Abstract
Nudaurelia ω capensis virus (NωV) capsids were previously characterized in two morphological forms, a T=4, 485-Å-diameter round particle with large pores and a tightly sealed 395-Å icosahedrally shaped particle with the same quasi-symmetric surface lattice. The large particle converts to the smaller particle when the pH is lowered from 7.6 to 5, and this activates an autocatalytic cleavage of the viral subunit at residue 570. Here we report that both 1-anilino-8 naphthalene sulfonate (ANS) and the covalent attachment of the thiol-reactive fluorophore, maleimide-ANS (MIANS), inhibit the structural transition and proteolysis at the lower pH. When ANS is exhaustively washed from the particles, the maturation proceeds normally; however, MIANS-modified particles are still inhibited after the same washing treatment, indicating that covalent attachment targets MIANS to a critical location for inhibition. Characterization of the low-pH MIANS product by electron cryo-microscopy (cryo-EM) and image reconstruction demonstrated a morphology intermediate between the two forms previously characterized. A pseudoatomic model of the intermediate configuration was generated by rigid body refinement of the X-ray structure of the subunits (previously determined in the assembled capsid) into the cryo-EM density, allowing a quantitative description of the inhibited intermediate and a hypothesis for the mechanism of the inhibition.
Protein capsids of many icosahedral viruses respond dynamically to environmental signals such as DNA or RNA packaging, receptor binding, and changes in pH and ionic composition. In so doing, they undergo conformational changes that involve the concerted reorganization of hundreds of protein subunits and nucleic acids. In many viruses, large-scale conformational transformations occur during assembly and maturation. Such transformations have been observed in several bacteriophages (7, 9, 11, 16, 19, 23, 30), herpesvirus (15, 33), and a single-stranded RNA insect virus (1, 4). Icosahedral virus particles with T numbers higher than 1 apparently do not always directly assemble into a highly stable form. Rather, capsid proteins initially organize into procapsid shells that place subunits into proper relative locations but with, in many cases, sparse interfacial contacts (4, 9, 16). These precursor particles are poised for reorganization into the stable mature particle when the proper signal is received by the protein or nucleo-protein aggregate (for examples, see references 4 and 9). Nudaurelia ω capensis virus (NωV), an insect tetravirus, was the first single-stranded RNA animal virus for which three-dimensional structures were determined for the procapsid and mature capsid conformations (4, 22). The procapsid structure was discovered for NωV virus-like particles (VLPs) expressed in a recombinant baculovirus system. These particles contain heterologous cellular RNA in a ratio to protein that is similar to that of authentic virions (4). A cryo-electron microscopy (cryo-EM) reconstruction at 26 Å revealed a nearly spherical protein shell perforated by large holes at all symmetry axes (4). These holes were absent in the mature, authentic virion structure, determined by crystallography at 2.8-Å resolution (4, 22). Maturation of procapsid nucleoprotein VLPs induced by acidification to pH 5 resulted in icosahedral capsid particles indistinguishable at 26-Å resolution from authentic virions (4). This transformation has also been monitored using small-angle X-ray scattering experiments, which reported a decrease in average diameter, from 485 to 395 Å, when procapsids were converted to capsids (5). The reorganization activates an autoproteolytic cleavage between residues 570 and 571 of the 70-kDa α protein into 62- and 8-kDa β and γ polypeptides. The resulting mature structure contains 240 copies of the β and γ polypeptides and the same nucleic acid composition as procapsid (4, 22). Comparison of the procapsid and capsid structures with the X-ray coordinates as a reference demonstrated that the conversion depends largely on quaternary reorganization of subunits and not large-scale refolding of tertiary or secondary structure (4, 5). The transition occurs in a highly cooperative manner over a 0.7 pH unit range between pH 6.7 and 6.0 (5). Above and below this range, particles exist as stable procapsid and capsid, respectively.
In order to further characterize the conformational transition, we labeled NωV procapsid particles with the thiol-reactive fluorophore 2-(4′-maleimidylanilino)naphthalene-6-sulfonate (MIANS). MIANS exhibits a pronounced sensitivity to its local environment, fluorescing strongly only when sequestered at hydrophobic surfaces (2, 18). We reasoned that the covalently attached MIANS situated at subunit interfaces might report on the conformational transition as it is buried during maturation. Instead, we found that upon acidification, the labeled particles were trapped at an intermediate stage of maturation and that the autocatalytic cleavage, believed to be essential for infectivity, was virtually inhibited.
The results presented in this communication provide support for the strategy of targeting subunit interfaces with antiviral compounds as a means of disrupting the viral life cycle (24, 32, 34). Most previous studies of small-compound inhibition of other viruses have focused on interrupting polymerization of capsid subunits. For example, a compound that is structurally related to MIANS, 4,4′-dianilino-1,1′-binaphthyl-5,5′-disulfonate (bis-ANS), was observed to inhibit in vitro assembly of hepatitis B virus capsids (34). In that case, the fluorophore was found to prevent assembly of complete capsids by binding to soluble dimers. It was postulated that bis-ANS acts as a molecular “wedge” that prevents formation of correct capsid geometry. Similarly, bis-ANS was observed to inhibit procapsid assembly in the bacteriophage P22 system (32) and to contribute to inactivation of vesicular stomatitis virus (3). Capsid-targeted compounds from the heteroaryldihydropyrimidine family also have been demonstrated to inhibit hepatitis B virus core particle formation (10); in vivo, this is accompanied by an increase in core protein degradation. Similarly, a recent study of human immunodeficiency virus type 1 assembly demonstrated that a small molecule (CAP-1) binds to the human immunodeficiency virus capsid protein and inhibits infectivity in a dose-dependent manner (28). The compound gives rise to abnormal core particles by interfering with interactions between capsid proteins that are critical for proper maturation. By contrast, our results demonstrate that a later stage of virus maturation, after the protein shell has fully polymerized, can be effectively disrupted by small compounds.
MATERIALS AND METHODS
Purification of NωV procapsid VLPs.
