Abstract
Insertional leukemogenesis represents the major risk factor of hematopoietic stem cell (HSC) based gene therapy utilizing integrating viral vectors. To develop a pre-clinical model for the evaluation of vector-related genotoxicity directly in the relevant human target cells, cord blood CD34+ HSCs were transplanted into immunodeficient NOD.SCID.IL2rg−/− (NSG) mice after transduction with an LTR-driven gammaretroviral vector (GV). Furthermore, we specifically investigated the effect of prolonged in vitro culture in the presence of cytokines recently described to promote HSC expansion or maintenance. Clonality of human hematopoiesis in NSG mice was assessed by high throughput insertion site analyses and validated by insertion site-specific PCR depicting a GV typical integration profile with insertion sites resembling to 25% those of clinical studies. No overrepresentation of integrations in the vicinity of cancer-related genes was observed, however, several dominant clones were identified including two clones harboring integrations in the ANGPT1 and near the ANGPT2 genes associated with deregulated ANGPT1- and ANGPT2-mRNA levels. While these data underscore the potential value of the NSG model, our studies also identified short-comings such as overall low numbers of engrafted HSCs, limited in vivo observation time, and the challenges of in-depth insertion site analyses by low contribution of gene modified hematopoiesis.
Keywords: clonal dominance, insertional mutagenesis, murine xenotransplant model, retroviral vector
Introduction
Hematopoietic gene therapy employing autologous hematopoietic stem cells (HSCs) constitutes an attractive strategy for the treatment of congenital disorders of the hematopoietic system. For severe diseases, it represents the only curative alternative to allogeneic bone marrow transplantation (BMT), particularly if a suitable HLA-matched donor is not available. Current approaches utilize integrating retroviral vectors for the transfer of a functional therapeutic gene copy into autologous HSCs in vitro before they get reinfused into the patient. Groundbreaking clinical studies in life-threatening hematological disorders such as primary immunodeficiencies (reviewed in ref. 1) have demonstrated the therapeutic efficacy of hematopoietic gene therapy showing reconstitution of the respective blood lineages with functionally corrected cells, clearance of infections, or independence from replacement therapies. However, in four independent studies, patients developed hematopoietic malignancies following therapy.2,3,4,5 A causal link between the gene therapeutic intervention and these malignancies was established by the demonstration of the transcriptional activation of known proto-oncogenes like LMO2, CCND2, BMI1, PRDM16, and MDS-EVI1 by retroviral vector integrations close to or in these genes. However, besides insertional mutagenesis, additional factors such as the preconditioning chemotherapy or the ex vivo culture of the transplanted cells in the presence of cytokines may have contributed to the induction of these malignancies.
In addition to the ex vivo culture of HSCs during gene therapy approaches, the expansion of transplantable HSCs in vitro represents a highly attractive goal given the limited numbers of available donor cells in allogeneic stem cell transplantations, particularly when single cord blood units are used as donor material. Therefore, a plethora of different strategies including the use of novel cytokines,6 co-culture systems,7,8 or small molecules9 have been evaluated for the expansion of long-term engrafting HSCs. However, prolonged culture with increased proliferation of hematopoietic stem and progenitor cells might raise new safety concerns in the context of gene therapy as cell clones harboring integrations near critical genes may proliferate overly and accumulate additional chromosomal aberrations already in vitro, thereby increasing the overall risk of adverse events for the patients.10 In this respect, thorough long-term monitoring of integrations in or near critical genes following the reinfusion of gene modified cells is of utmost importance. Linear amplification mediated (LAM)-PCR11 and derived methods to detect viral integrations12,13,14 in combination with high throughput sequencing technologies allow to comprehensively monitor the clonal repertoire of the gene modified hematopoiesis in the patients, but in the end cannot prevent clonal outgrowth. Thus, highly sensitive and well characterized preclinical models are needed to test integrating viral vectors prior to clinical application. In this context, several murine in vitro15 and in vivo16,17 as well as large animal models10,18,19,20 have been established and allow for long-term observations. Because the occurrence of insertional leukemias in the murine syngeneic model is a rare event, the development of clonal dominance serves as readout for the comparison of gene therapy vectors.21,22,23 However, given the inherent differences in the transformation potential between human and rodent cells,24,25 the generation of models evaluating the vector-related genotoxicity directly in the relevant human target cells appears highly warranted. Xenotransplantation of gene modified human CD34+ cells into immune-deficient mouse strains represent an attractive tool to achieve this aim.26,27,28 Although the xenotransplant model allows the evaluation of gene transfer efficacy,29 there is so far no report on clonal dominance or leukemia induction caused by retroviral insertional mutagenesis in human cells in this model. Therefore, in this study we explored the xenotransplant model for the assessment of genotoxicity after retroviral gene transfer into human HSCs subjected to different in vitro transduction and expansion protocols.
Results
In vitro expansion of CB-CD34+ cells in different cytokine conditions
Pilot experiments (n = 4) were performed to establish the in vitro expansion protocol. In these studies, 1.1–2.0 × 105 human CB-CD34+ cells were transduced and expanded in four different cytokine conditions (Table 1) for a total of 10 days. The combination of the cytokines SCF, THPO, and FLT3-L (referred to as “STF”) represented the baseline standard. The second approach evaluated the combination of G-CSF with STF (referred to as “GCSF”).19 In addition, two recently proposed HSC expansion protocols using either SCF, THPO, FGF1, IGFBP2, and Angiopoietin-like-5 (referred to as “Angptl5”)6 or the cytokines SCF, THPO, FLT3-L, IL-6, and the small molecule StemRegenin (referred to as “SR1”)9 were investigated. Cultivation in the GCSF cytokine combination yielded the highest proliferation of total cells (121 ± 48 fold), while Angptl5-cultured cells proliferated the least (41 ± 18 fold; Figure 1a). Likewise the total number of CD34+ cells increased between 8- and 40-fold with the highest expansion observed in the SR1-containing medium (Figure 1b). Although the relative contribution of CD34+ cells dropped substantially during the 10 days of in vitro culture, it remained highest in the SR1 medium (35.6% ± 1.5% vs. 16.6% ± 2.9% STF, 8.6% ± 1.1% GCSF, 13.7% ± 1.6% Angptl5; Figure 1c,d) and here also higher CD34 expression levels per cell were observed as measured by the mean fluorescence intensity (Figure 1e). In agreement with the expansion of CD34+ cells, also the highest number of colony forming cells was present in the SR1 cultures after 10 days. However, the potential of colony formation per cell decreased with increased culture time. In this comparison also SR1 cultured cells had the highest CFU potential, which was significantly higher than in GCSF cultures (Figure 1f,g).
Table 1. Cytokine conditions for in vitro culture of human CD34+ cells.

Figure 1.
