Abstract
Tumors with somatic mutations in the proofreading exonuclease domain of DNA polymerase epsilon (POLE-exo*) exhibit a novel mutator phenotype, with markedly elevated TCT→TAT and TCG→TTG mutations and overall mutation frequencies often exceeding 100 mutations/Mb. Here, we identify POLE-exo* tumors in numerous cancers and classify them into two groups, A and B, according to their mutational properties. Group A mutants are found only in POLE, whereas Group B mutants are found in POLE and POLD1 and appear to be nonfunctional. In Group A, cell-free polymerase assays confirm that mutations in the exonuclease domain result in high mutation frequencies with a preference for C→A mutation. We describe the patterns of amino acid substitutions caused by POLE-exo* and compare them to other tumor types. The nucleotide preference of POLE-exo* leads to increased frequencies of recurrent nonsense mutations in key tumor suppressors such as TP53, ATM, and PIK3R1. We further demonstrate that strand-specific mutation patterns arise from some of these POLE-exo* mutants during genome duplication. This is the first direct proof of leading strand-specific replication by human POLE, which has only been demonstrated in yeast so far. Taken together, the extremely high mutation frequency and strand specificity of mutations provide a unique identifier of eukaryotic origins of replication.
Damage to cellular DNA repair systems leads to a high frequency of mutations in all species (Kunkel and Erie 2005; Reha-Krantz 2010) and predisposes them to the development of cancer (Loeb 2011). The best-characterized mutator phenotype in cancer patients involves mismatch repair genes, principally MLH1, which is transcriptionally silenced by methylation of the promoter, leading to microsatellite instability (MSI) (Kane et al. 1997; Vilar and Gruber 2010). MSI is most prevalent in cancers of the colon and rectum (15%), cancers of the stomach (20%), and endometrium (30%). Rare occurrences of MSI are observed in a variety of other cancers, such as pancreatic adenocarcinoma, glioblastoma multiforme, and renal cell chromophobe tumors. Frequent insertion and deletion mutations (indels), especially at homopolymer and dinucleotide repeats, define MSI tumors. Whereas microsatellite stable (MSS) tumors exhibit mutation frequencies ranging from one to 10 mutations per Mb, MSI tumors from the same cancer are hypermutated, ranging from 10 to 100 mutations per Mb. MSI tumors also feature reduced chromosome instability compared to MSS tumors from the same organ (Ciriello et al. 2013).
Recently, large-scale genetic and genomic studies have revealed tumors mutated in the exonuclease (proofreading) domain of DNA polymerase epsilon (POLE), which is encoded by POLE, one of two major DNA polymerases responsible for replication of the eukaryotic nuclear genome. Approximately 3% of colorectal and 7% of endometrial cancers are reported with mutations in the proofreading domain of POLE (POLE-exo*) (The Cancer Genome Atlas Network 2012; Church et al. 2013; Palles et al. 2013; Kane and Shcherbakova 2014; Meng et al. 2014; Valle et al. 2014; Zou et al. 2014). The mutation incidence in tumors with this defect often exceeds 100 mutations/Mb. The spectrum of nucleotide changes in POLE-exo* tumors is distinct from the mutation spectrum in MSI and MSS tumors, characterized mainly by a prevalence of TCT→TAT and TCG→TTG mutations (see also Supplemental Fig. S1; The Cancer Genome Atlas Network 2012, 2013; Alexandrov et al. 2013; Kane and Shcherbakova 2014).
Studies in yeast based on selected, well-characterized origins of replication (ORI) demonstrated that POLE synthesizes the leading strand (Pursell et al. 2007b; Miyabe et al. 2011), whereas DNA polymerase delta (POLD1), which is encoded by POLD1, synthesizes the lagging strand (Nick McElhinny et al. 2008).
Although some colorectal and endometrial tumors have been reported with POLD1 exonuclease mutations (POLD1-exo*) (Palles et al. 2013; Valle et al. 2014), their relationship to the mutational properties of those tumors is not clear. Preliminary data on survival of patients harboring mutations in POLE suggest they have significantly better outcomes (The Cancer Genome Atlas Network 2013; Meng et al. 2014) and therefore may require less aggressive treatment. Thus, careful characterization of POLE-exo* and POLD1-exo* mutants is important to shed light on mutational processes leading to cancer, to impact therapeutic decision making, and even to yield clues into the fundamental DNA replicative processes in higher eukaryotes. In this study, we present a detailed analysis of POLE-exo* mutations in endometrial and colorectal tumors. Our results show that the novel patterns of somatic mutation described above are strand-specific and uniquely associated with a subset of POLE-exo* tumors. Moreover, these distinctive patterns confirm that POLE synthesizes the leading strand in humans.
Results
Classification of POLE-exo* mutants
Since the first report of a POLE-exo* mutation in human colorectal cancer (Yoshida et al. 2010), 50 mutations have been reported in the exonuclease domain of POLE in human cancers, mainly through large-scale sequencing studies (Table 1). In addition to colorectal and endometrial cancers, POLE-exo* was observed in <1% of glioblastoma multiforme, low grade glioma, lung adenocarcinoma, pancreatic ductal adenocarcinoma, breast invasive carcinoma, bladder adenocarcinoma, renal cell chromophobe carcinoma, head and neck cancer, and stomach cancers, representing a screen of 3719 patients in 12 cancers (data retrieved from The Cancer Genome Atlas data portal). As in colorectal and endometrial cancers, these POLE-exo* tumors exhibited ultrahigh substitution mutation frequencies, over half of which exceed 100 mutations/Mb (Table 1; Supplemental Table S1A). Overall, almost half the POLE-exo* occurred in either of two hot spots, encoding amino acid substitutions in P286R, H or S (15) and V411L (11). Four other amino acid substitutions were observed in at least two tumors: S297F, F367S, A456P, and S459F (Supplemental Table S1A).
Table 1.
Exonuclease domain mutations in replicative DNA polymerases in TCGA data

In most POLE-exo* tumors (40/50), C→A mutations regardless of immediate sequence context accounted for >20% of all substitutions, whereas all MSI or MSS tumors from these same cancers have C→A mutation frequencies <20% (Fig. 1; see Table 2). The frequencies of all other base substitution mutations and indels were similar to those observed in the MSS tumors (Fig. 1; Supplemental Fig. S2C). The extraordinary mutation frequencies of POLE-exo* tumors were accounted for almost exclusively by increased numbers of TCT→TAT and TCG→TTG substitutions (Fig. 2A). Together, TCG→TTG and TCT→TAT mutations account for >50% of the mutations found in POLE-exo* mutated tumors.
Figure 1.
Mutation spectra of POLE and POLD1 exonuclease domain-mutated tumors. The color code for the type of mutation is shown in the key above the histogram. Tumors are grouped first by polymerase and then by affected residue with C→A frequencies decreasing from left to right. The darker blue designates Group A, medium royal blue Group B, and for comparison, white designates wild-type polymerase in MSI and MSS. Black horizontal dotted line demarks 20% C→A, the provisional threshold for classifying Group A. Black vertical line separates Group A from Group B, and wild-type polymerase categories. In addition, the horizontal bar at the top of the histogram shows the group designation. Cancer abbreviations are the following: (CRC) TCGA COAD/READ colorectal cancers; (EEC) endometrial cancer; (STAD) stomach adenocarcinoma; (GBM) glioblastoma; (LGG) low grade glioma; (BRCA) breast carcinomas; (PAAD) pancreatic adenocarcinoma; (HNSC) head and neck cancer; (KICH) kidney chromophobe. The MSI status, type of cancer, and mutation frequency range are color coded as shown in the key below the histogram. Other abbreviations are as follows: (MSS) microsatellite stable; (MSI) microsatellite instable; (nt) not tested. The white asterisk in the cancer track demarks a pancreatic tumor that had both a POLE-exoP286R and POLD1-exoR311H. See structural features of these mutations in Supplemental Figure S1.
