Abstract
Alternative splicing (AS) of precursor RNAs is responsible for greatly expanding the regulatory and functional capacity of eukaryotic genomes. Of the different classes of AS, intron retention (IR) is the least well understood. In plants and unicellular eukaryotes, IR is the most common form of AS, whereas in animals, it is thought to represent the least prevalent form. Using high-coverage poly(A)+ RNA-seq data, we observe that IR is surprisingly frequent in mammals, affecting transcripts from as many as three-quarters of multiexonic genes. A highly correlated set of cis features comprising an “IR code” reliably discriminates retained from constitutively spliced introns. We show that IR acts widely to reduce the levels of transcripts that are less or not required for the physiology of the cell or tissue type in which they are detected. This “transcriptome tuning” function of IR acts through both nonsense-mediated mRNA decay and nuclear sequestration and turnover of IR transcripts. We further show that IR is linked to a cross-talk mechanism involving localized stalling of RNA polymerase II (Pol II) and reduced availability of spliceosomal components. Collectively, the results implicate a global checkpoint-type mechanism whereby reduced recruitment of splicing components coupled to Pol II pausing underlies widespread IR-mediated suppression of inappropriately expressed transcripts.
Alternative splicing (AS) is a widespread process by which splice sites in primary transcripts are differentially utilized to produce multiple mRNA and protein isoforms (Pan et al. 2008; Wang et al. 2008; Nilsen and Graveley 2010). It is regulated by the complex interplay of cis- and trans-acting factors that serve to promote or repress the assembly of productive splicing complexes, referred to as spliceosomes. Alternative “cassette” exon splicing is thought to represent the most frequent type of AS in animals and has been implicated in the control of diverse aspects of normal and disease biology (Kalsotra and Cooper 2011; Irimia and Blencowe 2012). In contrast, intron retention (IR), the process by which specific introns remain unspliced in polyadenylated transcripts, is thought to represent the least prevalent form of AS in animals (Galante et al. 2004; Sakabe and de Souza 2007; Wang et al. 2008), whereas it is the most frequent form in plants, fungi, and unicellular eukaryotes (Ner-Gaon et al. 2004; Marquez et al. 2012; Sebé-Pedrós et al. 2013).
Previous studies have shown that IR functions in the homeostatic control of the expression of some RNA processing and export factors (Kalyna et al. 2006; Li et al. 2006; Lareau et al. 2007; Ge and Porse 2013). More recently, it has emerged that IR also controls the expression of developmentally regulated genes in plants and animals (Kalyna et al. 2012; Yap et al. 2012; Wong et al. 2013). For example, a set of retained introns in a murine neuroblastoma cell line was shown to negatively regulate genes with neural-associated functions. Several of these introns were linked to nuclear retention and exosome-mediated RNA turnover of the host transcripts (Yap et al. 2012). In contrast, another set of IR events was found to control the levels of transcripts important for granulocyte maturation (Wong et al. 2013), largely through the process of nonsense-mediated mRNA decay (NMD). These recent studies suggest that different IR events control gene expression through distinct mechanisms. However, the extent to which IR operates across different primary cells and tissues to regulate gene expression via these and possibly additional mechanisms is not known.
By applying a new pipeline for IR detection to high coverage poly(A)+ RNA-seq data from more than 40 diverse human and mouse cell and tissue types, we have performed the most comprehensive analysis of IR to date in mammals. We find that IR is far more frequent in mammals than previously appreciated, affecting transcripts from most genes. A set of distinct cis features reliably discriminates this greatly expanded set of retained introns from introns that undergo constitutive splicing. These retained introns comprise three main classes with distinct evolutionary origins. We further provide evidence that retained introns act widely to functionally “tune” transcriptomes by further reducing the expression of relatively low abundance transcripts that often lack physiological relevance to the cells and tissues they are detected in. Finally, we show that IR is tightly linked to the increased occupancy of RNA Pol II over the corresponding intronic sequence at the genomic level and to reduced levels of core splicing factors. Our results thus suggest that bidirectional cross-talk between Pol II and IR as a consequence of reduced splicing factor recruitment is an important and widespread mechanism that contributes to the functional tuning of mammalian cell and tissue transcriptomes.
Results
High prevalence of intron retention in mammalian transcriptomes
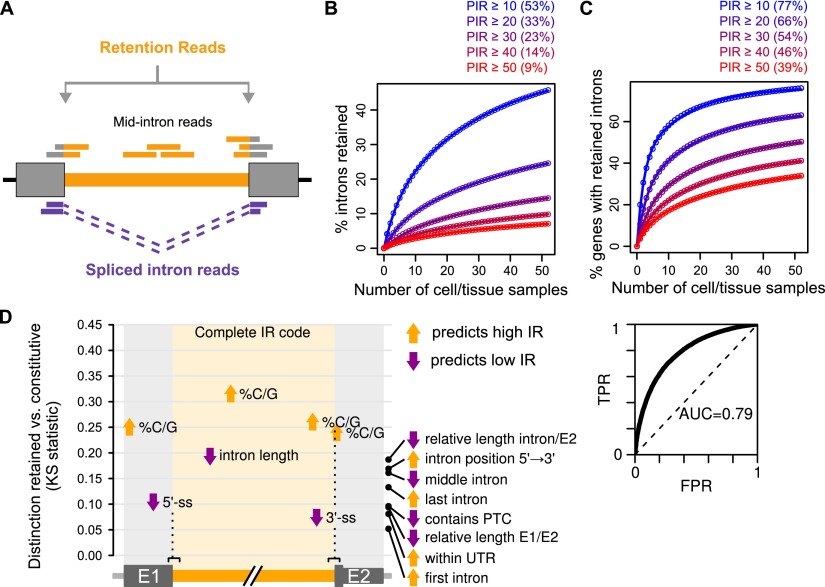
To systematically detect IR in mammals, we discriminated retained from constitutively spliced introns using alignments of reads from poly(A)+ RNA-seq data from ∼40 cell and tissue types from human and mouse (Supplemental Table S1). Reads were aligned to a comprehensive set of reference sequences comprising exon–intron junctions, intron midpoint sequences, and exon–exon junctions formed by intron removal. The degree of IR was represented using the metric percent intron retention (PIR), the percentage of transcripts in which a given intron is retained. In brief, PIR was calculated as the percentage of the average number of reads mapping to the 5′ and 3′ exon–intron junctions, over the sum of the average of the exon–intron junction reads plus the exon–exon junction reads. Additionally, we required a balanced accumulation of reads mapping to 5′ and 3′ exon–intron junctions and to intron midpoint sequences (Fig. 1A). These and additional steps were also used to distinguish IR from alternative transcription initiation/termination, alternative 5′/3′ splice-site selection, and/or overlap with transcripts from other genes including antisense loci (see Methods). IR was detected to variable extents between different cell and tissue types. In general, a higher proportion of introns were found to be retained in neural and immune cell types, whereas IR was detected less often in ES and muscle cells (Supplemental Fig. S1A,B; see below). These differences in IR frequency are unlikely due to differences in read coverage, since the majority of analyzed RNA-seq data sets had a comparable degree of read depth (Supplemental Table S1). RT-PCR validation experiments on 25 representative examples of IR detected by our analysis pipeline, using cell and tissue RNA samples from independent sources from those used to generate the RNA-seq data, confirmed the presence of IR in all cases (Methods; Supplemental Fig. S2). Moreover, measurements of cell/tissue-differential PIR by RNA-seq correlated well with corresponding measurements of PIR by RT-PCR (r = 0.63, P < 2.2 × 10−16, Pearson correlation) (Supplemental Fig. S2).
Figure 1.
Detection and prevalence of intron retention. (A) Intron retention was detected by aligning RNA-seq reads to comprehensive sets of exon–intron and exon–exon junctions. Reads mapping to mid-intron sequences and balanced counts of reads aligning to upstream and downstream exon–intron sequences were used to discriminate IR from other forms of transcript variation. IR levels were measured using percent intron retention (PIR): 100× mean retention reads over the sum of retention and spliced intron reads (see Methods for details). (B) Percentage of total human introns detected as retained at different PIR thresholds as increasing numbers of cell and tissue samples are randomly sampled. Numbers in parentheses are estimates for percentages of total introns retained at different PIR thresholds, as derived from extrapolation. (Circles) Means from 1,000 iterations; (lines) fitted function used for extrapolation (see Methods). Data for mouse introns in Supplemental Figure S1C. (C) Percentage of total human genes with retained introns at different PIR thresholds as increasing numbers of cell and tissue samples are randomly sampled. Numbers in parentheses are estimates for percentages of total genes with retained introns at different PIR thresholds, as derived from extrapolation. Circles and lines as in B; data for mouse genes in Supplemental Figure S1D. (D) Cis-acting features predictive of IR in human. The main graph quantifies, using the Kolmogorov-Smirnov statistic, how well individual features or a logistic regression model comprising 136 features (“Complete IR code”) (Supplemental Table S2) distinguish retained (PIR ≥ 10) from constitutive (PIR < 2) introns in neural tissues. See Methods for details. The graph on the upper right shows the receiver operating characteristic of the complete IR code with area under the curve (AUC) indicated. (TPR) True positive rate; (FPR) false positive rate. E1/E2 are 5′/3′ exons, respectively.
To estimate the total proportions of human and mouse introns that may be subjected to retention, we determined the frequency of IR detection after randomly sampling increasing numbers of cell and tissue RNA-seq data sets. Remarkably, ∼53% and 51% of all human and mouse introns, respectively, have the potential to be retained in poly(A)+ transcripts at a PIR ≥ 10 in at least one of the cell or tissue types, and ∼9% and 8% have the potential to be retained at a PIR ≥ 50 (Fig. 1B; Supplemental Fig. S1C). Moreover, ∼77% of human and mouse multiexonic genes contain one or more retained introns with a PIR ≥ 10, and 35% contain one or more retained introns with a PIR ≥ 50% (Fig. 1C; Supplemental Fig. S1D). These results reveal that the frequency of IR compares with the frequency of cassette exon AS (Pan et al. 2008) and is far more prevalent in mammalian cells and tissues than previously detected using lower coverage RNA-seq data (Wang et al. 2008).