In vitro characteristics of expanded CD34+ cells. (a) Cord blood-derived CD34+ cells were expanded with four different cytokine conditions for 10 days, the total cell numbers counted and the fold expansion of total cells calculated (mean ± SD, n = 4). (b) Expansion of CD34 marker positive cells after 10 days of culture in the four different cytokine conditions (mean ± SD, n = 3). (c) Representative flow cytometric analysis of CD34 marker expression after four (STF d4) and 10 days of culture in the different cytokine conditions compared to the expression on CD34+ enriched and uncultured cord blood cells. (d) Contribution (%) of CD34+ cells in four and 10 days expanded cells in comparison to day 0 cells (mean ± SD, n = 3). (e) CD34 expression levels, as measured by the Mean fluorescence intensity (MFI), on cultured cells at days 4 and 10 correlated to the expression level (MFI) in cells cultured with STF at the respective day of analysis (this value set to one, mean ± SD, n = 3). (f) Number of colonies of transduced and expanded CB-CD34+ cells plated after 4 and 10 days of culture (mean ± SD), compared to cells plated without in vitro culture (black column). Colony numbers per 1 × 103 cells plated cells/assay, three experiments were analyzed each, in triplicate assays. (g) Fold increase of colony forming cells in the cultures during the 4 and 10 days of culture. *P < 0.05; **P < 0.01; ns, not significant (n = 3, in triplicate assays each).
In vitro expanded cells engraft long-term in NOD.SCID.IL2rg−/− mice
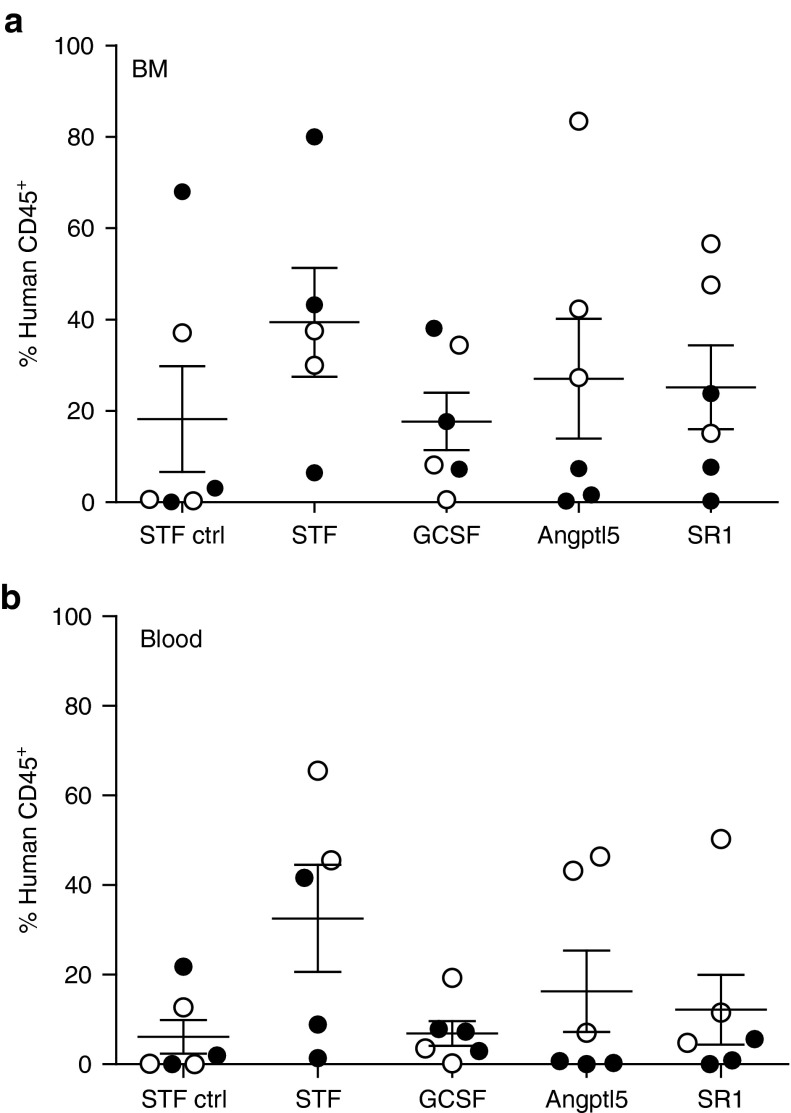
Next, we investigated the potential of in vitro transduced and expanded CB-CD34+ cells to engraft and maintain hematopoiesis in NOD.SCID.IL2rg−/− (NSG) mice. Per mouse 5 × 104 cells transduced with the gammaretroviral (GV) vector RSF91.eGFP.pre* and cultured for a total of 4 (STF-ctrl) or 10 days were transplanted (Supplementary Materials and Methods). Two independent experiments were carried out resulting in n = 5–6 mice per experimental group (Table 2). In both experiments, the transduction efficiency was beyond 90%, independent of the cytokine conditions employed during transduction (Supplementary Figure S1). In a control group, NSG mice were transplanted with cells transduced and cultured only for 4 days in STF medium (STF-ctrl; n = 6 mice). In all groups, the contribution of human CD45+ leukocytes in the peripheral blood increased over time indicating effective engraftment (Supplementary Figure S2). After 24 weeks, animals were sacrificed and the human engraftment and lineage contribution in the different hematopoietic tissues were determined by FACS analysis (Figure 2 and Supplementary Figure S3). In all groups, contribution of human cells to overall hematopoiesis varied substantially. There was a tendency towards lower BM engraftment and less human cell contribution to the blood in mice of the GCSF and STF-ctrl groups (Figure 2a,b); however, this was not significant by Kruskal–Wallis analysis across the groups, P = 0.5; (pairwise test in the blood: STF-ctrl versus STF P = 0.08, GCSF versus STF P = 0.1, Mann–Whitney test). In these mice, also human T-cell contribution was low (Supplementary Figure S4) in line with an insufficient thymus repopulation as well as peripheral blood mobilization observed in these groups. In the reconstituted thymi, human T-cell development was normal with human CD4/CD8 double- and single-positive T-cells detectable (Supplementary Figure S4).
Table 2. Experimental groups.

Figure 2.
Characterization of the human hematopoiesis in NSG mice. Contribution of human cells to the (a) BM and (b) blood. BM and blood cells were analyzed for the expression of human CD45 (each dot represents a transplanted mouse, filled circle: mice in the first experiment, open circle: mice in the second experiment; mean ± SEM).
The contribution of retrovirally transduced cells to hematopoiesis was variable but higher in the second experiment (Supplementary Figure S5). The number of GFP positive cells detected by FACS in individual mice correlated well with the detected vector copy number as measured by quantitative PCR (Supplementary Table S1). Over the course of our study, blood cell counts stayed well within the physiological range without any signs of abnormalities (Supplementary Figure S6). Furthermore, no major hematological abnormalities were detected by histopathological analysis of mouse organs (Supplementary Figure S7). In summary, engraftment of gene modified human CB-CD34+ cells was variable between mice but there was a tendency of improved engraftment after in vitro expansion in STF, Angptl5 or SR1 as compared to GCSF conditions or transplantation of unexpanded cells.