Purification of VLPs was performed following previously described procedures (12, 31). Briefly, Sf21 cells were infected with a recombinant baculovirus vector containing the NωV wild-type (WT) coat protein. After 4 days of incubation at 27°C, cells were lysed with 0.5% (vol/vol) NP-40 detergent. Supernatant and debris were separated by centrifugation. Supernatant containing the VLPs was treated with 10 μg of RNase A/ml in order to degrade ribosomal material that would otherwise copurify with the VLPs. VLPs were pelleted through a 30% (wt/vol) sucrose cushion at pH 7.6. The high-speed pellets were resuspended in 400 μl of pH 7.6 buffer containing 250 mM NaCl-50 mM Tris (buffer A). The soluble resuspended material was then sedimented through a 10-to-40% sucrose gradient (wt/vol) prepared using pH 7.6 buffer A. Standard centrifugation conditions were 113,000 × g for 1.25 h at 11°C in a Beckman SW41 rotor. Fractions were collected using an ISCO fractionator running at 0.75 min/fraction with continuous measurement of absorbance at 280 nm.
The primary sequence of the capsid protein subunit used in these studies differs from the sequence deposited in GenBank. The differences are isolated to three point mutations. Specifically, in our clone residue 37 is cysteine (rather than lysine), residue 235 is valine (rather than cysteine), and residue 283 is valine (rather than glutamate).
Labeling reaction.
NωV procapsid particles were labeled at a 50-fold molar excess concentration of MIANS (Molecular Probes) to capsid protein ([MIANS] = 700 μM; [capsid protein] = 14 μM) in buffer A with 10% dimethyl sulfoxide (DMSO). NωV particles mature normally and are insensitive to DMSO concentrations in this range. The reaction mixture was incubated at 4°C for 18 h. Excess dye was removed by centrifugal filtration through 100-kDa MWCO membranes (Millipore Corp.). The first three washes were performed using a mixture of 10% DMSO and 90% buffer A. This step was essential for removing all excess, unconjugated dye. Typically, five additional washes with only buffer A were done to remove all DMSO. Samples were protected from exposure to ambient light throughout labeling and purification and during storage.
Quantitation of labeling efficiency involved accurate determination of concentrations of both covalently bound fluorophore and capsid protein. Determination of capsid protein concentration is not trivial, however, due to the contributions of the nucleic acid to absorbance measurements at 280 nm. Moreover, the extinction coefficient available in the literature for NωV of A260 = 5 corresponding to 1 mg/ml has never been rigorously verified, and this relation presumably gives total VLP concentration (including RNA and protein) rather than protein capsid concentration alone. The relative mass fractions of RNA and protein have never been established for this virus, however, and thus it is not possible to directly determine protein concentration by using the A260 = 5 value.
In order to accurately determine capsid protein concentration and correlate A260 with actual capsid protein concentration, we developed a fluorescence assay that relates the intrinsic fluorescence intensity of a VLP sample to a known capsid protein concentration based upon a standard curve determined for pure NωV capsid protein in the absence of nucleic acid and under denaturing conditions. Because RNA does not fluoresce, the fluorescence intensity measured for a VLP sample containing both the capsid protein and encapsidated RNA denatured in 6 M guanidine HCl reports on the protein concentration alone. By this assay, we have found that an A260 value of 2.8 corresponds to a capsid protein concentration of 1 mg/ml. For virus samples labeled with MIANS, the A260 was further corrected to account for absorbance by MIANS, thus yielding the actual capsid protein concentration. The concentration of covalently bound MIANS was determined by measuring the absorbance at 325 nm (the absorbance maximum for MIANS shifts from 443 to 325 nm upon conjugation to sulfhydryl groups).
Cleavage maturation assayed by gel electrophoresis.
Purified procapsids in buffer A were converted to capsids by rapid dilution into pH 5.0 buffer B (250 mM NaCl, 50 mM Na-acetate; pH 5.0). Maturation was allowed to progress for a minimum of 4 h at 25°C. Previous studies have demonstrated that cleavage is essentially complete within 1 h under these conditions (1, 5). Identical conditions were used in attempting to mature MIANS-conjugated NωV (M-NωV) particles. Maturation inhibition by noncovalently bound 1-anilino-8 naphthalene sulfonate (ANS) was examined using solutions containing 0.1, 0.3, and 1.0 mM ANS (Sigma) made with buffer B. After 4 h of incubation at pH 5.0, half of each sample was removed and washed by centrifugal filtration (100-kDa MWCO membrane; Millipore Corp.). Three washes were performed using a 10% DMSO-90% buffer B, ANS-free solution followed by three additional washes with buffer B. Both washed and unwashed samples were allowed to incubate for an additional 2 h at pH 5.0 at 25°C. All samples were analyzed for cleavage maturation by running them on 4-to-12% denaturing acrylamide gradient gels (Invitrogen) according to standard procedures.
Velocity sedimentation of labeled VLPs.
M-NωV VLPs and unlabeled WT NωV capsid were sedimented at 113,000 × g for 1.25 h through a 10-to-40% (wt/vol) sucrose gradient prepared with pH 5.0 buffer B. Unlabeled WT NωV procapsid was sedimented at 113,000 × g for 1.25 h through a 10-to-40% (wt/vol) sucrose gradient prepared with pH 7.6 buffer A. Sedimentation conditions and fractionation methods were performed as described above for particle purification.
EM and image reconstruction.
Samples for negative-stain transmission EM were prepared at 0.5 mg/ml. Staining and microscopy were performed as described previously (31) using 1% uranyl-acetate solutions. Preliminary three-dimensional image reconstruction studies were carried out with negative-stained samples, and final reconstructions were calculated with frozen hydrated specimens. We note that negative stain and cryo-EM reconstructions of the M-NωV particles at pH 5.0 showed very good agreement with each other, underscoring the apparent robustness of the labeled particles even under the relatively harsh conditions of negative staining (data not shown). Three-hundred-mesh copper grids were used for making the holey film. Five-microliter samples at the concentration of 0.5 mg/ml were applied onto the holey grid before plunge-freezing in liquid ethane. A Gatan cryo-holder was used to insert the specimen into the electron microscope. The micrographs were taken at a magnification of ×45,000 on a Philip CEM 120 electron microscope equipped with decontaminating blades. Images were digitized using a ZEISS flat bed scanner with a step size of 21 μm corresponding to 4.67 Å/pixel. After digitizing the micrographs, virus particles were selected with the boxing program X3D from James Conway (8). The reconstructions were calculated with SPIDER (14). The resolution of the reconstruction (899 selected particles) was estimated with a Fourier shell correlation coefficient of 27 Å.