Distinguishing features and classes of retained introns
Consistent with previous reports (Galante et al. 2004; Sakabe and de Souza 2007), retained introns are on average shorter, more C/G rich, and associated with weaker splice sites than are constitutive introns (all tests P < 0.001, one-sided Mann-Whitney U test) (Supplemental Fig. S3). We used logistic regression to determine whether these and additional cis features, or an “IR code,” can be used to reliably discriminate retained from constitutively spliced introns, as detected by our analysis pipeline. The logistic model confirmed that retained introns are significantly associated with elevated C/G content, reduced length, and relatively weak 5′ and 3′ splice sites. However, additional features are also predicted to be important, including elevated C/G content in flanking exonic sequences, the ratios of the lengths of the intron and upstream exon to the length of the downstream exon, and the location of the intron within the gene body. Remarkably, although many of these features are highly correlated with each other, when combined, they more reliably discriminate retained from constitutively spliced introns (ROC AUC = 0.79, P < 1 × 10−300) (Fig. 1D; Supplemental Table S2). These results indicate that introns detected as retained in poly(A)+ RNA by our pipeline are associated with a set of hallmark features that distinguishes them from general introns.
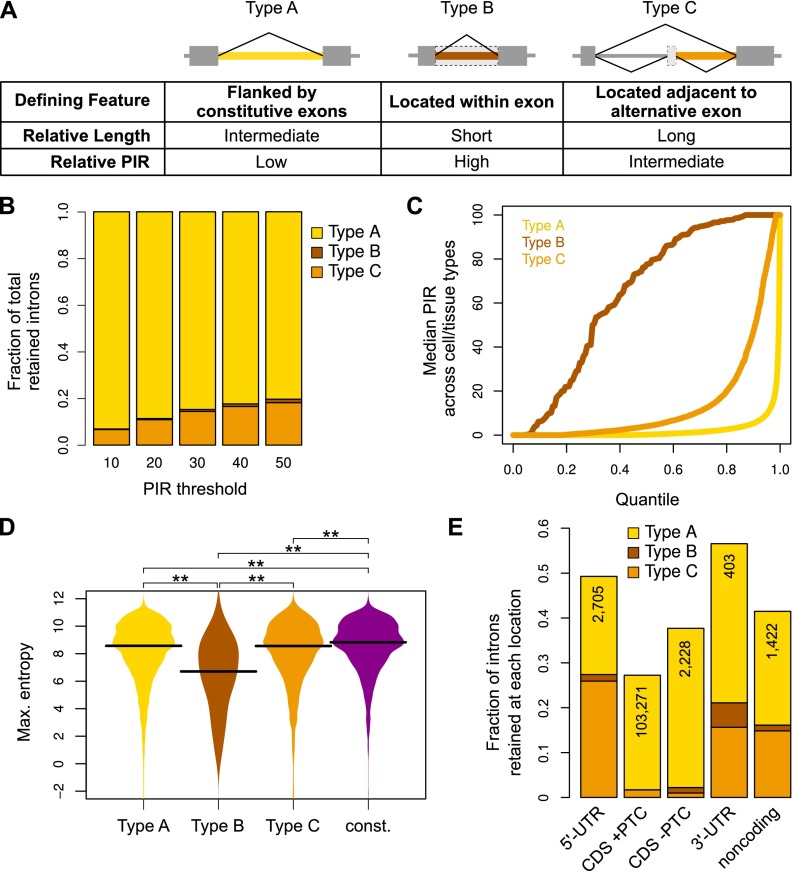
To further investigate cis features that define IR events, we considered that retained introns comprise three distinct types with different evolutionary origins, designated below as Types A–C (Fig. 2A; see Methods). Type A are ancestral introns flanked by constitutive exons, Type B arose by “intronization” of ancestral exonic sequence (Irimia et al. 2008), and Type C are located adjacent to one or more alternative exons that may or may not be conserved between species. Separating retained introns according to this classification reveals markedly different features. Type A introns are the most frequent and, relative to Type B and C, have an intermediate C/G content and length, and are the most weakly retained (Figs. 2B–D; Supplemental Fig. S4A–C). Type B represent the smallest fraction of retained introns and, consistent with their intra-exonic location, have the highest C/G content, the shortest length, the weakest splice sites, and the highest PIR values (Figs. 2B–D; Supplemental Fig. S4A–D). Type C are intermediate in number, have the lowest C/G content, and are longer than Type A and B introns, which suggest a relatively recent evolution that may be tied to the flanking alternative exons (Figs. 2B–D; Supplemental Fig. S4A–C). Retained introns thus comprise a heterogeneous group harboring distinct features and PIR levels that likely reflect different evolutionary origins and functional properties.
Figure 2.
Distinct types of retained introns and associated properties. (A) Classification of retained introns and associated properties. (B) Fractions of total human retained introns belonging to each evolutionary type at different PIR thresholds. Represented are introns that could be assigned as Type A–C and that are retained at the indicated PIR thresholds in ≥10% of the samples. (C) Cumulative distribution of median PIR levels for each retained intron type in human cells and tissues. Only introns where PIR could be determined in ≥10% of the samples are represented. (D) Comparison of donor splice site strength (measured using maximum entropy; see Methods) of human retained introns and of constitutively spliced introns. Retained introns compared have PIR ≥ 10 in ≥10% of the samples where PIR could be determined; constitutively spliced introns have PIR < 2 in all samples where PIR could be determined. (Asterisks) P < 0.001 in two-sided Mann-Whitney U test. (E) Fraction of all human introns in each genic region that is retained with PIR ≥ 10 in ≥10% of the samples where PIR could be determined. (UTR) Untranslated region; (CDS) coding region of gene; (PTC) premature termination codon that can elicit nonsense-mediated mRNA decay. The total number of retained introns in each region is indicated.
Evolutionary conservation of IR across vertebrate species
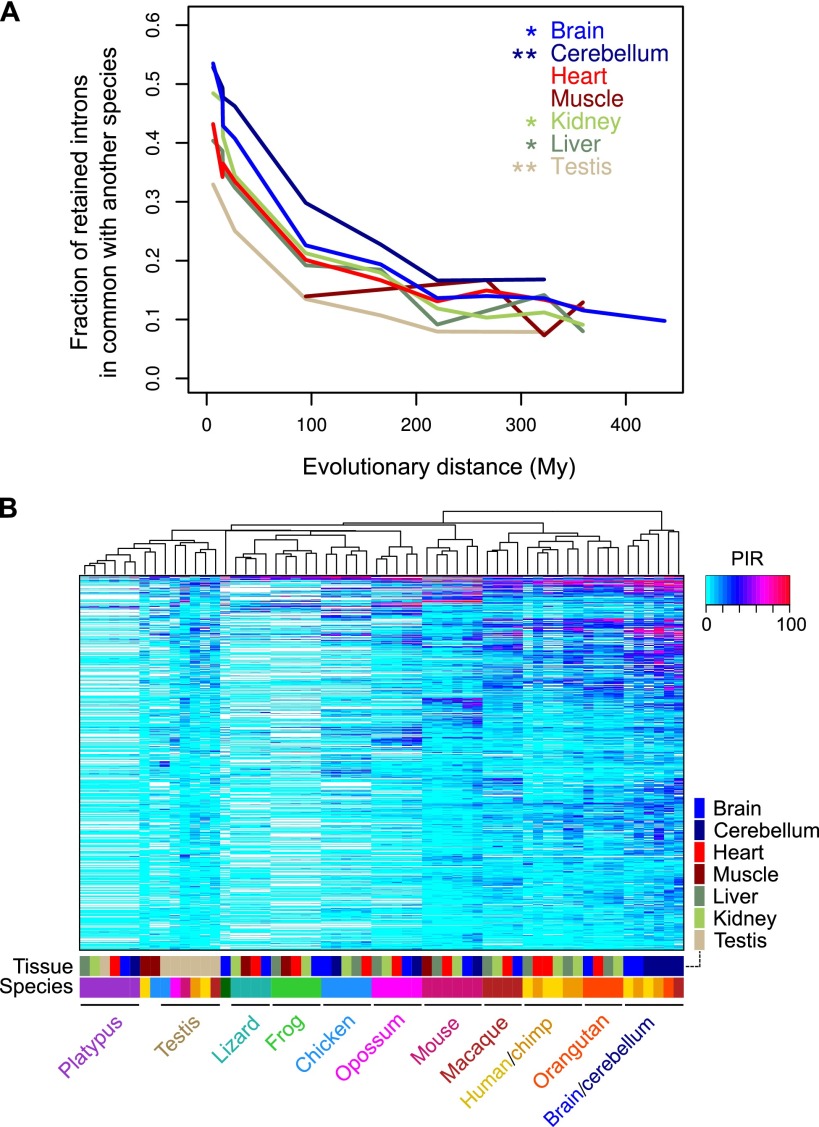
To gain insight into which cell and tissue differential IR events may have conserved functions, we next examined the extent to which IR events are conserved between the equivalent organs (whole brain, cerebellum, heart, muscle, liver, kidney, and testis) from up to 11 vertebrate species spanning ∼440 million years of evolution (see Fig. 3A). Similar to cassette exon AS (Barbosa-Morais et al. 2012; Merkin et al. 2012), IR has diverged rapidly in all tissues during vertebrate evolution, although to a considerably lesser extent in the brain than in organs such as testis, which shows the most divergent IR profiles (Fig. 3A; Supplemental Fig. S5A). Hierarchically clustering tissue samples by their transcriptome-wide PIR profiles results in the segregation of most brain tissues from primate species, whereas other samples generally cluster according to species; testis is an exception due to its high heterogeneity of PIR patterns (Fig. 3B, see legend). Similar results were obtained using principal component analysis (Supplemental Fig. S5B). These results thus suggest that IR, although having diverged rapidly in all analyzed vertebrate organs, more often provides conserved functions in the nervous system than in other tissues.
Figure 3.
Tissue-specific evolutionary conservation of IR across vertebrates. (A) Proportion of total orthologous introns retained (PIR ≥ 10) in an organ of one species that are also retained in the same organ of another of 11 vertebrate species being compared (see main text and panel B). Lines connect average values for each evolutionary distance. Asterisks indicate significance of differences between each organ and the average of all other organs (*) P < 0.05; (**) P < 0.001 (see Methods for details). (B) Hierarchical clustering of the same vertebrate species’ tissue samples in A, based on comparison of PIR values. Only introns with an intra-species PIR range ≥ 10 in at least three species are compared (n = 4835). (White) Missing data for nonconserved introns.