Table 2.
Exonuclease mutation categories

Figure 2.
Mutation propensities and structural relationships in POLE- and POLD1-exo* mutants. (A) Frequency of base substitutions with 5′ and 3′ contexts in colorectal (CRC) and endometrial (EEC) cancers in the most common mutation classes. (Upper) TCT→TAT; (lower) TCG→TTG; (left) CRC; (right) EEC. Individual dots represent mutation frequency in individual tumors. Group assignments are represented by colors. (Blue) POLE-exo* Group A; (green) MSI; (yellow) MSS (see also Supplemental Table S1A,B; Supplemental Figs. S1, S2). (B) Distance in angstroms of POLE-exo* mutations to the exonuclease catalytic residues. POLE-exo* Group A, POLE-exo* Group B, and POLD1-exo* Group B mutant residues were mapped to the yeast 3D structure, POLE PDB accession 4M8O (Hogg et al. 2014), and to POLD1-exo DNA PDB accession 3IAY (Swan et al. 2009) by homology using PyMOL (http://www.pymol.org). PyMol was used to estimate the distance from the mutated residue to either catalytic site residue. Top panels are the distances to the catalytic glutamate (Glu) residue of either polymerase; bottom panels are the distances to the catalytic aspartate (Asp) residue of either polymerase. The Group A or B category is defined in Tables 1, 2 (see also Supplemental Table S5). The polymerase group is indicated on the x-axis and the distance to either catalytic residue on the y-axis. (Asp) Exonuclease active site residues D275 (POLE) or D316 (POLD1); (exo) exonuclease catalytic residues E277 (POLE) and E318 (POLD1). Note that no Group A or B mutations have yet been found with 11 Å of the exonuclease catalytic site of POLD1. (See also Supplemental Fig. S1A,B.)
A group of 10 POLE-exo* mutants did not exhibit elevated context-dependent C→A and C→T mutation frequencies. These were observed in MSI or MSS tumors (Table 1; Supplemental Table S1) and had correspondingly lower mutation rates. Therefore, we divided the POLE-exo* mutants into two groups: Group A tumors were predominantly ultramutated, with context-dependent C→A frequency greater than 0.2, and contained several recurrent mutation sites within the POLE exonuclease domain. Group B tumors were predominantly MSI or MSS, with low frequencies of C→A mutations (<0.2) (Table 2) and no recurrent mutation sites within the POLE exonuclease domain. The Group B POLE-exo* mutants appear to be nonfunctional.
Among the same 3719 patients, we identified POLD1-exo* mutations in 13 samples (Supplemental Table S1B), all of which had mutation profiles similar to the ones observed in Group B POLE-exo* mutants. One exception was an ultramutated pancreatic tumor with POLD1-exoR311H with a high mutation frequency and C→A frequency (421/Mb; 0.21); but this tumor was also mutated in POLE-exoP286R, which is sufficient to explain the Group A phenotype we observed.
One gastric tumor with a POLD1-exoD316N mutation had a mutation frequency of 286/Mb, but its C→A frequency was low (0.1). The D316 residue in the exonuclease active site participates in the catalytic reaction. Mutation of the yeast homolog of POLD1-exoD316 abolishes exonuclease activity (Jin et al. 2001; Shevelev and Hübscher 2002). The phenotype of this mutation was unique among POLD1- and POLE-exo* and was left unclassified (see Tables 1, 2; Supplemental Table S1B).
Group A POLE-exo* mutations clustered nearer the exonuclease catalytic residues (POLE, D275 and E277; POLD1 D316 and E318). In contrast, Group B POLE-exo* and POLD1-exo* mutations were distributed around the exonuclease domain, more often found on the periphery, further away from the catalytic residues (Fig. 2B; see Supplemental Fig. S2AB).
Mutation of POLE-exo impairs 3′-5′ exonuclease activity
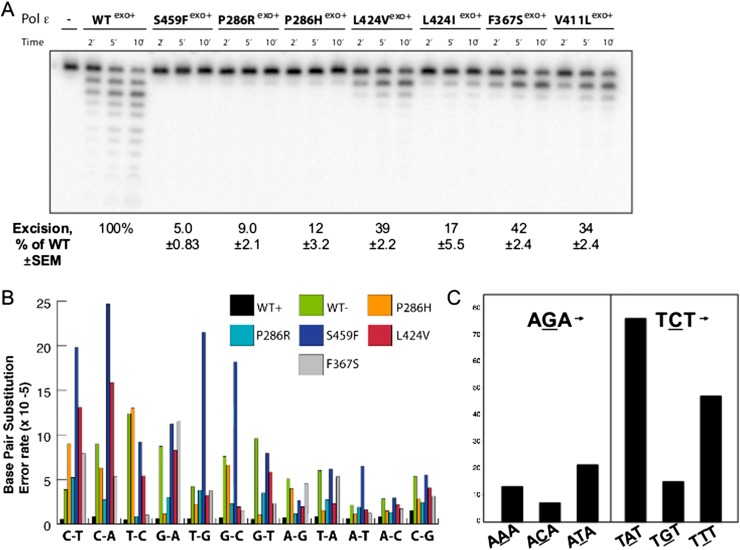
We used site-directed mutagenesis to introduce several CRC and endometrial cancer mutations into our previously described POLE expression construct (Korona et al. 2011; Goksenin et al. 2012), containing all six conserved motifs necessary for 3′-5′ exonuclease and DNA synthesis activities (Supplemental Fig. S3A). Although this construct encodes the N-terminal 140 kDa of the catalytic subunit, the equivalent yeast POLE construct was shown to have essentially identical replication fidelity to the four-subunit POLE holoenzyme and is therefore a useful substitute for in vitro replication fidelity assays (Pursell et al. 2007a). The amino acid substitutions in the POLE-exo domain had only marginal effects on DNA polymerase activity (Supplemental Fig. S3B); however, each mutation dramatically reduced 3′-5′ exonuclease activity relative to the wild-type POLE (Fig. 3A). Interestingly, the effects on exonuclease activity varied. The S459F substitution, which maps to the Exo III motif, reduced proofreading to a similar extent as the D275A/E277A amino acid substitutions, which essentially inactivate proofreading (Korona et al. 2011). Substitution of P286, a conserved Exo I residue, with either H or R caused similar reduction in excision. Interestingly, the F367S substitution, which is at a highly conserved residue immediately adjacent to the catalytic D368 in the Exo II motif, severely reduced, but did not completely abolish, exonuclease activity. The V411L variant, which is recurrent in CRC and endometrial tumors and maps to a conserved residue in between the Exo II and Exo III motifs, also reduced exonuclease activity without completely abolishing it (Fig. 3A). In contrast to Group A variants, the A428T Group B variant, which is found on the periphery of the active site, did not affect exonuclease function (Supplemental Fig. S4).
Figure 3.
