SUMMARY
The MUC1 heterodimeric transmembrane glycoprotein is aberrantly overexpressed by diverse human carcinomas. Galectin-3 is a β-galactoside binding protein that has also been associated with the development of human cancers. The present results demonstrate that MUC1 induces galectin-3 expression by a posttranscriptional mechanism. We show that the MUC1 C-terminal subunit is glycosylated on Asn-36 and that this modification is necessary for upregulation of galectin-3. N-glycosylated MUC1-C increases galectin-3 mRNA levels by suppressing expression of the microRNA miR-322 and thereby stabilizing galectin-3 transcripts. The results show that, in turn, galectin-3 binds to MUC1-C at the glycosylated Asn-36 site. The significance of the MUC1-C-galectin-3 interaction is supported by the demonstration that galectin-3 forms a bridge between MUC1 and the epidermal growth factor receptor (EGFR) and that galectin-3 is essential for EGF-mediated interactions between MUC1 and EGFR. These findings indicate that MUC1 and galectin-3 function as part of a miR-322-dependent regulatory loop.
INTRODUCTION
The MUC1 mucin-type, transmembrane glycoprotein is expressed on the apical borders of normal secretory epithelial cells (Kufe et al., 1984). With transformation and loss of polarity, MUC1 is aberrantly overexpressed on the entire cell surface in carcinomas of the breast, prostate, lung, and other epithelia (Kufe et al., 1984). The MUC1 polypeptide undergoes autoproteolysis in the endoplasmic reticulum with the generation of two subunits that in turn form a stable heterodimer (Levitin et al., 2005; Macao et al., 2006). The MUC1 N-terminal subunit (MUC1-N) consists in large part of variable numbers of 20 amino acid tandem repeats that are subject to extensive O-glycosylation. MUC1-N is tethered to the cell surface through noncovalent binding to the transmembrane MUC1 C-terminal subunit, which consists of a 58 amino acid extracellular domain, a 28 amino acid transmembrane domain, and a 72 amino acid cytoplasmic tail. MUC1-N extends beyond the cell glycocalyx as part of a physical barrier that protects epithelial cells from damage induced by free radicals, low pH, toxins, and other forms of stress that occur at the interface with the external environment. MUC1-N is also shed into this protective barrier, leaving MUC1-C at the cell surface as a putative receptor for signaling the presence of stress to the interior of the cell. Overexpression of MUC1 in transformed cells is also associated with accumulation of MUC1-C in the cytosol and targeting of this subunit to the nucleus (Li et al., 2003a, 2003b, 2003c) and mitochondria (Ren et al., 2004, 2006). In support of a role for MUC1-C in signal transduction, the cytoplasmic domain functions as a substrate for c-Src (Li et al., 2001a), c-Abl (Raina et al., 2006), glycogen synthase kinase 3β (Li et al., 1998), and protein kinase Cδ (Ren et al., 2002). Moreover, the MUC1-C cytoplasmic domain interacts directly with the Wnt pathway effector, β-catenin (Huang et al., 2005; Li et al., 2003b), the p53 tumor suppressor (Wei et al., 2005), and estrogen receptor α (Wei et al., 2006). Other studies have demonstrated that overexpression of MUC1 is sufficient to confer anchorage-independent growth and tumorigenicity (Li et al., 2003b; Raina et al., 2004; Ren et al., 2004; Wei et al., 2005) and resistance to stress-induced apoptosis (Ren et al., 2004; Yin and Kufe, 2003).
Galectins are a family of lectins that contain conserved carbohydrate-recognition domains (CRDs) of about 130 amino acids with specificity for β-galactosides found on both N- and O-linked glycans (Liu and Rabinovich, 2005). Of the 15 known mammalian galectins, the widely expressed galectin-3 is a structurally unique member with an N-terminal domain (ND) of repetitive sequences rich in proline, glycine, and tyrosine residues upstream to the CRD. The ND lacks a carbohydrate-binding function but is essential for the biologic activity of galectin-3 (Seetharaman et al., 1998). Cytosolic galectin-3 is targeted to the plasma membrane and released into the extracellular space, where it functions in the regulation of cell migration and adhesion (Ochieng et al., 2004). Galectin-3 is also targeted to the nucleus, where it plays a role in regulating pre-mRNA splicing (Patterson et al., 2004) and contributes to the activation of diverse transcription factors (Lin et al., 2002; Walzel et al., 2002). As found with MUC1 (Huang et al., 2005), galectin-3 binds directly to β-catenin and induces the transcriptional activity of Tcf-4 (Shimura et al., 2004). In addition and like MUC1 (Ren et al., 2004; Wei et al., 2005), galectin-3 blocks the apoptotic response of breast cancer cells to genotoxic anticancer agents (Takenaka et al., 2004a). The antiapoptotic activity of galectin-3 is regulated by casein kinase 1-mediated phosphorylation of nuclear galectin-3 on Ser-6 (Yoshii et al., 2002) and thereby its export to the cytoplasm (Takenaka et al., 2004b). Translocation of galectin-3 to mitochondria is associated with inhibition of cytochrome c release and contributes in part to protection against the induction of apoptosis (Matarrese et al., 2000; Yu et al., 2002). Galectin-3 also contains an Asn-Trp-Gly-Arg (NWGR) motif that is conserved in the BH1 domain of the Bcl-2 family members and is of importance to the galectin-3 antiapoptotic function (Akahani et al., 1997). Other studies have demonstrated that galectin-3 induces transformation (Nangia-Makker et al., 1995; Takenaka et al., 2003) and is upregulated in diverse carcinomas (Takenaka et al., 2004a; van den Brule et al., 2004). Notably, however, little is known about the regulation of galectin-3 expression in cancer cells.
The present results demonstrate that the MUC1-C subunit is modified by N-glycosylation of Asn-36. The N-glycosylated MUC1-C suppresses expression of microRNA miR-322 and thereby upregulates galectin-3. In turn, galectin-3 binds to MUC1 and physically integrates MUC1 with EGFR signaling. These findings support a model in which MUC1 and galectin-3 function in a microRNA-dependent regulatory loop.
RESULTS
MUC1 Upregulates Galectin-3 Expression in Carcinoma Cells
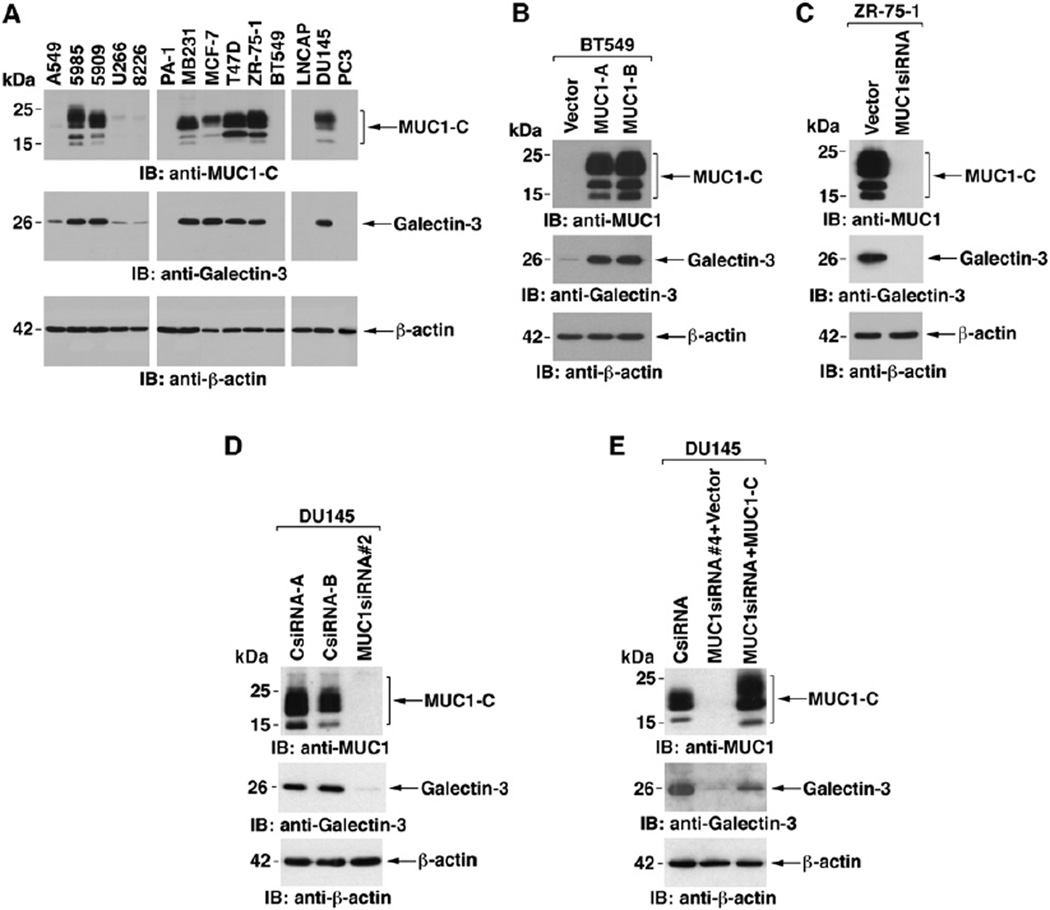
MUC1 and galectin-3 are both widely expressed in human carcinomas. To assess the coexpression of MUC1 and galectin-3, we analyzed lysates from diverse human carcinoma cells. The results indicated that MUC1 expression is associated with upregulation of galectin-3 (Figure 1A). The relationship between MUC1 and galectin-3 expression was further assessed in BT549 breast cancer cells, which are null for MUC1 and have low levels of galectin-3 (Figure 1B). Stable expression of MUC1, but not the empty vector, was associated with increases in galectin-3 levels (Figure 1B). MUC1-dependent regulation of galectin-3 expression was also assessed in human cancer cells that express endogenous MUC1. Stable silencing of MUC1 in ZR-75-1 breast cancer cells with a MUC1siRNA (Ren et al., 2004) was associated with decreases in galectin-3 (Figure 1C and Figure S1A in the Supplemental Data available with this article online). Transient silencing of MUC1 with a pool of MUC1 siRNAs also resulted in downregulation of galectin-3 (Figure S1B). Moreover, stable silencing of MUC1 in DU145 prostate cancer cells was associated with downregulation of galectin-3 (Figure 1D). To exclude off-target effects of the MUC1siRNA#2, which is directed against sequences encoding MUC1-C, we used MUC1-siRNA#4. Silencing MUC1 with MUC1siRNA#4 similarly resulted in downregulation of galectin-3 expression (Figure 1E). MUC1siRNA#4 targets sequences in the MUC1-N coding region. Consequently, we transfected these cells to express MUC1-C (Figure 1E). Notably, rescue of MUC1-C expression was associated with increases in galectin-3 (Figure 1E). These findings indicate that MUC1-C upregulates galectin-3 expression.
