Abstract
African women frequently acquire several genetically distinct human immunodeficiency virus type 1 (HIV-1) variants from a heterosexual partner, whereas the acquisition of multiple variants appears to be rare in men. To determine whether newly infected individuals in other risk groups acquire genetically diverse viruses, we examined the viral envelope sequences in plasma samples from 13 women and 4 men from the United States infected with subtype B viruses and 10 men from Kenya infected with non-subtype B viruses. HIV-1 envelope sequences differed by more than 2% in three U.S. women, one U.S. man, and one Kenyan man near the time of seroconversion. These findings suggest that early HIV-1 genetic diversity is not exclusive to women from Africa or to infection with any particular HIV-1 subtype.
Viral genetic variation, especially in the gene encoding the antigenic envelope protein, is one of the hallmarks of chronic human immunodeficiency virus type 1 (HIV-1) infection (18). Viruses in a long-term-infected individual are characterized by the presence of a complex swarm of quasispecies that may differ by up to 10% in the envelope gene (5). In contrast, relatively homogeneous envelope sequences with less than 1% diversity have been observed in many individuals, especially men, during primary HIV-1 infection. However, only a small number of men from different risk groups and different regions have been examined (1, 3, 32, 34, 35). Limited viral diversity was documented in some newly infected subjects even in cases in which the index partner was known to harbor a swarm of variants, suggesting that there was a selective barrier during transmission (32, 34, 35). This selective bottleneck is not as restrictive in women from Africa, of whom approximately 60% were observed to have multiple HIV-1 variants that in some cases differed by more than 5% in the envelope gene (8, 10, 21, 23). It is unknown whether there are other risk groups for which this selective bottleneck is also less restrictive.
Detection of viral diversity prior to seroconversion, along with other lines of evidence, suggested that the presence of multiple HIV-1 variants at the time of infection was not due to rapid evolution of the virus within the newly infected host (10, 21, 24). Phylogenetic analysis strongly suggested that multiple viruses were transmitted to women from one sexual partner (10, 21). Surprisingly, there was no viral diversity detected in any of the 10 men from Africa examined in our earlier studies, leading us to conclude that the transmission of multiple viral variants is much more common in women than in men (10). However, these studies focused on subjects at high risk of acquiring HIV-1 through heterosexual contact and thus may not be relevant to all groups.
To determine whether infection by multiple variants occurs in other risk groups, plasma samples were obtained near the time of infection from 11 women and 4 men who were in the U.S. HIVNET Vaccine Preparedness cohort and from 2 women who were monitored at a U.S. hospital clinic. The individuals selected for the study from the U.S. HIVNET cohort were those who had samples available within 6 months after documented seroconversion and within 1 year of an HIV-1-negative serological test. For the 13 women in the study, the median interval between the last seronegative date and the day of collection of the sample analyzed was 118 days (range, 6 to 247 days), while for the 4 men the median interval was 165 days (range, 142 to 250 days) (Table 1). The reported risk factors for HIV-1 acquisition were exclusively heterosexual contact for six women, both injection drug use (IDU) and heterosexual contact for four women, exclusively IDU for three women and two men, and homosexual contact for two men (Table 1).
TABLE 1.
