Abstract
Cancer cells are distinguished from each other and from healthy cells by features that drive clonal evolution and therapy resistance. New advances in high-dimensional flow cytometry make it possible to systematically measure mechanisms of tumor initiation, progression, and therapy resistance on millions of cells from human tumors. Here we describe flow cytometry techniques that enable a ‘single-cell systems biology’ view of cancer. High-dimensional techniques like mass cytometry enable multiplexed single-cell analysis of cell identity, clinical biomarkers, signaling network phospho-proteins, transcription factors, and functional readouts of proliferation, cell cycle status, and apoptosis. This capability pairs well with a signaling profiles approach that dissects mechanism by systematically perturbing and measuring many nodes in a signaling network. Single-cell approaches enable study of cellular heterogeneity of primary tissues and turn cell subsets into experimental controls or opportunities for new discovery. Rare populations of stem cells or therapy resistant cancer cells can be identified and compared to other types of cells within the same sample. In the long term, these techniques will enable tracking of minimal residual disease (MRD) and disease progression. By better understanding biological systems that control development and cell-cell interactions in healthy and diseased contexts, we can learn to program cells to become therapeutic agents or target malignant signaling events to specifically kill cancer cells. Single-cell approaches that provide deep insight into cell signaling and fate decisions will be critical to optimizing the next generation of cancer treatments combining targeted approaches and immunotherapy.
1 Introduction
Single-cell approaches reveal the heterogeneity inherent in primary tissues and tumors and provide the means to characterize complex phenotypes, isolate rare populations, and dissect underlying mechanisms. Especially critical for cancer research is the ability to track mutations and epigenetic events that confer hallmark attributes required for aggressive growth, malignancy, and therapeutic resistance (Hanahan and Weinberg, 2011). These changes impact network architecture and confer signatures that can be associated at the single-cell level with clinical features of each patient's disease (Irish et al., 2006a). Nearly all cellular features relevant for cancer research can now be measured on a per-cell basis (Table 1). A major advantage of a multidimensional, single-cell approach is that it allows determination of whether an abnormal trait in cancer, such as oncogenic signaling or a gene mutation, exists in all cells or is restricted to a cell subset (Fig. 1). As each piece of knowledge added per cell can dramatically improve the power to understand an experimental result (Krutzik et al., 2004), there has been a drive to expand the number of simultaneous per-cell measurements that can be made (Perfetto et al., 2004, Bendall et al., 2011). The creation of single-cell network profiling techniques has led to important breakthroughs in blood cancer, where flow cytometry techniques are straightforward to apply (Irish et al., 2006a). There is an urgent need now to apply these tools to the challenges of early detection and analysis of solid tumor cell signaling, tumor immunity, transformation to aggressive disease, and metastasis. High-dimensional flow cytometry approaches complement rapidly developing multiplex imaging cytometry tools (Gerner et al., 2012, Gerdes et al., 2013) and single-cell genetic tools (Kalisky and Quake, 2011, Wu et al., 2014). The promise of these techniques for precision medicine is great, but they also create the challenge of integrating results from multiple high-dimensional, single-cell quantitative techniques. Here we provide a primer for applying high-dimensional, single-cell flow cytometry in translational cancer research.
Table 1. Detecting cancer hallmarks in single cells.
| Cell property* | Example flow cytometry method (and referenced use in cancer) |
|---|---|
| Differentiation / Lineage | Antibodies against c-KIT (Wozniak and Kopec-Szlezak, 2004), CD34 (stem cells) (Holyoake et al., 1999, Wozniak and Kopec-Szlezak, 2004, Robillard et al., 2005); antibodies against CD38 (Robillard et al., 2005) or CD20 (Robillard et al., 2005, Irish et al., 2010) and other cluster of differentiation (CD) antigens in human (Mason et al., 2002, van Dongen et al., 2012, Amir el et al., 2013) and mouse (Van Meter et al., 2007, Mayle et al., 2013) tumor and blood cancer tissue samples |
| DNA Content (aneuploidy, DNA fragmentation) | Propidium iodide (PI)(O'Brien and Bolton, 1995), ethidium monoazide (O'Brien and Bolton, 1995), or 7-actinomycin D (7-AAD) (O'Brien and Bolton, 1995, Holyoake et al., 1999) staining of DNA; flow cytometry and FISH to evaluate telomere length (Baerlocher et al., 2006);γH2AX foci indicating DNA double-strand break repair (Huang et al., 2003, Bourton et al., 2012); rhodium and iridium metal intercalators (Ornatsky et al., 2008) |
| RNA Content (quiescence) | Pyronin Y (Holyoake et al., 1999) staining of RNA |
| Cell Cycle Stage | Antibodies against cyclin D (Holyoake et al., 1999), cyclin A (Juan et al., 1998), cyclin B1 (Juan et al., 1998), cyclin E (Erlanson and Landberg, 1998); phosphorylated histone H3 (M phase) (Juan et al., 1998); all cell cycle stages (Behbehani et al., 2012) |
| Proliferation | Bromo-deoxyuridine staining for newly replicated DNA (Robillard et al., 2005); antibodies against proliferating cell nuclear antigen (PCNA) (Castillo et al., 2000), antibodies against Ki67 (Holyoake et al., 1999, Castillo et al., 2000); carboxy-fluoresce in diacetate succinimidyl ester (CFSE) dye (Cooperman et al., 2004) |
| Oncogene Expression | Antibodies against BCL2 (Laane et al., 2005, Robillard et al., 2005, Irish et al., 2010), c-MYC (Morkve et al., 1992), RAS (Andreeff et al., 1986) |
| Mutations | Antibodies against mutant p53 (Zheng et al., 1999), H-Ras-Val12 (Carney et al., 1986) |
| Tumor Suppressor Activity | Antibodies against p53 protein (Zheng et al., 1999, Krutzik et al., 2004) or p21/Waf1 promoter activity driving GFP (p53R-GFP system) (Ohtani et al., 2004)‡; antibodies against phosphorylated p53 (Krutzik et al., 2004, Irish et al., 2007) or phosphorylated Rb (Behbehani et al., 2012); |
| Apoptotic Cell Death | Antibodies against Caspase 3 cleavage products (Belloc et al., 2000) |
| Cell Membrane Changes, Viability,& Necrosis | Annexin V (Belloc et al., 2000) staining for extracellular phosphatidylserine exposure, which occurs on apoptotic cells; detection of membrane permeability by PI dye exclusion (Nicoletti et al., 1991) or Alexa dye exclusion (Table 1); cisplatin exclusion (Fienberg et al., 2012) |
| Metabolism & Redox State | Dichlorofluoresceine diacetate (DCF-DA) staining (Armstrong et al., 2002), a measure of oxidation; monobromobimane (MBrB) staining (Chow and Hedley, 1995), a measure of glutathione; lipophilic fluorochrome dihexaoxacarbocyanine iodide (DiOC6 (3)) (Belloc et al., 2000), a measure of mitochondrial membrane potential; mitochondria peroxy yellow 1 (MitoPY1), a fluorescent probe to quantify hydrogen peroxide levels in living cells (Dickinson and Chang, 2008) |
| Tumor Antigens | Antibodies against B (Timmerman et al., 2002) or T (Maecker and Levy, 1989) cell receptor idiotype; tetramers against tumor antigen (e.g., tyrosinase) specific T cells (Lee et al., 1999) |
| Signaling Activity | Antibodies against phosphorylated STAT and MAPK proteins (Irish et al., 2004b, Van Meter et al., 2007, Kotecha et al., 2008), phosphorylated NF-κB, AKT, S6, Src family kinases (SFKs), and many more (Irish et al., 2010, Bendall et al., 2011); response to drug treatment (Krutzik et al., 2008, Bodenmiller et al., 2012); Indo-1 staining for Ca++ flux (Trentin et al., 2004); antibodies against IL-12 (Panoskaltsis et al., 2003), IFN-γ (Lee et al., 1999) or other cytokines |
Deep profiling enables >36 of such features to be measured on single cells (Bendall and Nolan, 2012, Bendall et al., 2012). Adapted from (Irish et al., 2006a).
Fig. 1.
