machinery BamCD protein complex. J Biol Chem. 2011;286:39116–39121. doi: 10.1074/jbc.M111.298166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knowles TJ, Browning DF, Jeeves M, Maderbocus V, Rajesh S, Sridhar P, et al. Structure and function of BamE within the outer membrane and the β-barrel assembly machine. EMBO J. 2011;12:123–128. doi: 10.1038/embor.2010.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rigel NW, Schwalm J, Ricci DP, Silhavy TJ. BamE modulates the Escherichia coli β-barrel assembly machine component BamA. J Bacteriol. 2012;194:1002–1008. doi: 10.1128/JB.06426-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robert V, Volokhina EB, Senf F, Bos MP, Gelder P van, Tommassen J. Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS Biol. 2006;11:1984–1995. doi: 10.1371/journal.pbio.0040377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim KH, Aulakh S, Paetzel M. The bacterial outer membrane β-barrel assembly machinery. Prot Sci. 2012;21:751–768. doi: 10.1002/pro.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ricci DP, Hagan CL, Kahne D, Silhavy TJ. Activation of the Escherichia coli β-barrel assembly machine (Bam). is required for essential components to interact properly with substrate. Proc Natl Acad Sci USA. 2012;109:3487–3491. doi: 10.1073/pnas.1201362109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rhodius VA, Suh WC, Nonaka G, West J, Gross CA. Conserved and variable functions of the σE stress response in related genomes. PLoS Biol. 2006;4:43–59. doi: 10.1371/journal.pbio.0040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gerken H, Leiser OP, Bennion D, Misra R. Involvement and necessity of the Cpx regulon in the event of aberrant β-barrel outer membrane protein assembly. Mol Microbiol. 2010;75:1033–1046. doi: 10.1111/j.1365-2958.2009.07042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stegmeier JF, Andersen C. Characterization of pores formed by YaeT (Omp85) from Escherichia coli. J Biochem. 2006;140:275–283. doi: 10.1093/jb/mvj147. [DOI] [PubMed] [Google Scholar]
- 47.Fardini Y, Trotereau J, Bottreau E, Souchard C, Velge P, Virlogeux-Payant I. Investigation of the role of the BAM complex and SurA chaperone in outer-membrane protein biogenesis and type III secretion system expression in Salmonella. Microbiol. 2009;155:1613–1622. doi: 10.1099/mic.0.025155-0. [DOI] [PubMed] [Google Scholar]
- 48.D'Andrea LD, Regan L. TPR proteins, the versatile helix. Trends Biochem Sci. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 49.Struyvé M, Moons M, Tommassen J. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J Mol Biol. 1991;218:141–148. doi: 10.1016/0022-2836(91)90880-f. [DOI] [PubMed] [Google Scholar]
- 50.Gentle IE, Burri L, Lithgow T. Molecular architecture and function of the Omp85 family of proteins. Mol Microbiol. 2005;58:1216–1225. doi: 10.1111/j.1365-2958.2005.04906.x. [DOI] [PubMed] [Google Scholar]
- 51.Waizenegger T, Habib SJ, Lech M, Mokranjac D, Paschen SA, Hell K, et al. Tob38, a novel essential component in the biogenesis of β-barrel proteins of mitochondria. EMBO Reports. 2004;5:704–709. doi: 10.1038/sj.embor.7400183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Milenkovic D, Kozjak V, Wiedemann N, Lohaus C, Meyer HE, Guiard B, et al. Sam35 of the mitochondrial protein sorting and assembly machinery is a peripheral outer membrane protein essential for cell viability. J Biol Chem. 2004;279:22781–22785. doi: 10.1074/jbc.C400120200. [DOI] [PubMed] [Google Scholar]
- 53.Ishikawa D, Yamamoto H, Tamura Y, Moritoh K, Endo T. Two novel proteins in the mitochondrial outer membrane mediate β-barrel protein assembly. J Cell Biol. 2004;166:621–627. doi: 10.1083/jcb.200405138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kutik S, Stojanovski D, Becker L, Becker T, Meinecke M, Krüger V, et al. Dissecting membrane insertion of mitochondrial β-barrel proteins. Cell. 2008;132:1011–1024. doi: 10.1016/j.cell.2008.01.028. [DOI] [PubMed] [Google Scholar]
- 55.Chan NC, Lithgow T. The peripheral membrane subunits of the SAM complex function codependently in mitochondrial outer membrane biogenesis. Mol Biol Cell. 2008;19:126–136. doi: 10.1091/mbc.E07-08-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Silhavy TJ, Berman ML, Enquist LW. Experiments with gene fusions. CSHL Press; Cold Spring Harbor, NY: 1984. [Google Scholar]
- 57.Casadaban MJ. Transposition and fusion of the lac genes to select promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976;141:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.