Fitting models into cryo-EM density.
The crystal structure of the mature NωV model was docked into the electron density calculated from the M-NωV particles. Because this particle is larger than the mature virus, the coordinates were moved radially outwards to fit the reconstruction. Then, the four subunits were treated as rigid bodies to fit the electron density map calculated from the cryo-EM reconstruction. Small portions of the subunits were not used in the pseudoatomic modeling, because these residues exhibit the greatest subunit-to-subunit differences in the capsid structure and are likely to be in substantially different conformations in procapsid and M-NωV structures, which exhibit less subunit differentiation (4). Only residues 61 to 599, 44 to 590, 42 to 591, and 44 to 590 out of 644 residues in each subunit were used in the modeling for the A, B, C, and D subunits, respectively. The fitting was optimized by a modified version (for cryo-EM application) (29) of the real-space refinement program Rsref (6).
The four subunits (A, B, C, and D) in the NωV T=4 capsid asymmetric unit were analyzed individually to determine the movement during the conformational transition from procapsid to the M-NωV and then to the mature capsid (see Table 1). Each subunit in the procapsid model was compared with its corresponding subunit in the M-NωV to find the translational and rotational transformation for superimposing them by the program LSQMAN from the Uppsala Software Factory. Similarly, the translational and rotational transformations from M-NωV to the matured capsid were analyzed for each subunit by LSQMAN.
TABLE 1.
Translation of subunits between procapsid, M-NωV (pH 5.0), and capsid
| Subunit | Particle dimension (Å)
|
|
|---|---|---|
| Procapsid → M-NωV | M-NωV → capsid | |
| A | 15 | 18 |
| B | 19 | 19 |
| C | 19 | 20 |
| D | 22 | 21 |
RESULTS
MIANS labeling of NωV procapsid.
MIANS reacts with the viral capsid protein by forming a covalent thio-ether linkage with the thiol group of cysteine residues. In order to quantify the efficiency of labeling, we determined the number of MIANS molecules attached to labeled procapsid VLPs by measuring absorbances at 325 and 260 nm (see Materials and Methods). We consistently obtained an average (± standard deviation) of 4.2 ± 0.8 MIANS molecules conjugated per subunit. Labeling of each subunit was inferred to be uniform, because all four quasi-equivalent subunit environments in the procapsid were similar (4). Labeling studies with maleimide-functionalized propionic acid followed by proteolytic digestion and matrix-assisted laser desorption ionization-mass spectrometry showed that the three most reactive cysteines are CYS-135, CYS-507, and CYS-552 (B. Bothner and J. E. Johnson, unpublished data). It is not clear whether the bulkier, aromatic MIANS probe accesses the same sites to the same degree. Mass spectrometry has not proven feasible for identifying the residues labeled with MIANS, possibly due to the influence of the negatively charged fluorophore upon peptide ionization. However, based on the known subunit structure in which two cysteines (CYS-247 and CYS-507) are buried in the β-sandwich domain, we infer that these cysteines may be inaccessible to the MIANS dye. Furthermore, since the reaction conditions should have saturated the available sites, we believe the figure of four MIANS per subunit that we measured indicates that all other cysteines (37, 135, 516, and 552) were completely labeled.
Autoproteolytic cleavage is greatly diminished.
NωV subunits normally undergo an autoproteolytic cleavage between residues 570 and 571 (1, 22); this converts the 70-kDa α protein into 62-kDa β and 8-kDa γ components. The reaction has a half-life on the order of an hour and only is initiated once the compact capsid state has been attained (5). Based on the structural similarities with nodaviruses such as Flock house virus, cleavage of the γ peptide in NωV is believed to be essential for infection (26). NωV VLPs labeled with MIANS showed almost no cleavage after 4 h of incubation in the pH 5.0 buffer at 25°C, while WT unlabeled VLPs were cleaved (Fig. 1a). Similarly, cleavage in unlabeled NωV VLPs incubated at pH 5.0 in the presence of a 1 mM concentration of the non-thiol-reactive MIANS analog ANS was significantly inhibited (see Fig. 3); lower concentrations of ANS, however, were not as effective at inhibiting maturation and preventing cleavage. Once the ANS was exhaustively washed away, the particles matured fully, exhibiting normal levels of cleavage and thus demonstrating that the ANS itself was not disrupting or irreversibly altering the particles and cleavage mechanism.
FIG. 1.