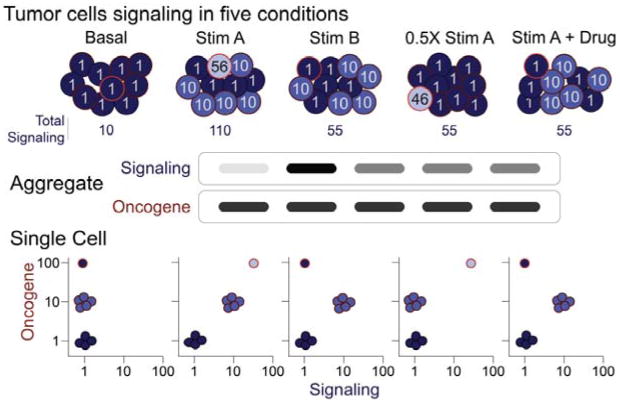
Multidimensional single-cell analysis pinpoints tumor cell signaling. In this example of 10 representative tumor cells analyzed under five stimulation conditions, oncogene expression marks three distinct populations of cells with contrasting signaling responses. In the top row, the number in each cell indicates the level of signaling in that cell under each condition. These values lead to the results shown as “Signaling”. An aggregate analysis might mistakenly be interpreted to suggest that three of the conditions (Stim B, 0.5× Stim A, and Stim A + Drug) elicited the same signaling responses. However, the single-cell view reveals key subset-specific signaling differences. For example, the signal from Stim B is not half as effective as Stim A. Stim B is completely effective at stimulating one subset and ineffective at stimulating another. The oncogene-high cells are hypersensitive to Stim A and non-responsive to Stim B. Similarly, the partial effect of the Drug is due to complete inhibition of one subset and no inhibition of another. Adapted from (Krutzik et al., 2004).
2 Single-Cell Quantification of Cancer Hallmarks
A vast array of cellular features can now be detected by flow cytometry (Table 1). Using mass cytometry and other high-dimensional techniques enables sets of 30 or more of these features to be measured at the single-cell level simultaneously. Each new feature measured brings the potential to better dissect the cellular heterogeneity of a tumor (Fig 1). These features can be generally categorized as markers of cell identity, surrogate markers, and effectors. Effectors differ from surrogate markers in that they directly measure a mechanistically important part of a cellular process such as signaling (MEK phosphorylation), apoptosis (caspase 3 cleavage), or proliferation (cyclin D). Surrogate markers have been shown to correlate with an outcome under some circumstances but they are not thought to be effectors of that outcome. An example of a surrogate marker of cancer stem cells is CD133: CD133 does not confer stem-ness but rather tends to enrich for cancer stem cells. High-dimensional single-cell analysis allows simultaneous quantification of many effectors of different cellular processes in all major cell types present in a sample.
In addition to measuring extracellular antigens or using live-cell permeable, non-toxic reagents, cytometry can quantify intracellular molecules and signaling activity in fixed and permeabilized cells, allowing targets in the cytoplasm and nucleus to be detected. Examples of intracellular targets include proteins with roles in metabolic potential (Armstrong et al, 2002, Chow and Hedley, 1995, and Belloc et al., 2000), phosphorylation induced signal transduction (Irish et al, 2004), and cytokine secretion (Panoskaltsis et al, 2003 and Lee et al, 1999).
As the technology to measure signaling has developed, it has aided in the development of computational modeling of biological networks in cancerous and healthy cells (Sachs et al., 2005). With the ability to quantitatively measure large sets of features simultaneously, this could lead to the systematic identification of clinically relevant signaling targets in a precision medicine setting where therapy is matched to the exact changes observed in the patient's cells. A single-cell view is critical to this, as drug responses in cell subsets are obscured when populations are analyzed in aggregate (Fig. 1).
Although a number of techniques can be used to measure certain features of cells, pragmatic concerns direct choice in many experiments. The detection techniques available to measure these features vary greatly in the amount of crosstalk that will be observed when measuring these features in combinations. A central challenge going forward is to quantitatively measure large sets of features in ways so that crosstalk between the measured channels is minimized. For example, loss of cell membrane integrity – a common surrogate for cell viability (Table 1) – should be routinely included and can be measured many different ways that have different impacts on experiment design. In traditional flow cytometry, exclusion of fluorescent molecules like propidium iodide (PI) (Nicoletti et al., 1991), 7-aminoactinomycin D (7-AAD) (Schmid et al., 1992), and Alexa dye succinimidyl esters (SE) (Krutzik and Nolan, 2006) is commonly used to detect cells lacking an intact membrane. However, PI has very broad excitation and emission spectra that greatly limits the use of additional fluorochromes detected at >550 nm.
As an alternative to PI or 7-AAD, Alexa 700 SE (Ax700-SE) can be used as a viability test (Box 1) in a manner analogous to the fluorescent cell barcoding protocol previously described (Krutzik and Nolan, 2006). The Alexa dyes can be used to minimize crosstalk from the viability detection channel into other channels or to allow staining for other targets of interest on specific channels occupied by PI or 7-AAD. Sequential use of spectrally distinct Alexa dyes can be used to track changes in viability over time. In mass cytometry, a rhodium or iridium nucleic acid intercalator (Ornatsky et al., 2008) or cisplatin (Fienberg et al., 2012) can be used in a similar manner to detect cells lacking an intact plasma membrane. Detection of dead cells is especially critical when working with necrotic tumor tissue and samples from patients undergoing therapy. While centrifugation at ∼180 × g is typical for live cells, centrifugation at ∼830 × g is recommended to effectively pellet dead and fixed cells.
Box 1.
Guidelines for titrating antibodies
Titrate antibodies in house using actual experimental conditions.
Mix positive and negative cells to create a signature pattern for titrations.
Use well characterized cells for titrations (not rare cells of interest).
Select optimal instrument channels for titrating reagents.
It may be necessary to titrate multiple clones under multiple perm conditions for intracellular epitopes that have not been widely studied.
It is often useful to measure cellular features that maintain or oppose tumor growth, such as proliferation, apoptosis, and cell cycle status (Table 1). Detection of bromo-deoxyuridine (BrdU) incorporation into newly replicated DNA (Robillard, 2005) and Ki67 (Holyoake et al., 1999 and Castillo et al., 2000), a protein strictly associated with proliferation (Scholzen and Gerdes, 2000), remain common indicators of proliferation. Apoptotic cell death is frequently measured by activation of the cleaved caspase 3 or by analysis of cell membrane changes like phosphatidylserine exposure (Belloc et al, 2000). In addition, experimental drug studies with chemotherapeutics and ionizing radiation have shown that cell cycle status play critical roles in maintaining tumor homeostasis. Cytometry has explored the therapeutic implications of cells in various states of the cell cycle by revealing quiescent cells kept in a drug-tolerant state. These cells can be identified by pyronin Y staining of RNA or by the abundance of cyclins that regulate cell cycle status (Holyoake et al, 1999, Juan et al, 1998, and Erlanson and Landberg 1998). To delineate cell cycle stages by mass cytometry, 5-iodo-2-deoxyuridine (IdU) is used to mark cells in S phase and G0/G1 cells are detected using antibodies against retinoblastoma protein (Rb) phosphorylated at serines 807 and 811 (Behbehani et al., 2012).
3 Dissecting Abnormal Signaling Networks
Genetic and epigenetic alterations in cancer cells lead to sustained changes in basal signaling and signaling responses (Fig. 2). The vast majority of driver mutations in cancer effect profound changes in cell signaling networks (Irish et al., 2006a). These observations indicate that differential activation of signaling pathways play critical roles in determining a cell's chance for survival or death. Epigenetic changes are also a potent force in shaping the structure of signaling networks in healthy development and cancer. Gain or loss of intercellular signaling interactions, activation of receptors whose signaling controls cell identity, and drug treatments can all trigger sustainable patterns of signaling that persist through cell division or isolation of those cells in culture. Epigenetic reprogramming of signaling networks is a primary mechanism of patterning in healthy development. B and T lymphocytes are an exception in that genetic changes are a mechanism driving healthy development and differentiation. As tools to sequence DNA and RNA continue to improve in speed, read depth, and single-cell precision (Marcy et al., 2007, Dalerba et al., 2011, Powell et al., 2012, Wu et al., 2014), genomic and proteomic tools for studying signaling network activity, transcription factor binding, and DNA methylation typically require tens of millions of cells for one test and are restricted to aggregate analysis (Fig. 1).
