Abstract
FXYD2 is a membrane protein responsible for regulating the function of the Na,K-ATPase in mammalian kidney epithelial cells. Here we report the structure of FXYD2b, one of two splice variants of the protein, determined by NMR spectroscopy in detergent micelles. Solid-state NMR characterization of the protein embedded in phospholipid bilayers indicates that several arginine side chains may be involved in hydrogen bond interactions with the phospholipid polar head groups. The structure and the NMR data suggest that FXYD2b could regulate the Na,K-ATPase by modulating the effective membrane surface electrostatics near the ion binding sites of the pump.
1. INTRODUCTION
The critical balance of Na+ and K+ ion concentrations across cell membranes is maintained primarily by the Na/K-ATPase, an integral membrane enzyme complex that hydrolyses intracellular ATP to generate the energy required to exchange three Na+ ions for two K+ ions across the plasma membrane [1–3]. The enzyme’s activity is regulated by its three subunits - a catalytic αsubunit, an auxiliary βsubunit and a regulatory γsubunit, also known as FXYD protein due to a conserved amino acid sequence in its N-terminus. The expression of FXYD genes is tissue-specific, cell-specific and developmentally regulated. The proteins are prevalent in the early stages of fetal life, in tissues that specialize in fluid or solute transport or that are electrically excitable. Their association with the Na,K-ATPase induces specific changes in the enzyme’s kinetics and affinity for Na+, K+ and ATP [3–6]. Several FXYD family members have been linked with major human diseases, including heart failure (FXYD1) [7], hypomagnesemia (FXYD2) [8], cancer (FXYD3, FXYD5) [9, 10], and schyzophrenia (FXYD6) [11], and represent attractive targets for therapeutic development.
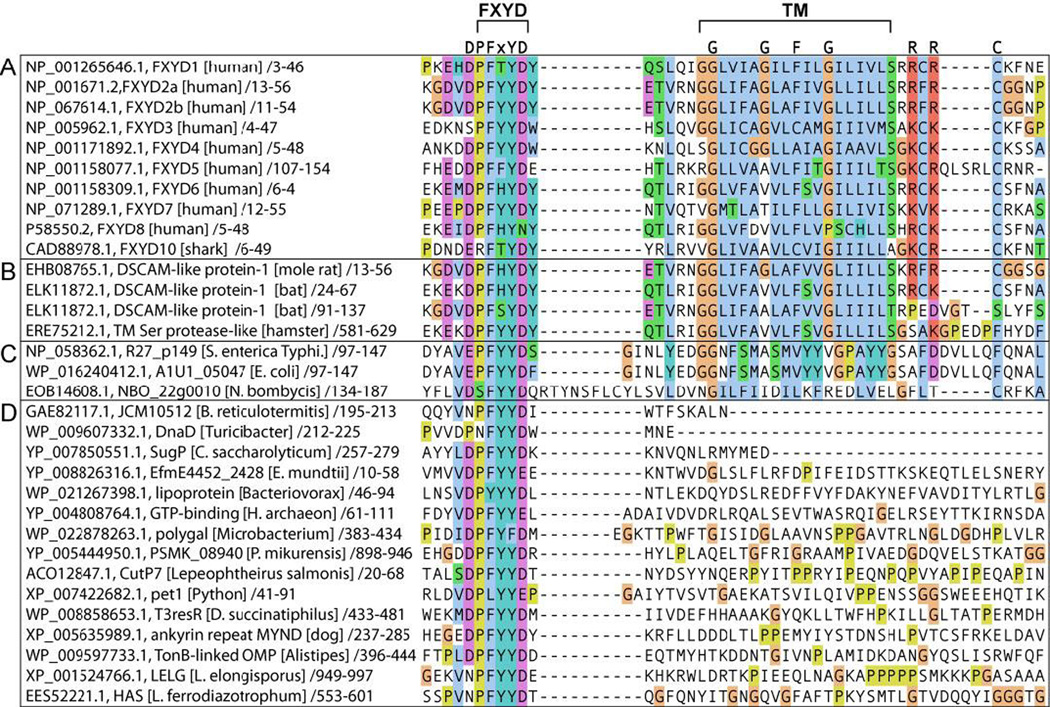
Although the FXYD proteins typically possess a single transmembrane helix and are relatively small, they are all encoded by genes with as many as nine exons [5]. Interestingly, the protein structures mirror the intron-exon arrangements of their corresponding genes, suggesting that discrete structured domains may have evolved to confer different functional properties in various physiological settings [12]. The family members share a core homology of 35 conserved amino acids, in and around a single transmembrane segment (Fig. 4A). The short signature motif, PFXYD (Pro, Phe, X, Tyr, Asp) is highly conserved in all known mammalian proteins, with residue X usually occupied by Tyr, but also Thr, Glu, or His. Conserved basic residues flank the transmembrane region, the extracellular N-termini are acidic, and the cytoplasmic C-termini are basic. However, outside of this homology region there is little sequence conservation among the family members. The distinct functionalities of FXYD proteins on the Na,K-ATPase's rate constant and affinities for Na+, K+ and ATP are largely ascribed to differences in their cytoplasmic domains, whose sequences vary widely among the family members.
Figure 4. Amino acid sequence alignment of FXYD homology domains.
(A) FXYD family proteins. (B-D) Proteins obtained by BLAST search of the NCBI database against the DVDPYYD polypeptide sequence. (B) Membrane proteins containing entire FXYD protein sequences, including conserved FXYD and transmembrane (TM) motifs. The Down syndrome cell adhesion molecule-like protein (DSCAM, ELK11872) contains two FXYD protein sequences from residues 24–67 and 91–137. (C) Other membrane proteins with a FXYD motif preceding a TM sequence. (D) Soluble protein sequences with a conserved FXYD motif. Alignments were generated with ClustalW [57] in Jalview [58], and rendered with ClustalW coloring. Transmembrane sequences were identified with TMHMM [56]. FASTA-formatted alignments are provided in Supporting materials.
FXYD2, the first FXYD protein to be identified as an accessory component of the Na,K-ATPase [13, 14], inhibits the activity of the pump by increasing its apparent affinity for both Na+ and K+ [15–20]. Its function is modulated by post-translational modification [16, 18], as well as by gene splicing and RNA editing, which respectively govern the expression of two splice variants, FXYD2a and FXYD2b, and of a truncated form of FXYD2b that is not exported to the plasma membrane [5, 13, 14, 21]. The two splice variants of FXYD2 have identical amino acid sequences, except in their N-terminal segments, encoded by the first exon [22, 23]. Both FXYD2a and FXYD2b are expressed primarily in kidney, albeit in distinct nephron segments, with FXYD2a in proximal and FXYD2b distal convoluted tubules [23–26]. The function of FXYD2 has been implicated in embryonic development [27], while misrouting of the protein due to the Gly41Arg mutation in the transmembrane segment has been linked with familial hypomagnesemia, a disease characterized by renal or intestinal Mg2+ loss [8]. The transmembrane span of FXYD2 has been implicated in modulation of the Na,K-ATPase's affinity for Na+ [28], while the N- and C-termini have been implicated in modulation of the Na+ and K+ affinities [25].
The first crystal structure, determined for pig kidney Na,K-ATPase confirmed that FXYD2 is an integral component of the enzyme complex and showed that its transmembrane helix associates with αM9, the ninth transmembrane helix of the αsubunit [29]. The α-FXYD association is stabilized by the amino acid sequence of αM9, which is highly conserved among all αsubunit isoforms [4], and by a set of highly conserved FXYD protein residues (Gly28, Gly33, Phe36 and Gly39 in FXYD2b) that form a distinctive "notch-peg-notch" shape along the helix length [30]. This intimate α-FXYD transmembrane association is also observed in the more recent crystal structure of Na,K-ATPase from pig kidney [31–34] and from shark [35, 36], and may be important for modulating the activity of the enzyme's Na+ binding sites [33].