Typical GV integration pattern without selection of clones with integrations near cancer-associated genes in vivo
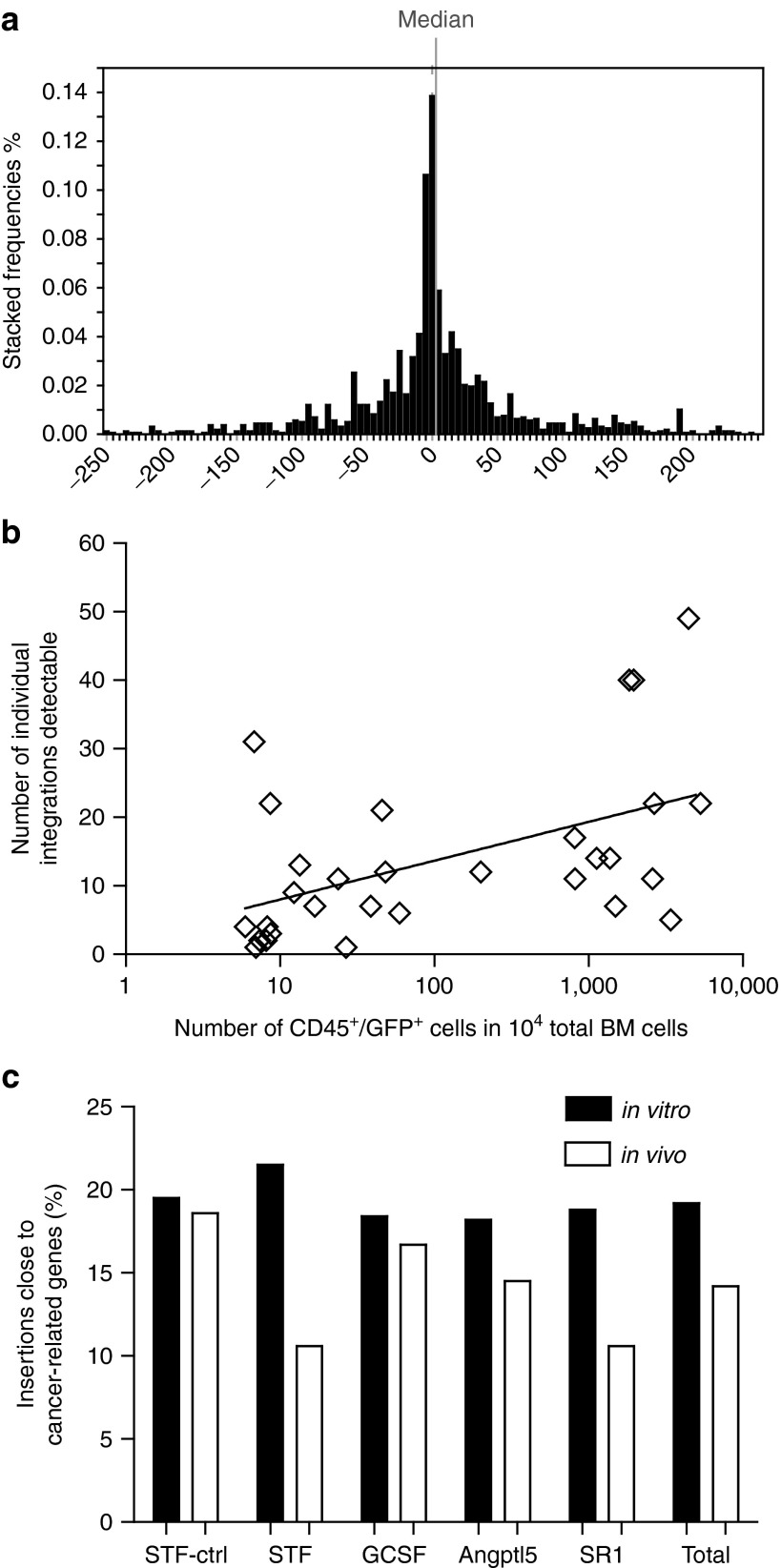
Linear amplification-mediated (LAM) PCR11 and 454-pyrosequencing were performed to detect GV vector integration sites. A combination of three restriction enzymes was used for LAM-PCR to detect insertion sites in the BM of 29 transplanted mice and 6 in vitro samples collected before transplantation (pre-TX). A total of 1615 integrations (1,197 in pre-TX, 418 in BM samples) were retrieved displaying an insertion profile typical of a GV vector with integrations clustering around the transcription start sites (TSS; Figure 3a). Median distance to the TSS was 0.7 kb and the frequency of intronic integrations was 62.8% with no significant difference between the pre-TX and the in vivo samples for these parameters. We did observe, however, a significantly lower number of insertions within a 10 kb distance to the TSS for the in vivo samples arguing against an in vivo selection for clones with insertions close to the TSS. Between 1 and 48 individual integration sites were recovered per mouse (Supplementary Table S2) and the number of integrations correlated directly to the human CD45+GFP+ cell engraftment in the BM (Spearman r = 0.52, P = 0.003; Figure 3b).
Figure 3.
Integration characteristics of the gammaretroviral vector RSF91.eGFP.pre. (a) The gammaretriviral vector integrations retrieved from all NSG mice and the pre-TX samples clustered around the TSS (0) of cellular genes; the median distance from the TSS was +0.7 kb indicated by the grey line. (b) The number of retrieved integrations per sample correlated with the amount of gene modified human cells (Spearman r = 0.52, P = 0.003). The numbers of gene modified human cells are determined by the percentages of GFP+/CD45+ cells within 10,000 total bone marrow cells. (c) Percentages of integrations retrieved from the pre-TX samples (black bar) and transplanted mice (white bar) in or close to cancer-related genes (cytokine condition as indicated) compared to genes listed in the Network of Cancer-related Genes Database (NCG), Retroviral Tagged Cancer Gene Database (RTCGD) and the Cancer Gene List assembled by the Bushman lab.
As the use of restriction enzymes may be problematic for the detection of certain insertion sites and could potentially interfere with the quantification by sequence reads,30 we also performed non-restricted (nr)LAM13 and re-free LAM PCRs.12 Due to the lower sensitivity of these methods, this analysis was only performed in mice with high numbers of gene modified cells in the BM. Here, 31 insertions in six mice were recovered by the re-free LAM PCR and 55 insertions in three mice by the nrLAM-PCR method (Supplementary Table S3). These 86 insertions were allocated to 74 unique genes, out of which 18 (24%) overlapped with those detected by the conventional LAM-PCR, but only three (4%) were identified by both non-restricted methods. Based on eight mice analyzed by repeated LAM and non-restricted LAM-PCRs, the overall pool size was calculated to 135 insertions per mouse using Chapman estimation (Supplementary Figure S8).
