ABSTRACT
Persistent human cytomegalovirus (HCMV) infection has been linked to several diseases, including atherosclerosis, transplant vascular sclerosis (TVS), restenosis, and glioblastoma. We have previously shown that factors secreted from HCMV-infected cells induce angiogenesis and that this process is due, at least in part, to increased secretion of interleukin-6 (IL-6). In order to identify the HCMV gene(s) responsible for angiogenesis promotion, we constructed a large panel of replication-competent HCMV recombinants. One HCMV recombinant deleted for UL1 to UL10 was unable to induce secretion of factors necessary for angiogenesis. Fine mapping using additional HCMV recombinants identified UL7 as a viral gene required for production of angiogenic factors from HCMV-infected cells. Transient expression of pUL7 induced phosphorylation of STAT3 and ERK1/2 MAP kinases and production of proangiogenic factors, including IL-6. Addition of recombinant pUL7 to cells was sufficient for angiogenesis and was again associated with increased IL-6 expression. Analysis of the UL7 structure revealed a conserved domain similar to the immunoglobulin superfamily domain and related to the N-terminal V-like domain of carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1). Our report therefore identifies UL7 as a novel HCMV-encoded molecule that is both structurally and functionally related to cellular CEACAM1, a proangiogenic factor highly expressed during vasculogenesis.
IMPORTANCE
A hallmark of cytomegalovirus (CMV) infection is its ability to modulate the host cellular machinery, resulting in the secretion of factors associated with long-term diseases such as vascular disorders and cancer. We previously demonstrated that HCMV infection alters the types and quantities of bioactive proteins released from cells (designated the HCMV secretome) that are involved in the promotion of angiogenesis and wound healing. A key proangiogenic and antiapoptotic factor identified from a proteomic-based approach was IL-6. In the present report, we show for the first time that HCMV UL7 encodes a soluble molecule that is a structural and functional homologue of the CEACAM1 proangiogenic cellular factor. This report thereby identifies a critical component of the HCMV secretome that may be responsible, at least in part, for the vascular dysregulation associated with persistent HCMV infection.
INTRODUCTION
Human cytomegalovirus (HCMV) is a β-herpesvirus that has been epidemiologically linked to several diseases that require angiogenesis (AG), including atherosclerosis, restenosis following angioplasty, transplant vascular sclerosis (TVS) associated with chronic allograft rejection, and glioblastoma (1–4). The best evidence that CMV is directly involved in these pathogenic processes comes from solid-organ transplantation models (5–7). Specifically, rat CMV (RCMV) was shown to decrease the mean time to allograft failure as well as increase the degree of TVS in the allograft vessels in a rat heart transplantation model (5–7). Treatment of allograft recipient rats with a CMV DNA polymerase inhibitor, ganciclovir, resulted in prolonged survival of the allograft, indicating that CMV replication was a requirement for acceleration of disease (8, 9). Microarray analysis of the RCMV-infected heart allograft transcriptome indicated that a significant number of RCMV-induced genes, including those encoding angiopoietin, vascular endothelial growth factor (VEGF), transforming growth factor beta (TGF-β), chemokines, members of the tumor necrosis factor receptor (TNFR) superfamily, and cytokines such as interleukin-6 (IL-6), were associated with AG and wound healing (WH) (10). Together, these results suggest that CMV-mediated acceleration of TVS occurs through direct or indirect alteration of AG and WH processes.
HCMV infection of cells in vitro alters the production of bioactive proteins released from infected cells into the supernatant (HCMV secretome) that, when applied to endothelial cell cultures, induce an angiogenic phenotype (11). Many of these factors have important roles in vascular disease, and we have hypothesized that a major role of CMV infection in the acceleration of TVS occurs through the increased production of AG/WH factors within the allograft. Mass spectrometry analysis of HCMV-infected and mock-infected secretomes identified more than 1,200 soluble proteins, more than 1,000 of which were specific to or highly enriched in the HCMV secretome. Pathway analysis indicates that a large number of angiogenic molecules, i.e., proteins involved in TGF-β and other factor signaling pathways, cytokines, and chemokines, as well as factors involved in extracellular matrix remodeling, are present in the HCMV secretome. Importantly, we demonstrated that the HCMV secretome contains high levels of IL-6 that promote AG and long-term survival of endothelial cells (EC). In EC, IL-6 prevented apoptosis by blocking caspase 3 and 7 activation through the induction of survivin (12). Although the identity of the HCMV gene(s) responsible for AG is unknown, this function was shown to be due to an HCMV late gene, since a drug that blocks the viral polymerase function also inhibits the ability of viral supernatants to induce neovessel formation.