Fig. 2.
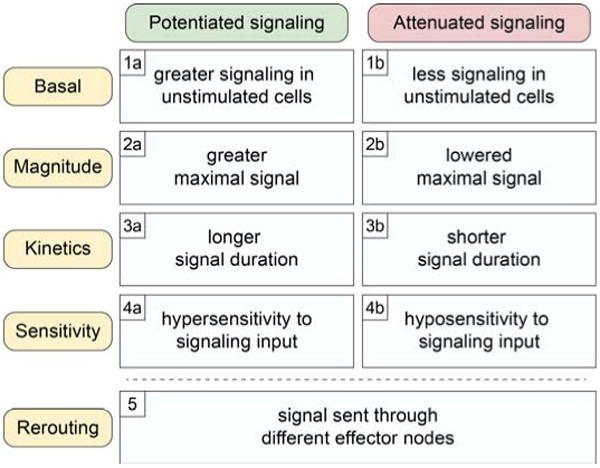
Abnormal signaling in cancer cell networks. Gains and losses of signaling drive oncogenesis and tumor progression. This figure classifies commonly observed signaling alterations according to direction (potentiated or attenuated) and mechanism. Basal signaling disruptions are commonly observed in cancer cells, and the signaling networks of the most negative prognostic cells typically display altered responses to environmental cues. Refer to (Irish et al., 2006a) for example cancer hallmark signaling changes conferred by gene mutations.
High-dimensional flow cytometry addresses this critical technology gap by quantifying single-cell epigenetic changes encoded by altered signaling mechanisms that transform cell function and fate (Fig. 2). Abnormal signaling in cancer can be viewed as changes in the function of signaling nodes within a network (Irish et al., 2006a). These changes are encoded by mechanisms such as constitutive basal activation of an oncogenic kinase (Fig. 2, 1a), loss of a tumor suppressor phosphatase (Fig. 2, 1b), or hypersensitivity to growth factor stimulation (Fig. 2, 4a). The signaling event can be either potentiated (strengthened) or attenuated (weakened), and these changes can have dramatic impacts on the overall function of the signaling network and the cell. Example signaling alterations in cancer that represent these mechanisms are highlighted in the following sections of this chapter.
Fig. 4.
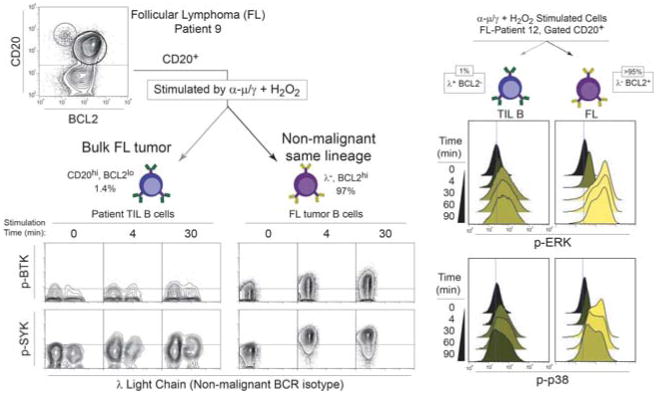
Identifying contrasting signaling in cancer and non-malignant cells of same lineage cell within a tumor. In this example, non-malignant tumor infiltrating lymphocyte (TIL) B cells are detected within follicular lymphoma B cell tumors from two patients. On the left, non-tumor cells were identified by the expression of the “wrong light chain” – a B cell receptor immunoglobulin light chain of a different isotype from the clonal tumor – combined with high CD20 expression and a lack of BCL2 expression. Here we can see that these cells have a distinct SYK and BTK signaling profile that contrasts with the bulk tumor. The histogram overlays on the right show potentiated magnitude and kinetics of ERK and p38 phosphorylation in lymphoma B cells (right side, BCL2+) vs. TIL B cells (left side, identified as λ+ non-tumor light chain and BCL2-).
To understand changes in regulation of signaling it is important to determine how signaling responses differ in cancer cells. A starting point to consider before analyzing a cell's entire signaling network is to identify signaling inputs that individually activate signaling nodes. In this method, cells treated with a stimulus typically serve as positive controls for signaling activity, whereas cells in a basal state function as negative controls. For constitutively active pathways, use of signaling node inhibitors may be necessary. Attention to inhibitor specificity and concentration should considered, as the signaling response may be the result of off-target effects in a signaling network (Bodenmiller et al., 2012). With this methodology, it is possible to reveal clinically relevant signaling profiles by comparing signaling networks among patients with different clinical outcomes (Fig. 3).
Fig. 3.
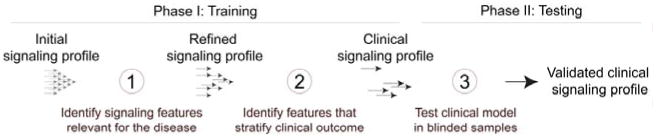
Discovery and validation of a clinical signaling profile. During the training phase, many hypotheses are tested as signaling is assessed at many nodes under a large number of conditions (basal, various signaling activators, doses, time points, drugs, and combinations). The signaling profile is then refined by determining which features differed in the experimental group (cancer) relative to controls (healthy). This feature selection step is based on the biosignature hypothesis (Irish et al., 2004a), which proposes that features that vary as much in the control group as they do in the experimental group are not likely to productively contribute to unsupervised stratification because they are not specific to the experimental group. Models based on one or more features are then built, and it is determined whether they stratify a feature of interest such as clinical outcome. This clinical signaling profile is then tested in a new set of samples comparable to the first and balanced for potential confounders. Ideally the test is performed by a new investigator or a computer algorithm that is blinded to the outcomes.
There are two main phases in the generation of a validated signaling profile (Fig. 3): (1) the training phase, which has the goal of hypothesis generation and new discovery and (2) the testing phase, which is a focused challenge of a small number of hypotheses identified during testing. Development of a signaling profile begins with assembling a list of measurable features and deciding how to organize the staining panels to maximize the information gained while minimizing issues like channel crosstalk. Features are then selected according to the biosignature hypothesis, refined for clinical relevance, and tested in a new set of samples (Fig. 3).
4 Single Cell Detection, Diagnosis, and Prognosis
The ability to measure multiple biomarkers per cell is particularly valuable in the study of genetically unstable tumors where new cell subsets continue to arise over time. Furthermore, cancer cells may resemble non-malignant tumor infiltrating cells of the same lineage (Fig. 4), and detection of multiple features per cell can help clarify each cell's identity. In this example from B cell follicular lymphoma, expression of CD20, the oncogene BCL2, and BCR light chain isotype (κ or λ) were all used to distinguish tumor B cells from non-tumor host B cells. Normally B cells exhibit a mixture of κ and λ light chains, but in lymphoma it is common for >95% of B cells to be a clonal expansion of a cancer cell with just one isotype. In a simple four-color panel it is possible to detect three identity markers and one phospho-protein signaling event (Fig. 4). Here, greater than normal ERK, BTK, SYK, and p38 signaling responses were identified specifically in the tumor B cells. Along with a greater magnitude of signaling potential, tumor cells sustained signaling for a significantly longer period (Irish et al., 2006b). This and other studies of BCR signaling in cancer have highlighted BCR signaling as a target for therapeutic discovery (Rickert, 2013). Recently, targeting BTK has shown great promise in B cell malignancies (Byrd et al., 2013, Wang et al., 2013).
A key advantage of mass cytometry is that many surface and signaling markers can be simultaneously detected. In the fluorescence example (Fig.5), different individual signaling readouts were repeated paired with the same three cell identity markers across four redundant staining panels in order to measure four phospho-proteins. A critical problem with this approach is that one cannot compare signaling vs. signaling in the same cell – the comparison must be made at the population level. With mass cytometry, 20 markers of identity can be paired with 14 phospho-proteins in a 34-dimensional panel. This removes redundant staining panels, conserves sample, and creates higher quality data. In cases where altered signaling distinguishes cancer cells from healthy cells (Fig. 4 and Fig. 5), mass cytometry may make it possible to quickly and accurately diagnose patients based on a flow cytometry signaling profile.
Fig. 5.
