Abstract
Tumor necrosis factor-induced protein 3 (TNFAIP3; also known as A20) negatively regulates NF-κB and MAPK signals to control inflammatory responses. TNFAIP3 also protects against TNF-induced cell death. Intestinal epithelial cell (IEC) expression of TNFAIP3 improves barrier function and tight junction integrity and prevents dextran sulfate sodium (DSS)-induced IEC death and colitis. We therefore investigated the effects of TNFAIP3 expression in IEC on immune homeostasis in the intestines of immune-compromised mice. Villin-TNFAIP3 (v-TNFAIP3) transgenic mice were interbred with IL-10−/− mice (v-TNFAIP3 × IL-10−/−) and incidence, onset, and severity of colitis was assessed. v-TNFAIP3 × IL-10−/− mice displayed severe, early onset, and highly penetrant colitis that was not observed in IL-10−/− or v-TNFAIP3 mice. V-TNFAIP3 mice displayed altered expression of mucosal cytokines, increased numbers of mucosal regulatory T cells, and altered expression of mucosal antimicrobial peptides (AMPs). Microbial colonization of the inner mucus layer of v-TNFAIP3 mice was observed, along with alterations in the microbiome, but this was not sufficient to induce colitis in v-TNFAIP3 mice. The relative sterility of the inner mucus layer observed in wild-type and IL-10−/− mice was lost in v-TNFAIP3 × IL-10−/− mice. Thus IEC-derived factors, induced by signals that are inhibited by TNFAIP3, suppress the onset of inflammatory bowel disease in IL-10−/− mice. Our results indicate that IEC expression of TNFAIP3 alters AMP expression and allows microbial colonization of the inner mucus layer, which activates an IL-10-dependent anti-inflammatory process that is necessary to prevent colitis.
Keywords: A20, Reg3, Ang4, NK-κB, intestinal mucus
tumor necrosis factor-induced protein 3 (TNFAIP3, also known as A20) is an ubiquitin-modifying enzyme that inhibits activation of NF-κB and MAPK by TNF receptor, Toll-like receptor, and other innate immune receptors (3, 16, 26). TNFAIP3 also inhibits apoptosis and some forms of necrotic cell death (3, 16, 26). The essential requirement for TNFAIP3 in preventing inflammation is evident by the profound inflammation and morbidity in TNFAIP3−/− mice (23). Lineage-specific deletion of TNFAIP3 in dendritic cells, B cells, intestinal epithelial cells (IEC), or macrophages and granulocytes results in inflammation mimicking human autoimmune and inflammatory diseases (12, 22, 28, 36, 38). Notably deletion of TNFAIP3 in each of these lineages results in distinct phenotypes, indicating cell-specific roles for TNFAIP3 in vivo. Although TNFAIP3 is broadly viewed as an enzyme that controls ubiquitin-dependent signaling to prevent inflammation, the essential function of TNFAIP3 is to control receptor-mediated cell signaling. Thus the role of TNFAIP3 in inflammation is dependent on the cellular context and the nature of the signal being controlled. This is exemplified by the phenotype of mice with a lineage-specific deletion of TNFAIP3 in keratinocytes that do not develop epidermal inflammation but rather display ectodermal abnormalities, reflecting a role for TNFAIP3 in controlling NF-κB-dependent homeostasis in epidermal tissue (24). Similarly, somatic mutations that delete TNFAIP3 in B cells are associated with human lymphomas, reflecting a key role for TNFAIP3 as a suppressor of NF-κB-induced B cell survival and tumorgenesis (4, 19). Thus the role of TNFAIP3 in tissue homeostasis is likely complex and parallels the complex and distinct roles that ubiquitin-mediated NF-κB, MAPK, and cell death signals play in different cell and tissue types.
Many genetic studies have implicated TNFAIP3 in human disease (3, 26). Polymorphisms in or near the TNFAIP3 gene locus are associated with altered risk for inflammatory bowel disease and many other human autoimmune or inflammatory disorders (18). Variants that reduce the function of TNFAIP3 are associated with systemic lupus erythematosus and B cell lymphomas (4, 19, 25, 29). These genetic associations with inflammatory disease and the fact that loss of TNFAIP3 in many cell types in mice results in inflammation have contributed to the idea that enhanced TNFAIP3 expression or function might have therapeutic potential in human disease. However, it is not known whether inflammatory bowel disease (IBD)-associated TNFAIP3 polymorphisms, or polymorphisms associated with other autoimmune diseases, reduce or enhance TNFAIP3 function. Given the cell and tissue context-dependent roles of TNFAIP3, it is of importance to determine how enhanced TNFAIP3 expression or function in distinct cell types might impact homeostasis.
TNFAIP3 is expressed in and inhibits NF-κB activation and TNF-induced cell death in IEC (38, 39). Lineage-specific deletion of TNFAIP3 in IEC does not result in spontaneous colitis but makes mice more susceptible to dextran sulfate sodium (DSS)-induced intestinal inflammation, potentially driven by TNF receptor-induced IEC death (38). Consistent with this, transgenic expression of TNFAIP3 in IEC ameliorates DSS-induced colitis and reduces DSS-induced IEC death (33). These phenotypes are not likely to result directly from inhibition of NF-κB signaling by TNFAIP3 in IEC because NF-κB signaling in IEC prevents inflammation and protects against DSS-induced colitis (31, 41). Instead, the prosurvival effects of TNFAIP3 in IEC may explain how IEC expression of TNFAIP3 prevents intestinal inflammation in these models. Additionally, TNFAIP3 may have distinct functions in IEC, as it promotes intestinal barrier function, regulates the ubiquitination of the tight junction protein occludin, and inhibits Wnt signaling and colon carcinogenesis (21, 35). The role of TNFAIP3 expression in IEC during inflammation may be context dependent, as mice expressing TNFAIP3 in IEC were not protected from 4,6-trinitrobenzene sulfonic acid (TNBS)-induced colitis and instead displayed a worse course of disease in that model (33). Therefore, we examined the effect of IEC expression of TNFAIP3 on intestinal homeostasis and inflammation in other mouse models of colitis.
Here we report that transgenic expression of TNFAIP3 in IEC alters the production of antimicrobial peptides (AMPs) in the intestinal mucosa and causes a loss of the normal spatial separation between IEC and lumenal microbes. Despite this altered microbial localization, v-TNFAIP3 mice do not develop colitis. In the context of IL-10 deficiency, IEC expression of TNFAIP3 causes severe early-onset Th-1-type colitis with 100% penetrance. These results suggest that enhanced expression of TNFAIP3 may not be anti-inflammatory in all circumstances and that genetic associations between TNFAIP3 and human disease should not be considered a priori as an indication of reduced TNFAIP3 expression or function in those diseases.
MATERIALS AND METHODS
Animal studies.
Animals were housed in specific pathogen-free conditions with investigation approved by the University of Chicago IACUC, under protocols 71661 and 72089. Villin-TNFAIP3 transgenic (Tg) mice and wild-type (WT) littermates were generated as previously described (21). For IL-10 knockout (KO) studies, these mice were bred onto the IL-10 KO C57BL/6 background and genotyped by tail clipping. Mouse weight was recorded weekly between weaning and death (6–8 wk), and occurrences of euthanasia resulting from mice exceeding IACUC-approved endpoints (weight loss exceeding 20% of maximum body weight, persistent positive fecal blood test or rectal bleeding, hunched posture, rectal prolapse) were recorded. Colons were surgically removed, weighed, and measured for length analysis. Histological examination was performed on tissue fixed in 10% formalin, paraffin embedded, and sectioned longitudinally. Sections (5 μm) were stained with hematoxylin and eosin and scored by a pathologist blinded to the genotype using previously described criteria (33).
ELISAs and immunoblotting.