Genetic diversity in subjects' samples early in infection
| Subject | PNSa | Risk factor(s)b | No. of samples sequenced | Nucleotide distancec | Amino acid difference (%)d | Insertions/deletionse |
|---|---|---|---|---|---|---|
| U.S. women | ||||||
| F14 | 91 | IDU, HSC | 7 | 0.86-3.25 | 2.00-7.49 | 18 bp in V4 |
| F2 | 148 | IDU | 4 | 0.77-2.71 | 2.02-5.02 | 30 bp in V1 |
| F15 | 49 | IDU | 6 | 0.67-2.34 | 1.15-4.60 | 3 bp in V2 and V5 |
| F7 | 247 | HSC | 4 | 0.59-1.68 | 1.17-3.81 | 39 bp in C3 |
| F3 | 191 | HSC | 4 | 0.54-1.54 | 1.18-3.81 | 39 bp in V1 |
| F9 | 234 | IDU | 5 | 0.62-1.08 | 1.17-2.62 | 66 bp in C2 |
| F5 | 114 | IDU, HSC | 4 | 0.57-1.03 | 0.86-2.22 | 27 bp in V1 |
| F10 | 118 | HSC | 4 | 0.56-1.09 | 0.83-2.71 | 24 bp in V1 |
| F16 | 99 | IDU, HSC | 5 | 0.09-0.54 | 0.00-1.08 | 3 bp in V2 |
| F4 | 126 | IDU, HSC | 2 | 0.49 | 1.16 | 16 bp in C2 |
| F11 | 165 | HSC | NAf | NA | NA | NA |
| MEM | 6 | HSC | NA | NA | NA | NA |
| K-C | 10 | HSC | NA | NA | NA | NA |
| U.S. men | ||||||
| M3 | 249 | IDU | 4 | 2.33-4.32 | 4.50-7.60 | 21 bp in V1 |
| M1 | 142 | MSM | 7 | 0.38-1.52 | 0.56-3.67 | 21 bp in V1 |
| M4 | 250 | IDU | 3 | 0.41-0.92 | 0.61-2.45 | 50 bp in C4 |
| M2 | 188 | MSM | NA | NA | NA | NA |
| Kenyan men | ||||||
| T135 | 230 | HSC | 5 | 0.51-3.20 | 1.51-6.67 | 15 bp in V2 |
| T263 | 283 | HSC | 2 | 0.97 | 2.31 | 15 bp in V4 |
| T199 | 322 | HSC | 3 | 0.38-0.88 | 0.86-2.35 | 15 bp in V4 |
| T158 | 139 | HSC | NA | NA | NA | NA |
| T387 | 280 | HSC | NA | NA | NA | NA |
| T457 | 28 | HSC | NA | NA | NA | NA |
| T518 | 280 | HSC | NA | NA | NA | NA |
| T594 | 133 | HSC | NA | NA | NA | NA |
| T645 | 273 | HSC | NA | NA | NA | NA |
| T650 | 161 | HSC | NA | NA | NA | NA |
PNS (post-negative serology), number of days between the last negative HIV-1 serology test and the time of sample collection.
Suspected route of HIV-1 acquisition. IDU, injection drug use; HSC, heterosexual contact; MSM, men having sex with men.
Range of Kimura two-parameter pairwise substitution rates per 100 bases, ignoring insertions and deletions
Range of pairwise percentage differences, ignoring insertions and deletions.
Largest insertion or deletion between sequence pairs and the envelope region.
NA, not applicable because no heteroduplexes were detected by HMA and therefore sequence analysis was not performed.
The methods used for plasma sample viral RNA isolation and reverse transcription (RT)-PCR have been described previously (7). To ensure that similar numbers of variants were analyzed for all subjects, the cDNA copy number was obtained by using real-time PCR (C. M. Rousseau, R. W. Nouat, B. A. Richardson, G. C. John-Stewart, D. Mbori-Ngacha, J. K. Kreiss, and J. Overbaugh, submitted for publication). A minimum of 10 and a maximum of 50 cDNAs were used in two independent RT-PCR amplifications of the V1-through-V5 envelope region. In cases in which the cDNA copies could not be quantified, a RNA dilution 10-fold higher than the lowest dilution that yielded a RT-PCR product (for subjects F3, F7, F14, and M4) was used for amplification of the envelope region.
The heteroduplex mobility assay (HMA) was used as a rapid screen to assess diversity. Because variants that have at least 2% genetic difference and/or insertions and deletions (indels) generally appear as heteroduplexes in the HMA (6, 29), we classified subjects as being infected with viruses with homogeneous envelope sequences without indels if no heteroduplexes were observed in the HMA of the two PCRs alone and in combination, as discussed previously (23). Three women (subjects F11, MEM, and K-C) and one man (subject M2) were found to have no detectable heteroduplexes when their plasma samples were analyzed by HMA (data not shown).