Global regulation of mRNA levels through IR
Retained introns, relative to constitutive introns, are enriched in untranslated regions (UTRs) and noncoding RNAs compared to protein-coding regions of genes (Figs. 1D, 2E). Moreover, retained introns that are predicted not to introduce an NMD-eliciting premature termination codon (PTC) are detected at a significantly higher frequency in coding-overlapping regions than are retained introns that introduce a PTC (P < 4.2 × 10−16, Fisher’s exact test). IR is also detected with increasing frequency toward the 3′ ends of transcripts (Supplemental Fig. S6A). Using ENCODE RNA-seq data generated from nuclear and cytoplasmic poly(A)+ RNA (Tilgner et al. 2012), we observe that this distribution pattern is detected in transcripts isolated from both cellular fractions (Supplemental Fig. S6B), indicating that it is not simply a consequence of reduced kinetics of splicing of retained introns in nascent transcripts. Moreover, PIR levels are lower, overall, in cytoplasmic compared to nuclear transcripts (Supplemental Fig. S6C). Taken together with the detection of prevalent IR (Fig. 1), these results suggest that cytoplasmic levels of transcripts containing retained introns may be reduced in diverse cells and tissues through nuclear restriction and NMD.
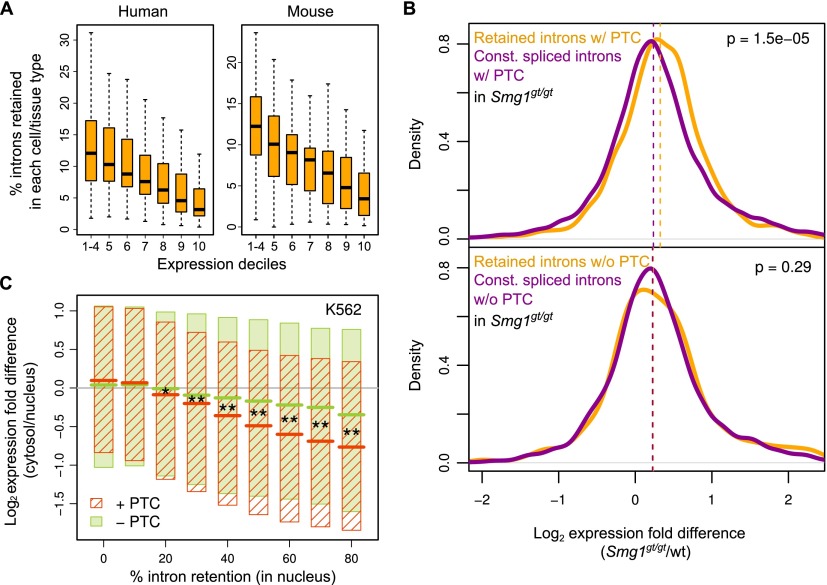
Supporting this possibility, we observed a significant negative relationship between IR detection and steady-state transcript levels (Fig. 4A; Supplemental Fig. S7A). To confirm whether intron retention leading to NMD has a significant global impact on transcripts, we analyzed transcript levels using RNA-seq data from mouse embryonic fibroblasts (MEFs) derived from wild-type mice and mice homozygous for a gene-trap (gt) insertion that disrupts Smg1 (McIlwain et al. 2010), a kinase that is critical for NMD activity (Fig. 4B). Transcripts with retained introns that introduce PTCs are expressed at significantly higher steady-state levels in the Smg1gt/gt than wild-type MEFs, compared to transcripts with PTC-containing introns that are not retained (P = 1.5 × 10−5, one-sided Mann-Whitney U test) (Fig. 4B, upper panel). Moreover, ∼10% of retained introns predicted to introduce PTCs display a pronounced (i.e., ≥15%) increase in PIR in Smg1gt/gt compared to wild-type MEFs, whereas only 2% displayed a comparable decrease. In contrast, a significant difference in overall steady-state transcript level changes between wild-type and Smg1gt/gt MEFs was not observed when comparing transcripts harboring either retained or constitutively spliced introns that are not predicted to introduce PTCs (Fig. 4B, lower panel). These results show that retained introns harboring PTCs contribute significantly to the global down-regulation of transcript levels via NMD.
Figure 4.
Global regulation of mRNA levels through IR. (A) Box plots showing distributions of percentages of total human and mouse introns detected as retained (PIR ≥ 10) in transcripts sorted into 10 different expression level bins (deciles, with deciles 1–4 averaged). (B) Distributions of expression difference between Smg1gt/gt and wild-type MEFs for transcripts harboring introns predicted to introduce a PTC that can trigger NMD upon retention (top), and for transcripts harboring introns that are predicted not to introduce a PTC (bottom). In each panel, retained (PIR ≥ 10) and constitutively (PIR < 2) spliced introns are compared. P-value indicates significance of expression change difference (one-sided Mann-Whitney U test). (C) Expression difference between cytosolic and nuclear fractions of K562 cells for transcripts that do or do not contain PTC-introducing introns, as measured at different PIR thresholds in the nuclear fraction (see also Supplemental Fig. S6C). Shaded boxes indicate upper and lower quartiles of distributions of expression level differences, and colored lines indicate median values. Expression-level differences for transcripts harboring retained introns that do or do not introduce PTCs are indicated by red and green, respectively. Asterisks indicate significance of difference between median values for expression level differences for transcripts with and without PTC-introducing retained introns (*) P < 0.05; (**) P < 0.001 (one-sided Mann-Whitney U tests after Bonferroni correction). See also Supplemental Figure S7B.
We next asked to what extent IR-mediated nuclear restriction versus NMD impacts steady-state transcript levels in the cytoplasm. Accordingly, we compared ratios of cytoplasmic to nuclear poly(A)+ mRNA for transcripts that harbor retained introns that are, or are not, predicted to introduce PTCs (Fig. 4C). These ratios were measured across a range of increasing nuclear PIR thresholds to determine how increased IR contributes to reduced levels of cytoplasmic transcripts. Importantly, as the PIR threshold increases in the nucleus, the levels of transcripts in the cytoplasm relative to the nucleus progressively decrease. This relationship is observed when the retained introns do not introduce a PTC (indicative of nuclear retention and turnover), but it is significantly enhanced when the retained introns introduce a PTC (Fig. 4C; Supplemental Fig. S7B). Intron retention thus results in global-scale reductions in cytoplasmic transcript levels through additive contributions from both nuclear restriction and NMD.
IR down-regulates nonphysiologically relevant transcripts
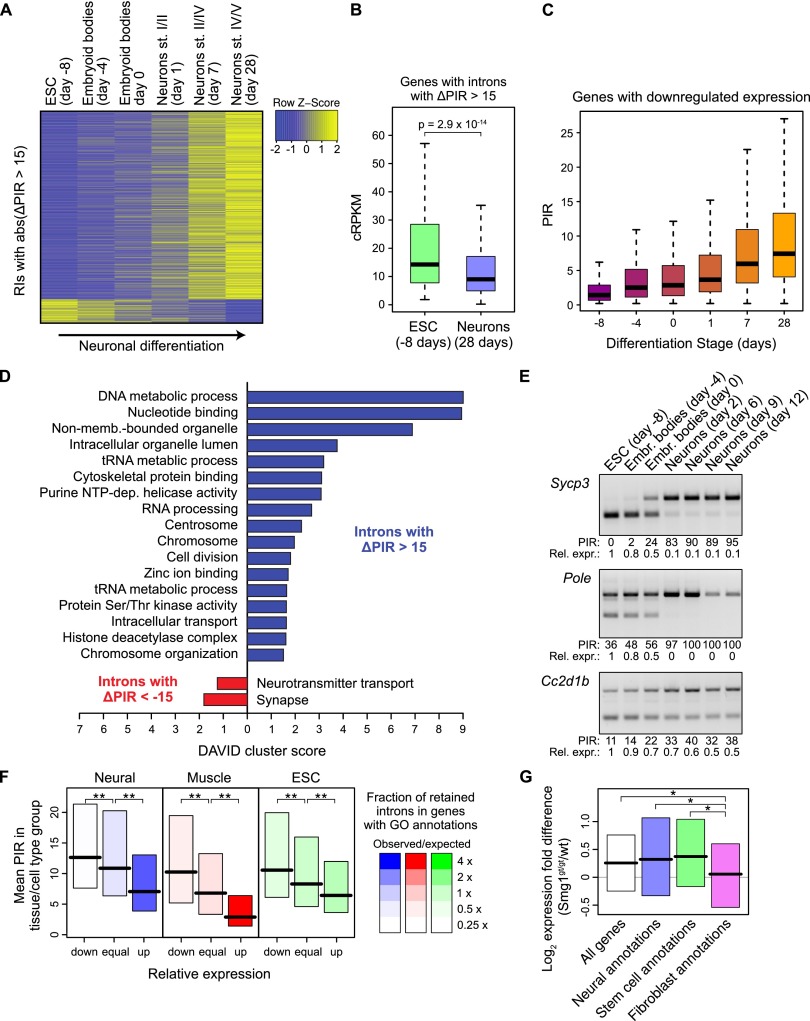
To investigate the potential physiological roles of IR during cell differentiation, we analyzed alternative retained introns using RNA-seq data generated across a time series of differentiation of cortical glutamatergic neurons from murine embryonic stem (ES) cells (Hubbard et al. 2013). Strikingly, the vast majority (88.7%) of detected differentially retained introns between ES and mature neurons display a progressive increase in retention during differentiation (Fig. 5A). Consistent with this observation and a global regulatory role for IR in the suppression of gene expression, transcripts with increased IR in mature neurons are, on average, expressed at significantly lower steady-state levels than are transcripts with increased IR in ES cells (Fig. 5B). Moreover, among the total set of genes that display progressively decreased steady-state levels as neuronal differentiation proceeds, overall PIR levels progressively increase (Fig. 5C). As a group, genes that contain introns with higher retention in differentiated neurons compared to ES cells are significantly enriched in multiple GO terms, including terms related to the cell cycle (Fig. 5D). Examination of individual genes that follow the same profile of IR regulation revealed examples that possess effector functions relevant to non-neural tissues, pluripotency, and DNA replication and repair (see below). Conversely, genes with introns whose retention is lower in differentiated neurons compared to ES cells are significantly enriched in GO terms that are relevant to neuronal biology, specifically, “Neurotransmitter transport” and “Synapse” (Fig. 5E). Taken together, these observations suggest that increased IR during neuronal differentiation primarily functions to down-regulate transcripts from genes, the expression of which is either not required or less required for the biology of mature neurons compared to ES or neural progenitor cells. It is also possible that IR down-regulates specific genes that, if otherwise expressed, could interfere with the specification of glutamatergic neuronal cell identity. To further investigate and confirm these possibilities, we used RT-PCR assays to validate individual events detected by RNA-seq that display concomitant increases in PIR and decreases in expression during neuronal differentiation. Of 11 tested IR events, all were confirmed to have increased levels of retention and decreased levels of processed mRNA expression (Fig. 5E; Supplemental Fig. S8). Among the validated examples are genes that function in meiosis in germ cells (Sycp3), DNA replication and repair (Mutyh, Pole), synthesis of organic acids concentrated in bile (Hsd3b7, Csad), smooth muscle and spleen biology (Fhod1), glomerulus integrity (Wtip), serotonergic synapses (Cc2d1b), astrocyte differentiation (Arhgap17), and the cytoskeleton organization and proliferation of neural progenitor cells (Flna). An additional example is an IR event in Ssrp1, a component of the “Facilitates Chromatin Transcription” (FACT) chromatin remodeling complex, that functions in transcription, replication, and DNA repair. Consistent with our observation that increased IR reduces Ssrp1 mRNA expression during neuronal differentiation, it has been previously reported that FACT is specifically up-regulated in proliferating cells relative to differentiated cells (Garcia et al. 2011). Our results thus reveal a mechanism by which FACT is down-regulated during differentiation.