Colons were removed and rinsed, and mucosal scrapings were performed, as previously described (33). Briefly, colons were opened longitudinally and rinsed, and the lumen was scraped using a glass slide. The scraping was collected in 500 μl buffer (10 mM Tris, 5 mM MgSO4, 250 μg DNAse2, 1× protease inhibitor cocktail from Roche), lysed, and sonicated. Normalization of samples was effected using a BCA kit (Pierce). ELISAs were performed using equal amounts of protein per sample (100 μg) according to manufacturer's protocols and IFN-γ, TNF, IL-4, IL-10, IL-6, mouse ELISA-kits (OptIEA BD Biosciences), IL-17, IL-25/17E, IL-22, IL-23, IL-12/23p40, and thymic stromal lymphopoietin (TSLP) mouse ELISA kits (DuoSet, eBioscience). For immunoblotting, samples were diluted 1:1 in Laemmli buffer (60 mM Tris·HCl pH 6.8, 2% SDS, 10% glycerol, 5% β-mercaptoethanol, 0.01% bromophenol blue), resolved by SDS-PAGE, transferred onto activated PVDF membrane, and probed using mouse-specific antibodies diluted in Li-COR (Odyssey), Ang4 (Santa Cruz Biotechnology clone M-18), Reg3β (R&D Systems clone P35230), biotin-labeled CD103 (Biolegend clone 2E7), myeloperoxidase (Thermo Scientific), Muc2 (Santa Cruz Biotechnology clone H-300), and actin (Santa Cruz Biotechnology clone C-11).
Immunofluorescent, immunohistochemical, and FISH staining.
Colons were removed and fixed in 10% formalin or Carnoy's solution (70% methanol, 20% chloroform, and 10% acetic acid) followed by paraffin embedding. Longitudinal sections (5 μm) were made, and antigen retrieval was performed as necessary. Briefly, sections to be stained were deparaffinized in 100% xylene followed by rehydration in decreasing concentrations of ethanol. Sections were then boiled in 10 mM sodium citrate and left to cool at room temperature for 25–30 min, followed by blocking with Li-COR buffer (Odyssey) and incubation with mouse-specific antibodies, Ang4 (Santa Cruz Biotechnology clone M-18), Reg3β (R&D Systems clone P35230), biotin-labeled CD103 (Biolegend clone 2E7), myeloperoxidase (Thermo Scientific), and Muc2 (Santa Cruz Biotechnology clone H-300) at 4°C overnight. NF-κB activity in epithelial cells was assessed by immunostaining as described with phospho-NF-κB antibody (Santa Cruz) followed by donkey anti-rabbit-horseradish peroxidase, development with diaminobenzidine, and counterstaining with Fast green. For fluorescence in situ hybridization (FISH) analyses, sections were deparaffinized in a similar manner, followed by incubation of FISH probe (EUB338 bacterial 16S rRNA) in hybridization buffer (35% formamide) at 50°C overnight, followed by counterstain with Hoescht (Invitrogen). Alexa Fluor-conjugated secondary antibodies, anti-rabbit 488 or 555, anti-goat 488 or 555, anti-sheep 555 (Invitrogen), or FITC-conjugated neutravidin (Invitrogen) were incubated for 1 h in the dark at room temperature followed by washing and mounting of slides with ProLong Gold anti-fade reagent (Invitrogen). Images were captured on an Olympus DSU spinning disk confocal microscope, collated in SlideBook, and analyzed using ImageJ.
Multicolor flow cytometry.
Spleen, mesenteric, and peripheral lymph nodes were surgically removed, and single-cell lymphocyte suspensions were generated. Briefly, nodes were homogenized using 0.3-μm gauze followed by washing with fluorescence-activated cell sorting (FACS) buffer (5% FBS in PBS). Samples were red blood cell lysed as required by addition of lysis buffer (155 mM NH4Cl, 12 mM NaHCO3, 0.1 mM EDTA) and filtered using gauze. Lipoprotein lipase preparations were generated by removal of colons followed by rinsing in cold calcium- and magnesium-free (CMF)-HEPES. Tissues were chopped into small pieces and washed in CMF-FBS-EDTA four times for 15 min at room temperature at 220 revolution/min. EDTA was removed by washing in complete RPMI followed by collagenase and DNAse treatment of samples in RPMI at 37°C with shaking for 1 h. Cells were filtered using 0.3-μm gauze, washed, and resuspended in FACS buffer. Cells were then counted, and an aliquot was made for antibody staining. For extracellular antigens, antibodies (BD Biosciences) were diluted in FACS buffer and incubated on ice for 1 h in the dark followed by washing and analyses. Intracellular staining was effected by fixation and permeabilized overnight at 4°C in the dark using the fixation/permeabilization kit (eBioscience). Samples were run on a FACSCalibur flow cytometer (BD Bioscience).
Microarray.
Whole colon pieces were removed from mice and frozen on dry ice. Samples were homogenized, and RNA was extracted per manufacturer's protocol using Trizol (Invitrogen). RNA was purified following manufacturer's protocol using the RNAeasy kit (Qiagen). Samples were hybridized onto Illumina MouseRef8 chips at the Functional Genomics Core at University of Chicago. Subsequent analyses were performed using the R statistical programming package and the InGeneuity and OntoExpress online tool. Statistical analyses were also performed in Prism (GraphPad) and Microsoft Excel.
Microbiome analysis.
Colons were removed from mice and lumens washed with 500 μl PBS, and flow through was collected; the same colons were then cut longitudinally and mucosal scrapings performed, with samples collected into TNES DNA extraction buffer (50 mM Tris·HCl, pH 7.4, 400 mM NaCl, 100 mM EDTA, pH 8.0, and 0.5% SDS) with Proteinase K (20 mg/ml). Samples were mixed using zircon beads (BioSpec Products) in a bead beater (BioSpec Products) and incubated overnight at 55°C with shaking. DNA was precipitated with a solution of phenol, chloroform, and isoamyl alcohol and precipitated with ethanol and washed, and the concentration was measured using a NanoDrop spectrophotometer (Thermo Scientific). MiSeq analyses were then performed; briefly, PCR primers used were specific for the 515–806 base pair (bp) region of the 16S rRNA-encoding gene (338F: 5′-GTGCCAGCMGCCGCGGTAA-3′ and 806R: 5′- GGACTACHVGGGTWTCTAAT-3) and contained Illumina 3′adapter sequences as well as a 12-bp barcode. Sequencing was performed by the Next Generation Sequencing Core at Argonne National Laboratory using an Illumia MiSeq. Sequences were then trimmed and classified with the QIIME toolkit. Using the QIIME wrappers, operational taxonomical units (OTUs) were picked at 97% sequence identity using uclust, and a representative sequence was then chosen for each OTU by selecting the most abundant sequence in that OTU. These representative sequences were classified and assigned a taxonomic string using the RDP Classifier. The PyNAST-aligned sequences were also used to build a phylogenetic tree with FastTree for calculation of unweighted/weighted UniFrac distances (as well as other measures of diversity) for ecological analyses, including visualization via ordination (e.g., principal coordinates analysis).
Statistical analyses.
Analyses were performed using Prism software from GraphPad, including t-tests and one-way ANOVA with multiple comparisons, and values of P ≤ 0.05 were considered significant.