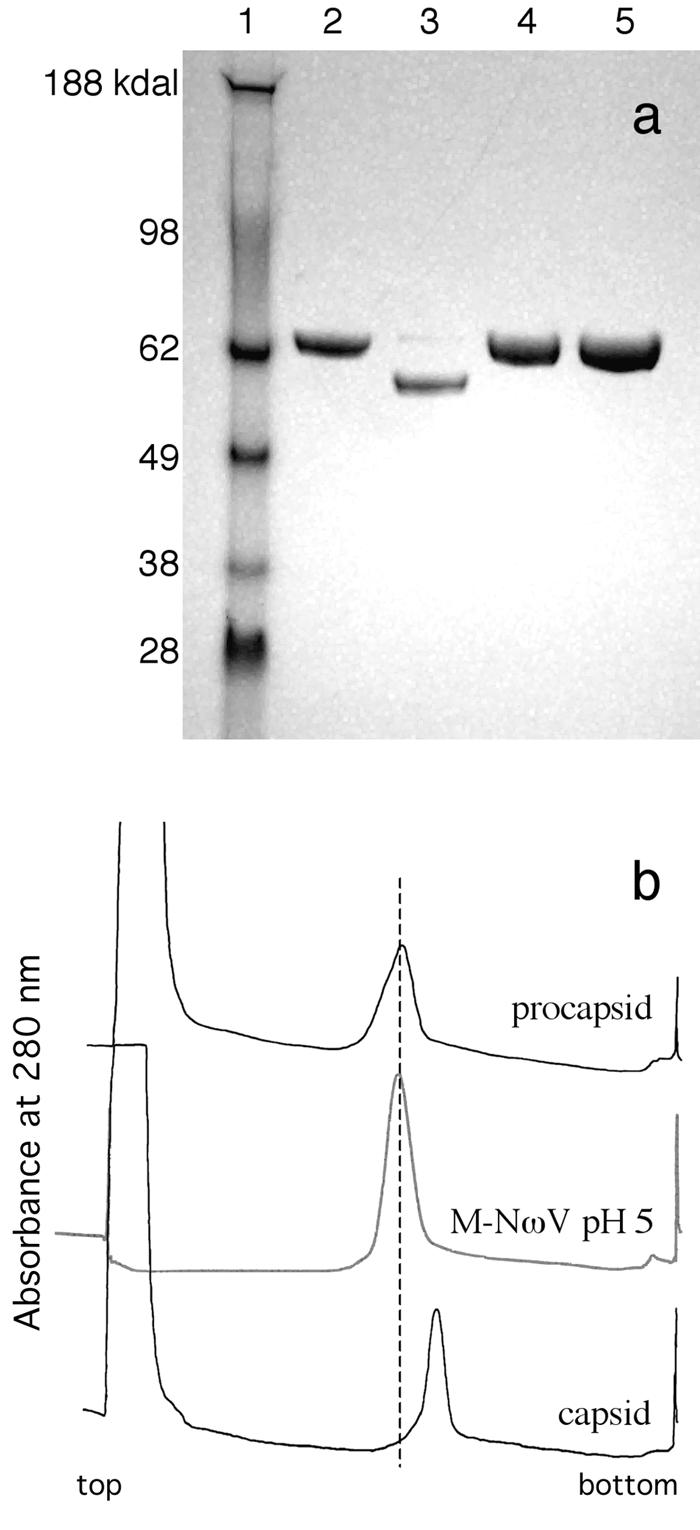
Labeled particles do not exhibit cleavage and sediment similarly to procapsid NωV during velocity sedimentation. (a) Labeled and unlabeled NωV samples run on a 4-to-12% denaturing acrylamide gradient gel. Lane assignment from left to right: 1, Invitrogen SeeBlue Plus 2 molecular weight standards; 2, WT procapsid pH 7.6; 3, WT capsid pH 5.0; 4, M-NωV pH 7.6; 5, M-NωV pH 5.0. (b) Fractionation of WT procapsid, capsid, and M-NωV particles sedimented through a 10-to-40% sucrose gradient. Ten-to-40% sucrose gradients (top of gradient on the left) prepared with 250 mM NaCl, pH 7.6 buffer for WT procapsid (buffer A) and 250 mM NaCl, pH 5.0 buffer (buffer B) for M-NωV and WT capsid. The curves presented are continuous measurements of the A280 for gradient fractionations.
FIG. 3.
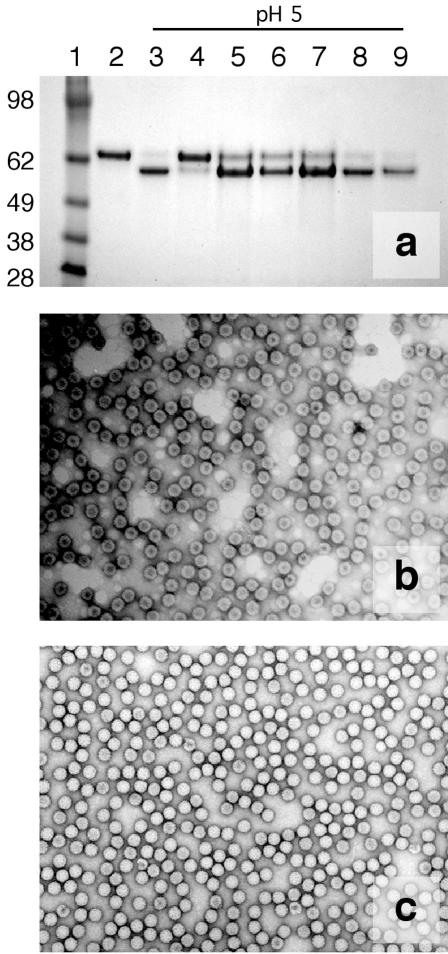
Noncovalently bound 1,8-ANS can inhibit maturation at pH 5.0 when present in high concentrations. If particles are washed with a 10% DMSO-90% buffer B solution, the ANS is removed and the particles mature normally. (a) Four-to-12% denaturing acrylamide gradient gel assay for subunit cleavage. Lanes from left to right: 1, Invitrogen SeeBlue Plus 2 molecular weight markers (sizes indicated in kilodaltons); 2, WT procapsid; 3, WT capsid (no ANS); 4, 1.0 mM ANS unwashed; 5, 1.0 mM ANS washed; 6, 0.3 mM ANS unwashed; 7, 0.3 mM ANS washed; 8, 0.1 mM unwashed; and 9, 0.1 mM washed. (b) Negative-stain EM micrograph of particles in 1.0 mM ANS at pH 5.0, revealing that the particles are intact but penetrable by uranyl-acetate stain. (c) Negative-stain EM micrograph of particles in 0.3 mM ANS at pH 5.0, revealing that particles are intact and appear to have matured to capsid.
M-NωV sediments similarly to unlabeled procapsid.
Velocity sedimentation through sucrose gradients provides a sensitive assay of hydrodynamic character for an entire population of particles. Sedimentation of unlabeled procapsid and capsid through a 10-to-40% sucrose gradient at 113,000 × g for 1.25 h consistently results in procapsid at fractions 17 to 18 and capsid at fractions 19 to 20 (31). Sedimentation of labeled M-NωV particles, at pH 5.0, generated a single peak at fractions 17 and 18 (Fig. 1b). By this assay, the labeled particles exhibited hydrodynamic properties similar to those of unlabeled procapsid, even at acidic pH.
M-NωV particles are permeable to negative stain at neutral and acidic pH.
