Abstract
BACKGROUND
Clonal evolution is frequently detected in patients developing resistance to imatinib. The outcome of patients with clonal evolution treated with second generation tyrosine kinase inhibitors is not known.
METHODS
The authors analyzed the outcome of 177 CML patients after second tyrosine kinase inhibitor therapy.
RESULTS
Ninety-five patients were in chronic phase, 30 had clonal evolution, 28 were in accelerated phase (AP), and 24 were in AP plus clonal evolution. Major cytogenetic response rates were 58%, 54%, 28%, and 13%; 2-year overall survival (OS) rates were 86%, 73%, 68%, and 33%; and 2-year event-free survival (EFS) rates were 69%, 67%, 31%, and 8%, respectively. The hematologic and cytogenetic response rates, OS, and EFS were no different between patients in chronic phase with clonal evolution and patients with chronic phase and no clonal evolution. However, clonal evolution had a significant adverse impact when associated with other features of AP. On multivariate analysis, clonal evolution had no independently significant effect on achieving major cytogenetic response on the second generation tyrosine kinase inhibitors. The factors predicting increasing major cytogenetic response to second generation tyrosine kinase inhibitors were prior achievement of major cytogenetic response with imatinib, higher hemoglobin levels, no splenomegaly, lower percentage of Philadelphia chromosome-positive metaphases, and no prior chemotherapy.
CONCLUSIONS
Clonal evolution constitutes a heterogeneous entity with variable outcome with second generation tyrosine kinase inhibitors, with trisomy 8, chromosome 17, and complex abnormalities having the worst outcome, regardless of the number of metaphases involved. The molecular events behind these abnormalities and potential therapeutic approaches directed at them need to be defined.
Keywords: clonal expression, clonal evolution, chronic myeloid leukemia, dasatinib, nilotinib
Chronic myeloid leukemia (CML) is a clonal myeloproliferative disorder that typically evolves through 3 clinical phases: chronic phase, accelerated phase (AP), and blastic phase. The hallmark of CML, the Philadelphia (Ph) chromosome, is an abnormally short chromosome 22 that results from a balanced translocation t(9;22) (q34;q11.2). The translocation leads to the formation of a hybrid gene BCR-ABL that encodes for the fusion protein Bcr-Abl with its characteristic deregulated tyrosine kinase activity.1
Clonal evolution in CML denotes the presence of a variety of additional, nonrandom chromosomal abnormalities besides the Ph chromosome. Clonal evolution occurs in approximately 30% of patients in AP and about 80% patients in blastic phase.2,3 Although clonal evolution may involve any abnormality within any chromosome, the most commonly reported abnormalities include double Ph, chromosome 17 abnormalities, and trisomy 8. Clonal evolution has been considered a criterion for AP of CML, particularly when it appears during the course of therapy. In this setting, clonal evolution is associated with a poor prognosis.3–11 The prognostic significance of clonal evolution may vary with different therapeutic modalities. The outcome after allogeneic stem cell transplant may not be adversely affected by clonal evolution, with a reported long-term survival rate of 60% if clonal evolution was the only feature of AP.12,13 Interferon (IFN)-α therapy caused complete resolution of clonal evolution in 46% patients, with a better response rate if clonal evolution was not associated with other features of AP, although patients with abnormalities of chromosome 17 had a worse outcome compared with those with other abnormalities.8 In patients treated with IFN-α and low-dose Ara-C, 3-year survival rates of 67% were observed if clonal evolution was the only feature of AP disease, compared with 22% if other features of AP were present (P < .01).14 With imatinib, the outcome of patients with clonal evolution is significantly inferior compared with those without clonal evolution.7
Patients who develop resistance to imatinib frequently develop clonal evolution as a mechanism of resistance (or associated with resistance).15–17 The second generation tyrosine kinase inhibitors dasatinib and nilotinib are effective in patients with CML after failure of imatinib.18,19 The significance of different chromosomal abnormalities associated with clonal evolution for the outcome after therapy with second generation tyrosine kinase inhibitors is unknown. Herein, we analyzed the response to therapy and long-term outcome after second generation tyrosine kinase inhibitor therapy in patients with CML with clonal evolution after imatinib failure.
MATERIALS AND METHODS
Patients
All patients with Ph+ CML in chronic phase or AP treated with dasatinib or nilotinib at The University of Texas M. D. Anderson Cancer Center between November 2003 and August 2007 were included in this analysis. Criteria for AP included the presence of any of the following10: 1) 15% to 29% peripheral or bone marrow blasts, 2) ≥20% peripheral or marrow basophils, 3) ≥30% peripheral or marrow blasts plus promyelocytes, 4) platelets <100 × 109/L unrelated to therapy, or 5) clonal evolution. Loss of chromosome Y, complex or variant Ph abnormalities, or chromosomal changes in Ph− cells were not considered clonal evolution. All patients were treated with either dasatinib or nilotinib after imatinib failure as part of a series of phase 1 and phase 2 studies at various doses as previously reported. These studies were approved by the institutional review board (IRB), and all patients signed IRB-approved informed consents.