Figure 5.
IR-mediated tuning of gene expression. (A) Heatmap of Z-scores for introns differentially retained during differentiation of ES cells into mature glutamatergic neurons. Z-scores are shown for retained introns with a change in PIR (ΔPIR) > 15 between ES cells and mature neurons (day 28). (B) Distribution of expression values (in cRPKM) in ES cells and mature neurons for genes that contain introns with increased retention during differentiation. (C) Distributions of PIR of introns in genes whose expression is down-regulated (more than fivefold between day −8 and day 28) at different time points of neuronal differentiation. (D) DAVID cluster analysis of enriched GO annotations for genes that contain introns with increased (blue) or decreased (red) retention during neuronal differentiation. (E) RT-PCR validation of RNA-seq–detected events with increasing PIR and decreasing spliced mRNA expression during differentiation of glutamatergic neurons from mouse ES cells. Quantification of PIR and the mRNA expression are shown beneath each panel. For relative expression, the spliced band was quantified and normalized to Gapdh, and day −8 was set to 1. See Supplemental Figure S8 for the full set of 11 tested events. (F) Relationship between the degree of IR and cell type-specific expression of genes annotated with cell/tissue-specific functions. Bars show the upper and lower quartile, and black lines the medians, of the PIR difference between neural and other cell/tissue types (left), muscle and other cell/tissue types (middle), and ESCs and other cell/tissue types (right). Intron PIR was measured in sets of genes assigned to equal-sized bins (three bars) based on having decreased (down), equivalent (equal), or increased (up) expression, as compared to the median expression values for the other cell types. Shading indicates the degree of enrichment of GO categories related to the biology of neural, muscle, and stem cells, respectively (see Methods for details). Asterisks indicate P < 0.001 in one-sided Mann-Whitney U tests after Bonferroni correction. (G) Expression difference between Smg1gt/gt and wild-type MEFs for transcripts harboring retained introns predicted to introduce a PTC that can trigger NMD, in genes associated with GO categories related to the biology of neural cells, stem cells, and fibroblasts. Asterisks indicate a significant difference between median values for expression level differences (P < 0.05 in one-sided Mann-Whitney U tests after Bonferroni correction).
To investigate whether IR is also involved in the down-regulation of transcripts that may be less physiologically relevant in other biological contexts, we next examined global relationships between IR and the expression levels of genes with varying degrees of functional specificity relevant to neural, muscle and ES cells. Genes were binned according to whether they are (1) down-regulated, (2) equally expressed, or (3) up-regulated in a specified group of tissue samples relative to the rest of the samples, and the degree of enrichment of IR and specific GO terms was analyzed within each bin. As expected (Miki et al. 2001; Zhang et al. 2004), GO terms reflecting cell/tissue-specific functions are increasingly enriched among the subsets of genes with increased cell/tissue-specific expression (Supplemental Table S3). Moreover, PIR levels are significantly lower in transcripts from these up-regulated, GO-enriched genes, and they are significantly higher in transcripts from genes with lower levels of cell/tissue-specific expression and GO enrichment (P < 0.001, one-sided Mann-Whitney U tests) (Fig. 5F). Extending the results shown in Figure 5A–E, these observations indicate that IR acts widely to reduce levels of transcripts from genes with functions that are not relevant or less relevant to the cell or tissue type in which IR is detected.
Finally, to assess whether this “transcriptome tuning” property of IR is linked to its ability to trigger NMD, we asked whether genes with increased transcript levels in Smg1gt/gt versus wild-type MEFs (see Fig. 4) are more often linked to annotations associated with neural or stem cell functions (i.e., less relevant or potentially detrimental to MEF biology) compared to fibroblast-associated functions. Remarkably, reduced NMD activity in the Smg1gt/gt MEFs indeed results in significant increases in transcript levels for genes associated with neural and ESC biology compared to fibroblast biology (P < 0.05, one-sided Mann-Whitney U tests after Bonferroni correction) (Fig. 5G). Collectively, these results indicate that IR-mediated down-regulation of transcripts acts widely to functionally tune cell type-specific gene expression profiles.
Mechanism of intron retention
To investigate mechanisms underlying IR-mediated gene regulation, we considered contributions both from cis- and trans-acting factors. Most RNA processing occurs cotranscriptionally, and there is increasing evidence for extensive cross-talk between splicing, transcription, and chromatin regulation (Braunschweig et al. 2013; Kornblihtt et al. 2013). Accordingly, because retained introns are associated with suboptimal splicing signals and negative cis-acting elements (Figs. 1D, 2D; Supplemental Fig. S3B; Galante et al. 2004; Sakabe and de Souza 2007; Yap et al. 2012; Wong et al. 2013), IR events may be particularly sensitive to kinetic coupling effects involving RNA Pol II; and conversely, IR may impact events acting at the level of chromatin and transcription that normally depend on efficient splicing. For example, it has been demonstrated that disruption of splicing in nascent RNA leads to proximal, intragenic pausing of RNA Pol II (Fong and Zhou 2001; Alexander et al. 2010; Chathoth et al. 2014) (see below).
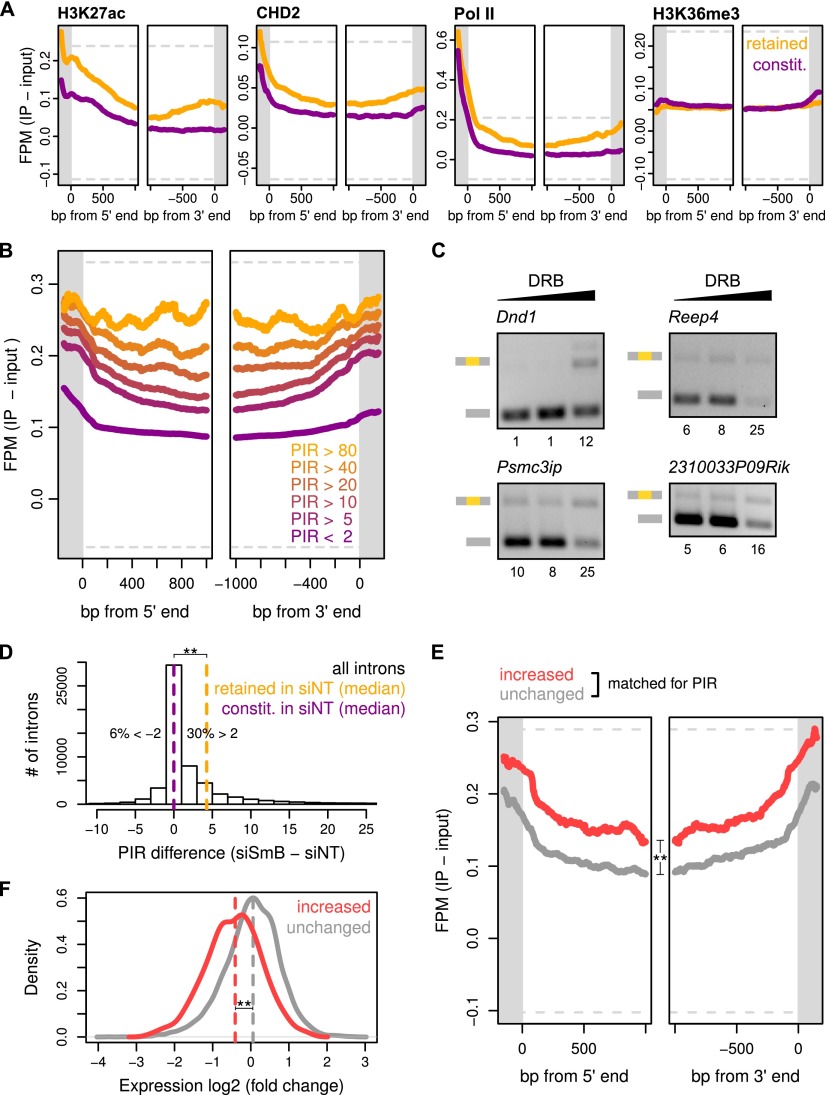
To investigate these possibilities, we analyzed ENCODE ChIP-seq data for POLR2A, the largest subunit of RNA Pol II, 128 additional transcription and chromatin components from the human K562 hematopoietic cell line, and 40 additional components from the mouse CH12 B lymphoma cell line (The ENCODE Project Consortium 2012), for which we measured IR levels using matching RNA-seq data. This analysis revealed that RNA Pol II, specific chromatin modifications (e.g., H3K27ac), and chromatin regulators (e.g., CHD2) are significantly enriched over retained compared to constitutive introns (Fig. 6A; Supplemental Fig. S9A). Moreover, enrichment of these and other specific transcription and chromatin components over retained introns is largely distinct from patterns of enrichment detected over exons. For example, H3K36me3 is significantly enriched over exons (Schwartz et al. 2009; Spies et al. 2009; Tilgner et al. 2009), but it is not enriched over retained introns when steady-state expression levels of the corresponding transcripts are controlled for (Fig. 6A; Supplemental Fig. S9A). These results thus reveal that, at the genomic level, retained introns are associated with specific chromatin and transcription components.
Figure 6.