In the current study, mutational analysis of the HCMV genome identified UL7 as the viral gene responsible for induction of AG factors during HCMV infection. UL7 is a late gene encoding a transmembrane protein that is cleaved to produce a small soluble protein secreted from the cell (13). We show that UL7 is sufficient to induce the AG phenotype and is structurally and functionally related to carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1), a cellular protein that also induces AG.
RESULTS
HCMV-induced angiogenesis is mediated by the UL7 late gene.
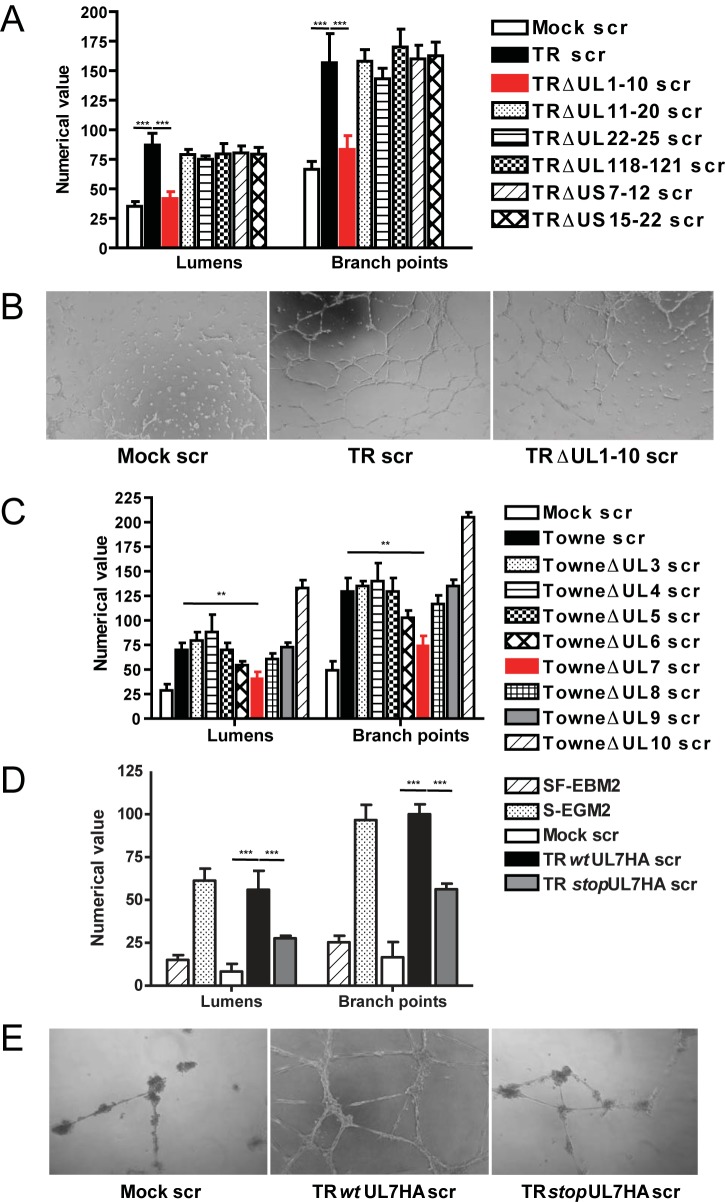
In order to identify the HCMV gene involved in AG, we generated a library of 6 recombinant viruses with large-scale deletions of nonessential open reading frames (ORFs) that include HCMV TR-bacterial artificial chromosome (BAC) ΔUL1 to UL10 (ΔUL1–10), ΔUL11–20, ΔUL22–25, ΔUL118–121, ΔUS7–12, and ΔUS15–22. Analysis of secretomes from these HCMV deletion mutants in a Matrigel angiogenesis assay indicated that only deletion of UL1–10 significantly decreased tubulogenesis in human aortic endothelial cells (HAEC) (70%) (Fig. 1A). The microscopy pictures (Fig. 1B) show some differences between the mock secretome and the UL1–10 mutant; however, the structures present in the TRΔUL1–10 secretome are mainly branching cells rather than lumens delimited by intact capillary-like tubule structures. As secretomes do not contain infectious virus, this finding suggests a direct or indirect role of HCMV protein(s) from the UL1–10 region in release of soluble proangiogenic factors.
FIG 1 .