Evaluation of Patients
All patients had a history and physical exam, complete blood counts, and blood chemistry before the start of therapy and every month for the first 3 months, then every 3 months until 12 months from the start of therapy, and then every 6 months. Cytogenetic response was assessed by G-banding assessed in the bone marrow with at least 20 metaphases counted. Molecular response was assessed by real-time polymerase chain reaction as previously reported.20 Both cytogenetic and molecular response assessments were performed at baseline, every 3 months for the first 12 months, and then every 6 months. Response and relapse criteria were as previously reported.21,22
Statistical Analysis
Overall survival (OS) was determined from the start of therapy with second generation tyrosine kinase inhibitors to death from any cause or last follow-up. Event-free survival (EFS) was measured from the start of each therapy until loss of complete hematologic response or major cytogenetic response, progression to the AP or blastic phase, or death from any cause during treatment. Chisquare test, Wilcoxon rank sum test, and Spearman rank correlation coefficient were used as appropriate to determine the inter-relationship of various parameters for this study. Survival curves were built using the Kaplan and Meier method, and differences in survival of subgroups were assessed by the log-rank test.23 Multivariate analyses were done to analyze the independent prognostic significance of various chromosomal abnormalities.
RESULTS
Among a total of 177 patients treated during this period, 54 (31%) had clonal evolution at the start of therapy with second generation tyrosine kinase inhibitors. Twenty-four (44%) of the 54 had clonal evolution in addition to other AP criteria, and 30 (56%) had clonal evolution as the only feature of AP. Among the others, 95 were in chronic phase, and 28 had other features of AP but no clonal evolution. All patients were previously treated with imatinib unsuccessfully. The median CML duration at the start of therapy of second generation tyrosine kinase inhibitors was 69 months (range, 4–241 months), and median follow-up after start of second generation tyrosine kinase inhibitors was 28.5 months (range, 5–53 months). Table 1 summarizes the characteristics of all patients.
Table 1.
Patient Characteristics by CML Phases
| Patient Characteristics | CP n = 95 |
CE alone (CP+CE), n = 30 |
AP (No CE), n = 28 |
CE+other AP features, n = 24 |
|---|---|---|---|---|
| Age, y (range) | 56 (21–83) | 57 (25–76) | 61 (28–81) | 56 (28–7) |
| Time from CML Dx to 2nd TKI, mo (range) | 67 (4–241) | 68 (8–207) | 80 (13–179) | 80 (4–191) |
| Follow-up after start of 2nd TKI, mo (range) | 28 (6–53) | 31 (18–46) | 29 (5–45) | 26 (8–35) |
| Prior therapy, No. (%) | ||||
| Imatinib | 95 (100) | 30 (100) | 28 (100) | 24 (100) |
| IFN | 54 (57) | 16 (53) | 17 (61) | 17 (71) |
| SCT | 1 (1) | 2 (7) | 2 (7) | 2 (8) |
| Other chemotherapy | 18 (19) | 7 (23) | 11 (39) | 9 (38) |
| Best imatinib response, No. (%) | ||||
| MCyR | 34 (36) | 12 (40) | 11 (39) | 2 (8) |
| Minor CyR | 13 (14) | 4 (13) | 3 (11) | 0 |
| CHR only | 35 (37) | 9 (30) | 4 (14) | 5 (21) |
| Resistant | 4 (4) | 1 (3) | 3 (11) | 5 (21) |
| Intolerable | 6 (6) | 2 (7) | 2 (7) | 3 (12) |
| Not known | 3 (3) | 2 (7) | 5 (18) | 9 (38) |
| BCR-ABL/KD mutations, No. (%) | ||||
| Yes | 34 (36) | 10 (33) | 12 (43) | 11 (46) |
| No | 28 (29) | 13 (43) | 3 (11) | 6 (25) |
| Not done | 33 (35) | 7 (23) | 13 (46) | 7 (29) |
CML indicates chronic myeloid leukemia; CP, chronic phase; CE, clonal evolution; AP, accelerated phase; Dx, diagnosis; TKI, second generation tyrosine kinase inhibitor; IFN, interferon; SCT, stem cell transplant; MCyR, major cytogenetic response; CyR, cytogenetic response; CHR, complete hematologic response; KD, kinase domain.
In 30 of 54 (56%) patients with clonal evolution (22 of 30 with clonal evolution alone and 8 of 24 with clonal evolution plus other AP features), there was a single additional chromosomal abnormality (besides the Ph chromosome) identified, with double Ph being the most common finding (Tables 2 and 3). The other 24 (44%) patients had >1 chromosomal abnormality besides the Ph chromosome, usually involving multiple abnormalities (median 2 abnormalities; range, 2–17).
Table 2.