Figure 1. MUC1 Upregulates Galectin-3 Expression.
(A) Lysates from the indicated cells were immunoblotted with anti-MUC1-C, anti-galectin-3, and anti-β-actin.
(B) The empty pIRES-puro vector and pIRES-puro-MUC1 were stably transfected into BT549 breast cancer cells. Lysates from the transfectants were immunoblotted with the indicated antibodies. The BT549/MUC1-A and -B cells represent two separately isolated clones.
(C) Lysates from ZR-75-1/vector-A and ZR-75-1/MUC1siRNA#2-A cells were immunoblotted with the indicated antibodies.
(D) Lysates from DU145 prostate cancer cells stably expressing the pRNA-U6.1/Neo CsiRNA or MUC1siRNA#2 were immunoblotted with the indicated antibodies.
(E) DU145 cells stably expressing MUC1siRNA#4 were transfected with pIRES-puro2 or pIRES-puro2-MUC1-C. Lysates were immunoblotted with the indicated antibodies. DU145/CsiRNA cells were included for comparison.
MUC1 Stabilizes Galectin-3 Transcripts
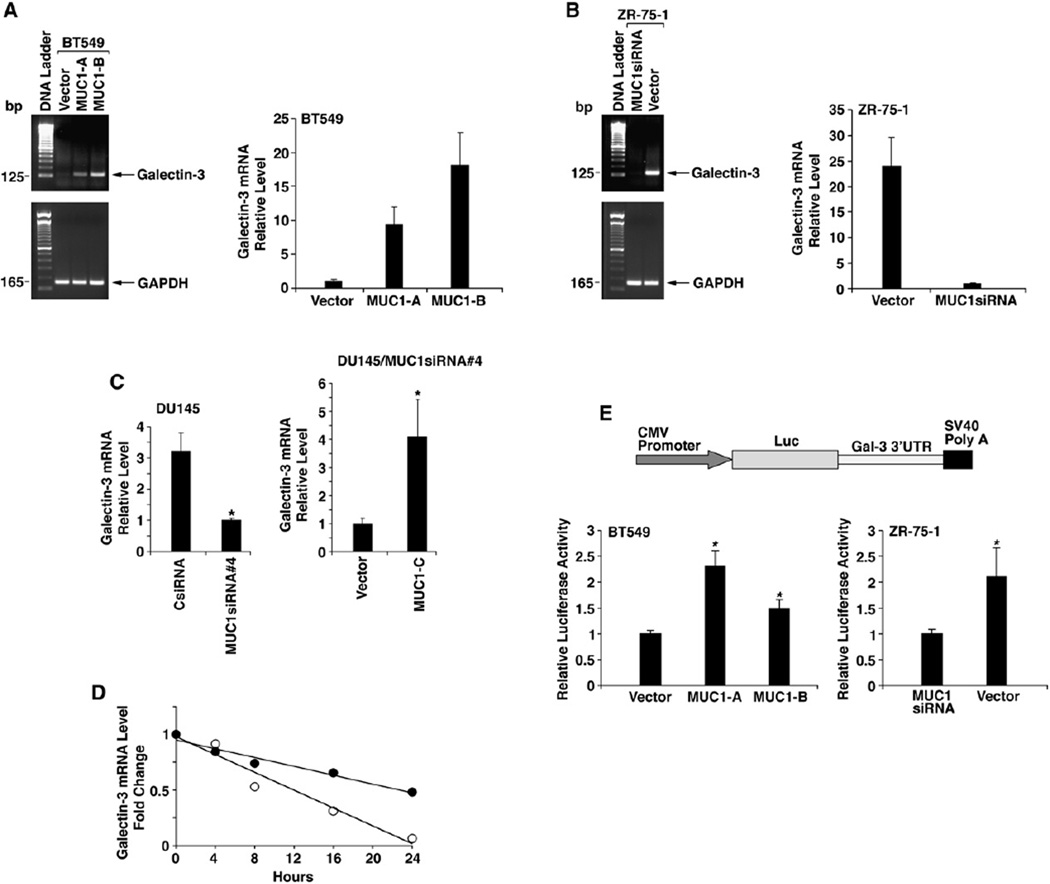
To define how MUC1 regulates galectin-3 expression, we performed semiquantitative RT-PCR analysis of galectin-3 mRNA levels. BT549 cells stably expressing MUC1 exhibited increases in galectin-3 transcripts as compared to that in BT549/vector cells (Figure 2A, left). MUC1-dependent upregulation of galectin-3 mRNA levels in BT549 cells was confirmed by quantitative RT-PCR (Figure 2A, right). In concert with these results, stable silencing of MUC1 in ZR-75-1 cells decreased galectin-3 mRNA levels as determined by semiquantitative (Figure 2B, left) and quantitative (Figure 2B, right) RT-PCR. Stable silencing of MUC1 in DU145 cells also decreased galectin-3 mRNA levels (Figure 2C, left). Moreover, expression of MUC1-C in DU145/MUC1siRNA#4 cells was associated with increases in galectin-3 transcripts (Figure 2C, right). To determine whether MUC1 activates galectin-3 gene transcription, we ligated regions (−3000 to +141 and −836 to +141) of the galectin-3 promoter upstream to a luciferase (Luc) reporter. The results obtained from transfecting pGal-3(−3000/+141)-Luc into BT549/vector and BT549/MUC1 cells indicated that MUC1 has little if any effect on activation of the galectin-3 promoter (Figure S2A). MUC1 also had no apparent effect on activation of the pGal-3(−3000/+141)-Luc or pGal-3(−836/+141)-Luc constructs in ZR-75-1 cells (Figures S2B and S2C), indicating that MUC1 upregulates galectin-3 mRNA levels by a posttranscriptional mechanism. To extend this analysis to the endogenous galectin-3 gene, we performed nuclear run-on assays. The rate of galectin-3 gene transcription in BT549 cells was unaffected by stable expression of MUC1 (Figure S2D, left). In addition, silencing of MUC1 in ZR-75-1 cells had little if any effect on transcription of the galectin-3 gene (Figure S2D, right). These findings indicate that MUC1 increases galectin-3 mRNA levels by a posttranscriptional mechanism. To define the mechanism by which MUC1 increases galectin-3 mRNA levels, we first analyzed stability of galectin-3 transcripts in BT549/vector and BT549/MUC1 cells. Galectin-3 and GAPDH mRNA levels were assayed by quantitative RT-PCR after inhibiting transcription with actinomycin D. The rate of galectin-3 mRNA degradation in BT549/vector cells was increased as compared to that in BT549/MUC1 cells, indicating that MUC1 stabilizes galectin-3 transcripts (Figure 2D). In this regard, the half-lives of galectin-3 transcripts were 22.8 and 12.0 hr in the presence and absence of MUC1, respectively. To determine if the galectin-3 mRNA 3′ untranslated region (3′UTR) is regulated by MUC1, the 3′UTR was ligated downstream to the luciferase gene in the pMIR reporter plasmid (Figure 2E). Transfection of BT549 cells with pMIR-Gal-3(3′UTR) demonstrated that MUC1 increases expression of the luciferase reporter (Figure 2E, left). Expression of pMIR-Gal-3(3′UTR) was also increased in ZR-75-1 cells by a MUC1-dependent mechanism (Figure 2E, right). These findings indicate that MUC1 stabilizes galectin-3 transcripts by a mechanism involving the 3′UTR.
Figure 2. MUC1 Stabilizes Galectin-3 Transcripts.
(A and B) BT549/vector and BT549/MUC1 cells (A) or ZR-75-1/vector and ZR-75-1/MUC1siRNA (stably silenced for MUC1) cells (B) were analyzed for galectin-3 and GAPDH mRNA levels by semiquantitative RT-PCR (left) and quantitative RT-PCR (right). The quantitative RT-PCR results (mean ± SD from three replicates) are expressed as the relative galectin-3 mRNA levels (normalized to GAPDH) compared to that obtained with BT549/vector (A) and ZR-75-1/MUC1siRNA(B) cells (assigned a value of 1).
(C) DU145/CsiRNA and DU145/MUC1siRNA#4 cells were analyzed for galectin-3 and GAPDH mRNA levels by quantitative RT-PCR (left). DU145/ MUC1siRNA#4 cells were transfected with pIRES-puro2 or pIRES-puro2-MUC1-C and then analyzed by quantitative RT-PCR (right). The results (mean ± SD from three replicates) are expressed as the relative galectin-3 mRNA levels (normalized to GAPDH) compared to that obtained with DU145/MUC1siRNA#4 (left) and DU145/MUC1siRNA#4 transfected with pIRES-puro2 (right) cells (assigned a value of 1). The asterisk (*) denotes a significant difference at p < 0.05 as compared to control.
(D) BT549/vector and BT549/MUC1 cells were treated with actinomycin D and then harvested at the indicated times. RNA was analyzed for galectin-3 and GAPDH mRNA levels by quantitative RT-PCR. The results are expressed as relative galectin-3 mRNA levels for BT549/vector (○) and BT549/ MUC1 (●) cells.
