Abstract
Studies of gene rearrangements and the consequent oncogenic fusion proteins have laid the foundation for targeted cancer therapy. To identify oncogenic fusions associated with glioma progression, we catalogued fusion transcripts by RNA-seq of 272 gliomas. Fusion transcripts were more frequently found in high-grade gliomas, in the classical subtype of gliomas, and in gliomas treated with radiation/temozolomide. Sixty-seven in-frame fusion transcripts were identified, including three recurrent fusion transcripts: FGFR3-TACC3, RNF213-SLC26A11, and PTPRZ1-MET (ZM). Interestingly, the ZM fusion was found only in grade III astrocytomas (1/13; 7.7%) or secondary GBMs (sGBMs, 3/20; 15.0%). In an independent cohort of sGBMs, the ZM fusion was found in three of 20 (15%) specimens. Genomic analysis revealed that the fusion arose from translocation events involving introns 3 or 8 of PTPRZ and intron 1 of MET. ZM fusion transcripts were found in GBMs irrespective of isocitrate dehydrogenase 1 (IDH1) mutation status. sGBMs harboring ZM fusion showed higher expression of genes required for PIK3CA signaling and lowered expression of genes that suppressed RB1 or TP53 function. Expression of the ZM fusion was mutually exclusive with EGFR overexpression in sGBMs. Exogenous expression of the ZM fusion in the U87MG glioblastoma line enhanced cell migration and invasion. Clinically, patients afflicted with ZM fusion harboring glioblastomas survived poorly relative to those afflicted with non-ZM-harboring sGBMs (P < 0.001). Our study profiles the shifting RNA landscape of gliomas during progression and reveled ZM as a novel, recurrent fusion transcript in sGBMs.
The paradigm of oncogene addiction is predicated on the premise that some oncogenes perform essential and irreplaceable functions required for the survival of cancer cells (Weinstein 2002). The aggregate of studies spanning the past two decades, however, reveal that very few oncogenes actually fulfill this criterion (Torti and Trusolino 2011). Most oncogenic pathways appear dynamic, with highly redundant circuitry (Stommel et al. 2007; Nitta et al. 2010). One of the notable exceptions to these observations involves fusion proteins (Ren 2005). These fusion proteins typically resulted from chromosomal translocations (Nambiar et al. 2008) and executed novel functions that cannot be reconstituted by the expression of either parental protein (Ren 2005; Singh et al. 2012). Importantly, these novel functions reprogrammed the cellular circuitry to a state of exquisite addiction (Ren 2005; Sasaki et al. 2010). Some of the most promising clinical results have arisen from targeted inhibition of these fusion proteins, including the BCR–ABL1 fusion (Ren 2005) and the EML4–ALK fusion (Sasaki et al. 2010).
As a first step toward the identification of novel fusion proteins, we performed RNA sequencing of 272 WHO grade II, III, and IV gliomas. Gliomas are the most common form of brain cancer and can be classified grade I to grade IV based on histologic features (Louis et al. 2007; Wang and Jiang 2013). Grade II gliomas are also known as low-grade gliomas, whereas grade III and IV tumors are frequently termed high-grade gliomas (Wen and Kesari 2008). The term glioblastoma (GBM) is synonymous with grade IV glioma (Wen and Kesari 2008). GBM is one of the deadliest of human cancers, with median survival of 14 mo after maximal surgical resection, chemotherapy, and radiation therapy (Stupp et al. 2005). Based on clinical history, GBMs can generally be classified into two subtypes (Ohgaki and Kleihues 2009). Primary GBM (pGBM) refers to the vast majority of GBMs, which are thought to form de novo in the elderly. On the other hand, secondary GBMs (sGBMs) typically progress from lower-grade tumors and affect younger patient populations. While pGBMs and sGBMs are indistinguishable histologically (Louis et al. 2007), emerging genomic profiling revealed a distinct genetic landscape between these two tumor types (Ohgaki and Kleihues 2009). For instance, mutation in the metabolic enzyme isocitrate dehydrogenase (IDH) is found almost exclusively in the sGBMs (Yan et al. 2009)
Our efforts unveiled a novel, recurrent fusion transcript involving the PTPRZ1 and the MET gene (ZM) that was found in 15% of sGBMs. sGBMs harboring this fusion exhibit aggressive clinical behavior and are associated with a poor prognosis for afflicted patients.
Results
RNA-seq of 272 gliomas
Two hundred seventy-two freshly frozen glioma samples were collected for the initial exploratory analysis using RNA-seq. The demographics of this patient population can be found in Supplemental Table 1. Central pathology reviews of these specimens were performed by independent board-certified neuropathologists and graded based on the 2007 WHO classification (Louis et al. 2007). All samples were collected based on the Cancer Genome Atlas Research Network (2008) criteria, such that collected specimens contained at least 80% viable GBM tissue, were frozen within 5 min of resection, and were subjected to RNA-seq analysis. In total, 1386 Gbs of 101-bp paired-end reads were generated, with an average of 50 million reads per sample. The sequencing data were mapped to the human reference gene set and reference genome RefSeq (hg19) using BWA (Li and Durbin 2009).
The TopHat-Fusion (Kim and Salzberg 2011) and deFuse (McPherson et al. 2011) algorithms were used for fusion detection. Only fusions that scored positively on both algorithms were subsequently analyzed (see Methods). In total, 214 fusion transcripts were detected in 38% of the samples (n = 104) (Supplemental Table 2). With the exception of the fusion transcripts that fell in GC-rich regions (4.7%), validation of all fusion transcripts was performed using conventional PCR amplification followed by Sanger sequencing.