Chromosomal Abnormalities Observed in Patients With CE Alone (n=30) in Ph+ Cells
| Types of Abnormalities in CEa |
Number (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Total pts | Trisomy 8 | Chr 17 Abnormality |
Double Ph+ | Other Translocations |
Other Chr Abnormality |
Complex | |||
| 1 | ≥2 | 1 | ≥2 | ||||||
| Total pts | 30 (100) | 2 (6) | 3 (10) | 15 (50) | 9 (30) | 1 (3) | 8 (27) | 1 (3) | 2 (6) |
| Trisomy 8 | 2 (6) | 1 (3)b | — | 1 (3) | — | — | — | — | — |
| Chr 17 abnormality | 3 (10) | — | 1 (3)b | 2 (6) | — | — | — | — | — |
| Double Ph+ | 15 (50) | 1 (3) | 2 (6) | 8 (27)b | 3 (10) | — | 2 (6) | 1 (3) | 2 (6) |
| Other translocation | |||||||||
| 1 | 9 (30) | — | — | 3 (10) | 6 (20)b | 1 (3) | 1 (3) | 1 (3) | 2 (6) |
| ≥2 | 1 (3) | — | — | — | — | — | — | — | — |
| Other Chr abnormality | |||||||||
| 1 | 8 (27) | — | — | 2 (6) | 1 (3) | — | 6 (20)b | — | 1 (3) |
| ≥2 | 1 (3) | — | — | 1 (3) | 1 (3) | — | — | — | 1 (3) |
| Complexc | 2 (6) | — | — | 2 (6) | 2 (6) | 1 (3) | 1 (3) | 1 (3) | 2 (6) |
CE indicates clonal evolution; Ph+, Philadelphia positive; pts, patients; Chr, chromosome.
In all categories, some patients had >2 chromosomal abnormalities; therefore, they were counted ≥2 times.
Patients with single chromosomal abnormalities.
Complex abnormalities: ≥3 additional types of chromosomal abnormalities besides the Ph chromosome.
Table 3.
Chromosomal Abnormalities Observed in Patients With CE Plus Other AP Features (n=24) in Ph+ Cells
| Types of Abnormalities in CEa |
Number (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Total pts | Trisomy 8 | Chr 17 Abnormality |
Double Ph+ |
Other Translocations |
Other Chr Abnormality |
Complex | |||
| 1 | ≥2 | 1 | ≥2 | ||||||
| Total pts | 24 (100) | 7 (29) | 10 (42) | 13 (54) | 8 (33) | — | 6 (25) | 4 (17) | 10 (42) |
| Trisomy 8 | 7 (29) | — | 3 (13) | 5 (21) | — | — | 2 (8) | 1 (4) | 4 (17) |
| Chr 17 abnormality | 10 (42) | 3 (13) | 3 (13)b | 4 (17) | 3 (13) | — | 2 (8) | 1 (4) | 5 (21) |
| Double Ph+ | 13 (54) | 5 (21) | 4 (17) | 3 (13)b | 4 (17) | — | 2 (8) | 2 (8) | 6 (25) |
| Other translocation | |||||||||
| 1 | 8 (33) | — | 3 (13) | 4 (17) | 1 (4)b | — | 3 (13) | 2 (8) | 5 (21) |
| ≥2 | — | — | — | — | — | — | — | — | — |
| Other Chr abnormality | |||||||||
| 1 | 6 (25) | 2 (8) | 2 (8) | 2 (8) | 3 (13) | — | 1 (4)b | — | 4 (17) |
| ≥2 | 4 (17) | 1 (4) | 1 (4) | 2 (8) | 2 (8) | — | — | 1 (4) | 4 (17) |
| Complexc | 10 (42) | 4 (17) | 5 (21) | 6 (25) | 5 (21) | — | 4 (17) | 4 (17) | 10 (42) |
CE indicates clonal evolution; AP, accelerated phase; Ph+, Philadelphia positive; pts, patients; Chr, chromosome.
In all categories, some patients had >2 chromosomal abnormalities; therefore, they were counted ≥2 times.
Patients with single chromosomal abnormalities.
Complex abnormalities: ≥3 additional types of chromosomal abnormalities besides the Ph chromosome.