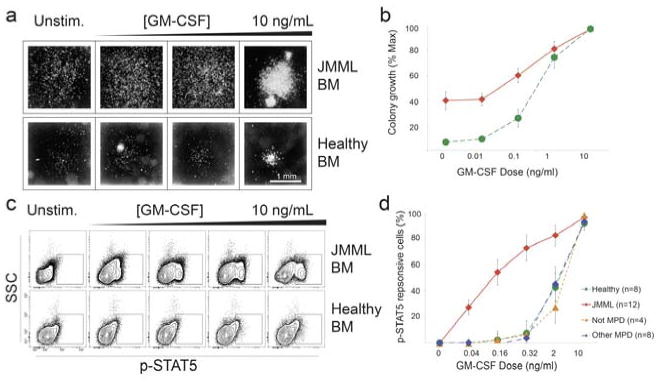
Hypersensitivity to a signaling input is diagnostic for JMML. a Previously, 3-4 weeks were required to confirm a suspected diagnosis of JMML with a granulocyte-macrophage colony-forming units (CFU-GM) assay. In the CFU assay, bone marrow cells from healthy donors (green curve) and patients have different responses to GM-CSF. b Plot of colony growth vs. GM-CSF dose in healthy volunteers (green) and patients (red).c By flow cytometry, a hypersensitive population of JMML cells is detected in cancerous bone marrow compared to the normal control. d A dose-dependent increase in hypersensitive activity of p-STAT5 uniquely distinguished JMML from other myeloproliferative disorders as well as healthy patients. Adapted from (Kotecha et al., 2008).
Juvenile myelomonocytic leukemia (JMML) has historically diagnosed and confirmed with a granulocyte-macrophage colony forming units (CFU-GM) assay ((Emanuel et al., 1991) and Fig. 5). The disadvantage of this approach is that 3-4 weeks are required to confirm the diagnosis when the potentially curative therapy for JMML is an early allogeneic stem cell transplant. While it had become clear that RAS signaling dysregulation occurs in at least 75% of JMML (Flotho et al., 2007), the role of STAT5 activation had not been investigated. In a study that used single-cell profiling of JMML patient blood and bone marrow samples, a small proportion of CD33+, CD14+, CD38dim cells exhibited hypersensitive p-STAT5 responses in response to sub-maximal concentrations of GM-CSF (Kotecha et al., 2008). This diagnostic approach was recently independently validated (Hasegawa et al., 2013). Thus, phospho-flow cytometry provides a precise readout for the aberrant signaling in JMML that distinguishes JMML from both healthy subjects and from patients with other myeloproliferative disorders. Analysis of cell subpopulations associated with disease opens opportunities for quick detection of minimal residual disease (MRD) and has potential to assess therapeutic resistance (Kotecha, 2008). The application to MRD is especially important in the clinical setting of cancer chemotherapy, and a vital need exists for flow cytometry tools that track and automatically identify MRD using surface markers or signaling events (Amir el et al., 2013).
5 Predicting Therapy Response and Tracking Evolution
Surface and signaling-based single-cell analysis can track the abundance of malignant cells at diagnosis and spot the emergence of drug-resistant cells over time during treatment. An example of this is the detection of a clinically significant tumor cell subset of lymphoma cells defined by altered BCR signaling (Fig. 6). Following α-BCR stimulation, several phospho-epitopes had impaired BCR signaling responses in a subset of cells termed lymphoma negative prognostic (LNP) cells. The presence of BCR-insensitive LNP cells was negatively correlated with overall patient survival and LNP cells increased in abundance following treatment and disease progression (Irish et al., 2010). These results indicate that BCR-insensitive LNP cells may have a selective survival advantage compared with bulk tumor B cells (Fig. 6). The close associations between the signaling profiles and risk of death strongly suggest that these cells are therapy insensitive due to specific changes to cell signaling. Perturbing cells with an input stimulus to observe differential activation of signaling networks in cancer has repeatedly been shown to stratify survival (Irish et al., 2004a, Irish et al., 2010).
Fig. 6.
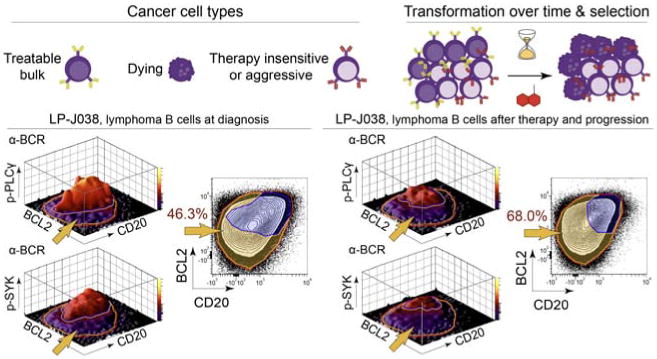
Emergence of a negative prognostic subset over time following treatment. In this example, LNP tumor cells from lymphoma patient J038 are distinguished by abnormal SYK and PLCγ signaling and differential BCL2 and CD20 expression (gold arrow). At the time of diagnosis, LNP cells constituted only 46.3% of the tumor cells. After therapy and disease progression, LNP cells increased to 68% of the tumor. Each 1% increase in LNP cells is associated with a 2.5% increased risk of death in the following year (p < 0.000005, z-score = 4.68). Adapted from (Irish et al., 2010).
Targeted cancer therapies have advanced rapidly as our understanding of cancer cell-specific signaling alterations has increased (Irish et al., 2006a). Genomic technologies can now identify patterns of gene expression or detect the presence of novel point mutations on a case-by-case basis. This has led to the identification of tumor subclasses and improved understanding of disease biology for appropriate therapies. For example, targeting the overexpression of HER2 with lapatinib or trastuzumab in breast cancer has benefited patients (Schnitt, 2010). However, it can be difficult to target newly discovered mutations, and separating drivers from passengers can be challenging when normal, pre-, and post-treatment sample sets are not available. In contrast, the signaling events measured in phospho-flow panels are typically highly targetable, and in many cases there are drugs available that are already being used in the clinic in other settings.
An alternative strategy is to measure deregulation of an oncogenic pathway by measuring active kinase signaling and a cell networks signaling potential when perturbed (Fig. 7). For example, signaling alterations that predict therapy outcome are observed in acute myeloid leukemia (AML) patient samples. Increased activity of STAT5 and STAT3 activity is known to induce the expression of genes for survival and proliferation. Interferon γ treatment activates STAT1 activity, which can oppose survival by activation of genes involved in antigen presentation to the immune system. Cells from patients who did not respond to induction chemotherapy shared a profile including a critical failure to phosphorylate STAT1 in response to interferon γ (Fig. 7, Therapy-resistant AML cells). Instead of activating STAT1, these cells have rerouted IFNγ signaling to phosphorylate oncogenic STAT5. These results provide a rationale for the investigation of STAT5 inhibition in therapy-resistant AML to improve the outcome of patients with this resistant subset. Thus, a key promise of the signaling profile approach is that observed cancer-specific signaling disruptions are required for cancer cell survival or aggressive behavior.
Fig. 7.
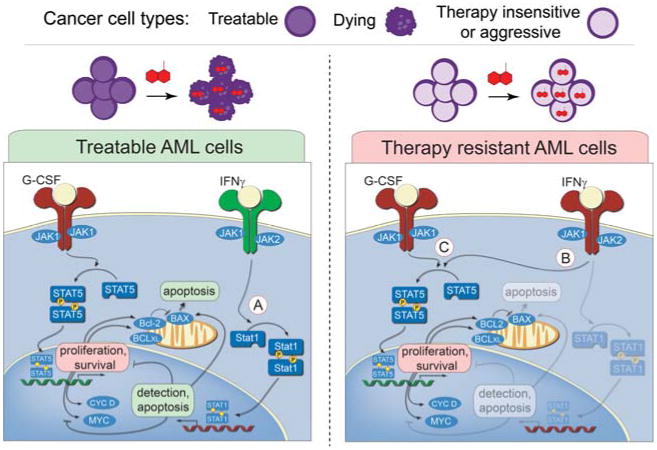
A hallmark mechanism of AML therapy resistance is rewired JAK/STAT signaling. In this example, signaling profiles of two different AML cancer cells are shown. In treatable AML cells, G-CSF signaling through JAK1 and induction of STAT5 phosphorylation mediates transcription of pro-survival and proliferation genes. Conversely, IFNγ signaling through JAK2 results in induction of STAT1 phosphorylation that mediates cell cycle arrest and apoptosis. In the signaling network of the therapy resistant AML cell, the response to IFNγ has become rerouted to STAT5, which, like G-CSF, mediates transcription of pro-survival and proliferation genes. The lack of functional STAT1 activation, which activates cell cycle arrest induced apoptosis, explains why patients with these cancer cells are often resistant to DNA-damage-induction therapy. Inhibition of JAK2/STAT5 signaling in therapy resistant AML cells could potentially improve the outcome of patients with this resistant subset.