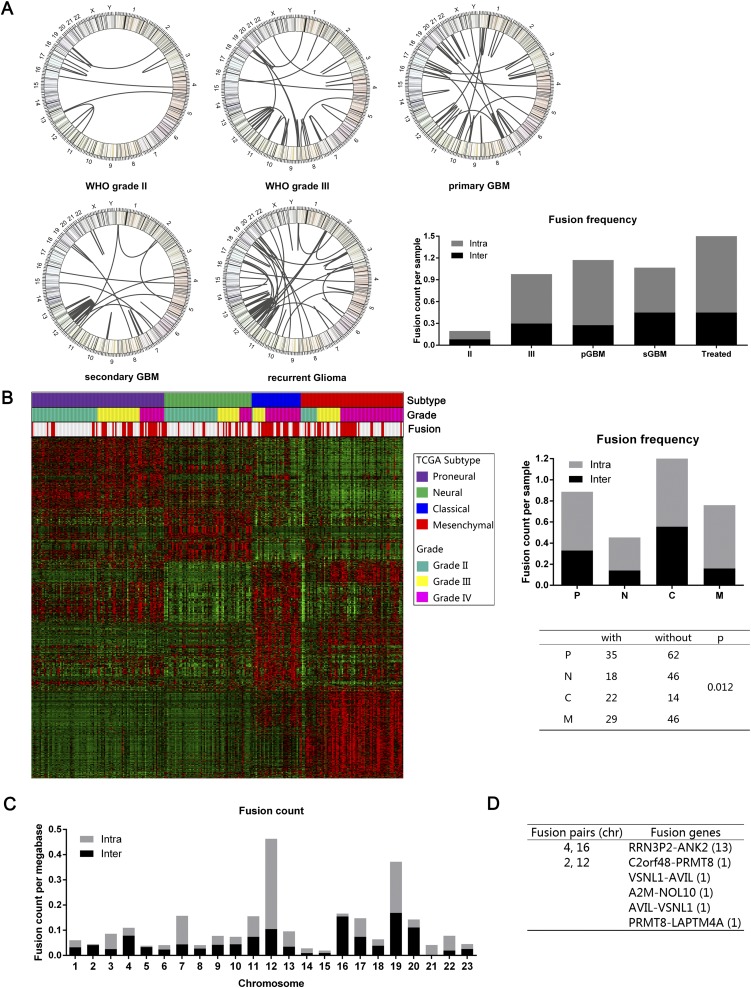
In general, fusion transcripts were more frequent in high-grade gliomas. Only 18.0% of grade II gliomas harbored fusion transcripts. In contrast, nearly half of the high-grade gliomas (42.5% of grade III glioma and 55.6% grade IV glioma) harbored fusion transcripts (Fig. 1A). These results were generally consistent with the progressive increase in genomic instability during advancing tumor grade (Negrini et al. 2010). The highest numbers of fusion transcripts were found in gliomas that recurred after radiation and/or temozolomide treatment (Fig. 1A), suggesting that DNA damage accumulation contributes to fusion transcript formation. The number of fusion transcripts detected did not significantly differ between pGMBs and sGBMs. However, the classical subtype of the gliomas (Verhaak et al. 2010) was more likely to harbor fusion transcripts relative to the other Cancer Genome Atlas–defined transcriptional subtypes (P < 0.03) (Fig. 1B)
Figure 1.
Fusion distribution depending on WHO classification, Cancer Genome Atlas subtypes, or chromosomes. (A) Circos plot of genomic distribution of fusion genes in grade II, grade III, primary GBM, secondary GBM, and recurrent gliomas. (B) Fusion distribution in the four Cancer Genome Atlas subtypes. There was a distinct higher proportion of patients with fusion in a classical subtype (P = 0.012). (C) Genomic distribution of fusion genes, indicating that chromosome 12 was a hot spot for intrachromosome fusion. (D) For the interchromosome fusion detection, chromosomes 4 and 6 and chromosomes 2 and 12 were the fusion pairs with the most interchromosome fusion frequency.
The most common form of fusion transcripts (75.7%) arose from the joining of sequences from the same chromosome (Fig. 1C). These fusion transcripts most commonly mapped to chromosomes 12 and 19, suggesting these chromosomes may represent “hot-spots” for deletional instability. Fusion transcripts involving sequences from distinct chromosomes constituted 24.3% of all fusion transcripts. These events most commonly involved exchanges between chromosomes 4 and 16, suggesting that these chromosomes may be located in physical proximity in a three-dimensional chromatin structure (Fig. 1D; Sajan and Hawkins 2012). The ratio of intrachromosomal and interchromosomal translocations was consistent with those reported in breast cancer (Kangaspeska et al. 2012).
Functional annotation of the in-frame fusion transcripts
Of the 214 fusion transcripts, 147 were out-of-frame and 67 were in-frame (sequences found in Supplemental Table 2). The distribution of these fusion transcripts as a function of age, sex, and glioma grade is shown in Supplemental Figure 1A. Of these in-frame fusions, 55 arose from the fusion of sequences located on the same chromosome and 12 arose from joining of sequences derived from different chromosomes.
Functional annotation of the fusion transcripts revealed the following. We identified 14 fusion transcripts containing sequences of genes involved in the canonical GBM signaling pathways (The Cancer Genome Atlas Research Network 2008) including the RTK/PIK3CA, RB1, and TP53 signaling pathways: CBL–FBXO2, FGFR3–TACC3, PTPRZ1–MET (ZM), VHL–BRK1, EGFR–VSTM2A, JAK1–HIVEP3, PTEN–COL17A1, CDK4–TSFM, IL1RAP–FGF12, NFATC3–CPNE2, PLA2G6–CRYBB1, APEH–SHISA5, TCF7L1–KIF1B, and TGFB1–SAE1 (Supplemental Fig. 1B). Additionally, we identified 11 fusion transcripts containing sequences of genes with metabolic function: CCM2–OGDH, FRMD4A–PFKP, TPT1–AADAT, AHCYL2–TMEM178B, PTN–DGKI, ZMIZ1–MAT1A, MTAP–C9orf92, CD81–SPAG6, CDK17–KCNC2, BCR–LZTR1, and AP2A2–SBF2. We found five fusion transcripts containing sequences encoding kinase domains: FGFR3–TACC3, PTPRZ1–MET (ZM), ST7–CTTNBP2, PLAGL2–HCK, and NEK6–RXRA. Finally, we identified two fusion transcripts containing sequences from genes implicated in chromatin remodeling: IFT80–MLH1 and MLL3–CHGB.