RESULTS
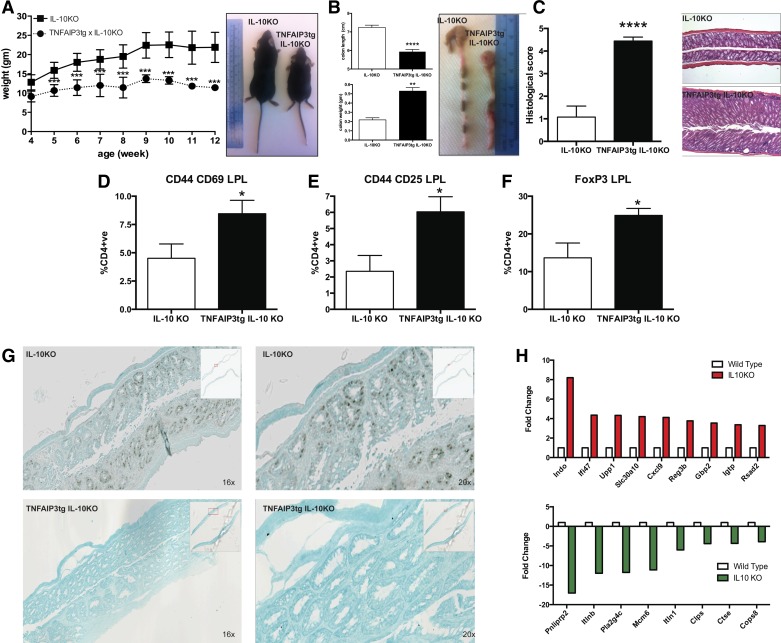
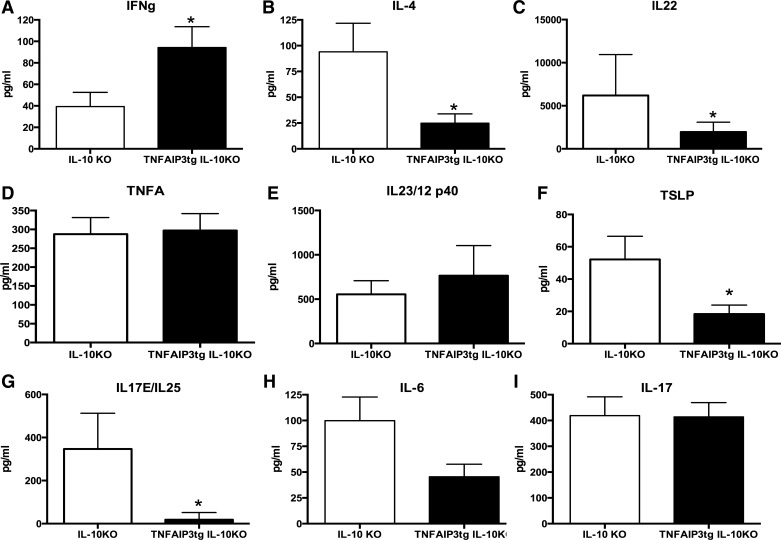
We have previously found that v-TNFAIP3 mice display enhanced barrier function and more stable tight junctions than WT mice (21). In addition, we found that v-TNFAIP3 mice are protected from DSS-induced IEC death and colitis but not from TNBS-induced colitis (33). This suggested that IEC might play distinct roles in T cell-mediated colitis that are regulated by epithelial expression of TNFAIP3. Colitis in IL-10−/− mice requires T cells and the presence of microbes and is preceded by increased intestinal permeability and IEC apoptosis (5, 8, 27, 34). Activated NF-κB was evident in the epithelium of IL-10−/− mice, but, consistent with the effects of TNFAIP3 expression in other tissues, we observed reduced activation of NF-κB in the epithelium of v-TNFAIP3 × IL-10−/− mice (Fig. 1). We therefore tested whether expression of TNFAIP3 in IEC alters the course or severity of colitis in IL-10−/− mice. In our colony, all mice (including v-TNFAIP3 mice) are on a C57Bl/6 background under specific pathogen-free conditions. We typically observe a low penetrance of colitis only at an advanced age (10–20% at 16–20 wk) in IL-10−/− mice under these conditions. Although overt colitis was not observed in IL-10−/− mice, changes in gene expression consistent with those reported in IL-10−/− mice with colitis were observed (13). These included increased expression of indolamine 2,3 dioxygenase (INDO), immunity-related GTPase family M protein (IRGM) (ifi47), uridine phosphorylase 1, and chemokine ligand 9 (CXCL9), indicative of a response to interferon-γ in the tissue of IL-10−/− mice. In addition, decreased expression of pancreatic lipase-related protein 2 (pnliprp2) was observed, consistent with prior studies in IL-10−/− mice with colitis. Together, these data suggest that the mucosa of IL-10−/− mice has altered gene expression consistent with a Th1-type colitis even though colitis is not manifest in these mice. However, when crossed to v-TNFAIP3 mice, we observed severe colitis with very early age of onset in 100% of IL-10−/− mice. This was characterized by failure of v-TNFAIP3 × IL-10−/− mice to gain weight compared with their IL-10−/− littermates (Fig. 1). The colitis in these mice was manifest as colonic shortening and increased histological scores for inflammation owing to immune cell infiltration, crypt hypertrophy, and abscesses (Fig. 1). Cytokine profiles from mucosal scrapings showed elevated levels of IFN-γ, reduced levels of IL-4, and no alteration of IL-17, indicative of a Th1-type colitis (Fig. 2). We did not observe significant changes in the levels of IL-12/23 p40 or IL-23 associated with colitis in these mice, indicating that colitis can occur in this model without elevated levels of these cytokines (Fig. 2). These findings indicate that expression of TNFAIP3 in IEC markedly exacerbates Th1-type colitis in IL-10−/− mice.
Fig. 1.
Epithelial expression of tumor necrosis factor-induced protein 3 (TNFAIP3) suppresses NF-κB activation and induces colitis in IL-10 knockout (KO) mice. Villin-TNFAIP3 transgenic mice (TNFAIP3tg) were interbred with IL-10−/− mice, and littermates were assessed for the activation of epithelial cell NF-κB and incidence and severity of colitis. TNFAIP3tg × IL-10KO, compared with IL-10KO mice, exhibited reduced body weight and failure to thrive apparent by 4 wk, significant by 5 wk of age (A); reduced length, increased weight, and visible turgidity of the colon (B); increased histological signs of inflammation (C); increased levels of activated CD4 T cells (D and E); and increased levels of FoxP3+ CD4 T cells in the colonic mucosa (F). G: reduced immunostaining for activated NF-κB (phospho-NF-κB) in the nuclei of epithelial cells. H: altered gene expression in the intestinal mucosa of noncolitic IL-10−/− mice. *P < 0.05, **P < 0.01, ***P < 0.005, ****P < 0.001.
Fig. 2.
Epithelial expression of TNFAIP3 induces a Th1-type colitis in IL-10KO mice. ELISAs were performed on the colonic mucosa from IL-10KO vs. TNFAIP3tg × IL-10KO mice, and these revealed elevated levels of IFN-γ (A) and reduced levels of IL-4 (B) typical of a Th1-type of inflammation. There were also significantly reduced levels of IL-22 (C), thymic stromal lymphopoietin (TSLP) (F), and IL-25 (G) and a trend toward reduced IL-6 (H), in the mucosa of TNFAIP3tg × IL-10KO mice, compared with IL-10KO mice. No differences were observed in the mucosal levels of TNF (D), IL-12/23 (E), or IL-17 (I). *P < 0.05.
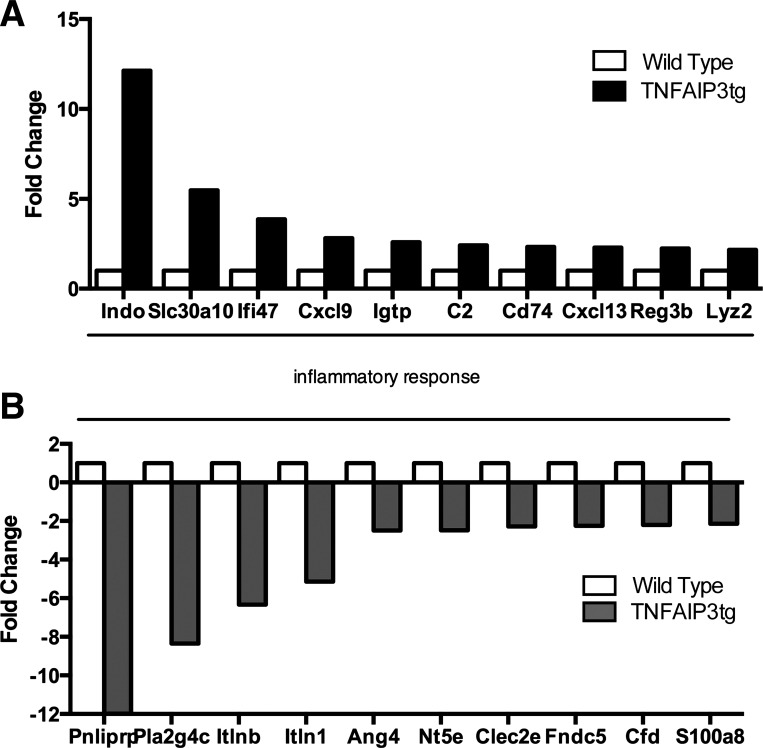
The enhanced colitis in IL-10−/− × v-TNFAIP3 mice compared with IL-10−/− mice suggests that IEC actively suppress colitis in immune-compromised mice. To interrogate this further, we examined the mucosal gene expression in v-TNFAIP3 mice (Fig. 3). We have not observed colitis or inability to thrive in v-TNFAIP3 mice compared with WT littermates. To investigate the effect of IEC expression of TNFAIP3 on intestinal homeostasis, we performed a gene microarray analysis on v-TNFAIP3 mice and their WT littermates. Following microarray, gene ontology analysis (GO) was performed using registered GO terms from OntoExpress and a cut off of fold change expression alteration of 2.0. Significant altered expression was found for GO terms controlling the inflammatory response, including those involved in antibacterial humoral responses (Fig. 3). Upregulated genes included INDO1, involved in dendritic and neutrophil function, Igtp, an interferon-induced gene, and Reg3β, an antimicrobial c-type lectin (Fig. 3). Downregulated genes also grouped into similar GO classes, specifically those involved in the inflammatory response (Fig. 3). These downregulated genes included the intelectin family members, Itlnb and Itln1, and the AMP Ang4. Thus expression of TNFAIP3 in IEC does not lead to colitis but does alter mucosal gene expression in the intestine.