The signature FXYD motif has been proposed to adopt an extended hook-like conformation that stabilizes the extracellular region of the β chain through hydrogen bond and salt bridge contacts [34–36]. However, the extramembrane regions of the FXYD and βchains are incompletely and/or poorly defined by the electron densities in the crystal structures of Na,K-ATPase. The NMR structures of FXYD1 and FXYD4, determined in micelles, separate from the Na,K-ATPase complex, show that the FXYD motif adopts a short loosely helical conformation in FXYD1 and a long well-formed helix in FXYD4 [30, 37]. In both structures, the transmembrane helix is followed by a short helical extension of conserved basic residues that anchor it near the membrane-water interface, and a ~10-residue amphipathic C-terminal helix that associates with the membrane surface. This helix contains the phosphorylation sites of FXYD1 and could modulate the electrostatic potential at the membrane surface, near the Na+/K+ ion binding sites of the Na,K-ATPase αsubunit [38].
Here we describe the NMR structure of FXYD2b determined in micelles. Solid-state NMR data, obtained for FXYD2b in magnetically aligned lipid bilayers, reflect the transmembrane orientation of the protein and show that several arginine side chains are immobilized, possibly due to hydrogen-bonded interactions with the phospholipid polar head groups. Such interactions could contribute to the membrane surface electrostatics and assist the recruitment of specific negatively charged phospholipids important for Na,K-ATPase stability and function.
2. MATERIALS AND METHODS
2.1 Sample preparation
Human FXYD2a and FXYD2b were prepared as described previously [39]. Briefly, the proteins were each expressed as C-terminal fusions to His9-TrpΔLE-Met and, purified by Ni-affinity chromatography and then separated from the fusion partner by CNBr chemical cleavage. Two mutations Met1Leu and Cys50Ser (in FXYD2b) or Cys52Ser (in FXYD2a) were introduced to facilitate cleavage and purification. Uniform 13C/15N isotopic labeling was obtained by supplementing minimal M9 growth media with 13C-glucose and (15NH4)2SO4 (Cambridge Isotope Laboratories, Andover, MA).
Samples for solution NMR spectroscopy were prepared by dissolving pure lyophilized protein in 300 uL of buffer (20 mM sodium citrate at pH 5, 10 mM DTT, 10% D2O) containing 500 mM sodium dodecyl sulfate (SDS) or 150 mM dodecyl-phosphocholine (DPC). For residual dipolar coupling (RDC) measurements, the samples were weakly aligned in 6.5% polyacrylamide gels by means of vertical compression, as described previously [12].
Magnetically aligned lipid bilayer samples for solid-state NMR spectroscopy were prepared by dissolving 2–3 mg of pure lyophilized protein in 1,2-O-dihexyl-sn-glycero-3-phosphocholine (6-O-PC) and then adding the resulting solution to a dispersion of the longer chain lipid 1,2-O-ditetradecyl-sn-glycero-3-phosphocholine (14-O-PC) to obtain a molar ratio 3.2 for long-chain to short-chain lipid. The final sample, containing 300 mM 14-O-PC in a volume of 180 µL at pH 6.7, was transferred to a flat-bottomed, 5 mm outer diameter NMR tube (New Era Enterprises, Vineland, NJ) for NMR studies.
2.2 NMR experiments
Solution NMR experiments were performed at 40°C on a Bruker AVANCE 600 MHz spectrometer equipped with a 1H/15N/13C triple-resonance cryoprobe. Solid-state NMR experiments were performed on a Bruker AVANCE 500 MHz spectrometer with a home-built 1H/15N double-resonance 5 mm solenoid coil probe. The NMR data were processed using NMRPipe [40] and analyzed using Sparky [41].
The 1H/15N/13C two- and three-dimensional solution NMR experiments, for backbone resonance assignments, measurements of resonance intensity and RDC measurements, were performed as described previously for FXYD1 [30]. Chemical shifts were referenced to the H2O resonance set to its expected position at 40°C [42]. Dihedral angles were derived from analysis of the NMR chemical shifts with the program TALOS+ [43]. The 1H/15N one- and two-dimensional oriented sample (OS) solid-state NMR experiments were performed as described previously [44].
To probe the association of FXYD2b with the micelle environment, we examined the paramagnetic relaxation enhancement (PRE) broadening effect of Mn2+ on the 1H/15N HSQC spectrum of the protein. PRE restraints were obtained by measuring the resonance intensities in the 1H/15N HSQC spectrum of FXYD2b after addition of increasing amounts of 1.8 mM MnCl2 to the micelle solution.
2.3 Computational methods
Structure calculations were performed with XPLOR-NIH [45, 46]. The structure coordinates and NMR restraints have been deposited in the Protein Data Bank (PDB ID: 2MKV) and structure statistics are reported in Table 1.
Table 1.
NMR structure statistics for FXYD2b.
| Number of experimental NMR restraints | |
| Dihedral angles | |
| phi | 38 |
| psi | 38 |
| Residual dipolar couplings (RDC) | |
| 1H-15N RDC | 60 |
| Distances | |
| CO-HN hydrogen bonds | 38 |
| Plane distances | |
| HN-plane | 8 |
| Structure statisticsa | |
| Violations (mean ± s.d.) | |
| Dihedral angle restraints (°) | 1.145 ± 0.111 |
| 1H-15N RDC restraints (Hz) | 0.767 ± 0.029 |
| distance restraints (Å) | 0.144 ± 0.003 |
| plane distance restraints (Å) | 1.311 ± 0.087 |
| Deviations from idealized geometry | |
| Bond lengths (Å) | 0.002 ± 0.000 |
| Bond angles (°) | 0.461 ± 0.008 |
| Impropers (°) | 0.460 ± 0.011 |
| Average pairwise r.m.s.d. (Å) | |
| Backbone | 0.181 ± 0.059 |
| Heavy | 1.038 ± 0.106 |
| Ramachandran Plot Statisticsb | |
| residues in most favored regions (%) | 86.3 |
| residues in additional allowed regions (%) | 11.2 |
| residues in generously allowed regions (%) | 2.2 |
| residues in disallowed regions (%) | 0.4 |
Evaluated for 10 lowest energy structures out of a total 100 calculated structures.
Evaluated with the program PROCHECK [49].
Two conventional simulated annealing protocols were used [47], the first for folding 100 structures from an initially extended conformation and the second for subsequent refinement of 100 structures from the lowest energy structure of the first protocol. Experimental restraints included dihedral angles derived from chemical shifts, hydrogen bond distances derived from 1H/2H exchange, and amide NH bond orientations derived from RDCs (Table 1). The knowledge-based statistical torsion angle potential torsionDB was implemented as described previously [47].
Plane distance restraints, derived from the Mn2+ PRE data, were implemented in the refinement stage, using the plane distance potential recently developed for XPLOR-NIH [48], instead of the harmonic coordinate plane restraints that we used previously for structure determination of FXYD1 [30]. Based on the data, backbone N atoms from residues Trp4, Tyr5 and Leu29 were restrained to one common plane representative of the extracellular membrane surface, and N atoms from Lys55-Arg57 and Ser50 were restrained to another plane representative of the cytoplasmic surface. The planes were defined to be perpendicular to the long axis of the transmembrane helix and the plane distance restrains were implemented loosely (± 3 Å).