(E) Construction of the galectin-3 3′UTR-luciferase reporter plasmid. BT549/vector and BT549/MUC1 cells (left) or ZR-75-1/vector and ZR-75-1/ MUC1siRNA cells (right) were transfected with the pMIR-Gal-3(3′UTR) and, as a control, pMIR-β-galactosidase plasmids. The cells were analyzed for luciferase and β-gal activities at 48 hr after transfection. The results (mean ± SD of three experiments) are expressed as the relative luciferase activity (normalized to β-gal) compared to that in BT549/vector (left) or ZR-75-1/MUC1siRNA (right) cells (assigned a value of 1). The asterisk (*) denotes a significant difference at p < 0.05 as compared to control.
MUC1 Suppresses MiR-322 Expression and Thereby Increases Stability of Galectin-3 mRNA
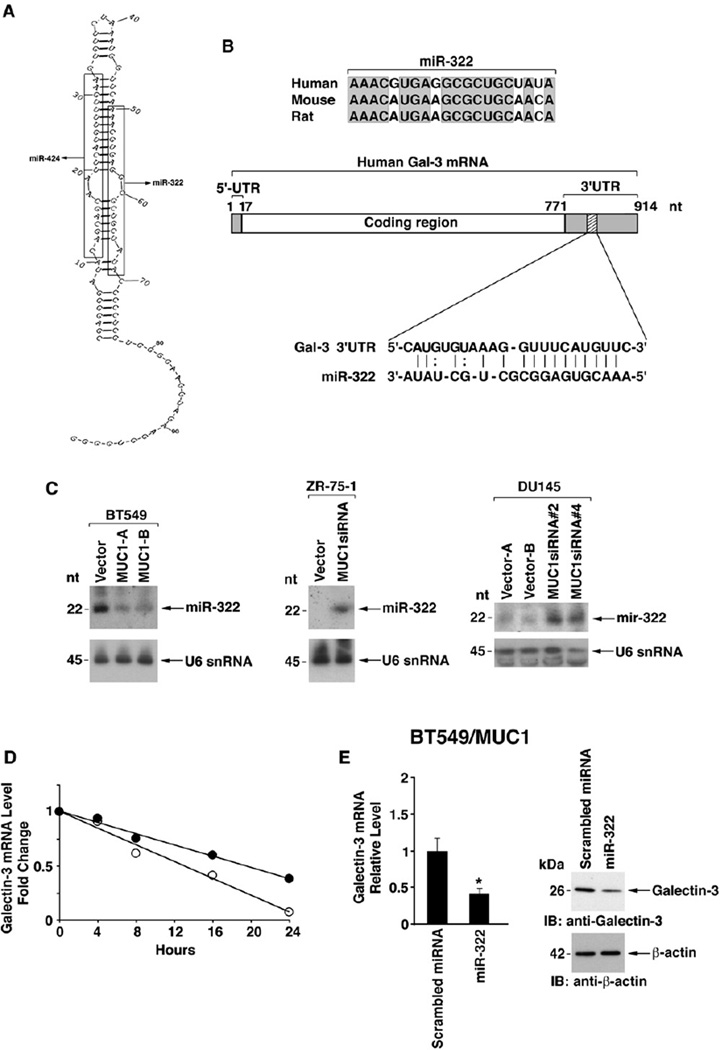
The galectin-3 3′UTR has no identifiable AU-rich elements that function in the regulation of mRNA stability. Consequently, we asked if the galectin-3 3′UTR contains sequences that could be targeted by a microRNA. A search of the Sanger miRNA registry (http://microrna.sanger.ac.uk) identified miR-322 as a possible candidate. miR-322 is expressed from a stem-loop structure (pre-miR-322) at which miR-424 also originates (Figure 3A). Expression of miR-424 has been demonstrated in human cells (Kasashima et al., 2004); however, there is no available evidence for a human miR-322. Alignment of mouse, rat, and the putative human miR-322 revealed conservation of the sequences (Figure 3B). In addition, human miR-322 has recognition sequences for the human galectin-3 3′UTR (Figure 3B). Northern blot analysis of RNA from BT549/ vector cells demonstrated expression of miR-322 (Figure 3C, left). By contrast, we found that miR-322 levels are decreased in BT549 cells that express MUC1 (Figure 3C, left). Studies with ZR-75-1 cells further showed that miR-322 expression is suppressed by a MUC1-dependent mechanism (Figure 3C, middle). Moreover, silencing MUC1 in DU145 cells with MUC1siRNA#2 or MUC1 siRNA#4 increased miR-322 expression (Figure 3C, right).
Figure 3. MUC1 Downregulates MiR-322 and Thereby Increases Stability of Galectin-3 mRNA.
(A) Model of the pre-miR stem-loop structure encoding miR-424 and miR-322.
(B) Sequence alignment of human miR-322 with mouse and rat miR-322 and with the galectin-3 3′UTR.
(C) Northern blot analyses of RNA from the indicated BT549, ZR-75-1, and DU145 cells probed for miR-322 (upper panels) and U6 snRNA as a loading control (lower panels).
(D) BT549/vector cells were transfected with an antisense 2′-O-methyl oligoribonucleotide targeted against miR-322 or a scrambled 2′-O-methyl oligoribonucleotide as a control for 48 hr. The cells were treated with actinomycin D and harvested at the indicated times. Total RNA was analyzed for galectin-3 and GAPDH mRNA levels by quantitative RT-PCR. The results are expressed as relative galectin-3 mRNA levels for BT549 cells transfected with the scrambled oligo (○) or the anti-miR-322 (●).
(E) BT549/MUC1 cells were transfected with a pre-miR-322 or a scrambled miRNA and selected in the presence of blasticidin. Cells were assayed for galectin-3 mRNA (left) and protein (right). The results (mean±SD for three replicates) are expressed as the relative galectin-3 mRNA levels (normalized to GAPDH) compared to that obtained with the scrambled miRNA (assigned a value of 1) (left). The asterisk (*) denotes a significant difference at p < 0.01 as compared to control (left).
These results thus demonstrate that MUC1 suppresses miR-322 expression. To determine whether miR-322 regulates galectin-3 expression, BT549/vector cells were transfected with an antisense 2′-O-methyl oligoribonucleotide targeted against miR-322 or, as a control, with a scrambled 2′ -O-methyl oligoribonucleotide. We found that anti-miR-322, and not the scrambled oligo, upregulates galectin-3 expression at the mRNA (Figure S3A, left) and protein levels (Figure S3A, right). Stability of galectin-3 transcripts was also determined by quantitative RT-PCR after actinomycin D treatment. Degradation of galectin-3 mRNA was increased in cells transfected with the scrambled oligo compared to that obtained with anti-miR-322 (Figure 3D), indicating that miR-322 decreases galectin-3 mRNA stability. The half-lives of galectin-3 transcripts were 13.0 and 19.5 hr in cells transfected with the scrambled oligo and anti-miR-322, respectively. As another approach, BT549/MUC1 cells were transfected with a pre-miR-322 or a scrambled miRNA. Ectopic miR-322 had no detectable effect on galectin-3 gene transcription as determined by run-on assays (Figure S3B). Moreover, analysis of galectin-3 expression demonstrated that the ectopic miR-322 decreases galectin-3 mRNA and protein levels (Figure 3E). These findings indicate that miR-322 decreases stability of galectin-3 transcripts.