Coupling between IR and RNA Pol II elongation. (A) Average ChIP-seq signals across introns that are retained (orange, PIR ≥ 10) or not retained (purple, PIR < 2) in K562 cells, for four of 129 analyzed human chromatin features. Retained and not retained introns analyzed are in genes with matched expression levels. (Gray bars) Flanking exons; (y-axes) input-subtracted average ChIP fragment density per million reads (FPM-input) at each aligned bp; (gray dashed lines) 2nd and 98th percentiles of all values in the plot region before averaging to give an indication of the dynamic range. See Supplemental Figure S9A for similar analysis in mouse CH12 cells. (B) Average ChIP-seq signal of Ser2-phosphorylated RNA Pol II over introns with different PIR thresholds in K562 cells. Labels and plots as in A. See Supplemental Figure S9B for similar analysis in mouse CH12 cells. (C) RT-PCR assays of IR events in CGR8 mouse ES cells with and without treatment with the RNA Pol II elongation inhibitor DRB. Retained introns with low PIR in ES cells relative to other cell types were analyzed. (Lane 1) 0 μg/mL DRB; (lane 2) 10 μg/mL; (lane 3) 25 μg/mL. Quantitations are shown beneath each panel. See Supplemental Figure S10 for the full set of tested introns. (D) Number of introns with PIR changes in HeLa cells transfected with siRNAs targeting SNRPB, which codes for SmB/B′, relative to cells transfected with a control, nontargeting siRNA (Saltzman et al. 2011). (Dashed lines) Medians of retained (PIR ≥ 10) and constitutive (PIR < 2) introns in control treated cells; (asterisks) P < 0.001 between these groups (one-sided Mann-Whitney U test). (E) RNA Pol II occupancy over introns (in untreated HeLa cells) that become more retained (PIR difference > 15, red) or that do not change (PIR difference between −2 and +2, gray) after SNRPB knockdown. Because RNA Pol II occupancy is dependent on absolute PIR, groups of introns with matching PIR distributions in control treated cells and >2 kb away from the transcription start site are shown. (Asterisks) P < 0.001 for difference in median Pol II-Ser2p occupancy (see Methods for test details). (F) Expression change between SNRPB and control knockdown for genes containing introns that become more retained (PIR difference > 15, red) or that do not change (PIR difference between −2 and +2, gray). (Asterisks) P < 0.001 between these groups (one-sided Mann-Whitney U test).
Of all the factors analyzed, Pol II hyperphosphorylated on its C-terminal domain (CTD) at Ser2 (Pol II-Ser2p), the form of Pol II associated with transcription elongation, displays the strongest enrichment over retained introns compared to constitutive introns (Fig. 6B). This enrichment pattern is observed irrespective of the location of retained introns in transcripts, although occupancy of Pol II is highest near transcription start and termination sites (Supplemental Fig. S9A). Importantly, we further observe a strong positive correlation between the level of Pol II-Ser2p enrichment over retained introns and the level of IR (Fig. 6B; Supplemental Fig. S9B). These results thus indicate that retained introns are sites of significant accumulation of the elongating form of RNA Pol II, and the levels of this accumulation appear to be tightly coupled with IR levels.
To further explore the relationship between IR and Pol II elongation, we asked whether drug-induced inhibition of elongation results in increased IR. Previously, we showed that treatment of cells with the elongation inhibitor 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole (DRB), which inhibits the kinase activity of the positive transcription elongation factor (P-TEFb), leads to cassette exon splicing changes and increased occupancy of Pol II at specific sites adjacent to the affected exons (Ip et al. 2011), in addition to its known role in causing increased accumulation of Pol II at transcription start sites (Marshall et al. 1996). Following mock- or DRB-treatment of CGR8 ES cells, we used RT-PCR assays to analyze PIR of introns that are highly retained in differentiated mouse cells relative to mouse ES cells. We observed that 13 of 18 analyzed introns showed detectable increases in retention, with four increasing by PIR ≥ 10, whereas only two showed decreases and the remaining three either no detectable change, or else a band corresponding to the size of the intron-retained isoform could not be detected (P ≈ 0.01, one-sided Mann-Whitney U test for PIR difference) (Fig. 6C; Supplemental Fig. S10). Collectively, these results provide evidence that localized Pol II pausing over retained introns in genes is coupled to increased IR levels.
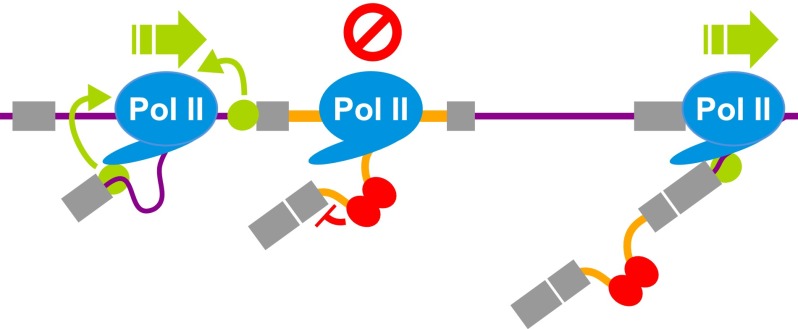
Finally, we hypothesized that if Pol II accumulation at genomic locations coinciding with retained introns is due, at least in part, to the inefficient recruitment of splicing factors (see Discussion), further reducing the levels of core spliceosome components would preferentially increase PIR levels of introns that are already associated with increased Pol II occupancy. To test this hypothesis, we analyzed poly(A)+ RNA-seq data (Saltzman et al. 2011) generated from HeLa cells following knockdown of spliceosomal snRNP components. As expected, we observed a general increase in PIR for retained versus constitutive introns (Fig. 6D). Importantly, introns that display the largest increases in PIR upon snRNP depletion are associated with significantly higher levels of Pol II occupancy than are introns that have comparable PIR levels in untreated cells but show no increase in retention upon snRNP depletion (Fig. 6E). Furthermore, the levels of transcripts containing introns that show increased retention upon snRNP depletion are significantly reduced compared to the levels of transcripts with introns that do not show an increased PIR (Fig. 6F). Collectively, although we cannot exclude possible indirect effects of snRNP depletion contributing to increases in IR, these observations further support the conclusion that IR controls mammalian gene expression via a global, bidirectional cross-talk mechanism, in which low levels of transcription lead to impaired recruitment of core splicing factors, which in turn results in localized pausing of RNA Pol II, further intron retention, and ultimately transcript turnover (Fig. 7).
Figure 7.
Mechanistic model for gene regulation via coupling between IR and RNA Pol II elongation. Inaccurate cell/tissue-specific transcription leads to low levels of expression and reduced recruitment of splicing factors to nascent transcripts. Weak splice sites and/or other cis features associated with retained introns leads to their retention. Binding of basal splicing components such as U1 snRNP (green circle) to the 5′ splice site of constitutive introns promotes Pol II elongation (Fong and Zhou 2001; Alexander et al. 2010), whereas the absence of recruitment of such factors promotes IR and reduces RNA Pol II elongation. Reduced Pol II elongation may further promote and commit introns to retention by favoring binding of splicing repressive factors (red ovals).
Discussion
In this study, we show that IR is a widespread regulatory mechanism that contributes to the functional tuning of mammalian transcriptomes. Consistent with emerging evidence for IR controlling cell type-specific and developmentally regulated gene expression (Yap et al. 2012; Wong et al. 2013), we observe that IR globally impacts gene expression in mammalian cells and tissues by negatively regulating cytoplasmic transcript levels. Although IR-mediated nuclear restriction is important, the majority of IR-dependent regulation appears to operate through NMD (Fig. 4B,C). Strikingly, IR is detected at higher levels in transcripts expressed at relatively low levels and annotated with gene functions that are not relevant or less relevant to the cell type or developmental stage in which the transcripts are detected. As we have shown from analyzing IR during neuronal differentiation, this tuning function acts to reduce the expression of genes that are more required for the biology of ES and progenitor cells than the biology of mature neurons. Similarly, this function of IR may act widely to suppress the expression of spurious transcripts that arise as a consequence of the inability of the cell to fully shut off inappropriate transcription.
Our results provide a model in which the functional tuning of gene expression is achieved through a global “checkpoint” mechanism by which IR regulates transcript levels via cross-talk with RNA Pol II. We show that IR is closely linked to the stalling of RNA Pol II, since increased occupancy of Pol II over retained introns in genomic DNA correlates strongly with increased IR levels (Fig. 6B; Supplemental Fig. S9B). IR is also highly correlated with specific sequence features including reduced intron length, elevated C/G content, intron position within the transcript, and splice site strength. The increased pausing of Pol II over retained introns may in part be due to the increased C/G content of these sequences, as increased C/G content of genes has recently been shown to negatively correlate with Pol II elongation rate (Veloso et al. 2014). Moreover, it is also possible that intron retention is related to differences in the relative G/C content of intron versus flanking intron sequences, which has been shown to impact whether splicing proceeds via an intron or exon definition-type mechanism (Amit et al. 2012).
Given previous extensive evidence of splicing factor recruitment to sites of active transcription through coupling mechanisms that involve chromatin and transcription factors (Braunschweig et al. 2013; Kornblihtt et al. 2013), including splicing activators and the Pol II carboxyl-terminal domain (David and Manley 2011), it is possible that reduced transcriptional levels, for example arising from low frequency spurious promoter activation, result in insufficient recruitment of splicing factors to promote efficient intron removal in nascent RNA. Since retained introns generally have weaker splice sites compared to constitutive introns (this study; Sakabe and de Souza 2007), they are particularly sensitive to reduced splicing factor concentrations (Fig. 6; Wong et al. 2013). Moreover, because Pol II elongation in turn depends on productive splicing (Fong and Zhou 2001; Alexander et al. 2010; Chathoth et al. 2014), retained introns likely become sites of increased pausing of Pol II because of their inefficient splicing. As also supported by the results of DRB treatment, which led to increased retention of most analyzed introns (Fig. 6C), reduced elongation of Pol II over retained introns appears to be mechanistically linked to increased intron retention levels. The kinetic delay in Pol II elongation could lead to increased binding of repressive splicing factors, such as the hnRNP splicing regulator PTBP1 (Yap et al. 2012), and as a consequence further contribute to establishing IR in otherwise processed and polyadenylated RNA. These nuclear events ultimately culminate in the turnover of retained intron-containing poly(A)+ RNA in the nucleus and cytoplasm and contribute to the down-regulation of gene expression.
Methods
Data sets
This study used poly(A)+ RNA-seq data generated as part of the Illumina Human BodyMap 2.0 Project and as described in Brawand et al. (2011) and Barbosa-Morais et al. (2012), as well as multiple other data sets described in the literature (Supplemental Table S1). ENCODE ChIP-seq data were downloaded from the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra; The ENCODE Project Consortium 2012). GEO accessions of all analyzed ChIP-seq data sets are provided in Supplemental Table S4.
