ABSTRACT
The actin cytoskeleton and its network of associated proteins constitute a physical barrier that viruses must circumvent to gain entry into cells for productive infection. The mechanisms by which the physical signals of infection are sensed by the host to activate an innate immune response are not well understood. The antiviral endoribonuclease RNase L is ubiquitously expressed in a latent form and activated upon binding 2-5A, a unique oligoadenylate produced during viral infections. We provide evidence that RNase L in its inactive form interacts with the actin-binding protein Filamin A to modulate the actin cytoskeleton and inhibit virus entry. Cells lacking either RNase L or Filamin A displayed increased virus entry which was exacerbated in cells lacking both proteins. RNase L deletion mutants that reduced Filamin A interaction displayed a compromised ability to restrict virus entry, supporting the idea of an important role for the RNase L-Filamin A complex in barrier function. Remarkably, both the wild type and a catalytically inactive RNase L mutant were competent to reduce virus entry when transfected into RNase L-deficient cells, indicating that this novel function of RNase L is independent of its enzymatic activity. Virus infection and RNase L activation disrupt its association with Filamin A and release RNase L to mediate its canonical nuclease-dependent antiviral activities. The dual functions of RNase L as a constitutive component of the actin cytoskeleton and as an induced mediator of antiviral signaling and effector functions provide insights into its mechanisms of antiviral activity and opportunities for the development of novel antiviral agents.
IMPORTANCE
Cells constantly face and sample pathogens on their outer surface. The actin cytoskeleton and interacting proteins associate with the cell membrane and constitute a barrier to infection. Disruption of the actin cytoskeleton allows viruses to enter the cell and induces innate immune responses to clear infections. The molecular mechanisms that link virus-induced physical perturbations to host defense pathways remain unclear. Our studies identified a novel interaction between the antiviral endoribonuclease RNase L and the actin-binding protein Filamin A that enhances host defense by preventing viral entry into naive cells. This role for RNase L is independent of its enzymatic function. Virus infection alters actin dynamics, disrupts the RNase L-Filamin A complex, and releases RNase L to mediate antiviral signaling and effector functions via its established nucleolytic activities. These dual roles for RNase L provide an efficient strategy to protect cells from infection and rapidly respond upon pathogen exposure.
INTRODUCTION
Viral infection is initiated by attachment to its cellular receptors. The subsequent activation of receptor-mediated endocytosis results in actin clustering and perturbation of the actin cytoskeleton (1). The disruption of barrier function provided by the actin cytoskeleton is a central component of infection by diverse pathogens and exposes viral pathogen-associated molecular patterns (PAMPs) to host pattern recognition receptors (PRRs) that activate an innate immune response (2–4). Pathogen recognition by PRRs results in the induction of cytokines and chemokines, including type I interferon (IFN), that mediate pleiotropic activities to clear viral infections (4). Consistent with a key role for disruption of the actin cytoskeleton in virus infection, viruses have evolved strategies to perturb this barrier by targeting actin-binding proteins. For instance, the actin-binding protein Rac1 GTPase is required for actin remodeling to facilitate entry, virus production, and release of dengue virus and coxsackievirus (5, 6). Influenza virus interacts with the actin cytoskeleton in all stages of the viral life cycle, including entry, replication, antiviral signaling, and intracellular trafficking, for egress in polarized cells (7). Gelsolin, an actin-severing protein, restricts human immunodeficiency virus type 1 (HIV-1) infection in a prefusion step and is inhibited by the IFN-regulated antiviral RNA-dependent protein kinase PKR (8, 9). Recent studies have implicated AMP-related protein kinase (AMPK) activity in actin polymerization that increased entry of both vaccinia virus and Ebola virus (10, 11). Thus, while pathogen targeting of actin-binding proteins as a strategy to promote infection is well established, less is known about how the physical disruption of the actin cytoskeleton is sensed by the host to activate established innate immune effectors.
Filamin A is a multifunctional actin-binding protein which cross-links actin filaments into a network through its N-terminal actin-binding domain (ABD). The C-terminal β-sheets form immunoglobulin-like domains that provide interfaces for protein-protein interactions. In this manner, Filamin A serves as a scaffold to connect the actin cytoskeleton with over 60 functionally diverse cellular proteins, including membrane receptors, signaling molecules, and DNA repair proteins. In the context of pathogen-host interactions, Filamin A serves as an adaptor protein that links HIV-1 receptors to the actin cytoskeleton to promote viral entry (12, 13) and associates with viral NS3 and NS5A proteins in chronic hepatitis C virus (HCV) infection (14). Filamin A binds to Integrin β1 (15), and integrins are receptors for adenoviruses and coxsackievirus. These viruses use Filamin A as an adaptor to connect receptor binding to the actin cytoskeleton that is required for their life cycles (16, 17). Filamin A thus functions in infection by diverse viral pathogens; however, little is known about its interactions with components of the host innate immune response.
RNase L is a unique endoribonuclease that is activated during viral infections or in cells treated with IFN (18–20). IFN induces transcription of IFN-stimulated genes (ISGs), including 2′-5′-oligoadenylate synthetase (OAS). OAS proteins are activated by viral double-stranded RNA (dsRNA) PAMPs to produce 2-5A [px5′A(2′p5′A)n, where x = 1 to 3 and n = ≥2] from cellular ATP, which in turn binds specifically to the latent endoribonuclease, RNase L (21). To date, the only well-established function of 2-5A is activation of RNase L. 2-5A binding promotes dimerization of RNase L and converts it to an active enzyme (21, 22). Activated RNase L cleaves single-stranded viral and host RNAs to mediate its antiviral and antiproliferative activities (23, 24). Thus, in the context of virus infection, RNase L is thought to function after release of viral nucleic acids and interferon production to activate OAS and synthesize 2-5A from ATP. Structure-function analysis of RNase L revealed that the R667A substitution inhibits endoribonuclease activity (nuclease dead) and that the R462Q substitution impairs the ability of RNase L to dimerize (25–28). In addition to the direct cleavage of viral RNA to inhibit replication, the products of RNase L cleavage include small RNA with duplex structures which can signal through the RIG-I-like helicases RIG-I (retinoic acid-inducible gene I) and MDA5 (melanoma differentiation-associated protein 5) to amplify the production of IFN-β (29).
In addition to the established roles of RNase L as an antiviral protein that require its endoribonuclease activity, RNase L interacts with several cellular proteins that may provide alternative mechanisms by which it mediates biological functions. For example, RNase L interacts with IQGAP1 (an IQ [isoleucine-glutamine] motif containing GTPase activating protein 1) (30), which functions as an assembly scaffold for the organization of a multimolecular complex and could interface with incoming signals to induce reorganization of the actin cytoskeleton. The interaction with IQGAP1 may thus position RNase L to respond to viral pathogens that target the actin cytoskeleton. Recent studies have identified interactions of RNase L with extracellular matrix (ECM) and cytoskeletal proteins (31). In this regard, RNase L has been shown to regulate tight-junction proteins and maintain barrier integrity in intestinal epithelial cells during enteropathogenic Escherichia coli infection (32). Relatedly, RNase L interacts with the androgen receptor (AR) (33) that, in turn, interacts with Filamin A (34). These observations suggested that RNase L may modulate the cytoskeleton. Therefore, we investigated a potential interaction of RNase L with Filamin A that may influence the cellular actin network. Here we identify Filamin A as a novel RNase L interacting protein under resting, uninfected conditions. This interaction functioned to limit viral entry via a mechanism that is independent of RNase L enzymatic activity and virus-induced production of 2-5A. Consistent with this role, activation of RNase L by 2-5A or viral infections disrupted the interaction, leading to canonical RNase L activity in antiviral signaling. Cells lacking either RNase L or Filamin A or lacking both proteins exhibited an altered actin cytoskeleton, enhanced entry of virus, and compromised antiviral activity, supporting the idea of an important function for this interaction. Together, these studies identified a previously unrecognized nonenzymatic role for RNase L to inhibit virus entry through interaction with Filamin A.
RESULTS
RNase L interacts with Filamin A and dissociates upon activation.