6 Experimental and Clinical Considerations
Cytometry can be used to quantitate multiple properties per cell that can then be correlated with biological processes or disease progression. With the ability to simultaneously measure more targets and with the ever-increasing sizes of datasets, experimental design and data processing have become critical aspects of these experiments.
A primary challenge in high-dimensional profiling of heterogeneous cells is optimization of a staining protocol that facilitates the detection of extracellular and intracellular targets of the cells. A target's localization should be considered and a range of appropriate reagents tested in order to develop a protocol that balances speed, reproducibility, and sensitivity. Optimizing signal to noise remains a central goal in fluorescent flow cytometry (Maecker and Trotter, 2006) and mass cytometry (Bendall et al., 2012). This may involve titrating the detection of the target on live cells, after para-formaldehyde fixation, and/or after permeabilization (e.g., methanol, ethanol, saponin, Triton X-100) of the cell membrane (Krutzik and Nolan, 2003). Panels that measure all features except one are a classic flow cytometry control termed ‘fluorescence minus one’ (FMO), described in detail by Maecker and Trotter (Maecker and Trotter, 2006). For mass cytometry, a comparable ‘mass minus one’ (MMO) control is equally valuable for determining what level of signal can reliably be considered positive.
When creating multi-step staining protocols for detection of extracellular and intracellular epitopes, a key advantage of small molecule dyes and the polymer metal chelators used in mass cytometry is that they are not sensitive to the common permeabilization agents. This contrasts with large protein fluorophores; fluorescence of protein fluorophores can be harmed by harsh alcohol treatments used in storage of fixed samples and during permeabilization. In mass cytometry, a multi-step staining protocol is a common alternative to seeking epitope unmasking staining conditions that work well for a variety of epitopes that are localized in different cellular compartments and differentially dependent on three-dimensional conformation. In a typical signaling experiment, surface marker staining occurs after the cells have been fixed so that detection of cell identity does not alter signaling. However, since many surface marker target epitopes are no longer detectable following harsh permeabilization conditions, surface staining occurs immediately following the short fix step that stops signaling in the phospho-flow protocol (1.6% paraformaldehyde for 5 minutes at room temperature). Thus, surface staining occurs following stimulation/fixation and prior to methanol permeabilization. For more information, see Table I (Krutzik et al., 2005) and Fig. 2 (Krutzik and Nolan, 2003). For certain intracellular targets – especially transcription factors – permeabilization with saponin or Triton X-100 can yield superior staining. Usually a short formaldehyde fix (≤ 10 minutes) does not destroy target epitopes and detection of surface markers is decreased by an acceptable ∼10% of the original signal, although there are exceptions.
Antibody titration and staining optimization should follow well-established guidelines (Box 1). It is critical to titrate antibodies in the exact conditions that they will be used and to include populations of positive and negative control cells at known ratios. The stain index between positive and negative cells allows verification of the subset pattern. It is not sufficient to titrate an antibody on a uniform positive population while using unstained or isotype control stained cells as a comparison point. It is acceptable for the positive and negative cells to be in different tubes, but the advantage of staining all the cells simultaneously in multidimensional cytometry is lost. With intracellular work: less is more. Problems are typically due to over-staining, which leads to non-specific background signal (see Figure 2 in (Krutzik and Nolan, 2003)). Antibodies that work well by immunofluorescence nearly always are suitable for fluorescent flow and mass cytometry when the same fixation, permeabilization, and staining conditions are used.
For all types of cytometry, internal biological control populations are ideal controls. Intracellular controls transform the cellular heterogeneity that confounds aggregate approaches (Fig. 1) into a distinct advantage of single-cell approaches. Markers of stem-ness, such as CD34 (Woziniak and Kopec-Szlezak, 2004), and lineage-restricted molecules expressed during differentiation (Mason et al., 2002) help determine the identity of tumor cells. However, developmental programs can be aberrantly activated or suppressed in both the cancer cells and the surrounding microenvironment. Because phenotypic plasticity characterizes cancer, it is especially valuable to have multiple markers that are expected to be positive and negative on each major tumor and host-cell population. A general rule is to include two positive markers and one negative marker for each major tumor and host cell type (Irish et al., 2010). Negative markers help rule out artifacts. In immune cancers, markers of clonality can be used to confirm cancer cell identity or dissect cancer cell lineage (Irish et al., 2006b, Sachen et al., 2012, Green et al., 2013). Cell isolation by fluorescence-activated cell sorting followed by sequencing for oncogenic mutations can confirm the identity of cancer cells or be used to identify underlying driver and passenger mutations (Green et al., 2013). Ultimately, the more features detected (Table 1), the more confidence one has in the identity and biology of a given population during the discovery or training phases of a project (Fig. 3). Cytometry provides the toolkit for tracking and characterizing the ubiquitous heterogeneity of cancer.
The dysregulated intracellular signaling observed in cancer cells contrasts greatly from signaling in normal cells. Challenging of the cancer cells with perturbation reagents can reveal divergent response patterns. Even when the mechanism is not directly inferable, analysis of multiple signaling events can identify the point in a cellular system that is dysregulated. To develop a protocol that profiles signaling responses, comparison to a healthy population of cells, such as peripheral blood mononuclear cells or a tractable genetically modified cell line, often establishes a comparison point for how intracellular systems should behave.
After acquiring a large dataset, data interpretation can be a challenging hurdle in high-dimensional experiments. Traditional multi-parameter techniques like flow cytometry have relied upon two-dimensional plots to visualize the data to understand correlations between the parameters. Unfortunately, as the number of parameters increase, the number of two-dimensional plots increase to create an overwhelming visualization problem. Analytical approaches developed to tackle this complexity include dimensional reduction tools such as SPADE (Bendall et al., 2011, Qiu et al., 2011) and viSNE (Amir el et al., 2013). To achieve a greater understanding of tumor proteins and signaling, these tools can be used to then computationally compare this new view of cancer across patients and tumor subtypes.
7 Future Perspectives
Going forward, the field must address a number of challenges in data analysis and platform integration raised by the increased power to simultaneously detect many features of single cells. Four key areas are:
Data analysis, storage, and sharing with collaborative teams.
Cross-platform comparisons with other systems biology techniques.
Cross-scale data integration, especially between single cells and aggregates.
Comparisons across time, especially in clinical studies.
Technical tools and experimental designs have far outpaced the existing computational tools. Many are working to address this need, but it is important to go beyond the basic challenge of clustering groups of cells by similar features. Tools for identification of populations within single-cell datasets have increased dramatically in sophistication and speed (Pyne et al., 2009, Qiu et al., 2011, Aghaeepour et al., 2013, Amir el et al., 2013), and now there is an urgent need for tools that model the populations and derive biological meaning from the markers used to find populations. It is critical to make sure that tools do not find populations in such a way that they are limited to a particular dataset. This is vital for reproducibility as well as for clinical application. In the end, it is critical to define the difference metric in terms of the underlying biological mechanisms and to refine the model to the minimal parts for clinical testing.
How do we connect measurements made at the single-cell level with knowledge gained using other aggregate analysis tools? Single-cell techniques have essentially been developing independently of aggregate analysis tools because it is unclear as to how to connect the information gained at such different scales. Thus, approaches to span experimental platform and biological scale are sorely needed for the next generation of single-cell opportunities in cancer biology (Fig. 8).
Fig. 8.