3. RESULTS AND DISCUSSION
3.1 Structure of FXYD2b in micelles
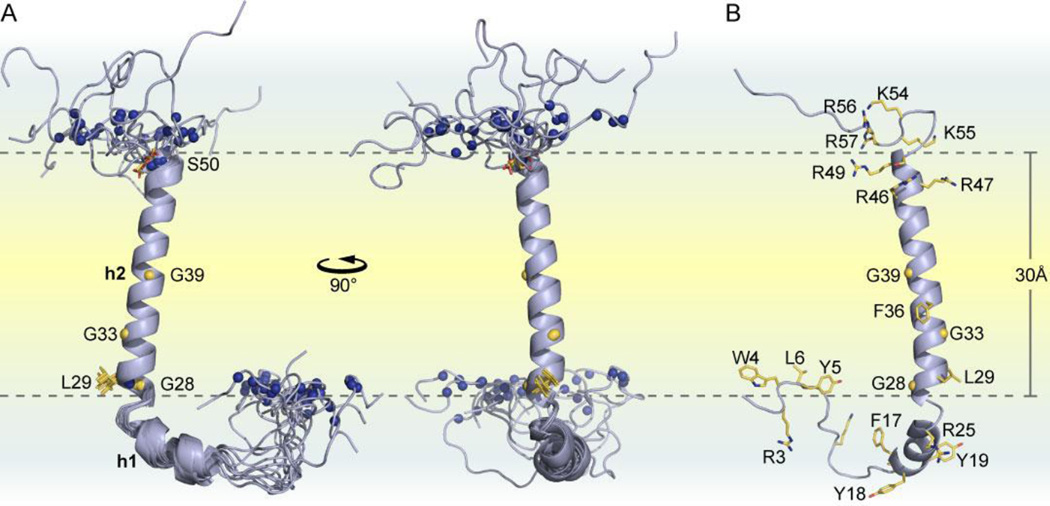
The structure of human FXYD2b is shown in Fig. 1. The transmembrane helix (h2), spanning residues Gly27-Leu44, is clearly defined by backbone amide hydrogens that are protected from exchange with bulk water (Fig. 2A). As observed in the structures of human FXYD1 and rat FXYD4 [30, 37], a set of conserved amino acids form a long groove parallel to the length of the transmembrane helix, delineating the binding interface with the Na,K-ATPase αM9 helix. The groove is lined by conserved glycines (Gly28, Gly33, Gly39) and interrupted by the aromatic ring of Phe36, which protrudes from the transmembrane helix like a key ready to engage its lock. Helix h2 is preceded by a short helical segment (h1) that spans residues Phe17-Arg25 and includes the FXYD signature motif (FYYD in FXYD2).
Figure 1. Structure of FXYD2b determined in micelles.
(A) Orthogonal views of the ten lowest energy NMR structures. Amide N atoms whose positions were restricted by plane distance restraints, derived from the Mn2+ PRE data, are shown as blue spheres. The two planes coincide with the micelle-water interface and each contain the amide N atoms of L29 or S50. (B) Lowest energy structure of FXYD2b showing side chains for arginines and other key residues that associate with the micelle interior.
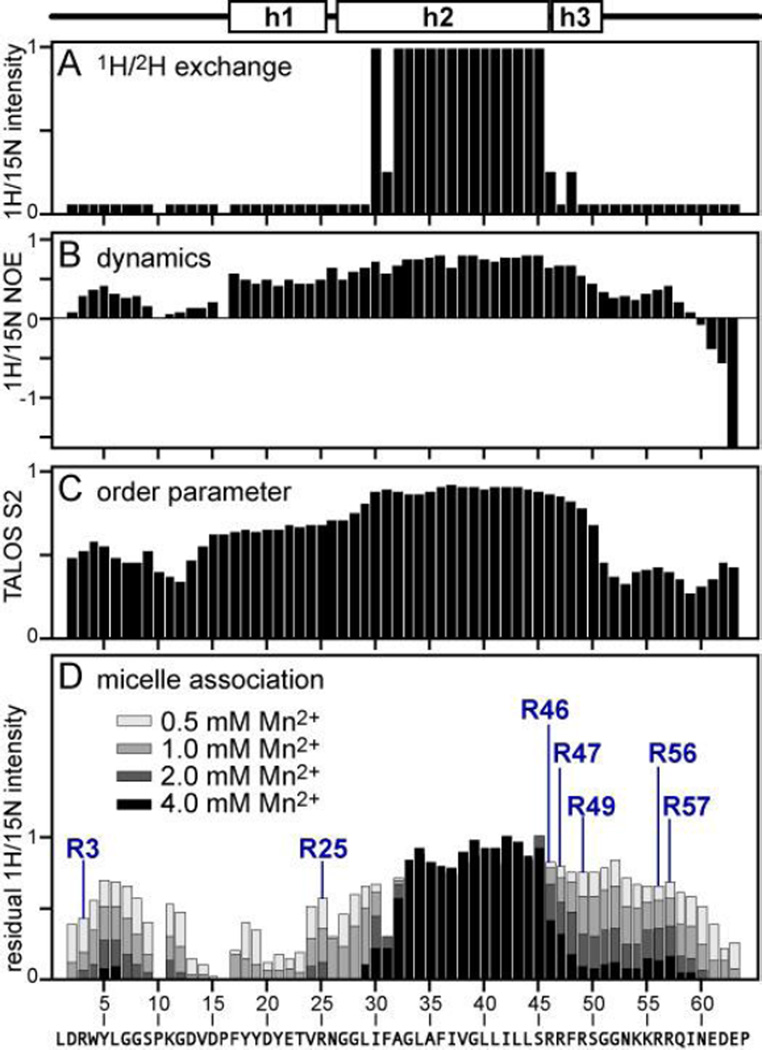
Figure 2. Summary of structural and dynamic features of FXYD2b in micelles.
The helical structure is outlined above the graphs. (A) Amide 1H/2H exchange profile. (B) Heteronuclear 1H/15N NOEs. (C) Order parameters (S2) derived from TALOS+ analysis of the chemical shifts. (D) Mn2+ PRE profile shwoing residual 1H/15N NMR intensity measured after addition of increasing concentrations of MnCl2. Peak intensities were measured in the presence of increasing concentrations of Mn2+ (see legend). Residual normalized peak intensity is the ratio of the intensity measured without Mn2+ to that measured in the presence of Mn2+. Data for the seven Arg backbone sites are labeled.
A short helix (h3), spanning residues Ser45-Ser50, extends beyond the hydrophobic portion of the transmembrane helix and contains the conserved arginine-rich sequence RRFR of the protein. Helices h2 and h3 of FXYD2b form a single extended transmembrane helix, while those of FXYD1 and FXYD4 are separated by a kink that changes helix direction slightly. Furthermore, FXYD2b lacks the well-defined cytoplasmic amphipathic helix observed in FXYD1 and FXYD4, and has somewhat less structural order in this region. Beyond the helical regions, the backbone of extracellular Trp4, Tyr5 and Leu6 and cytoplasmic Lys55, Arg56, and Arg57 associate with the micelle-water interface.
Overall, helices h1, h2, and h3 have similar backbone dynamics, with similar values of 1H/15N heteronuclear NOEs (Fig. 2B), order parameters (Fig. 2C) and 1H/15N HSQC peak intensities (Fig. 3B). Notably, residues Trp4-Gly7 in the N-terminus of the protein and residues Lys55-Arg57 in the C-terminal cytoplasmic region, also exhibit restricted dynamics, while Pro10-Pro16 and the C-terminus are significantly more flexible, with negative values of the 1H/15N NOE and greater peak intensities.