Fig. 3.
Altered gene expression in the intestinal mucosa of TNFAIP3tg mice. Total RNA was extracted from the intestinal mucosa of wild-type (WT) or villin-TNFAIP3 transgenic (TNFAIP3tg) littermates and assessed for gene expression by array. Selected mRNA levels from the complete set of data are shown graphically here. Genes with higher expression in TNFAIP3tg mucosa include indolamine 2,3 dioxygenase (INDO), solute carrier family 30, member 10 (SLC30A10), interferon-γ-inducible protein 47 (ifi47, also known as IRG-47), chemokine ligand 9 (CxCl9), and interferon-γ-induced GTPase (IgtP, also known as IRGM3). Genes with lower expression in TNFAIP3tg mice include pancreatic lipase-related protein precursor (Pnliprp), phospholipase A2 group VIC (Pla2g4c or cytosolic PLA2), intelectin b (itlnb), intelectin 1 (itln1), angiogenin-4 (Ang4), and 5′ nucleotides, ecto (Nt5e or CD73).
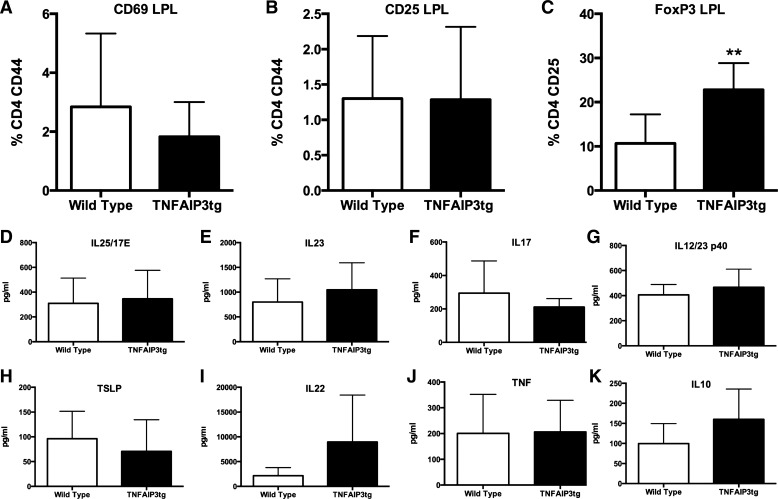
The changes in gene expression observed in v-TNFAIP3Tg mice included altered levels of INDO1 and Igtp (IRGM3) in the colonic mucosa. This led us to assess T-cell phenotypes in the intestinal mucosa and secondary lymphoid organs. We determined that there was no significant increase in the number of CD4+CD44+CD69+ or CD4+CD44+CD25+ activated T-lymphocytes in the lamina propria (LP) or in the spleen, peripheral lymph nodes (PLN), or mesenteric lymph nodes (MLN) of v-TNFAIP3Tg mice (Fig. 1A). Significant increases were identified however, in the CD4+CD25+FoxP3+ population of T cells in the LP but not in spleen, PLN, or MLN of v-TNFAIP3Tg mice, demonstrating that IEC expression of TNFAIP3 leads to increased numbers of Tregs particularly in the colonic mucosa (Fig. 4). Examination of cytokine levels in the mucosa of v-TNFAIP3Tg mice revealed no significant differences in the levels of IL25/17E, TSLP, IL-10, IL23, IL12/23p40, IL-17, TNF, or IL-22 although the latter showed a trend toward increased expression in v-TNFAIP3Tg mice compared with WT littermates (Fig. 4). Thus expression of TNFAIP3 in IEC does not significantly alter levels of cytokines or the activation state of T cells but does result in an increase in the number or Tregs in the intestinal mucosa. This, combined with the lack of colitis in these mice, suggested that IEC expression of TNFAIP3 was leading to changes in gut mucosal homeostasis but that compensatory changes, possibly increased numbers of Tregs, were preventing spontaneous inflammation in v-TNFAIP3 mice.
Fig. 4.
Increased Tregs in the colonic mucosa of TNFAIP3tg mice. Colonic lamina propria lymphocytes were isolated and analyzed for the presence of activated CD4 T-lymphocytes (A and B) and FoxP3+ CD4 T lymphocytes (C). Colonic mucosal tissue was assessed for the level of IL-25 (D), IL-23 (E), IL-17 (F), IL-12/23 (G), TSLP (H), IL-22 (I), TNF (J), and IL-10 (K). **P < 0.01.
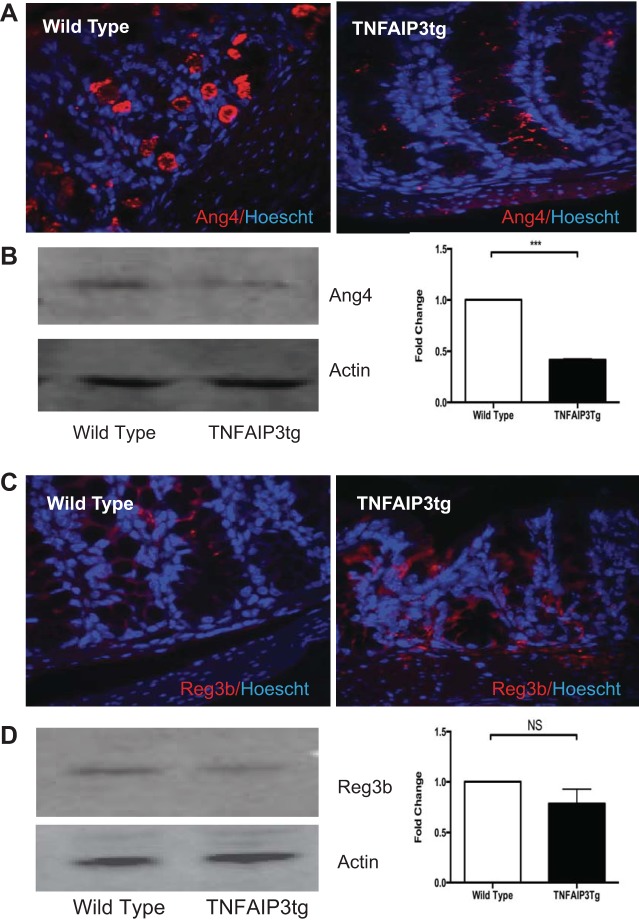
Given the significant alteration of expression of genes involved in antimicrobial defense, we focused on the functional outcomes of their altered expression. Reg3β is a member of the Reg3 family of C-type lectins that protect the epithelial cell surface by preventing bacterial translocation and colonization of the relatively sterile inner mucus layer (10). Ang4 is induced upon colonization of the gut by Bacteroidetes thetaiotaomicron and has antimicrobial activity against Listeria monocytogenes and Enterococcus faecalis but is selective, as it has no or reduced activity against Listeria innocua, some strains of Escherichia coli and Bacteroides thetaiotaomicron itself (10, 15). The gene expression changes were validated at the protein level using immunofluorescent staining of Carnoy's fixed tissue and immunoblot analysis of mucosal scrapings (Fig. 5). v-TNFAIP3 mice displayed markedly less Ang4 staining within the inner mucus layer of the colon, compared with WT littermates. Conversely, v-TNFAIP3 mice displayed increased expression of Reg3β in the colon, and these results were validated by immunoblots for Ang4 and Reg3β (Fig. 5). Thus expression of TNFAIP3 in IEC alters the expression of AMP in the mucosa, with some AMPs exhibiting increased expression and others decreased expression.