Sequence annotation
Full genomic sequences for the 11 species analyzed in this study were downloaded from the UCSC Genome Browser database (Dreszer et al. 2012) (assemblies listed in Supplemental Table S5). Full transcriptomic sequences for all species were downloaded from Ensembl (Flicek et al. 2013) (transcriptome versions listed in Supplemental Table S5). For each gene, the representative transcript was selected for gene expression (GE) analysis as in Barbosa-Morais et al. (2012). Intron annotations (including genomic coordinates) for intron retention (IR) analysis were derived from tables downloaded from the UCSC Genome Browser database. The selected tables for each species are listed in Supplemental Table S5.
Orthology definition
The comparison of GE levels between species relied on orthology relationships provided by Ensembl. The analysis was restricted to 1:1 orthology relationships between any given pair of species. The orthology relationships between introns for cross-species comparative IR analyses were obtained by converting the genomic coordinates between genomes using the liftOver tool from Galaxy (Giardine et al. 2005; Blankenberg et al. 2010; Goecks et al. 2010) and selecting 1:1 overlaps.
Gene expression estimation
Gene expression levels were determined using the cRPKM metric, i.e., reads per thousand unique-mappable positions of target transcript sequence per million of mapped reads, as described in Labbe et al. (2012).
Percent intron retention (PIR) estimation
We considered every intron a potential retained intron. Each putative IR event was delineated by the adjacent 5′ and 3′ exons (E1 and E2, respectively) and the intron itself (I). For each event, we define two retention junctions, E1I (connecting exon E1 and the intron) and IE2 (connecting the intron and exon E2), and one constitutive (i.e., no retention) junction, E1E2 (connecting exons E1 and E2). For each species and each read length, k, we assembled all unique retention and constitutive junction sequences for subsequent poly(A)+ RNA-seq alignments. These junction sequences were constructed such that there is a minimum overlap of 8 nt between the reads and each of the exons or introns involved (i.e., not exceeding 2k − 16 in length) and such that only sequences from those two exons/intron are aligned (i.e., the length of the alignment is reduced if any of the exons or the intron are less than k − 8).
For each junction, we then determined the effective number of uniquely mappable positions. We extracted the L − k + 1 (L being the junction length) k-mers from each junction sequence and then aligned the full set of k-mers against the respective genome plus all exon–exon junctions, using Bowtie (Langmead et al. 2009) and allowing for a maximum of two mismatches. k-mers with a single alignment (thus potential junction reads) were then aligned back to the full, nonredundant set of junction sequences. The number of such k-mers with one unique alignment mapping to a junction was counted. This corresponds to the junction’s effective number of uniquely mappable positions for k-mer poly(A)+ RNA-seq reads.
For each sample, the corresponding poly(A)+ RNA-seq data were aligned against the respective genome plus all exon–exon junctions, using Bowtie and allowing for a maximum of two mismatches. Reads with a single mapping were then aligned to the full nonredundant set of junction sequences and, for each junction, the number of reads with one unique alignment mapping to it were counted. For each junction, the read count was normalized for mappability by multiplying it by the ratio between the maximum number of mappable positions (i.e., k − 15) and its effective number of uniquely mappable positions (as defined above).
To control for the possibility of reads mapping to exon–intron junctions reflecting alternative 5′ and/or 3′ splice sites and not bona fide IR events, we also aligned the poly(A)+ RNA-seq data against intron body sequences. To optimize processing time and disk space usage, for each intron we selected its middle 200 nt (or the full intron, if shorter) as the sample window. Selecting midpoint intronic sequences for quantification purposes further has the advantage of avoiding situations where unannotated alternative 5′ and 3′ splice sites may affect the accuracy of intron level measurements.
For each intronic window, we determined the effective number of uniquely mappable positions. We extracted the L − k + 1 (L being the intronic window length) k-mers from each intronic window and then aligned the full set of k-mers against the respective genome, using Bowtie and allowing for a maximum of two mismatches. k-mers with a single alignment (thus potential intronic reads) were then aligned back to the full, nonredundant set of intron sample sequences. The number of such k-mers with one unique alignment mapping to an intron was counted. This corresponds to the intron sample sequence’s effective number of uniquely mappable positions for k-mer poly(A)+ RNA-seq reads.
For each sample, the corresponding poly(A)+ RNA-seq data were aligned against the respective genome, using Bowtie and allowing for a maximum of two mismatches. Reads with a single mapping were then aligned to the full nonredundant set of intron sample sequences and, for each sequence, the number of reads with one unique alignment mapping to it were counted. For each intronic sequence, the read count was normalized for mappability and made directly comparable with the corresponding junction read counts (see coverage and balance motivations below) by multiplying it by the ratio between the junctions’ maximum number of mappable positions (i.e., k−15) and the intronic sequence’s effective number of uniquely mappable positions (as defined above).
The PIR value for each intron was defined as follows: PIR = 100 × average(#E1I, #IE2)/(#E1E2 + average(#E1I, #IE2)), where #E1I, #IE2 and #E1E2 are the normalized read counts for the associated junctions.
PIR values were then filtered according to the following empirical criteria:
PIRmin* ≤ 95;
median(#EI1, #IE2, #I) + #E1E2 > 10;
- P-value(binomial{M = min(#E1I, #IE2, #I),
- N = min(#E1I, #IE2, #I) + max(#E1I, #IE2, #I),
- P = 1/3.5,
- alternative = lower
- }) ≥ 0.05;
The first and last bp of the intron have no overlap with annotated exons either in the same or a different gene annotated in UCSC knownGene.
#I is the normalized read count for the associated intron sample sequence. PIRmin* is the minimum PIR value across all the samples in which the intron fulfills the second and third criteria. Binomial is an exact binomial test. In this case, M is the number of successes, N is the number of trials, P is the hypothesized probability of success, and alternative indicates the alternative hypothesis. In our case, the alternative is a proportion of successes lower than the null probability. We actually keep the events for which there is not enough evidence that the ratio between M and N is lower than P.
The goal of the first criterion was to exclude the possibility of measuring constitutive retention of a false intron due to misannotation issues. The goal of the second criterion (coverage) was to ensure sufficient read evidence for IR detection and enough coverage for sufficient precision and resolution in the estimation of PIR levels. The goal of the third criterion (balance) was to exclude events in which there is a high imbalance in read counts among the two exon–intron junctions and the intron body sequence. Such imbalances can arise from neighboring alternative 5′ and/or 3′ splice sites or overlapping genes, confound PIR estimates, and lead to the false detection of IR. The goal of the last criterion was to avoid falsely interpreting exonic reads from an alternative transcript isoform or another gene as junction reads supporting IR. The resulting PIR calls were robust with respect to sequencing depth, which was tested by randomly sampling between 1.25% and 80% of the reads in the original sample and recalculating PIR as described above (data not shown).
Mouse and human introns considered analyzed in this study are provided in Supplemental Tables S6, S7. PIR values for each intron in each tissue/cell type/treatment are provided in Supplemental Tables S8–S13. The thresholds assigned for PIR differed between analyses, as indicated in the main text.
Logistic model of IR in pooled human neural samples (“IR code”)
Aligned reads from six human neural samples (see Supplemental Table S14) were pooled, and PIR was calculated for the pool as described above. Then, of the 123.042 introns with PIR values that passed the quality control criteria (see above), we labeled all introns with PIRs ≤ 2 “constitutive” (71,188 introns) and all introns with PIRs ≥ 10 “retained” (17,185 introns). We learned a logistic regression model using a total of 136 features that describe length, sequence composition and dinucleotide frequency of introns and flanking exons, gene architecture, and splice site strength (Supplemental Table S2). Learning was done using fivefold cross-validation. The area under the ROC curve of the predictions of unseen data was AUC = 0.79 (P < 1 × 10−300, n = 88,373), and the KS statistic was 0.433 (P < 1 × 10−300). Moreover, when only predicting the most reliable introns (sorted based on PIR; top half of retained and bottom half of constitutive introns), the AUC increased to 0.85 and the KS statistic to 0.483. For Figure 1D, we elected to display only features that are significant in a reduced model with 19 lowly correlated features but which retains an AUC of 0.76 (Supplemental Table S2).
Estimation of the total number of retained introns
To estimate the total number of retained introns at a given PIR threshold, t, the order of all n samples was randomized, and the total number of introns with PIR ≥ t in any of the first 1, 2,..., n samples was determined. This procedure was iterated 1000 times, and the mean of each number of samples was calculated. A function with two exponential terms, f(x) = p1 (1 − e−p2 x) + p3 (1 − e−p4 x), was fitted to these means. Fitting using two terms afforded a substantial improvement over fitting using a single exponential term. The limit of the fitted function, given by p1 + p3, was used as the estimation of the total fraction of introns that are potentially retained at a given PIR threshold in one or more cell and tissue type. Estimation of the total fraction of genes with retained introns was performed in a similar manner, in this case by determining the total fraction of genes in which any intron is retained at a given PIR threshold.
Location of IR and intron PTC status
The location of introns within the 5′-UTR, CDS, 3′-UTR, or within a noncoding transcript, was inferred from the CDS coordinate information in annotation tables downloaded from the UCSC Genome Browser. Introns located precisely between two regions were disregarded. We used custom Perl scripts to annotate whether or not CDS introns introduce a PTC upon retention, essentially as described for cassette alternative exons in Saltzman et al. (2008). For the analysis of 5′–3′ bias, we converted the relative intron ranks (intron rank within the transcript divided by number of introns in the transcript) to 10 equal-sized bins.
Conservation of IR in vertebrates
The fraction of retained introns in species S (PIR ≥ 10) that are also retained in another species P, was determined considering all introns where a PIR could be calculated in both species. For each of the major organs, IR was compared in all pairwise combinations of species (S, P). Note that the fraction in S also retained in P need not be the same as the fraction in P also retained in S. In Figure 3A, these fractions were plotted against the evolutionary distance.
Significance of PIR differences between each organ and the average of all other organs was determined as follows: For each organ, a table was created in which the rows corresponded to all pairwise species comparisons, and columns contained the fraction of retained introns in the given organ and the average in all other organs; paired two-sided Wilcoxon signed-rank tests were performed on these tables, and derived P-values were Bonferroni-corrected.
Clustering of organ samples from different species based on PIR values was performed using Euclidean distance (complete linkage). Introns that have a minimum differential PIR of 10 in at least three species were clustered (n = 4835). For plotting purposes only, missing values were imputed using an R implementation of the DINEOF procedure (Beckers and Rixen 2003) (http://menugget.blogspot.ca/2012/10/dineof-data-interpolating-empirical.html). Rows (introns) were then clustered by column Euclidean distance using Ward’s linkage, and both rows and columns were reordered according to mean PIR within the constraints of the dendrograms.