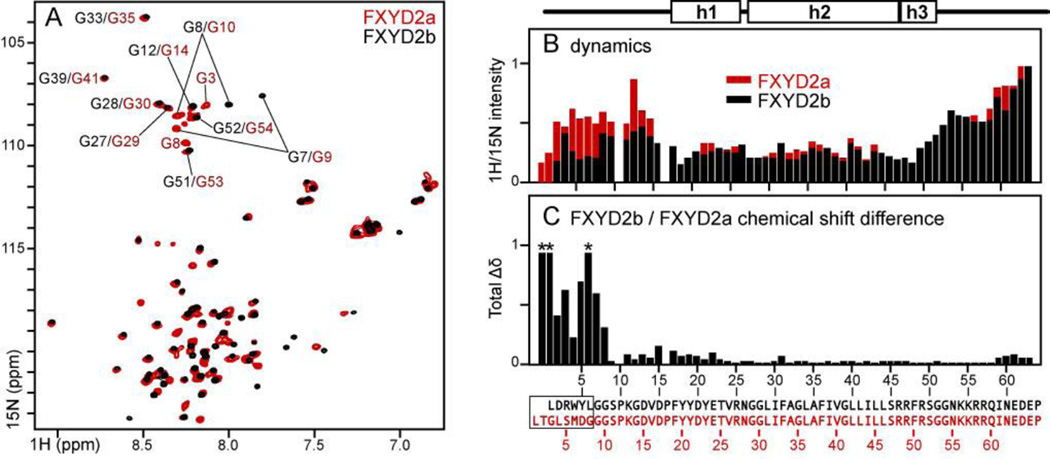
Figure 3. Comparison between FXYD2b and FXYD2a in micelles.
(A) 1H/15N HSQC spectra of FXYD2a (red) and FXYD2b (black) in SDS. (B) Normalized 1H/15N HSQC peak intensities measured for FXYD2b (black) or FXYD2a (red). (C) Total difference in 1H and 15N chemical shifts between FXYD2a and FXYD2b (Δδtot=[(ΔδH)2+(ΔδN/5)2]1/2). ΔδN is scaled by 1/5 to account for the 5-fold difference between the chemical shift dispersions of 15N and 1H. Red asterisks denote cross peaks with Δδtot >1 ppm. Amino acid sequences and numbers at the bottom correspond to FXYD2b (black) or FXYD2a (red). Differences in the N-termini of the two splice variants are enclosed in the box.
FXYD2a, the longer splice variant, is structurally similar to FXYD2b, as evidenced by the similarity of their HSQC cross peak chemical shifts (Fig. 3A). Significant chemical shift changes are observed only in the N-termini where the amino acid sequences of the two proteins differ (Fig. 3C). Notably, however, the slightly longer N-terminus of FXYD2a is significantly more dynamic all the way to Gly14, as evidenced by much greater HSQC peak intensities (Fig. 3B). This feature could be important for the different physiological functions of FXYD2a and FXYD2b.
In FXYD2b, the FYYD-spanning helix is oriented at a ~90° angle from the transmembrane helix. The arrangement of the FYYD motif helix relative to the micelle-water interface is consistent with the propensity of Phe and Tyr to associate with lipids. By contrast, the FXYD motifs of shark FXYD10 and pig FXYD2 have been proposed to adopt an extended hook-like structure forming extensive interactions with the extracellular regions of the βsubunit of their respective Na,K-ATPase [34–36]. These regions of the complex are incompletely and/or poorly defined by the crystallographic data, especially in the case of pig kidney Na,K-ATPase containing FXYD2 [29, 31–34]. However, it is possible that the extramembrane regions of FXYD proteins adopt different structures in their isolated states separate from the Na,K-ATPase complex. Several studies indicate that FXYD proteins can exist separate from the Na,K-ATPase [50] and associate with different partners, including the Na+/Ca2+ exchanger and some Ca2+ channels [51, 52]. The NMR data for FXYD2b (Fig. 2B) and other FXYD proteins [30, 37, 38, 53] are consistent in showing that the extramembrane regions of FXYD proteins are more dynamic, a property that could enable them to adopt different conformations in different settings or in the presence of their different partners. Structure determination of the whole FXYD protein within the enzyme complex will be needed to understand the precise conformations and functions of the extramembrane regions with respect to Na,K-ATPase regulation.
No conclusive function has been assigned to the signature FXYD sequences. The FXYD motifs of FXYD2 and FXYD10 have been proposed to stabilize the Na,K-ATPase through interactions with the extracellular regions of the αand βchains [34–36]. However, recent data argue against involvement of the FXYD, βand αextramembrane regions in stabilizing the Na,K-ATPase, and instead, attribute the stabilizing influence of the FXYD proteins on the Na,K-ATPase primarily to the FXYD transmembrane helices and to the effects of FXYD proteins in promoting the interactions of the enzyme with specific phospholipids, such as negatively charged phosphatidylserine [54, 55]. It is also possible that the FXYD motif helps stabilize the lipid bilayer membrane structure surrounding the enzyme complex and/or constitutes a localization signal.
A BLAST (Basic Local Alignment Search Tool) search of the NCBI protein sequence database, seeded with the short polypeptide sequence DVDPFYYD, identifies this sequence in numerous proteins from a wide variety of organisms, including prokaryotes (Fig. 4). Notably, the identification of entire FXYD protein sequences within other multi-domain membrane proteins (identified with TMHMM [56]) further indicates that they can have functions beyond Na,K-ATPase regulation.
Interesting examples include the 2174-residue DSCAM protein (Down syndrome cell adhesion molecule-like protein 1), which contains an entire FXYD protein sequence spanning residues 13–56, the shorter 343-residue DSCAM, which contains two contiguous entire FXYD protein sequences spanning residues 24–67 and 91–137, and the 894-residue serine protease, which also contains an entire FXYD protein sequence from residues 13–56 (Fig. 4B). These may be classified as members of the FXYD family since they share sequence homology across the FXYD motif as well as the conserved features of the transmembrane helix. The FXYD motif is also found in other membrane proteins (Fig. 4C) and several soluble proteins (Fig. 4D) with various functions, including GTP-binding proteins, lipoproteins and ankyrin repeat proteins. Functional insights may be also attained by careful analysis of other FXYD motif containing proteins as their sequences and structures become available in the databases.
3.2 Association of FXYD2b with the micelle
The depth of micelle insertion of FXYD2b backbone amide sites is reflected by the Mn2+ PRE profile (Fig. 2D). Distance-dependent broadening is observed for peaks from solvent-exposed sites, while residues associated with the hydrophobic regions of the micelle are less or not affected. The data mirror the hydrophobic character of the micelle-associated residues. Addition of MnCl2 results in substantial line broadening and disappearance of peaks from the N- and C-termini as well as from helix h1. By contrast, peaks from helix h2 retain close to their full intensity in the presence of Mn2+, consistent with the membrane-embedded topology of this segment. The PRE data also mirror the profile of backbone dynamics (Fig. 2B) with micelle-associated sites exhibiting more restricted mobility than water-exposed sites, and water-exposed regions (Pro10-Pro16 and the C-terminus) exhibiting enhanced mobility.
Notably, cross peaks from residues Trp4-Gly7 and Lys55-Arg57 also retain appreciable intensity at high Mn2+ concentrations. These sites appear protected from interaction with aqueous Mn2+ to a similar extent as Leu29 and Ser50 flanking the transmembrane helix, indicating that they are buried at a similar depth within the micelle. The data effectively restrain the backbone of extracellular Trp4, Tyr5 and Leu6 and cytoplasmic Lys55, Arg56, and Arg57 to the micelle-water interface. The aliphatic side chains of arginines and lysines are frequently found embedded in membranes with their positively charge groups reaching towards the membrane-water interface where they can hydrogen bond with the lipid phosphate head groups [59]. The 1H/15N HSQC peak from the indole group of Trp4 also resists complete obliteration by Mn2+, consistent with its insertion in the hydrophobic micelle, and the orientation of the hydrophobic side chain of Leu6 reflects its propensity for membrane insertion.