We identified three recurrent fusion transcripts: FGFR3–TACC3, RNF213–SLC26A11, and the ZM fusion. The FGFR3–TACC3 fusion was previously reported by Singh et al. (2012). This fusion transcript was found in three of 59 pGBMs (5.1%) (Supplemental Fig. 1C). The RNF213–SLC26A11 fusion transcript was previously identified in a chronic myeloid leukemia specimen (Zhou et al. 2013). This fusion transcript was detected in one pGBM (one of 59 or 1.7%) and a recurrent grade III glioma (one of 13, or 7.7%) (Supplemental Fig. 1D). The ZM fusion has not been previously reported and was detected in one grade III glioma and three sGBMs (Supplemental Fig. 1E). The four fusion transcripts involved four different breakpoints within the PTPRZ1 coding sequence. In contrast, the breakpoints in the MET gene were located at the same junction.
Validation of the PTPRZ1–MET as a recurrent fusion transcript
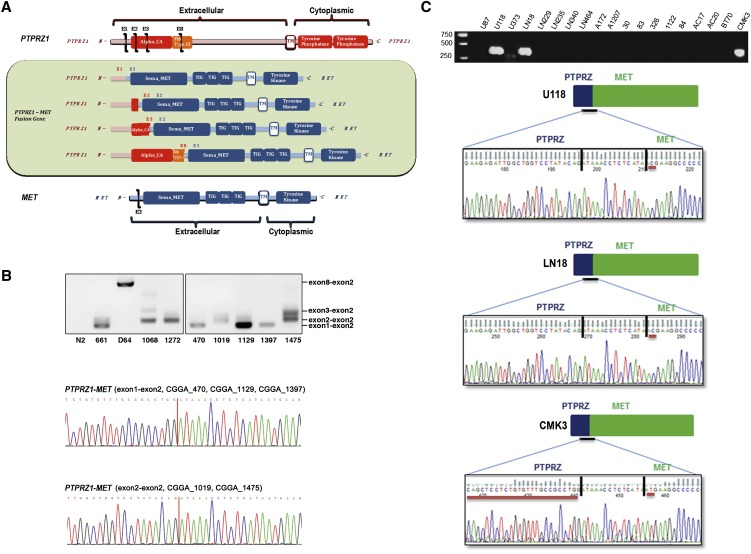
Three fusion transcripts contained sequences encoding the carbonic anhydrase (CA) domain and fibronectin type III (Fib III) domain of PTPRZ1 (Mohebiany et al. 2013) fused to the dimerization domain, immunoglobulin-like domains, transmembrane domain, and the tyrosine kinase domain of MET (Fig. 2A; Organ and Tsao 2011). To confirm the recurrent nature of the ZM fusion, we collected an additional 192 glioma samples (Supplemental Table 3) and screened for the presence of the fusion transcript in these samples using fusion-specific PCR primers (Supplemental Table 4). Of the samples tested, the ZM fusion transcript was detected in five additional specimens (Fig. 2B). These fusion breakpoints were confirmed by direct Sanger sequencing. Importantly, a variety of fusion junctions were detected, rendering cross-contamination as the reason for fusion detection unlikely. Interestingly, six out of 40 (15%) specimens in total were derived from patients afflicted with sGBM.
Figure 2.
ZM fusion in training and validation sets. (A) Schematic of PTPRZ1, MET, and the resulting PTPRZ1-MET fusion proteins. (B) PCR and Sanger sequencing validation of the positive fusion samples in the training and validation sets. (C) ZM fusion screening in 19 glioma cell lines. Three cell lines (U118, LN18, and CMK3) showed as ZM fusion positive.
To further confirm the recurrent nature of the ZM fusion, we screened 19 GBM cell lines using fusion-specific PCR primers and detected the fusion sequence in three cell lines (two long-term passaged lines [U118 and LN18] and one primary neurosphere line [CMK3]) (Fig. 2C). In U118 and LN18, a T→C transition mutation was found in the second nucleotide of the MET sequence, changing methionine into threonine. In CMK3, additional mutations were found in addition to the T→C transition. A stretch of nucleotide changes between nucleotide 73 to 123 of the PTPRZ1 coding sequence changed the stretch of amino acids from Tyr-Leu-Lys-Arg-Phe-Leu-Ala-Cys-Ile-Gln-Leu-Leu-Leu-Cys-Val-Cys-Arg-Leu to Tyr-Tyr-Arg-Gln-Gln-Arg-Lys-Leu-Val-Glu-Glu-Ile-Gly-Try-Ser-Tyr-Thr. These results together confirmed the recurrent nature of ZM fusion in GBMs.
Genomic translocation events that give rise to ZM fusion transcripts
We wished to characterize the genomic translocation that gave rise to the ZM fusion transcript. To this end, we extracted DNA from two ZM-harboring GBM specimens: CGGA_D64 and CGGA_1068. CGGA_D64 harbored a ZM fusion that fused exon 8 of PTPRZ to exon 2 of MET (Supplemental Fig. 2A). PCR amplification and Sanger sequencing of the genomic breakpoint in CGGA_D64 revealed a translocation fusing DNA sequences from intron 8 of PTPRZ1 and intron 1 of MET (Supplemental Fig. 2B). Splicing of the fuse intron is expected to give rise to the fusion transcript observed in CGGA_D64. CGGA_1068 harbored a RNA transcript fusing exon 2 of PTPRZ to exon 2 of MET (Supplemental Fig. 2C). Sanger sequencing of the genomic breakpoint in CGGA_1068 revealed a translocation fusing DNA sequences from intron 1 of PTPRZ1 and intron 1 of MET (Supplemental Fig. 2D). Splicing of the fuse intron is expected to give rise to the fusion transcript observed in CGGA_1068. Of note, intact PTPRZ transcripts can be detected in both specimens (Supplemental Fig. 2E), suggesting tandem duplication of the region of PTPRZ involved in ZM fusion.