Direct Binding of the MUC1-C Extracellular Domain and Galectin-3
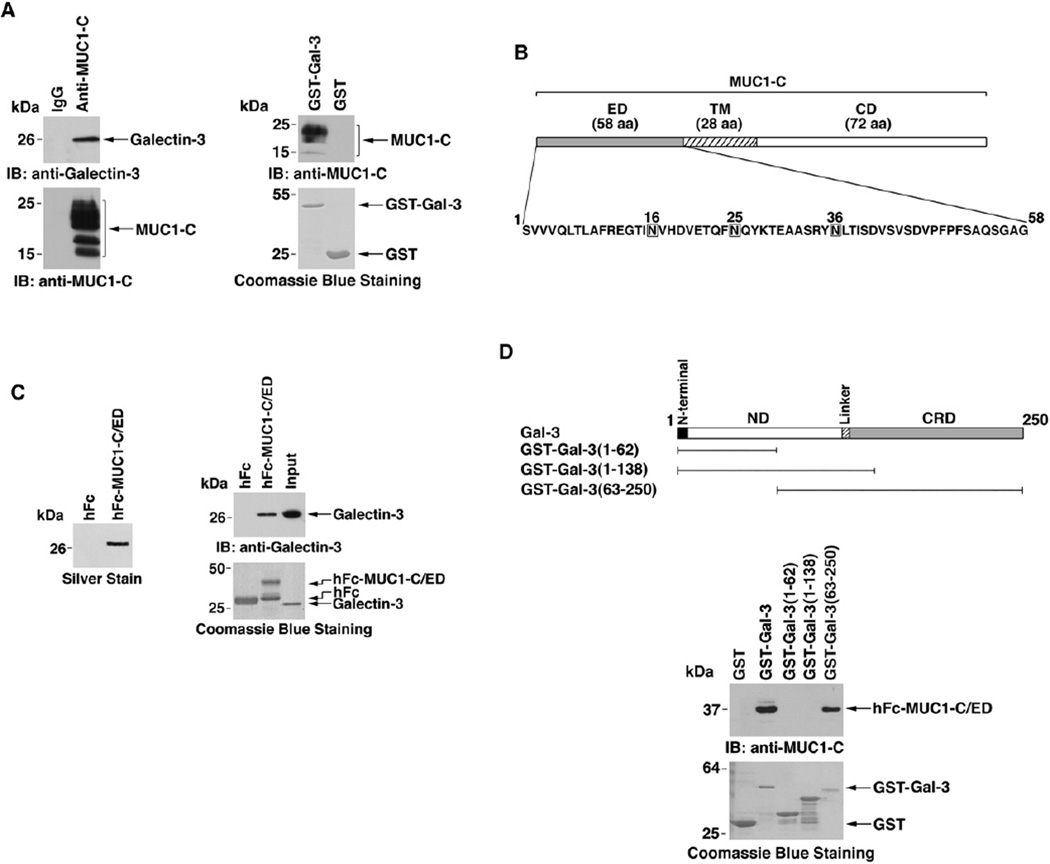
Galectin-3 binds to glycans on cell surface molecules like MUC1. To determine if the upregulation of galectin-3 expression is associated with binding of MUC1 and galectin-3, lysates from ZR-75-1 cells were immunoprecipitated with anti-MUC1-C or, as a control, IgG. Galectin-3 was detectable in the anti-MUC1-C, and not the IgG, precipitates (Figure 4A, left). Incubation of a GST-galectin-3 fusion protein with ZR-75-1 cell lysates in pull-down experiments further demonstrated binding of MUC1-C and galectin-3 (Figure 4A, right). MUC1-C consists of a 58 amino acid extracellular domain (ED), a 28 amino acid transmembrane domain (TM), and a 72 amino acid cytoplasmic domain (CD) (Figure 4B). The MUC1-C extracellular domain (MUC1-C/ED) contains three potential N-glycosylation sites that could confer the interaction between MUC1-C and galectin-3 (Figure 4B). To determine if galectin-3 associates with the MUC1-C/ED, we fused the MUC1-C/ED to human Fc (hFc-MUC1-C/ED) (Figure 4B). hFc and hFc-MUC1-C/ED bound to agarose beads were incubated with supernatants from ZR-75-1 breast cancer cells. Analysis of adsorbed proteins by SDS-PAGE and silver staining demonstrated binding of a ~26 kDa protein to hFc-MUC1-C/ED and not hFc (Figure 4C, left). Digestion of the adsorbed protein with trypsin and mass spectroscopy analysis of the tryptic peptides supported identity with galectin-3 (data not shown). Immunoblot analysis of the adsorbed protein also confirmed the association of MUC1-C/ED and galectin-3. To determine if MUC1-C/ED and galectin-3 interact directly, we incubated hFc or hFc-MUC1-C/ED with purified recombinant galectin-3. Analysis of the adsorbates by immunoblotting with anti-galectin-3 demonstrated binding of galectin-3 to hFc-MUC1-C/ED and not hFc (Figure 4C, right). To define the region of galectin-3 that confers binding to MUC1-C/ ED, we prepared GST fusion proteins with full-length galectin-3 or certain deletion mutants (Figure 4D). MUC1-C/ED was pulled down with GST-galectin-3 and GST-galectin-3(63–250) that contains the CRD (Figure 4D). By contrast, there was no detectable binding of MUC1-C/ ED and GST-galectin-3(1–62) or GST-galectin-3(1–138) that include the ND (Figure 4D). The kinetics of the interaction between MUC1-C/ED and galectin-3 were assessed by immobilizing galectin-3 to a sensor chip and assaying for binding of MUC1-C/ED in a BIAcore (Figure S4A). MUC1-C/ED bound to galectin-3 with a dissociation constant (KD) of 11.1 nM. Similar kinetics (KD = 9.8 nM) were obtained for binding of MUC1-C/ED to galectin-3(63–250) (Figure S4B). These findings indicate that MUC1-C/ ED and galectin-3 bind directly and that this interaction is mediated by the galectin-3 CRD.
Figure 4. Galectin-3 Associates with the MUC1-C Extracellular Domain.
(A) Lysates from ZR-75-1 cells were immunoprecipitated with anti-MUC1-C and, as a control, a nonspecific IgG (left). The precipitates were immunoblotted with the indicated antibodies. ZR-75-1 cell lysates were incubated with GST or GST-galectin-3 bound to glutathione beads (right). The adsorbates were immunoblotted with anti-MUC1-C. Input of the GST and GST-galectin-3 proteins was assessed by Coomassie blue staining.
(B) Schematic representation of the transmembrane MUC1-C subunit and amino acid sequence of the extracellular domain (MUC1-C/ED).
(C) Left, ZR-75-1 cells were grown in 0.1% FBS for 2 days. Cell surface binding proteins were released in salt solution, diluted, and passed through hFc or hFc-MUC1-C/ED columns. The adsorbed proteins were eluted and analyzed by SDS-PAGE and silver staining. The 26 kDa protein was subjected to trypsin digestion. Analysis of the tryptic peptides by LC-MS demonstrated identity with galectin-3. Right, hFc and hFc-MUC1-C/ED were incubated with purified galectin-3 and precipitated with protein G-Sepharose. The adsorbates were immunoblotted with anti-galectin-3 (upper panel). The input proteins were stained with Coomassie blue (lower panel).
(D) Schema of galectin-3 is shown with the regions expressed as GST fusion proteins. hFc-MUC1-C/ED was incubated with GST or the indicated GST-galectin-3 fusion proteins. Adsorbates to glutathione beads were immunoblotted with anti-MUC1-C/ED. Input proteins were stained with Coomassie blue.
MUC1-C Glycosylation Is Necessary for the Galectin-3 Interaction
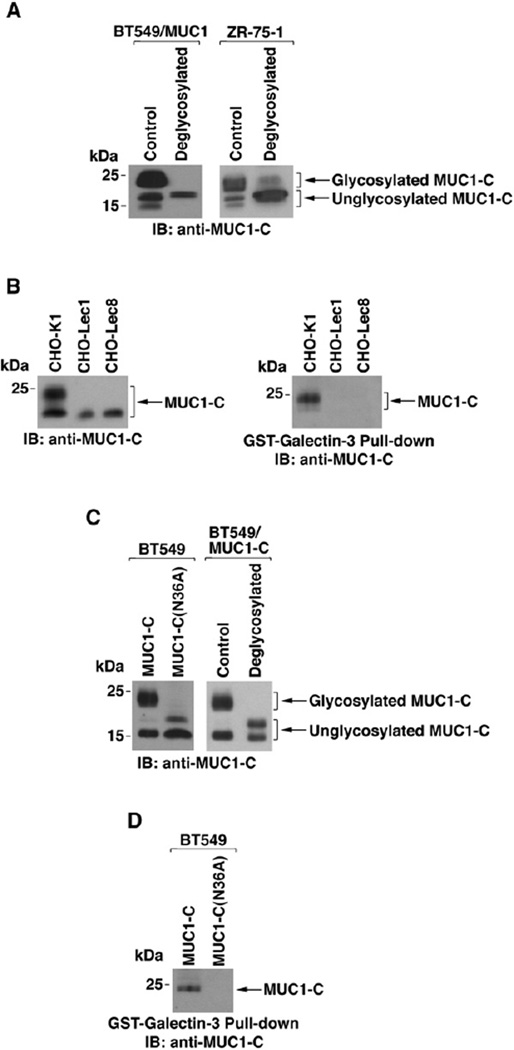
Galectin-3 binds to both β-galactoside-containing glycoproteins and nonglycosylated proteins (Shimura et al., 2004). MUC1-C is expressed as 25–20, 17, and 15 kDa species (Figure 5A); however, it is not known if any of these species are subject to glycosylation. To assess MUC1-C glycosylation, anti-MUC1-C precipitates from BT549/MUC1 cells were incubated in the absence and presence of N-glycosidases. Deglycosylation was associated with a decrease in the broad anti-MUC1-C band at 25–20 kDa and an increase in the 17 kDa band (Figure 5A, left), consistent with the presence of N-linked glycans. Similar results were obtained when endogenous MUC1-C from ZR-75-1 cells was incubated with N-glycosidases (Figure 5A, right). To extend this analysis, we expressed MUC1-C in wild-type and glycosylation-deficient CHO cells. Expression of MUC1-C in CHO-Lec1 cells, which are deficient in N-glycosylation, was associated with loss of the 25–20 kDa band (Figure 5B, left). A similar pattern of MUC1-C expression was observed in CHO-Lec8 cells, which are deficient for incorporation of β-galactosides (Figure 5B, left). Notably, GST-galectin-3 pull-downs showed binding only to the 25–20 kDa MUC1-C expressed in CHO-K1 cells (Figure 5B, right), consistent with interaction of galectin-3 and N-linked glycans on MUC1-C. The MUC1-C extracellular domain contains three asparagine residues (positions 16, 25, and 36; see Figure 4B), one of which (NLT) conforms to a predicted N-glycosylation site. Consequently, we stably expressed wild-type MUC1-C or MUC1-C with an N36A mutation in human BT549 cells. Expression of wild-type MUC1-C was associated with anti-MUC1-C reactivity that was predominant at 25–20 and 15 kDa (Figure 5C, left). By contrast, expression of MUC1-C(N36A) resulted in anti-MUC1-C reactivity at 17 and 15 kDa (Figure 5C, left), consistent with loss of the glycosylated 25–20 kDa species. As a control, deglycosylation of wild-type MUC1-C was also associated with reactivity at 17 and 15 kDa (Figure 5C, right). Importantly, binding of galectin-3 was detectable with wild-type MUC1-C, but not with MUC1-C(N36A) (Figure 5D). These findings indicate that galectin-3 binds to MUC1-C glycosylated at the Asn-36 site.
Figure 5. Glycosylation of MUC1-C on Asn-36 Is Necessary for Galectin-3 Binding.
(A) Lysates from BT549/MUC1 (left) and ZR-75-1 (right) cells were immunoprecipitated with anti-MUC1-C. The precipitates were left untreated or digested with N-glycosidases and then immunoblotted with anti-MUC1-C.
(B) MUC1-C was transiently expressed in wild-type CHO-K1 cells or the glycosylation-deficient Lec1 and Lec8 variants. Lysates were immunoblotted with anti-MUC1-C (left). The lysates were also incubated with GST-galectin-3 and the precipitates immunoblotted with anti-MUC1-C (right).
(C) Left, lysates from BT549cells stably expressing MUC1-Cor MUC1-C(N36A) were immunoblotted with anti-MUC1-C. Right, lysates from BT549/MUC1-C cells were immunoprecipitated with anti-MUC1-C. The precipitates were left untreated or digested with N-glycosidases and immunoblotted with anti-MUC1-C.