3.3 Characterization of the arginine side chains
FXYD2b contains seven arginines: Arg3 just before the Trp4-Leu6 micelle-associated segment, Arg25 located after the FYYD motif near the extracellular membrane-water interface, Arg46, Arg47 and Arg49 located at the end of the transmembrane helix near the cytoplasmic membrane-water interface, and Arg56 and Arg57 in the micelle-associated region of the C-terminus. All of these may be expected to interact appreciably with the membrane since their backbone amide sites exhibit 1H/15N HSQC signals with protection from Mn2+ PRE broadening (Fig. 2D).
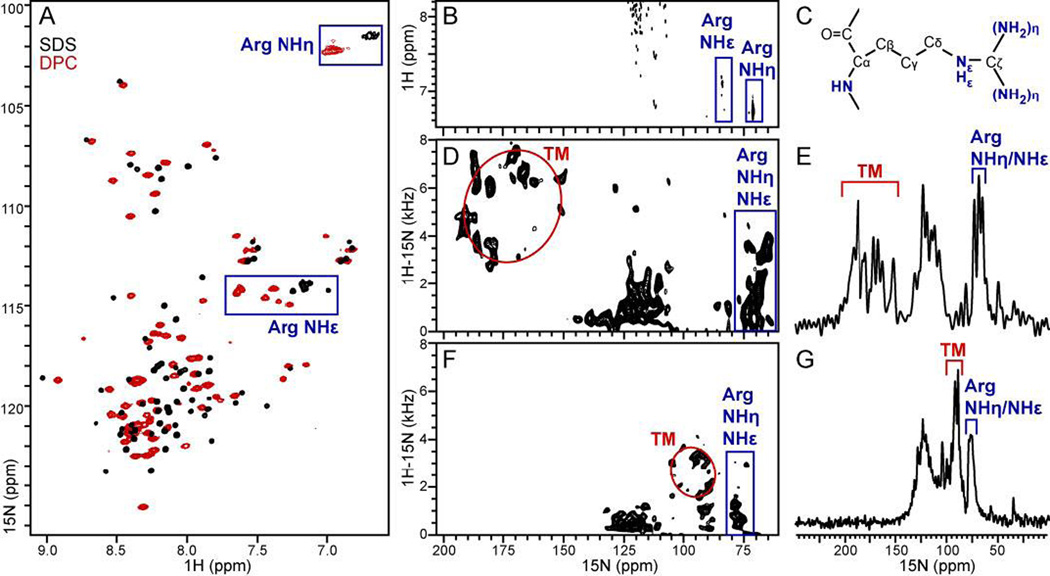
The HSQC spectra of FXYD2b in micelles display a broad arginine guanidinium NHη signal intensity, consistent with intermediate rotational dynamics of the Arg side chains at this temperature (Fig. 5A). In the spectra of FXYD2b in SDS, cross peaks from arginine guanidinium NHε groups are clustered in a region between 7.0 and 7.2 ppm (Fig. 5A; black). However, the spectra obtained for the protein in DPC exhibit six well-resolved NHε cross peaks between 7.2 and 7.7 ppm (Fig. 5A; red). Two of these peaks appear significantly shifted downfield, as might be expected for hydrogen-bonded guanidinium groups [60, 61], suggesting that these arginine side chains interact with the DPC phosphates. This result is substantiated by the solid-state NMR spectra obtained for uniformly 15N-labeled FXYD2b in magnetically aligned phospholipid bilayers (Fig. 5D-G), where significant resonance intensity is observed at 15N frequencies (70–90 ppm) associated with arginine side chains [60–62].
Figure 5. Solution and solid-state NMR spectra of uniformly 15N labeled FXYD2b in detergent micelles and phospholipid bilayers.
(A, B) Solution NMR 1H/15N HSQC spectra in SDS (black) and DPC (red) micelles. Peaks from the seven arginine guanidinium groups are enclosed in blue boxes. (A) The 15N frequencies from Arg side chains peaks are not actual but reflect the reduced 15N spectral width used to measure the spectra. (B) The spectrum was obtained with full 15N spectral width to observe the correct 1H and 15N frequencies of peaks from arginine guanidinium NH sites. (C) Molecular structure of arginine. (D-G) One-and two-dimensional 1H/15N OS solid-state NMR spectra spectra of FXYD2b in lipid bilayers aligned with the membrane perpendicular (D, E) or parallel (F, G) to the magnetic field. Peaks from the transmembrane helix (TM) trace wheel-like patterns (red circles). Peaks assigned to arginine guanidinium NH groups are enclosed in blue boxes.
The one-dimensional cross polarization (CP) spectra and the two-dimensional 1H/15N separated local field (SLF) spectra exhibit remarkable resolution. Resonances from amide sites in the transmembrane helix (TM) conform to wheel-like patterns in the spectral regions expected for an α-helix crossing the membrane at an angle of ~20° [63, 64], with 15N frequencies of 150–200 ppm in the spectra from bilayers aligned perpendicular to magnetic field (Fig. 5D, E), or 85–100 ppm in the spectra from parallel membranes (Fig. 5F, G). For both membrane alignments, peaks in the central region of the 15N spectrum (100–130 ppm) are assigned to sites with sufficient mobility to cause isotropic averaging of the 15N chemical shift and 1H-15N dipolar coupling, as might be expected for the non-helical regions of the protein.
In the case of fully rigid guanidinium groups, each of the seven arginines in FXYD2b would be expected to contribute three peaks to the solution and solid-state NMR spectra, one from NHε and two from NHη groups. However, in the absence of hydrogen bonding or other immobilizing influences, rapid flip rates around the Nε-Cζ bond average the NHη NMR signals [60, 61]. Resonance overlap in the solid-state NMR spectra of FXYD2b precludes identification of the number of peaks from arginine side chains. However, since these signals display observable 1H-15N dipolar couplings in the SLF spectra, we infer that they must emanate from sites that do not undergo rapid isotropic reorientation.
Notably, both the 1H-15N dipolar couplings and the 15N chemical shift frequencies of these peaks change with the overall magnetic alignment of the lipid bilayer membrane in the magnetic field, indicating that the Arg side chains themselves adopt preferred orientations relative to the lipid bilayer. Since FXYD2b contains no other N-bearing side chains that would resonate in this region of the spectrum, we conclude that at least some of the arginine side chains are sufficiently immobilized to yield orientation-dependent solid-state NMR signals. Such immobilization could result from hydrogen bond formation between arginine side chains and the lipid phosphate or polar head groups.