POLE-exo* cancer mutations severely and differentially inhibit 3′→5′ exonuclease activity in a strand-specific manner. (A) The relative 3′-5′ exonuclease activities of the POLE cancer mutant variants were compared to those of the wild-type POLE enzyme (WTexo+). Reactions were carried out at 37°C and initiated by the addition of the enzyme. Aliquots were removed at the indicated times and resolved by 12% denaturing PAGE (see Methods for reaction details). Exonuclease activity was quantified and expressed as a percentage of wild-type activity (±SEM). (B) POLE cancer variants selectively affect specific replication errors in vitro. Error rates for each of the 12 possible individual base pair substitutions were calculated using a lacZ forward mutation assay as described by Bebenek and Kunkel (1995) and Korona et al. (2011). (WT) A previously characterized exonuclease-inactivating mutant (D275A/E277A). (C) POLE-exo* P286R exonuclease mutants exhibit a preference for TCT→TAT mutations in a cell-free reaction. Purified POLE containing the P286R substitution, three equally spaced oligonucleotide primers, and an M13mp2 single-stranded DNA template were used to generate a sequence product that was directly sequenced using Illumina paired end protocol. In the resulting collection of sequenced POLE replication errors, the count of TCT→TAT mutations is higher than the count of the complementary AGA→ATA mutations. (See also Supplemental Fig. S3.)
POLE-exo* mutants generate a unique pattern of replication errors
Amino acid substitutions in replicative polymerases that suppress or inactivate proofreading also cause an increase in extension from misincorporated nucleotides and subsequent increases in error rates (Shcherbakova et al. 2003; Fortune et al. 2005; Schmitt et al. 2009; Korona et al. 2011). We measured the effects on replication fidelity of the mutant POLE variants using a lacZ forward mutation assay (Bebenek and Kunkel 1995). The mutant POLE variants all increased the mutation frequency over the wild-type, exonuclease-proficient POLE (Supplemental Fig. S3C) in proportion to their relative impairments to proofreading. The S459F mutant increased the mutation frequency beyond that of the catalytic inactivation mutant. The mutation frequency of the D275A/E277A/S459F (100 × 10−4) triple mutant was identical to the D275A/E277A double mutant, suggesting a role for S459 only when the metal ion is properly coordinated by D275 and E275 (Yang 2011).
It is possible that the mutation spectrum observed in tumors with POLE-exo* results not only from increased mutation frequency of the polymerase, but also from the action of the mismatch repair system. To investigate the relative contribution of the mutant polymerase, we sequenced individual lacZ mutant clones from the forward mutation assay and calculated error rates for each individual mispairing. Although error rates varied for each mutant and each mispairing, only two errors were strongly elevated in every one of the POLE cancer mutants: C→A transversions and C→T transitions (Fig. 3B) irrespective of the context of flanking bases. Several POLE variants do show elevated error rates for a subset of different mispairings. In particular, several variants show increased T→C and G→A transitions. Both involve the formation of a G•T mispairing, which is the single most efficiently corrected mispairing by mismatch repair (Kunkel and Erie 2005), suggesting that any made by POLE mutants are likely readily removed in vivo. The S459F POLE variant shows high error rates for two additional errors, T→G and G→C transversions, which may indicate a more complex effect on the exonucleolytic mechanism for this mutant. Transversion errors are the most poorly corrected group of errors by mismatch repair (Schaaper and Dunn 1991; Lujan et al. 2012), suggesting that a POLE mutant-mediated increase in the C•dT mispairing-induced C→A errors could more easily saturate or circumvent the already less efficient mismatch repair correction. C→T transitions can arise in the lacZ forward mutation assay due to dA misinsertion opposite an undamaged C or opposite the uracil formed as a product of cytosine deamination. This shared elevation in error rate may reflect uracil bypass as well as true C•dA misincorporation (Barnes and Lindahl 2004).
In our lacZ system, there is no TCT→TAT change that permits direct observation and error rate calculation at this motif. An alternate way of determining the effects of sequence context on the specificity of POLE-induced TCT→TAT transversion errors is to directly sequence products of DNA synthesis. We used M13mp2 ssDNA, the same template DNA from the lacZ fidelity assay, as a template primed with three equally spaced oligonucleotide primers to generate products of DNA synthesis that could then be processed and sequenced directly. Our results using POLE-exoP286R show a preference for TCT→TAT transversions over AGA→ATA, confirming strand specificity of POLE (Fig. 3C). The analogous P286R substitution in yeast was recently shown to drive a strong mutator phenotype, supporting the idea that POLE replication errors may play a role in tumorigenesis (Kane and Shcherbakova 2014).
Genome-wide mutation patterns show replication strand bias
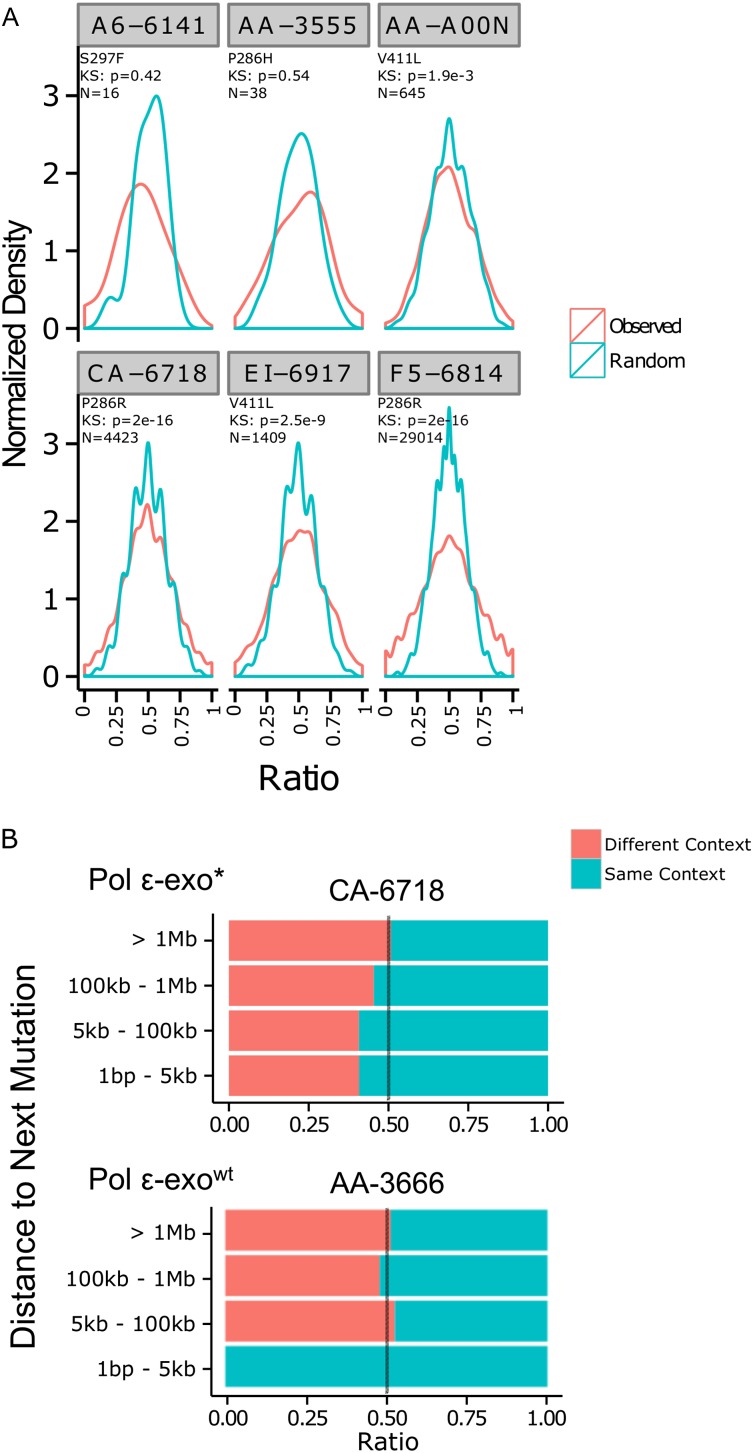
To ask what fraction of the genome exhibits strand bias, we tallied all TCT→TAT and AGA→ATA mutations in consecutive 50-kb windows across all chromosomes. The ratios of C→A to the total number of C→A plus G→T mutations in each window is shown as a frequency distribution in Figure 4A, compared to the distribution of ratios obtained from drawing an equivalent number of C→A and G→T mutations at random in each window. The POLE-exo* tumors exhibited increased accumulation of windows on the tails of the distribution (<0.2 and >0.8), and decreased accumulation in the center (0.5). The frequency distribution of C→A ratios differed from the random expectation in four of six tumors (Komolgrov Smirnov [KS] test P-value range 0.0019–2 × 10−16). The two tumors, A6-6141 and AA-3555, with KS P > 0.05, had relatively low mutation frequencies and less than 50 eligible windows across the genome (see Methods). In all cases, between 10% and 20% of the genome was found in 50-kb windows with <0.2 or >0.8 ratios (data not shown).