The HMAs of the plasma samples from the remaining 10 women and 3 men showed distinct heteroduplexes. For these subjects, the independent RT-PCR product that showed the largest number of heteroduplexes was cloned into a Topo TA vector (Invitrogen, Carlsbad, Calif.). For each subject, 12 to 50 different clones were obtained. The inserts from the clones were amplified by using PCR conditions described previously (17, 21). An arbitrarily chosen reference envelope and any envelope insert that in combination with the reference sequence showed a unique heteroduplex mobility were sequenced. In addition, the products from all of the envelope inserts with sequence data were combined and evaluated by HMA to show that, in aggregate, the inserts with sequence data replicated the heteroduplex pattern of the initial plasma sample. For most subjects, the HMA pattern obtained by combining all of the variants with sequence data was similar to the HMA pattern of the original plasma sample, implying that most of the major envelope variants had been cloned and sequenced (Fig. 1A). In some cases (for subjects F3, F10, F15, M1, and M3), new heteroduplexes were detected when the combination of the products of the individual clones was compared to the HMA of the original plasma sample (Fig. 1B). This finding may represent variants that were present at a low frequency in the parent sample and thus could not be visualized in the original HMA pattern.
FIG. 1.
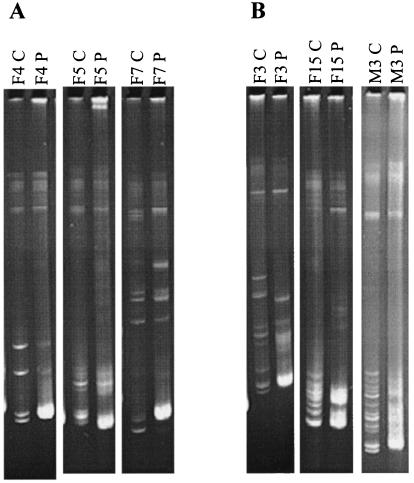
Examples of the HMA of the V1-through-V5 envelope region in cases in which there were detectable heteroduplexes. A subject's identification number is shown above each lane of the gel and is followed by a “P” or a “C.” P lanes show the combination of two PCR amplifications of 10 HIV-1 genomes from the original plasma sample. C lanes show the combination of the PCR products from envelope inserts. These inserts were those with sequence data from the cloned variants from the subject. (A) Examples of HMA in which the combination of all of the variants with sequence data matched the HMA pattern of the original plasma sample. (B) Examples of HMA in which new heteroduplexes were detected in the combination of the individual clones when this combination was compared to the original plasma sample.
Three of the 10 women and 1 of the 3 men were infected with viral variants that had a maximum nucleotide pairwise difference of more than 2% and indels in the envelope gene (Table 1). The variants from the remaining seven women and two men with heteroduplexes had indels but nucleotide variation of less than 2%. Indels, which presumably can result from a single error during RT, were also observed in recently infected African subjects, but they were usually also accompanied by numerous nucleotide differences (10, 21). Hypermutated sequences with a predominance of unidirectional G-to-A mutations were not observed in any of the cloned envelopes (3, 19, 30). The amino acid sequence differences were as high as 7.6% between sequences within a patient (Table 1), and the majority of the amino acid changes clustered in the defined envelope variable domains (data not shown) (27). These differences in the nucleotide and amino acid sequences are lower than the 15 and 10% nucleotide differences observed in the V1-V2 (20, 26, 31) and C2-V5 (5, 25) envelope regions, respectively, in some long-term-infected subjects. Thus, it is possible that in cases in which multiple viruses are acquired, specific HIV-1 variants are selected for transmission, similar to what has been proposed for subjects infected with a homogeneous virus population (32-35). However, it is also possible that the range of viruses infecting subjects with multiple HIV-1 variants may be similar to what is present in the index case if the source partner is within the first few years after infection. A detailed study of the transmission of viral variants in couples discordant for HIV-1 infection would help address these possibilities.
The median intervals from the last seronegative date to the day of collection of the sample assayed for diversity were not significantly different between those subjects infected with HIV-1 variants with more than 2% genetic diversity (4 subjects; median interval, 119.5 days) and the other individuals (13 subjects; median interval, 142 days) (P = 0.7; Mann-Whitney U test). Thus, there was no significant difference in the times the virus had to evolve within the host between the two groups of subjects.