Principal component analysis (PCA) was carried out with the “princomp” function in R (R Core Team 2014), following standardization without scaling (“stdize” function, CRAN package “pls”), using the same introns as for clustering, replacing missing values with imputed ones.
Assignment of retained intron types
Three types of introns were defined based on their evolutionary origin. Type A are ancestral introns flanked by constitutive exons; Type B arose by “intronization” of ancestral exonic sequence (Irimia et al. 2008); and Type C are located adjacent to one or more alternative exons that may or may not be conserved between species.
Retained introns were classified as Type A if the intron in question was conserved between the human and mouse genomes (but not necessarily retained in both species), with 9-bp precision after coordinate conversion using the UCSC liftOver tool (parameter -inMatch=0.10). Classification as Type A also required that there were no overlapping RefSeq-annotated exons from the same gene overlapping the intron and that no introns overlapped the exons.
To identify Type B retained introns in human and mouse, we mapped the genomic coordinates for each human and mouse intron (“probed introns”) to the following species’ genome assemblies using the liftOver tool (parameters: -minMatch=0.10 -multiple -minChainT=200 -minChainQ=200): Anolis carolinensis (anoCar2), Bos taurus (bosTau6), Canis familiaris (canFam3, for human, and canFam2, for mouse), Gallus gallus (galGal3), Loxodonta africana (loxAfr3), Monodelphis domestica (monDom5), Ornithorhynchus anatinus (ornAna1), Rattus norvegicus (rn5), Sus scrofa (susScr2), plus human (hg19, for mouse), and mouse (mm9, for human). Next, we downloaded Gene Transfer Format annotations for these species from Ensembl (ftp://ftp.ensembl.org), and intersected the lifted-over coordinates of each probed intron with the corresponding annotated exonic and intronic coordinates using BEDTools “intersect” (Quinlan and Hall 2010). We then asked which probed introns fully overlapped with annotated exons but not with introns in at least two other species. For this, we required that (1) the length of the intron-exon intersect was equal to the length of the lifted-over intron, and (2) that the length of the lifted-over intron was at least half of the probed intron. Finally, in the case of probed introns with partial intersections with both annotated exons and introns in other species, we discarded those for which there were more species with longer overlap to introns compared to exons than vice versa. Using this approach, we defined 327 and 141 Type B introns in human and mouse, respectively.
Type C introns in human and mouse were defined as follows: At least one of the flanking exons was required to overlap entirely with both a RefSeq-annotated intron in the same orientation in the same species and similarly to an intron lifted-over from the other species (mouse/human) with 9-bp precision.
Type annotations for each intron are provided in Supplemental Tables S6 and S7.
Analysis of splice-site strength
MaxEntScan (Yeo and Burge 2004) was used to calculate maximum entropy scores for 9-bp 5′ splice sites and 20-bp 3′ splice sites.
Correlation between IR and gene expression
Spearman rank correlations of PIR and expression (cRPKM) of the corresponding gene (see above) were calculated for all introns in which there were at least two samples in which both PIR and expression could be calculated, and where no more than one sample had a PIR = 0. The latter was required in order to exclude introns in which most of the observed variance in expression was due to other mechanisms of gene regulation.
We also investigated the association between PIR and gene expression using groups of genes with increasing expression levels. For each sample in human (52 in total) and in mouse (65 in total), we divided genes into deciles according to their cRPKM value in that specific sample and calculated the fraction of retained introns (defined as PIR ≥ 10, PIR ≥ 20, or PIR ≥ 50) in each decile. Lower deciles (1–4) were merged to increase the number of data points due to their inherently lower read coverage. Genes with cRPKM < 2 across all samples were discarded.
Analysis of IR during in vitro neuronal differentiation
PIR and gene expression cRPKM were calculated as described above. For the introns with enough read coverage and balance for the six analyzed time points, we asked which had an absolute PIR difference of more than 15 between ES cells (−8 d) and fully mature neurons (28 d). We identified 825 introns with a dPIR > 15 and 105 with a dPIR < −15. A heatmap of Z-scores of PIRs for these introns was plotted using the heatmap.2 function in R. GO (Ashburner et al. 2000) and KEGG Pathway (Kanehisa et al. 2004) enrichment analysis for the genes containing the two groups of introns was performed in the Database for Annotation, Visualization and Integrated Discovery (DAVID) (Huang da et al. 2009) (using raw P-value < 0.005 for at least one term within the cluster as cutoff).
Analysis of functional annotations associated with IR in ES, muscular, neural, and fibroblast cells
Median PIR in neural, muscle, and ES cells, and in complementary sets of “other” cell/tissue types, was calculated for selections of samples in which assignment to “biological type” was unambiguous (i.e., samples such as precursor cells and cell lines were excluded) (see Supplemental Table S14). Only events in which PIR values had been determined in at least two samples of the respective tissue group and in at least five “other” samples were considered. Analysis of differential gene expression relied on the B-statistic, i.e., the empirical Bayes log-odds of differential expression (Smyth 2004). Genes were considered to be differentially expressed between the sample group of interest (neural/muscle/ES cells) and the “other” cell/tissue types if B > log2(19), corresponding to 95% odds of differential expression.
Broad functional categories of genes were constructed as follows: The full table of GO categories, downloaded on September 15, 2013, was searched with the following search terms:
Neural categories: “brain” OR “retina” OR “synapse” AND NOT “immuno”
Muscle categories: “muscle” OR “myo” OR “sarco” OR “contractile”
Stem cell categories: “stem cell”
Fibroblast-related categories: “fibroblast”
Genes were then labeled with the broad categories if they were annotated with any of the found GO categories. Lists of all considered categories and categories found associated with genes in human and mouse are provided in Supplemental Table S3.
Selection of candidate IR events for RT-PCR validation
Mouse IR events that, by RNA-seq analysis, displayed differential retention in mouse and human neural, muscle, or ES cells/tissues, or had a flat retention profile in both species, were selected for validation by RT-PCR. Events selected for analysis also had a PIR of 0 or 100 in less than half of the samples assayed. Finally, the selected events were also those predicted to be expressed in at least 10 analyzed RNA samples, had intron and flanking exon lengths amenable to RT-PCR, and that did not overlap other genes.
Analysis of ChIP-seq data
Reads were converted to FASTQ format, mapped to the human (hg19) or mouse (mm9) genomes using Bowtie with settings–best -n 2 -k 1, and duplicate reads were removed. Reads falling into an arbitrary genomic region of 2 Mb were used to estimate ChIP fragment length by determining the distance at which the cross-correlation of the numbers of reads mapping to each bp on the + and − strands was maximal. All reads were then extended to that fragment length or 120 bp, whichever was higher. Pileups—the number of fragments overlapping each genomic bp—were calculated, and were normalized by million mappable reads in the ChIP-seq library. Normalized pileups from replicate experiments were then averaged, and matched input samples were subtracted, creating input-subtracted FPM (fragments per million reads). Input subtraction was deemed necessary because without it, ChIP-seq profiles routinely displayed enrichment or depletion on average across introns, exons, or transcription start sites. For alignment plots, sets of introns with their flanking exons were defined as outlined in the figures, and input-subtracted (unless indicated otherwise), normalized FPM were averaged per bp such that the grouped introns were superimposed by their splice donor or acceptor sites (left and right half of plots, respectively). Values for each aligned intronic (white plot area) or exonic (gray plot area) bp were averaged at any one position. These within-group averages were plotted directly without smoothing.
Statistical tests
All statistical analyses were performed in R (R Core Team 2014), a free software environment for statistical computing and graphics, making use of packages from the Comprehensive R Archive Network (CRAN) and Bioconductor (http://www.bioconductor.org/, tools for the analysis of high-throughput genomic data). Where applicable and not indicated otherwise, P-values were corrected for multiple testing with the Bonferroni method.
To test the significance of Pol II-Ser2p ChIP-seq signal differences between introns with unchanged or increased PIR after knockdown of SNRPB in HeLa cells, groups of introns were first matched for comparable PIR in control-treated cells, giving rise to 1442 pairs of introns. Median ChIP signals were then calculated from normalized, control-subtracted ChIP signals (see above) in both introns of each pair. To avoid confounding possible factors due to different ChIP signals near the ends versus in the interior of introns, the whole intron was considered for the shorter intron, but for the longer intron only equal-sized regions from the 5′ and 3′ end that together were of the same length as the shorter intron were considered. The resulting median value pairs were subjected to a paired, one-sided Mann-Whitney U test.
Cell culture
For in vitro differentiation of ES cells into neurons, CGR8 mouse ES cells were maintained at subconfluent conditions on gelatin-coated plates and were differentiated into neurons as previously described (Hubbard et al. 2013). RNA was extracted at different time points during the differentiation protocol using the RNeasy Mini Kit (Qiagen) as recommended by the manufacturer.
For treatment with DRB, CGR8 mouse ES cells were seeded at 50% confluence and treated for 24 h with 10 or 25 μg/mL DRB or DMSO as a control.
RT-PCR validation
RT-PCR assays were performed essentially as previously described (Calarco et al. 2007). In each reaction, 3–10 ng total RNA (40 ng for validation of events during neuronal differentiation) or 0.3–1 ng poly(A)+ RNA was used as input, and cDNA synthesis and amplification were performed using the OneStep RT-PCR kit (Qiagen) following the manufacturer’s recommendations. For assays in DRB-treated CGR8 cells, total RNA was purified and DNase treated using Qiagen’s RNeasy kit, and 40 ng were used for cDNA synthesis and amplified by PCR. For short introns (<400 bp), the assay used two primers, one in each flanking exon. For long introns (>2 kb), the assay was designed with one reverse primer in the flanking downstream exon and two mutually exclusive forward primers: one in the flanking upstream exon, which could only amplify a spliced product in the given amplification time, and one in the intron itself, which could only amplify a retained product. The number of amplification cycles varied from 29 to 37, depending on the transcript and the sample analyzed; 24 cycles were used for Gapdh. Reaction products were resolved using 1.5%–3% TAE-agarose gels stained with ethidium bromide and imaged using the Gel Doc XR System (Bio-Rad). ImageJ software was used to measure the intensity of bands representing the retained and spliced isoforms, and these values were normalized by product size. PIR levels of retained introns were then calculated as the amount of retained isoform divided by the sum of spliced and retained isoforms.