Figure 4.
Propensity of of POLE-exo* Group A mutations to cluster on DNA strands. (A) POLE-exo* context mutations (TCT→TAT and AGA→ATA) were counted in 50-kb windows across POLE-exo* tumors F5-6814, AA-A00N, EI-6917, CA-6718, AA-3555, and A6-6141 from TCGA. Normalized density of TCT→TAT mutations divided by total POLE-exo* context is shown in red, and randomized contexts are shown in blue. POLE-exo* tumors with high mutation rates show enrichment for extreme ratios of TCT→TAT mutations indicating clusters of same-strand mutation within the 50-kb windows. Four of six significantly deviate from random expectation by Kolmogorov-Smirnov (KS) P-value < 0.05. Two with KS P-value > 0.05 had low mutation frequencies and too few windows available (N) for a valid test (see Methods). (B) POLE-exo* context mutations were paired such that for each mutation, the partner was the next mutation at the indicated minimum genomic distance upstream. The POLE-exo* (CA-6718) tumor showed an enrichment for the same context (TCT→TAT, TCT→TAT or AGA→ATA, AGA→ATA) versus different context (TCT→TAT, AGA→ATA or AGA→ATA, TCT→TAT). This enrichment was not observed in the POLE-exowt (AA-3666) tumor. The blue bar in the 1-bp to 5-kb range of the POLE-wt only had 10 observations across the entire genome and may be a statistical outlier due to the limited number of observations. (See also Supplemental Fig. S5 for analysis of other patients.)
In a second test, for each POLE-exo* mutation we scanned ahead to the next POLE-exo* mutation that was within 1 kb, 5 kb, 100 kb, and 1 Mb away. We then classified these mutation pairs as either same-strand pairs or opposite-strand pairs and compared the ratios of same-strand pairs to opposite-strand pairs at the chosen distance intervals (Fig. 4B; Supplemental Fig. S5). We found that POLE-exo* tumors had consistent enrichment of same-strand mutations when close together, but became random at greater distance (1 Mb away). This enrichment was not observed in non-POLE-exo* tumors. These data indicate that POLE-exo* mutations are strand enriched in a spatially dependent manner.
Strand-specific changes correlate with origins of replication
We hypothesized that C→A mutations would be enriched on the leading strand near origins of replication in the genomes of POLE-exo* tumors, given the proposed role of POLE in replication in yeast. Whole-genome sequencing (WGS) data from six CRC POLE-exo* tumors were available for analysis. Because the leading strand is the plus-strand heading in one direction from an origin and the minus strand in the other direction, with all mutations mapped to the plus strand we should see consecutive G→T or C→A mutations, depending on which side of the origin is the leading strand.
To visualize the distribution of the six Group A POLE-exo* WGS tumors individually, we uploaded genome-wide mutations in TCT→TAT and AGA→ATA to the UCSC Genome Browser (Fig. 5A). Indeed, each chromosome contained many loci where strand biases could be observed in the distribution of these mutations. Regions of alternating TCT→TAT and AGA→ATA mutations on chromosome 17, consistent with a pattern of strand-specific DNA synthesis by POLE, can be seen in Figure 5A. Interestingly, in some cases regions exhibiting alternating strand-specific mutations were shared across all six tumors, suggesting these various tumors execute their replication program with a high degree of consistency.
Figure 5.
Mutations of POLE-exo* Group A mutants in the UCSC Genome Browser. The TCT→TAT and AGA→ATA mutations were taken from six whole-genome sequences of POLE-exo* mutant tumors from TCGA patients (F5-6814, AA-A00N, EI-6917, CA-6718, AA-3555, A6-6141). (A) POLE-exo* mutants TCT→TAT (red) or AGA→ATA (blue) from chromosome 17 (top tracks) and a representative segment zoomed in to show clustering of same-strand mutations. TCGA patient ID is to the left of the track. Note alignment of red and blue clusters preserved across multiple patients and clusters predominating in red and blue (boxes). Ordering of patient tracks is the same on top and bottom. (B) POLE-exo* mutations mapped to LMNB2 ORI. The ORI position is chromosome 19:2367825 to 2368428. (Top) Schematic of a replication origin showing leading strand and expected mutations from POLE-exo* mutants mapped to the reference strand. The turquoise bar indicates ORI location. (Bottom) C→A (red hash marks) predominate to the left of the ORI (17/26); C→A (16/60) are the minority to the right (χ2 P = 0.00164). (See also Supplemental Fig. S6 for well-characterized ORI at TOP1 and DHFR.)
To seek evidence linking the strand bias in POLE-exo*, we examined the mutation pattern around known human ORIs. The promoter of LAMB2 is one of the best-characterized human origins of replication (Giacca et al. 1994; Abdurashidova et al. 2000; Vaara et al. 2012). As shown in Figure 5B, the mutation pattern is asymmetrical with respect to the origin. On the plus strand, C→A predominates in the 5′ direction relative to the LAMB2 ORI (65%, 17 of 26 mutations), whereas in the 3′ direction, C→A mutations are less abundant (26%, 16 of 60 mutations), in which the opposite strand is now leading (Fig. 5B) (χ2 P = 0.001635). We also examined the mutational pattern near the TOP1 ORI (Keller et al. 2002) and in an adjacent region to the DHFR ORI (Looney and Hamlin 1987; Altman and Fanning 2001). We found alternating bands of C→A (Supplemental Fig. S6, red) adjacent to G→T mutations (Supplemental Fig. S6, blue), indicating strand switching. In the case of the DHFR ORI, red and blue bands are also found upstream of the reported origin. This may not be surprising because the mapping of mammalian origins is complicated by many factors, with variability based on methods used. Mapping of the DHFR ORI is particularly complicated by tandem origins (β, β′, γ) involved in replicating nearby genes (Hamlin et al. 2010). In addition, origins can be constitutive, flexible, or inactive and are influenced by replication timing events and stress (Mechali 2010), all factors that make their location harder to define. Clearly, in the vicinity of three well-characterized origins of replication, there is strand bias, consistent with the observation in yeast that POLE plays a major role in the synthesis of the leading strand. This correlation supports the notion that the regions of alternating strand bias we observed were marking origins of replication.