It has been estimated that HIV-1 diversifies at a rate of approximately 1% per year in the C2-V5 envelope region (25), which makes it unlikely that the infecting virus would accumulate enough point mutations to be more than 2% different in the 119.5 days from the last seronegative date to the day when the sequences were analyzed. The interval between the day of infection and the collection day for the sample analyzed was less than 150 days in all cases except one (Table 1). In the one case in which we observed diversity at a relatively later time (subject M3, 249 days), there were HIV-1 variants that were up to 4.3% different at the nucleotide level. This difference is considerably greater than what we would predict based on the rates of diversification. However, we cannot say with certainty that this is true in all cases because diversification rates can vary greatly among subjects (4, 11, 14, 16) and in different envelope regions, especially the V1-V2 region (22; M. Sagar and J. Overbaugh, unpublished observations). The RT and nested PCR used to amplify the plasma sequences from subjects with homogeneous and heterogeneous viruses would be predicted to introduce two nucleotide changes in a 1,000-bp template, assuming an error rate of 10−3 for a reverse transcriptase and 10−5 for a DNA polymerase (12, 13). Although it is possible that the viral variants early in infection may arise due to de novo diversification or sampling methodology, studies showing the presence of the same variants before and after HIV-1 seroconversion strongly suggest that multiple variants are acquired at the time of infection and are not due to the rapid mutation of a single variant (10).
The U.S. men examined here acquired HIV-1 through either homosexual contact or IDU, whereas the 10 men from Kenya, described in our previous studies, had acquired HIV-1 through heterosexual contact (10). Recent studies suggest that some men who acquire HIV-1 through homosexual contact and/or IDU appear to have multiple HIV-1 variants at the time of infection, similar to subject M3 examined here (2, 9). To further test whether men exposed through heterosexual contact could also acquire genetically diverse variants, we examined viral diversity in an additional 10 men from Kenya early in infection. Sample collection and monitoring have been described previously (10, 15). The median interval between the last seronegative date and the day of collection of the sample analyzed was 251 days (range, 28 to 322 days) (Table 1). Seven of the 10 men had no heteroduplexes, as assessed by HMA, and thus were classified as being infected with viruses with a genetically homogeneous envelope sequence without indels. Heteroduplexes were observed in the HMAs of the plasma samples from three men, and thus envelope variants were cloned and sequenced by using methods described above. One of the three men was infected with viral variants that had up to 3.2% genetic diversity and also had indels in the envelope gene (Table 1). Two of the 10 men had viral variants with indels but with less than 1% genetic diversity. The results for these 2 men were similar to those for 3 of 12 women from Africa and 1 of 10 men from Africa examined in our previous studies, who were classified as having a homogeneous virus population because they had rare deletion variants with minimal genetic differences (10).
These results show that infection with multiple HIV-1 variants is not specific to women from Africa. In 3 of 13 women and 1 of 4 men from the United States, we observed distinct variants early in infection that differed by more than 2% in the envelope sequence. Phylogenetic analysis suggests that all of the different genetic variants isolated from one subject were acquired from a single source and were not due to infection from multiple donors or laboratory contamination (Fig. 2). In this study, all of the U.S. individuals were infected with HIV-1 subtype B, whereas in this and our previous studies the Kenyan subjects were infected with subtypes A, C, and D (10). Together, these findings suggest that the acquisition of multiple envelope genotypes does not occur only during the transmission of specific HIV-1 subtypes.
FIG. 2.
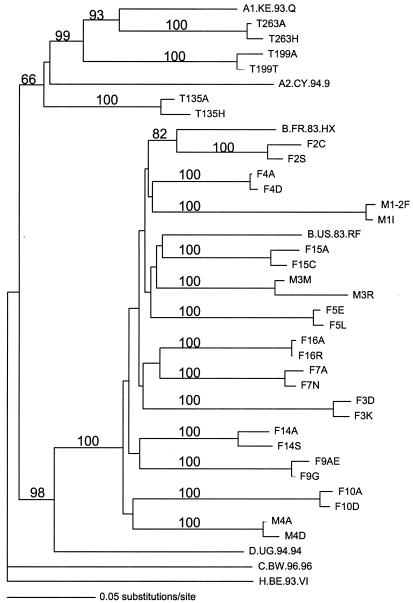
Phylogram of the two most divergent V1-through-V5 envelope gene sequences from each subject. The two most divergent sequences were selected based on pairwise differences. The sequence designation includes the subject identification number followed by a letter designating the clone. Sequences were aligned by using ClustalX with manual adjustment by use of MacClade (version 4.01). A phylogenetic tree was generated by using a distance-based method, neighbor joining, in the software package Phylogenetic Analysis Using Parsimony and Other Methods (PAUP 4.02b2a) (28). For these analyses, a two-parameter distance matrix, the Kimura two-parameter distance correction, was applied. H.BE.93.VI was selected as an outgroup to root the tree because it was likely to be the least related to the other sequences. Numbers at the nodes represent the bootstrap values for those nodes from 100 bootstrap resamplings. The results of this analysis were confirmed by using parsimony and maximum-likelihood methods.