Deletion of UL7 within the UL1–10 region impaired tubulogenesis. (A) Quantitative measures of angiogenesis consisting of the numbers of lumens and branch points observed in the presence of secretomes generated from mock infections (Mock scr), TR secretome (TR scr), TRΔUL1–10 secretome (TR ΔUL1–10 scr), TRΔUL11–20 secretome (TR ΔUL11–20 scr), TRΔUL22–25 secretome (TR ΔUL22–25 scr), TRΔUL118–121 secretome (TR ΔUL118–121 scr), TRΔUS7–12 secretome (TR ΔUS7–12 scr), and TRΔUL15-12 secretome (TR ΔUS5-12 scr). Three independent experiments were performed. Data are expressed as means ± SEM of the results of three independent biological replicates for each viral construct assayed in triplicate. Statistical significance is represented with three asterisks (***; P < 0.001). (B) Representative examples of mock secretome (Mock scr), HCMV TR secretome (TR scr), and HCMV TR ΔUL1–10 secretome (TR ΔUL1–10 scr) are shown as a low-power image (magnification, ×10). (C) HFFF cells were infected with wild-type Towne (wt Towne), Towne ΔUL3, Towne ΔUL4, Towne ΔUL5, Towne ΔUL6, Towne ΔUL7, Towne ΔUL8, Towne ΔUL9, and Towne ΔUL10 or were mock infected. Secretomes were collected at 96 hpi and tested in a Matrigel assay using HAEC. Three independent experiments were performed. Data are expressed as means ± SEM of the results of three independent biological replicates for each viral construct assayed in triplicate. Statistical significance is represented with two asterisks (**; P < 0.01). (D) HAEC were infected with TR wtUL7HA or TR stopUL7HA and secretomes collected at 96 hpi. Statistical significance is represented with three asterisks (***; P < 0.001). (E) Representative examples of mock secretome (Mock scr), TR wtUL7HA secretome (TR wtUL7HA scr), and TR stopUL7HA secretome (TR stopUL7HA scr) are shown as a high-power image (magnification, ×20).
In order to determine which of the HCMV genes encoded in the TR ΔUL1–10 mutant were responsible for HCMV-induced AG, a library of HCMV Towne mutants with transposon mutations in UL3, UL4, UL5, UL6, UL7, UL8, UL9, or UL10 was used to generate secretomes in fibroblasts (14). The UL2 mutant was not included in these studies since this virus exhibits a growth defect (14). The role of UL1 was also not examined since the parental HCMV Towne strain contains a spontaneous deletion in this gene that does not affect the ability of the virus to induce a secretome that stimulates AG (15). Analysis of HCMV mutant secretomes for induction of AG indicated that mutation of UL7 resulted in a 50% reduction of tubulogenesis (Fig. 1C). As further confirmation that UL7 was important for HCMV-induced AG, an HCMV UL7 mutant was generated in the clinical strain TR (TR stopUL7HA) by insertion of two stop codons as well as an influenza virus hemagglutinin (HA) epitope tag in frame at the 3′ end of the UL7 ORF. A correlate HCMV TR revertant virus (TR wild-type UL7HA [wtUL7HA]) was also constructed by replacing the stopUL7-HA cassette with a functional UL7-HA insertion (see Fig. S1 in the supplemental material). The replication kinetics of TR wtUL7HA and TR stopUL7HA were similar to those seen with the TR wt (see Fig. S2). Expression of UL7 in TR wtUL7HA- or TR stopUL7HA-infected cells was determined by immunofluorescence using antibodies to the HA epitope, with results indicating that only the revertant virus expressed UL7 (see Fig. S3). Subsequently, analysis of secretomes obtained from TR stopUL7HA- and TR wtUL7HA-infected HAEC indicated that only the revertant virus and not the UL7 mutant induced AG (Fig. 1D and E). These results demonstrate that UL7 is the gene within the UL1–10 region required for HCMV-induced AG.
UL7 is a novel CEACAM-like protein and is sufficient to induce angiogenesis.
UL7 is a member of the HCMV RL11 gene family (15, 16). Previous studies indicated that UL7 is expressed as an early-late gene and encodes a 222-amino-acid type I glycoprotein, which is proteolytically cleaved to release a heavily glycosylated ectodomain (13). The late gene expression of UL7 was confirmed by Northern blotting (see Fig. S4 in the supplemental material) and is consistent with the previous observation that HCMV-induced AG is due to expression of a viral late gene (11). Although pUL7 was previously reported to have homology to the CD229 lymphocyte-activating molecule receptor (13), a query of the HCMV TR UL7 protein against NCBI GenBank conserved domains revealed a similarity to the first Ig-like domain of the carcinoembryonic antigen (CEA)-related cell adhesion molecule (CEACAM) protein subfamily (cd, 05741; E value, 3.90e-04). A BLASTp alignment performed with the four members of the CEACAM family (CEACAM1, CEACAM3, CEACAM5, and CEACAM6) displayed 39% similarity and 29% amino acid identity between the UL7 Ig-like domain and the N-terminal V-like domain of CEACAM1 (Fig. 2). Interestingly, the CEACAM1 molecule is highly expressed during vasculogenesis (17), and soluble recombinant CEACAM1 added to endothelial cell cultures has been shown to stimulate significant tubule formation (18).