Amino acid substitution patterns unique to POLE-exo* Group A mutants
The nucleotide substitutions resulting from POLE-exo* mutants gave rise to a shift in amino acid change frequencies compared to MSS and MSI tumors. In coding regions, the POLE-exo* preferential sites, TCT and TCG, encode Ser; and their reverse complements, AGA and CGA, encode Arg. In the Group A mutants, Ser codons should mutate to Tyr, Leu by a C→A and C→T mutation, respectively; whereas the Arg codons should mutate to Ile and Gln by a G→T and G→A substitution, respectively. We therefore expect an increased proportion of these amino acid changes in POLE-exo* tumors relative to MSS and MSI. Inspection of the codon change tables confirms that the relative proportions of these changes increases significantly in the POLE-exo* mutant tumors (Supplemental Tables S2, S3) compared to MSI and MSS proportions for the same changes. For example, the relative proportion of Ser→Tyr in POLE-exo* (0.36) is three times higher than in MSI tumors (0.12) and six times higher than in MSS tumors (0.06). Arg→Ile is 13 times higher in POLE-exo* than in MSS, and eight times higher than MSI. The percentage of Ser→Tyr and Leu and Arg→Ile and Gln yields eight comparisons (POLE-exo* mut to MSI and to MSS). Across two diseases (CRC and EEC), there are a total of 16 comparisons, and all exhibit similar increased frequencies. We have performed χ2 analysis on all 16 comparisons, and only one, Ser→Leu in endometrial cancer, falls shy of statistical significance (see Supplemental Table S2). Moreover, for Ser and Arg, these four amino acid changes are among the highest proportion of all changes in POLE-exo* mutants. Thus, the POLE-exo* mutant profoundly skews the amino acid replacement frequencies.
Interestingly, the most frequent amino acid change of all was mutation of Glu codons to nonsense codons (Supplemental Tables S2, S3). Glu codons (GAG and GAA) can form a POLE-exo* context site if preceded by any codon with A in the third position (AGAG and AGAA), to form a POLE-exo* AGA, thereby causing mutation of the first codon position G to T and creating a stop codon. Thus, both Glu codons contribute to the dramatic increase in nonsense mutations in POLE-exo* Group A tumors. In contrast, in MSI and MSS tumors, Arg codons are the most frequently mutated, including the most frequent contributors of nonsense mutation (de Beer et al. 2013). For both of those amino acids’ codons, nonsense mutations are relatively rare compared to missense mutations (Supplemental Table S3).
Several cancer-related tumor suppressor genes are affected by the POLE-exo* specific nonsense codon changes in these patients, including key tumor suppressors TP53, PIK3R1, and ATM.
Strand-specific mutation patterns in tumor suppressor genes
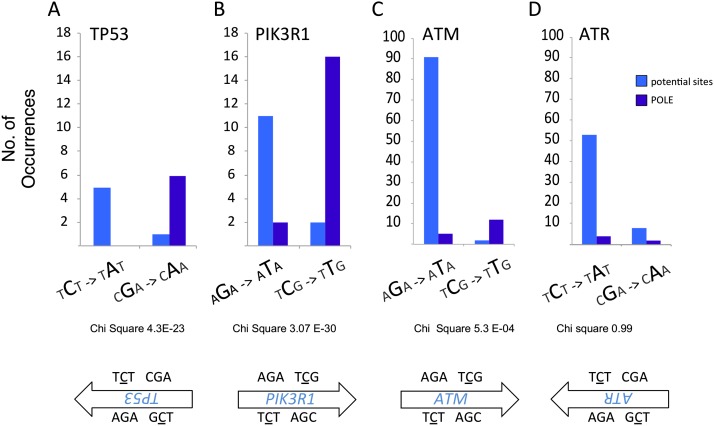
Group A POLE-exo* tumors are enriched in nonsense mutations in TP53 at R213X relative to tumors from MSI and MSS patients (Fig. 6). This is a Glu to Stop mutation, which also occurs at a POLE-exo* hotspot (TCG to TTG) with a flanking A (TCGA to TTGA). Surprisingly, TP53 has five other sites that could mutate to a nonsense codon through a POLE-exo* context-specific site, by a G→T mutation on the antisense strand at TCT hotspot motifs (Fig. 6). However, the R213X, TCG→TTG occurs six times, whereas the five other nonsense codons, which require a TCT→TAT change have not yet been observed (P < 10−4, χ2) (Fig. 6). This same pattern of strand specificity was also seen in PIK3R1 and ATM. In each of these cases, potential nonsense sites from both hotspots are available from both strands, but only one occurs in this data set. These recurrent nonsense mutations can be explained if POLE replicates on only one strand in the neighborhood of these genes in a manner similar to that shown in yeast (Pursell et al. 2007b).
Figure 6.
POLE-exo* mutations lead to enrichment of nonsense mutations at tumor suppressor genes in endometrial and colorectal tumors. Comparison of potential nonsense sites that occur in POLE-exo* context (TCT/AGA →TAT/ATA and TCG/CGA →TTG/CAA) to actual nonsense mutations found in POLE-exo* tumors. Selected tumor suppressor genes are shown. The number of potential nonsense mutation sites are shown in light blue; the nonsense mutation sites that actually occurred in POLE-exo* tumors are in dark blue. (A) TP53 (χ2 P = 4.3 × 10−23); (B) PIK3R1 (χ2 P = 3.07 × 10−30); (C) ATM (χ2 P = 5.3 × 10−4); (D) ATR; the POLE-exo* mutations are evenly distributed among the potential sites (χ2 P = 0.99). (See also Supplemental Tables S2, S3 for full amino acid change tables.)
Discussion
A detailed analysis of the properties of POLE-exo* reveals that they can be classified into two groups. In addition to an ultrahigh mutation frequency dominated by C→A substitutions (>20%), Group A mutants exhibit a very low relative rate of somatic indel mutation (<2%) and arise from mutation of a limited number of amino acid positions in the exonuclease domain. The cell-free experiments with purified human wild-type and mutant POLE demonstrate that mutation of the exonuclease domain of the polymerase can account for the nucleotide preference and mutation frequencies found in the tumors. Group B mutants exhibit mutation properties of MSI tumors, and indeed, they are found almost always in tumors classified as MSI (Table 2). Group B mutants appear to be nonfunctional; however, two of the Group B sites are observed more than once, which raises the possibility that at least some of the POLD1 are functional in the tumor. Resolution of this question awaits subsequent in vitro functional studies.
Group A mutants are found at nine sites in 38 patients, mapping close to the exonuclease catalytic residues, all but one have been seen recurrently, suggesting most alleles that can give rise to the Group A phenotype are now known. Group B are observed in 10 POLE and 13 POLD1 and map further from the catalytic site. These observations raise the interesting possibility that in POLD1-exo* mutant tumors, mutations near the catalytic residues are under negative selection in human tumors, except for an exonuclease catalytic reside, the aforementioned D316 itself. In contrast, mutation of a catalytic residual of POLE was not observed. Mutation of POLD1-exoD316 was associated with an ultrahigh mutation rate, but it exhibited none of the other Group A properties.
Only the Group A mutants are demonstrably carcinogenic, not just through increased mutation rate, but also through specificity of the predominant C→A and C→T nucleotide changes, which increase nonsense mutation rates in key tumor suppressor genes. Specific recurrent mutations were found in many other genes with known involvement in human cancer, such as MSH6, MSH2, PTEN, and PIK3R1 (data not shown) but require functional studies to assess their impact on the cancers. Thus, further analysis of all the recurrent POLE-exo* site-specific changes may reveal important additional features of tumor initiation and maintenance in these patients.