Our previous studies, which showed that the likelihood of being infected by multiple HIV-1 variants was associated with the presence of exogenous factors present at the time of infection (24), suggest that individuals in different populations may differ in their risks of acquiring multiple HIV-1 variants. For high-risk women, these factors included the use of hormonal contraceptives (HCs) and the presence of genital tract infections (GTIs) (24). Thus, we speculate that the low percentage of women from the United States who are infected with multiple variants may reflect in part a lower prevalence of HC use or GTIs at the time of infection. Unfortunately, data on HC use and GTIs was not available at the time of HIV-1 acquisition for most of the study population. It is noteworthy that in each of the four cases in which we observed genetically heterogeneous viruses at primary infection in the U.S. subjects, IDU was reported as a risk factor. Thus, it is possible that IDU increases the likelihood that a person will acquire multiple viruses, although larger studies are needed to test this association.
Infection by genetically diverse viruses has been linked to a higher level of viral replication and a faster decline in CD4+-T-cell counts (23). Here, we showed that infection with multiple genotypic variants occurs in multiple risk groups, albeit at different frequencies. Studies of factors that promote the acquisition of multiple viruses may provide important information on modifiable factors present at the time of infection that could impact HIV-1 pathogenesis.
Nucleotide sequence accession numbers.
All sequences reported in this publication have been submitted to GenBank (accession numbers AY525444 to AY525503).
Acknowledgments
We thank Geoffrey Gottlieb and Stephanie Rainwater for help with the phylogenetic analysis and the HIVNET program for providing samples for study.
This study was supported by NIH grant AI38518 (J.O.), NIH grants AI43638 and RR00425 (E.S.D.), and a mentored clinical scientist development award (KO8) to M.S. (AI52759).
REFERENCES
- 1.Bernardin, F., B. L. Herring, L. Peddada, and E. L. Delwart. 2003. Primary infection of a male plasma donor with divergent HIV variants from the same source followed by rapid fluctuations in their relative frequency and viral recombination. AIDS Res. Hum. Retrovir. 19:1009-1015. [DOI] [PubMed] [Google Scholar]
- 2.Carneiro, M., X. F. Yu, C. Lyles, A. Templeton, A. E. Weisstein, M. Safaeian, H. Farzadegan, D. Vlahov, and R. B. Markham. 1999. The effect of drug-injection behavior on genetic evolution of HIV-1. J. Infect. Dis. 180:1025-1032. [DOI] [PubMed] [Google Scholar]
- 3.Delwart, E., M. Magierowska, M. Royz, B. Foley, L. Peddada, R. Smith, C. Heldebrant, A. Conrad, and M. Busch. 2002. Homogeneous quasispecies in 16 out of 17 individuals during very early HIV-1 primary infection. AIDS 16:189-195. [DOI] [PubMed] [Google Scholar]
- 4.Delwart, E. L., H. Pan, H. W. Sheppard, D. Wolpert, A. U. Neumann, B. Korber, and J. I. Mullins. 1997. Slower evolution of human immunodeficiency virus type 1 quasispecies during progression to AIDS. J. Virol. 71:7498-7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delwart, E. L., H. W. Sheppard, B. D. Walker, J. Goudsmit, and J. I. Mullins. 1994. Human immunodeficiency virus type 1 evolution in vivo tracked by DNA heteroduplex mobility assays. J. Virol. 68:6672-6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delwart, E. L., E. G. Shpaer, J. Louwagie, F. E. McCutchan, M. Grez, H. Rubsamen-Waigmann, and J. I. Mullins. 1993. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science 262:1257-1261. [DOI] [PubMed] [Google Scholar]