Fig. 5.
Reduced expression of angiogenin-4 in the intestinal mucosa of TNFAIP3tg mice. Immunohistochemical localization (red) (A), immunoblots of Ang4 (B), and Reg3β protein in WT and TNFAIP3tg colon (C and D), showing decreased Ang4 expression in the colon of TNFAIP3tg mice.
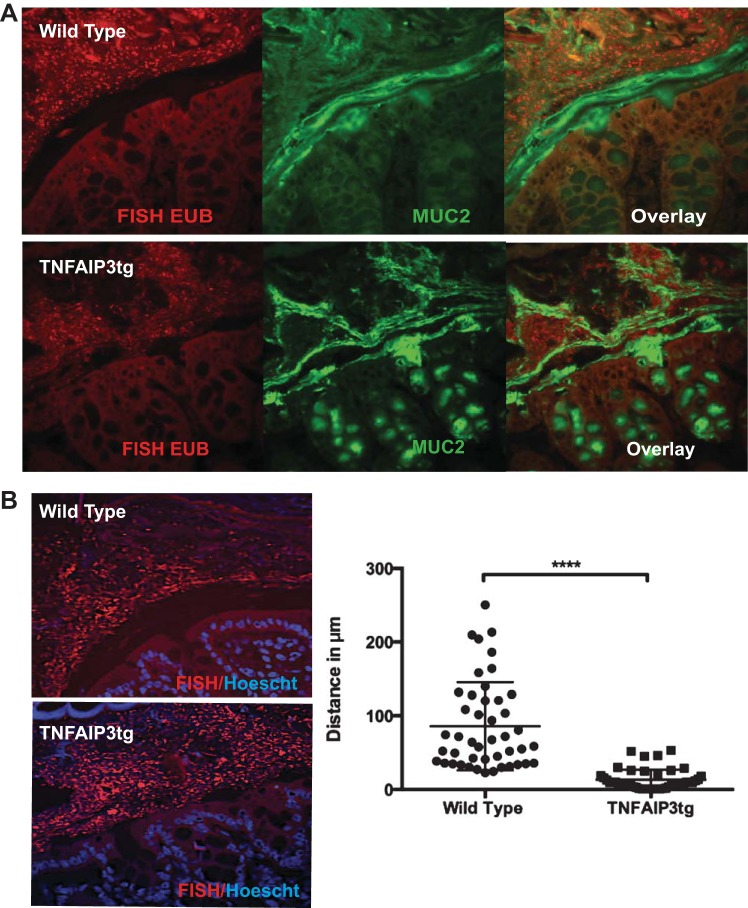
To assess the functional consequences of this AMP expression alteration, we examined the possibility of altered microbial localization in the colon of v-TNFAIP3 mice. Compared with their WT littermates, v-TNFAIP3 mice displayed a loss of the spatial separation between the intestinal epithelium and luminal bacteria (Fig. 6). The inner mucus layer of the large intestine is largely composed of a heavily glycosylated layer of the Mucin-2 protein (17). Although we did not observe a change in Muc2 expression in v-TNFAIP3 mice, we did find that bacteria colonized the inner mucus layer in a way that we did not visualize in WT littermates. Thus we observed that expression of TNFAIP3 in IEC alters AMP expression in the mucosa and allows luminal microbes to invade the inner mucus layer of the gut. These microbes are likely to be resistant to Reg3-type AMPs and potentially sensitive to Ang4 or other lectins that are reduced in v-TNFAIP3 mice. We interpret this to indicate that microbial invasion of the inner mucus layer occurs when IEC express TNFAIP3, and, although this may cause alterations in immune homeostasis, such as increased numbers of Tregs in the mucosa, it is insufficient to cause colitis in immune competent hosts.
Fig. 6.
Microbial invasion of the intestinal inner mucus layer in villin-TNFAIP3 transgenic mice. A: fluorescence in situ hybridization (FISH) with EUB338 probe for bacterial 16S rRNA (EUB) (red) and immunohistochemistry for Muc-2 (green) in Carnoy's-fixed paraffin-embedded colonic tissue. B: FISH and Hoescht (blue) staining to detect the spatial separation of bacteria from epithelial cells with quantification of the distance (μM) between the epithelial surface and the bacterial population. ****P < 0.001.
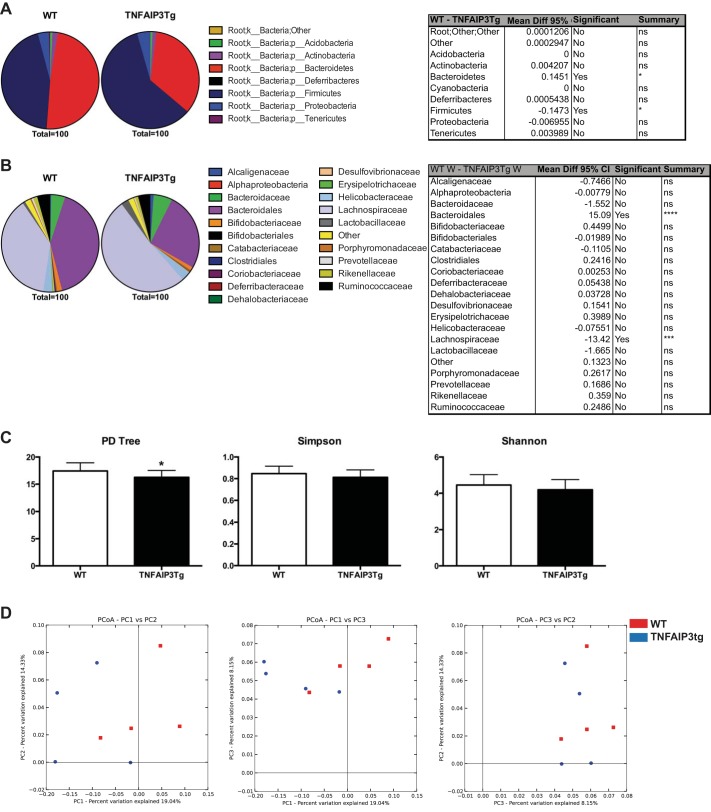
Given that the localization of bacteria is altered in v-TNFAIP3 mice compared with littermate controls, we next investigated whether the structure of the intestinal bacterial populations were altered. To do so, we performed microbiome analyses of colonic mucosa and compared v-TNFAIP3 mice with WT littermate controls. Statistically significant differences existed at the phylum taxonomical level (Fig. 7), displaying a reduction in the proportion of Bacteroidetes and an increase in the proportion of Firmicutes in colonic washes of v-TNFAIP3 mice compared with controls. Furthermore at the deeper order taxonomical level, proportional differences were observed in the Bacteriodales (contraction) and Lachnospiraceae (expansion) (Fig. 7), mirroring those observed at the phylum level. Metrics for α-diversity show imperceptible but statistically significant differences in groups, at analysis four across five iterations for the overall phylogenetic diversity tree (Fig. 7). However, the more robust Simpson and Shannon metrics of α-diversity for both weighted and unweighted data show no such significant difference. β-Diversity as assessed by both weighted and unweighted principal component analyses also showed no significant clustering comparing between all principal components P1, P2, and P3 (Fig. 7). Taken together these results warrant further exploration of the exact microbes present and their localization, using a similar 16S RNA clone library approach to that used to identify the Bilophila wadsworthia pathobiont by Devkota et al. (6).
Fig. 7.
Altered microbiome of colonic mucosal samples in v-TNFAIP3 compared with littermate controls. Comparison between taxonomical population proportions of v-TNFAIP3 vs. littermate controls at both the phylum (A) and order (B) levels. Pie charts depict average proportions for N = 4 mice per group; significant differences were observed between Bacteroidetes and Firmicutes (A) and Bacteriodales and Lachnospiraceae (B), respectively. Tabular results of a 2-way ANOVA with multiple comparisons are also shown. α-Diversity metrics, phylogenetic diversity (PD) tree, Shannon and Simpson are also shown comparing between groups (C) as is β-diversity as assessed by principal coordinate analysis (PCoA) (D).