(D) Lysates from BT549/MUC1-C and BT549/MUC1-C(N36A) cells were incubated with GST-galectin-3, and the adsorbates were immunoblotted with anti-MUC1-C.
MUC1-C Subunit Induces Galectin-3 Expression
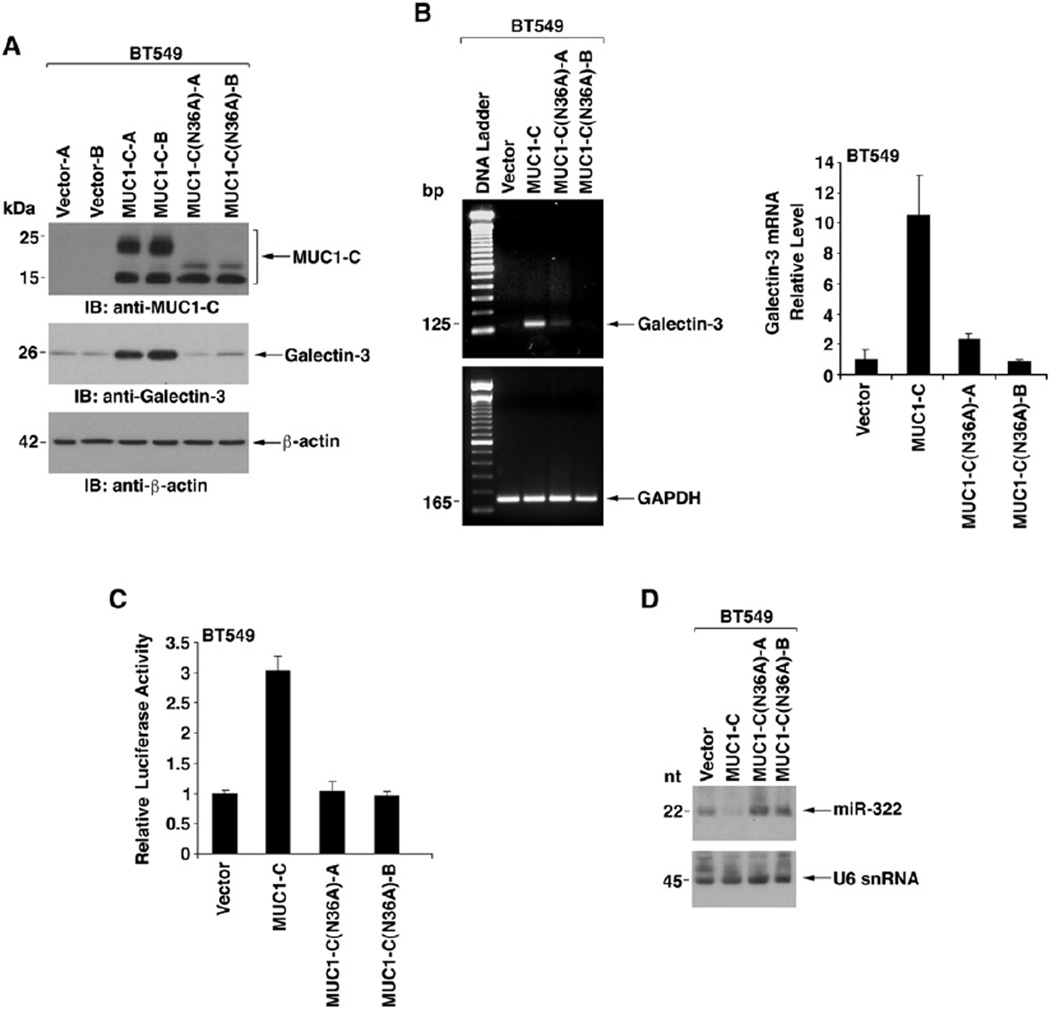
To further define the interaction between MUC1-C and galectin-3, BT549 cells stably transfected with MUC1-C or MUC1-C(N36A) were analyzed for induction of galectin-3 (Figure 6A). The results demonstrate that MUC1-C is sufficient to upregulate galectin-3 expression (Figure 6A). By contrast, MUC1-C(N36A) had little effect on galectin-3 levels (Figure 6A). Semiquantitative and quantitative RT-PCR also demonstrated that MUC1-C-induced upregulation of galectin-3 mRNA levels is substantially attenuated with MUC1-C(N36A) (Figure 6B). As found with MUC1, there was no apparent effect of MUC1-C or MUC1-C(N36A) on activation of the galectin-3 promoter (data not shown). However, MUC1-C, but not MUC1-C(N36A), was effective in increasing expression of the pMIR-Gal-3(3′UTR) reporter (Figure 6C). In concert with these results, we found that MUC1-C-induced suppression of miR-322 is also abrogated by the N36A mutation (Figure 6D). These findings indicate that glycosylation of MUC1-C Asn-36 is necessary for suppression of miR-322 expression and upregulation of galectin-3.
Figure 6. Glycosylation of MUC1-C on Asn-36 Is Associated with Upregulation of Galectin-3 Expression.
(A) Lysates from BT549 cells stably expressing the empty vector, MUC1-C, or MUC1-C(N36A) were immunoblotted with the indicated antibodies.
(B) The indicated BT549 cells were analyzed for galectin-3 and GAPDH mRNA levels by semiquantitative RT-PCR (left) and real-time RT-PCR (right). The real-time RT-PCR results (mean ± SD from three replicates) are expressed as the relative galectin-3 mRNA levels (normalized to GAPDH) compared to that obtained with BT549/vector cells (assigned a value of 1).
(C) The indicated BT549 cells were transfected with pMIR-Gal-3(3′UTR) and pMIR-β-gal plasmids. The cells were assayed for luciferase and β-gal activities at 48 hr after transfection. The results (mean ± SD of three experiments) are expressed as the relative luciferase activity (normalized to β-gal) compared to that in BT549/vector cells.
(D) Northern blot analysis of RNA from the indicated BT549 cells probed for miR-322 and U6 snRNA.
Galectin-3 Confers the Interaction between MUC1 and EGFR
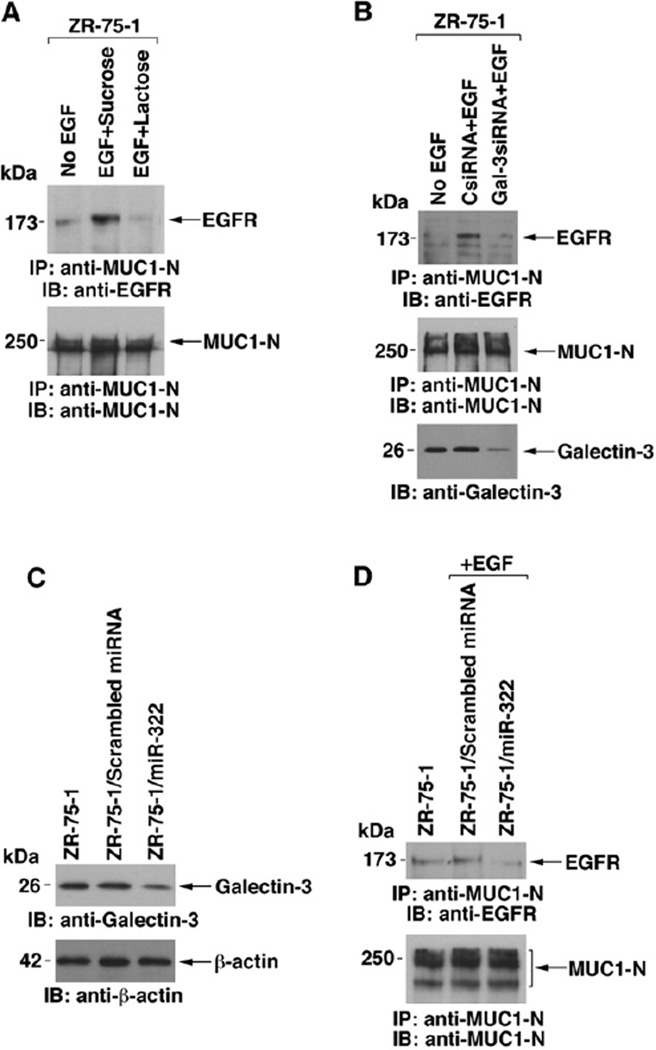
Previous work has demonstrated that MUC1 associates with EGFR constitutively and that this interaction is stimulated by EGF treatment (Li et al., 2001b). Other studies have shown that EGFR is crosslinked at the cell surface by galectin-3 (Partridge et al., 2004). To determine if galectin-3 functions in the interaction between MUC1 and EGFR, we treated ZR-75-1 cells with lactose, a competitive inhibitor of galectin-3 binding and, as a control, with sucrose. EGF-stimulated binding of MUC1 and EGFR was attenuated by lactose and not sucrose, indicating that galectin-3 may facilitate this interaction (Figure 7A). To extend the analysis, the ZR-75-1 cells were transfected with a pool of galectin-3-specific siRNAs or a control siRNA. The results demonstrate that silencing galectin-3 blocks the EGF-induced association of MUC1 and EGFR (Figure 7B). To relate these studies to the downregulation of galectin-3 by miR-322, we transfected ZR-75-1 cells with pre-miR-322 or a scrambled miRNA. As found in BT549/MUC1 cells (Figure 3E), ectopic miR-322 decreased galectin-3 expression (Figure 7C). Moreover, downregulation of galectin-3 by ectopic miR-322 attenuated the EGF-induced interaction between MUC1 and EGFR (Figure 7D). EGF stimulation is associated with increased colocalization of MUC1 and EGFR at the cell membrane (Li et al., 2001b). As shown previously by confocal microscopy (Li et al., 2001b), EGFR and MUC1 are uniformly distributed over the cell membrane of control ZR-75-1 cells (Figure S5A). After EGF stimulation, EGFR and MUC1 cluster in patches (Li et al., 2001b). The colocalization of EGFR and MUC1 in clusters was blocked by lactose and not sucrose (Figure S5A). Consistent with involvement of galectin-3, EGF-induced clustering of EGFR and MUC1 was also blocked by silencing galectin-3 (Figure S5B). These findings indicate that MUC1 suppression of miR-322 and thereby upregulation of galectin-3 is of importance to the interaction between MUC1 and EGFR.