- 7.Hajjar, A. M., P. F. Lewis, Y. Endeshaw, J. Ndinya-Achola, J. K. Kreiss, and J. Overbaugh. 1998. Efficient isolation of human immunodeficiency virus type 1 RNA from cervical swabs. J. Clin. Microbiol. 36:2349-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kampinga, G. A., A. Simonon, P. Van de Perre, E. Karita, P. Msellati, and J. Goudsmit. 1997. Primary infections with HIV-1 of women and their offspring in Rwanda: findings of heterogeneity at seroconversion, coinfection, and recombinants of HIV-1 subtypes A and C. Virology 227:63-76. [DOI] [PubMed] [Google Scholar]
- 9.Learn, G. H., D. Muthui, S. J. Brodie, T. Zhu, K. Diem, J. I. Mullins, and L. Corey. 2002. Virus population homogenization following acute human immunodeficiency virus type 1 infection. J. Virol. 76:11953-11959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Long, E. M., H. L. Martin, Jr., J. K. Kreiss, S. M. Rainwater, L. Lavreys, D. J. Jackson, J. Rakwar, K. Mandaliya, and J. Overbaugh. 2000. Gender differences in HIV-1 diversity at time of infection. Nat. Med. 6:71-75. [DOI] [PubMed] [Google Scholar]
- 11.Lukashov, V. V., and J. Goudsmit. 1997. Founder virus population related to route of virus transmission: a determinant of intrahost human immunodeficiency virus type 1 evolution? J. Virol. 71:2023-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malet, I., M. Belnard, H. Agut, and A. Cahour. 2003. From RNA to quasispecies: a DNA polymerase with proofreading activity is highly recommended for accurate assessment of viral diversity. J. Virol. Methods 109:161-170. [DOI] [PubMed] [Google Scholar]
- 13.Mansky, L. M. 2000. In vivo analysis of human T-cell leukemia virus type 1 reverse transcription accuracy. J. Virol. 74:9525-9531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Markham, R. B., W. C. Wang, A. E. Weisstein, Z. Wang, A. Munoz, A. Templeton, J. Margolick, D. Vlahov, T. Quinn, H. Farzadegan, and X. F. Yu. 1998. Patterns of HIV-1 evolution in individuals with differing rates of CD4 T cell decline. Proc. Natl. Acad. Sci. USA 95:12568-12573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin, H. L., P. M. Nyange, B. A. Richardson, L. Lavreys, K. Mandaliya, D. J. Jackson, J. O. Ndinya-Achola, and J. Kreiss. 1998. Hormonal contraception, sexually transmitted diseases, and the risk of heterosexual transmission of HIV-1. J. Infect. Dis. 178:1053-1059. [DOI] [PubMed] [Google Scholar]
- 16.McDonald, R. A., D. L. Mayers, R. C. Chung, K. F. Wagner, S. Ratto-Kim, D. L. Birx, and N. L. Michael. 1997. Evolution of human immunodeficiency virus type 1 env sequence variation in patients with diverse rates of disease progression and T-cell function. J. Virol. 71:1871-1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Overbaugh, J., R. J. Anderson, J. O. Ndinya-Achola, and J. K. Kreiss. 1996. Distinct but related human immunodeficiency virus type 1 variant populations in genital secretions and blood. AIDS Res. Hum. Retrovir. 12:107-115. [DOI] [PubMed] [Google Scholar]
- 18.Overbaugh, J., and C. R. Bangham. 2001. Selection forces and constraints on retroviral sequence variation. Science 292:1106-1109. [DOI] [PubMed] [Google Scholar]
- 19.Overbaugh, J., S. M. Jackson, M. D. Papenhausen, and L. M. Rudensey. 1996. Lentiviral genomes with G-to-A hypermutation may result from Taq polymerase errors during polymerase chain reaction. AIDS Res. Hum. Retrovir. 12:1605-1613. [DOI] [PubMed] [Google Scholar]
- 20.Palmer, C., P. Balfe, D. Fox, J. C. May, R. Frederiksson, E. M. Fenyo, and J. A. McKeating. 1996. Functional characterization of the V1V2 region of human immunodeficiency virus type 1. Virology 220:436-449. [DOI] [PubMed] [Google Scholar]