Group A has a better prognosis in endometrial cancer, where a sufficient number of POLE-exo* patients have been accumulated (The Cancer Genome Atlas Network 2013; Meng et al. 2014). Whether or not this clinical feature will be realized in the other cancers is now a critical question since some POLE-exo* mutations occur in cancers where no cures are available (e.g., glioblastoma multiforme and pancreatic adenocarcinoma). The need for a clinical test to uncover these mutants underscores the importance of distinguishing Group A from Group B mutations.
Group A mutants generate a significant strand-specific pattern of mutation on the genome consistent with the role of POLE in leading strand synthesis observed in yeast. These strand-specific patterns often show alternating regions of C→A followed by G→T stretching across thousands of bases in a manner consistent with localization of an ORI. We observe these clustered and alternating patterns in ∼10%–20% of the tumor genome, including loci at or near ORI well characterized in other cell culture systems, for example at ORI in the promoter regions of LMNB2, TOP1, and DHFR. The mutational signatures we report are the first from a higher eukaryote that are in agreement with the concept, demonstrated in yeast, that POLE is involved in leading strand synthesis.
It is well known that mammalian ORI fire inconsistently, or shift location, from one round of replication to the next (for review, see Mechali 2010). Indeed most studies of mammalian ORI use cell cultures in synchronized replication to avoid this feature of the replication system (Hamlin et al. 2010; Valenzuela 2012; Aparicio 2013). In these tumors, we are observing the accumulation of mutations over many rounds of cell division, so any inconsistencies in the firing or location of the cells’ ORI could obscure the mutational pattern by randomly changing which strand is the leading strand. Thus, the 10%–20% of the genome we observe in clustered C→A and G→T must represent consistent, or “constitutive” ORI across all cells in the tumor through all rounds of division, across multiple individuals (Mechali 2010). The observation of these putative ORI were remarkably similar across the six tumors, suggesting that cancer cells with Group A mutations may offer an ideal system for delving into the basic biology of genome replication.
Methods
Identification of POLE-exo* mutants
Mutational data for colorectal cancer was obtained by sequencing 497 tumor and normal pairs of colon and rectal cancer. The study design and sequencing approaches were reported (The Cancer Genome Atlas Network 2012). POLE mutation data in other cancers was obtained from The Cancer Genome Atlas Data Portal (https://tcga-data.nci.nih.gov/tcga/dataAccessMatrix.htm), Genome Data Analysis Center (GDAC) (https://confluence.broadinstitute.org), and the cBio Portal for cancer genomics (http://www.cbioportal.org). Other mutations affecting the POLE- and POLD1-exo domains are reported in COSMIC (http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/) but were not evaluated in the rest of this study because (1) they were not accompanied with overall mutation frequencies and nucleotide change frequencies, or (2) their validation status was not clear.
Cancers in TCGA that were found to contain POLE-exo* mutations are colorectal cancers (CRC), endometrial cancer (EEC), stomach adenocarcinoma (STAD), glioblastoma (GBM), low grade glioma (LGG), breast carcinomas (BRCA), bladder carcinomas (BLCA), pancreatic adenocarcinoma (PAAD), head and neck cancer (HNSC), kidney chromophobe (KICH), and lung adenocarcinoma (LUAD). All publicly available TCGA tumor data comply with U.S. law protecting patient confidentiality and other ethical standards.
Measurement of microsatellite instability
The MSI status was initially obtained from The Cancer Genome Atlas Data Portal (https://tcga-data.nci.nih.gov/tcga/dataAccessMatrix.htm). TCGA MSI status was determined using a panel of four mononucleotide and three dinucleotide repeats, previously described (The Cancer Genome Atlas Network 2012). The MSI status was reevaluated using a panel of 48 genes with mononucleotide runs in our sequence based homopolymer analysis, previously described (The Cancer Genome Atlas Network 2012). The majority of MSI calls matched between the two analyses; however, there were exceptions. TCGA-AP-A059 and TCGA-D1-A103 were judged MSI using the BAT-25 panel, but they were reclassified as MSS using our homopolymer analysis. In addition, TCGA-AP-A056, TCGA-D1-A16Y, and TCGA D1-A17Q were classified as MSI-L by TCGA and reclassified as MSS by our homopolymer analysis. Because of the low frequency of insertion and deletion mutations and high percentage of C→A substitutions, we reclassified these samples as MSS (Supplemental Fig. S7).
Mutation context clustering and enrichment
Whole-genome data from TCGA patients TCGA-F5-6814, TCGA-AA-A00N, TCGA-EI-6917, TCGA-CA-6718, TCGA-AA-3555, and TCGA-A6-6141 were analyzed as follows: To test for clustering of mutations, windows 50 kb in width were generated across the genomic space and the ratio of TCT→TAT to TCT→TAT plus AGA→ATA was calculated. Windows were analyzed further if they contained 10 or greater mutations. To generate a random mutation model, at each window a random sampling from either TCT→TAT or AGA→ATA was taken for each observed mutation within that window, and the ratio of TCT→TAT to total was reported. To test for correlation, for each TCT→TAT and AGA→ATA mutation, the distance (in genomic space) was calculated to the next (higher chromosomal coordinate) mutation subject to a minimum distance of 1 bp, 5 kb, 100 kb, or 1 Mb. These mutations were paired and classified as same context if they were identical and different context if they were inverse mutations. Distances and orientation configurations for the contexts (same orientation or reverse compliment) were binned for analysis.
For annotation of mutation classes in the UCSC Genome Browser, mutation data for specific contexts were extracted from MAF files for each subject. Mutations were uploaded to the browser and colored depending on strand orientation. TCT→TAT are colored red, and AGA→ATA mutations are blue.
Expression and purification of recombinant cancer-associated human POLE constructs
An expression vector encoding amino acid residues 1–1189 of the exonuclease-proficient catalytic subunit of human POLE (POLE-N140) was used as described (Korona et al. 2011; Goksenin et al. 2012). Site-directed mutagenesis was used to introduce the POLE-exo* mutations encoding: P286H, F367S, and S459F substitutions. Expression vectors were cotransformed into Rosetta cells (EMD Millipore) along with the pRK603 vector, which drives coexpression of the TEV protease along with the GSTTEV-POLE-exoN140 construct. This allows for intracellular cleavage of the N-terminal GST tag.
An aliquot of an overnight culture of Rosetta cells containing the POLE-exoN140 expression vector was added to 6 L of LB media to a final A600nm of 0.1. Cells were grown at 37°C with shaking at 220 rpm to an OD600 of 0.6, when IPTG was added to a final concentration of 4 µM. Expression was induced for 4 h at room temperature, then cells were harvested by centrifugation. Cells were resuspended in lysis buffer [300 mM Tris, 100 mM NaCl, 20 mM K2SO4, 0.5 mM EDTA, pH 7.8, protease inhibitor tablets (Roche)] at 1.5 mL per 1 g cells. Cells were lysed by several passages through a French press, and lysate was homogenized with a 20.5-gauge needle. Cell lysates were purified using a HisTrapFF column (GE Healthcare). Columns were equilibrated with 10 column volumes of wash buffer (150 mM Tris, 200 mM NaCl, 20 mM K2SO4, and 2 mM DTT, pH 7.8). Bound protein was washed with 25 c.v. of wash buffer, then 10 c.v. of wash buffer containing 75 mM imidazole. The sample was eluted in 150 mM imidazole and dialyzed overnight into SEC buffer (150 mM Tris, 200 mM NaCl, 20 mM K2SO4, and 2mM DTT at pH 7.8). The sample was concentrated to 100 μL in a spin concentrator and resolved by a Superdex 200 10/300 GL column (GE Healthcare). Peak fractions were monitored by UV and by Western blot, pooled, and quick frozen in liquid N2 and stored at −80°C.