Figure 7. Interaction between MUC1 and EGFR Is Mediated by Galectin-3.
(A) ZR-75-1 cells were incubated with sucrose or lactose in complete medium for 24 hr and then with sucrose and lactose for an additional 24hr in 0.1% serum. The cells were then stimulated with EGF for5 min.
(B) ZR-75-1 cells were transfected with a control siRNA or a galectin-3 siRNA pool for 48hr and then grown in the presence of 0.1% serum for 24 hr. The cells were then stimulated with EGF for 5 min. Anti-MUC1-N precipitates were immunoblotted with the indicated antibodies (A and B). Lysates were also directly immunoblotted with anti-galectin-3 (B).
(C and D) ZR-75-1 cells were transfected with a pre-miR-322 or a scrambled miRNA and selected in blasticidin. Lysates were immunoblotted with the indicated antibodies (C). Anti-MUC1-N precipitates were immunoblotted with the indicated antibodies (D).
DISCUSSION
MUC1 C-Terminal Subunit Is Expressed as N-Glycosylated and Unglycosylated Forms
MUC1 is over expressed in human malignancies as a heterodimer of the MUC1-N and MUC1-C subunits. MUC1-N is the large mucin component of the molecule with the characteristic variable numbers of tandem repeats that are extensively modified by O-linked glycans. The MUC1-C subunit was believed to function primarily in tethering MUC1-N at the cell membrane. However, the findings that MUC1-C interacts with diverse signaling molecules (Huang et al., 2005; Li et al., 2001a; Raina et al., 2006), localizes to the nucleus (Li et al., 2003a, 2003b, 2003c) and mitochondria (Ren et al., 2004, 2006), and induces transformation (Huang et al., 2005; Li et al., 2003b) have generated interest in the structure and function of this subunit. The present studies demonstrate that MUC1-C is endogenously expressed as a 25–20 kDa form that is glycosylated on Asn-36. Treatment of MUC1-C with N-glycosidase, expression of MUC1 in N-glycosylation-deficient CHO cells, and mutation of MUC1-C at Asn-36 were each associated with loss of the 25-20 kDa species. MUC1-C is a 158 amino acid protein with a predicted molecular mass of 17 kDa. In this regard, MUC1-C is also detectable as a 17 kDa species, the electrophoretic mobility of which is not affected by N-glycosidases or expression in glycosylation-deficient cells. Consistent with the MUC1-C 25-20 kDa form being a glycosylated product of the 17 kDa backbone, digestion with N-glycosidases was associated with conversion of the 25-20 kDa glycoprotein to the 17 kDa protein. MUC1-C is also expressed as a ~15 kDa species, which is presently under study as a putative cleavage product of the 17 kDa protein. These findings indicate that MUC1-C as found endogenously in diverse types of carcinoma cells (see Figure 1A) is expressed as N-glycosylated and unglycosylated forms.
MUC1 Upregulates Galectin-3 Expression by a Posttranscriptional Mechanism
Galectin-3 is a unique member of the galectin family that, in addition to the CRD, has an N-terminal region capable of forming oligomers (Ahmad et al., 2004). Overexpression of galectin-3 has been associated with the development of human malignancies (Califice et al., 2004; Liu and Rabinovich, 2005; van den Brule et al., 2004). Overexpression of MUC1 is also found in diverse human cancers (Kufe et al., 1984); however, there has been no evidence supporting a relationship between MUC1 and galectin-3. In the present work, studies in different types of human cancer cell lines suggested that MUC1 and galectin-3 expression could be coordinately regulated. Moreover, use of cells with gain and silencing of MUC1 demonstrated that MUC1 upregulates galectin-3 expression at the mRNA level. The human galectin-3 gene (LGALS3), located on chromosome 14q21–22, includes a promoter region that contains putative elements for activation by the SP1, AP-1, CREB, and NF-κB transcription factors (Kadrofske et al., 1998). In this regard, c-Jun, CREB, and NF-κB have been implicated in activation of the galectin-3 gene (Dumic et al., 2006; Liu and Rabinovich, 2005). Of potential relevance, MUC1-C has been shown to function as a coactivator of β-catenin, p53-, and estrogen receptor α-mediated gene transcription (Huang et al., 2005; Wei et al., 2005, 2006), indicating that MUC1 might function in coactivation of galectin-3 expression. However, our studies with galectin-3 promoter-Luc reporter constructs and with nuclear run-on assays demonstrated that MUC1 has no apparent effect on transcription of the galectin-3 gene. In contrast, assessment of galectin-3 mRNA stability and results obtained with the galectin-3 3′UTR ligated into the pMIR-Luc reporter indicated that MUC1 increases galectin-3 expression by stabilizing the galectin-3 transcript. We also found that MUC1-C-in-duced activation of pMIR-Luc-gal-3(3′UTR) and upregulation of galectin-3 mRNA levels are substantially attenuated by expressing MUC1-C that is defective as a substrate for N-glycosylation. These findings indicate that MUC1-C induces galectin-3 expression by a posttranscriptional mechanism and that N-glycosylation of MUC1-C Asn-36 is necessary for this response.
MUC1-C Suppresses MiR-322 in the Regulation of Galectin-3 Expression
miRNAs are noncoding RNAs of about 22 nucleotides that posttranscriptionally regulate gene expression. Base pairing of the miRNA with complementary sequences in an mRNA 3′UTR leads to suppression of translation or decreases in mRNA stability (Bartel, 2004). Translational suppression is a more commonly described mechanism of miRNA action; however, there are now increasing reports of miRNA-mediated mRNA degradation (Bagga et al., 2005; Krutzfeldt et al., 2005; Lim et al., 2005; Wu and Belasco, 2005). Binding of the miRNA is associated with accelerated deadenylation of the mRNA (Wu et al., 2006). In the present work, we identified miR-322 as a putative regulator of galectin-3 expression and demonstrated that miR-322 is expressed in human cells. Alignment of miR-322 sequences with the galectin-3 3′UTR showed incomplete complementarity and G-U base pairing typically found for binding of miRNAs to their targets (Esquela-Kerscher and Slack, 2006). The precursor stem-loop sequence for miR-322 is located at chromosome Xq26.3. The genomic annotation specifies overlapping of the miR-322 precursor sequence with the 5′UTR of a hypothetical gene, designated MGC16121, indicating that both the pre-miRNA and the MGC16121 mRNA may originate from the same transcript. In this regard, like miR-322, MUC1 suppresses MGC16121 mRNA levels (data not shown). Upstream to the MGC16121 5′UTR is a putative promoter with a TATA box and, within 2000 bp, potential binding sites for 39 different transcription factors. Our results demonstrate that (1) MUC1 suppresses miR-322 expression and (2) MUC1-C is sufficient for the suppression. Importantly, and in concert with the effects of MUC1-C on both miR-322 and galectin-3, antisense inhibition of miR-322 resulted in upregulation of galectin-3 expression. Ectopic expression of pre-miR-322 in BT549/MUC1 cells was also associated with downregulation of galectin-3 mRNA and protein. Moreover, consistent with the demonstration that glycosylation of MUC1-C on Asn-36 is necessary for upregulation of galectin-3, the MUC1-C(N36A) mutant had little if any effect on miR-322 levels. Whereas the MUC1-C(N36A) mutant disrupts binding with galectin-3, the effects of MUC1-C on galectin-3 mRNA stability could be secondary to regulation of the galectin-3 protein. However, the results obtained from expression of exogenous MUC1-C or MUC1-C(N36A) in BT549 cells indicate that the effects of MUC1-C on galectin-3 expression are mediated through suppression of miR-322.
Galectin-3 Binds to the MUC1-C Extracellular Domain
Galectin-3 is unique member of the galectin family that, in addition to the conserved CRD, contains an ND that functions in the formation of oligomers (Dumic et al., 2006). At the cell surface, galectin-3 binds to N-glycans on EGF, TGFβ, and T cell receptors (Demetriou et al., 2001; Partridge et al., 2004). Multimerization of galectin-3, in turn, generates crosslinking and lattice formation among these receptors and potentially other cell surface molecules. Our results demonstrate that glycosylation of MUC1-C on Asn-36 functions as a binding site for galectin-3. Deglycosylation of MUC1-C abrogates binding of MUC1-C and galectin-3, indicating that their association is not mediated by protein-protein interactions. Other work has shown that the MUC1 heterodimer forms complexes with EGFR (Li et al., 2001b; Schroeder et al., 2001); however, the basis for this interaction was not known. The present findings support a model in which galectin-3 functions in crosslinking MUC1 to EGFR and possibly other cell surface receptors. Thus, blocking galectin-3 binding with lactose, downregulating galectin-3 with ectopic miR-322, or silencing galectin-3 with siRNA was associated with inhibition of MUC1-EGFR complex formation in the response to EGF stimulation. Activation of EGFR also induces binding of the MUC1-C cytoplasmic domain to β-catenin (Huang et al., 2005; Li et al., 2001b), and this response is dependent on the galectin-3-mediated interaction between MUC1 and EGFR (data not shown). Our results also indicate that MUC1-C associates with galectin-3 intracellularly. Like MUC1-C, galectin-3 binds directly to β-catenin (Shimura et al., 2004). Moreover, like MUC1-C, galectin-3 localizes to the nucleus and coactivates β-catenin/Tcf4-mediated gene transcription (Shimura et al., 2004). MUC1-C and galectin-3 also both localize to mitochondria and block activation of the intrinsic apoptotic pathway (Dumic et al., 2006; Ren et al., 2004; Ren et al., 2006). Further study will be needed to determine whether MUC1-C and galectin-3 associate in the nucleus and mitochondria and whether their interaction regulates gene transcription and the apoptotic response.