FIG 2 .

UL7 is a homologue of secreted CEACAM1. An alignment of the sequences of the V-set Ig domain of CEACAM1 and UL7 is shown. Asterisks, colons, and dots indicate identical amino acid residues, conserved amino acid residues, and semiconserved amino acid residues, respectively.
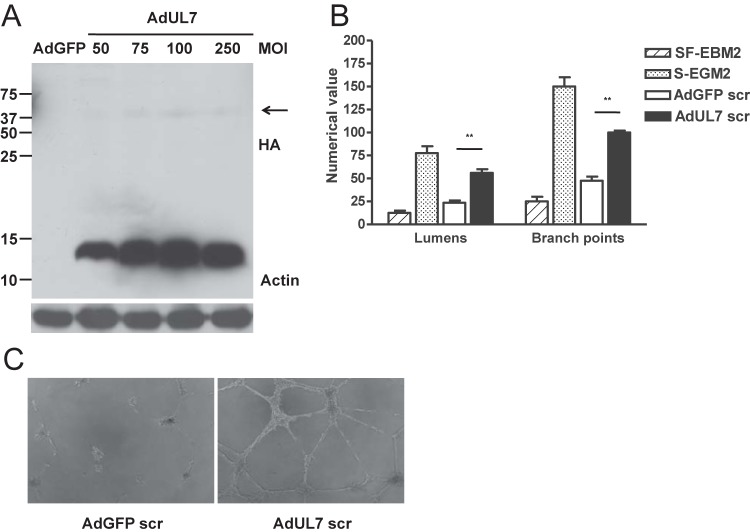
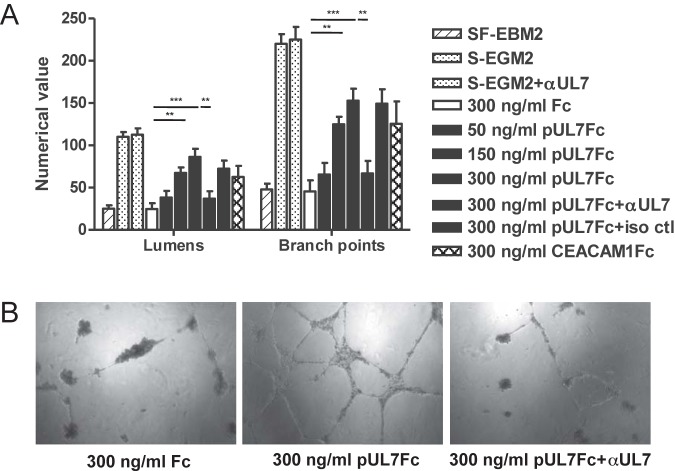
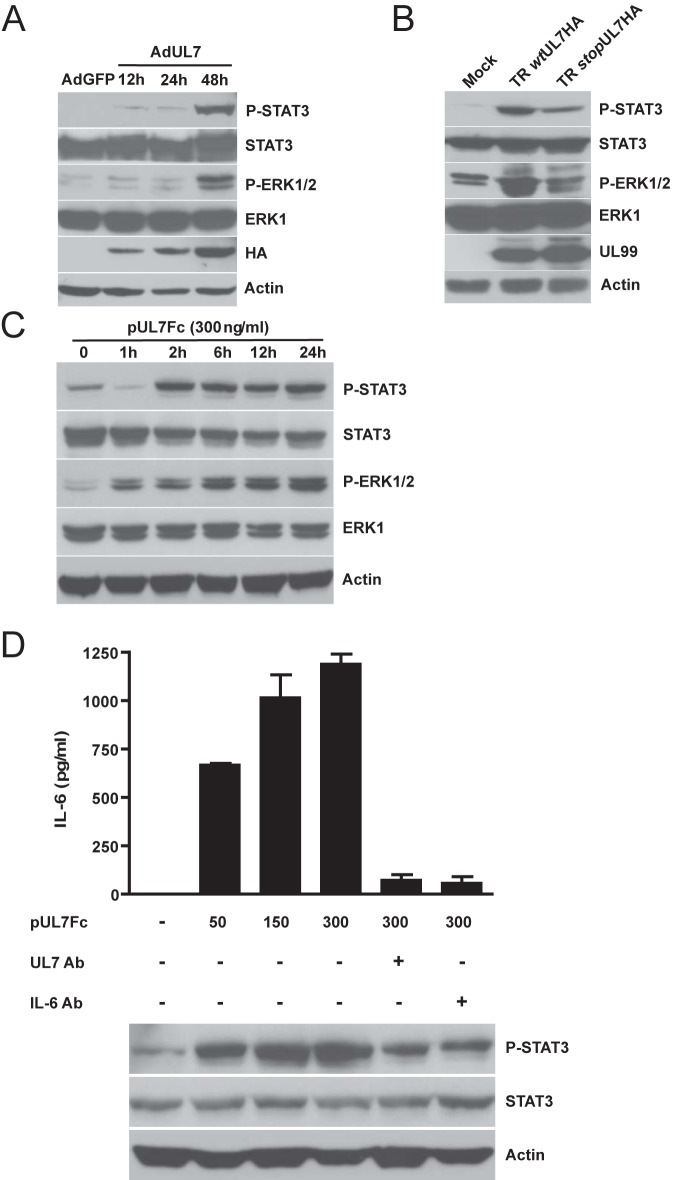
To confirm that UL7 is cleaved from the cell surface, HEK293 cells were transfected with a plasmid expressing pUL7 tagged at the C terminus with a HA epitope. After 48 h, immunoblot analyses of cell lysates by the use of an antibody specific for the HA epitope and of supernatants by the use of a specific polyclonal UL7 antibody were performed. Two prominent bands of 75 and 12 kDa, corresponding to the UL7 precursor and the cytoplasmic tail, were detected in the pUL7HA-expressing cells. PNGaseF treatment led to a reduction of the UL7 precursor to the predicted molecular mass of 25 kDa (see the left panel of Fig. S5 in the supplemental material). In the supernatant of the transfected cells, only the 75-kDa ectodomain was present, demonstrating that pUL7 is partially cleaved in HEK293 cells (see Fig. S5, right panel). To allow us to express detectable levels of pUL7 in HAEC, we constructed an adenoviral vector expressing pUL7HA (designated AdUL7). Western blotting of cell lysate from HAEC transduced with AdUL7 showed the 12-kDa protein, corresponding to cytoplasmic tail regions of UL7, and a faint band around 75 kDa (Fig. 3A), suggesting that the ectodomain of pUL7 in EC is mainly released from the cells. Secretomes were harvested from cells transduced with AdUL7 or adenovirus construct expressing green fluorescent protein (AdGFP) and examined for the ability to induce AG. As shown in Fig. 3B and C, only the secretome from HAEC transduced with AdUL7 was capable of inducing a consistent network of interconnecting tubules. To determine whether the ectodomain of pUL7 was sufficient to induce AG, we assessed the angiogenic properties of a recombinant protein containing only the extracellular domain of UL7 fused to the Fc region of the mouse IgG heavy chain. As shown in Fig. 4, stimulation of HAEC with pUL7Fc, but not with Fc alone, resulted in a