To identify a potential selection of integrations in or close to known proto-oncogenes we compared the integrations in mice and preTx samples with genes listed in the Retroviral Tagged Cancer Gene Database (RTCGD),31 the Network of Cancer-related Genes (NCG)32 and a compiled Cancer Gene List from the Bushman lab (www.bushmanlab.org/links/genelists). There were no significant difference between the individual in vitro (χ2 P = 0.8979) or in vivo groups (χ2 P = 0.4587). In all groups, a reduction of hits close to cancer related genes was observed in vivo; however, this is not significantly different for any of the groups (Fisher's exact test, P = 0.0666 (Figure 3c)). This was also true for the fold changes in hits close to oncogenes for each mouse calculated separately (Supplementary Figure S9). These data argue against a general enrichment of clones with insertions close to proto-oncogenes in vivo or during in vitro expansion.
Insertion sites in the NSG model recapitulate data from clinical trials
We further compared the insertions identified in our study with the integration data from five clinical trials and three preclinical studies using GV reported by Deichmann and colleagues.33 We found a total overlap of 217 (21.4%) insertions with the Deichmann study. In addition, among all insertions recovered in our study, 45 common insertion sites (CIS) of 4th or higher order, as defined by Wu and colleagues34 were detected (Table 3) with nine of these CIS observed near or in cancer-related genes. For these CIS again a considerable overlap with insertions reported by the Deichmann study (13/45, 29%) was observed. Furthermore, the ANGPT1 locus was identified as a CIS of higher order (6th in our analysis; 5th in the Deichmann study). These results underline the fact, that clinically relevant insertion sites can be retrieved in the humanized mouse model.
Table 3. Common insertions sites.

Human dominant clones arise in NSG mice
As we expected higher sequence reads of integration sites in dominant clones compared to clones of low abundance, we analyzed the occurrence of high read insertions in the mice. Although high read insertions (>100 reads) were detected in all transplant groups (23/418 total integrations for LAM-PCR analysis (Figure 4a) and 16/86 integrations for nrLAM and referee LAM-PCR) there was no significant difference in their occurrence between the groups (Supplementary Figure S10).
Figure 4.
Contribution of individual integrations quantified by insertion site-specific qPCR. (a) Number of sequence reads displayed for each integration from all mice of the indicated groups collected by LAM-PCR analysis. The black line marks the 100-reads threshold. (b) Human engraftment (identified by hCD45 expression), contribution of gene modified cells (GFP expression) detected by flow cytometric analysis and contribution of specific insertion sites measured by insertion site-specific (IS)-qPCR to the total BM cells. (*this insertion site was detected by the non-restricted LAM PCR). One further integration was detected in the PB (24 weeks), but not BM, of Mouse M77 at a contribution of 0.4% by IS-qPCR and not included in the graph. Below the x-axis the identity of the mice with the specific insertion sites are indicated. (c) Comparison of the contribution of clones with specific integration sites in the indicated genes quantified by the sequencing read counts (white bars) with the contribution of the clones as measured by insertion-site specific qPCR (black bars) performed on DNA isolated from BM cells at the end of the observation time. The measurements are displayed as percentages of the gene modified BM cells. (d–h) Contribution of integrations as indicated by closed black symbols to the blood at different time points. (d) ANGPT1 and EHD1 integrations in mouse M69. (e) ANGPT2 in mouse M53. (f) PMVK and TXNDC2 integrations in M65. (g) C16orf7 in mouse M65. (h) ERMP1 integration in mouse M63. (d–h) All insertion sites were quantified by insertion site specific qPCRs and displayed as contribution to total blood leukocytes. The contributions of the human cells in the blood are shown with open squares and the GFP positive blood cells by open circles. Please note the different scales of the y-axis in the different blots. In (f), almost all human cells were GFP positive and, therefore, the lines are blotted on top of each other.
To verify the presence and quantify the contribution of clones with specific insertion sites, we performed insertion site-specific (IS) quantitative (q)PCRs on BM-DNA of six mice with varying human engraftment but good contribution of transgenic cells (Figure 4b). We selected eleven integrations of which five had particularly high read counts. We confirmed four of the high read insertions by IS-qPCR in the BM (ANGPT1, ANGPT2, EHD1, TXNDC2). One insertion site was detected as dominant clone although the read counts did not predict this (PMVK). In three cases, the estimation of abundance by insertion site analysis was >30-fold different than by IS-qPCR (ERMP1, CD3EAP, ARHGEF, Supplementary Table S4). In general, IS-qPCR detected higher contributions to the transduced hematopoiesis in two and lower in nine cases (Figure 4c and Supplementary Table S4), indicating that the sequence reads mostly overestimated the abundance of clones.
In a next step, we tried to quantify these insertions in the peripheral blood over time by IS-qPCR. Here, in mouse M69 a clone with an integration in the fourth intron of ANGPT1 became progressively dominant in the gene-modified human hematopoiesis (2.1–29.4%; week 9–24, Figure 4d). In the same mouse, we further detected a clone with an insertion site close to EHD1 with almost the same kinetics as the ANGPT1 clone indicating that both insertions probably were contained in the same cell clone. A dominant clone harboring an integration in reverse orientation 5.8 kb upstream of the ANGPT2 gene progressively contributed to human hematopoiesis in mouse M53 (Figure 4e). In mouse M65, a clone with an integration 387 bp downstream of the TSS of PMVK with increasing blood contribution was quantifiable as well as a low abundance clone with an insertion close to TXND2 which was dominant in the BM (Figure 4f). Two additional integrations—C16orf7 (mouse M70) and ERMP1 (mouse M63)—were tracked over time in blood samples albeit their contribution to overall human hematopoiesis was low (Figure 4g,h). Thus, we were able to validate integration sites by IS-qPCR and to track clones by their insertion site in the blood. We also identified dominant clones with increasing contribution to gene modified hematopoiesis over time.
Viral vector integrations caused deregulated expression of human cellular genes
To investigate whether the occurrence of dominant clones led to deregulation of gene expression due to vector integration, we analyzed expression of ANGPT1, ANGPT2, and PMVK in the three animals harboring these integrations. The mRNA levels of ANGPT1 in mouse M69 were significantly reduced in comparison to other transplanted NSG mice not containing this integration (Figure 5a, left panel) and CD34+ control cells. The integration site was located in the fourth intron in sense orientation (Figure 5b), which was also a CIS in our study, and may have interfered with cellular splicing; however, we were not able to detect fusion transcripts between the vector and gene. Sequencing of ANGPT1 mRNA detected alternative splice forms lacking exon5 coding sequence, arguing for altered splicing. In contrast, in mouse M53 the GV integration caused a significant upregulation of the ANGPT2 mRNA levels (~50 times, Figure 5a, middle panel) which was in agreement with the typical GV integration close to the promoter, favoring enhancer-mediated interactions between the vector enhancer and the cellular promoter (Figure 5c). PMVK expression in mouse M65 was not significantly deregulated compared to the other mice (Figure 5a, right panel). The inability to detect altered gene expression in this case may in part be due to the low contribution of the human hematopoiesis in this specific mouse, although the clone was dominant within the human transduced cells.
Figure 5.