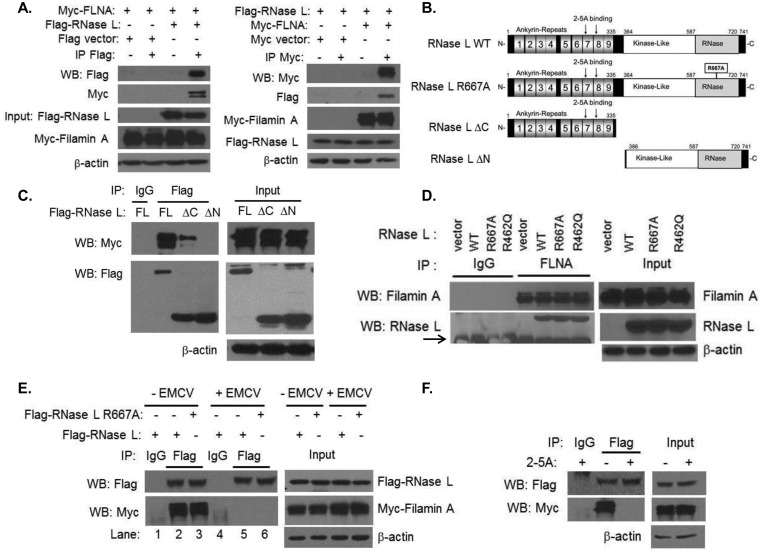
RNase L was reported to interact with the androgen receptor (AR) which also interacts with Filamin A, suggesting that RNase L may interact, directly or indirectly, with Filamin A. To determine if these proteins associate in cells, HEK293T cells were transfected with Flag-RNase L and Myc-Filamin A. Immunoprecipitation (IP) of either RNase L or Filamin A from transfected cells demonstrated the presence of Filamin A or RNase L in IP complexes, respectively (Fig. 1A). An N-terminal proteolytic product of Filamin A is also detected as has been reported in other cells (34, 35). HEK293T cells do not express androgen receptor, suggesting that the interaction is independent of the presence of the AR. We further confirmed the interaction of RNase L and Filamin A in HT1080 cells (data not shown). RNase L has a modular structure with an N-terminal ankyrin repeat region that contains the sites for binding of its activator, 2-5A, a C-terminal nuclease domain, and a kinase-like domain in the middle of the protein (Fig. 1B). To identify the region of RNase L that interacts with Filamin A, we used RNase L deletion constructs that comprised N-terminal residues 1 to 335 (ΔC, lacking nuclease domain) or C-terminal residues 386 to 741 (ΔN, lacking ankyrin repeats) in coimmunoprecipitation experiments with full-length Myc-Filamin A. Interaction of the ΔC RNase L protein with Filamin A was slightly reduced compared to that seen with full-length RNase L; however, deletion of the N-terminal residues, including the ankyrin repeats in the ΔN construct, abrogated Filamin A binding (Fig. 1C; see also Fig. S2 in the supplemental material). These results suggested that the N-terminal half of RNase L is essential for Filamin A interaction and that, while they are not required, the C-terminal residues may impact the interaction by affecting protein folding. To determine if endogenous Filamin A can bind to RNase L, we used HeLa cells which express very small amounts of RNase L protein (36) that were reconstituted with a vector alone, wild-type (WT) RNase L, or RNase L point mutants R667A and R462Q, which render the enzyme nuclease dead and dimerization deficient, respectively (36, 37). Filamin A coimmunoprecipitated with WT RNase L and to comparable levels with mutant forms of the protein, suggesting that the interaction did not require the enzymatic or dimerization capacities of RNase L (Fig. 1D). RNase L is expressed as an inactive monomer in the absence of its 2-5A activator that is produced in response to virus infection or IFN treatment. To analyze the impact of RNase L activation on its interaction with Filamin A in the context of viral infection, cells expressing Filamin A and WT or nuclease-dead (R667A) RNase L were infected with encephalomyocarditis virus (EMCV), which is known to activate RNase L (29, 38, 39), or left uninfected. Interestingly, the interaction between Filamin A and either WT or nuclease-dead RNase L decreased significantly following virus infection (Fig. 1E). This finding suggested that RNase L activation during viral infection results in its dissociation from a Filamin A complex. To directly demonstrate that 2-5A-induced activation of RNase L results in its dissociation from Filamin A in cells, HEK293T cells expressing Flag-RNase L and Myc-Filamin A were transfected with 2-5A. Immunoprecipitation of RNase L indicated that its interaction with Filamin A was indeed reduced in the presence of 2-5A compared to the results seen with untreated cells (Fig. 1F). These results suggested that RNase L interacts with Filamin A under basal, unstimulated conditions but that the interaction is disrupted when RNase L is activated during viral infections.
FIG 1 .
RNase L interacts with Filamin A in uninfected cells and dissociates upon activation. (A) HEK293T cells expressing Flag-RNase L and Myc-Filamin A or Flag and Myc tag vector alone were immunoprecipitated with Flag-M2 agarose beads, Myc-EZview agarose beads, or control IgG. The samples were separated on SDS-PAGE, and the presence of interacting proteins was determined by immunoblot analysis. WB, Western blot analysis. (B) Domain organization and key features of RNase L and mutants used in this study. (C) Myc-Filamin A and Flag-RNase L (full length [FL]), Flag-RNase L (ΔC), or Flag-RNase L (ΔN) were immunoprecipitated using Flag-M2 agarose beads or control IgG. Blots were probed with anti-Myc or anti-Flag antibodies. For the IP analysis whose results are presented in panels C to F, 5% of the immunoprecipitate was loaded in blots probed with the antibodies indicated. (D) HeLa M cells stably expressing pcDNA3 vector, RNase L WT, RNase L R667A, or RNase L R462Q were immunoprecipitated with anti-Filamin A antibodies or control IgG. Endogenous Filamin A and coimmunoprecipitated RNase L proteins were detected on immunoblots using specific antibodies. IgG heavy chain is indicated by an arrow. (E) HEK293T cells expressing Flag-RNase L or Flag-RNase L R667A and Myc-Filamin A were infected with EMCV (MOI = 1.0) for 1 h or left uninfected. Cell lysates were immunoprecipitated with Flag-M2 agarose beads or control IgG. Samples were subjected to immunoblot analysis using anti-Flag or anti-Myc antibodies. (F) HEK293T cells expressing Flag-RNase L and Myc-Filamin A were transfected with 10 µM 2-5A for 1 h followed by immunoprecipitation and immunoblot analysis performed as described above. Protein expression in all lysates was normalized to β-actin levels. Data are representative of the results of 3 independent experiments.
Actin dynamics are altered in cells lacking RNase L.
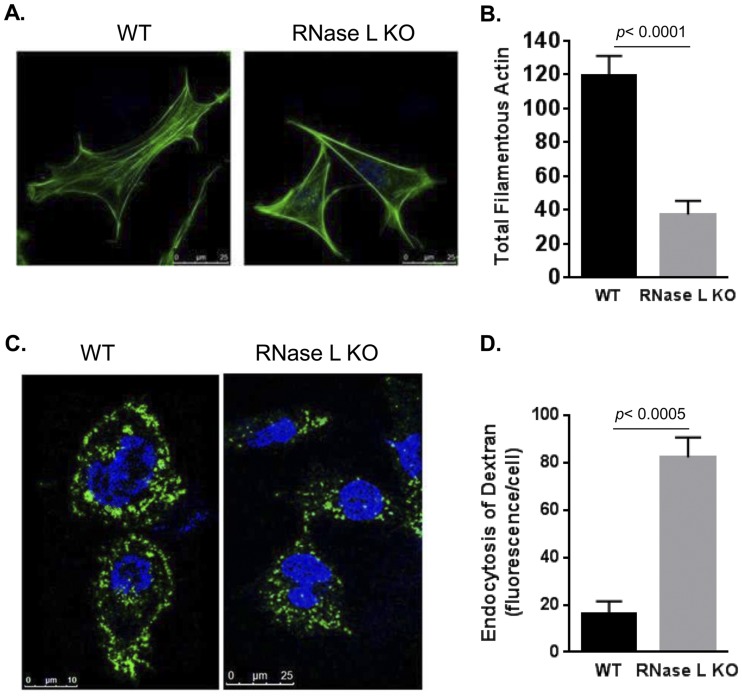
Actin networks define cell shape, mediate motility and migration, and are an important target for invading pathogens (1, 2). Filamin A modulates actin dynamics, and its interaction with RNase L suggested that RNase L deficiency may impact the actin cytoskeleton. Consistent with this prediction, WT and RNase L-knockout (KO) mouse embryonic fibroblasts (MEFs) exhibited distinct morphologies that may reflect alterations in the actin cytoskeleton. Therefore, we examined the organization of filamentous actin in RNase L KO and WT MEFs by staining with Alexa 488-labeled phalloidin. The immunofluorescence staining was analyzed using a Leica TCS SP5 multiphoton laser scanning confocal microscope (Fig. 2A). Quantification of the mean fluorescence intensity of stained cells normalized to surface area revealed a significant (30%) reduction in the total number of actin filaments in RNase L KO compared to WT MEFs (Fig. 2B). Alterations in the actin cytoskeleton are associated with functional changes, including macropinosome formation and nonselective particle uptake, that also serve as mechanisms for viral entry (40–42). To determine if the altered actin network observed in RNase L KO MEFs led to the induction of macropinocytosis, we compared endocytic activity levels in WT RNase L and KO MEFs by monitoring the uptake of soluble fluorescein isothiocyanate (FITC)-labeled dextran. RNase L KO MEFs showed a 5-fold increase in endocytic uptake as evidenced by the appearance of FITC-dextran granules in the cytoplasm, whereas most of the staining remained on the periphery of the WT cells (Fig. 2C and D). These results suggested that RNase L plays a role in regulating actin dynamics and actin-mediated cellular processes and that these functions may be mediated, in part, by its interaction with Filamin A.
FIG 2 .
RNase L regulates actin dynamics. (A) WT and RNase L KO primary MEFs were stained with Alexa 488 phalloidin to detect actin filaments (F-actin). (B) Total F-actin amounts were analyzed from 10 fields of MEFs from three independent experiments stained as described above and quantitated using ImageJ software. (C) Fluid-phase uptake of FITC-dextran (10 kDa) was performed in primary WT and RNase L KO MEFs starved in medium containing 1% serum. (D) The amount of fluorescent dextran internalized was quantified by marking the cell boundary, and the mean fluorescence inside the cell was normalized to the cell surface area using ImageJ and graphed. Representative images from triplicate experiments are shown. Quantitation data in panels A and B are from the results of three independent experiments and represent means ± standard errors (SE). P values from comparisons performed using Student’s t test are as shown.
RNase L enzymatic activity is not affected by Filamin A or disruption of the actin cytoskeleton.