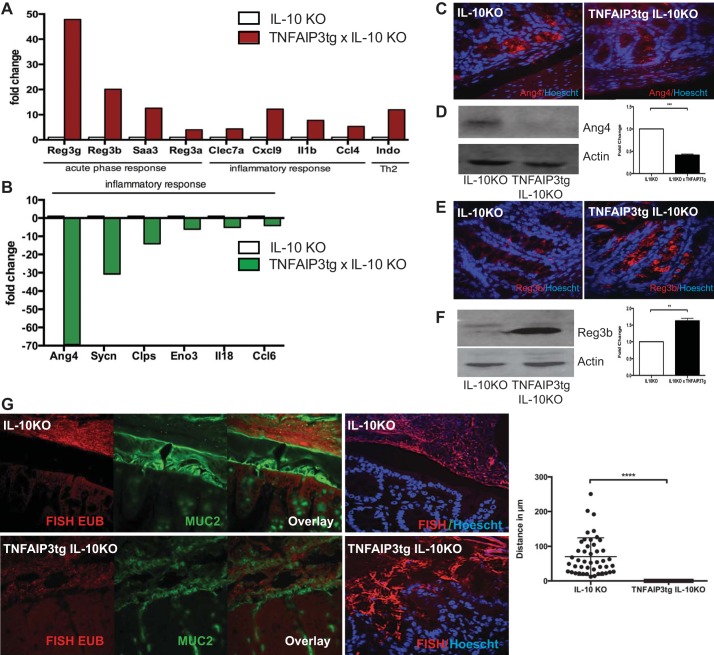
To investigate whether TNFAIP3 expression in IEC alters gene expression in IL-10−/− mice, we performed a gene array analysis of intestinal mucosa from IL-10−/− vs. IL-10−/− × v-TNFAIP3tg littermates. Significant differences in AMP expression were observed in IL-10−/− × v-TNFAIP3tg mice compared with IL-10−/− mice, most notably increased levels of the Reg3 family of genes and markedly decreased Ang4 expression (Fig. 8). Altered expression of Reg3β and Ang4 were validated by immunoblot and immunohistochemical analyses (Fig. 8). We next characterized whether control of bacterial burden was also altered as a result of altered AMP expression. Similar to changes observed in v-TNFAIP3 mice, we observed a loss of sterility of the inner mucus layer and an encroachment of bacteria to the epithelial cell surface in IL-10−/− × v-TNFAIP3 mice (Fig. 8). Because IL-10−/− × v-TNFAIP3 mice have chronic colitis, it is possible that the loss of sterility of the inner mucus layer is a reflection of the ongoing inflammation in these mice. However, the encroachment of bacteria in v-TNFAIP3 mice, which do not develop colitis, supports the idea that expression of TNFAIP3 in IEC reduces the spatial separation of host and microbes as a primary effect, which contributes to early and severe onset of IBD in IL-10−/− mice.
Fig. 8.
Altered production of antimicrobial peptides and invasion of the inner mucus layer in villin-TNFAIP3 × IL-10KO mice. Representative gene expression profile from an expression array showing mRNA that are increased (A) or decreased (B) in the colonic mucosa of TNFAIP3tg × IL-10KO mice, compared with IL-10KO mice. Immunohistochemistry and immunoblots showing decreased angiogenin-4 (C and D) and increased Reg3b protein (E and F) in the colonic mucosa of TNFAIP3tg × IL-10KO mice. G: FISH, Muc-2, and Hoescht staining showing microbial invasion of the colonic inner mucus layer and loss of spatial separation between the epithelial surface and bacteria in the intestines of TNFAIP3tg × IL-10KO mice, compared with IL-10KO mice. **P < 0.01, ***P < 0.005, ****P < 0.001.
DISCUSSION
We have found that IL-10−/− mice develop severe and early onset of colitis when TNFAIP3 is expressed in the intestinal epithelium. This is consistent with our previous finding that IEC expression of TNFAIP3 results in worse intestinal inflammation in the TNBS colitis model (33). This proinflammatory effect of TNFAIP3 expression was not expected because TNFAIP3 is typically described as an anti-inflammatory protein. TNFAIP3 inhibits NF-κB and MAPK signaling induced by TNF, NOD2, Toll-like receptor ligands, IL-17, and other factors (2, 11, 14, 23, 26, 37). We interpret our present findings with the hypothetical model that inhibition of NF-κB and MAPK signals in IEC leads to reduced production of a subset of AMPs, such as Ang4 and intelectins, that normally contribute to the relative sterility of the inner mucus layer of the colon. This leads to microbial contact with the intestinal mucosa, resulting in increased production of AMPs that do not depend on NF-κB or MAPK signals in IEC. These include Reg3-type peptides that rely on STAT3 signals in IEC for their production (40). In addition, microbial invasion induces regulatory T cells in the mucosa, leading to homeostatic control and prevention of colitis. In v-TNFAIP3 × IL-10−/− mice, which lack these regulatory T cells, microbial contact with the mucosa leads to severe and early onset of colitis. It is likely that the microbes invading the inner mucus layer are a subset of the gut microbiome that are normally controlled by Ang4, intelectins, and other AMPs that are reduced in mice expressing TNFAIP3 in IEC. These microbes are also likely to be resistant to Reg3-type and other AMPs that are increased in v-TNFAIP3 mice. It will be of interest to identify the types of microbes that invade the inner mucus layer in v-TNFAIP3 mice, as they have the capacity to cause severe colitis when the mucosal immune system is compromised. Typically microbial contact with the intestinal epithelium should elicit induction of protective cytokines like IL-22, TSLP, and IL-25, but we observed lower levels of these cytokines in v-TNFAIP3 × IL-10−/− mice. The failure to produce these protective cytokines may reflect the capacity of TNFAIP3 to block Toll-like receptor and TNF-induced NF-κB and MAPK activation in IEC and may have contributed to the inflammation seen in v-TNFAIP3 × IL-10−/− mice. However, we did not observe altered expression of these cytokines in v-TNFAIP3 mice without inflammation, and so it remains possible that the decreased expression observed in v-TNFAIP3 × IL-10−/− mice was the result of the inflammation in those mice, rather than a primary cause of the inflammation.
TNFAIP3 also regulates cell death in a manner that is cell and context dependent. In B cells or keratinocytes, loss of TNFAIP3 leads to excessive NF-κB activation, leading to hyperproliferation and, in the case of B cells, cellular transformation (4, 19, 24). Conversely, loss of TNFAIP3 in fibroblasts, T cells, and IEC leads to increased TNF-induced cell death (23, 38). TNFAIP3 also protects against DSS-induced IEC death, and mice deficient for, or overexpressing, TNFAIP3 in IEC are sensitive or resistant to DSS colitis, respectively (33, 38). Lastly, TNFAIP3 has unique functions in IEC, including the regulation of Wnt signaling and support of tight junction integrity to suppress carcinogenesis and promote barrier function, respectively (21, 35). The present studies do not allow us to definitively rule out these other functions of TNFAIP3 in IEC as potential contributors to the increased IBD in v-TNFAIP3 × IL-10−/− mice. However, the prevention of IEC death by TNFAIP3 is unlikely to explain our findings because increased IEC apoptosis in IL-10−/− mice contributes to the pathogenesis of colitis. Similarly, enhanced barrier function resulting from TNFAIP3 expression in IEC would be expected to prevent colitis in IL-10−/− mice, which are known to display increased gut permeability before the onset of colitis (27). Conversely, increased gut permeability can be protective in DSS colitis, as it increases the priming of mucosal regulatory T cells (20). However, we observed increased regulatory T cells in the mucosa of v-TNFAIP3 mice, suggesting that the increased barrier function in this model does not prevent Treg priming and that increased barrier function is unlikely to explain the more severe colitis in v-TNFAIP3 × IL-10−/− mice.