Unlabeled procapsid and capsid NωV VLPs are distinguishable by negative-stain transmission EM (Fig. 2). Procapsids exhibit a round shape and are permeable to stain, whereas capsid particles are polyhedral and are mostly impermeable to stain. Significant populations of procapsids break apart, presumably due to the low-pH shock produced by uranyl-acetate staining, while capsid particles maintain their integrity. Figure 2 also shows that M-NωV particles were permeable to stain at both neutral and acidic pH. Yet, compared with the fragile NωV procapsid particles (Fig. 2c), the labeled particles appeared regular and robust at low pH (Fig. 2b). M-NωV particles at high pH also appeared to be somewhat less prone to breaking apart than the unlabeled procapsid.
FIG. 2.
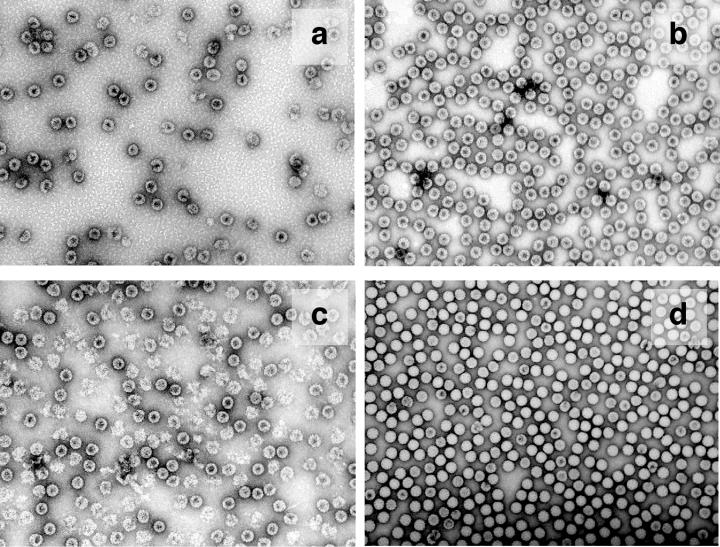
Negative-stain EM micrographs. (a) M-NωV in pH 7.6 buffer A; (b) M-NωV in pH 5.0 buffer B; (c) WT NωV pH 7.6 buffer A; (d) WT NωV pH 5.0 buffer B. Magnification, ×52,000.
Particles incubated in 1 mM ANS at pH 5.0 likewise were permeable to the stain, whereas particles in 0.3 mM ANS appeared to have matured fully to form mostly stain-impermeable capsids (Fig. 3). These results corroborate the findings that VLPs in 1 mM ANS are unable to undergo autoproteolytic cleavage maturation (Fig. 3), while those in 0.3 mM ANS mature normally. Negative-stain EM micrographs also confirmed that ANS does not break apart the particles.
EM reconstructions and pseudoatomic model of M-NωV at pH 5.
The three-dimensional cryo-EM reconstruction of labeled VLPs at pH 5.0 is shown in Fig. 4, with reconstructions of unlabeled NωV procapsid (pH 7.6) and capsid (pH 5.0) adapted from Canady et al. (4) for comparison. The unlabeled procapsid and capsid exhibit distinct surface morphologies due to different configurations of the immunoglobulin (Ig) domains and lack of close-packed β-sandwich interfaces in procapsid (4). Ig domains in the procapsid form dumbbell-shaped surface protrusions corresponding to dimer pairs between the A and B and the C and D subunits. The dimers differentiate into two categories of trimer (ABC and DDD) in mature capsid, dramatically altering the particle shape and surface morphology. The internal surface of the procapsid reconstruction exhibits a trefoil-shaped density at the three-fold and quasi-three-fold axes. This density is absent in the capsid and may denote a region of the subunit that refolds during the transition (4).
FIG. 4.
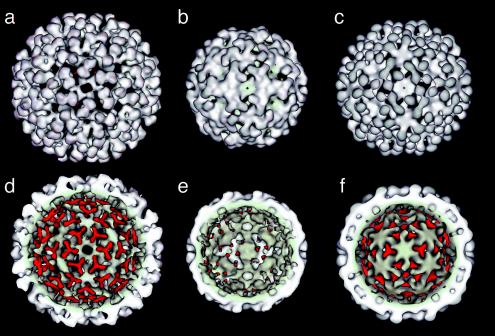
Cryo-EM reconstructions for procapsid, capsid, and M-NωV at pH 5.0 particles. (a, b, and c) Outer surface view of the procapsid, capsid, and M-NωV particle, respectively. (d, e, and f) Inner surface view with the trefoil feature highlighted in red. Capsid and procapsid reconstructions were adapted from Canady et al. (4).
The reconstruction of M-NωV particles at pH 5.0 (Fig. 4) resembles procapsid despite the fact that at pH 5.0 unlabeled NωV exists in the compact capsid form. Closer examination revealed that M-NωV at pH 5.0 was intermediate in size between procapsid and capsid, with an approximate diameter of 440 Å. The size determination by EM contrasted with the results of a sedimentation assay that suggested the M-NωV particles at pH 5 were similar in size to procapsid. This indicates that the sucrose gradient sedimentation assay is more sensitive to the hydrodynamic surface properties and porous nature of the particles than to the relatively small change in particle diameter. Indeed, the exterior of the labeled particles at pH 5 was very procapsid-like (Fig. 4). The Ig domains were still dimeric dumbbells, and close packing of interfaces had not yet occurred. The trefoil densities observed on the inner surface of procapsid persisted, but densities at three-fold and quasi-three-fold axes differed, with the latter showing a weaker density, suggesting that subunits are differentiated to a greater extent than in procapsid. Holes present in the procapsid structure at quasi-six-fold and five-fold axes were closed, but pores still penetrated at the three-fold and quasi-three-fold axes (see Fig. 5). The existence of the open pores likely was responsible for the penetration of negative stain into M-NωV particles (Fig. 2). The NωV subunit X-ray coordinates were docked into the cryo-EM density and refined as four rigid bodies in the icosahedral asymmetric unit. The excellent fit to the density was consistent with limited refolding between the capsid-based X-ray structure and the inhibited, low-pH, procapsid-like structure (Fig. 4).
FIG. 5.