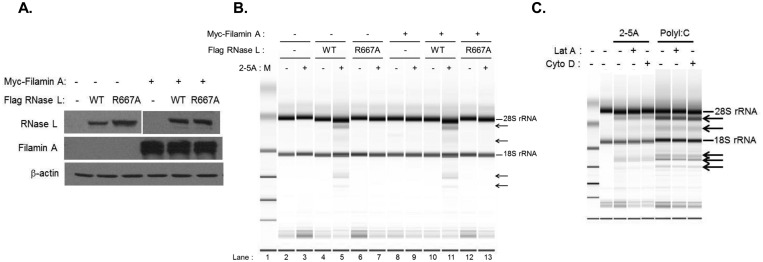
Our data indicate that RNase L associates with Filamin A under basal conditions and dissociates upon 2-5A-mediated activation (Fig. 1). To determine if the presence of Filamin A impacts RNase L enzymatic activity, we utilized the M2 melanoma cell line that lacks Filamin A mRNA and protein expression (43) and does not express detectable RNase L (Fig. 3A). To analyze RNase L activity in the presence and absence of Filamin A, M2 cell lines stably transfected with Filamin A and with either WT or catalytically inactive RNase L were generated and protein expression was validated by immunoblot analysis (Fig. 3A). Each cell line was then transfected with 2-5A to activate RNase L or mock transfected and analyzed for RNase L activity. 2-5A-induced activation of RNase L results in the cleavage of rRNA into discrete fragments that can be quantified as a measure of RNase L activity in cells (29, 44, 45). 2-5A transfection of M2 cells expressing WT but not catalytically inactive RNase L induced characteristic rRNA cleavage products in a manner independent of Filamin A expression (compare lanes 5 and 11 in Fig. 3B). Thus, consistent with the 2-5A-induced dissociation of Filamin A and RNase L (Fig. 1F), Filamin A did not alter the enzymatic activity of RNase L. Since RNase L deficiency resulted in a reduction in the levels of filamentous actin (Fig. 2A and B) that might have occurred via its interaction with Filamin A, we next determined if perturbation of actin filaments impacted RNase L activity. HT1080 cells that express functional endogenous RNase L (see Fig. S1 in the supplemental material) were pretreated with Cytochalasin D (Cyto D), which inhibits actin polymerization, or with Latrunculin A (Lat A), which prevents F-actin assembly and inhibits polymerization, or were left untreated. Cells were then transfected with 2-5A or with poly(I·C) to activate OAS and produce endogenous 2-5A, and RNase L activity was measured by monitoring rRNA cleavage. RNase L activity in response to poly(I·C) and, to a lesser extent, 2-5A was observed that was independent of cytoskeletal disruption (Fig. 3C). Together, these data indicate that the presence of the actin-binding protein Filamin A, or perturbation of the actin network, does not affect RNase L activity. Moreover, RNase L interacts with Filamin A only in the absence of its activator, suggesting that RNase L-dependent modulation of the actin cytoskeleton does not require its enzymatic activity.
FIG 3 .
Effect of Filamin A and actin cytoskeleton on RNase L activity. M2 stable cell lines expressing Myc-Filamin A and/or Flag-RNase L WT or Flag-RNase L R667A were analyzed for expression of RNase L and Filamin A on immunoblots (A) and transfected with 10 µM 2-5A (B), and RNase L-mediated cleavage of rRNA (arrows) was analyzed on RNA chips using an Agilent 2100 Bioanalyzer. (C) HT1080 cells were pretreated with Cytochalasin D (Cyto D; 5 µM), Latrunculin A (Lat A; 0.5 µM), or vehicle (DMSO) for 1 h followed by transfection with 10 µM 2-5A or 2 µg/ml of poly(I·C). Characteristic RNase L-generated rRNA cleavage products (arrows) were detected as described for panel B. Representative images from three independent experiments are shown.
RNase L and Filamin A reduce virus entry into cells.
Many viruses such as dengue virus and influenza virus use macropinocytosis as a mechanism of viral entry into cells (40–42). We demonstrated that cells lacking RNase L show an increase in nonselective uptake of fluid-phase dextran by macropinocytosis (Fig. 2C), suggesting that RNase L may play a role in forming a barrier for viral entry into cells. Indeed, Filamin A functions as a scaffold for cytoskeletal rearrangement that is associated with virus infection (12, 13) and RNase L may contribute to this activity via its interaction with Filamin A. We therefore tested the hypothesis that RNase L and Filamin A function together to reduce virus entry.
Sendai virus (SeV) is a paramyxovirus that is known to induce rearrangement of the actin cytoskeleton upon infection (2, 46); we therefore compared the levels of entry of Sendai virus (SeV) into M2 and A7 cells that lack or express both RNase L and Filamin A. A7 cells are M2 cells reconstituted with Filamin A and are widely used as control cells for the study of Filamin A (43, 47, 48). We determined that A7 cells also express functional RNase L protein (see Fig. S1 in the supplemental material). Entry of SeV into cells was detected by immunofluorescence using antibodies against SeV that recognize all structural proteins (49) (Fig. 4A) and quantified by measuring the fluorescence in a defined region normalized to the cell surface area. This analysis was performed at 1 h postinfection to provide an indication of the impact of Filamin A and RNase L on virus entry prior to replication and activation of RNase L and other innate immune responses. In A7 cells that express Filamin A and RNase L, most of the virus was restricted to the cell membrane. In contrast, M2 cells that lack these proteins exhibited 2.5-fold-higher levels of intracellular SeV staining (Fig. 4A and C), suggesting that RNase L and Filamin A function to restrict virus entry. To determine if RNase L deficiency alone also led to increased virus entry, SeV entry was analyzed in WT and RNase L KO MEFs (Fig. 4B). Consistent with the enhanced uptake of dextran in RNase L KO MEFs (Fig. 2C), the intracellular SeV immunofluorescence level was increased 4-fold in RNase L KO compared to WT MEFs (Fig. 4B and D). Further quantification of SeV entry by quantitative real-time PCR revealed a 40% decrease in intracellular viral RNA levels in WT compared to KO MEFs (Fig. 4E). Together, these data suggest that RNase L, perhaps via its interaction with Filamin A, functions to inhibit virus entry as a novel mechanism of its antiviral activity. To dissect the relative contributions of RNase L and Filamin A in the inhibition of viral entry and assess the potential role of RNase L enzymatic activity in this function, we used RNase L- and Filamin A-deficient M2 cells that had been stably transfected with Filamin A, WT RNase L, or catalytically inactive (R667A) RNase L alone and in combination as shown in Fig. 3A. Cells were infected with SeV or EMCV for 1 h to allow viral entry and then washed with acidic glycine to inactivate extracellular viruses. Intracellular virus was quantified by quantitative real-time PCR for SeV RNA or plaque assay for EMCV (45). SeV entry into M2 cells expressing WT, R667A, or Filamin A proteins was inhibited by more than 3.5-log and EMCV entry was reduced by 2-log compared to the levels seen with M2 cells which lack both proteins (Fig. 4F and G). Strikingly, coexpression of Filamin A protein with either WT or R667A mutant RNase L resulted in a significant further inhibition of entry by both viruses (Fig. 4F and G). Similar effects of Filamin A and WT or R667A RNase L on virus titer were observed when the acidic glycine wash was omitted; however, intracellular EMCV titers were marginally higher, likely due to the prolonged exposure to viable virus (see Fig. S3 in the supplemental material). These findings demonstrate novel roles for RNase L and Filamin A in restricting virus entry. While expression of either protein resulted in a significant reduction in the level of virus entry, the presence of both proteins further decreased virus entry levels, suggesting that RNase L and Filamin A function as part of a mechanical barrier to virus entry. More remarkable is the observation that the inhibition of virus entry by RNase L occurs in a manner independent of its enzymatic activity, suggesting a novel structural mechanism by which RNase L mediates its antiviral activity. To evaluate the contribution of RNase L-Filamin A interaction to viral entry, M2 cells stably expressing Filamin A were transfected with vector alone, full-length RNase L (WT), or N- and C-terminal deletion mutants (ΔN and ΔC) that abolish and reduce Filamin A interactions, respectively (Fig. 1C). Cells were then infected with SeV to allow virus entry for 1 h, and intracellular virus was quantified as described above. Expression of WT RNase L resulted in a significant 52-fold reduction in virus entry compared to the level seen with cells lacking RNase L expression (Fig. 5, WT versus none). In contrast, expression of the ΔN RNase L mutant that does not bind Filamin A led to a dramatic 23-fold increase in virus entry compared to the level seen with WT RNase L (Fig. 5). The ΔC RNase L mutant, which exhibits only slightly reduced Filamin A binding compared to WT RNase L, resulted in a correspondingly modest 7-fold increase in SeV entry over that observed for WT RNase L (Fig. 5). These results link RNase L-Filamin A interaction with the restriction of viral entry and support an important role for this interaction in barrier function.
FIG 4 .
Regulation of virus entry by RNase L and Filamin A. (A and C) Immunofluorescence of A7 and M2 melanoma cells infected with SeV (40 HAU/ml) for 1 h. Unbound virus was removed, and cells were stained with antibody against SeV and detected using Alexa 633-conjugated secondary antibodies and imaged. Virus entry was quantitated by measuring the mean fluorescence intensity (in thousands) in cells normalized to the cell surface area (n = 10 from three independent experiments). (B and D) WT and RNase L KO MEFs were infected, stained, and quantitated as described for panel A (n = 10 from three independent experiments). (E) WT and RNase L KO MEFs were infected with SeV (40 HAU/ml) for 1 h. After unbound virus was removed, entry was quantitated by determining copy numbers of SeV genomic RNA strands by real-time quantitative PCR (qPCR) as described in Materials and Methods. (F and G) M2 stable cells expressing Myc-Filamin A and/or Flag-RNase L WT or Flag-RNase L R667A were infected with SeV (40 HAU/ml) (F) or EMCV (MOI = 1.0) (G) for 1 h and washed briefly with acidic glycine to remove unbound extracellular viruses. Intracellular EMCV titers were determined by a plaque assay using L929 cells, and copy numbers of SeV genomic RNA strands were determined by real-time qPCR and normalized to GAPDH mRNA levels. Student’s t test was used to determine P values (as shown) for comparisons of M2 cells expressing Filamin A to cells expressing Filamin A and either RNase L WT or R667A. Data are representative of the results of two independent experiments performed in triplicate ± standard deviations (SD).
FIG 5 .
Effect of RNase L and Filamin A interaction on viral entry. (A) M2 cells stably expressing Myc-Filamin A transfected with Flag-RNase L (full length), Flag-RNase L (ΔC), or Flag-RNase L (ΔN) or with vector (none) were infected with SeV (40 HAU/ml) for 1 h and washed briefly with acidic glycine to remove unbound extracellular viruses. Copy numbers of SeV genomic RNA strands were determined by real-time qPCR and normalized to GAPDH mRNA levels. Student’s t test was used to determine P values (as shown). (B) Fold change in viral entry was normalized to WT data. *, P < 0.01; **, P < 0.001 (for comparisons using Student’s t test).
Filamin A inhibits virus production.