Although v-TNFAIP3 mice do not develop overt inflammation, changes were observed in the gene expression and Treg populations in the mucosa of these mice. The increased expression of INDO, IRG-47, IRGM3, and CXCL9 in the mucosa of v-TNFAIP3 are consistent with an IFN-γ profile, suggesting that the mucosa is responding to the microbial contact with the epithelium with a subclinical or nonpathological Th1 type inflammatory response. Consistent with this, decreased expression of pancreatic lipase-related protein may reflect reduced levels of IL-4 in the mucosa of these mice. Proinflammatory cytokines induce the expression of c-PLA2, CD73, Ang4, and intelectins, and TNFAIP3 inhibits signaling by proinflammatory cytokines. The reduced expression of these genes in v-TNFAIP3 mice likely reflects the inhibition of inflammatory signals in the epithelium of these mice. Thus, although pathological inflammation does not spontaneously occur in v-TNFAIP3, there is altered homeostasis in the mucosa of these mice that may predispose them to colitis, given an additional challenge. We observed alterations in the microbiome of v-TNFAIP3 mice that included contraction in the Bacteriodales and expansion of Lachnospiraceae. It has previously been shown that, in human samples from inflamed intestines, there is a proportional contraction of both Bacteroidetes and Firmicutes in favor of an expansion of proteobacteria (9). It is possible that the loss of Bacteriodales, which is observed in inflammatory states, may be compensated by the concomitant expansion of Lachnospiraceae, contributing to the protective effect of v-TNFAIP3 expression in other mouse models of IBD (33). Additionally it suggests that the spatial alteration usually observed with these microbes (interfold vs. digesta) (30) is lost attributable to our observed alterations in AMP production. It will be of interest to determine how epithelial expression of TNFAIP3 alters specific microbial species and the incidence and severity of inflammation in other models of IBD.
Genetic variants in the TNFAIP3 locus have been implicated in a variety of human autoimmune disorders, including IBD (18, 26). In addition, TNFAIP3 expression is part of a signature profile that can predict responses to therapy in IBD (1). TNFAIP3 inhibits NF-κB and MAPK signals, and this accounts for its anti-inflammatory function in innate immune cells. However, targeted deletion of NF-κB and MAPK signaling components in IEC and innate immune cells have clearly demonstrated that these signaling pathways play distinct cell-type-specific roles that have, in some cases, opposing effects on intestinal inflammation (7, 31, 32, 41). Induction of TNFAIP3 in IEC vs. innate immune cells may also therefore have distinct effects on intestinal inflammation, as we have found in this study. For example, induction of TNFAIP3 in IEC by TNF could potentially lead to altered production of AMPs, allowing invasion of the inner mucus layer by a subset of microbes and induction of colitis in an immune-compromised individual. Some of the benefits of anti-TNF therapy may result from reducing TNFAIP3 expression in IEC and thus restoring the sterility of the inner mucus layer. Conversely, therapies aimed at increasing TNFAIP3 expression or activity may prove beneficial for the prevention of many types of inflammation, but care should be taken when considering the potential deleterious effects of increased IEC expression or activity of TNFAIP3 in the colon. Lastly, genetic studies that implicate the TNFAIP3 locus in inflammation should be mindful of the fact that variants leading to increased expression or activity of TNFAIP3 may also predispose to some types of inflammation.
GRANTS
This work was supported by AI083375-01, DK42086, Crohn's and Colitis Foundation of America 0-34493-1362, Broad Medical Research Foundation IBD-0259 (to D. Boone), F32DK082104 (to J. Messer), and F32AI077235 (to J. Lodolce).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
Author contributions: S.F.M., L.R., J.S.M., J.P.L., J.E.C., and D.L.B. conception and design of research; S.F.M., L.R., W.A.G., J.P.L., J.E.C., S.J.B., T.M.N., R.A.K., G.M., C.B., and L.E.K. performed experiments; S.F.M., L.R., W.A.G., C.R.W., J.S.M., and D.L.B. analyzed data; S.F.M., L.R., W.A.G., C.R.W., J.S.M., and D.L.B. interpreted results of experiments; S.F.M., L.R., W.A.G., and C.R.W. prepared figures; S.F.M. and D.L.B. drafted manuscript; S.F.M., J.S.M., and D.L.B. edited and revised manuscript; D.L.B. approved final version of manuscript.
REFERENCES
- 1.Arsenescu R, Bruno ME, Rogier EW, Stefka AT, McMahan AE, Wright TB, Nasser MS, de Villiers WJ, Kaetzel CS. Signature biomarkers in Crohn's disease: toward a molecular classification. Mucosal Immunol 1: 399–411, 2008. [DOI] [PubMed] [Google Scholar]
- 2.Boone DL, Turer EE, Lee EG, Ahmad RC, Wheeler MT, Tsui C, Hurley P, Chien M, Chai S, Hitotsumatsu O, McNally E, Pickart C, Ma A. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol 5: 1052–1060, 2004. [DOI] [PubMed] [Google Scholar]
- 3.Catrysse L, Vereecke L, Beyaert R, van Loo G. A20 in inflammation and autoimmunity. Trends Immunol 35: 22–31, 2014. [DOI] [PubMed] [Google Scholar]
- 4.Compagno M, Lim WK, Grunn A, Nandula SV, Brahmachary M, Shen Q, Bertoni F, Ponzoni M, Scandurra M, Califano A, Bhagat G, Chadburn A, Dalla-Favera R, Pasqualucci L. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature 459: 717–721, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davidson NJ, Leach MW, Fort MM, Thompson-Snipes L, Kuhn R, Muller W, Berg DJ, Rennick DM. T helper cell 1-type CD4+ T cells, but not B cells, mediate colitis in interleukin 10-deficient mice. J Exp Med 184: 241–251, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Devkota S, Chang EB. Diet-induced expansion of pathobionts in experimental colitis: implications for tailored therapies. Gut Microbes 4: 172–174, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eckmann L, Nebelsiek T, Fingerle AA, Dann SM, Mages J, Lang R, Robine S, Kagnoff MF, Schmid RM, Karin M, Arkan MC, Greten FR. Opposing functions of IKKbeta during acute and chronic intestinal inflammation. Proc Natl Acad Sci USA 105: 15058–15063, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edelblum KL, Yan F, Yamaoka T, Polk DB. Regulation of apoptosis during homeostasis and disease in the intestinal epithelium. Inflamm Bowel Dis 12: 413–424, 2006. [DOI] [PubMed] [Google Scholar]
- 9.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA 104: 13780–13785, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallo RL, Hooper LV. Epithelial antimicrobial defence of the skin and intestine. Nat Rev Immunol 12: 503–516, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garg AV, Ahmed M, Vallejo AN, Ma A, Gaffen SL. The deubiquitinase A20 mediates feedback inhibition of interleukin-17 receptor signaling. Sci Signal 6: ra44, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hammer GE, Turer EE, Taylor KE, Fang CJ, Advincula R, Oshima S, Barrera J, Huang EJ, Hou B, Malynn BA, Reizis B, DeFranco A, Criswell LA, Nakamura MC, Ma A. Expression of A20 by dendritic cells preserves immune homeostasis and prevents colitis and spondyloarthritis. Nat Immunol 12: 1184–1193, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansen JJ, Holt L, Sartor RB. Gene expression patterns in experimental colitis in IL-10-deficient mice. Inflamm Bowel Dis 15: 890–899, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hitotsumatsu O, Ahmad RC, Tavares R, Wang M, Philpott D, Turer EE, Lee BL, Shiffin N, Advincula R, Malynn BA, Werts C, Ma A. The ubiquitin-editing enzyme A20 restricts nucleotide-binding oligomerization domain containing 2-triggered signals. Immunity 28: 381–390, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hooper LV, Stappenbeck TS, Hong CV, Gordon JI. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat Immunol 4: 269–273, 2003. [DOI] [PubMed] [Google Scholar]