dose-dependent increase of tubule formation that was inhibited by preincubation of pUL7Fc with an anti-UL7 polyclonal antibody. Human recombinant CEACAM1Fc was used as a positive control. To verify the hypothesis that UL7 is binding a cellular receptor on the cell surface, we performed a flow cytometry-based binding assay with purified pUL7Fc or Fc alone, demonstrating the specific interaction of UL7 with CD31-positive HAEC (see Fig. S6). These data indicate that the pUL7 ectodomain is sufficient to induce the production of angiogenic factors in the cellular secretome.
FIG 3 .
pUL7 is secreted and sufficient to induce AG. HAEC were transduced with AdGFP and AdUL7, and the secretomes were collected at 48 h. (A) Cell lysates were analyzed by Western blotting with antibodies specific for HA and actin. Right margin, the arrowhead indicates the pUL7 high-molecular-weight form. (B) Quantitative measures of angiogenesis consisted of the numbers of lumens and branch points. Three independent experiments were performed. Data are expressed as means ± SEM of the results of three independent biological replicates for each adenoviral construct assayed in triplicate. Statistical significance of the results of comparisons with the AdGFP secretome is indicated with two asterisks (**; P < 0.01). (C) Representative examples of AdGFP and AdUL7 secretomes are shown as a high-power image (magnification, ×20).
FIG 4 .
Recombinant UL7 promotes HAEC tube formation. Increasing concentrations (50, 150, and 300 ng/ml) of pUL7Fc were directly added to serum-free basal media in the presence or absence of neutralizing antibodies against UL7 or the isotype control (iso ctl). Controls included HAEC stimulated with 300 ng/ml of Fc and 300 ng/ml of CEACAM1Fc. (A) Quantitative measures of angiogenesis consisted of the numbers of lumens and branch points. Three independent experiments were performed. Data are expressed as means ± SEM of the results of three independent biological replicates for each condition assayed in triplicate. Statistical significance is represented with three asterisks (***; P < 0.001) or with two asterisks (**; P < 0.01). (B) Representative examples of Fc, pUL7Fc, and pUL7Fc plus αUL7 (pUL7Fc+αUL7) are shown as a high-power image (magnification, ×20).