Vector integration alters cellular gene expression. Expression of the gene close to the insertion site was determined by qPCR: (a) ANGPT1 in BM samples of mouse M69 (left, arrow); ANGPT2 in BM samples of mouse M53 (middle); PMVK in BM samples of mouse M65 (right). Expression levels were normalized to mobilized peripheral blood CD34+ cells, CD8+ T-cells served as a negative control (mean ± SD). The expression was further compared to other mice of the study with different integration sites (mean ± SD). Student's t-test: **P < 0.01; n.d. = not detectable. (b) Insertion in the ANGPT1 gene took place in the fourth intron in forward orientation with a distance to the TSS of 192183 bp. This position in the ANGPT1 gene was also a CIS; other insertions detected in ANGPT1 in our study are indicated by flags. All other insertions were also in forward orientation. Exons are indicated as black boxes and labeled Ex1-Ex9. The vector integration and orientation is shown in detail in the blow up of the intron 4. (c) The insertion in ANGPT2 was located 5675 bp upstream of the transcription start site (TSS) in reverse orientation.
Discussion
In this study, we addressed the question whether we could detect genotoxic events in the humanized mouse model. For transduction, we employed a GV LTR vector with a known genotoxic potential expressing eGFP as a marker gene16,35 and expanded the cells under different cytokine conditions before transplantation. The potential development of clonal dominance was investigated by insertion site analysis in the BM at the end of the observation time of 6 months. The combination of the GV LTR vector and the prolonged culture with the risk of selection of dominant clones even before transplantation increased the chance to induce clonal imbalances in vivo. The experimental setup was, therefore, designed to test the potential of the humanized mouse model to read out gene therapy-related genotoxic events.
The engraftment with gene modified human cells was efficient but highly variable between mice and thus made the discrimination between the different media conditions used for in vitro expansion difficult. Specifically, the superior in vitro expansion of CD34+ cells by SR1 was not reflected by better engraftment in NSG mice. There was, however, a tendency of improved engraftment of cells expanded in STF, Angptl5, or SR1 condition in contrast to expansion in GCSF. While the transplantation of fewer cells may have yielded more distinct differences between expansion conditions,9 this was incompatible with the aim of our study, as in our model the assessment of genotoxicity relies on a high number of transduced and transplanted CD34+ cells.
Employing LAM-PCR, we recovered 418 in vivo-insertion sites with one to 48 per mouse, on the basis of an estimated pool of ~135 insertions per mouse. In comparison, Greene and colleagues reported 82, 38, and 27 unique insertion sites in three NSG mice,36 but in this study, four times more CD34 cells were transplanted. Based on limiting dilution assays37 or recent studies employing barcoding strategies in the NSG model that estimated the number of long-term engrafting HSCs to only 0.02% of cord blood CD34+ cells,38 we can estimate a number of 10–25 long-term repopulating HSCs per mouse in our experiments (not taking into account a potential effect of the in vitro culture on this number). As we identified the insertion sites 24 weeks after transplantation, we can assume that our analysis was focused on long-term repopulating HSCs.
We performed nrLAM- and referee LAM-PCR to correct for the bias introduced by the use of restriction enzymes.30 Although these methods have the disadvantage of lower sensitivity and a higher need for input DNA compared to the conventional LAM-PCR, we detected insertion sites not identified by conventional LAM-PCR (e.g. in ANGPT1). However, the overall number of individual insertion sites per mouse detected by the non-restricted methods was lower than from the conventional LAM-PCR. Importantly, only the use of insertion site-specific qPCR allowed us to unequivocally determine clone size. Thus, among the ten investigated insertion sites, only in half of them a dominant contribution to human hematopoiesis was confirmed. This recapitulates our recent results in the humanized mouse model using MGMT-expressing vector27 and highlights the technical limitations associated with quantitative analysis of clonality based on read counts also observed in other studies.39,40,41 Our data suggest, however, that the read count approach represents a valid selection criterion for further analysis.
Most importantly, our study provides clear evidence that clonal dominance within the human hematopoiesis can be detected and quantified in the xenograft model. This is in contrast to the published data for the evaluation of a clinical grade vector for the treatment of Wiskott Aldrich Syndrome42 and most likely reflects the increased mutagenic potential of our LTR-driven GV vector in combination with the prolonged in vitro expansion culture.
We detected dominant clones with an activating integration closely upstream of the TSS of ANGPT2 in one, and downregulation of ANGPT1 with the vector integration in the fourth intron in the other. Both angiopoietins bind to the Tie2 receptor which is expressed on long-term repopulating HSCs. A role of Tie2 in maintaining HSC quiescence and self-renewal was demonstrated in mouse models.41 The overexpression of ANGPT1 in the BM niche during steady-state-hematopoiesis increased the number of LSK cells.40 However, our experiments were based on the in vitro transduction, culture and transplantation of HSCs and their hematopoietic reconstitution after transplantation. The reciprocal regulation of the ANGPT genes as seen in our study may be benificial in this situation. However, the function of ANGPT2 in hematopoiesis is not well examined. Both ANGPTs are secreted from BM niche cells and only at very low levels expressed in HSCs under normal conditions (Supplementary Figure S11).
Following the contribution to the gene modified hematopoiesis in the blood, the ANGPT1, ANGPT2, EHD1, and PMVK clones progressively took over the gene modified compartment, indicating a growth advantage over the other clones. We, therefore, confirmed three dominant clones in thirty mice and calculated an incidence of clonal dominance of 2 × 10-6 per transplanted cell and 3.3 × 10-6 per vector insertion. We did not detect leukemic transformation of human cells in the xenograft model which in the murine BMT model using the same gammaretroviral vector occurred with an estimated frequency of <2 × 10–7 per transplanted cell43 and up to 10–6 after high dose transduction and in vitro expansion of transduced cells before transplantation.16 Clonal dominance in the murine model occurred in almost every mouse when using the gammaretroviral LTR vectors containing strong enhancer/promoter elements as used in our study;44,45 however, in general, more stem cells are transplanted in the murine model. From these estimations, it can be concluded that the occurrence of clonal dominance in the humanized mouse model was only slightly lower than in the murine model, but that the low number of initially transduced and transplanted CD34+ cells precluded a comprehensive comparison between our groups. Furthermore, we did not observe leukemic transformation of human cells. This can be explained by the inherent differences between human and murine cells in respect to transformation,24 but complicates the development of humanized genotoxicity models. In addition, although the observation time was long enough to detect clonal dominance, for leukemic transformation longer observation times, including secondary transplantations, may be needed. Taken together, our study sheds light on the current obstacles that need to be addressed for efficient use of humanized mouse models to assess genotoxicity. It further supports the cautious interpretation of gene therapy safety analysis in respect to genotoxicity in the humanized mouse model based on a similar study design.
Materials and Methods
Virus production. Human CD34+ cells were transduced with the gammaretroviral vector RSF91.eGFP.pre*46 pseudotyped with the modified feline endogenous virus glycoprotein RD114/TR.47 Infectious viral particles were produced with a split packaging system in human embryonic kidney 293T cells48 and concentrated by ultracentrifugation, resuspended in StemSpan (Stemcell Technologies, Vancouver, Canada) and stored at −80 °C.