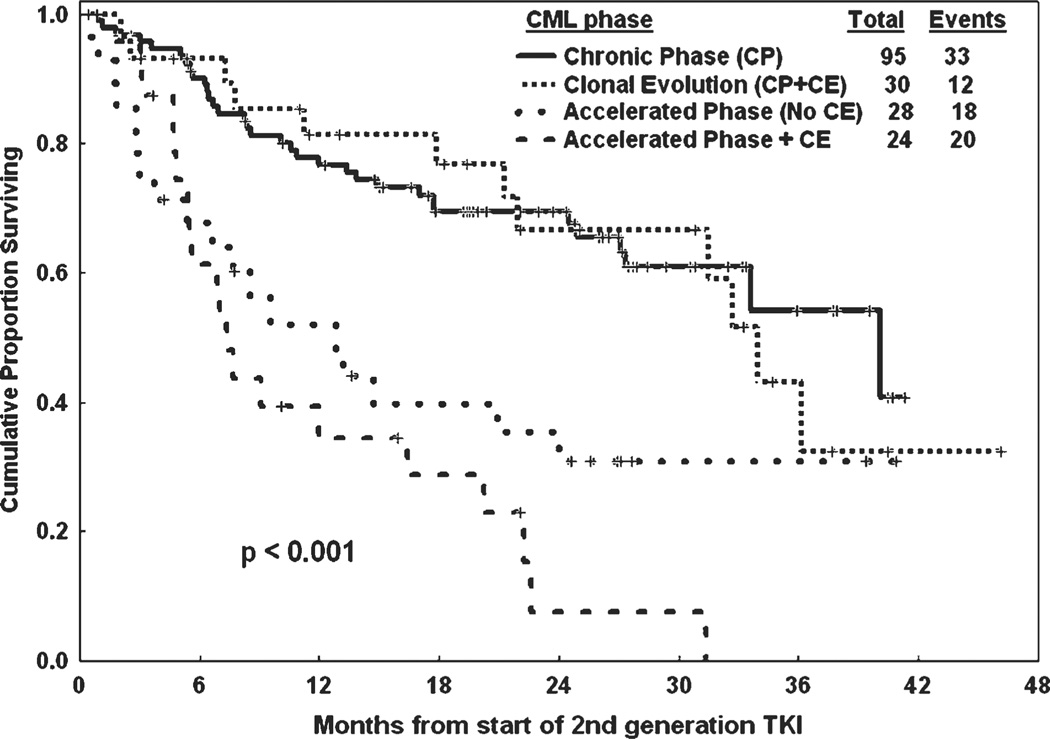
The absence or presence of clonal evolution in itself was a significant factor for response to treatment and survival; with major cytogenetic response rates of 53% versus 35% (P = .04), 2-year OS of 82% versus 56% (P < .001), and 2-year EFS of 60% versus 40% (P = .04), respectively, for patients without clonal evolution and with clonal evolution. Analyzing the outcome by individual groups, in the patients with active disease (ie, patients not in complete hematologic response at start of therapy), 60 of the 73 (82%) patients in chronic phase, 21 of the 23 (91%) with clonal evolution alone, 21 of the 28 (75%) in AP, and 13 of the 24 (54%) with clonal evolution plus other features of AP disease achieved a hematologic response with second generation tyrosine kinase inhibitors (P=.01), but the difference was not significant when subgroup comparison was done (Table 4). A major cytogenetic response was achieved in 55 of the 95 (58%) patients in chronic phase, 16 of the 30 (54%) with clonal evolution alone, 8 of the 28 (28%) in AP, and 3 of the 24 (13%) with clonal evolution plus other features of AP disease (P < .001), but the difference was not significant when subgroup comparison was done, as shown in Table 4. When comparing patients in chronic phase versus patients in chronic phase plus clonal evolution, the complete hematologic response rates were 79% versus 87% (P = .61), major cytogenetic response rates were 58% versus 54% (P = .82), and complete cytogenetic response rates were 53% versus 47% (P = .71), respectively. Similarly, comparing patients in AP without clonal evolution versus patients in AP with clonal evolution, the complete hematologic response rates were 71% versus 46% (P = .11), major cytogenetic response rates were 28% versus 13% (P = .28), and complete cytogenetic response rates were 21% versus 13% (P = .63), respectively. When comparing patients with chronic phase to those with clonal evolution alone, there was no significant difference in the 2-year OS and EFS (Table 4). Similarly, there was no difference in EFS between AP versus AP plus clonal evolution (Fig. 1). However, patients with AP plus clonal evolution had a significantly lower 2-year OS than those with AP alone (P = .004).
Table 4.
Response to Therapy by CML Phases
| Response, No. (%) | CP n = 95 |
CE alone (CP+CE), n = 30 |
AP (No CE), n = 28 |
CE+other AP features, n = 24 |
||
|---|---|---|---|---|---|---|
| Total/active diseasea | 95/73 | 30/23 | 28 | 24 | ||
| Hematologic response | ||||||
| CHR | 58 (79) | 20 (87) | 20 (71) | 11 (46) | ||
| P=.61 | P=.11 | |||||
| Partial HR | 2 (3) | 1 (4) | 1 (4) | 2 (8) | ||
| Overall | 60 (82) | 21 (91) | 21 (75) | 13 (54) | ||
| P=.47 | P=.20 | |||||
| Cytogenetic response | ||||||
| Overall | 69 (73) | 19 (63) | 12 (43) | 5 (21) | ||
| P=.45 | P=.25 | |||||
| CCyR | 50 (53) | 14 (47) | 6 (21) | 3 (13) | ||
| P=.71 | P=.63 | |||||
| PCyR | 5 (5) | 2 (7) | 2 (7) | 0 | ||
| MCyR, P=.82 | MCyR, P=.28 | |||||
| Minor CyR | 14 (15) | 3 (10) | 4 (14) | 2 (8) | ||
| Overall survival | ||||||
| Median, mo | NA | 43 | 45 | 18 | ||
| 2 year, % | 86 | 73 | 68 | 33 | ||
| P=.11 | P=.004 | |||||
| Event-free survival | ||||||
| Median, mo | 40 | 34 | 13 | 7 | ||
| 2 year, % | 69 | 67 | 31 | 8 | ||
| P=.87 | P=.18 | |||||
CML indicates chronic myeloid leukemia; CP, chronic phase; CE, clonal evolution; AP, accelerated phase; CHR, complete hematologic response; HR, hematologic response; CCyR, complete cytogenetic response; PCyR, partial cytogenetic response; MCyR, major cytogenetic response; CyR, cytogenetic response; NA, not available.