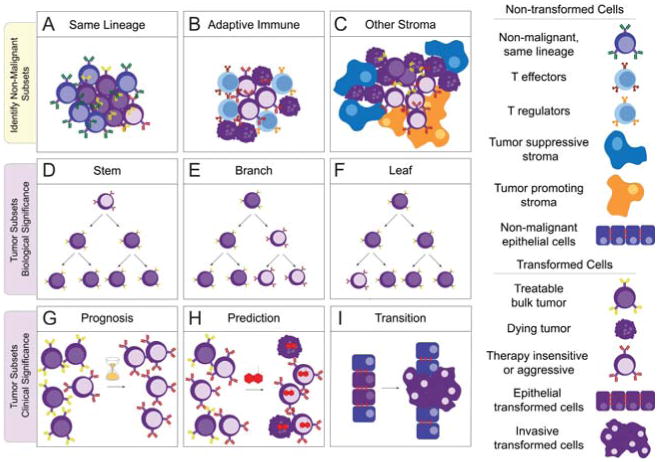
Key single-cell opportunities in cancer research. The first row depicts the opportunities of detecting non-malignant cells of the same lineage as the tumor (A, as in Fig. 5), tumor infiltrating immune responders (B, as in (Myklebust et al., 2013)), and other non-malignant stromal cells (C). It will be important to distinguish between abnormal signaling that promotes cancer, such as inflammation, and abnormal signaling that results from cancer, such as T cell suppression via PD-1 or generation of cancer associated fibroblasts (Barcellos-Hoff et al., 2013). In the second row, (D), (E), and (F) depict contrasting biological origins of an aggressive, therapy-insensitive tumor subpopulation that can be dissected with single-cell tools. A gatekeeper mutation conferring resistance to targeted therapy might be an apomorphy that distinguishes a rare ‘leaf’ subset (F). Alternatively, a slow cell cycle phenotype might distinguish a cancer stem cell (D)(Reya et al., 2001). A large, heterogeneous branch (E) observed at the time of diagnosis might need to be treated with a combination of therapies in order to kill all populations and obtain a clinical response. The third row depicts clinical single-cell opportunities, such as detecting negative prognostic subpopulations (G, as in Fig. 7), treatment insensitive subsets (H), and cellular transitions as would be observed when epithelial cancer cells become an invasive, metastatic population (I).
Increasingly, single-cell tools will need to take into account changes over long periods of time – such as in the case with samples obtained over time during treatment. The concepts of before and after treatment and of subset evolution, emergence, transformation, and metastasis must be considered. What are reliable markers of stable cellular identity and how do we track ‘a population’ of cells over time?
Table 2. Exclusion viability test using Alexa 700 succinimidyl ester (Ax700-SE).
| Step | Details |
|---|---|
| Ax700-SE 50,000× Stock | Dissolve 1 mg Ax700-SE in 0.5 mL dimethyl sulfoxide (DMSO) to achieve a 50,000× long term frozen stock of 2,000 μg/mL. Store frozen and protected from water. |
| Ax700-SE 500× Aliquots | Prepare 500× frozen stocks of 20 μg/mL Ax700-SE in DMSO. A 20-μL aliquot is sufficient to stain approximately 50 experimental samples in 200 μL. |
| Ax700-SE 50× Working | Dilute Ax700-SE in DMSO to prepare a 50× of 2 μg/mL. Do not store. |
| Stain† | Add 4 μ L of 50× working stock of Ax700-SE directly to cells in suspension to achieve a final concentration of 0.04 μg/mL. Stain for 10 min; titrate as needed. |
| Wash & Collect | Wash with 1× PBS§ containing 1% bovine serum albumin (BSA) or other carrier protein. Pellet cells by centrifugation and continue with other staining steps or collect. |
Typically, live cells are stained prior to stimulation and no apparent impact on biology is observed. For a mass cytometry version using cisplatin, refer to (Fienberg et al., 2012).
Sterile filtered phosphate buffered saline (PBS) without calcium or magnesium is recommended.
References
- Aghaeepour N, Finak G, Flow CAPC. Consortium D. Hoos H, Mosmann TR, et al. Critical assessment of automated flow cytometry data analysis techniques. Nat Methods. 2013;10:228–38. doi: 10.1038/nmeth.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir el AD, Davis KL, Tadmor MD, Simonds EF, Levine JH, Bendall SC, et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol. 2013;31:545–52. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeff M, Slater DE, Bressler J, Furth ME. Cellular ras oncogene expression and cell cycle measured by flow cytometry in hematopoietic cell lines. Blood. 1986;67:676–81. [PubMed] [Google Scholar]
- Armstrong JS, Steinauer KK, Hornung B, Irish JM, Lecane P, Birrell GW, et al. Role of glutathione depletion and reactive oxygen species generation in apoptotic signaling in a human B lymphoma cell line. Cell Death Differ. 2002;9:252–63. doi: 10.1038/sj.cdd.4400959. [DOI] [PubMed] [Google Scholar]
- Baerlocher GM, Vulto I, de Jong G, Lansdorp PM. Flow cytometry and FISH to measure the average length of telomeres (flow FISH) Nat Protoc. 2006;1:2365–76. doi: 10.1038/nprot.2006.263. [DOI] [PubMed] [Google Scholar]
- Barcellos-Hoff MH, Lyden D, Wang TC. The evolution of the cancer niche during multistage carcinogenesis. Nat Rev Cancer. 2013;13:511–8. doi: 10.1038/nrc3536. [DOI] [PubMed] [Google Scholar]
- Behbehani GK, Bendall SC, Clutter MR, Fantl WJ, Nolan GP. Single-cell mass cytometry adapted to measurements of the cell cycle. Cytometry A. 2012;81:552–66. doi: 10.1002/cyto.a.22075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belloc F, Belaud-Rotureau MA, Lavignolle V, Bascans E, Braz-Pereira E, Durrieu F, et al. Flow cytometry detection of caspase 3 activation in preapoptotic leukemic cells. Cytometry. 2000;40:151–60. doi: 10.1002/(sici)1097-0320(20000601)40:2<151::aid-cyto9>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Bendall SC, Nolan GP. From single cells to deep phenotypes in cancer. Nat Biotechnol. 2012;30:639–47. doi: 10.1038/nbt.2283. [DOI] [PubMed] [Google Scholar]
- Bendall SC, Nolan GP, Roederer M, Chattopadhyay PK. A deep profiler's guide to cytometry. Trends Immunol. 2012;33:323–32. doi: 10.1016/j.it.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendall SC, Simonds EF, Qiu P, Amir el AD, Krutzik PO, Finck R, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–96. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenmiller B, Zunder ER, Finck R, Chen TJ, Savig ES, Bruggner RV, et al. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat Biotechnol. 2012;30:858–67. doi: 10.1038/nbt.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourton EC, Plowman PN, Zahir SA, Senguloglu GU, Serrai H, Bottley G, et al. Multispectral imaging flow cytometry reveals distinct frequencies of gamma-H2AX foci induction in DNA double strand break repair defective human cell lines. Cytometry A. 2012;81:130–7. doi: 10.1002/cyto.a.21171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd JC, Furman RR, Coutre SE, Flinn IW, Burger JA, Blum KA, et al. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. N Engl J Med. 2013;369:32–42. doi: 10.1056/NEJMoa1215637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney WP, Petit D, Hamer P, Der CJ, Finkel T, Cooper GM, et al. Monoclonal antibody specific for an activated RAS protein. Proc Natl Acad Sci U S A. 1986;83:7485–9. doi: 10.1073/pnas.83.19.7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo R, Mascarenhas J, Telford W, Chadburn A, Friedman SM, Schattner EJ. Proliferative response of mantle cell lymphoma cells stimulated by CD40 ligation and IL-4. Leukemia. 2000;14:292–8. doi: 10.1038/sj.leu.2401664. [DOI] [PubMed] [Google Scholar]
- Chow S, Hedley D. Flow cytometric determination of glutathione in clinical samples. Cytometry. 1995;21:68–71. doi: 10.1002/cyto.990210113. [DOI] [PubMed] [Google Scholar]
- Cooperman J, Neely R, Teachey DT, Grupp S, Choi JK. Cell division rates of primary human precursor B cells in culture reflect in vivo rates. Stem Cells. 2004;22:1111–20. doi: 10.1634/stemcells.22-6-1111. [DOI] [PubMed] [Google Scholar]