CD34+ cell isolation, culture, and transduction. Cord blood (CB) samples were obtained from the Hannover Medical School following written consent of the donors as approved by the Hannover Medical School local ethics committee. Total nucleated cells were isolated by a Ficoll gradient followed by enrichment for CD34+ cells employing MACS purification (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany). Isolated cells were frozen until further usage. CB-CD34+ cells were cultured in StemSpan medium supplemented with 1% penicillin/streptomycin and one of four cytokine combinations using hSCF, hTHPO, hFLT3-L, hGCSF, hIGFBP2, hAngptl5, hFGF-1, and hIL6 (PeproTech, Rocky Hill, CT). The StemRegenin compound (Cellagen Technology, San Diego, CA) was used at a concentration of 1 µmol/l as described.9
5 × 104 CB-CD34+ cells/mouse were transduced two times (days 2 and 3) on Retronectin with an MOI of 20 each day. Cells were transplanted the next day (day 4) or expanded for further 6 days. For more detailed information, see Supplementary Materials and Methods.
Colony assays. Colony Assays were performed in triplicates using Methocult H4034 Optimum (Stem Cell Technologies). CB-CD34+ cells were plated at a density of 4-5 × 102 cells (uncultured cells; 80–90% purity), 0.5-1 × 103 cells (day 4), and 1 × 103 cells (day 10).
Mouse experiments. NOD.SCID.IL2rg−/− (NSG) mice49 aged 6–8 weeks received a sublethal irradiation dose of 3 Gy 24 h before transplantation (TX). Progenies of 5 × 104 CB-CD34+ cells were transplanted per mouse. The experiments were done in two cohorts with transplantation performed on different days. Each cohort was transplanted from the same pool of donor CB-CD34+ cells but two different pools were used for the two cohorts. Starting at 6 weeks post TX, blood samples were taken in intervals of 3 weeks for blood counts, flow cytometric analysis and DNA isolation. Mice were sacrificed at 24 weeks, cells from the peripheral blood, BM and thymus isolated and analyzed by flow cytometry using antibodies as specified in Supplementary Materials and Methods.
Linear amplification mediated (LAM)-PCRs and high throughput sequencing. Conventional, restriction enzyme-free (re-free) and non-restrictive (nr) linear amplification mediated (LAM)-PCR were performed as described earlier.11,12,13 For further information, see Supplementary Materials and Methods.
Insertion site-specific qPCR and qRT-PCR. Individual vector integrations were validated using reverse primers specific for the genomic region designed according to the 454-sequences, the NCBI BLAST search results and the NCBI reference sequences. The LTR primer located at the 5'end of the LTR (JM354) from the nested PCR step during the LAM-PCR served as the forward primer for validation of all integration sites. Subsequently, real-time TaqMan PCR (Life Technologies GmBH, Frankfurt, Germany) was carried out on all the PB samples collected at different time points using a StepOnePlus (Applied Biosystems, Carlsbad, CA). Additionally, the VCN was determined using primers detecting the woodchuck posttranscriptional element (PRE) and polypyrimidine tract-binding protein 2 (PTBP2) as an internal reference for both quantitative PCRs (qPCRs). All qPCRs were performed in triplicates. A plasmid standard harboring sequences for the residual part of the gammaretroviral LTR, PRE, and PTPB2 was used for the quantification (see Supplementary Materials and Methods for the list of all primers).
Quantitative RT-PCR was done to evaluate gene expression in total RNA isolated from the BM using Quantitect primer assays (QIAGEN) and quantified using SYBR Green on the StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, CA). Expression of beta-ACTIN served as control.
Bioinformatic analysis of viral integration sites. Post-sequencing data processing was carried out with customized PERL (Practical Extraction and Report Language) and EXCEL-based VBA (Visual Basic for Application) scripts as described before.27,39,50 For sequence alignment the freely accessible MAVRIC (Methods for Analyzing Viral Integration Clusters) online tool was employed (http://mavric.erasmusmc.nl/index.php).51 A detailed description of data procession and the basic parameters used for MAVRIC are provided in the Supplementary Materials and Methods.
Statistical tests. Statistical significance was determined by non-parametric t-test, two-way analysis of variance test and Spearman's rank correlation.
SUPPLEMENTARY MATERIAL Figure S1. Gammaretroviral vector and transduction efficiency. Figure S2. Development of the human chimerism and presence of transduced cells in the peripheral blood of transplanted NSG mice. Figure S3. Contribution of human hematopoiesis to the different blood cell lineages in the BM and blood. Figure S4. Thymus reconstitution in transplanted NSG mice. Figure S5. Number of GFP positive cells in the BM and blood. Figure S6. Peripheral blood cell counts. Figure S7. Histopathological analysis of different tissues. Figure S8. Pool size calculation by Chapman Estimation. Figure S9. Fold change of insertion sites close to oncogenes in each mouse. Figure S10. Number of reads per insertion site for each mouse. Figure S11. Expression of Tie1, Tie2, ANGPT1 and ANGPT2 in CD34+ cells and the mice M53 and M69. Table S1. Overview of transplanted cell numbers and vector copies per mouse. Table S2. Overview of the insertion site recovered by LAM-PCR in transplanted NSG mice. Table S3. Overview of the insertion site analysis using nrLAM and referee LAM PCR. Table S4. Correlation of sequence reads and quantification by IS-qPCR. Materials and Methods.
Acknowledgments
We thank Sabine Knoess, Doreen Lüttge, and Rena-Mareike Struss for their excellent technical support; Jannik Daudert for his help in developing scripts to evaluate integration analysis data (all from the Institute of Experimental Hematology, Hannover Medical School); and Jörg Frühauf for his assistance during irradiation of mice (Institute of Radiology, Hannover Medical School). This work was supported by grants from the Deutsche Forschungsgemeinschaft: Cluster of Excellence REBIRTH (Exc 62/1; T.M.) and SPP1230 grant MO 886/3–1 (U.M. and T.M.), the European Union (Cell-PID, M.R. and U.M.) and the LOEWE Center for Cell and Gene Therapy Frankfurt, funded by the Hessian Ministry of Higher Education, Research and the Arts (III L 4-518/17.004, 2013) (U.M.). R.P. was partially supported by a stipend from Hannover Biomedical Research School. The authors declare no conflict of interest.