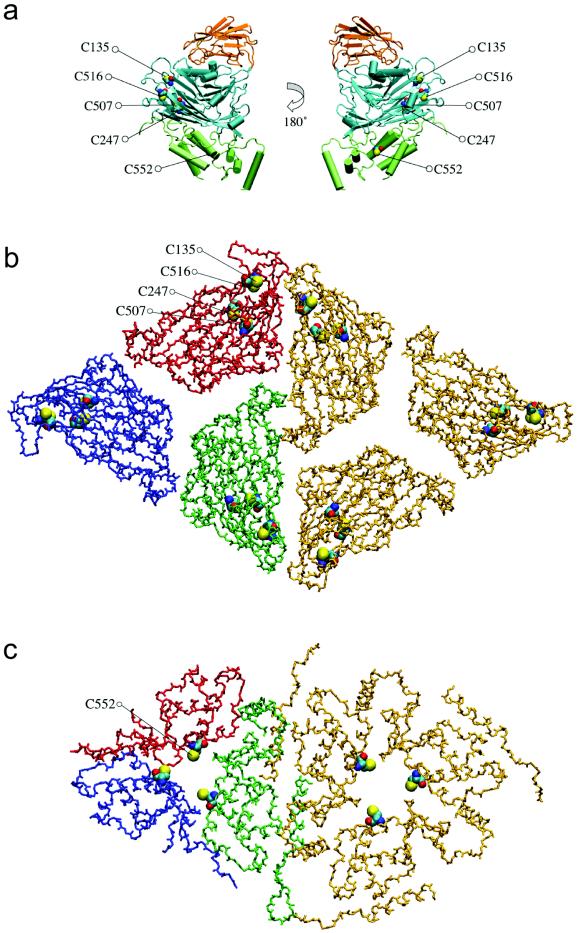
(a) Location of five of the six cysteine residues in the NωV D subunit; CYS-37 is not observed in the crystallographic structure. The Ig-like domain is orange, the β-sandwich domain is cyan, and the helical internal domain is green. (b) View of the pseudoatomic model for M-NωV at pH 5.0 down the threefold and quasi-threefold axes; only the polypeptide backbone trace for the β-sandwich domains is depicted. Cysteines 135, 247, 507, and 516 are rendered as space-filling models. Subunit interfaces are open and not tightly packed. (c) Same view as in panel b, but of the internal helical domain and CYS-552.
DISCUSSION
NωV morphogenesis and maturation involve three main stages: (i) an assembly event in which capsid protein subunits and nucleic acid coalesce to form a fragile particle with subunits loosely arranged on a T=4 quasi-equivalent lattice (procapsid), (ii) a pH-dependent conformational compaction in which subunits undergo a dramatic rearrangement to form the more robust, closed capsid, and (iii) an autoproteolytic cleavage event that locks the particles into their capsid states (4). Comparing particle dimensions and the refined pseudoatomic coordinates of M-NωV with those of the capsid and procapsid (Table 1), we estimate that M-NωV has progressed halfway along the trajectory between procapsid and capsid. Indeed, the pH 5.0 M-NωV reconstruction was consistent with data obtained from X-ray scattering experiments for a structural intermediate that becomes populated during the course of this transition (5). We conclude that M-NωV at pH 5.0 represents a trapped intermediate along the maturation pathway between procapsid and capsid. This result underscores the fact that labeling of NωV with MIANS did not disable the pH-triggering event that initiates the conformational compaction of procapsid. Instead, it appears that the particles were prevented from attaining their fully compact conformation that is required for autoproteolytic cleavage.
The crystal structure of the NωV capsid revealed that the structural control elements required for the formation of the T=4 lattice reside in the N- and C-terminal helical regions of the subunit and that these are internal in the particles (22). Canady et al. suggested that a trefoil of internal density located at icosahedral and quasi-three-fold symmetry axes in the procapsid but absent in the capsid may correspond to segments of the subunit that become unstructured at low pH (Fig. 4) (4). Taylor et al. have also suggested that this portion of the subunit domains may represent the conformational switch that directs the transformation as a whole (31). We attribute the trefoil-shaped density to the clustered internal domain helices of the NωV subunit (Fig. 5), which form trimeric interactions around the three-fold and quasi-three-fold vertices. The trefoils are present in the M-NωV intermediate structure determined at pH 5.0, although at reduced density around the quasi-three-fold axes. This reduction in trefoil density around the quasi-three-fold axes (A, B, and C subunits) is accompanied by the appearance of additional density at the five-fold vertex. This may indicate that the procapsid-capsid transition is initiated by movement of subunits around the five-fold vertex, namely the A subunit which is at the vertex and the B subunits that interact closely with the A subunits. Trefoils centered on the true three-fold axes (D subunits) are found with nearly the same density as in the procapsid structure, suggesting that these portions of the icosahedral assembly mature at a later stage.
The trefoil structures may persist at lower pH because the MIANS as well as ANS bind directly to portions of this domain and inhibit its dissolution by unfolding of helices or dissociation of paired helices at low pH. ANS and the closely related compound bis-ANS have been observed to influence protein conformation and oligomerization in many other systems. In some cases, binding of the fluorophore to proteins induces tightened conformational states, particularly when the proteins are initially denatured or only partly structured (17, 21). In NωV, binding of ANS or attachment of MIANS to the helical domain may stabilize the trefoil conformation, thus restraining the switching mechanism that drives the conformational maturation to completion. The EM reconstruction suggests this inhibition is most effective for the D subunits that ring the three-fold axes.
Based upon the pseudoatomic structure we have determined, the largest gaps between subunits in the maturation-arrested particles are located along the three-fold and quasi-three-fold axes (Fig. 5). Of the six cysteine residues present in each subunit, CYS-552 is the only residue located along the three-fold and quasi-three-fold axes. CYS-552 is also the only cysteine that is located on the internal helical domain of the subunit which, as described above, is believed to contribute to the trefoil density observed by EM. The locations of the other cysteines make them less likely to be involved in arresting the conformational change. CYS-247 and CYS-507 are buried in the β-sandwich domain of the virus; as such they are unlikely to be readily accessible to covalent modification by bulky molecules such as MIANS. CYS-37 is not crystallographically ordered, and thus its precise location is not known. However this residue is surrounded by several arginine and lysine residues that are believed to interact with the encapsidated RNA. CYS-135 is the only cysteine situated on the exterior surface of the mature capsid. While this residue sits near a subunit interface, compounds bound to this site probably are not sterically restrained, and thus they are probably incapable of preventing the subunit interfaces from sealing shut. CYS-516 is located along the face of the β-sandwich domain, relatively distant from the three-fold and quasi-three-fold vertices. Our working hypothesis is that labeling of CYS-552 may be critical for inhibition of the conformational maturation.