The absence of Filamin A led to enhanced viral entry, suggesting that it may also impact downstream virus production and release. To test this prediction, we first measured viral genomic RNA following infection of M2 and A7 cells with SeV for 8 and 24 h. M2 cells that lack both RNase L and Filamin A showed a greater than 6-fold increase in viral RNA levels compared to A7 cells which express both proteins (Fig. 6A). This observation correlated with a dramatically increased accumulation of SeV proteins in M2 compared to A7 cells at the 24-h time point (Fig. 6B). RNase L has been shown to exert antiviral effects that result in a decrease in SeV titers in vitro and in vivo (29, 45); therefore, the compromised antiviral effect observed in M2 cells could have been due to the lack of detectable RNase L in these cells. To specifically examine the antiviral effect of Filamin A, we knocked down Filamin A in HT1080 cells that express functional RNase L (Fig. 6F). SeV infection of Filamin A knockdown and control cells resulted in a significant 3-fold increase in the level of SeV genomic RNA in the supernatant of Filamin A knockdown compared to control cell results at both 8 and 24 h postinfection (Fig. 6C). To assess the effect of Filamin A on virus production and release, intracellular and extracellular SeV genomic RNA was quantified following infection of HT1080 stable cells with Filamin A knockdown and control cells (Fig. 6G) for 8 and 24 h. Filamin A knockdown resulted in an increase in the levels of both intracellular and released SeV at all the time points, suggesting that Filamin A impaired virus entry and led to a functional decrease in virus production (Fig. 6D and E).
FIG 6 .
Filamin A inhibits SeV production and release. (A) A7 or M2 cells were infected with SeV (40 HAU/ml), and viral yield levels at the indicated times were quantitated by determining SeV RNA copy numbers in culture supernatants by qPCR. Copy numbers were interpolated from a T7-transcribed RNA standard and are expressed as copy number per ml of supernatant. (B) Cell lysates of A7 and M2 cells infected with SeV as described for panel A were analyzed for viral protein expression at the indicated times using anti-SeV antibodies. β-Actin was used as a loading control. (C) HT1080 cells transfected with control small interfering RNA (Ctrl siRNA) or Filamin A siRNA were infected with SeV, and viral yield levels were determined as described for panel A. Student’s t test was used for comparisons. *, P < 0.001 (for comparisons to control shRNA cell results performed using Student’s t test). HT1080 shRNA Filamin A knockdown or control stable cell lines were infected with SeV (40 HAU/ml) for 1 h. Unbound virus was washed with PBS and replaced with complete growth medium. (D and E) Cell samples and supernatant were collected at the indicated times, and the intracellular (D) or extracellular (supernatant) (E) SeV genomic RNA copy number was determined as described for panel A. The intracellular viral copy number was normalized to GAPDH mRNA levels. (F and G) Knockdown of Filamin A using control (Ctrl) and Filamin A (FilA) siRNAs (F) or an shRNA-expressing stable cell line (G) is shown on immunoblots. Data are representative of the results of two independent experiments performed in triplicate ± SD. *, P < 0.01; **, P < 0.001 (for comparisons to control shRNA cell results performed using Student’s t test).
Filamin A potentiates the antiviral effect of RNase L.
Coexpression of RNase L and Filamin A potently inhibited virus entry at 1 h postinfection (Fig. 4F and G, Fig. 5). To evaluate the functional impact of RNase L and Filamin A expression on virus production, viral RNA or viral titers were measured at later time points following SeV or EMCV infection of M2 cells that stably express Filamin A and either WT or catalytically inactive (R667A) RNase L (Fig. 3A). WT RNase L dramatically reduced SeV and EMCV titers in the supernatant under all conditions tested. Expression of catalytically inactive RNase L or Filamin A alone resulted in a more modest diminution of viral titers (Fig. 7) and likely reflects the contribution of antiviral activity mediated by the enzymatic activity of WT RNase L which occurs post-virus entry. Coexpression of RNase L and Filamin A significantly reduced the levels of viral RNA or titer (7- to 10-fold for SeV RNA and 1- to 2.5-log for EMCV PFU) compared to the results seen with either protein alone (Fig. 7). However, the impact of expression of both proteins on virus production was less dramatic than its effect on virus entry (Fig. 4F and G), suggesting that Filamin A plays a more prominent role in restricting virus entry (see, e.g., Fig. 3A). Our findings support a model in which RNase L and Filamin A function together to provide a barrier to viral entry that does not require RNase L enzymatic activity (Fig. 8). Following viral infection, production of type I IFNs and 2-5A activates RNase L, which dissociates from Filamin A and mediates antiviral activity via its established enzymatic mechanisms (18).
FIG 7 .
Filamin A potentiates RNase L antiviral effect. M2 cells expressing Myc-Filamin A and/or Flag-RNase L WT or Flag-RNase L R667A were infected with SeV (40 HAU/ml) (A and B) or EMCV (MOI = 1.0) (C and D) for the indicated times. (A and B) Viral yields at 8 h (A) and 24 h (B) were quantitated by determining SeV RNA copy numbers in culture supernatants by qPCR. Copy numbers were interpolated from the T7-transcribed RNA standard and are expressed as copy number per ml of supernatant. (C and D) EMCV viral titers were determined by a plaque assay using L929 cells after 4 h (C) or 8 h (D). P values are as shown for comparisons performed using Student’s t test. Data are representative of the results of two independent experiments performed in triplicate ± SD.
FIG 8 .
Proposed antiviral mechanism of RNase L-Filamin A interaction. RNase L-Filamin A interaction prevents viral entry and promotes resistance under basal conditions by maintaining a barrier. Virus infection alters actin dynamics and disrupts RNase L-Filamin A interaction to release RNase L from the scaffold. dsRNAs produced during viral infections activate OAS to produce 2-5A, which in turn binds monomeric, inactive RNase L and converts it to an active dimer with potent endoribonuclease activity that mediates an antiviral response. CM, cell membrane; PRR, pattern recognition receptor.
DISCUSSION
Viral and bacterial pathogens interact with host plasma membrane proteins that act as receptors as a first step in infection. These receptors are linked to intercellular junctional proteins and the intracellular cytoskeletal network, which together comprise a physical barrier to pathogen entry (1). Disruption of this barrier is essential for a productive infection; however, the mechanisms by which the physical signals of infection are sensed by the host, and how these signals activate established innate immune mediators to initiate a host response, are not well understood. In this report, we provide the first evidence that RNase L, an established mediator of the innate immune response to viral and bacterial pathogens (18–20, 29, 50), interacts with the actin-binding protein Filamin A to modulate the actin cytoskeleton and inhibit virus entry into cells. Upon infection and activation of its enzymatic activity, RNase L dissociates from Filamin A to mediate its canonical antiviral signaling and effector functions. Our report thus identifies a novel mechanism by which RNase L exerts its antiviral activity via an interface with the actin cytoskeleton that is modulated in response to the physical perturbation of this network.
The interaction between RNase L and Filamin A was detected and mapped by coimmunoprecipitation to investigate their association in a cellular context. This analysis indicated that the N-terminal portion of RNase L was required for coimmunoprecipitation with Filamin A (Fig. 1C; see also Fig. S2 in the supplemental material). A small reduction in the IP of Filamin A with the ΔN RNase L mutant (Fig. 1C) but not in the reciprocal IP (see Fig. S2) was observed, suggesting that C-terminal residues may also influence the interaction. Additional work is required to identify the specific RNase L residues that mediate its association with a Filamin A complex. This information will permit the generation of interaction-deficient mutants to assess the role of this interaction in antiviral activities. Interestingly, despite the presence of the 8.5 ankyrin repeat domains in this region that are known to mediate protein-protein interactions, this is the first report of a protein that interacts with RNase L via its N terminus. The ankyrin repeat domains that comprise the RNase L N terminus contain sites that are critical to coordination of the binding of its activator, 2-5A (27, 28, 51). 2-5A is produced in cells following virus infection or IFN treatment, and its binding to RNase L induces conformational change and dimerization that convert RNase L into an active endoribonuclease (21, 51, 52). As predicted from the overlap between the 2-5A binding sites and the Filamin A interaction region, the RNase L-Filamin A interaction was disrupted following virus infection or direct treatment of cells with 2-5A (Fig. 1E and F). Thus, it is likely that Filamin A interacts with inactive monomeric RNase L. Conversely, the presence of Filamin A did not impair RNase L activation (Fig. 3B). These results may reflect a stronger affinity of RNase L for 2-5A compared to that seen with Filamin A and may involve distinct residues that mediate these interactions. Another potential impact of the association of RNase L with Filamin A via its N terminus is that the pseudokinase domain, which is critical for RNase L homodimerization and interaction with all heterologous RNase L binding partners reported to date, remains free (27, 28, 53). This orientation may function to anchor RNase L to the cytoskeletal complex while allowing the pseudokinase domain to interact with regulatory molecules. For example, the pseudokinase domain also contains a site that is required for 2-5A binding (27); thus, access to the pseudokinase domain may provide an initial docking site for 2-5A, thereby promoting subsequent dissociation from Filamin A and dimerization. Together, our data suggest that Filamin A, by virtue of its actin-binding ability, may sequester a pool of RNase L in the cytoskeleton that dissociates from this complex upon activation. In a related study, RNase L was isolated in association with cytoskeletal proteins in a detergent-insoluble cell fraction and was released to the cytosol upon stimulation with the phorbol ester mitogen 12-O-tetradecanoylphorbol-13-acetate (TPA). TPA treatment induced phosphorylation of RNase L, which led to its activation and dissociation from cytoskeletal components (54). Furthermore, proteomic analysis of RNase L-interacting proteins identified extracellular matrix (ECM) and cytoskeletal proteins as binding partners (31). These findings suggest that the association of RNase L with the cytoskeleton is both dynamic and context specific.