3.4 Implications for function
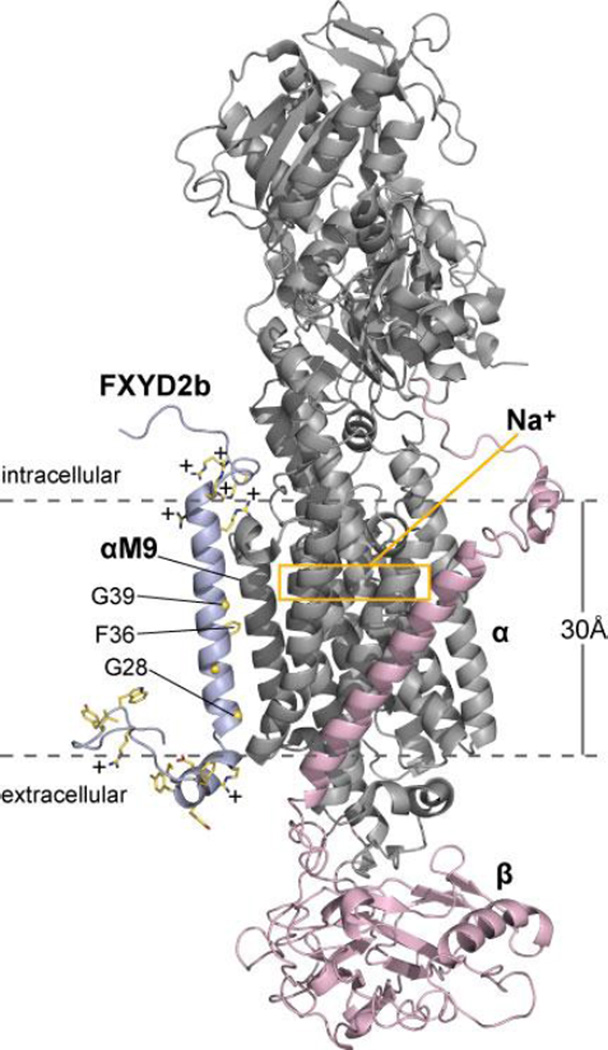
The crystal structures of pig kidney Na,K-ATPase highlight a highly electropositive region near the cytoplasmic membrane surface and the Na+/K+ ion binding sites of the αsubunit. This region is composed of arginine and lysine residues located at the cytoplasmic ends and connecting loops of the αsubunit's transmembrane helices, including three arginines that were proposed to constitute a voltage-sensing module [29]. To examine the location of FXYD2 arginines in the context of the Na,K-ATPase, we generated a model of the α/β/FXYD2b complex by replacing the coordinates of the transmembrane helix of endogenous FXYD2 in the recent crystal structure of Na,K-ATPase from pig kidney [34] with those of full-length FXYD2b determined by NMR in this study (Fig. 6). The model shows that the membrane surface location of Arg and Lys in FXYD2b could significantly contribute to the effective electrostatic potential near the Na,K-ATPase's Na+ binding sites.
Figure 6. Structural model of the α/β/FXYD2b complex.
The model was generated by replacing the coordinates of the transmembrane helix of endogenous FXYD2 in the crystal structure of Na,K-ATPase from pig kidney [34] with those of full-length FXYD2b determined by NMR. FXYD2b (light blue) associates with helix M9 of the αsubunit (gray). FXYD2b arginine and lysine side chains contribute significant positive charge (+) to the cytoplasmic membrane surface. The FYYD spanning helix of FXYD2b rests on the extracellular membrane surface near but without contacting the βsubunit (pink). The yellow box marks the location of the intramembrane binding sites for Na+.
Molecular dynamics simulations indicate that specific arginine side chains in channel voltage sensors can pair with one or more lipid phosphodiester groups at the membrane-water interface, dramatically influencing the local electric field [65]. This is consistent with the requirement of negatively charged lipid phosphodiester groups for channel function [66] and has provided an attractive explanation for the voltage sensor mechanism. Similar requirements for negatively charged phospholipids have been reported for optimal activity of the Na,K-ATPase activity (see [67, 68] and references therein).
Our data show that several arginines of FXYD2b appear to associate with the lipid bilayer interfacial regions. In the presence of phospholipids, they are sufficiently immobilized to yield both orientation-dependent solid-state NMR signals as well as shifted solution NMR signals. Their strategic positions in the structure of FXYD2b allows their guanidinium groups to form hydrogen bonds with the phosphate and polar head groups of membrane phospholipids, interactions that could be particularly significant in the presence of negatively charged lipids such as phosphatidylserine. This property could be important for the recruitment of specific phospholipids near the Na,K-ATPase, providing one potential explanation for the Na,K-ATPase stabilizing effect of FXYD proteins [54, 55], as well as for regulating the enzyme's activity.
Supplementary Material
HIGHLIGHTS.
The structure of FXYD2b is determined in micelles by NMR.
Several arginines yield orientation-dependent solid-state NMR signals.
Arginine side chains may hydrogen bond with lipid head groups.
The FXYD motif adopts a helical fold.
Additional FXYD-containing sequences are identified.
ACKNOWLEDGMENTS
This research was supported by grants from the National Institutes of Health (GM100265; AI074805; CA082864). The NMR studies utilized the NMR Facility at Sanford-Burnham Medical Research Institute, and the Resource for Molecular Imaging of Proteins at UCSD, each supported by grants from the National Institutes of Health (P30 CA030199; P41 EB002031).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Cornelius F, Mahmmoud YA. Functional modulation of the sodium pump: the regulatory proteins "Fixit". News Physiol Sci. 2003;18:119–124. doi: 10.1152/nips.01434.2003. [DOI] [PubMed] [Google Scholar]
- 2.Garty H, Karlish SJ. Role of fxyd proteins in ion transport. Annu. Rev. Physiol. 2006;68:431–459. doi: 10.1146/annurev.physiol.68.040104.131852. [DOI] [PubMed] [Google Scholar]
- 3.Geering K. FXYD proteins: new regulators of Na-K-ATPase. Am J Physiol Renal Physiol. 2006;290:F241–F250. doi: 10.1152/ajprenal.00126.2005. [DOI] [PubMed] [Google Scholar]
- 4.Blanco G, Mercer RW. Isozymes of the Na-K-ATPase: heterogeneity in structure, diversity in function. Am. J. Physiol. 1998;275:F633–F650. doi: 10.1152/ajprenal.1998.275.5.F633. [DOI] [PubMed] [Google Scholar]
- 5.Sweadner KJ, Rael E. The FXYD gene family of small ion transport regulators or channels: cDNA sequence, protein signature sequence, and expression. Genomics. 2000;68:41–56. doi: 10.1006/geno.2000.6274. [DOI] [PubMed] [Google Scholar]
- 6.Pihakaski-Maunsbach K, Vorum H, Locke EM, Garty H, Karlish SJ, Maunsbach AB. Immunocytochemical localization of Na,K-ATPase gamma subunit and CHIF in inner medulla of rat kidney. Ann. N. Y. Acad. Sci. 2003;986:401–409. doi: 10.1111/j.1749-6632.2003.tb07221.x. [DOI] [PubMed] [Google Scholar]
- 7.Shattock MJ. Phospholemman: its role in normal cardiac physiology and potential as a druggable target in disease. Curr Opin Pharmacol. 2009;9:160–166. doi: 10.1016/j.coph.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 8.Meij IC, Koenderink JB, van Bokhoven H, Assink KF, Groenestege WT, de Pont JJ, Bindels RJ, Monnens LA, van den Heuvel LP, Knoers NV. Dominant isolated renal magnesium loss is caused by misrouting of the Na(+),K(+)-ATPase gamma-subunit. Nat. Genet. 2000;26:265–266. doi: 10.1038/81543. [DOI] [PubMed] [Google Scholar]
- 9.Morrison BW, Moorman JR, Kowdley GC, Kobayashi YM, Jones LR, Leder P. Mat-8, a novel phospholemman-like protein expressed in human breast tumors, induces a chloride conductance in Xenopus oocytes. J. Biol. Chem. 1995;270:2176–2182. doi: 10.1074/jbc.270.5.2176. [DOI] [PubMed] [Google Scholar]
- 10.Nam JS, Hirohashi S, Wakefield LM. Dysadherin: a new player in cancer progression. Cancer Lett. 2007;255:161–169. doi: 10.1016/j.canlet.2007.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choudhury K, McQuillin A, Puri V, Pimm J, Datta S, Thirumalai S, Krasucki R, Lawrence J, Bass NJ, Quested D, Crombie C, Fraser G, Walker N, Nadeem H, Johnson S, Curtis D, St Clair D, Gurling HM. A genetic association study of chromosome 11q22-24 in two different samples implicates the FXYD6 gene, encoding phosphohippolin, in susceptibility to schizophrenia. Am. J. Hum. Genet. 2007;80:664–672. doi: 10.1086/513475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franzin CM, Yu J, Thai K, Choi J, Marassi FM. Correlation of Gene and Protein Structures in the FXYD Family Proteins. J. Mol. Biol. 2005;354:743–750. doi: 10.1016/j.jmb.2005.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forbush B, 3rd, Kaplan JH, Hoffman JF. Characterization of a new photoaffinity derivative of ouabain: labeling of the large polypeptide and of a proteolipid component of the Na, K-ATPase. Biochemistry. 1978;17:3667–3676. doi: 10.1021/bi00610a037. [DOI] [PubMed] [Google Scholar]