Is MIANS unique in its ability to inhibit particle maturation? We have demonstrated that noncovalently bound ANS alone will inhibit the transition at sufficient concentrations. However, we do not know the location of the specific site(s) at which ANS binds to the capsid protein; indeed, ANS may interact with capsid proteins through generalized, nonspecific association at numerous hydrophobic surface patches. The covalent attachment of MIANS is significant, because it localizes the inhibitory compounds to specific sites that also successfully arrest the transition. Preliminary studies involving a variety of other compounds conjugated to the cysteines in NωV also revealed inhibition of the maturation. These compounds include an uncharged, 252-Da spin label (TEMPO-maleimide), as well as bulkier fluorophores such as PyMPO and Oregon Green 488-maleimide that bear opposite electrostatic charges (K. K. Lee, B. Bothner, and J. E. Johnson, unpublished data). Although the role of charge burial cannot be ruled out (13, 27), one feature these compounds have in common with MIANS is a bulky apolar moiety. The specific structures of this moiety vary widely. This suggests that the protein-protein interactions that must undergo rearrangement during maturation are highly sensitive to perturbation at their interfaces, particularly when it is localized to the pressure points marked by the cysteine residues.
Whereas studies of disruption of virus function by capsid-directed compounds in other systems have demonstrated inhibition of proper assembly (24, 28, 32, 34), in the case of the M-NωV capsids the correct assembly has already been attained, and the interfering molecule does not break down the assembly. Instead, it appears that a later stage involving a large-scale conformational reorganization, required for maturation, has been inhibited by the introduction of the inhibitory compounds at interfaces. In this respect, the actions of the MIANS and ANS compounds on NωV maturation are similar to the action of the WIN compounds that inhibit picornavirus infectivity by binding to fully formed virus capsids and inhibiting dynamic quaternary breathing motions that are necessary for infection (20, 25). Clearly, viral capsids can be targeted at several stages of the viral life cycle: by disrupting subunit polymerization (24, 28, 32, 34) and by inhibiting dynamic capsid breathing (20, 25), as well as by preventing maturation of the already-assembled capsid.
Lastly, the covalently modified NωV particles appear to be stable and more robust than procapsid NωV. We are, thus, presented with an opportunity to characterize an alternative state of subunit organization in this virus by using moderate (EM) and high-resolution (X-ray crystallography) methods. This will provide insight into the mechanisms of large-scale conformational transformations in these massive macromolecular complexes.
Acknowledgments
This work was supported by NIH grant R01-GM54076. K. K. Lee is supported by a postdoctoral fellowship grant from the National Institutes of Health (F32-GM65013).
REFERENCES
- 1.Agrawal, D. K., and J. E. Johnson. 1995. Assembly of the T=4 Nudaurelia capensis omega virus capsid protein, post-translational cleavage, and specific encapsidation of its mRNA in a baculovirus expression system. Virology 207:89-97. [DOI] [PubMed] [Google Scholar]
- 2.Andley, U. P., J. N. Liang, and B. Chakrabarti. 1982. Spectroscopic investigations of bovine lens crystallins. 2. Fluorescent probes for polar-apolar nature and sulfhydryl group accessibility. Biochemistry 21:1853-1858. [DOI] [PubMed] [Google Scholar]
- 3.Bonafe, C. F., M. Glaser, E. W. Voss, G. Weber, and J. L. Silva. 2000. Virus inactivation by anilinonaphthalene sulfonate compounds and comparison with other ligands. Biochem. Biophys. Res. Commun. 275:955-961. [DOI] [PubMed] [Google Scholar]
- 4.Canady, M. A., M. Tihova, T. N. Hanzlik, J. E. Johnson, and M. Yeager. 2000. Large conformational changes in the maturation of a simple RNA virus, Nudaurelia capensis omega virus (NωV). J. Mol. Biol. 299:573-584. [DOI] [PubMed] [Google Scholar]
- 5.Canady, M. A., H. Tsuruta, and J. E. Johnson. 2001. Analysis of rapid, large-scale protein quaternary structural changes: time-resolved X-ray solution scattering of Nudaurelia capensis omega virus (NωV) maturation. J. Mol. Biol. 311:803-814. [DOI] [PubMed] [Google Scholar]
- 6.Chapman, M. S. 1995. Restrained real-space macromolecular atomic refinement using a new resolution-dependent electron density function. Acta Crystallogr. A 51:69-80. [Google Scholar]
- 7.Conway, J. F., R. L. Duda, N. Cheng, R. W. Hendrix, and A. C. Steven. 1995. Proteolytic and conformational control of virus capsid maturation: the bacteriophage HK97 system. J. Mol. Biol. 253:86-99. [DOI] [PubMed] [Google Scholar]
- 8.Conway, J. F., and A. C. Steven. 1999. Methods for reconstructing density maps of “single” particles from cryoelectron micrographs to subnanometer resolution. J. Struct. Biol. 128:106-118. [DOI] [PubMed] [Google Scholar]
- 9.Conway, J. F., W. R. Wikoff, N. Cheng, R. L. Duda, R. W. Hendrix, J. E. Johnson, and A. C. Steven. 2001. Virus maturation involving large subunit rotations and local refolding. Science 292:744-748. [DOI] [PubMed] [Google Scholar]
- 10.Deres, K., C. H. Schroder, A. Paessens, S. Goldmann, H. J. Hacker, O. Weber, T. Kramer, U. Niewohner, U. Pleiss, J. Stoltefuss, E. Graef, D. Koletzki, R. N. Masantschek, A. Reimann, R. Jaeger, R. Gross, B. Beckermann, K. H. Schlemmer, D. Haebich, and H. Rubsamen-Waigmann. 2003. Inhibition of hepatitis B virus replication by drug-induced depletion of nucleocapsids. Science 299:893-896. [DOI] [PubMed] [Google Scholar]