HCMV UL7 induces the production of IL-6 that is associated with increased STAT3 and extracellular signal-regulated kinase 1 (ERK1) and ERK2 (ERK1/2) phosphorylation.
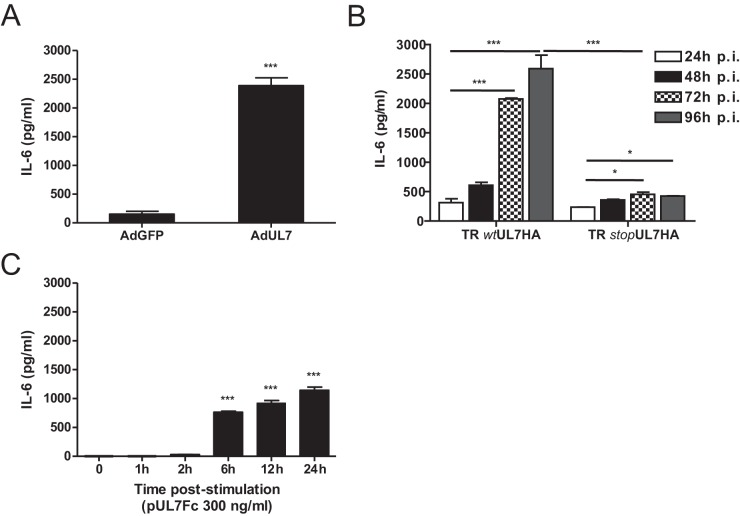
We previously reported that HCMV-induced AG is mediated by viral induction of IL-6 in the secretome (12). To determine whether pUL7 induced the production of IL-6, HAEC transduced with AdUL7 or AdGFP were analyzed for IL-6 production. As shown in Fig. 5A, transduction of cells with AdUL7 resulted in an ~12-fold increase in production of IL-6 in comparison to the AdGFP results. These observations are also consistent with the 10-fold-lower level of IL-6 release seen following infection of cells with the UL7 mutant (TR stopUL7HA) compared to the revertant virus TR wtUL7HA at 96 h postinfection (hpi) (Fig. 5B). To determine whether extracellular addition of UL7 induced production of IL-6, pUL7Fc was added to cells followed by quantification of the cytokine in supernatants. As shown in Fig. 5C, addition of pUL7Fc to HAEC was sufficient to induce secretion of IL-6 within the first 24 h.
FIG 5 .
UL7 upregulates IL-6 protein secretion. Data represent ELISA quantification of IL-6 levels in HAEC transduced with AdGFP and AdUL7 (A), infected with TR wtUL7HA or with TR stopUL7HA (B), and stimulated with recombinant pUL7Fc (C). IL-6 secretion is shown as the means ± SEM of the results of two independent experiments. ***, P < 0.001; *, P < 0.05.
Consistent with the release of IL-6 in the supernatant of AdUL7-transduced HAEC, we observed elevated levels of phospho-STAT3 and phospho-ERK1/2 at 48 hpi, compared to the control AdGFP-transduced cell results (Fig. 6A). To investigate the phosphorylation status of STAT3 and ERK1/2 during HCMV infection, we infected HAEC with TR wtUL7HA and TR stopUL7HA. Western blot analysis of cells infected with TR wtUL7HA for 96 h showed a 35-fold increase in phospho-STAT3 and a 10-fold increase in phospho-ERK1/2 levels compared to the mock-infected cell results. The level of phospho-STAT3 in HAEC infected with TRstopUL7HA was 12-fold higher than that seen with the mock infection, while the level of phospho-ERK1/2 was comparable to that seen with the mock infection (Fig. 6B). In order to determine whether soluble pUL7 binding to the cells induced IL-6 production, pUL7Fc was added to cultures of HAEC at increasing concentrations. As shown in Fig. 6D, addition of pUL7Fc resulted in IL-6 production, with optimal effects observed at 300 ng/ml. Moreover, pUL7Fc induced STAT3 and ERK1/2 phosphorylation (Fig. 6C). Treatment with neutralizing antibodies against pUL7 or IL-6 resulted in an 80% decrease in IL-6 secretion and a 40% reduction in STAT3 phosphorylation (Fig. 6D).
FIG 6 .
UL7 induces STAT3 and ERK1/2 phosphorylation. Cell lysates from HAEC transduced with AdGFP or AdUL7 (A), infected with TR wtUL7HA or TR stopUL7HA for 96 h (B), and stimulated with pUL7Fc (C) were subjected to SDS-PAGE followed by immunoblotting. The membranes were decorated with antibodies specific for phospho-STAT3 and phospho-ERK1/2. Immunodetection was performed with total STAT3, ERK1, and actin serving as the internal controls. (D) Cells were treated with increasing concentrations of recombinant pUL7Fc in the presence or absence of UL7 and IL-6 neutralizing antibodies (Ab) for 24 h. ELISA was used to quantify the amount of IL-6 secreted from HAEC. Cell lysates were analyzed by Western blotting with specific antibodies for phospho-STAT3, total STAT3, and actin.