- 21.Poss, M., H. L. Martin, J. K. Kreiss, L. Granville, B. Chohan, P. Nyange, K. Mandaliya, and J. Overbaugh. 1995. Diversity in virus populations from genital secretions and peripheral blood from women recently infected with human immunodeficiency virus type 1. J. Virol. 69:8118-8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poss, M., A. G. Rodrigo, J. J. Gosink, G. H. Learn, D. de Vange Panteleeff, H. L. Martin, Jr., J. Bwayo, J. K. Kreiss, and J. Overbaugh. 1998. Evolution of envelope sequences from the genital tract and peripheral blood of women infected with clade A human immunodeficiency virus type 1. J. Virol. 72:8240-8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sagar, M., L. Lavreys, J. M. Baeten, B. A. Richardson, K. Mandaliya, B. H. Chohan, J. K. Kreiss, and J. Overbaugh. 2003. Infection with multiple HIV-1 variants is associated with faster disease progression. J. Virol. 77:12921-12926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sagar, M., L. Lavreys, J. M. Baeten, B. A. Richardson, K. Mandaliya, J. O. Ndinya-Achola, J. K. Kreiss, and J. Overbaugh. 2003. Identification of modifiable factors that affect the genetic diversity of the transmitted HIV-1 population. AIDS 18:615-619. [DOI] [PubMed] [Google Scholar]
- 25.Shankarappa, R., J. B. Margolick, S. J. Gange, A. G. Rodrigo, D. Upchurch, H. Farzadegan, P. Gupta, C. R. Rinaldo, G. H. Learn, X. He, X. L. Huang, and J. I. Mullins. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 73:10489-10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shioda, T., S. Oka, X. Xin, H. Liu, R. Harukuni, A. Kurotani, M. Fukushima, M. K. Hasan, T. Shiino, Y. Takebe, A. Iwamoto, and Y. Nagai. 1997. In vivo sequence variability of human immunodeficiency virus type 1 envelope gp120: association of V2 extension with slow disease progression. J. Virol. 71:4871-4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Starcich, B. R., B. H. Hahn, G. M. Shaw, P. D. McNeely, S. Modrow, H. J. Wolf, E. S. Parks, W. P. Parks, S. F. Josephs, R. C. Gallo, and F. Wong-Staal. 1986. Identification and characterization of conserved and variable regions in the envelope gene of HTLV-III/LAV, the retrovirus of AIDS. Cell 45:637-648. [DOI] [PubMed] [Google Scholar]
- 28.Swofford, D. L. 1998. Phylogenetic analysis using parsimony, 4th ed. Sinauer Associates, Sunderland, Mass.
- 29.Upchurch, D. A., R. Shankarappa, and J. I. Mullins. 2000. Position and degree of mismatches and the mobility of DNA heteroduplexes. Nucleic Acids Res. 28:E69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vartanian, J.-P., A. Meyerhans, B. Åsjö, and S. Wain-Hobson. 1991. Selection, recombination, and G→A hypermutation of human immunodeficiency virus type 1 genomes. J. Virol. 65:1779-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, N., T. Zhu, and D. D. Ho. 1995. Sequence diversity of V1 and V2 domains of gp120 from human immunodeficiency virus type 1: lack of correlation with viral phenotype. J. Virol. 69:2708-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolfs, T. F., G. Zwart, M. Bakker, and J. Goudsmit. 1992. HIV-1 genomic RNA diversification following sexual and parenteral virus transmission. Virology 189:103-110. [DOI] [PubMed] [Google Scholar]
- 33.Wolinsky, S. M., C. M. Wike, B. T. Korber, C. Hutto, W. P. Parks, L. L. Rosenblum, K. J. Kunstman, M. R. Furtado, and J. L. Munoz. 1992. Selective transmission of human immunodeficiency virus type-1 variants from mothers to infants. Science 255:1134-1137. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, L. Q., P. MacKenzie, A. Cleland, E. C. Holmes, A. J. Brown, and P. Simmonds. 1993. Selection for specific sequences in the external envelope protein of human immunodeficiency virus type 1 upon primary infection. J. Virol. 67:3345-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu, T., H. Mo, N. Wang, D. S. Nam, Y. Cao, R. A. Koup, and D. D. Ho. 1993. Genotypic and phenotypic characterization of HIV-1 in patients with primary infection. Science 261:1179-1181. [DOI] [PubMed] [Google Scholar]