Active disease: Patients who were not in CHR. The gap in number is for the patients who were in CHR at the start of second generation tyrosine kinase inhibitor therapy.
Figure 1.
Event-free survival after treatment with second generation tyrosine kinase inhibitors (TKIs) in various phases of chronic myeloid leukemia (CML).
We then analyzed whether various features of clonal evolution had different prognostic impact. The OS and EFS were worse with increasing numbers of cells/metaphases showing Ph positivity (90% or more vs less). However, within the patients with clonal evolution, the percentage of metaphases involved with clonal evolution had no statistically significant impact on major cytogenetic response, OS, or EFS albeit with a trend for a worse outcome for patients with >90% metaphases with clonal evolution (Table 5). Regarding the different chromosomal abnormalities seen, when presenting as individual abnormalities, the 2-year survival probability was 11% for patients with chromosome 8 abnormalities, 35% for chromosome 17 involvement, 57% for double Ph+ cells, 52% for patients with other chromosomal translocations, 56% for other chromosomal abnormalities, 27% for complex chromosomal abnormalities, and 39% for ≥2 other chromosomal abnormalities (Table 6).
Table 5.
Response to Therapy and Survival by Presence or Absence of CE and by Percentage of Metaphases Involved With CE in CML
| Type of CE | Total No. of pts |
MCyR No. (%) |
OS | EFS | ||
|---|---|---|---|---|---|---|
| Median, mo |
2-Year % | Median, mo |
2-Year % | |||
| Clonal evolution | ||||||
| No | 120 | 63 (53) | 45 | 82 | 34 | 60 |
| Yes | 54 | 19 (35) | 27 | 56 | 21 | 40 |
| P = .04 | P <.001 | P = .04 | ||||
| Number of Ph+ cells, % | ||||||
| ≥90 | 32 | 25 (78) | NA | 84 | NA | 77 |
| >90 | 140 | 56 (40) | 43 | 71 | 24 | 49 |
| P <.001 | P = .07 | P = .007 | ||||
| Clonal evolution by % of metaphases involved | ||||||
| 1–25% | 14 | 5 (36) | NA | 71 | 33 | 50 |
| 26–50% | 7 | 4 (57) | NA | 54 | 32 | 63 |
| 51–90% | 7 | 4 (57) | 27 | 57 | 21 | 50 |
| >90% | 25 | 7 (28) | 20 | 46 | 12 | 25 |
| P = .13 | P = .23 | P = .14 | ||||
CE indicates clonal evolution; CML, chronic myeloid leukemia; MCyR, major cytogenetic response; OS, overall survival; EFS, event-free survival; Ph+, Philadelphia positive; NA, not available.
Table 6.
Response to Therapy and Survival by Type of CE in CML
| Type of CEa | Total No. of pts |
MCyR, No. (%) |
OS | EFS | ||
|---|---|---|---|---|---|---|
| Median, mo |
2-Year % | Median, mo |
2-Year % | |||
| All CEa | 54 | 19 (35) | 27 | 56 | 21 | 40 |
| Trisomy 8 | ||||||
| No | 45 | 17 (39) | 43 | 65 | 23 | 47 |
| Yes | 9 | 2 (22) | 6 | 11 | 5 | 0 |
| P=.35 | P <.001 | P=.001 | ||||
| Chr 17 abnormalities | ||||||
| No | 41 | 16 (40) | 32 | 62 | 23 | 47 |
| Yes | 13 | 3 (23) | 19 | 35 | 6 | 20 |
| P=.27 | P <.03 | P=.02 | ||||
| Double Ph+ | ||||||
| No | 26 | 10 (38) | 27 | 54 | 22 | 45 |
| Yes | 28 | 9 (32) | 29 | 57 | 20 | 32 |
| P=.70 | P=.75 | P=.73 | ||||
| Other Chr translocations | ||||||
| No | 36 | 15 (43) | 29 | 57 | 23 | 46 |
| Yes | 18 | 4 (22) | 27 | 52 | 21 | 28 |
| P=.14 | P=.85 | P=.41 | ||||
| Other Chr abnormalities | ||||||
| No | 35 | 12 (35) | 27 | 55 | 22 | 34 |
| Yes | 19 | 7 (37) | 32 | 56 | 21 | 49 |
| P=.91 | P=.80 | P=.64 | ||||
| Complex Chr abnormalitiesb | ||||||
| No | 42 | 17 (41) | 43 | 63 | 23 | 48 |
| Yes | 12 | 2 (17) | 8 | 27 | 7 | 12 |
| P=.12 | P=.02 | P=.004 | ||||
| No. of Chr abnormalities besides Ph Chr | ||||||
| 1 | 30 | 14 (47) | 43 | 67 | 32 | 52 |
| ≥2 | 24 | 5 (21) | 18 | 39 | 8 | 22 |
| P=.06 | P=.06 | P=.007 | ||||
| BCR-ABL/KD mutations | ||||||
| Yes | 21 | 6 (30) | 19 | 43 | 21 | 27 |
| No | 19 | 7 (37) | NA | 73 | 31 | 53 |
| Na | 14 | 6 (43) | 27 | 51 | 15 | 40 |
| P=.74 | P=.21 | P=.51 | ||||
CE indicates clonal evolution; CML, chronic myeloid leukemia; pts, patients; MCyR, major cytogenetic response; OS, overall survival; EFS, event-free survival; Chr, chromosome; Ph+, Philadelphia positive; KD, kinase domain; NA, not available.