- Dalerba P, Kalisky T, Sahoo D, Rajendran PS, Rothenberg ME, Leyrat AA, et al. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat Biotechnol. 2011;29:1120–7. doi: 10.1038/nbt.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson BC, Chang CJ. A targetable fluorescent probe for imaging hydrogen peroxide in the mitochondria of living cells. J Am Chem Soc. 2008;130:9638–9. doi: 10.1021/ja802355u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuel PD, Bates LJ, Castleberry RP, Gualtieri RJ, Zuckerman KS. Selective hypersensitivity to granulocyte-macrophage colony-stimulating factor by juvenile chronic myeloid leukemia hematopoietic progenitors. Blood. 1991;77:925–9. [PubMed] [Google Scholar]
- Erlanson M, Landberg G. Flow cytometric quantification of cyclin E in human cell lines and hematopoietic malignancies. Cytometry. 1998;32:214–22. [PubMed] [Google Scholar]
- Fienberg HG, Simonds EF, Fantl WJ, Nolan GP, Bodenmiller B. A platinum-based covalent viability reagent for single-cell mass cytometry. Cytometry A. 2012;81:467–75. doi: 10.1002/cyto.a.22067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flotho C, Kratz C, Niemeyer CM. Targeting RAS signaling pathways in juvenile myelomonocytic leukemia. Curr Drug Targets. 2007;8:715–25. doi: 10.2174/138945007780830773. [DOI] [PubMed] [Google Scholar]
- Gerdes MJ, Sevinsky CJ, Sood A, Adak S, Bello MO, Bordwell A, et al. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc Natl Acad Sci U S A. 2013;110:11982–7. doi: 10.1073/pnas.1300136110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerner MY, Kastenmuller W, Ifrim I, Kabat J, Germain RN. Histo-cytometry: a method for highly multiplex quantitative tissue imaging analysis applied to dendritic cell subset microanatomy in lymph nodes. Immunity. 2012;37:364–76. doi: 10.1016/j.immuni.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green MR, Gentles AJ, Nair RV, Irish JM, Kihira S, Liu CL, et al. Hierarchy in somatic mutations arising during genomic evolution and progression of follicular lymphoma. Blood. 2013;121:1604–11. doi: 10.1182/blood-2012-09-457283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hasegawa D, Bugarin C, Giordan M, Bresolin S, Longoni D, Micalizzi C, et al. Validation of flow cytometric phospho-STAT5 as a diagnostic tool for juvenile myelomonocytic leukemia. Blood Cancer J. 2013;3:e160. doi: 10.1038/bcj.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holyoake T, Jiang X, Eaves C, Eaves A. Isolation of a highly quiescent subpopulation of primitive leukemic cells in chronic myeloid leukemia. Blood. 1999;94:2056–64. [PubMed] [Google Scholar]
- Huang X, Traganos F, Darzynkiewicz Z. DNA damage induced by DNA topoisomerase I- and topoisomerase II-inhibitors detected by histone H2AX phosphorylation in relation to the cell cycle phase and apoptosis. Cell Cycle. 2003;2:614–9. [PubMed] [Google Scholar]
- Irish J, Hovland R, Krutzik P, Perez O, Bruserud O, Gjertsen B, et al. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell. 2004a;118:217–28. doi: 10.1016/j.cell.2004.06.028. [DOI] [PubMed] [Google Scholar]
- Irish J, Kotecha N, Nolan G. Innovation - Mapping normal and cancer cell signalling networks: towards single-cell proteomics. Nature Reviews Cancer. 2006a;6:146–55. doi: 10.1038/nrc1804. [DOI] [PubMed] [Google Scholar]
- Irish JM, Anensen N, Hovland R, Skavland J, Borresen-Dale AL, Bruserud O, et al. Flt3 Y591 duplication and Bc1-2 overexpression are detected in acute myeloid leukemia cells with high levels of phosphorylated wild-type p53. Blood. 2007;109:2589–96. doi: 10.1182/blood-2006-02-004234. [DOI] [PubMed] [Google Scholar]
- Irish JM, Czerwinski DK, Nolan GP, Levy R. Altered B-cell receptor signaling kinetics distinguish human follicular lymphoma. B cells from tumor-infiltrating nonmalignant B cells. Blood. 2006b;108:3135–42. doi: 10.1182/blood-2006-02-003921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish JM, Hovland R, Krutzik PO, Perez OD, Bruserud O, Gjertsen BT, et al. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell. 2004b;118:217–28. doi: 10.1016/j.cell.2004.06.028. [DOI] [PubMed] [Google Scholar]
- Irish JM, Myklebust JH, Alizadeh AA, Houot R, Sharman JP, Czerwinski DK, et al. B-cell signaling networks reveal a negative prognostic human lymphoma cell subset that emerges during tumor progression. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:12747–54. doi: 10.1073/pnas.1002057107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan G, Traganos F, James WM, Ray JM, Roberge M, Sauve DM, et al. Histone H3 Phosphorylation and Expression of Cyclins A and B1 Measured in Individual Cells During Their Progression Through G2 and Mitosis. Cytometry. 1998;32:71–7. doi: 10.1002/(sici)1097-0320(19980601)32:2<71::aid-cyto1>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Kalisky T, Quake SR. Single-cell genomics. Nat Methods. 2011;8:311–4. doi: 10.1038/nmeth0411-311. [DOI] [PubMed] [Google Scholar]
- Kotecha N, Floress NJ, Irish JM, Simonds EF, Sakai DS, Archambeault S, et al. Single-cell profiling identifies aberrant STAT5 activation in myeloid malignancies with specific clinical and biologic correlates. Cancer Cell. 2008;14:335–43. doi: 10.1016/j.ccr.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutzik PO, Crane JM, Clutter MR, Nolan GP. High-content single-cell drug screening with phosphospecific flow cytometry. Nat Chem Biol. 2008;4:132–42. doi: 10.1038/nchembio.2007.59. [DOI] [PubMed] [Google Scholar]
- Krutzik PO, Hale MB, Nolan GP. Characterization of the murine immunological signaling network with phosphospecific flow cytometry. J Immunol. 2005;175:2366–73. doi: 10.4049/jimmunol.175.4.2366. [DOI] [PubMed] [Google Scholar]
- Krutzik PO, Irish JM, Nolan GP, Perez OD. Analysis of protein phosphorylation and cellular signaling events by flow cytometry: techniques and clinical applications. Clin Immunol. 2004;110:206–21. doi: 10.1016/j.clim.2003.11.009. [DOI] [PubMed] [Google Scholar]
- Krutzik PO, Nolan GP. Fluorescent cell barcoding in flow cytometry allows high-throughput drug screening and signaling profiling. Nat Methods. 2006;3:361–8. doi: 10.1038/nmeth872. [DOI] [PubMed] [Google Scholar]
- Krutzik PO, Nolan GP. Intracellular phospho-protein staining techniques for flow cytometry: monitoring single cell signaling events. Cytometry A. 2003;55:61–70. doi: 10.1002/cyto.a.10072. [DOI] [PubMed] [Google Scholar]
- Laane E, Tani E, Bjorklund E, Elmberger G, Everaus H, Skoog L, et al. Flow cytometric immunophenotyping including Bcl-2 detection on fine needle aspirates in the diagnosis of reactive lymphadenopathy and non-Hodgkin's lymphoma. Cytometry B Clin Cytom. 2005;64:34–42. doi: 10.1002/cyto.b.20043. [DOI] [PubMed] [Google Scholar]
- Lee PP, Yee C, Savage PA, Fong L, Brockstedt D, Weber JS, et al. Characterization of circulating T cells specific for tumor-associated antigens in melanoma patients. Nat Med. 1999;5:677–85. doi: 10.1038/9525. [DOI] [PubMed] [Google Scholar]
- Maecker HT, Levy R. Prevalence of antigen receptor variants in human T cell lines and tumors. J Immunol. 1989;142:1395–404. [PubMed] [Google Scholar]
- Maecker HT, Trotter J. Flow cytometry controls, instrument setup, and the determination of positivity. Cytometry A. 2006;69:1037–42. doi: 10.1002/cyto.a.20333. [DOI] [PubMed] [Google Scholar]