Genetic landscape of ZM fusion containing sGBMs
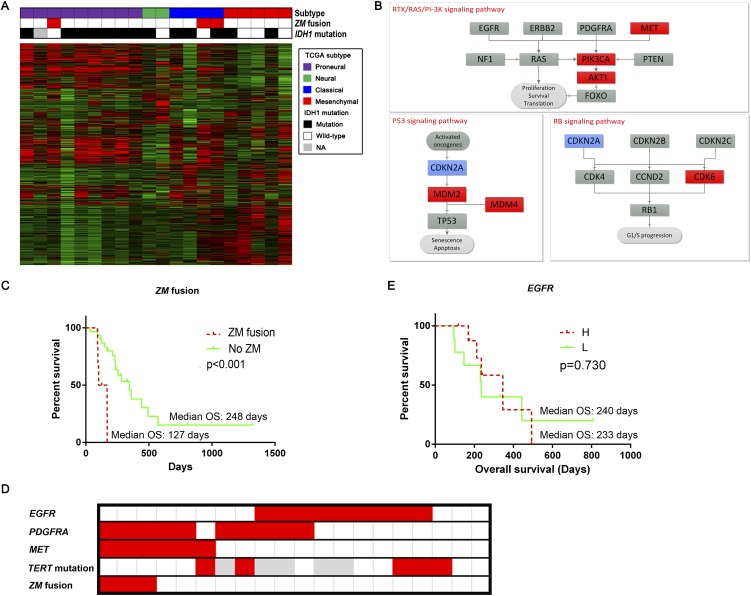
We wished to assess whether the genetic landscape of the ZM fusion containing sGBM differed from those of non-fusion-containing sGBMs. Given the emerging importance of IDH1 mutations in sGBM (Yan et al. 2009), we first tested whether the presence of ZM fusion transcripts coincided with the presence of IDH1 mutations. The ZM fusion transcripts were found in one IDH1 mutated GBM and two IDH wild-type GBMs (Fig. 3A). We next tested whether ZM-fusion transcripts were enriched in any particular Cancer Genome Atlas transcriptional subtypes (Verhaak et al. 2010). The fusion transcript was detected in a proneural subtype and two classical subtype GBMs. These results suggest that the ZM fusion was not tightly coupled to IDH1 mutation status or transcriptional subtype.
Figure 3.
PTPRZ1-MET likely confers unique function. (A) Fusion distribution of the Cancer Genome Atlas subtypes in the sGBM samples. The three ZM-fused sGBM samples consisted of one proneural and two classical subtypes. Due to the small sample size, ZM fusion did not show much association with IDH1 mutation. (B) Expression alteration in ZM-fused sGBMs compared with those without it. The former group showed higher expression of the MET-PIK3CA-AKT1 axis, as well as MDM2 and MDM4. (C) ZM-fused sGBM samples had a significantly shorter overall survival. (D) ZM-fused sGBM samples showed a higher proportion of MET overexpression and EGFR underexpression. (E) EGFR showed no prognostic value in the sGBM samples.
We next determined whether the expression of nodal genes involved in canonical GBM signaling pathways (PIK3CA, RB1, and TP53) (The Cancer Genome Atlas Research Network 2008) differed between sGBMs with or without ZM fusion transcripts. Of the key genes that mediate PIK3CA signaling, there was significant overexpression of the MET, PIK3CA, and AKT1 transcripts in the ZM fusion–bearing GBMs, suggesting hyperactivation of the PIK3CA pathway (Ng et al. 2012). Of the key genes mediating RB1 function, the expression of CDKN2A, a suppressor in this pathway (Foulkes et al. 1997), appeared significantly suppressed, while CDK6, an activator of this pathway (Deshpande et al. 2005), was overexpressed in the ZM fusion–harboring sGBMs. Of the key genes mediating TP53 function, MDM2 and MDM4 (Wade et al. 2013) appeared highly overexpressed in the fusion-bearing GBMs, implicating suppression of TP53-mediated DNA damage response (Bartkova et al. 2005, 2010; Bartek et al. 2007) as a key step in the pathogenesis of these tumors (Fig. 3B).
These results suggest that the ZM fusion–harboring tumors may exhibit a more aggressive phenotype. Clinically, this aggressive phenotype would manifest in the form of decreased overall survival (OS). Indeed, we found that the patients afflicted with ZM fusion–harboring sGBMs fared particularly poorly, with significantly compromised OS relative to those afflicted with sGBMs without the ZM fusion (median OS with ZM fusion vs. without ZM fusion: 127 d vs. 248 d, P < 0.001, log-rank test) (Fig. 3C).
Mutual exclusivity of EGFR expression and ZM fusion in sGBMs
Epidermal growth factor receptor (EGFR) is a receptor tyrosine kinase that is frequently amplified in GBMs (Wen and Kesari 2008) and signals through the RTK/PIK3CA cascade. Our analysis suggested that this pathway is hyperactive in ZM fusion–harboring sGBMs. Since genetic events that are functionally redundant (Ciriello et al. 2012) frequently demonstrate mutually exclusive patterns in genomic analysis, we tested whether the expression of ZM fusion and EGFR overexpression were mutually exclusive in sGBMs. Consistent with our hypothesis, overexpression of EGFR and the presence of the ZM fusion transcript appeared mutually exclusive (Fig. 3D).