Supplementary Material
References
- Candotti F. Gene transfer into hematopoietic stem cells as treatment for primary immunodeficiency diseases. Int J Hematol. 2014;99:383–392. doi: 10.1007/s12185-014-1524-z. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Garrigue A, Wang GP, Soulier J, Lim A, Morillon E, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe SJ, Mansour MR, Schwarzwaelder K, Bartholomae C, Hubank M, Kempski H, et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J Clin Invest. 2008;118:3143–3150. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein S, Ott MG, Schultze-Strasser S, Jauch A, Burwinkel B, Kinner A, et al. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nat Med. 2010;16:198–204. doi: 10.1038/nm.2088. [DOI] [PubMed] [Google Scholar]
- Braun CJ, Boztug K, Paruzynski A, Witzel M, Schwarzer A, Rothe M, et al. Gene therapy for Wiskott-Aldrich syndrome–long-term efficacy and genotoxicity. Sci Transl Med. 2014;6:227ra33. doi: 10.1126/scitranslmed.3007280. [DOI] [PubMed] [Google Scholar]
- Zhang CC, Kaba M, Iizuka S, Huynh H, Lodish HF. Angiopoietin-like 5 and IGFBP2 stimulate ex vivo expansion of human cord blood hematopoietic stem cells as assayed by NOD/SCID transplantation. Blood. 2008;111:3415–3423. doi: 10.1182/blood-2007-11-122119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JM, Nolan DJ, Vertes EL, Varnum-Finney B, Kobayashi H, Hooper AT, et al. Endothelial cells are essential for the self-renewal and repopulation of Notch-dependent hematopoietic stem cells. Cell Stem Cell. 2010;6:251–264. doi: 10.1016/j.stem.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lima M, McNiece I, Robinson SN, Munsell M, Eapen M, Horowitz M, et al. Cord-blood engraftment with ex vivo mesenchymal-cell coculture. N Engl J Med. 2012;367:2305–2315. doi: 10.1056/NEJMoa1207285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boitano AE, Wang J, Romeo R, Bouchez LC, Parker AE, Sutton SE, et al. Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science. 2010;329:1345–1348. doi: 10.1126/science.1191536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers S, Gomes TJ, Larochelle A, Lopez R, Adler R, Krouse A, et al. Ex vivo expansion of retrovirally transduced primate CD34+ cells results in overrepresentation of clones with MDS1/EVI1 insertion sites in the myeloid lineage after transplantation. Mol Ther. 2010;18:1633–1639. doi: 10.1038/mt.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Schwarzwaelder K, Bartholomae C, Zaoui K, Ball C, Pilz I, et al. High-resolution insertion-site analysis by linear amplification-mediated PCR (LAM-PCR) Nat Methods. 2007;4:1051–1057. doi: 10.1038/nmeth1103. [DOI] [PubMed] [Google Scholar]
- Wu C, Jares A, Winkler T, Xie J, Metais JY, Dunbar CE. High efficiency restriction enzyme-free linear amplification-mediated polymerase chain reaction approach for tracking lentiviral integration sites does not abrogate retrieval bias. Hum Gene Ther. 2013;24:38–47. doi: 10.1089/hum.2012.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paruzynski A, Arens A, Gabriel R, Bartholomae CC, Scholz S, Wang W, et al. Genome-wide high-throughput integrome analyses by nrLAM-PCR and next-generation sequencing. Nat Protoc. 2010;5:1379–1395. doi: 10.1038/nprot.2010.87. [DOI] [PubMed] [Google Scholar]
- Brady T, Roth SL, Malani N, Wang GP, Berry CC, Leboulch P, et al. A method to sequence and quantify DNA integration for monitoring outcome in gene therapy. Nucleic Acids Res. 2011;39:e72. doi: 10.1093/nar/gkr140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modlich U, Bohne J, Schmidt M, von Kalle C, Knöss S, Schambach A, et al. Cell-culture assays reveal the importance of retroviral vector design for insertional genotoxicity. Blood. 2006;108:2545–2553. doi: 10.1182/blood-2005-08-024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modlich U, Kustikova OS, Schmidt M, Rudolph C, Meyer J, Li Z, et al. Leukemias following retroviral transfer of multidrug resistance 1 (MDR1) are driven by combinatorial insertional mutagenesis. Blood. 2005;105:4235–4246. doi: 10.1182/blood-2004-11-4535. [DOI] [PubMed] [Google Scholar]
- Montini E, Cesana D, Schmidt M, Sanvito F, Ponzoni M, Bartholomae C, et al. Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nat Biotechnol. 2006;24:687–696. doi: 10.1038/nbt1216. [DOI] [PubMed] [Google Scholar]
- Beard BC, Keyser KA, Trobridge GD, Peterson LJ, Miller DG, Jacobs M, et al. Unique integration profiles in a canine model of long-term repopulating cells transduced with gammaretrovirus, lentivirus, or foamy virus. Hum Gene Ther. 2007;18:423–434. doi: 10.1089/hum.2007.011. [DOI] [PubMed] [Google Scholar]
- Neff T, Beard BC, Peterson LJ, Anandakumar P, Thompson J, Kiem HP. Polyclonal chemoprotection against temozolomide in a large-animal model of drug resistance gene therapy. Blood. 2005;105:997–1002. doi: 10.1182/blood-2004-08-3169. [DOI] [PubMed] [Google Scholar]
- Seggewiss R, Pittaluga S, Adler RL, Guenaga FJ, Ferguson C, Pilz IH, et al. Acute myeloid leukemia is associated with retroviral gene transfer to hematopoietic progenitor cells in a rhesus macaque. Blood. 2006;107:3865–3867. doi: 10.1182/blood-2005-10-4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kustikova O, Fehse B, Modlich U, Yang M, Düllmann J, Kamino K, et al. Clonal dominance of hematopoietic stem cells triggered by retroviral gene marking. Science. 2005;308:1171–1174. doi: 10.1126/science.1105063. [DOI] [PubMed] [Google Scholar]
- Cornils K, Lange C, Schambach A, Brugman MH, Nowak R, Lioznov M, et al. Stem cell marking with promotor-deprived self-inactivating retroviral vectors does not lead to induced clonal imbalance. Mol Ther. 2009;17:131–143. doi: 10.1038/mt.2008.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maetzig T, Brugman MH, Bartels S, Heinz N, Kustikova OS, Modlich U, et al. Polyclonal fluctuation of lentiviral vector-transduced and expanded murine hematopoietic stem cells. Blood. 2011;117:3053–3064. doi: 10.1182/blood-2010-08-303222. [DOI] [PubMed] [Google Scholar]
- Rangarajan A, Hong SJ, Gifford A, Weinberg RA. Species- and cell type-specific requirements for cellular transformation. Cancer Cell. 2004;6:171–183. doi: 10.1016/j.ccr.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Mestas J, Hughes CC. Of mice and not men: differences between mouse and human immunology. J Immunol. 2004;172:2731–2738. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- Biffi A, Bartolomae CC, Cesana D, Cartier N, Aubourg P, Ranzani M, et al. Lentiviral vector common integration sites in preclinical models and a clinical trial reflect a benign integration bias and not oncogenic selection. Blood. 2011;117:5332–5339. doi: 10.1182/blood-2010-09-306761. [DOI] [PubMed] [Google Scholar]