In vitro primer polymerization and excision assays
A 20-mer DNA oligo, 5′-CCTCTTCGCTATTACGCCAG-3′, was 5′ end-labeled with 32P by incubating with T4 polynucleotide kinase (Invitrogen) and ATP Gamma 32P (Perkin Elmer). Primers used in polymerization assays were annealed to a complementary 45-mer DNA oligonucleotide, 5′-TTGCAGCACATCCCCCTTTCGCCAGCTGGCGTAATAGCGAAGAGG-3′. DNA polymerization reaction (30 μL) contained 50 mM Tris, pH 7.4, 8 mM MgCl2, 1 mM DTT, 10% glycerol, 25 μM each dNTP, and 100 nM DNA primer-template. Reactions were started with the addition of 1 nM enzyme and carried out at 37°C. Reactions were stopped by addition of an equal volume of stop buffer (95% formamide, 5 mM EDTA, 0.1% xylene cylanol, and 0.1% bromphenol blue). Exonuclease reactions were identical except dNTPs were withheld, and the 20-mer primer ended in T to form a terminal mismatch. Enzyme was added at 2 nM to initiate reactions. Products were resolved by 12% denaturing PAGE. Gels were dried and exposed to phosphorimager cassettes and quantified using a phosphorimager (GE Healthcare) and ImageQuantTL software (GE Healthcare).
In vitro error rate measurements
The in vitro lacZ forward mutation assay was performed essentially as described previously (Bebenek and Kunkel 1995). Briefly, double-stranded M13mp2 DNA containing a 407-nt ssDNA gap was used as a substrate in reactions containing 0.15 nM DNA, 50 mM Tris-Cl, pH 7.4, 8 mM MgCl2, 2 mM DTT, 100 μg/mL BSA, 10% glycerol, 250 μM dNTPs, and 1.5 nM purified POLE at 37°C. Complete gap-filling was monitored by gel electrophoresis. Completely filled product was then transformed into E. coli cells, which were used to determine the frequency of light blue and colorless plaques on plates containing X-gal. Filled product containing no replication errors yields dark blue plaques, whereas replication errors introduced during gap-filling DNA synthesis yield light blue or colorless mutant plaques. lacZ mutant frequencies were calculated from combining at least two independent experiments. DNA from mutant plaques was purified, and the lacZ gene was sequenced. Error rates were calculated as described (Bebenek and Kunkel 1995).
Direct sequencing of DNA polymerization reactions
To generate the substrate for the direct sequencing of POLE synthesis products, three approximately equally spaced DNA primers were hybridized to single-strand M13mp2 DNA. The primers were located at positions 1324, 4012, and 6434 of the 7216-nt M13mp2 template. Primer sequences were 1324: 5′-AGCAACGGCTACAGAGGCT-3′; 4012: 5′-TTTTTAACCTCCGGCTTAGG-3′; 6434: 5′-GATCGCACTCCAGCCAGC-3′. Primers were added to the template on ice at 1.2-fold molar excess and hybridized by heating for 5 min at 80°C followed by slow cooling to room temperature. Reactions were assembled containing 40 nM DNA, 50 mM Tris-Cl, pH 7.4, 8 mM MgCl2, 2 mM DTT, 100 μg/mL BSA, 10% glycerol, and 250 μM dNTPs. Reactions were initiated by the addition of 40 nM POLE and incubated for 30 min at 37°C. DNA synthesis was measured in parallel in separate identical reactions in which dATP Alpha 32P (3000 Ci/mmol; Perkin Elmer) was added, followed by separation by denaturing PAGE.
Libraries were prepared from the reactions lacking radio-labeled dATP by the following method: Single-stranded product was removed from the synthesis reactions using mung bean nuclease to generate double-stranded blunt-end DNA molecules. The double-stranded product was then formed into next-generation sequencing libraries using a ThruPLEX-FD kit (Rubicon Genomics). Libraries were individually barcoded and pooled. Pooled libraries were sequenced on a single MiSeq lane (Illumina) using a 2 × 150 paired-end protocol.
Data access
Sequences obtained from the POLE synthesis products from single-strand M13mp2 DNA have been submitted to NCBI’s BioSample database (http://www.ncbi.nlm.nih.gov/biosample) under accession number SAMN03023853.
Acknowledgments
This work was funded by U54HG003273 from the National Human Genome Research Institute to Baylor College of Medicine; NCI grant U24 CA143843 to D.A.W.; NCI-U24CA143840 and SU2C-AACR-DT0209 from the Stand Up To Cancer Dream Team Translational Research Grant, a Program of the Entertainment Industry Foundation to Memorial Sloan Kettering Cancer Center; and National Institutes of Health (ES016780, RR020152) to Z.F.P.
Author contributions: E.S. and D.A.W. discovered POLE-exo* mutations in ultramutated CRC patients, performed bioinformatics analysis, and wrote the paper; N.W. and N.S. correlated exonuclease mutations to mutation spectrum, performed statistical and bioinformatics analysis, and prepared figures; E.E.H. and A.Y.G. performed enzyme purifications, polymerization, excision reactions, and lacZ mutant frequency plaque assays; H.C., H.D., and D.M.M. performed direct sequencing of the DNA polymerization reactions; K.R.C. performed bioinformatics and statistical analysis; and Z.F.P., C.S., and D.A.W. provided scientific oversight.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.174789.114.