The intricate network of actin filaments associated with the plasma membrane presents a physical barrier that viruses must disrupt to gain entry into the cytoplasm. The interaction of RNase L with Filamin A suggested that RNase L may impact cellular processes regulated by actin dynamics. In support of this prediction, we observed distinct morphologies in MEFs lacking RNase L that correlated with reduced levels of filamentous actin and increased dextran uptake by macropinocytosis (Fig. 2). In contrast, RNase L KO bone marrow-derived macrophages (BMMs) were previously reported to exhibit reduced uptake of FITC-dextran compared to WT BMMs, whereas phagocytic activity was not affected (55). The different results reported in these studies may reflect fundamental differences in pinocytic activity and innate immune responses between MEFs and BMMs. In line with the observed increase in macropinocytosis, RNase L KO MEFs showed increased entry of SeV and EMCV, suggesting that RNase L serves as a barrier for virus entry. In a similar role, RNase L inhibited translocation of enteropathic E. coli across intestinal epithelial cells by regulating tight-junction proteins and barrier integrity (32). Together, these findings suggest a novel barrier function for RNase L in pathogen exclusion.
Our finding that RNase L inhibits virus entry and interacts with Filamin A suggested that Filamin A may also function in inhibiting viral entry into cells. Consistent with this prediction, infection of Filamin A-deficient and -competent M2 melanoma cells with either SeV or EMCV demonstrated increased virus entry in the absence of Filamin A (Fig. 4A, F, and G). Furthermore, M2 cells lacking either RNase L or Filamin A displayed significantly more virus entry and cells lacking both proteins showed a further increase in virus entry (Fig. 4A, F, and G). This finding indicated roles for both RNase L and Filamin A in limiting virus entry. In support of a critical role for the interaction of RNase L and Filamin A in restricting virus entry, RNase L deletion mutants that abolished or reduced Filamin A interaction resulted in a proportional decrease in barrier function and increase in virus entry (Fig. 5). Interestingly, the presence of either the N- or C-terminal regions of RNase L reduced viral entry compared to that observed in RNase L-deficient cells, suggesting that this activity is mediated by multiple regions and interacting proteins. Thus, Filamin A may serve as the primary cytoskeletal contact for the RNase L N terminus, with other cytoskeletal proteins interacting with the internal and C-terminal regions. Consistent with this interpretation, cytoskeletal proteins were recently identified as a major class of candidate RNase L interactors (31) and we validated RNase L interaction with a cytoskeletal protein through its C terminus (B. A. Hassel and H. J. Ezelle, unpublished data). The extent to which RNase L interaction with Filamin A and other cytoskeletal proteins enhances barrier function requires additional studies to map and mutate the interaction sites and to generate interaction-deficient mutants. Our work further demonstrated that both WT and catalytically inactive RNase L were competent to reduce virus entry when transfected into RNase L-deficient cells. To our knowledge, this observation of the novel role of RNase L in limiting virus entry represents the first report of its antiviral function that is independent of enzymatic activity. Analysis of intracellular and extracellular viral genomic RNA at later times postinfection demonstrated a significant, but more modest, combined effect of RNase L and Filamin A on viral replication and release from cells compared to entry (Fig. 6D and E and 7). This activity exhibited greater dependence on RNase L enzymatic function, which is an important component of antiviral mechanisms post-entry and replication. Taken together, our results support a model in which RNase L and Filamin A form a structural complex to restrict viral entry in naive cells (Fig. 8). Invading viruses alter the actin cytoskeleton, in part by targeting actin-binding proteins to facilitate their entry. These alterations, in turn, disrupt the RNase L-Filamin A interaction to release RNase L from the barrier complex and promote downstream antiviral signaling.
Innate immune sensors and effectors that have historically been thought to function only in the context of pathogen infection have recently been reported to play key roles in host homeostatic activities in the absence of pathogen threat. Specifically, several IFN-stimulated genes (ISG) have been shown to modulate the actin cytoskeleton and enhance barrier function as a baseline defense against pathogen infection. For example, the PKR antiviral protein kinase binds to and inhibits an actin-severing protein, gelsolin, to alter actin dynamics limiting viral entry and to maintain a basal innate immune defense (8). IFITM is an ISG that is part of the tight-junction complex and promotes barrier function from HCV entry in response to IFN (56). RIG-I is a PRR that senses viral RNAs, including the products of RNase L cleavage (29), to activate signaling pathways that induce type I IFNs (57, 58) and is associated with the actin cytoskeleton (59, 60). The localization of RIG-I, and other innate immune signaling proteins, including RNase L and PKR, close to the plasma membrane-proximal cytoskeleton is thought to facilitate the formation of viral RNA granules that act as platforms for virus sensing and signaling upon viral entry (61). Furthermore, the close association of viral RNA with RNase L and RIG-I provides a mechanism by which the viral RNA products of RNase L cleavage can activate RIG-I. In addition, the ISG15 ubiquitin-like protein that mediates diverse IFN-induced antiviral functions also modulates the actin cytoskeleton and cell architecture and promotes cell migration by stabilizing proteins that promote migration and invasion (62). The dual roles these ISGs serve in the absence and presence of pathogens provide an efficient strategy to protect against future infection and to rapidly respond upon pathogen exposure. Our report thus identifies RNase L as the latest entry in a growing list of innate immune effectors that mediate critical host defense activities under basal conditions and following pathogen infection. Virtually all viruses remodel the actin network to optimize multiple aspects of their infection cycle; therefore, an understanding of the host proteins and mechanisms underlying these regulatory nodes may provide opportunities for the development of broad-spectrum antiviral agents that target these activities.
MATERIALS AND METHODS
Cells, reagents, and plasmids.
HEK293T (provided by Fan Dong, University of Toledo), HT1080, and L929 cells were cultured in Dulbecco’s modified minimal essential medium (DMEM) with 10% fetal bovine serum (Invitrogen, Carlsbad, CA) and 100 µg/ml penicillin/streptomycin. WT and RNaseL−/− (RNase L KO) MEFs, primary and transformed with simian virus 40 (SV40) large T antigen (a gift from Robert Silverman, Cleveland Clinic), were grown in RPMI 1640 supplemented with streptomycin (100 µg/ml), penicillin (100 units/ml), 2 mmol/liter glutamine, and 10% fetal bovine serum (Invitrogen, Carlsbad, CA). HeLa M cells expressing pcDNA3 vector, WT RNase L, RNase L R667A (nuclease dead), or RNase L R462Q (dimerization defective and with 3-fold-reduced enzyme activity) kindly provided by Robert Silverman, Cleveland Clinic, were maintained in DMEM with 10% fetal bovine serum (Invitrogen, Carlsbad, CA) and 100 µg/ml penicillin-streptomycin supplemented with 250 µg/ml G418 (Life Technologies, Carlsbad, CA). M2 melanoma cells lacking Filamin A and A7 cells derived from M2 cells by stably expressing Filamin A (provided by J. H. Hartwig, Harvard Medical School, Boston, MA) were cultured in Dulbecco’s modified minimal essential medium with 10% fetal bovine serum (Invitrogen, Carlsbad, CA) and 100 µg/ml penicillin-streptomycin. A7 cells were maintained in complete DMEM with 0.5 mg/ml G418 (Life Technologies, Carlsbad, CA). All chemicals, unless indicated otherwise, were from Sigma-Aldrich (St. Louis, MO). Antibody to Flag tag (monoclonal and polyclonal), Flag-M2 agarose beads, Myc-EZview beads, and β-actin were from Sigma (St. Louis, MO), Myc tag (monoclonal and polyclonal) was from Cell Signaling, Inc. (Danvers, MA), Filamin A (monoclonal) was from Neomarkers (Fremont, CA), and polyclonal antibody was from Santa Cruz Biotechnology (Santa Cruz, CA). Goat anti-Sendai virus antibody raised against the Cantell strain which recognizes all structural antigens was used for immunofluorescence (Meridian Life Sciences). Rabbit anti-SeV antibody from MBL (Woburn, MA) was used for immunoblots. RNase L monoclonal antibody was kindly provided by Robert Silverman (Cleveland Clinic). Anti-mouse IgG and anti-rabbit IgG horseradish peroxidase (HRP)-linked secondary antibodies were from Cell Signaling, Inc. (Danvers, MA), and ECL reagents were from GE Healthcare (Piscataway, NJ). FITC-labeled dextran (10 kDa), Alexa 488-labeled phalloidin, and Alexa Fluor 633 donkey anti-goat IgG were from Life Technologies. Cytochalasin D and Latrunculin A were obtained from Cayman Chemicals (Ann Arbor, MI) and used at 5 µM and 0.5 µM dissolved in dimethyl sulfoxide (DMSO). Sendai virus (Cantell strain; Charles River Laboratories), EMCV, and preparation and transfection of 2-5A have been described previously (45). Plasmids Flag-RNase L, Flag-RNase L R667A, Flag-RNase L (residues 1 to 335; ΔC), Flag-RNase L (residues 386 to 741; ΔN) (kindly provided by Robert Silverman, Cleveland Clinic), and Myc-Filamin A (Addgene) were transfected using Lipofectamine 2000 per the manufacturer’s instructions.
Stable cell lines.
Filamin A silencing and nonsilencing small harpin RNAs (shRNAs) were generated as suggested by the manufacturer using a GIPZ-lentiviral shRNA system, and knockdown HT1080 cells or controls were selected with 1 µg/ml of puromycin (Open Biosystems/Thermo Scientific, Pittsburgh, PA). M2 cells transfected with Myc-Filamin A were selected in the presence of 0.5 mg/ml of G418 to generate stable M2 Filamin A (FLNA) cells. M2 cells or M2 FLNA cells were transfected with Flag-CMV2 vector, Flag-RNase L, or Flag-RNase LR667A along with pbabe-puro plamid and selected in the presence of 1 µg/ml of puromycin to generate stable cell lines. Cells with comparable levels of expression of Filamin A and/or RNase L were selected and used.
Dextran uptake assays.
WT and RNase L KO MEFs were grown on glass coverslips and replaced with RPMI medium containing 1% serum for 18 h. Cells were incubated with 25 µg/ml of FITC-dextran (10 kDa) for 1 h at 37°C, washed twice in phosphate-buffered saline (PBS), fixed, and mounted in Vectashield with DAPI (4′,6-diamidino-2-phenylindole) (Vector Laboratories, Burlingame, CA). Cells were imaged by the use of a Leica TCS SP5 multiphoton laser scanning confocal microscope, and fluorescence was quantitated using NIH ImageJ software. A minimum of 10 fields from triplicate experiments were analyzed.
Virus infection and virus assays.