- 16.Hymowitz SG, Wertz IE. A20: from ubiquitin editing to tumour suppression. Nat Rev Cancer 10: 332–341, 2010. [DOI] [PubMed] [Google Scholar]
- 17.Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci USA 105: 15064–15069, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, Essers J, Mitrovic M, Ning K, Cleynen I, Theatre E, Spain SL, Raychaudhuri S, Goyette P, Wei Z, Abraham C, Achkar JP, Ahmad T, Amininejad L, Ananthakrishnan AN, Andersen V, Andrews JM, Baidoo L, Balschun T, Bampton PA, Bitton A, Boucher G, Brand S, Buning C, Cohain A, Cichon S, D'Amato M, De Jong D, Devaney KL, Dubinsky M, Edwards C, Ellinghaus D, Ferguson LR, Franchimont D, Fransen K, Gearry R, Georges M, Gieger C, Glas J, Haritunians T, Hart A, Hawkey C, Hedl M, Hu X, Karlsen TH, Kupcinskas L, Kugathasan S, Latiano A, Laukens D, Lawrance IC, Lees CW, Louis E, Mahy G, Mansfield J, Morgan AR, Mowat C, Newman W, Palmieri O, Ponsioen CY, Potocnik U, Prescott NJ, Regueiro M, Rotter JI, Russell RK, Sanderson JD, Sans M, Satsangi J, Schreiber S, Simms LA, Sventoraityte J, Targan SR, Taylor KD, Tremelling M, Verspaget HW, De Vos M, Wijmenga C, Wilson DC, Winkelmann J, Xavier RJ, Zeissig S, Zhang B, Zhang CK, Zhao H, International IBD. Genetics Consortium, Silverberg MS Annese V Hakonarson H Brant SR Radford-Smith G Mathew CG Rioux JD Schadt EE Daly MJ Franke A Parkes M Vermeire S Barrett JC Cho JH Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491: 119–124, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato M, Sanada M, Kato I, Sato Y, Takita J, Takeuchi K, Niwa A, Chen Y, Nakazaki K, Nomoto J, Asakura Y, Muto S, Tamura A, Iio M, Akatsuka Y, Hayashi Y, Mori H, Igarashi T, Kurokawa M, Chiba S, Mori S, Ishikawa Y, Okamoto K, Tobinai K, Nakagama H, Nakahata T, Yoshino T, Kobayashi Y, Ogawa S. Frequent inactivation of A20 in B-cell lymphomas. Nature 459: 712–716, 2009. [DOI] [PubMed] [Google Scholar]
- 20.Khounlotham M, Kim W, Peatman E, Nava P, Medina-Contreras O, Addis C, Koch S, Fournier B, Nusrat A, Denning TL, Parkos CA. Compromised intestinal epithelial barrier induces adaptive immune compensation that protects from colitis. Immunity 37: 563–573, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolodziej LE, Lodolce JP, Chang JE, Schneider JR, Grimm WA, Bartulis SJ, Zhu X, Messer JS, Murphy SF, Reddy N, Turner JR, Boone DL. TNFAIP3 maintains intestinal barrier function and supports epithelial cell tight junctions. PLoS One 6: e26352, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kool M, van Loo G, Waelput W, De Prijck S, Muskens F, Sze M, van Praet J, Branco-Madeira F, Janssens S, Reizis B, Elewaut D, Beyaert R, Hammad H, Lambrecht BN. The ubiquitin-editing protein A20 prevents dendritic cell activation, recognition of apoptotic cells, and systemic autoimmunity. Immunity 35: 82–96, 2011. [DOI] [PubMed] [Google Scholar]
- 23.Lee EG, Boone DL, Chai S, Libby SL, Chien M, Lodolce JP, Ma A. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science 289: 2350–2354, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lippens S, Lefebvre S, Gilbert B, Sze M, Devos M, Verhelst K, Vereecke L, Mc Guire C, Guerin C, Vandenabeele P, Pasparakis M, Mikkola ML, Beyaert R, Declercq W, van Loo G. Keratinocyte-specific ablation of the NF-kappaB regulatory protein A20 (TNFAIP3) reveals a role in the control of epidermal homeostasis. Cell Death Diff 18: 1845–1853, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lodolce JP, Kolodziej LE, Rhee L, Kariuki SN, Franek BS, McGreal NM, Logsdon MF, Bartulis SJ, Perera MA, Ellis NA, Adams EJ, Hanauer SB, Jolly M, Niewold TB, Boone DL. African-derived genetic polymorphisms in TNFAIP3 mediate risk for autoimmunity. J Immunol 184: 7001–7009, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma A, Malynn BA. A20: linking a complex regulator of ubiquitylation to immunity and human disease. Nat Rev Immunol 12: 774–785, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madsen KL, Malfair D, Gray D, Doyle JS, Jewell LD, Fedorak RN. Interleukin-10 gene-deficient mice develop a primary intestinal permeability defect in response to enteric microflora. Inflamm Bowel Dis 5: 262–270, 1999. [DOI] [PubMed] [Google Scholar]
- 28.Matmati M, Jacques P, Maelfait J, Verheugen E, Kool M, Sze M, Geboes L, Louagie E, Mc Guire C, Vereecke L, Chu Y, Boon L, Staelens S, Matthys P, Lambrecht BN, Schmidt-Supprian M, Pasparakis M, Elewaut D, Beyaert R, van Loo G. A20 (TNFAIP3) deficiency in myeloid cells triggers erosive polyarthritis resembling rheumatoid arthritis. Nat Genet 43: 908–912, 2011. [DOI] [PubMed] [Google Scholar]
- 29.Musone SL, Taylor KE, Lu TT, Nititham J, Ferreira RC, Ortmann W, Shifrin N, Petri MA, Kamboh MI, Manzi S, Seldin MF, Gregersen PK, Behrens TW, Ma A, Kwok PY, Criswell LA. Multiple polymorphisms in the TNFAIP3 region are independently associated with systemic lupus erythematosus. Nat Genet 40: 1062–1064, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nava GM, Friedrichsen HJ, Stappenbeck TS. Spatial organization of intestinal microbiota in the mouse ascending colon. ISME J 5: 627–638, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nenci A, Becker C, Wullaert A, Gareus R, van Loo G, Danese S, Huth M, Nikolaev A, Neufert C, Madison B, Gumucio D, Neurath MF, Pasparakis M. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature 446: 557–561, 2007. [DOI] [PubMed] [Google Scholar]
- 32.Otsuka M, Kang YJ, Ren J, Jiang H, Wang Y, Omata M, Han J. Distinct effects of p38alpha deletion in myeloid lineage and gut epithelia in mouse models of inflammatory bowel disease. Gastroenterology 138: 1255–1265; e1251–e1259, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rhee L, Murphy SF, Kolodziej LE, Grimm WA, Weber CR, Lodolce JP, Chang JE, Bartulis SJ, Messer JS, Schneider JR, Paski S, Nero TM, Boone DL. Expression of TNFAIP3 in intestinal epithelial cells protects from DSS- but not TNBS-induced colitis. Am J Physiol Gastrointest Liver Physiol 303: G220–G227, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sellon RK, Tonkonogy S, Schultz M, Dieleman LA, Grenther W, Balish E, Rennick DM, Sartor RB. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun 66: 5224–5231, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shao L, Oshima S, Duong B, Advincula R, Barrera J, Malynn BA, Ma A. A20 restricts Wnt signaling in intestinal epithelial cells and suppresses colon carcinogenesis. PLoS One 8: e62223, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tavares RM, Turer EE, Liu CL, Advincula R, Scapini P, Rhee L, Barrera J, Lowell CA, Utz PJ, Malynn BA, Ma A. The ubiquitin modifying enzyme A20 restricts B cell survival and prevents autoimmunity. Immunity 33: 181–191, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turer EE, Tavares RM, Mortier E, Hitotsumatsu O, Advincula R, Lee B, Shifrin N, Malynn BA, Ma A. Homeostatic MyD88-dependent signals cause lethal inflammation in the absence of A20. J Exp Med 205: 451–464, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vereecke L, Sze M, Mc Guire C, Rogiers B, Chu Y, Schmidt-Supprian M, Pasparakis M, Beyaert R, van Loo G. Enterocyte-specific A20 deficiency sensitizes to tumor necrosis factor-induced toxicity and experimental colitis. J Exp Med 207: 1513–1523, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J, Ouyang Y, Guner Y, Ford HR, Grishin AV. Ubiquitin-editing enzyme A20 promotes tolerance to lipopolysaccharide in enterocytes. J Immunol 183: 1384–1392, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Willson TA, Jurickova I, Collins M, Denson LA. Deletion of intestinal epithelial cell STAT3 promotes T-lymphocyte STAT3 activation and chronic colitis following acute dextran sodium sulfate injury in mice. Inflamm Bowel Dis 19: 512–525, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zaph C, Troy AE, Taylor BC, Berman-Booty LD, Guild KJ, Du Y, Yost EA, Gruber AD, May MJ, Greten FR, Eckmann L, Karin M, Artis D. Epithelial-cell-intrinsic IKK-beta expression regulates intestinal immune homeostasis. Nature 446: 552–556, 2007. [DOI] [PubMed] [Google Scholar]