TERT promoter mutations are often associated with EGFR amplification or overexpression (Nonoguchi et al. 2013; Appin and Brat 2014). We thus, hypothesize that TERT mutations will cosegregate with EGFR overexpression and will be mutually exclusive to the presence of a ZM fusion transcript. To test this hypothesis, we performed Sanger sequencing of the TERT promoter region in 20 sGBMs (three samples harboring ZM fusion transcripts and 17 ZM-negative samples). TERT mutation was found in approximately a third of the ZM-negative sGBMs but none of the ZM-harboring sGBMs (Fig. 3D), further confirming our hypothesis.
Importantly, EGFR overexpression was not associated with changes in the OS of sGBM patients (median OS with high EGFR expression vs. low EGFR expression: 233 d vs. 240 d, P = 0.730, log-rank test) (Fig. 3E), suggesting that ZM fusion protein likely mediates activities not attributed to EGFR.
Expression of the ZM fusion protein
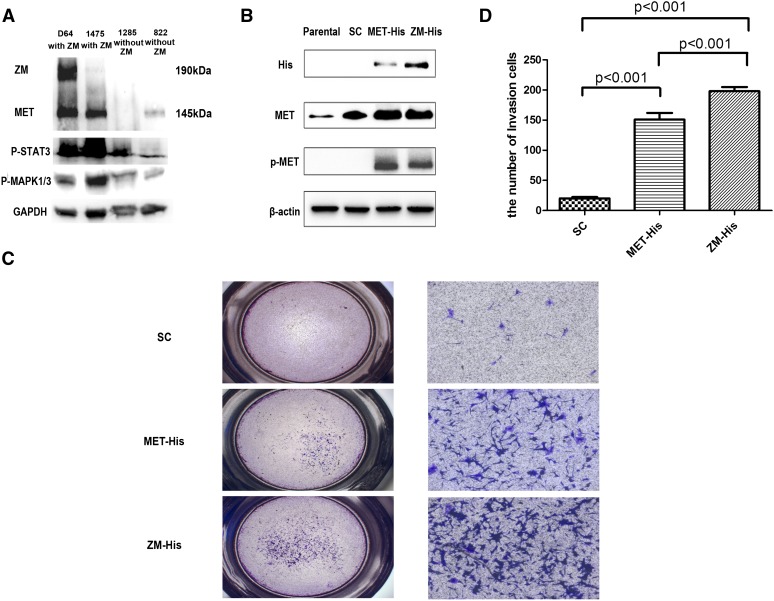
We characterized MET expression at the protein level in two ZM-harboring sGBM specimens with sufficient quantity for immunoblotting analysis (CGGA_1475 and CGGA_D64) and two ZM-negative samples (CGGA_822 and CGGA_1285). CGGA_D64 harbored a ZM fusion that fused exon 8 of PTPRZ to exon 2 of MET. The expected molecular weight of this fusion protein is ∼190 kDa. This prediction is born out when the D64 ZM fusion protein was exogenously expressed in 293 T (Supplemental Fig. 2). Indeed, when protein extract from CGGA_64 was probed with an anti-MET antibody, a 190-kDa band was observed in addition to the 145-kDa band. These results suggest that the ZM fusion transcript was translated into protein (Fig. 4A).
Figure 4.
Immunoblot analysis and invasion assay used for oncogenic alteration detection of ZM fusion in vitro. (A) Immunoblot analysis of ZM-negative samples (822, 1285) and ZM-positive samples (1475, D64). Other than the band of wild-type (WT) MET at 145 kDa, D64 showed a distinct band at 190 kDa, in accordance with ZM fusion protein (ZM is 942 bp larger than WT MET at nucleotide level), which also hybridized with antibody to MET. STAT3 and the MAPK1/3 pathway were intensively activated in ZM-positive samples. (B) His tagged version of the CGGA_1475 ZM fusion was cloned into an adenovirus vector and stably expressed this protein in the U87MG GBM line. The MET endogenously expressed in U87MG is not phosphorylated at residue 1234/5. In contrast, exogenously expressed MET or ZM fusion harbors this phosphorylation. (C) Contrasted with scrambled, notably more U87 cells infected by ZM and MET adenovirus penetrated the Matrigel-coated transwell at 24 h after cells seeded. Meanwhile, the ZM group showed more an intensive invasion than did cells over expressing MET. (Left) 1×; (right) 10×, with scale bar, 200 μm. (D) The fold induction in migration relative to the SC (scramble control) group.
CGGA_1475 harbored a ZM fusion that fused exon 2 of PTPRZ to exon 2 of MET. The anticipated molecular weight of exons 1 and 2 of PTPRZ is 2.3 kDa and 2.7 kDa, respectively. The molecular weight of MET is ∼145 kDa. Exon 1 of MET encodes the 5′ untranslated sequence (394 bp). The anticipated molecular weight of the ZM fusion in CGGA_1475, where exons 1 and 2 of PTPRZ are fused to exon 2 of MET, therefore approximates that of the native MET (∼145 kDa). As such, these two species cannot be discriminated based on SDS-PAGE. This prediction was born out when the CGGA_1475 ZM fusion was overexpressed in 293T cells (Supplemental Fig. 3). Given this ambiguity, it is difficult to interpret whether the strong 145-kDa band in the CGGA_1475 extract represents MET or ZM fusion protein expression.
To explore the oncologic function of the ZM protein, we cloned a His tagged version of the CGGA_1475 ZM fusion into an adenovirus vector and stably expressed this protein in the U87MG GBM line. Stable expression of this ZM protein can be detected as evidenced by a 145-kDa band when probed with an anti-His tag antibody or an anti-MET antibody. Importantly, the MET endogenously expressed in U87MG is not phosphorylated at residue 1234/5 (Cooper et al. 1984; Eder et al. 2009). This phosphorylation event occurs upon dimerization and activation of MET (Wickramasinghe and Kong-Beltran 2005). In contrast, exogenously expressed MET or ZM fusion harbors this phosphorylation (Fig. 4B).