- 14.Collins JH, Leszyk J. The "gamma subunit" of Na,K-ATPase: a small, amphiphilic protein with a unique amino acid sequence. Biochemistry. 1987;26:8665–8668. doi: 10.1021/bi00400a026. [DOI] [PubMed] [Google Scholar]
- 15.Beguin P, Wang X, Firsov D, Puoti A, Claeys D, Horisberger JD, Geering K. The gamma subunit is a specific component of the Na,K-ATPase and modulates its transport function. EMBO J. 1997;16:4250–4260. doi: 10.1093/emboj/16.14.4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arystarkhova E, Wetzel RK, Asinovski NK, Sweadner KJ. The gamma subunit modulates Na(+) and K(+) affinity of the renal Na,K-ATPase. J. Biol. Chem. 1999;274:33183–33185. doi: 10.1074/jbc.274.47.33183. [DOI] [PubMed] [Google Scholar]
- 17.Pu HX, Cluzeaud F, Goldshleger R, Karlish SJ, Farman N, Blostein R. Functional role and immunocytochemical localization of the gamma a and gamma b forms of the Na,K-ATPase gamma subunit. J. Biol. Chem. 2001;276:20370–20378. doi: 10.1074/jbc.M010836200. [DOI] [PubMed] [Google Scholar]
- 18.Arystarkhova E, Donnet C, Asinovski NK, Sweadner KJ. Differential regulation of renal Na,KATPase by splice variants of the gamma subunit. J. Biol. Chem. 2002;277:10162–10172. doi: 10.1074/jbc.M111552200. [DOI] [PubMed] [Google Scholar]
- 19.Jones DH, Li TY, Arystarkhova E, Barr KJ, Wetzel RK, Peng J, Markham K, Sweadner KJ, Fong GH, Kidder GM. Na,K-ATPase from mice lacking the gamma subunit (FXYD2) exhibits altered Na+ affinity and decreased thermal stability. J. Biol. Chem. 2005;280:19003–19011. doi: 10.1074/jbc.M500697200. [DOI] [PubMed] [Google Scholar]
- 20.Zouzoulas A, Dunham PB, Blostein R. The effect of the gamma modulator on Na/K pump activity of intact mammalian cells. J. Membr. Biol. 2005;204:49–56. doi: 10.1007/s00232-005-0746-7. [DOI] [PubMed] [Google Scholar]
- 21.Sweadner KJ, Pascoa JL, Salazar CA, Arystarkhova E. Post-transcriptional control of Na,KATPase activity and cell growth by a splice variant of FXYD2 protein with modified mRNA. J. Biol. Chem. 2011;286:18290–18300. doi: 10.1074/jbc.M111.241901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuster B, Shainskaya A, Pu HX, Goldshleger R, Blostein R, Mann M, Karlish SJ. A new variant of the gamma subunit of renal Na,K-ATPase. Identification by mass spectrometry, antibody binding, and expression in cultured cells. J. Biol. Chem. 2000;275:18441–18446. doi: 10.1074/jbc.M001411200. [DOI] [PubMed] [Google Scholar]
- 23.Arystarkhova E, Wetzel RK, Sweadner KJ. Distribution and oligomeric association of splice forms of Na(+)-K(+)-ATPase regulatory gamma-subunit in rat kidney. Am J Physiol Renal Physiol. 2002;282:F393–F407. doi: 10.1152/ajprenal.00146.2001. [DOI] [PubMed] [Google Scholar]
- 24.Mercer RW, Biemesderfer D, Bliss DP, Jr, Collins JH, Forbush B., 3rd Molecular cloning and immunological characterization of the gamma polypeptide, a small protein associated with the Na,KATPase. J. Cell Biol. 1993;121:579–586. doi: 10.1083/jcb.121.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pu HX, Scanzano R, Blostein R. Distinct regulatory effects of the Na,K-ATPase gamma subunit. J. Biol. Chem. 2002;277:20270–20276. doi: 10.1074/jbc.M201009200. [DOI] [PubMed] [Google Scholar]
- 26.Pihakaski-Maunsbach K, Vorum H, Honore B, Tokonabe S, Frokiaer J, Garty H, Karlish SJ, Maunsbach AB. Locations, abundances, and possible functions of FXYD ion transport regulators in rat renal medulla. Am J Physiol Renal Physiol. 2006;291:F1033–F1044. doi: 10.1152/ajprenal.00086.2006. [DOI] [PubMed] [Google Scholar]
- 27.Jones DH, Davies TC, Kidder GM. Embryonic expression of the putative gamma subunit of the sodium pump is required for acquisition of fluid transport capacity during mouse blastocyst development. J. Cell Biol. 1997;139:1545–1552. doi: 10.1083/jcb.139.6.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindzen M, Aizman R, Lifshitz Y, Lubarski I, Karlish SJ, Garty H. Structure-function relations of interactions between Na,K-ATPase, the gamma subunit, and corticosteroid hormone-induced factor. J. Biol. Chem. 2003;278:18738–18743. doi: 10.1074/jbc.M213253200. [DOI] [PubMed] [Google Scholar]
- 29.Morth JP, Pedersen BP, Toustrup-Jensen MS, Sorensen TL, Petersen J, Andersen JP, Vilsen B, Nissen P. Crystal structure of the sodium-potassium pump. Nature. 2007;450:1043–1049. doi: 10.1038/nature06419. [DOI] [PubMed] [Google Scholar]
- 30.Teriete P, Franzin CM, Choi J, Marassi FM. Structure of the Na,K-ATPase regulatory protein FXYD1 in micelles. Biochemistry. 2007;46:6774–6783. doi: 10.1021/bi700391b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yatime L, Laursen M, Morth JP, Esmann M, Nissen P, Fedosova NU. Structural insights into the high affinity binding of cardiotonic steroids to the Na+,K+-ATPase. J. Struct. Biol. 2011;174:296–306. doi: 10.1016/j.jsb.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 32.Laursen M, Yatime L, Nissen P, Fedosova NU. Crystal structure of the high-affinity Na+K+- ATPase-ouabain complex with Mg2+ bound in the cation binding site. Proc. Natl. Acad. Sci. U. S. A. 2013;110:10958–10963. doi: 10.1073/pnas.1222308110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nyblom M, Poulsen H, Gourdon P, Reinhard L, Andersson M, Lindahl E, Fedosova N, Nissen P. Crystal structure of Na+, K(+)-ATPase in the Na(+)-bound state. Science. 2013;342:123–127. doi: 10.1126/science.1243352. [DOI] [PubMed] [Google Scholar]
- 34.Kanai R, Ogawa H, Vilsen B, Cornelius F, Toyoshima C. Crystal structure of a Na+-bound Na+,K+-ATPase preceding the E1P state. Nature. 2013;502:201–206. doi: 10.1038/nature12578. [DOI] [PubMed] [Google Scholar]
- 35.Ogawa H, Shinoda T, Cornelius F, Toyoshima C. Crystal structure of the sodium-potassium pump (Na+,K+-ATPase) with bound potassium and ouabain. Proc. Natl. Acad. Sci. U. S. A. 2009;106:13742–13747. doi: 10.1073/pnas.0907054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shinoda T, Ogawa H, Cornelius F, Toyoshima C. Crystal structure of the sodium-potassium pump at 2.4 A resolution. Nature. 2009;459:446–450. doi: 10.1038/nature07939. [DOI] [PubMed] [Google Scholar]