Includes patients with CE alone (n=30) and CE + other AP features (n=24).
The clonal evolution represents changes reported in Ph+ cells. Many patients had >1 type of abnormality. Each abnormality (whether alone or in combination) was considered individually in statistical methods.
Complex abnormalities: ≥3 additional types of chromosomal abnormalities besides the Ph chromosome.
BCR-ABL mutations were present in 67 (57%) of the 117 patients in whom sequencing was done, and mutations were almost equally distributed (33%–46%) in all 4 groups (chronic phase or clonal evolution alone or AP or clonal evolution + AP, P = .13, Table 1). The mutations were varied, and the most common observed were G250E mutation in 9 patients (6 in chronic phase and 3 in AP alone); M351T mutation in 4 patients in chronic phase; and T315I mutation in 3 patients in chronic phase. There were 3 patients in chronic phase and 2 patients in the clonal evolution alone group who had 2 coexisting mutations: M351T + F317L, T315I + M351T, and V299L + F486S in chronic phase patients, and F311L + E453K, and G250E + F359I in patients with clonal evolution alone. Within the 54 patients with clonal evolution (clonal evolution alone and clonal evolution plus other AP features), there was no statistically significant difference in major cytogenetic response (30% vs 37%, P = .74), OS (43% vs 73%, P = .21), or EFS (27% vs 53%, P = .51), respectively, between patients with and without mutations, but there was a trend for inferior survival for patients with mutations (Table 6).
On univariate analysis, other factors besides clonal evolution that were associated with inferior OS or EFS were presence of splenomegaly, hemoglobin <10 g/dL, platelet count <100 × 109/L), basophils >5% or blasts ≥5% in peripheral blood or bone marrow, stage of CML (AP), and hematological resistance to imatinib therapy. On multivariate analysis, clonal evolution did not show an independently significant effect on achieving major cytogenetic response on the second generation tyrosine kinase inhibitors. The factors predicting increasing major cytogenetic response to second generation tyrosine kinase inhibitors were identified as prior achievement of major cytogenetic response with imatinib, higher hemoglobin levels, no splenomegaly, lower percentage of Ph+ metaphases, and no prior chemotherapy (eg, cytarabine, decitabine). Prior achievement of major cytogenetic response to imatinib and lower number of marrow blasts were significantly associated with better EFS. For OS, the positive factors were higher hemoglobin, lower marrow blast percent, and no thrombocytopenia using a Cox proportional hazard model. Clonal evolution was not independently associated with an inferior EFS or OS.
DISCUSSION
The adverse prognosis conferred by the presence of clonal evolution has been demonstrated in patients treated with imatinib. O’Dwyer et al7 reported on 71 patients with CML in AP treated with imatinib, of whom 15 had clonal evolution alone, 32 had AP features without clonal evolution, and 24 had clonal evolution plus other features of AP. Major cytogenetic response was achieved with imatinib in 73%, 31% (P = .043), and 13% (P = .007), respectively. With a mean follow-up of 11.2 months, the 1-year probability of survival was 100%, 85%, and 67.5% (P = .01), respectively. In another series from our institution,5 498 patients with CML treated with imatinib were analyzed, of whom 295 were in chronic phase, 70 had clonal evolution alone, 82 had AP without clonal evolution, and 51 had clonal evolution plus other features of AP disease. The rates of major cytogenetic response were 66%, 54%, 40%, and 33%, (P = .5), respectively. After a median follow-up of 30 months, 2-year survival rates were 92%, 77%, 71%, and 46%, respectively. In a multivariate analysis, clonal evolution was not independently associated with the probability of achieving major cytogenetic response or complete cytogenetic response, but it was an independent poor prognostic factor for survival. In the present analysis, the presence of clonal evolution did not show similar prognostic impact for patients treated with second generation tyrosine kinase inhibitors mostly after failure of imatinib therapy.