- Marcy Y, Ouverney C, Bik EM, Losekann T, Ivanova N, Martin HG, et al. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc Natl Acad Sci U S A. 2007;104:11889–94. doi: 10.1073/pnas.0704662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason D, Andre P, Bensussan A, Buckley C, Civin C, Clark E, et al. CD antigens 2002. Blood. 2002;99:3877–80. doi: 10.1182/blood.v99.10.3877. [DOI] [PubMed] [Google Scholar]
- Mayle A, Luo M, Jeong M, Goodell MA. Flow cytometry analysis of murine hematopoietic stem cells. Cytometry A. 2013;83:27–37. doi: 10.1002/cyto.a.22093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morkve O, Halvorsen OJ, Stangeland L, Gulsvik A, Laerum OD. Quantitation of biological tumor markers (p53, c-myc, Ki-67 and DNA ploidy) by multiparameter flow cytometry in non-small-cell lung cancer. Int J Cancer. 1992;52:851–5. doi: 10.1002/ijc.2910520603. [DOI] [PubMed] [Google Scholar]
- Myklebust JH, Irish JM, Brody J, Czerwinski DK, Houot R, Kohrt HE, et al. High PD-1 expression and suppressed cytokine signaling distinguish T cells infiltrating follicular lymphoma tumors from peripheral T cells. Blood. 2013;121:1367–76. doi: 10.1182/blood-2012-04-421826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoletti I, Migliorati G, Pagliacci MC, Grignani F, Riccardi C. A rapid and simple method for measuring thymocyte apoptosis by propidium iodide staining and flow cytometry. Journal of Immunological Methods. 1991;139:271–9. doi: 10.1016/0022-1759(91)90198-o. [DOI] [PubMed] [Google Scholar]
- O'Brien MC, Bolton WE. Comparison of cell viability probes compatible with fixation and permeabilization for combined surface and intracellular staining in flow cytometry. Cytometry. 1995;19:243–55. doi: 10.1002/cyto.990190308. [DOI] [PubMed] [Google Scholar]
- Ohtani S, Kagawa S, Tango Y, Umeoka T, Tokunaga N, Tsunemitsu Y, et al. Quantitative analysis of p53-targeted gene expression and visualization of p53 transcriptional activity following intratumoral administration of adenoviral p53 in vivo. Mol Cancer Ther. 2004;3:93–100. [PubMed] [Google Scholar]
- Ornatsky OI, Lou X, Nitz M, Schafer S, Sheldrick WS, Baranov VI, et al. Study of cell antigens and intracellular DNA by identification of element-containing labels and metallointercalators using inductively coupled plasma mass spectrometry. Anal Chem. 2008;80:2539–47. doi: 10.1021/ac702128m. [DOI] [PubMed] [Google Scholar]
- Panoskaltsis N, Reid CD, Knight SC. Quantification and cytokine production of circulating lymphoid and myeloid cells in acute myelogenous leukaemia. Leukemia. 2003;17:716–30. doi: 10.1038/sj.leu.2402835. [DOI] [PubMed] [Google Scholar]
- Perfetto SP, Chattopadhyay PK, Roederer M. Seventeen-colour flow cytometry: unravelling the immune system. Nat Rev Immunol. 2004;4:648–55. doi: 10.1038/nri1416. [DOI] [PubMed] [Google Scholar]
- Powell AA, Talasaz AH, Zhang H, Coram MA, Reddy A, Deng G, et al. Single cell profiling of circulating tumor cells: transcriptional heterogeneity and diversity from breast cancer cell lines. PLoS One. 2012;7:e33788. doi: 10.1371/journal.pone.0033788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyne S, Hu X, Wang K, Rossin E, Lin TI, Maier LM, et al. Automated high-dimensional flow cytometric data analysis. Proc Natl Acad Sci U S A. 2009;106:8519–24. doi: 10.1073/pnas.0903028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu P, Simonds EF, Bendall SC, Gibbs KD, Jr, Bruggner RV, Linderman MD, et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol. 2011;29:886–91. doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–11. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- Rickert RC. New insights into pre-BCR and BCR signalling with relevance to B cell malignancies. Nat Rev Immunol. 2013;13:578–91. doi: 10.1038/nri3487. [DOI] [PubMed] [Google Scholar]
- Robillard N, Pellat-Deceunynck C, Bataille R. Phenotypic characterization of the human myeloma cell growth fraction. Blood. 2005;105:4845–8. doi: 10.1182/blood-2004-12-4700. [DOI] [PubMed] [Google Scholar]
- Sachen KL, Strohman MJ, Singletary J, Alizadeh AA, Kattah NH, Lossos C, et al. Self-antigen recognition by follicular lymphoma B-cell receptors. Blood. 2012;120:4182–90. doi: 10.1182/blood-2012-05-427534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs K, Perez O, Pe'er D, Lauffenburger DA, Nolan GP. Causal protein-signaling networks derived from multiparameter single-cell data. Science. 2005;308:523–9. doi: 10.1126/science.1105809. [DOI] [PubMed] [Google Scholar]
- Schmid I, Krall WJ, Uittenbogaart CH, Braun J, Giorgi JV. Dead cell discrimination with 7-amino-actinomycin D in combination with dual color immunofluorescence in single laser flow cytometry. Cytometry. 1992;13:204–8. doi: 10.1002/cyto.990130216. [DOI] [PubMed] [Google Scholar]
- Schnitt SJ. Classification and prognosis of invasive breast cancer: from morphology to molecular taxonomy. Mod Pathol. 2010;23 Suppl 2:S60–4. doi: 10.1038/modpathol.2010.33. [DOI] [PubMed] [Google Scholar]
- Scholzen T, Gerdes J. The Ki-67 protein: from the known and the unknown. J Cell Physiol. 2000;182:311–22. doi: 10.1002/(SICI)1097-4652(200003)182:3<311::AID-JCP1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Timmerman JM, Czerwinski DK, Davis TA, Hsu FJ, Benike C, Hao ZM, et al. Idiotype-pulsed dendritic cell vaccination for B-cell lymphoma: clinical and immune responses in 35 patients. Blood. 2002;99:1517–26. doi: 10.1182/blood.v99.5.1517. [DOI] [PubMed] [Google Scholar]
- Trentin L, Cabrelle A, Facco M, Carollo D, Miorin M, Tosoni A, et al. Homeostatic chemokines drive migration of malignant B cells in patients with non-Hodgkin lymphomas. Blood. 2004;104:502–8. doi: 10.1182/blood-2003-09-3103. [DOI] [PubMed] [Google Scholar]
- van Dongen JJ, Lhermitte L, Bottcher S, Almeida J, van der Velden VH, Flores-Montero J, et al. EuroFlow antibody panels for standardized n-dimensional flow cytometric immunophenotyping of normal, reactive and malignant leukocytes. Leukemia. 2012;26:1908–75. doi: 10.1038/leu.2012.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Meter MEM, Diaz-Flores E, Archard JA, Passegue E, Irish JM, Kotecha N, et al. K-Ras(G12D) expression induces hyperproliferation and aberrant signaling in primary hematopoietic stem/progenitor cells. Blood. 2007;109:3945–52. doi: 10.1182/blood-2006-09-047530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ML, Rule S, Martin P, Goy A, Auer R, Kahl BS, et al. Targeting BTK with ibrutinib in relapsed or refractory mantle-cell lymphoma. N Engl J Med. 2013;369:507–16. doi: 10.1056/NEJMoa1306220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak J, Kopec-Szlezak J. c-Kit receptor (CD117) expression on myeloblasts and white blood cell counts in acute myeloid leukemia. Cytometry B Clin Cytom. 2004;58:9–16. doi: 10.1002/cyto.b.10068. [DOI] [PubMed] [Google Scholar]
- Wu AR, Neff NF, Kalisky T, Dalerba P, Treutlein B, Rothenberg ME, et al. Quantitative assessment of single-cell RNA-sequencing methods. Nat Methods. 2014;11:41–6. doi: 10.1038/nmeth.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng A, Castren K, Saily M, Savolainen ER, Koistinen P, Vahakangas K. p53 status of newly established acute myeloid leukaemia cell lines. Br J Cancer. 1999;79:407–15. doi: 10.1038/sj.bjc.6690064. [DOI] [PMC free article] [PubMed] [Google Scholar]