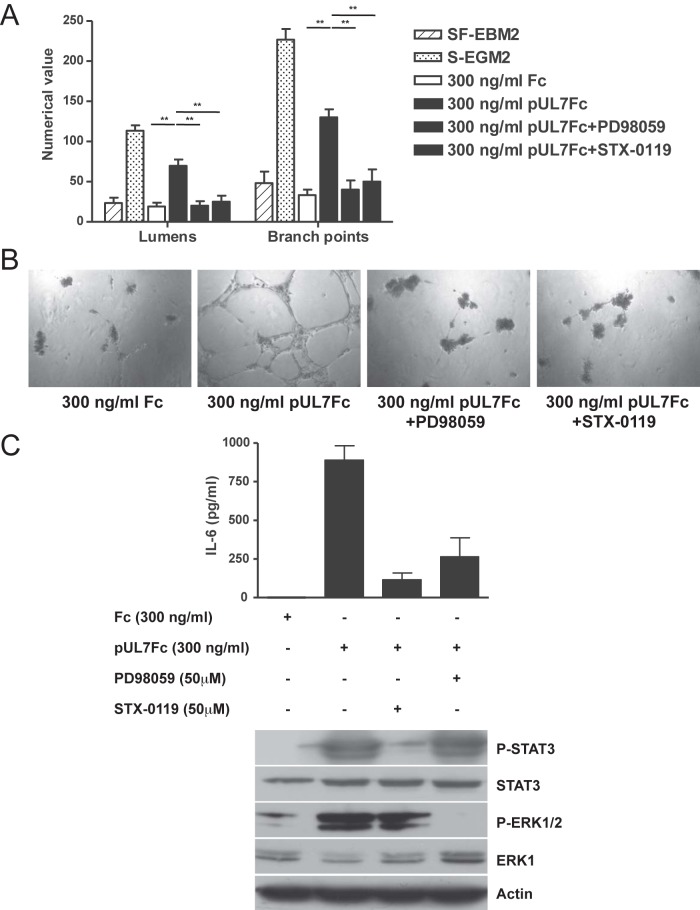
Finally, treatment of pUL7Fc-stimulated HAEC with either an ERK1/2 (PD98059) or a STAT3 (STX-0119) inhibitor resulted in decreased tubule formation and IL-6 secretion (Fig. 7).
FIG 7 .
Pharmacological inhibition of STAT3 or ERK1/2 phosphorylation impairs pUL7-induced AG and IL-6 secretion. HAEC were incubated in the presence of 300 ng/ml of Fc, pUL7Fc, pUL7Fc with 50 µM of ERK1/2 inhibitor PD98059, and pUL7Fc with 50 µM of STAT-3 inhibitor STX-0119. (A) Quantitative measures of angiogenesis consisted of the numbers of lumens and branch points. Three independent experiments were performed. Data are expressed as means ± SEM of the results of three independent biological replicates for each conditions assayed in triplicate. Statistical significance is represented with two asterisks (**; P < 0.01). (B) Representative examples of Fc, pUL7Fc, pUL7Fc+PD9859, and pUL7Fc+STX-0119 are shown as a high-power image (magnification, ×20). (C) ELISA quantification of IL-6 levels and immunoblotting on cell lysates with specific antibodies for phospho-STAT3, total STAT3, and actin.
Together, these observations indicate that binding of secreted pUL7 to an unknown cellular receptor on HAEC induces IL-6 through the activation of STATs and of ERK1/2-mitogen-activated protein kinase (MAPK) pathways.
DISCUSSION
In the present study, we demonstrated that the presence of HCMV pUL7 is sufficient to induce endothelial cell angiogenesis. Interestingly, pUL7 is a secreted viral protein and contains an Ig-like domain with structural and functional homology to CEACAM1, a molecule highly expressed during vasculogenesis. We found that UL7 expression stimulates an inflammatory response in EC, inducing the secretion of IL-6 and the activation of STATs and of MAPK pathways. Importantly, these findings identify a mechanism by which HCMV can influence and manipulate the vascular environment despite limited infection of the vessel wall.