As MET signaling is known to enhance GBM migration and invasion (The Cancer Genome Atlas Research Network 2008), we tested whether ZM fusion modulated these properties in U87MG. Consistent with prior reports (Brockmann et al. 2003), MET expression caused a 7.55-fold increase in the migratory activity of U87MG in a Matrigel-coated transwell assay relative to a vector control. Expression of the ZM fusion caused a 9.9-fold increase in the migratory activity of U87MG cells relative to a vector control. This represents an ∼30% increase in cellular migratory activity in ZM expressing U87MG relative to cells expressing wild-type MET (Fig. 4C). In sum, these results suggest that the recurrent nature of the ZM fusion is unlikely a statistical artifact and that the ZM fusion contributes to the oncologic function of GBMs.
Discussion
Our study is the first study to profile the shifting RNA landscape of gliomas as a function of tumor grade and the identification of ZM as a recurrent fusion gene in sGBMs. In total, 214 fusion transcripts were detected. We observed a notable increase in the proportion of gliomas harboring fusion transcripts during the transition from grade II to grade III. However, this proportion remained somewhat constant between the grade III or grade IV gliomas. These results suggest chromosomal instability as a phenotype associated with the transition from low-grade glioma (grade II) to high-grade glioma (grades III and IV). The highest number of fusion transcripts was found in GBMs that recurred after radiation/temozolomide treatment, suggesting that DNA damage accumulation contributed to fusion transcript formation. Supporting this hypothesis, the classical subtypes of GBM were more likely to harbor fusion transcripts relative to other subtypes. A hallmark of the classical subtype involved aberrant EGFR signaling (Verhaak et al. 2010), and this aberrant signaling has been shown to induce increased DNA damage accumulation and chromosomal instability (Nitta et al. 2010).
Two of the three recurrent fusion transcripts have been previously reported. The RNF213–SLC26A11 fusion transcript was found in a chronic myeloid leukemia patient (Zhou et al. 2013). The FGFR3–TACC3 fusion transcript was initially reported by Singh et al. (2012) as a recurrent fusion transcript in GBMs. The finding was subsequently confirmed by an independent group (Parker et al. 2013). Singh et al. (2012) reported transcripts fusing exon 16 of FGFR3 to exon 8 of TACC3. Soon after this initial report, Parker et al. (2013) reported fusion transcripts involving exons 18 or 19 of FGFR3 and exons 4, 10, or 11 of TACC3. Our analysis showed fusion involving exon 17 of FGFR3 and exons 8, 10, or 11 of TACC3. We found FGFR3–TACC3 fusion transcripts in 5% of pGBMs. This prevalence is largely consistent with those reported by Singh et al. (2012) and Parker et al. (2013).
In addition to the two previously reported fusion transcripts, we identified a novel recurrent fusion transcript that we termed the ZM fusion transcript. ZM is detected in ∼15% of sGBMs in independent cohorts, rendering it the most frequently recurring transcripts for sGBMs. This transcript arose as a result of translocation events between the introns of PTPRZ and the MET proto-oncogene, resulting in a fusion containing variable numbers of PTPRZ1 extracellular domains (including the CA domain and the fibronectin domain) and the entire intracellular domain of MET (Fig. 2A). MET encodes a well-studied proto-oncogene whose activation is triggered by dimerization of the intracellular domain upon binding of hepatocyte growth factor (HGF) (Stommel et al. 2007; Li et al. 2011). PTPRZ1 encodes a membrane-associated tyrosine phosphatase that is highly expressed in the central nervous system (Muller et al. 2003) and signals through beta-catenin–mediated functions (Diamantopoulou et al. 2012). The recurrent nature of the ZM fusion suggests that it plays an active role in GBM biology. Consistent with this hypothesis (1) stable expression of ZM fusion enhanced GBM migration and invasion, (2) ZM fusion harboring sGBM harbored overexpression of genes involved in PIK3CA signaling, and (3) the expression of the ZM fusion transcript was associated with worsened OS in sGBM patients (127 d vs. 248 d). It remains to be determined whether ZM-expressing GBMs are sensitive to MET inhibitors.
The heterogeneity of most fusion transcripts observed in this study and their nonrecurrent nature are highly reminiscent of RNA-seq efforts in other solid tumor types, including melanoma (Berger et al. 2010) and lung cancer (Seo et al. 2012). In our profiling of 272 gliomas, 67 in-frame fusion transcripts were identified. While many of these fusion proteins involve genes that participate in canonical GBM signaling pathways, including RTK/PIK3CA, RB1, and TP53 (The Cancer Genome Atlas Research Network 2008), most of the fusion transcripts identified here involved gene sequences that have not been well studied in GBMs. Characterization of these fusion sequences may unveil novel biologic insights.
In sum, we reported a comprehensive RNA-seq analysis that characterized the RNA fusion landscape during glioma progression. The study provided a catalog of novel fusion transcripts as potential GBM therapeutic targets and revealed ZM as a novel, recurrent fusion transcript in sGBMs.
Methods
Clinical specimen collection
All research performed was approved by the institutional review board (IRB) at Tiantan Hospital and was in accordance with the principles expressed at the declaration at Helsinki. A dedicated clinical research specialist obtained consent from each patient prior to collection. Written consent was obtained for each patient. The specimens were collected under IRB KY2013-017-01. For each patient, the following clinical information was collected: diagnosis, gender, age, WHO grades, and OS.
RNA-seq and quality control
The libraries were sequenced on an Illumina HiSeq 2000 platform using a 101-bp paired-end sequencing strategy. The original image data generated by the sequencing machine were converted into sequence data via base calling (Illumina pipeline CASAVA v1.8.2) and then subjected to standard quality control (QC) criteria to remove all of the reads that fit any of the following parameters: (1) reads that aligned to adaptors or primers with no more than two mismatches, (2) reads with >10% unknown bases (N bases), and (3) reads with >50% of low-quality bases (quality value ≤5) in one read. Finally, 1308.3 Gb (94.4%) of filtered reads were left for further analysis after QC.