MUC1 and Galectin-3 Function in a Positive Regulatory Loop
In summary, the present results support a model in which MUC1-C is glycosylated on Asn-36 and upregulates galectin-3 expression. In turn, galectin-3 binds to MUC1-C at the glycosylated Asn-36 site and integrates MUC1 with EGFR signaling. Of central importance to this MUC1-galectin-3 regulatory loop is miR-322, which blocks galectin-3 expression and is suppressed by MUC1-Cthat is glycosylated on Asn-36. Integration of this positive MUC1-C-galectin-3 regulatory loop by miR-322 differs from reports of miRNAs acting in a double-negative feedback loop to control neuronal fate decisions (Johnston et al., 2005) and in a negative feedback loop to prevent the accumulation of E2F transcription factors (Sylvestre et al., 2007). In the present model, overexpression of MUC1-C as found in human cancers suppresses miR-322 and thereby upregulates galectin-3, an effector of cell surface, nuclear, and mitochondrial signaling that, like MUC1-C, induces transformation and is present at increased levels in diverse carcinomas.
EXPERIMENTAL PROCEDURES
Cell Culture
Human breast cancer (ZR-75-1, BT549) and lung cancer (CRL-5985, CRL-5909) cells were grown in RPMI1640 medium supplemented with 10% fetal bovine serum (FBS), 100U/ml penicillin, and 100 µg/ml streptomycin. The human breast cancer (MD-MB-231, MCF-7), prostate cancer (LnCAP, DU145, PC3), lung cancer (A549), kidney epithelial (293), and Chinese hamster ovary (CHO-K1) cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS, antibiotics, and 1 mM L-glutamine. The glycosylation-deficient CHO-Lec1 and CHO-Lec8 cells (ATCC) were grown in alpha minimum essential medium containing 10% FBS and antibiotics. In certain studies, cells were cultured in medium with 0.1 % FBS for 24 hr and then stimulated with 100ng/ml EGF (Calbiochem-Novabiochem) for 5 min at 37°C.
Immunoblotting and Precipitates
Lysates were prepared from subconfluent cells as described (Li et al., 2001b). Immunoblot analysis was performed with anti-MUC1-C (Ab5; Neomarkers), anti-galectin-3 (Abcam), and anti-β-actin (Sigma). Immune complexes were prepared as described with anti-MUC1-C and anti-MUC1-N (Ren et al., 2004, 2006). For pull-down assays, GST or GST-galectin-3 was incubated with cell lysates for 2 hr at 4°C and precipitated with glutathione-Sepharose 4B. Precipitates were subjected to immunoblotting with anti-MUC1-C, anti-galectin-3 (Santa Cruz), anti-MUC1-N (DF3) (Ren et al., 2004), or anti-EGFR (Santa Cruz Biotechnology).
In Vitro Binding Assays
hFc and hFc-MUC1-C/ED were incubated with purified galectin-3 in lysis buffer for 1 hr at 4°C. The reaction products were precipitated with protein G-Sepharose and immunoblotted with anti-galectin-3. In other studies, hFc-MUC1-C/ED was incubated with GST-galectin-3 deletion mutants. The reaction products were precipitated with glutathione-Sepharose 4B and immunoblotted with anti-MUC1-C/ED (Ren et al., 2004).
Luciferase Assays
Cells were transfected with pGL3-galectin-3 promoter constructs pGal-3(−3000/+141)-Luc or pGal-3(−836/+141)-Luc and pcDNA-lacZ using Fugene6. Galectin-3 promoter activity was measured 24 hr after transfection. To assess function of the galectin-3 3′UTR, cells were transfected with pMIR-Gal-3(3′UTR) and pMIR-β-galactosidase using Fugene6 and assayed at 48 hr after transfection. Luciferase assays were performed with the luciferase assay system (Promega Corp.). The results were normalized to β-galactosidase activity and presented as relative luciferase activity.
Analysis of MiR-322 Expression
Northern blotting for detection of miRNAs was performed as described (Johnson et al., 2005; Lee and Ambros, 2001). Total cellular RNA (40 µg) isolated with TRIzol reagent (Invitrogen) was size fractionated in 15% urea-acrylamide gels. The RNA was transferred to positively charged nylon membranes (BrightStar-Plus, Ambion) by electroblotting. After UV crosslinking and prehybridization, the RNA was hybridized to miR-322 (5′-GCAGCGCCTCACGTTT-3′) and U6 snRNA (5′-GCGTGTCATCCTTGCGCAG-3′) 32P-labeled probes (Starfire Labeling System; Integrated DNA Technologies).
Antisense Inhibition of MiR-322
An antisense 2′-O-methyl oligoribonucleotide (5′-GGUAUAGCA GCGCCUCACGUUUUG-3′) against miR-322 and a scrambled 2′-O-methyl oligoribonucleotide (5′-CACGGAUCUAGGUCGUAACGUAGG-3′) (Integrated DNA Technologies) were transfected into BT549 cells (50 pmoles/10,000 cells) in the presence of Lipofectamine 2000. At 72 hr after transfection, the cells were analyzed for galectin-3 expression.
Ectopic Expression of Pre-MiR-322
Pre-miR constructs encoding mature miR-322 (5′-AAACGUGAGGCG CUGCUAUA-3′) or a scrambled control sequence (5′-GUCAGGUCA AACGGCCUAGAU-3′) were cloned into pcDNA 6.2-GW/miR vector (BLOCK-iT Pol II miR RNAi Expression Vector Kit; Invitrogen). The constructed clones were introduced into cells by Fugene6. The transfected cells were selected in 10 µg/ml blasticidin.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Grants CA97098 and CA100707 awarded by the National Cancer Institute. The authors acknowledge Kamal Chauhan for technical support and thank Daniel Tom, Harvard Center for Neurodegeneration and Repair, for assistance with the confocal studies. D.F. is a founder of Genus Oncology and a consultant to the company.
Footnotes
Supplemental Data
Supplemental Data include Supplemental Experimental Procedures, Supplemental References, and five figures and can be found with this article online at http://www.molecule.org/cgi/content/full/27/6/992/DC1/.
REFERENCES
- Ahmad N, Gabius HJ, Andre S, Kaltner H, Sabesan S, Roy R, Liu B, Macaluso F, Brewer CF. Galectin-3 precipitates as a pentamer with synthetic multivalent carbohydrates and forms heterogeneous cross-linked complexes. J. Biol. Chem. 2004;279:10841–10847. doi: 10.1074/jbc.M312834200. [DOI] [PubMed] [Google Scholar]
- Akahani S, Nangia-Makker P, Inohara H, Kim HR, Raz A. Galectin-3: a novel antiapoptotic molecule with a functional BH1 (NWGR) domain of Bcl-2 family. Cancer Res. 1997;57:5272–5276. [PubMed] [Google Scholar]
- Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Califice S, Castronovo V, Van Den Brule F. Galectin-3 and cancer. Int. J. Oncol. 2004;25:983–992. [PubMed] [Google Scholar]
- Demetriou M, Granovsky M, Quaggin S, Dennis JW. Negative regulation of T-cell activation and autoimmunity by Mgat5 N-glycosylation. Nature. 2001;409:733–739. doi: 10.1038/35055582. [DOI] [PubMed] [Google Scholar]
- Dumic J, Dabelic S, Flogel M. Galectin-3: an open-ended story. Biochim. Biophys. Acta. 2006;1760:616–635. doi: 10.1016/j.bbagen.2005.12.020. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Huang L, Chen D, Liu D, Yin L, Kharbanda S, Kufe D. MUC1 oncoprotein blocks GSK3β-mediated phosphorylation and degradation of β-catenin. Cancer Res. 2005;65:10413–10422. doi: 10.1158/0008-5472.CAN-05-2474. [DOI] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Johnston RJ, Jr., Chang S, Etchberger JF, Ortiz CO, Ho-bert O. MicroRNAs acting in a double-negative feedback loop to control a neuronal cell fate decision. Proc. Natl. Acad. Sci. USA. 2005;102:12449–12454. doi: 10.1073/pnas.0505530102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadrofske MM, Openo KP, Wang JL. The human LGALS3 (galectin-3) gene: determination of the gene structure and functional characterization of the promoter. Arch. Biochem. Biophys. 1998;349:7–20. doi: 10.1006/abbi.1997.0447. [DOI] [PubMed] [Google Scholar]
- Kasashima K, Nakamura Y, Kozu T. Altered expression profiles of microRNAs during TPA-induced differentiation of HL-60 cells. Biochem. Biophys. Res. Commun. 2004;322:403–410. doi: 10.1016/j.bbrc.2004.07.130. [DOI] [PubMed] [Google Scholar]
- Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Kufe D, Inghirami G, Abe M, Hayes D, Justi-Wheeler H, Schlom J. Differential reactivity of a novel monoclonal antibody (DF3) with human malignant versus benign breast tumors. Hybridoma. 1984;3:223–232. doi: 10.1089/hyb.1984.3.223. [DOI] [PubMed] [Google Scholar]
- Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- Levitin F, Stern O, Weiss M, Gil-Henn C, Ziv R, Prokocimer Z, Smorodinsky NI, Rubinstein DB, Wreschner DH. The MUC1 SEA module is a self-cleaving domain. J. Biol. Chem. 2005;280:33374–33386. doi: 10.1074/jbc.M506047200. [DOI] [PubMed] [Google Scholar]