Cells (2 × 105 in 12-well plates) were grown overnight, washed two times with PBS, and infected with EMCV (strain K) at a multiplicity of infection (MOI) of 1.0 or with Sendai virus (SeV) at 40 hemagglutination units (HAU)/ml in media without serum. After 1 h, media were removed, cells were washed with PBS, and complete media were added until they were harvested. EMCV titers in supernatants were determined by a plaque assay after 4 h and 8 h in serial dilutions using L929 cells. SeV was quantified after 8 h and 24 h from supernatants by RNA isolation and real-time PCR, and copy numbers were interpolated from a T7-transcribed RNA standard and expressed as copy numbers per ml of supernatant as previously described (45). To determine the effect on SeV production and release, control or Filamin A shRNA-expressing stable HT1080 cells were infected with SeV (40 HAU/ml) for 8 h or 24 h. Intracellular SeV RNA copy numbers were determined by reverse transcription-PCR (RT-PCR) and normalized to GAPDH (glyceraldehyde-3-phosphate dehydrogenase), and extracellular RNA levels were determined for the same samples from culture supernatants. For assays of viral entry, 1 × 106 cells were infected with EMCV (MOI = 1.0) or SeV (40 HAU/ml) at 37° C for 1 h. The inoculum of virus was removed, and the cells were treated with acid glycine (pH 3.0) for 2 min to inactivate unbound extracellular virus (5, 63–67). M2 cells stably expressing Filamin A were transiently transfected with Flag-RNase L (full length), Flag-RNase L, Flag-RNase L (residues 1 to 335; ΔC), Flag-RNase L (residues 386 to 741; ΔN), or Flag vector alone (1 × 105 cells in 12-well dishes) and infected with SeV (40 HAU/ml) as described above to monitor virus entry. Following virus attachment, virus internalization was allowed to proceed for 1 h. Cell samples were collected, and the intracellular titer of virus in each sample was determined by a plaque assay for EMCV after repeat freeze-thaw cycles or by determining SeV RNA copy numbers normalized to GAPDH by RT-PCR. Experiments were performed two times, and data are shown as standard deviations (SD) for experiments performed in triplicate.
Monitoring RNase L activity in intact cells.
Cells were transfected with 10 µM 2-5A or 2 µg/ml of synthetic dsRNA [poly(I·C)], using Lipofectamine 2000. In some experiments, cells were pretreated with Cytochalasin D (5 µM) or Latrunculin A (0.5 µM) or vehicle (DMSO) for 1 h followed by 2-5A transfection. At the indicated times, the total RNA was isolated from transfected cells using Trizol reagent (Invitrogen/Life Technologies) and quantitated by measuring absorbance at 260 nm using a NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE). RNAs (2 µg) were separated on RNA chips and analyzed with a 2100 Bioanalyzer (Agilent Technologies) to monitor characteristic rRNA cleavage products generated by RNase L activity as described previously (44).
Coimmunoprecipitation and immunoblotting.
HEK293T cells expressing Myc-Filamin A along with various Flag-RNase L constructs (WT, R667A, residues 1 to 335 [ΔC] or residues 386 to 741[ΔN]) were left untreated or transfected with 10 µM 2-5A or infected with EMCV (MOI = 1.0) for 1 h and harvested. Cells expressing Myc or Flag tag alone were used as controls. Cells were washed with ice-cold PBS and lysed in buffer containing 0.5% NP-40, 90 mM KCl, 5 mM magnesium acetate, 20 mM Tris (pH 7.5), 5 mM β mercaptoethanol, 0.1 M phenylmethylsulfonyl fluoride (PMSF), 0.2 mM sodium orthovanadate, 50 mM NaF, 10 mM glycerophosphate, and protease inhibitor (Roche Diagnostics, Indianapolis, IN) on ice for 20 min. The lysates were clarified by centrifugation at 10,000 × g (at 4°C for 20 min). Clarified cell lysates were precleared and mixed with control IgG, FlagM2-agarose, or Myc-EZview beads and rotated end to end of 1 h or overnight at 4°C. The beads were collected and washed five times in lysis buffer. Cell lysates (2 mg) of HeLa M cells expressing RNase L (WT) or the RNase L R667A mutant or the RNase L R462Q mutant were subjected to immunoprecipitation using Filamin A polyclonal antibodies (Bethyl Laboratories, Montgomery, TX) and control IgG antibodies. The immunoprecipitated proteins were dissociated by boiling in Laemmli sample buffer, separated on 8% or 10% SDS-polyacrylamide gels, transferred to nitrocellulose membrane (Bio-Rad, Hercules, CA), and subjected to immunoblotting. Membranes were probed with different primary antibodies according to the manufacturer’s protocols. Membranes were washed with Tris-buffered saline (TBS) with 1% Tween 20 and incubated with goat anti-mouse or goat anti-rabbit antibody tagged with horseradish peroxidase (Cell Signaling, Danvers, MA) for 1 h. Proteins in the blots were detected by enhanced chemiluminescence (GE Healthcare).
Immunofluorescence assay (IFA).
Cells were seeded on glass coverslips 24 h prior to use for IFA. Infected or uninfected cells were rinsed in cold PBS and fixed with 4% paraformaldehyde (EM Sciences, Hatfield, PA) for 15 min and permeabilized with 0.2% Triton X-100 for 5 min. After washing and blocking in 1% bovine serum albumin (BSA) PBS, the cells were reacted with primary antibodies (1:300, overnight at 4°C) reactive against all structural SeV antigens followed by washing and incubation with fluorescent dye-conjugated secondary antibodies and Alexa 633 donkey anti-goat IgG (Molecular Probes) (1:100; 1 h at 4°C). F-actin was visualized by staining with Alexa 488 phalloidin for 1 h at 37°C according to the manufacturer’s instructions (Molecular Probes). Cells were mounted in Vectashield with DAPI to stain the nucleus (Vector Laboratories, Burlingame, CA). Fluorescence and confocal microscopy assessments were performed with a Leica CS SP5 multiphoton laser scanning confocal microscope (Leica Microsystems, Weitzler, Germany) and quantitated using ImageJ software (National Institutes of Health). Images were processed using Adobe Photoshop CS4 (Adobe, San Jose, CA). More than 10 cells (from two coverslips) were analyzed for each condition.
Statistical analysis.
Student’s t tests were used for determining the statistical significance of the results of comparisons between groups. P values were obtained from a two-tailed, unpaired Student’s t test and are shown for all analyses. P < 0.05 was considered significant in all cases.
SUPPLEMENTAL MATERIAL
(A) Expression levels of endogenous RNase L and Filamin A in A7, M2 melanoma, and HT1080 cells normalized to β-actin levels. (B) M2 and A7 melanoma cells were transfected with 10 µM 2-5A or 2 µg/ml of poly(I·C), and specific cleavage of rRNA (shown by arrows) was analyzed on RNA chips using an Agilent 2100 Bioanalyzer. Download
(A) HEK293T cells expressing Myc-Filamin A or Myc tag and Flag-RNase L (full length, WT), Flag-RNase L (ΔC), or Flag-RNase L (ΔN) were immunoprecipitated using Myc-EZview agarose beads or control IgG beads. Samples were subjected to immunoblot analysis using anti-Flag or anti-Myc antibodies. For IP analysis, 5% of the immunoprecipitate was loaded in blots probed with the antibodies indicated. (B) Protein expression in cell lysates normalized to β-actin levels. Download
M2 stable cells expressing Myc-Filamin A and/or Flag-RNase L WT or Flag-RNase L R667A were infected with EMCV (MOI = 1.0) for 1 h and washed briefly with PBS to remove unbound extracellular viruses. Intracellular EMCV titers were determined by a plaque assay using L929 cells. Student’s t test was used to determine P values (as shown) of M2 cells expressing Filamin A compared to cells expressing Filamin A and either RNase L WT or R667A. P values determined using Student’s t test are as shown. Data are representative of the results of experiments performed in triplicate ± SD. Download
M2 cells stably expressing Myc-Filamin A and transiently transfected with Flag-RNase L (WT, full length), Flag-RNase L (ΔC), or Flag-RNase L (ΔN) or with vector alone were subjected to immunoblot analysis using anti-Flag antibodies. Protein expression in cell lysates was normalized to β-actin levels. *, nonspecific band. Download
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health (NIH) grant AI089518 (K.M.) and startup funds from the University of Toledo (K.M.) and, in part, by the NIH (NIAID grant AI077556 to B.A.H. and NIDDK grant DK084460-01A2 to A.Z.).
We thank Robert Silverman for providing WT and RNase L Ko MEFs and HeLa M stable cells as well as the RNase L plasmids used in this study. We thank Douglas Leaman (Department of Biological Sciences, University of Toledo) for valuable discussions through the course of this work.
Footnotes
Citation Malathi K, Siddiqui MA, Dayal S, Naji M, Ezelle HJ, Zeng C, Zhou A, Hassel BA. 2014. RNase L interacts with Filamin A to regulate actin dynamics and barrier function for viral entry. mBio 5(6):e02012-14. doi:10.1128/mBio.02012-14.