Supplementary Material
Acknowledgments
We thank Andrew Delong and Eduardo Eyras for helpful discussions and insights, and members of the Blencowe laboratory for valuable comments on the manuscript. This work was supported by grants from the Canadian Institutes of Health Research and Canadian Cancer Society (B.J.B.); EMBO long-term fellowships (U.B. and T.G.-P.); Human Frontier Science Program Organization long-term fellowships (U.B. and M.I.); an OSCI fellowship (T.G.-P.); CIHR postdoctoral and Marie Curie IOF fellowships (N.L.B.-M.); and an NSERC studentship (E.N.).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.177790.114.
References
- Alexander RD, Innocente SA, Barrass JD, Beggs JD. 2010. Splicing-dependent RNA polymerase pausing in yeast. Mol Cell 40: 582–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amit M, Donyo M, Hollander D, Goren A, Kim E, Gelfman S, Lev-Maor G, Burstein D, Schwartz S, Postolsky B, et al. 2012. Differential GC content between exons and introns establishes distinct strategies of splice-site recognition. Cell Rep 1: 543–556 [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter C, Watt S, Colak R, et al. 2012. The evolutionary landscape of alternative splicing in vertebrate species. Science 338: 1587–1593 [DOI] [PubMed] [Google Scholar]
- Beckers JM, Rixen M. 2003. EOF calculations and data filling from incomplete oceanographic datasets. J Atmos Ocean Technol 20: 1839–1856 [Google Scholar]
- Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J. 2010. Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol 89: 19.10.1–19.10.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunschweig U, Gueroussov S, Plocik AM, Graveley BR, Blencowe BJ. 2013. Dynamic integration of splicing within gene regulatory pathways. Cell 152: 1252–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawand D, Soumillon M, Necsulea A, Julien P, Csárdi G, Harrigan P, Weier M, Liechti A, Aximu-Petri A, Kircher M, et al. 2011. The evolution of gene expression levels in mammalian organs. Nature 478: 343–348 [DOI] [PubMed] [Google Scholar]
- Calarco JA, Xing Y, Cáceres M, Calarco JP, Xiao X, Pan Q, Lee C, Preuss TM, Blencowe BJ. 2007. Global analysis of alternative splicing differences between humans and chimpanzees. Genes Dev 21: 2963–2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chathoth KT, Barrass JD, Webb S, Beggs JD. 2014. A splicing-dependent transcriptional checkpoint associated with prespliceosome formation. Mol Cell 53: 779–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David CJ, Manley JL. 2011. The RNA polymerase C-terminal domain: a new role in spliceosome assembly. Transcription 2: 221–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreszer TR, Karolchik D, Zweig AS, Hinrichs AS, Raney BJ, Kuhn RM, Meyer LR, Wong M, Sloan CA, Rosenbloom KR, et al. 2012. The UCSC Genome Browser database: extensions and updates 2011. Nucleic Acids Res 40: D918–D923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium . 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, et al. 2013. Ensembl 2013. Nucleic Acids Res 41: D48–D55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong YW, Zhou Q. 2001. Stimulatory effect of splicing factors on transcriptional elongation. Nature 414: 929–933 [DOI] [PubMed] [Google Scholar]
- Galante PA, Sakabe NJ, Kirschbaum-Slager N, de Souza SJ. 2004. Detection and evaluation of intron retention events in the human transcriptome. RNA 10: 757–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia H, Fleyshman D, Kolesnikova K, Safina A, Commane M, Paszkiewicz G, Omelian A, Morrison C, Gurova K. 2011. Expression of FACT in mammalian tissues suggests its role in maintaining of undifferentiated state of cells. Oncotarget 2: 783–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y, Porse BT. 2013. The functional consequences of intron retention: alternative splicing coupled to NMD as a regulator of gene expression. Bioessays 36: 236–243 [DOI] [PubMed] [Google Scholar]
- Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, et al. 2005. Galaxy: a platform for interactive large-scale genome analysis. Genome Res 15: 1451–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J. 2010. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11: R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Hubbard KS, Gut IM, Lyman ME, McNutt PM. 2013. Longitudinal RNA sequencing of the deep transcriptome during neurogenesis of cortical glutamatergic neurons from murine ESCs. F1000 Res 2: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip JY, Schmidt D, Pan Q, Ramani AK, Fraser AG, Odom DT, Blencowe BJ. 2011. Global impact of RNA polymerase II elongation inhibition on alternative splicing regulation. Genome Res 21: 390–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M, Blencowe BJ. 2012. Alternative splicing: decoding an expansive regulatory layer. Curr Opin Cell Biol 24: 323–332 [DOI] [PubMed] [Google Scholar]
- Irimia M, Rukov JL, Penny D, Vinther J, Garcia-Fernandez J, Roy SW. 2008. Origin of introns by ‘intronization’ of exonic sequences. Trends Genet 24: 378–381 [DOI] [PubMed] [Google Scholar]
- Kalsotra A, Cooper TA. 2011. Functional consequences of developmentally regulated alternative splicing. Nat Rev Genet 12: 715–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyna M, Lopato S, Voronin V, Barta A. 2006. Evolutionary conservation and regulation of particular alternative splicing events in plant SR proteins. Nucleic Acids Res 34: 4395–4405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyna M, Simpson CG, Syed NH, Lewandowska D, Marquez Y, Kusenda B, Marshall J, Fuller J, Cardle L, McNicol J, et al. 2012. Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res 40: 2454–2469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. 2004. The KEGG resource for deciphering the genome. Nucleic Acids Res 32: D277–D280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt AR, Schor IE, Alló M, Dujardin G, Petrillo E, Muñoz MJ. 2013. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat Rev Mol Cell Biol 14: 153–165 [DOI] [PubMed] [Google Scholar]
- Labbe RM, Irimia M, Currie KW, Lin A, Zhu SJ, Brown DD, Ross EJ, Voisin V, Bader GD, Blencowe BJ, et al. 2012. A comparative transcriptomic analysis reveals conserved features of stem cell pluripotency in planarians and mammals. Stem Cells 30: 1734–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. 2007. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature 446: 926–929 [DOI] [PubMed] [Google Scholar]
- Li Y, Bor YC, Misawa Y, Xue Y, Rekosh D, Hammarskjöld ML. 2006. An intron with a constitutive transport element is retained in a Tap messenger RNA. Nature 443: 234–237 [DOI] [PubMed] [Google Scholar]
- Marquez Y, Brown JW, Simpson C, Barta A, Kalyna M. 2012. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res 22: 1184–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall NF, Peng J, Xie Z, Price DH. 1996. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem 271: 27176–27183 [DOI] [PubMed] [Google Scholar]
- McIlwain DR, Pan Q, Reilly PT, Elia AJ, McCracken S, Wakeham AC, Itie-Youten A, Blencowe BJ, Mak TW. 2010. Smg1 is required for embryogenesis and regulates diverse genes via alternative splicing coupled to nonsense-mediated mRNA decay. Proc Natl Acad Sci 107: 12186–12191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkin J, Russell C, Chen P, Burge CB. 2012. Evolutionary dynamics of gene and isoform regulation in mammalian tissues. Science 338: 1593–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki R, Kadota K, Bono H, Mizuno Y, Tomaru Y, Carninci P, Itoh M, Shibata K, Kawai J, Konno H, et al. 2001. Delineating developmental and metabolic pathways in vivo by expression profiling using the RIKEN set of 18,816 full-length enriched mouse cDNA arrays. Proc Natl Acad Sci 98: 2199–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ner-Gaon H, Halachmi R, Savaldi-Goldstein S, Rubin E, Ophir R, Fluhr R. 2004. Intron retention is a major phenomenon in alternative splicing in Arabidopsis. Plant J 39(6):877–885 [DOI] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR. 2010. Expansion of the eukaryotic proteome by alternative splicing. Nature 463: 457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40: 1413–1415 [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . 2014. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/
- Sakabe NJ, de Souza SJ. 2007. Sequence features responsible for intron retention in human. BMC Genomics 8: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltzman AL, Kim YK, Pan Q, Fagnani MM, Maquat LE, Blencowe BJ. 2008. Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Mol Cell Biol 28: 4320–4330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltzman AL, Pan Q, Blencowe BJ. 2011. Regulation of alternative splicing by the core spliceosomal machinery. Genes Dev 25: 373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Meshorer E, Ast G. 2009. Chromatin organization marks exon-intron structure. Nat Struct Mol Biol 16: 990–995 [DOI] [PubMed] [Google Scholar]
- Sebé-Pedrós A, Irimia M, Del Campo J, Parra-Acero H, Russ C, Nusbaum C, Blencowe BJ, Ruiz-Trillo I. 2013. Regulated aggregative multicellularity in a close unicellular relative of metazoa. eLife 2: e01287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3. [DOI] [PubMed] [Google Scholar]
- Spies N, Nielsen CB, Padgett RA, Burge CB. 2009. Biased chromatin signatures around polyadenylation sites and exons. Mol Cell 36: 245–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilgner H, Nikolaou C, Althammer S, Sammeth M, Beato M, Valcárcel J, Guigó R. 2009. Nucleosome positioning as a determinant of exon recognition. Nat Struct Mol Biol 16: 996–1001 [DOI] [PubMed] [Google Scholar]
- Tilgner H, Knowles DG, Johnson R, Davis CA, Chakrabortty S, Djebali S, Curado J, Snyder M, Gingeras TR, Guigo R. 2012. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome Res 22: 1616–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veloso A, Kirkconnell KS, Magnuson B, Biewen B, Paulsen MT, Wilson TE, Ljungman M. 2014. Rate of elongation by RNA polymerase II is associated with specific gene features and epigenetic modifications. Genome Res 24: 896–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JJ, Ritchie W, Ebner OA, Selbach M, Wong JW, Huang Y, Gao D, Pinello N, Gonzalez M, Baidya K, et al. 2013. Orchestrated intron retention regulates normal granulocyte differentiation. Cell 154: 583–595 [DOI] [PubMed] [Google Scholar]
- Yap K, Lim ZQ, Khandelia P, Friedman B, Makeyev EV. 2012. Coordinated regulation of neuronal mRNA steady-state levels through developmentally controlled intron retention. Genes Dev 26: 1209–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G, Burge CB. 2004. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol 11: 377–394 [DOI] [PubMed] [Google Scholar]
- Zhang W, Morris QD, Chang R, Shai O, Bakowski MA, Mitsakakis N, Mohammad N, Robinson MD, Zirngibl R, Somogyi E, et al. 2004. The functional landscape of mouse gene expression. J Biol 3: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.