Our report represents the first systematic analysis of the significance of clonal evolution in the outcome after therapy with second generation tyrosine kinase inhibitors. Our results suggest that for CML patients with clonal evolution but no other features of AP treated with a second generation tyrosine kinase inhibitor after imatinib failure, the hematologic and cytogenetic response rates, OS, and EFS are no different than for patients with chronic phase and no clonal evolution. Clonal evolution, however, has a significant adverse impact when associated with other features of AP. However, in a multivariate analysis, clonal evolution does not have an independent prognostic impact on the outcome of patients. This suggests that outcome after second generation tyrosine kinase inhibitors is driven by BCR-ABL inhibition, and the molecular events associated with clonal evolution have relatively little impact on the outcome after therapy, at least in the chronic phase. In contrast, in the AP, the presence of clonal evolution adversely affects outcome, particularly OS. Although the response rate and EFS are not statistically significant in AP, the trends in cytogenetic response rate and EFS suggest an adverse impact of clonal evolution. These are probably not statistically significant because of the small sample size. The molecular mechanisms associated with clonal evolution remain to be identified, and they are likely to be heterogeneous as a variety of different chromosomal abnormalities are associated with clonal evolution, frequently coexisting in the same patient. Unraveling the different molecular mechanisms involved in this process may help us better understand the pathophysiology of CML progression.
We also found that trisomy 8, chromosome 17, or complex chromosomal abnormalities had worse 2-year OS and EFS, whereas double Ph chromosome, other translocations, other clonal abnormalities, or the percentage of cells showing clonal evolution had no significant impact on EFS/OS. This might be an oversimplification of the complicated process that is clonal evolution, as there were 44% patients who had >1 chromosomal abnormality, and we here are considering each chromosomal abnormality as if it existed alone. Yet it gives us an idea that chromosome 8 abnormalities have worst outcome followed by complex abnormalities, chromosome 17 abnormalities, and abnormalities involving >1 chromosome.
It has been previously reported that mutations may occur more frequently among patients with clonal evolution.24 This association might represent further manifestation of the genomic instability that has been reported in patients with CML.25 Interestingly, in our series the presence of kinase domain mutations had little prognostic impact on the response to second generation tyrosine kinase inhibitors, although there is a trend for inferior OS and EFS.
In conclusion, clonal evolution in CML constitutes a heterogeneous entity with variable outcome with second generation tyrosine kinase inhibitor therapy. As a group, patients with clonal evolution have inferior outcome with second generation tyrosine kinase inhibitors, and the presence of other features of AP further worsens the outcome. However, other clinical features are mainly responsible for the inferior outcome seen among patients with clonal evolution. The molecular events behind the different clonal evolution categories and the potential therapeutic approaches directed at them need to be defined.
Acknowledgments
CONFLICT OF INTEREST DISCLOSURES
Supported by a research grant from Novartis and BMS (H.K. and J.C.).
Footnotes
D.V. wrote the manuscript, analyzed data, and approved it. J.C. designed the study, managed the patients, analyzed data, and reviewed and approved the manuscript. J.S. collected data and assisted with data analysis. H.K., S.O., Z.E., G.G.M., C.K., and G.B. approved the manuscript and managed the patients.
REFERENCES
- 1.Quintas-Cardama A, Cortes J. Chronic myeloid leukemia: diagnosis and treatment. Mayo Clin Proc. 2006;81:973–988. doi: 10.4065/81.7.973. [DOI] [PubMed] [Google Scholar]
- 2.Anastasi J, Feng J, Le Beau MM, Larson RA, Rowley JD, Vardiman JW. The relationship between secondary chromosomal abnormalities and blast transformation in chronic myelogenous leukemia. Leukemia. 1995;9:628–633. [PubMed] [Google Scholar]
- 3.Cortes J, O’Dwyer ME. Clonal evolution in chronic myelogenous leukemia. Hematol Oncol Clin North Am. 2004;18:671–684. doi: 10.1016/j.hoc.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 4.Fabarius A, Haferlach C, Muller MC, et al. Dynamics of cytogenetic aberrations in Philadelphia chromosome positive and negative hematopoiesis during dasatinib therapy of chronic myeloid leukemia patients after imatinib failure. Haematologica. 2007;92:834–837. doi: 10.3324/haematol.11064. [DOI] [PubMed] [Google Scholar]