REFERENCES
- 1. Delorme-Axford E, Coyne CB. 2011. The actin cytoskeleton as a barrier to virus infection of polarized epithelial cells. Viruses 3:2462–2477. 10.3390/v3122462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cudmore S, Reckmann I, Way M. 1997. Viral manipulations of the actin cytoskeleton. Trends Microbiol. 5:142–148. 10.1016/S0966-842X(97)01011-1. [DOI] [PubMed] [Google Scholar]
- 3. Taylor MP, Koyuncu OO, Enquist LW. 2011. Subversion of the actin cytoskeleton during viral infection. Nat. Rev. Microbiol. 9:427–439. 10.1038/nrmicro2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Akira S. 2009. Innate immunity to pathogens: diversity in receptors for microbial recognition. Immunol. Rev. 227:5–8. 10.1111/j.1600-065X.2008.00739.x. [DOI] [PubMed] [Google Scholar]
- 5. Wang JL, Zhang JL, Chen W, Xu XF, Gao N, Fan DY, An J. 2010. Roles of small GTPase Rac1 in the regulation of actin cytoskeleton during dengue virus infection. PLoS Negl. Trop. Dis. 4:e809. 10.1371/journal.pntd.0000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Coyne CB, Shen L, Turner JR, Bergelson JM. 2007. Coxsackievirus entry across epithelial tight junctions requires occludin and the small GTPases Rab34 and Rab5. Cell Host Microbe 2:181–192. 10.1016/j.chom.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sun X, Whittaker GR. 2007. Role of the actin cytoskeleton during influenza virus internalization into polarized epithelial cells. Cell. Microbiol. 9:1672–1682. 10.1111/j.1462-5822.2007.00900.x. [DOI] [PubMed] [Google Scholar]
- 8. Irving AT, Wang D, Vasilevski O, Latchoumanin O, Kozer N, Clayton AH, Szczepny A, Morimoto H, Xu D, Williams BR, Sadler AJ. 2012. Regulation of actin dynamics by protein kinase R control of gelsolin enforces basal innate immune defense. Immunity 36:795–806. 10.1016/j.immuni.2012.02.020. [DOI] [PubMed] [Google Scholar]
- 9. García-Expósito L, Ziglio S, Barroso-González J, de Armas-Rillo L, Valera MS, Zipeto D, Machado JD, Valenzuela-Fernández A. 2013. Gelsolin activity controls efficient early HIV-1 infection. Retrovirology 10:39. 10.1186/1742-4690-10-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Moser TS, Jones RG, Thompson CB, Coyne CB, Cherry S. 2010. A kinome RNAi screen identified AMPK as promoting poxvirus entry through the control of actin dynamics. PLoS Pathog. 6:e1000954. 10.1371/journal.ppat.1000954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kondratowicz AS, Hunt CL, Davey RA, Cherry S, Maury WJ. 2013. AMP-activated protein kinase is required for the macropinocytic internalization of ebolavirus. J. Virol. 87:746–755. 10.1128/JVI.01634-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cooper J, Liu L, Woodruff EA, Taylor HE, Goodwin JS, D’Aquila RT, Spearman P, Hildreth JE, Dong X. 2011. Filamin A protein interacts with human immunodeficiency virus type 1 gag protein and contributes to productive particle assembly. J. Biol. Chem. 286:28498–28510. 10.1074/jbc.M111.239053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jiménez-Baranda S, Gómez-Moutón C, Rojas A, Martínez-Prats L, Mira E, Ana Lacalle R, Valencia A, Dimitrov DS, Viola A, Delgado R, Martínez-A C, Mañes S. 2007. Filamin-A regulates actin-dependent clustering of HIV receptors. Nat. Cell Biol. 9:838–846. 10.1038/ncb1610. [DOI] [PubMed] [Google Scholar]
- 14. Ghosh S, Ahrens WA, Phatak SU, Hwang S, Schrum LW, Bonkovsky HL. 2011. Association of filamin A and vimentin with hepatitis C virus proteins in infected human hepatocytes. J. Viral Hepat. 18:e568–e577. 10.1111/j.1365-2893.2011.01487.x. [DOI] [PubMed] [Google Scholar]
- 15. Calderwood DA, Huttenlocher A, Kiosses WB, Rose DM, Woodside DG, Schwartz MA, Ginsberg MH. 2001. Increased filamin binding to beta-integrin cytoplasmic domains inhibits cell migration. Nat. Cell Biol. 3:1060–1068. 10.1038/ncb1201-1060. [DOI] [PubMed] [Google Scholar]
- 16. Kälin S, Amstutz B, Gastaldelli M, Wolfrum N, Boucke K, Havenga M, DiGennaro F, Liska N, Hemmi S, Greber UF. 2010. Macropinocytotic uptake and infection of human epithelial cells with species B2 adenovirus type 35. J. Virol. 84:5336–5350. 10.1128/JVI.02494-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shafren DR, Bates RC, Agrez MV, Herd RL, Burns GF, Barry RD. 1995. Coxsackieviruses B1, B3, and B5 use decay accelerating factor as a receptor for cell attachment. J. Virol. 69:3873–3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chakrabarti A, Jha BK, Silverman RH. 2011. New insights into the role of RNase L in innate immunity. J. Interferon Cytokine Res. 31:49–57. 10.1089/jir.2010.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liang SL, Quirk D, Zhou A. 2006. RNase L: its biological roles and regulation. IUBMB Life 58:508–514. 10.1080/15216540600838232. [DOI] [PubMed] [Google Scholar]
- 20. Brennan-Laun SE, Ezelle HJ, Li XL, Hassel BA. 2014. RNase-L control of cellular mRNAs: roles in biologic functions and mechanisms of substrate targeting. J. Interferon Cytokine Res. 34:275–288. 10.1089/jir.2013.0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Silverman RH. 2007. A scientific journey through the 2-5A/RNase L system. Cytokine Growth Factor Rev. 18:381–388. 10.1016/j.cytogfr.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Silverman RH. 2007. Viral encounters with 2′,5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. J. Virol. 81:12720–12729. 10.1128/JVI.01471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Silverman RH, Cayley PJ, Knight M, Gilbert CS, Kerr IM. 1982. Control of the ppp(a2’p)nA system in HeLa cells. Effects of interferon and virus infection. Eur. J. Biochem. 124:131–138. [DOI] [PubMed] [Google Scholar]
- 24. Wreschner DH, James TC, Silverman RH, Kerr IM. 1981. Ribosomal RNA cleavage, nuclease activation and 2-5A(ppp(A2’p)nA) in interferon-treated cells. Nucleic Acids Res. 9:1571–1581. 10.1093/nar/9.7.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dong B, Silverman RH. 1999. Alternative function of a protein kinase homology domain in 2′, 5′-oligoadenylate dependent RNase L. Nucleic Acids Res. 27:439–445. 10.1093/nar/27.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xiang Y, Wang Z, Murakami J, Plummer S, Klein EA, Carpten JD, Trent JM, Isaacs WB, Casey G, Silverman RH. 2003. Effects of RNase L mutations associated with prostate cancer on apoptosis induced by 2′,5′-oligoadenylates. Cancer Res. 63:6795–6801. [PubMed] [Google Scholar]
- 27. Huang H, Zeqiraj E, Dong B, Jha BK, Duffy NM, Orlicky S, Thevakumaran N, Talukdar M, Pillon MC, Ceccarelli DF, Wan LC, Juang YC, Mao DY, Gaughan C, Brinton MA, Perelygin AA, Kourinov I, Guarné A, Silverman RH, Sicheri F. 2014. Dimeric structure of pseudokinase RNase L bound to 2-5A reveals a basis for interferon-induced antiviral activity. Mol. Cell 53:221–234. 10.1016/j.molcel.2013.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Han Y, Donovan J, Rath S, Whitney G, Chitrakar A, Korennykh A. 2014. Structure of human RNase L reveals the basis for regulated RNA decay in the IFN response. Science 343:1244–1248. 10.1126/science.1249845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Malathi K, Dong B, Gale M, Jr, Silverman RH. 2007. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature 448:816–819. 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sato A, Naito T, Hiramoto A, Goda K, Omi T, Kitade Y, Sasaki T, Matsuda A, Fukushima M, Wataya Y, Kim HS. 2010. Association of RNase L with a Ras GTPase-activating-like protein IQGAP1 in mediating the apoptosis of a human cancer cell-line. FEBS J. 277:4464–4473. 10.1111/j.1742-4658.2010.07833.x. [DOI] [PubMed] [Google Scholar]
- 31. Gupta A, Rath PC. 2014. Expression of mRNA and protein-protein interaction of the antiviral endoribonuclease RNase L in mouse spleen. Int. J. Biol. Macromol. 69:307–318. 10.1016/j.ijbiomac.2014.04.042. [DOI] [PubMed] [Google Scholar]
- 32. Long TM, Nisa S, Donnenberg MS, Hassel BA. 2014. Enteropathogenic Escherichia coli inhibits type I interferon- and RNase-L-mediated host defense to disrupt intestinal epithelial cell barrier function. Infect. Immun. 82:2802–2814. 10.1128/IAI.00105-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bettoun DJ, Scafonas A, Rutledge SJ, Hodor P, Chen O, Gambone C, Vogel R, McElwee-Witmer S, Bai C, Freedman L, Schmidt A. 2005. Interaction between the androgen receptor and RNase L mediates a cross-talk between the interferon and androgen signaling pathways. J. Biol. Chem. 280:38898–38901. 10.1074/jbc.C500324200. [DOI] [PubMed] [Google Scholar]
- 34. Loy CJ, Sim KS, Yong EL. 2003. Filamin-A fragment localizes to the nucleus to regulate androgen receptor and coactivator functions. Proc. Natl. Acad. Sci. U. S. A. 100:4562–4567. 10.1073/pnas.0736237100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang Y, Kreisberg JI, Bedolla RG, Mikhailova M, deVere White RW, Ghosh PM. 2007. A 90 kDa fragment of filamin A promotes Casodex-induced growth inhibition in Casodex-resistant androgen receptor positive C4-2 prostate cancer cells. Oncogene 26:6061–6070. 10.1038/sj.onc.1210435. [DOI] [PubMed] [Google Scholar]