References
- Abdurashidova G, Deganuto M, Klima R, Riva S, Biamonti G, Giacca M, Falaschi A. 2000. Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science 287: 2023–2026 [DOI] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SAJR, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børresen-Dale AL, et al. 2013. Signatures of mutational processes in human cancer. Nature 500: 415–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman AL, Fanning E. 2001. The Chinese hamster dihydrofolate reductase replication origin β is active at multiple ectopic chromosomal locations and requires specific DNA sequence elements for activity. Mol Cell Biol 21: 1098–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio OM. 2013. Location, location, location: it’s all in the timing for replication origins. Genes Dev 27: 117–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes DE, Lindahl T. 2004. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet 38: 445–476 [DOI] [PubMed] [Google Scholar]
- Bebenek K, Kunkel TA. 1995. Analyzing fidelity of DNA polymerases. Methods Enzymol 262: 217–232 [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Network . 2012. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487: 330–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Network . 2013. Integrated genomic characterization of endometrial carcinoma. Nature 497: 67–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church DN, Briggs SEW, Palles C, Domingo E, Kearsey SJ, Grimes JM, Gorman M, Martin L, Howarth KM, Hodgson SV, et al. 2013. DNA polymerase ɛ and δ exonuclease domain mutations in endometrial cancer. Hum Mol Genet 22: 2820–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriello G, Miller ML, Aksoy BA, Senbabaoglu Y, Schultz N, Sander C. 2013. Emerging landscape of oncogenic signatures across human cancers. Nat Genet 45: 1127–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Beer TA, Laskowski RA, Parks SL, Sipos B, Goldman N, Thornton JM. 2013. Amino acid changes in disease-associated variants differ radically from variants observed in the 1000 genomes project dataset. PLoS Comput Biol 9: e1003382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortune JM, Pavlov YI, Welch CM, Johansson E, Burgers PM, Kunkel TA. 2005. Saccharomyces cerevisiae DNA polymerase δ: high fidelity for base substitutions but lower fidelity for single- and multi-base deletions. J Biol Chem 280: 29980–29987 [DOI] [PubMed] [Google Scholar]
- Giacca M, Zentilin L, Norio P, Diviacco S, Dimitrova D, Contreas G, Biamonti G, Perini G, Weighardt F, Riva S. 1994. Fine mapping of a replication origin of human DNA. Proc Natl Acad Sci 91: 7119–7123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goksenin AY, Zahurancik W, LeCompte KG, Taggart DJ, Suo Z, Pursell ZF. 2012. Human DNA polymerase ɛ is able to efficiently extend from multiple consecutive ribonucleotides. J Biol Chem 287: 42675–42684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamlin JL, Mesner LD, Dijkwel PA. 2010. A winding road to origin discovery. Chromosome Res 18: 45–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg M, Osterman P, Bylund GO, Ganai RA, Lundström EB, Sauer-Eriksson AE, Johansson E. 2014. Structural basis for processive DNA synthesis by yeast DNA polymerase ɛ. Nat Struct Mol Biol 21: 49–55 [DOI] [PubMed] [Google Scholar]
- Jin YH, Obert R, Burgers PM, Kunkel TA, Resnick MA, Gordenin DA. 2001. The 3′→5′ exonuclease of DNA polymerase δ can substitute for the 5′ flap endonuclease Rad27/Fen1 in processing Okazaki fragments and preventing genome instability. Proc Natl Acad Sci 98: 5122–5127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane DP, Shcherbakova PV. 2014. A common cancer-associated DNA polymerase ɛ mutation causes an exceptionally strong mutator phenotype, indicating fidelity defects distinct from loss of proofreading. Cancer Res 74: 1895–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, Jessup JM, Kolodner R. 1997. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res 57: 808–811 [PubMed] [Google Scholar]
- Keller C, Ladenburger EM, Kremer M, Knippers R. 2002. The origin recognition complex marks a replication origin in the human TOP1 gene promoter. J Biol Chem 277: 31430–31440 [DOI] [PubMed] [Google Scholar]
- Korona DA, LeCompte KG, Pursell ZF. 2011. The high fidelity and unique error signature of human DNA polymerase. Nucleic Acids Res 39: 1763–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA, Erie DA. 2005. DNA mismatch repair. Annu Rev Biochem 74: 681–710 [DOI] [PubMed] [Google Scholar]
- Loeb LA. 2011. Human cancers express mutator phenotypes: origin, consequences and targeting. Nat Rev Cancer 11: 450–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looney JE, Hamlin JL. 1987. Isolation of the amplified dihydrofolate reductase domain from methotrexate-resistant Chinese hamster ovary cells. Mol Cell Biol 7: 569–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujan SA, Williams JS, Pursell ZF, Abdulovic-Cui AA, Clark AB, Nick McElhinny SA, Kunkel TA. 2012. Mismatch repair balances leading and lagging strand DNA replication fidelity. PLoS Genet 8: e1003016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechali M. 2010. Eukaryotic DNA replication origins: many choices for appropriate answers. Nat Rev Mol Cell Biol 11: 728–738 [DOI] [PubMed] [Google Scholar]
- Meng B, Hoang LN, McIntyre JB, Duggan MA, Nelson GS, Lee CH, Köbel M. 2014. POLE exonuclease domain mutation predicts long progression-free survival in grade 3 endometrioid carcinoma of the endometrium. Gynecol Oncol 134: 15–19 [DOI] [PubMed] [Google Scholar]
- Miyabe I, Kunkel TA, Carr AM. 2011. The major roles of DNA polymerases ɛ and δ at the eukaryotic replication fork are evolutionarily conserved. PLoS Genet 7: e1002407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PMJ, Kunkel TA. 2008. Division of labor at the eukaryotic replication fork. Mol Cell 30: 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palles C, Cazier JB, Howarth KM, Domingo E, Jones AM, Broderick P, Kemp Z, Spain SL, Guarino E, Salguero I, et al. 2013. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet 45: 136–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pursell ZF, Isoz I, Lundström EB, Johansson E, Kunkel TA. 2007a. Regulation of B family DNA polymerase fidelity by a conserved active site residue: characterization of M644W, M644L and M644F mutants of yeast DNA polymerase ɛ. Nucleic Acids Res 35: 3076–3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pursell ZF, Isoz I, Lundström EB, Johansson E, Kunkel TA. 2007b. Yeast DNA polymerase ɛ participates in leading-strand DNA replication. Science 317: 127–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reha-Krantz LJ. 2010. DNA polymerase proofreading: multiple roles maintain genome stability. Biochim Biophys Acta 1804: 1049–1063 [DOI] [PubMed] [Google Scholar]
- Schaaper RM, Dunn RL. 1991. Spontaneous mutation in the Escherichia coli lacI gene. Genetics 129: 317–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt MW, Matsumoto Y, Loeb LA. 2009. High fidelity and lesion bypass capability of human DNA polymerase δ. Biochimie 91: 1163–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbakova PV, Pavlov YI, Chilkova O, Rogozin IB, Johansson E, Kunkel TA. 2003. Unique error signature of the four-subunit yeast DNA polymerase ε. J Biol Chem 278: 43770–43780 [DOI] [PubMed] [Google Scholar]
- Shevelev IV, Hübscher U. 2002. The 3′–5′ exonucleases. Nat Rev Mol Cell Biol 3: 364–376 [DOI] [PubMed] [Google Scholar]
- Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK. 2009. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase δ. Nat Struct Mol Biol 16: 979–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaara M, Itkonen H, Hillukkala T, Liu Z, Nasheuer HP, Schaarschmidt D, Pospiech H, Syvaoja JE. 2012. Segregation of replicative DNA polymerases during S phase: DNA polymerase ε, but not DNA polymerases α/δa, are associated with lamins throughout S phase in human cells. J Biol Chem 287: 33327–33338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela MS. 2012. Initiation of DNA replication in the human genome. Hereditary Genet Supp 1: 4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle L, Hernández-Illán E, Bellido F, Aiza G, Castillejo A, Castillejo MI, Navarro M, Seguí N, Vargas G, Guarinos C, et al. 2014. New insights into POLE and POLD1 germline mutations in familial colorectal cancer and polyposis. Hum Mol Genet 23: 3506–3512 [DOI] [PubMed] [Google Scholar]
- Vilar E, Gruber SB. 2010. Microsatellite instability in colorectal cancer—the stable evidence. Nat Rev Clin Oncol 7: 153–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. 2011. Nucleases: diversity of structure, function and mechanism. Q Rev Biophys 44: 1–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida R, Miyashita K, Inoue M, Shimamoto A, Yan Z, Egashira A, Oki E, Kakeji Y, Oda S, Maehara Y. 2010. Concurrent genetic alterations in DNA polymerase proofreading and mismatch repair in human colorectal cancer. Eur J Hum Genet 19: 320–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Liu FY, Liu H, Wang F, Li W, Huang MZ, Huang Y, Yuan XQ, Xu XY, Huang OP, et al. 2014. Frequent POLE1 p.S297F mutation in Chinese patients with ovarian endometrioid carcinoma. Mutat Res 761: 49–52 [DOI] [PubMed] [Google Scholar]