- 11.Dokland, T., and H. Murialdo. 1993. Structural transitions during maturation of bacteriophage lambda capsids. J. Mol. Biol. 233:682-694. [DOI] [PubMed] [Google Scholar]
- 12.Dong, X. F., P. Natarajan, M. Tihova, J. E. Johnson, and A. Schneemann. 1998. Particle polymorphism caused by deletion of a peptide molecular switch in a quasiequivalent icosahedral virus. J. Virol. 72:6024-6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fitch, C. A., D. A. Karp, K. K. Lee, W. E. Stites, E. E. Lattman, and E. B. Garcia-Moreno. 2002. Experimental pKa values of buried residues: analysis with continuum methods and role of water penetration. Biophys. J. 82:3289-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frank, J., M. Radermacher, P. Penczek, J. Zhu, Y. Li, M. Ladjadj, and A. Leith. 1996. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116:190-199. [DOI] [PubMed] [Google Scholar]
- 15.Heymann, J. B., N. Cheng, W. W. Newcomb, B. L. Trus, J. C. Brown, and A. C. Steven. 2003. Dynamics of herpes simplex virus capsid maturation visualized by time-lapse cryo-electron microscopy. Nat. Struct. Biol. 10:334-341. [DOI] [PubMed] [Google Scholar]
- 16.Jiang, W., Z. Li, Z. Zhang, M. L. Baker, P. E. Prevelige, Jr., and W. Chiu. 2003. Coat protein fold and maturation transition of bacteriophage P22 seen at subnanometer resolutions. Nat. Struct. Biol. 10:131-135. [DOI] [PubMed] [Google Scholar]
- 17.Kamen, D. E., and R. W. Woody. 2001. A partially folded intermediate conformation is induced in pectate lyase C by the addition of 8-anilino-1-naphthalenesulfonate (ANS). Protein Sci. 10:2123-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lakowicz, J. R. 1999. Principles of fluorescence spectroscopy, 2nd ed. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 19.Lata, R., J. F. Conway, N. Cheng, R. L. Duda, R. W. Hendrix, W. R. Wikoff, J. E. Johnson, H. Tsuruta, and A. C. Steven. 2000. Maturation dynamics of a viral capsid: visualization of transitional intermediate states. Cell 100:253-263. [DOI] [PubMed] [Google Scholar]
- 20.Lewis, J. K., B. Bothner, T. J. Smith, and G. Siuzdak. 1998. Antiviral agent blocks breathing of the common cold virus. Proc. Natl. Acad. Sci. USA 95:6774-6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matulis, D., C. G. Baumann, V. A. Bloomfield, and R. E. Lovrien. 1999. 1-Anilino-8-naphthalene sulfonate as a protein conformational tightening agent. Biopolymers 49:451-458. [DOI] [PubMed] [Google Scholar]
- 22.Munshi, S., L. Liljas, J. Cavarelli, W. Bomu, B. McKinney, V. Reddy, and J. E. Johnson. 1996. The 2.8 Å structure of a T=4 animal virus and its implications for membrane translocation of RNA. J. Mol. Biol. 261:1-10. [DOI] [PubMed] [Google Scholar]
- 23.Prasad, B. V., P. E. Prevelige, E. Marietta, R. O. Chen, D. Thomas, J. King, and W. Chiu. 1993. Three-dimensional transformation of capsids associated with genome packaging in a bacterial virus. J. Mol. Biol. 231:65-74. [DOI] [PubMed] [Google Scholar]
- 24.Prevelige, P. E., Jr. 1998. Inhibiting virus-capsid assembly by altering the polymerisation pathway. Trends Biotechnol. 16:61-65. [DOI] [PubMed] [Google Scholar]
- 25.Reisdorph, N., J. J. Thomas, U. Katpally, E. Chase, K. Harris, G. Siuzdak, and T. J. Smith. 2003. Human rhinovirus capsid dynamics is controlled by canyon flexibility. Virology 314:34-44. [DOI] [PubMed] [Google Scholar]
- 26.Schneemann, A., W. Zhong, T. M. Gallagher, and R. R. Rueckert. 1992. Maturation cleavage required for infectivity of a nodavirus. J. Virol. 66:6728-6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schutz, C. N., and A. Warshel. 2001. What are the dielectric “constants” of proteins and how to validate electrostatic models? Proteins 44:400-417. [DOI] [PubMed] [Google Scholar]
- 28.Tang, C., E. Loeliger, I. Kinde, S. Kyere, K. Mayo, E. Barklis, Y. Sun, M. Huang, and M. F. Summers. 2003. Antiviral inhibition of the HIV-1 capsid protein. J. Mol. Biol. 327:1013-1020. [DOI] [PubMed] [Google Scholar]
- 29.Tang, J., D. W. Taylor, and K. A. Taylor. 2001. The three-dimensional structure of alpha-actinin obtained by cryoelectron microscopy suggests a model for Ca2+-dependent actin binding. J. Mol. Biol. 310:845-858. [DOI] [PubMed] [Google Scholar]
- 30.Tao, Y., N. H. Olson, W. Xu, D. L. Anderson, M. G. Rossmann, and T. S. Baker. 1998. Assembly of a tailed bacterial virus and its genome release studied in three dimensions. Cell 95:431-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor, D. J., N. K. Krishna, M. A. Canady, A. Schneemann, and J. E. Johnson. 2002. Large-scale, pH-dependent, quaternary structure changes in an RNA virus capsid are reversible in the absence of subunit autoproteolysis. J. Virol. 76:9972-9980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teschke, C. M., J. King, and P. E. Prevelige, Jr. 1993. Inhibition of viral capsid assembly by 1,1′-bi(4-anilinonaphthalene-5-sulfonic acid). Biochemistry 32:10658-10665. [DOI] [PubMed] [Google Scholar]
- 33.Trus, B. L., F. P. Booy, W. W. Newcomb, J. C. Brown, F. L. Homa, D. R. Thomsen, and A. C. Steven. 1996. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J. Mol. Biol. 263:447-462. [DOI] [PubMed] [Google Scholar]
- 34.Zlotnick, A., P. Ceres, S. Singh, and J. M. Johnson. 2002. A small molecule inhibits and misdirects assembly of hepatitis B virus capsids. J. Virol. 76:4848-4854. [DOI] [PMC free article] [PubMed] [Google Scholar]