- 37.Franzin CM, Teriete P, Marassi FM. Structural similarity of a membrane protein in micelles and membranes. J. Am. Chem. Soc. 2007;129:8078–8079. doi: 10.1021/ja0728371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teriete P, Thai K, Choi J, Marassi FM. Effects of PKA phosphorylation on the conformation of the Na,K-ATPase regulatory protein FXYD1. Biochim. Biophys. Acta. 2009;1788:2462–2470. doi: 10.1016/j.bbamem.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crowell KJ, Franzin CM, Koltay A, Lee S, Lucchese AM, Snyder BC, Marassi FM. Expression and characterization of the FXYD ion transport regulators for NMR structural studies in lipid micelles and lipid bilayers. Biochim. Biophys. Acta. 2003;1645:15–21. doi: 10.1016/s1570-9639(02)00473-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 41.Goddard TD, Kneller DG. SPARKY 3. San Francisco: University of California; 2004. [Google Scholar]
- 42.Cavanagh J, Fairbrother WJ, Palmer AG, Skelton NJ. Protein NMR spectroscopy : principles and practice. San Diego: Academic Press; 1996. [Google Scholar]
- 43.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mahalakshmi R, Marassi FM. Orientation of the Escherichia coli outer membrane protein OmpX in phospholipid bilayer membranes determined by solid-State NMR. Biochemistry. 2008;47:6531–6538. doi: 10.1021/bi800362b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 46.Schwieters CD, Kuszewski JJ, Marius Clore G. Using Xplor-NIH for NMR molecular structure determination. Prog. Nucl. Magn. Reson. Spectrosc. 2006;48:47–62. [Google Scholar]
- 47.Bermejo GA, Clore GM, Schwieters CD. Smooth statistical torsion angle potential derived from a large conformational database via adaptive kernel density estimation improves the quality of NMR protein structures. Protein Sci. 2012;21:1824–1836. doi: 10.1002/pro.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu C, Gagnon E, Call ME, Schnell JR, Schwieters CD, Carman CV, Chou JJ, Wucherpfennig KW. Regulation of T cell receptor activation by dynamic membrane binding of the CD3epsilon cytoplasmic tyrosine-based motif. Cell. 2008;135:702–713. doi: 10.1016/j.cell.2008.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 50.Wypijewski KJ, Howie J, Reilly L, Tulloch LB, Aughton KL, McLatchie LM, Shattock MJ, Calaghan SC, Fuller W. A separate pool of cardiac phospholemman that does not regulate or associate with the sodium pump: multimers of phospholemman in ventricular muscle. J. Biol. Chem. 2013;288:13808–13820. doi: 10.1074/jbc.M113.460956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang XQ, Wang J, Carl LL, Song J, Ahlers BA, Cheung JY. Phospholemman regulates cardiac Na+/Ca2+ exchanger by interacting with the exchanger's proximal linker domain. Am J Physiol Cell Physiol. 2009 doi: 10.1152/ajpcell.00196.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang X, Gao G, Guo K, Yarotskyy V, Huang C, Elmslie KS, Peterson BZ. Phospholemman modulates the gating of cardiac L-type calcium channels. Biophys. J. 2010;98:1149–1159. doi: 10.1016/j.bpj.2009.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Franzin CM, Yu J, Thai K, Choi J, Marassi FM. Correlation of gene and protein structures in the FXYD family proteins. J. Mol. Biol. 2005;354:743–750. doi: 10.1016/j.jmb.2005.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mishra NK, Peleg Y, Cirri E, Belogus T, Lifshitz Y, Voelker DR, Apell HJ, Garty H, Karlish SJ. FXYD proteins stabilize Na,K-ATPase: amplification of specific phosphatidylserine-protein interactions. J. Biol. Chem. 2011;286:9699–9712. doi: 10.1074/jbc.M110.184234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beguin P, Crambert G, Guennoun S, Garty H, Horisberger JD, Geering K. CHIF, a member of the FXYD protein family, is a regulator of Na,K-ATPase distinct from the gamma-subunit. EMBO J. 2001;20:3993–4002. doi: 10.1093/emboj/20.15.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 57.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 59.Schow EV, Freites JA, Cheng P, Bernsel A, von Heijne G, White SH, Tobias DJ. Arginine in membranes: the connection between molecular dynamics simulations and translocon-mediated insertion experiments. J. Membr. Biol. 2011;239:35–48. doi: 10.1007/s00232-010-9330-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pascal SM, Yamazaki T, Singer AU, Kay LE, Forman-Kay JD. Structural and dynamic characterization of the phosphotyrosine binding region of a Src homology 2 domain--phosphopeptide complex by NMR relaxation, proton exchange, and chemical shift approaches. Biochemistry. 1995;34:11353–11362. doi: 10.1021/bi00036a008. [DOI] [PubMed] [Google Scholar]
- 61.Henry GD, Sykes BD. Determination of the rotational dynamics and pH dependence of the hydrogen exchange rates of the arginine guanidino group using NMR spectroscopy. J. Biomol. NMR . 1995;6:59–66. doi: 10.1007/BF00417492. [DOI] [PubMed] [Google Scholar]
- 62.Petkova AT, Hu JG, Bizounok M, Simpson M, Griffin RG, Herzfeld J. Arginine activity in the proton-motive photocycle of bacteriorhodopsin: solid-state NMR studies of the wild-type and D85N proteins. Biochemistry. 1999;38:1562–1572. doi: 10.1021/bi981968z. [DOI] [PubMed] [Google Scholar]
- 63.Marassi FM, Opella SJ. A solid-state NMR index of helical membrane protein structure and topology. J. Magn. Reson. 2000;144:150–155. doi: 10.1006/jmre.2000.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang J, Denny J, Tian C, Kim S, Mo Y, Kovacs F, Song Z, Nishimura K, Gan Z, Fu R, Quine JR, Cross TA. Imaging membrane protein helical wheels. J. Magn. Reson. 2000;144:162–167. doi: 10.1006/jmre.2000.2037. [DOI] [PubMed] [Google Scholar]
- 65.Jogini V, Roux B. Dynamics of the Kv1.2 voltage-gated K+ channel in a membrane environment. Biophys. J. 2007;93:3070–3082. doi: 10.1529/biophysj.107.112540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schmidt D, Jiang QX, MacKinnon R. Phospholipids and the origin of cationic gating charges in voltage sensors. Nature. 2006;444:775–779. doi: 10.1038/nature05416. [DOI] [PubMed] [Google Scholar]
- 67.Cornelius F, Mahmmoud YA. Modulation of FXYD Interaction with Na,K-ATPase by Anionic Phospholipids and Protein Kinase Phosphorylation. Biochemistry. 2007 doi: 10.1021/bi062239j. [DOI] [PubMed] [Google Scholar]
- 68.Lifshitz Y, Petrovich E, Haviv H, Goldshleger R, Tal DM, Garty H, Karlish SJ. Purification of the human alpha2 Isoform of Na,K-ATPase expressed in Pichia pastoris. Stabilization by lipids and FXYD1. Biochemistry. 2007;46:14937–14950. doi: 10.1021/bi701812c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.