- 36. Chakrabarti A, Ghosh PK, Banerjee S, Gaughan C, Silverman RH. 2012. RNase L triggers autophagy in response to viral infections. J. Virol. 86:11311–11321. 10.1128/JVI.00270-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Han JQ, Townsend HL, Jha BK, Paranjape JM, Silverman RH, Barton DJ. 2007. A phylogenetically conserved RNA structure in the poliovirus open reading frame inhibits the antiviral endoribonuclease RNase L. J. Virol. 81:5561–5572. 10.1128/JVI.01857-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hassel BA, Zhou A, Sotomayor C, Maran A, Silverman RH. 1993. A dominant negative mutant of 2-5A-dependent RNase suppresses antiproliferative and antiviral effects of interferon. EMBO J. 12:3297–3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhou A, Paranjape J, Brown TL, Nie H, Naik S, Dong B, Chang A, Trapp B, Fairchild R, Colmenares C, Silverman RH. 1997. Interferon action and apoptosis are defective in mice devoid of 2′,5′-oligoadenylate-dependent RNase L. EMBO J. 16:6355–6363. 10.1093/emboj/16.21.6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mercer J, Helenius A. 2012. Gulping rather than sipping: macropinocytosis as a way of virus entry. Curr. Opin. Microbiol. 15:490–499. 10.1016/j.mib.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 41. Mercer J, Helenius A. 2009. Virus entry by macropinocytosis. Nat. Cell Biol. 11:510–520. 10.1038/ncb0509-510. [DOI] [PubMed] [Google Scholar]
- 42. Krzyzaniak MA, Zumstein MT, Gerez JA, Picotti P, Helenius A. 2013. Host cell entry of respiratory syncytial virus involves macropinocytosis followed by proteolytic activation of the F protein. PLoS Pathog. 9:e1003309. 10.1371/journal.ppat.1003309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cunningham CC, Gorlin JB, Kwiatkowski DJ, Hartwig JH, Janmey PA, Byers HR, Stossel TP. 1992. Actin-binding protein requirement for cortical stability and efficient locomotion. Science 255:325–327. 10.1126/science.1549777. [DOI] [PubMed] [Google Scholar]
- 44. Malathi K, Paranjape JM, Bulanova E, Shim M, Guenther-Johnson JM, Faber PW, Eling TE, Williams BR, Silverman RH. 2005. A transcriptional signaling pathway in the IFN system mediated by 2′-5′-oligoadenylate activation of RNase L. Proc. Natl. Acad. Sci. U. S. A. 102:14533–14538. 10.1073/pnas.0507551102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Siddiqui MA, Malathi K. 2012. RNase L induces autophagy via c-Jun N-terminal kinase and double-stranded RNA-dependent protein kinase signaling pathways. J. Biol. Chem. 287:43651–43664. 10.1074/jbc.M112.399964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Miazza V, Mottet-Osman G, Startchick S, Chaponnier C, Roux L. 2011. Sendai virus induced cytoplasmic actin remodeling correlates with efficient virus particle production. Virology 410:7–16. 10.1016/j.virol.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 47. Cunningham CC. 1992. Actin structural proteins in cell motility. Cancer Metastasis Rev. 11:69–77. 10.1007/BF00047604. [DOI] [PubMed] [Google Scholar]
- 48. Muriel O, Echarri A, Hellriegel C, Pavón DM, Beccari L, Del Pozo MA. 2011. Phosphorylated filamin A regulates actin-linked caveolae dynamics. J. Cell Sci. 124:2763–2776. 10.1242/jcs.080804. [DOI] [PubMed] [Google Scholar]
- 49. Loo YM, Fornek J, Crochet N, Bajwa G, Perwitasari O, Martinez-Sobrido L, Akira S, Gill MA, García-Sastre A, Katze MG, Gale M., Jr. 2008. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 82:335–345. 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li XL, Ezelle HJ, Kang TJ, Zhang L, Shirey KA, Harro J, Hasday JD, Mohapatra SK, Crasta OR, Vogel SN, Cross AS, Hassel BA. 2008. An essential role for the antiviral endoribonuclease, RNase-L, in antibacterial immunity. Proc. Natl. Acad. Sci. U. S. A. 105:20816–20821. 10.1073/pnas.0807265105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dong B, Silverman RH. 1995. 2-5A-dependent RNase molecules dimerize during activation by 2-5A. J. Biol. Chem. 270:4133–4137. 10.1074/jbc.270.8.4133. [DOI] [PubMed] [Google Scholar]
- 52. Dong B, Niwa M, Walter P, Silverman RH. 2001. Basis for regulated RNA cleavage by functional analysis of RNase L and Ire1p. RNA 7:361–373. 10.1017/S1355838201002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hetz C, Bernasconi P, Fisher J, Lee AH, Bassik MC, Antonsson B, Brandt GS, Iwakoshi NN, Schinzel A, Glimcher LH, Korsmeyer SJ. 2006. Proapoptotic BAX and BAK modulate the unfolded protein response by a direct interaction with IRE1alpha. Science 312:572–576. 10.1126/science.1123480. [DOI] [PubMed] [Google Scholar]
- 54. Tnani M, Aliau S, Bayard B. 1998. Localization of a molecular form of interferon-regulated RNase L in the cytoskeleton. J. Interferon Cytokine Res. 18:361–368. 10.1089/jir.1998.18.361. [DOI] [PubMed] [Google Scholar]
- 55. Yi X, Zeng C, Liu H, Chen X, Zhang P, Yun BS, Jin G, Zhou A. 2013. Lack of RNase L attenuates macrophage Functions. PLoS One 8:e81269. 10.1371/journal.pone.0081269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wilkins C, Woodward J, Lau DT, Barnes A, Joyce M, McFarlane N, McKeating JA, Tyrrell DL, Gale M., Jr. 2013. IFITM1 is a tight junction protein that inhibits hepatitis C virus entry. Hepatology 57:461–469. 10.1002/hep.26066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Takeuchi O, Akira S. 2008. MDA5/RIG-I and virus recognition. Curr. Opin. Immunol. 20:17–22. 10.1016/j.coi.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 58. Takeuchi O, Akira S. 2007. Recognition of viruses by innate immunity. Immunol. Rev. 220:214–224. 10.1111/j.1600-065X.2007.00562.x. [DOI] [PubMed] [Google Scholar]
- 59. Ohman T, Rintahaka J, Kalkkinen N, Matikainen S, Nyman TA. 2009. Actin and RIG-I/MAVS signaling components translocate to mitochondria upon influenza A virus infection of human primary macrophages. J. Immunol. 182:5682–5692. 10.4049/jimmunol.0803093. [DOI] [PubMed] [Google Scholar]
- 60. Mukherjee A, Morosky SA, Shen L, Weber CR, Turner JR, Kim KS, Wang T, Coyne CB. 2009. Retinoic acid-induced gene-1 (RIG-I) associates with the actin cytoskeleton via caspase activation and recruitment domain-dependent interactions. J. Biol. Chem. 284:6486–6494. 10.1074/jbc.M807547200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Onomoto K, Jogi M, Yoo JS, Narita R, Morimoto S, Takemura A, Sambhara S, Kawaguchi A, Osari S, Nagata K, Matsumiya T, Namiki H, Yoneyama M, Fujita T. 2012. Critical role of an antiviral stress granule containing RIG-I and PKR in viral detection and innate immunity. PLoS One 7:e43031. 10.1371/journal.pone.0043031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Desai SD, Reed RE, Burks J, Wood LM, Pullikuth AK, Haas AL, Liu LF, Breslin JW, Meiners S, Sankar S. 2012. ISG15 disrupts cytoskeletal architecture and promotes motility in human breast cancer cells. Exp. Biol. Med. (Maywood) 237:38–49. 10.1258/ebm.2011.011236. [DOI] [PubMed] [Google Scholar]
- 63. Weiss C, Clark HF. 1985. Rapid inactivation of rotaviruses by exposure to acid buffer or acidic gastric juice. J. Gen. Virol. 66:2725–2730. 10.1099/0022-1317-66-12-2725. [DOI] [PubMed] [Google Scholar]
- 64. Suksanpaisan L, Susantad T, Smith DR. 2009. Characterization of dengue virus entry into HepG2 cells. J. Biomed. Sci. 16:17. 10.1186/1423-0127-16-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hung SL, Lee PL, Chen HW, Chen LK, Kao CL, King CC. 1999. Analysis of the steps involved in dengue virus entry into host cells. Virology 257:156–167. 10.1006/viro.1999.9633. [DOI] [PubMed] [Google Scholar]
- 66. Thongtan T, Cheepsunthorn P, Chaiworakul V, Rattanarungsan C, Wikan N, Smith DR. 2010. Highly permissive infection of microglial cells by Japanese encephalitis virus: a possible role as a viral reservoir. Microbes Infect. 12:37–45. 10.1016/j.micinf.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 67. Helander A, Silvey KJ, Mantis NJ, Hutchings AB, Chandran K, Lucas WT, Nibert ML, Neutra MR. 2003. The viral sigma1 protein and glycoconjugates containing alpha2-3-linked sialic acid are involved in type 1 reovirus adherence to M cell apical surfaces. J. Virol. 77:7964–7977. 10.1128/JVI.77.14.7964-7977.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Expression levels of endogenous RNase L and Filamin A in A7, M2 melanoma, and HT1080 cells normalized to β-actin levels. (B) M2 and A7 melanoma cells were transfected with 10 µM 2-5A or 2 µg/ml of poly(I·C), and specific cleavage of rRNA (shown by arrows) was analyzed on RNA chips using an Agilent 2100 Bioanalyzer. Download
(A) HEK293T cells expressing Myc-Filamin A or Myc tag and Flag-RNase L (full length, WT), Flag-RNase L (ΔC), or Flag-RNase L (ΔN) were immunoprecipitated using Myc-EZview agarose beads or control IgG beads. Samples were subjected to immunoblot analysis using anti-Flag or anti-Myc antibodies. For IP analysis, 5% of the immunoprecipitate was loaded in blots probed with the antibodies indicated. (B) Protein expression in cell lysates normalized to β-actin levels. Download
M2 stable cells expressing Myc-Filamin A and/or Flag-RNase L WT or Flag-RNase L R667A were infected with EMCV (MOI = 1.0) for 1 h and washed briefly with PBS to remove unbound extracellular viruses. Intracellular EMCV titers were determined by a plaque assay using L929 cells. Student’s t test was used to determine P values (as shown) of M2 cells expressing Filamin A compared to cells expressing Filamin A and either RNase L WT or R667A. P values determined using Student’s t test are as shown. Data are representative of the results of experiments performed in triplicate ± SD. Download
M2 cells stably expressing Myc-Filamin A and transiently transfected with Flag-RNase L (WT, full length), Flag-RNase L (ΔC), or Flag-RNase L (ΔN) or with vector alone were subjected to immunoblot analysis using anti-Flag antibodies. Protein expression in cell lysates was normalized to β-actin levels. *, nonspecific band. Download