Read mapping
Hg19 RefSeq (RNA sequences, GRCh37) was downloaded from the UCSC Genome Browser (http://genome.ucsc.edu).
Candidate gene fusion identification
We used two algorithms, deFuse (deFuse-0.6.1) (McPherson et al. 2011) and TopHat-Fusion (TopHatFusion-0.1.0) (Kim and Salzberg 2011), to detect gene fusion based on the paired-end reads in different samples. The candidates simultaneously detected by both deFuse and TopHat-Fusion were regarded as reliable candidate gene fusions, which were carried forward for further analysis.
Expression analysis of RefSeq genes
The gene expression was calculated using the RPKM (reads per kilobase transcriptome per million reads) method (Audic and Claverie 1997; Mortazavi et al. 2008). The RPKM method is able to remove the influence of different gene lengths and sequencing discrepancies from the calculation of gene expression. Therefore, the calculated gene expression can be directly used to compare the differences in gene expression among samples.
RNA extraction and PCR validation
First-strand cDNA was synthesized from 500–1000 ng of total RNA with random hexamer primers (Promega) using SuperScript III reverse transcriptase (Invitrogen). Reverse transcription was performed for 60 min at 55°C followed by 15 min at 70°C to inactivate the reaction. Escherichia coli RNase H (New England Biolabs) was added to remove RNA complementary to cDNA. Primers (Supplemental Table 3) were designed as flanking fusion points. PCR products were purified using a QIAquick PCR purification kit (Qiagen) and cloned into pGEM-T easy vector (Promega) and then sequenced by a ABI Prism 3730×l DNA sequencer (Applied Biosystems). Sixty-four out of 67 (95.5%) fusion genes were confirmed by sequencing.
Statistical analysis
OS time was calculated from the date of diagnosis until death or the last follow-up. The survival curve was calculated with the Kaplan-Meier method, and the difference was analyzed using a two-sided log-rank test. A P-value < 0.05 was considered statistically significant. All the data analysis was performed in GraphPad Prism and R.
Data access
The raw sequencing data for 272 gliomas have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE48865.
Acknowledgments
This work was supported by grants from the National High Technology Research and Development Program (no. 2012AA02A508), International Science and Technology Cooperation Program (no. 2012DFA30470), National Natural Science Foundation of China (no. 91229121), Beijing Science and Technology Plan (no. Z131100006113018), and National Key Technology Research and Development Program of the Ministry of Science and Technology of China (no. 2013BAI09B03). C.C.C. is supported by the Doris Duke Charitable Foundation, Sontag Foundation, Burroughs Wellcome Fund, Forbeck Foundation, and Kimmel Foundation.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.165126.113.
References
- Appin CL, Brat DJ. 2014. Molecular genetics of gliomas. Cancer J 20: 66–72 [DOI] [PubMed] [Google Scholar]
- Audic Sp, Claverie JM. 1997. The significance of digital gene expression profiles. Genome Res 7: 986–995 [DOI] [PubMed] [Google Scholar]
- Bartek J, Bartkova J, Lukas J. 2007. DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene 26: 7773–7779 [DOI] [PubMed] [Google Scholar]
- Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, et al. 2005. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434: 864–870 [DOI] [PubMed] [Google Scholar]
- Bartkova J, Hamerlik P, Stockhausen MT, Ehrmann J, Hlobilkova A, Laursen H, Kalita O, Kolar Z, Poulsen HS, Broholm H, et al. 2010. Replication stress and oxidative damage contribute to aberrant constitutive activation of DNA damage signalling in human gliomas. Oncogene 29: 5095–5102 [DOI] [PubMed] [Google Scholar]
- Berger MF, Levin JZ, Vijayendran K, Sivachenko A, Adiconis X, Maguire J, Johnson LA, Robinson J, Verhaak RG, Sougnez C, et al. 2010. Integrative analysis of the melanoma transcriptome. Genome Res 20: 413–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockmann MA, Ulbricht U, Gruner K, Fillbrandt R, Westphal M, Lamszus K. 2003. Glioblastoma and cerebral microvascular endothelial cell migration in response to tumor-associated growth factors. Neurosurgery 52: 1391–1399 [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network . 2008. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455: 1061–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriello G, Cerami E, Sander C, Schultz N. 2012. Mutual exclusivity analysis identifies oncogenic network modules. Genome Res 22: 398–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper CS, Park M, Blair DG, Tainsky MA, Huebner K, Croce CM, Vande Woude GF. 1984. Molecular cloning of a new transforming gene from a chemically transformed human cell line. Nature 311: 29–33 [DOI] [PubMed] [Google Scholar]
- Deshpande A, Sicinski P, Hinds PW. 2005. Cyclins and cdks in development and cancer: a perspective. Oncogene 24: 2909–2915 [DOI] [PubMed] [Google Scholar]
- Diamantopoulou Z, Kitsou P, Menashi S, Courty J, Katsoris P. 2012. Loss of receptor protein tyrosine phosphatase β/ζ (RPTPβ/ζ) promotes prostate cancer metastasis. J Biol Chem 287: 40339–40349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eder JP, Vande Woude GF, Boerner SA, LoRusso PM. 2009. Novel therapeutic inhibitors of the c-Met signaling pathway in cancer. Clin Cancer Res 15: 2207–2214 [DOI] [PubMed] [Google Scholar]
- Foulkes WD, Flanders TY, Pollock PM, Hayward NK. 1997. The CDKN2A (p16) gene and human cancer. Mol Med 3: 5–20 [PMC free article] [PubMed] [Google Scholar]