- Li Y, Bharti A, Chen D, Gong J, Kufe D. Interaction of glycogen synthase kinase 3β with the DF3/MUC1 carcinoma-associated antigen and β-catenin. Mol. Cell. Biol. 1998;18:7216–7224. doi: 10.1128/mcb.18.12.7216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Kuwahara H, Ren J, Wen G, Kufe D. The c-Src tyrosine kinase regulates signaling of the human DF3/MUC1 carcinoma-associated antigen with GSK3β and β-catenin. J. Biol. Chem. 2001a;276:6061–6064. doi: 10.1074/jbc.C000754200. [DOI] [PubMed] [Google Scholar]
- Li Y, Ren J, Yu W-H, Li G, Kuwahara H, Yin L, Carraway KL, Kufe D. The EGF receptor regulates interaction of the human DF3/MUC1 carcinoma antigen with c-Src and β-catenin. J. Biol. Chem. 2001b;276:35239–35242. doi: 10.1074/jbc.C100359200. [DOI] [PubMed] [Google Scholar]
- Li Y, Chen W, Ren J, Yu W, Li Q, Yoshida K, Kufe D. DF3/MUC1 signaling in multiple myeloma cells is regulated by interleukin-7. Cancer Biol. Ther. 2003a;2:187–193. doi: 10.4161/cbt.2.2.282. [DOI] [PubMed] [Google Scholar]
- Li Y, Liu D, Chen D, Kharbanda S, Kufe D. Human DF3/MUC1 carcinoma-associated protein functions as an oncogene. Oncogene. 2003b;22:6107–6110. doi: 10.1038/sj.onc.1206732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Yu W-H, Ren J, Huang L, Kharbanda S, Loda M, Kufe D. Heregulin targets γ-catenin to the nucleolus by a mechanism dependent on the DF3/MUC1 protein. Mol. Cancer Res. 2003c;1:765–775. [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Micro-array analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lin HM, Pestell RG, Raz A, Kim HR. Galectin-3 enhances cyclin D(1) promoter activity through SP1 and a cAMP-responsive element in human breast epithelial cells. Oncogene. 2002;21:8001–8010. doi: 10.1038/sj.onc.1205820. [DOI] [PubMed] [Google Scholar]
- Liu FT, Rabinovich GA. Galectins as modulators of tumour progression. Nat. Rev. Cancer. 2005;5:29–41. doi: 10.1038/nrc1527. [DOI] [PubMed] [Google Scholar]
- Macao B, Johansson DG, Hansson GC, Hard T. Autoproteolysis coupled to protein folding in the SEA domain of the membrane-bound MUC1 mucin. Nat. Struct. Mol. Biol. 2006;13:71–76. doi: 10.1038/nsmb1035. [DOI] [PubMed] [Google Scholar]
- Matarrese P, Tinari N, Semeraro ML, Natoli C, Iacobelli S, Malorni W. Galectin-3 overexpression protects from cell damage and death by influencing mitochondrial homeostasis. FEBS Lett. 2000;473:311–315. doi: 10.1016/s0014-5793(00)01547-7. [DOI] [PubMed] [Google Scholar]
- Nangia-Makker P, Thompson E, Ochieng J, Raz A. Induction of tumorigenicity by gelectin-3 in a non-tumorigenic human breast carcinoma cell line. Int. J. Oncol. 1995;7:1079–1087. doi: 10.3892/ijo.7.5.1079. [DOI] [PubMed] [Google Scholar]
- Ochieng J, Furtak V, Lukyanov P. Extracellular functions of galectin-3. Glycoconj. J. 2004;19:527–535. doi: 10.1023/B:GLYC.0000014082.99675.2f. [DOI] [PubMed] [Google Scholar]
- Partridge EA, Le Roy C, Di Guglielmo GM, Pawling J, Cheung P, Granovsky M, Nabi IR, Wrana JL, Dennis JW. Regulation of cytokine receptors by Golgi N-glycan processing and endocytosis. Science. 2004;306:120–124. doi: 10.1126/science.1102109. [DOI] [PubMed] [Google Scholar]
- Patterson RJ, Wang W, Wang JL. Understanding the biochemical activities of galectin-1 and galectin-3 in the nucleus. Glycoconj. J. 2004;19:499–506. doi: 10.1023/B:GLYC.0000014079.87862.c7. [DOI] [PubMed] [Google Scholar]
- Raina D, Kharbanda S, Kufe D. The MUC1 oncoprotein activates the anti-apoptotic PI3K/Akt and Bcl-xL pathways in rat 3Y1 fibroblasts. J. Biol. Chem. 2004;279:20607–20612. doi: 10.1074/jbc.M310538200. [DOI] [PubMed] [Google Scholar]
- Raina D, Ahmad R, Kumar S, Ren J, Yoshida K, Kharbanda S, Kufe D. MUC1 oncoprotein blocks nuclear targeting of c-Abl in the apoptotic response to DNA damage. EMBO J. 2006;25:3774–3783. doi: 10.1038/sj.emboj.7601263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Li Y, Kufe D. Protein kinase C d regulates function of the DF3/MUC1 carcinoma antigen in β-catenin signaling. J. Biol. Chem. 2002;277:17616–17622. doi: 10.1074/jbc.M200436200. [DOI] [PubMed] [Google Scholar]
- Ren J, Agata N, Chen D, Li Y, Yu W-H, Huang L, Raina D, Chen W, Kharbanda S, Kufe D. Human MUC1 carcinoma-associated protein confers resistance to genotoxic anti-cancer agents. Cancer Cell. 2004;5:163–175. doi: 10.1016/s1535-6108(04)00020-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Bharti A, Raina D, Chen W, Ahmad R, Kufe D. MUC1 oncoprotein is targeted to mitochondria by heregulin-induced activation of c-Src and the molecular chaperone HSP90. Oncogene. 2006;25:20–31. doi: 10.1038/sj.onc.1209012. [DOI] [PubMed] [Google Scholar]
- Schroeder J, Thompson M, Gardner M, Gendler S. Transgenic MUC1 interacts with epidermal growth factor receptor and correlates with mitogen-activated protein kinase activation in the mouse mammary gland. J. Biol. Chem. 2001;276:13057–13064. doi: 10.1074/jbc.M011248200. [DOI] [PubMed] [Google Scholar]
- Seetharaman J, Kanigsberg A, Slaaby R, Leffler H, Barondes SH, Rini JM. X-ray crystal structure of the human galectin-3 carbohydrate recognition domain at 2.1-A resolution. J. Biol. Chem. 1998;273:13047–13052. doi: 10.1074/jbc.273.21.13047. [DOI] [PubMed] [Google Scholar]
- Shimura T, Takenaka Y, Tsutsumi S, Hogan V, Kikuchi A, Raz A. Galectin-3, a novel binding partner of beta-catenin. Cancer Res. 2004;64:6363–6367. doi: 10.1158/0008-5472.CAN-04-1816. [DOI] [PubMed] [Google Scholar]
- Sylvestre Y, De Guire V, Querido E, Mukhopadhyay UK, Bour-deau V, Major F, Ferbeyre G, Chartrand P. An E2F/miR-20a autoregulatory feedback loop. J. Biol. Chem. 2007;282:2135–2143. doi: 10.1074/jbc.M608939200. [DOI] [PubMed] [Google Scholar]
- Takenaka Y, Inohara H, Yoshii T, Oshima K, Nakahara S, Aka-hani S, Honjo Y, Yamamoto Y, Raz A, Kubo T. Malignant transformation of thyroid follicular cells by galectin-3. Cancer Lett. 2003;195:111–119. doi: 10.1016/s0304-3835(03)00056-9. [DOI] [PubMed] [Google Scholar]
- Takenaka Y, Fukumori T, Raz A. Galectin-3 and metastasis. Glycoconj. J. 2004a;19:543–549. doi: 10.1023/B:GLYC.0000014084.01324.15. [DOI] [PubMed] [Google Scholar]
- Takenaka Y, Fukumori T, Yoshii T, Oka N, Inohara H, Kim HR, Bresalier RS, Raz A. Nuclear export of phosphorylated galectin-3 regulates its antiapoptotic activity in response to chemo-therapeutic drugs. Mol. Cell. Biol. 2004b;24:4395–4406. doi: 10.1128/MCB.24.10.4395-4406.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brule F, Califice S, Castronovo V. Expression of galectins in cancer: a critical review. Glycoconj. J. 2004;19:537–542. doi: 10.1023/B:GLYC.0000014083.48508.6a. [DOI] [PubMed] [Google Scholar]
- Walzel H, Blach M, Hirabayashi J, Arata Y, Kasai K, Brock J. Galectin-induced activation of the transcription factors NFAT and AP-1 in human Jurkat T-lymphocytes. Cell. Signal. 2002;14:861–868. doi: 10.1016/s0898-6568(02)00035-9. [DOI] [PubMed] [Google Scholar]
- Wei X, Xu H, Kufe D. Human MUC1 oncoprotein regulates p53-responsive gene transcription in the genotoxic stress response. Cancer Cell. 2005;7:167–178. doi: 10.1016/j.ccr.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Wei X, Xu H, Kufe D. MUC1 oncoprotein stabilizes and activates estrogen receptor alpha. Mol. Cell. 2006;21:295–305. doi: 10.1016/j.molcel.2005.11.030. [DOI] [PubMed] [Google Scholar]
- Wu L, Belasco JG. Micro-RNA regulation of the mammalian lin-28 gene during neuronal differentiation of embryonal carcinoma cells. Mol. Cell. Biol. 2005;25:9198–9208. doi: 10.1128/MCB.25.21.9198-9208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl. Acad. Sci. USA. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Kufe D. Human MUC1 carcinoma antigen regulates intracellular oxidant levels and the apoptotic response to oxidative stress. J. Biol. Chem. 2003;278:35458–35464. doi: 10.1074/jbc.M301987200. [DOI] [PubMed] [Google Scholar]
- Yoshii T, Fukumori T, Honjo Y, Inohara H, Kim HR, Raz A. Galectin-3 phosphorylation is required for its anti-apoptotic function and cell cycle arrest. J. Biol. Chem. 2002;277:6852–6857. doi: 10.1074/jbc.M107668200. [DOI] [PubMed] [Google Scholar]
- Yu F, Finley RL, Jr., Raz A, Kim HR. Galectin-3 translocates to the perinuclear membranes and inhibits cytochrome c release from the mitochondria. A role for synexin in galectin-3 translocation. J. Biol. Chem. 2002;277:15819–15827. doi: 10.1074/jbc.M200154200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.