- 5.Cortes J, Talpaz M, Giles F, et al. Prognostic significance of cytogenetic clonal evolution in patients with chronic myelogenous leukemia on imatinib mesylate therapy. Blood. 2003;101:3794–3800. doi: 10.1182/blood-2002-09-2790. [DOI] [PubMed] [Google Scholar]
- 6.Marktel S, Marin D, Foot N, et al. Chronic myeloid leukemia in chronic phase responding to imatinib: the occurrence of additional cytogenetic abnormalities predicts disease progression. Haematologica. 2003;88:260–267. [PubMed] [Google Scholar]
- 7.O’Dwyer ME, Mauro MJ, Kurilik G, et al. The impact of clonal evolution on response to imatinib mesylate (STI571) in accelerated phase CML. Blood. 2002;100:1628–1633. doi: 10.1182/blood-2002-03-0777. [DOI] [PubMed] [Google Scholar]
- 8.Cortes J, Talpaz M, O’Brien S, et al. Suppression of cytogenetic clonal evolution with interferon alfa therapy in patients with Philadelphia chromosome-positive chronic myelogenous leukemia. J Clin Oncol. 1998;16:3279–3285. doi: 10.1200/JCO.1998.16.10.3279. [DOI] [PubMed] [Google Scholar]
- 9.Majlis A, Smith TL, Talpaz M, O’Brien S, Rios MB, Kantarjian HM. Significance of cytogenetic clonal evolution in chronic myelogenous leukemia. J Clin Oncol. 1996;14:196–203. doi: 10.1200/JCO.1996.14.1.196. [DOI] [PubMed] [Google Scholar]
- 10.Kantarjian HM, Dixon D, Keating MJ, et al. Characteristics of accelerated disease in chronic myelogenous leukemia. Cancer. 1988;61:1441–1446. doi: 10.1002/1097-0142(19880401)61:7<1441::aid-cncr2820610727>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 11.Sokal JE, Gomez GA, Baccarani M, et al. Prognostic significance of additional cytogenetic abnormalities at diagnosis of Philadelphia chromosome-positive chronic granulocytic leukemia. Blood. 1988;72:294–298. [PubMed] [Google Scholar]
- 12.Clift RA, Buckner CD, Thomas ED, et al. Marrow transplantation for patients in accelerated phase of chronic myeloid leukemia. Blood. 1994;84:4368–4373. [PubMed] [Google Scholar]
- 13.Przepiorka D, Thomas ED. Prognostic significance of cytogenetic abnormalities in patients with chronic myelogenous leukemia. Bone Marrow Transplant. 1988;3:113–119. [PubMed] [Google Scholar]
- 14.Kantarjian HM, Keating MJ, Estey EH, et al. Treatment of advanced stages of Philadelphia chromosome-positive chronic myelogenous leukemia with interferon-alpha and low-dose cytarabine. J Clin Oncol. 1992;10:772–778. doi: 10.1200/JCO.1992.10.5.772. [DOI] [PubMed] [Google Scholar]
- 15.Rosenhahn J, Weise A, Michel S, et al. Cytogenetic characterization and proteomic profiling of the imatinib-resistant cell line KCL22-R. Int J Oncol. 2007;31:121–128. [PubMed] [Google Scholar]
- 16.Lahaye T, Riehm B, Berger U, et al. Response and resistance in 300 patients with Bcr-Abl-positive leukemias treated with imatinib in a single center: a 4.5-year follow-up. Cancer. 2005;103:1659–1669. doi: 10.1002/cncr.20922. [DOI] [PubMed] [Google Scholar]
- 17.Hochhaus A, Kreil S, Corbin AS, et al. Molecular and chromosomal mechanisms of resistance to imatinib (STI571) therapy. Leukemia. 2002;16:2190–2196. doi: 10.1038/sj.leu.2402741. [DOI] [PubMed] [Google Scholar]
- 18.Guilhot F, Apperley J, Kim DW, et al. Dasatinib induces significant hematologic and cytogenetic responses in patients with imatinib-resistant or -intolerant chronic myeloid leukemia in accelerated phase. Blood. 2007;109:4143–4150. doi: 10.1182/blood-2006-09-046839. [DOI] [PubMed] [Google Scholar]
- 19.Kantarjian H, Giles F, Wunderle L, et al. Nilotinib in imatinib-resistant CML and Philadelphia chromosome-positive ALL. N Engl J Med. 2006;354:2542–2551. doi: 10.1056/NEJMoa055104. [DOI] [PubMed] [Google Scholar]
- 20.Cortes J, Talpaz M, O’Brien S, et al. Molecular responses in patients with chronic myelogenous leukemia in chronic phase treated with imatinib mesylate. Clin Cancer Res. 2005;11:3425–3432. doi: 10.1158/1078-0432.CCR-04-2139. [DOI] [PubMed] [Google Scholar]
- 21.Talpaz M, Silver RT, Druker BJ, et al. Imatinib induces durable hematologic and cytogenetic responses in patients with accelerated phase chronic myeloid leukemia: results of a phase 2 study. Blood. 2002;99:1928–1937. doi: 10.1182/blood.v99.6.1928. [DOI] [PubMed] [Google Scholar]
- 22.Kantarjian H, Sawyers C, Hochhaus A, et al. Hematologic and cytogenetic responses to imatinib mesylate in chronic myelogenous leukemia. N Engl J Med. 2002;346:645–652. doi: 10.1056/NEJMoa011573. [DOI] [PubMed] [Google Scholar]
- 23.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- 24.Jabbour E, Kantarjian H, Jones D, et al. Frequency and clinical significance of BCR-ABL mutations in patients with chronic myeloid leukemia treated with imatinib mesylate. Leukemia. 2006;20:1767–1773. doi: 10.1038/sj.leu.2404318. [DOI] [PubMed] [Google Scholar]
- 25.Skorski T. BCR/ABL regulates response to DNA damage: the role in resistance to genotoxic treatment and in genomic instability. Oncogene. 2002;21:8591–8604. doi: 10.1038/sj.onc.1206087. [DOI] [PubMed] [Google Scholar]