- Kangaspeska S, Hultsch S, Edgren H, Nicorici D, Murumagi A, Kallioniemi O. 2012. Reanalysis of RNA-sequencing data reveals several additional fusion genes with multiple isoforms. PLoS ONE 7: e48745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Salzberg SL. 2011. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol 12: R72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li A, Glas M, Lal B, Ying M, Sang Y, Xia S, Trageser D, Guerrero-Cazares H, Eberhart CG, et al. 2011. c-Met signaling induces a reprogramming network and supports the glioblastoma stem-like phenotype. Proc Natl Acad Sci 108: 9951–9956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. 2007. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 114: 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson A, Hormozdiari F, Zayed A, Giuliany R, Ha G, Sun MG, Griffith M, Heravi Moussavi A, Senz J, Melnyk N, et al. 2011. deFuse: an algorithm for gene fusion discovery in tumor RNA-Seq data. PLoS Comput Biol 7: e1001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohebiany AN, Nikolaienko RM, Bouyain S, Harroch S. 2013. Receptor-type tyrosine phosphatase ligands: looking for the needle in the haystack. FEBS J 280: 388–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Muller S, Kunkel P, Lamszus K, Ulbricht U, Lorente GA, Nelson AM, von Schack D, Chin DJ, Lohr SC, Westphal M, et al. 2003. A role for receptor tyrosine phosphataseζ in glioma cell migration. Oncogene 22: 6661–6668 [DOI] [PubMed] [Google Scholar]
- Nambiar M, Kari V, Raghavan SC. 2008. Chromosomal translocations in cancer. Biochim Biophys Acta 1786: 139–152 [DOI] [PubMed] [Google Scholar]
- Negrini S, Gorgoulis VG, Halazonetis TD. 2010. Genomic instability—an evolving hallmark of cancer. Nat Rev Mol Cell Biol 11: 220–228 [DOI] [PubMed] [Google Scholar]
- Ng K, Kim R, Kesari S, Carter B, Chen CC. 2012. Genomic profiling of glioblastoma: convergence of fundamental biologic tenets and novel insights. J Neurooncol 107: 1–12 [DOI] [PubMed] [Google Scholar]
- Nitta M, Kozono D, Kennedy R, Stommel J, Ng K, Zinn PO, Kushwaha D, Kesari S, Furnari F, Hoadley KA, et al. 2010. Targeting EGFR induced oxidative stress by PARP1 inhibition in glioblastoma therapy. PLoS ONE 5: e10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonoguchi N, Ohta T, Oh JE, Kim YH, Kleihues P, Ohgaki H. 2013. TERT promoter mutations in primary and secondary glioblastomas. Acta Neuropathol 126: 931–937 [DOI] [PubMed] [Google Scholar]
- Ohgaki H, Kleihues P. 2009. Genetic alterations and signaling pathways in the evolution of gliomas. Cancer Sci 100: 2235–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organ SL, Tsao MS. 2011. An overview of the c-MET signaling pathway. Ther Adv Med Oncol 3: S7–S19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker BC, Annala MJ, Cogdell DE, Granberg KJ, Sun Y, Ji P, Li X, Gumin J, Zheng H, Hu L, et al. 2013. The tumorigenic FGFR3-TACC3 gene fusion escapes miR-99a regulation in glioblastoma. J Clin Invest 123: 855–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren R. 2005. Mechanisms of BCR-ABL in the pathogenesis of chronic myelogenous leukaemia. Nat Rev Cancer 5: 172–183 [DOI] [PubMed] [Google Scholar]
- Sajan SA, Hawkins RD. 2012. Methods for identifying higher-order chromatin structure. Annu Rev Genomics Hum Genet 13: 59–82 [DOI] [PubMed] [Google Scholar]
- Sasaki T, Rodig SJ, Chirieac LR, Janne PA. 2010. The biology and treatment of EML4-ALK non-small cell lung cancer. Eur J Cancer 46: 1773–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo JS, Ju YS, Lee WC, Shin JY, Lee JK, Bleazard T, Lee J, Jung YJ, Kim JO, Yu SB, et al. 2012. The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Res 22: 2109–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D, Chan JM, Zoppoli P, Niola F, Sullivan R, Castano A, Liu EM, Reichel J, Porrati P, Pellegatta S, et al. 2012. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science 337: 1231–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stommel JM, Kimmelman AC, Ying H, Nabioullin R, Ponugoti AH, Wiedemeyer R, Stegh AH, Bradner JE, Ligon KL, Brennan C, et al. 2007. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science 318: 287–290 [DOI] [PubMed] [Google Scholar]
- Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, et al. 2005. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 352: 987–996 [DOI] [PubMed] [Google Scholar]
- Torti D, Trusolino L. 2011. Oncogene addiction as a foundational rationale for targeted anti-cancer therapy: promises and perils. EMBO Mol Med 3: 623–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. 2010. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17: 98–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade M, Li YC, Wahl GM. 2013. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer 13: 83–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Jiang T. 2013. Understanding high grade glioma: molecular mechanism, therapy and comprehensive management. Cancer Lett 331: 139–146 [DOI] [PubMed] [Google Scholar]
- Weinstein IB. 2002. Cancer. Addiction to oncogenes—the Achilles heal of cancer. Science 297: 63–64 [DOI] [PubMed] [Google Scholar]
- Wen PY, Kesari S. 2008. Malignant gliomas in adults. N Engl J Med 359: 492–507 [DOI] [PubMed] [Google Scholar]
- Wickramasinghe D, Kong-Beltran M. 2005. Met activation and receptor dimerization in cancer: a role for the Sema domain. Cell Cycle 4: 683–685 [DOI] [PubMed] [Google Scholar]
- Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, et al. 2009. IDH1 and IDH2 mutations in gliomas. N Engl J Med 360: 765–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JB, Zhang T, Wang BF, Gao HZ, Xu X. 2013. Identification of a novel gene fusion RNF213-SLC26A11 in chronic myeloid leukemia by RNA-Seq. Mol Med Rep 7: 591–597 [DOI] [PubMed] [Google Scholar]