- Phaltane R, Haemmerle R, Rothe M, Modlich U, Moritz T. Efficiency and safety of O6-methylguanine DNA methyltransferase (MGMT(P140K))-mediated in vivo selection in a humanized mouse model. Hum Gene Ther. 2014;25:144–155. doi: 10.1089/hum.2013.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S, Wang H, Bailey B, Ernstberger A, Juliar BE, Sinn AL, et al. Humanized bone marrow mouse model as a preclinical tool to assess therapy-mediated hematotoxicity. Clin Cancer Res. 2011;17:2195–2206. doi: 10.1158/1078-0432.CCR-10-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larochelle A, Dunbar CE. Hematopoietic stem cell gene therapy:assessing the relevance of preclinical models. Semin Hematol. 2013;50:101–130. doi: 10.1053/j.seminhematol.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel R, Eckenberg R, Paruzynski A, Bartholomae CC, Nowrouzi A, Arens A, et al. Comprehensive genomic access to vector integration in clinical gene therapy. Nat Med. 2009;15:1431–1436. doi: 10.1038/nm.2057. [DOI] [PubMed] [Google Scholar]
- Akagi K, Suzuki T, Stephens RM, Jenkins NA, Copeland NG. RTCGD: retroviral tagged cancer gene database. Nucleic Acids Res. 2004;32 Database issue:D523–D527. doi: 10.1093/nar/gkh013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Antonio M, Pendino V, Sinha S, Ciccarelli FD. Network of Cancer Genes (NCG 3.0): integration and analysis of genetic and network properties of cancer genes. Nucleic Acids Res. 2012;40 Database issue:D978–D983. doi: 10.1093/nar/gkr952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deichmann A, Brugman MH, Bartholomae CC, Schwarzwaelder K, Verstegen MM, Howe SJ, et al. Insertion sites in engrafted cells cluster within a limited repertoire of genomic areas after gammaretroviral vector gene therapy. Mol Ther. 2011;19:2031–2039. doi: 10.1038/mt.2011.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Luke BT, Burgess SM. Redefining the common insertion site. Virology. 2006;344:292–295. doi: 10.1016/j.virol.2005.08.047. [DOI] [PubMed] [Google Scholar]
- Modlich U, Navarro S, Zychlinski D, Maetzig T, Knoess S, Brugman MH, et al. Insertional transformation of hematopoietic cells by self-inactivating lentiviral and gammaretroviral vectors. Mol Ther. 2009;17:1919–1928. doi: 10.1038/mt.2009.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene MR, Lockey T, Mehta PK, Kim YS, Eldridge PW, Gray JT, et al. Transduction of human CD34+ repopulating cells with a self-inactivating lentiviral vector for SCID-X1 produced at clinical scale by a stable cell line. Hum Gene Ther Methods. 2012;23:297–308. doi: 10.1089/hgtb.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott SP, Eppert K, Lechman ER, Doedens M, Dick JE. Comparison of human cord blood engraftment between immunocompromised mouse strains. Blood. 2010;116:193–200. doi: 10.1182/blood-2010-02-271841. [DOI] [PubMed] [Google Scholar]
- Cheung AM, Nguyen LV, Carles A, Beer P, Miller PH, Knapp DJ, et al. Analysis of the clonal growth and differentiation dynamics of primitive barcoded human cord blood cells in NSG mice. Blood. 2013;122:3129–3137. doi: 10.1182/blood-2013-06-508432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugman MH, Suerth JD, Rothe M, Suerbaum S, Schambach A, Modlich U, et al. Evaluating a ligation-mediated PCR and pyrosequencing method for the detection of clonal contribution in polyclonal retrovirally transduced samples. Hum Gene Ther Methods. 2013;24:68–79. doi: 10.1089/hgtb.2012.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano FA, Sorg UR, Appelt JU, Lachmann N, Bleier S, Roeder I, et al. Clonal inventory screens uncover monoclonality following serial transplantation of MGMT P140K-transduced stem cells and dose-intense chemotherapy. Hum Gene Ther. 2011;22:697–710. doi: 10.1089/hum.2010.088. [DOI] [PubMed] [Google Scholar]
- Harkey MA, Kaul R, Jacobs MA, Kurre P, Bovee D, Levy R, et al. Multiarm high-throughput integration site detection: limitations of LAM-PCR technology and optimization for clonal analysis. Stem Cells Dev. 2007;16:381–392. doi: 10.1089/scd.2007.0015. [DOI] [PubMed] [Google Scholar]
- Scaramuzza S, Biasco L, Ripamonti A, Castiello MC, Loperfido M, Draghici E, et al. Preclinical safety and efficacy of human CD34(+) cells transduced with lentiviral vector for the treatment of Wiskott-Aldrich syndrome. Mol Ther. 2013;21:175–184. doi: 10.1038/mt.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum C, Modlich U, Göhring G, Schlegelberger B. Concise review: managing genotoxicity in the therapeutic modification of stem cells. Stem Cells. 2011;29:1479–1484. doi: 10.1002/stem.716. [DOI] [PubMed] [Google Scholar]
- Stein S, Scholz S, Schwäble J, Sadat MA, Modlich U, Schultze-Strasser S, et al. From bench to bedside: preclinical evaluation of a self-inactivating gammaretroviral vector for the gene therapy of X-linked chronic granulomatous disease. Hum Gene Ther Clin Dev. 2013;24:86–98. doi: 10.1089/humc.2013.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kustikova OS, Schiedlmeier B, Brugman MH, Stahlhut M, Bartels S, Li Z, et al. Cell-intrinsic and vector-related properties cooperate to determine the incidence and consequences of insertional mutagenesis. Mol Ther. 2009;17:1537–1547. doi: 10.1038/mt.2009.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schambach A, Bohne J, Chandra S, Will E, Margison GP, Williams DA, et al. Equal potency of gammaretroviral and lentiviral SIN vectors for expression of O6-methylguanine-DNA methyltransferase in hematopoietic cells. Mol Ther. 2006;13:391–400. doi: 10.1016/j.ymthe.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Sandrin V, Boson B, Salmon P, Gay W, Nègre D, Le Grand R, et al. Lentiviral vectors pseudotyped with a modified RD114 envelope glycoprotein show increased stability in sera and augmented transduction of primary lymphocytes and CD34+ cells derived from human and nonhuman primates. Blood. 2002;100:823–832. doi: 10.1182/blood-2001-11-0042. [DOI] [PubMed] [Google Scholar]
- Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, et al. A third-generation lentivirus vector with a conditional packaging system. J Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz LD, Lyons BL, Burzenski LM, Gott B, Chen X, Chaleff S, et al. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. J Immunol. 2005;174:6477–6489. doi: 10.4049/jimmunol.174.10.6477. [DOI] [PubMed] [Google Scholar]
- Rittelmeyer I, Rothe M, Brugman MH, Iken M, Schambach A, Manns MP, et al. Hepatic lentiviral gene transfer is associated with clonal selection, but not with tumor formation in serially transplanted rodents. Hepatology. 2013;58:397–408. doi: 10.1002/hep.26204. [DOI] [PubMed] [Google Scholar]
- Huston MW, Brugman MH, Horsman S, Stubbs A, van der Spek P, Wagemaker G. Comprehensive investigation of parameter choice in viral integration site analysis and its effects on the gene annotations produced. Hum Gene Ther. 2012;23:1209–1219. doi: 10.1089/hum.2011.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.