The CMV genome encodes 12 multigene families (16, 19), including the RL11 family, which contains the RL11–13, UL1, and UL4–11 genes (20). All members of the RL11 family are located adjacent to each other near the left terminus of the genome and are dispensable for virus growth in vitro (14). Among the RL11D genes, UL7 exhibits high sequence conservation across different CMV genotypes (97% to 100% intra- and 83% to 93% intergenotype conservation), suggesting a critical role of UL7 for viral fitness (15). Consistent with the literature data, we found that pUL7 is shed from HAEC cells and that the transcript is expressed in the late phase of the viral replication cycle. Upon further sequence inspection of pUL7, however, we identified homology with the first Ig-like domain of the carcinoembryonic antigen (CEA)-related cell adhesion molecule (CEACAM) protein subfamily. Interestingly, another HCMV RL11 protein, UL1, which is virion associated and required for efficient replication in epithelial cells, had previously been reported to share sequence homology with CEACAM1 (21). Because RL11 members are more likely the descendants of a common cellular ancestor that was incorporated into the HCMV genome, duplicated, and subsequently subjected to functional divergence during coevolution with the host, pUL7 may have evolved to mimic the angiogenic properties of soluble CEACAM1 whereas pUL1 has retained the receptor function contributing to HCMV access to host cells by interacting with members of the CEA family.
CEACAM1 is a transmembrane cell adhesion molecule that is abundantly expressed in epithelia, vessel endothelia, and hematopoietic cells (22). In primary EC culture, recombinant human CEACAM1 enhances the sprouting and migration of EC, acting synergistically with VEGF (18). EC produces both secreted and membrane-bound forms of CEACAM1. The soluble CEACAM1 form may stimulate proliferation and chemotaxis of EC, whereas the membrane-bound form may be involved in morphogenic events such as formation of capillary tubes. Moreover, lack of endothelial CEACAM1 expression in Ceacam1−/− mice leads to defective vascular remodeling in vivo, whereas endothelial overexpression of CEACAM1 in CEACAM1endo+ mice induces extensive vascular growth (23). Membrane-bound CEACAM1 can have a long (L) or a short (s) cytoplasmic domain (24). Both isoforms are coexpressed in most CEACAM1-expressing tissues, and the ratio between the two isoforms determines the signaling outcome (25, 26). The structural and functional homology of UL7 with CEACAM1 and the inter-strain-specific conservation level reflect the existence of a selective pressure against mutations in the protein sequence, supporting a relevant role of this viral product. The short cytoplasmic tail of pUL7 resembles the “s” isoform of CEACAM1, suggesting that pUL7 does not function as a signal receptor but more likely evolved to mimic the angiogenic properties of soluble CEACAM1. However, our studies presented here do not rule out a possible function for the membrane-bound form, in addition to the AG functions of the soluble form.
We have previously shown that one of the key cytokines present in the HCMV secretome is IL-6, which promotes AG and blocks apoptosis through increased levels of survivin (12). The inflammatory effects of IL-6 have been implicated as a factor in the failure of organ and tissue grafts, and elevated levels of the cytokine have been reported to accompany HCMV replication in transplanted lungs and bone marrow during episodes of inflammation or rejection (27–29). Interestingly, we found that binding of pUL7 stimulates an inflammatory response in vascular endothelial cells that leads to IL-6 secretion. Our data are therefore in contrast with the observations by Engel et al., who reported that activated monocyte-derived immature dendritic cells (DCs) and monocytic cell lines transiently expressing pUL7 secreted less IL-8, IL-6, and tumor necrosis factor (TNF) than the control cells (13). It is possible that the binding of pUL7 with its cellular receptor(s) triggers a pro- or anti-inflammatory response depending on the role played by different cell types in HCMV spread and persistence in the host. For example, CEACAM1-L, upon activation, can discriminate between binding of cytoplasmic protein tyrosine phosphate SHP-1/SHP-2 and binding of Src family tyrosine kinases in a manner that is regulated by and dependent on CEACAM1-L supramolecular organization. The discriminatory binding of SHP-1/SHP-2 and of Src is the basis for the different effects of signal regulation by CEACAM1, which have found to be both inhibitory and stimulatory (30).
Notably, the concentration of IL-6 was increased after AdUL7 transduction or addition of recombinant pUL7Fc protein (Fig. 5A and C). IL-6 is a potent, pleiotropic, inflammatory cytokine that mediates multiple physiological functions, including developmental differentiation of lymphocytes, cell proliferation, cell survival, and inhibition of apoptotic signals (31–34). IL-6 mRNA has also been detected in human atherosclerotic lesions, indicating that this cytokine is expressed at the site of injury (35). Engagement of IL-6 with the IL-6 receptor activates three main signaling pathways, the JAK/STAT, ERK1/2-MAPK, and phosphoinositol 3-kinase (PI3K)/Akt pathways (36, 37). Activation of the JAK-STAT pathway was reported to have short-term cardioprotective effects as well as long-term inflammation-promoting effects (38). ERK1/2-MAPK activation has been implicated in hypertrophy and cell survival (39). IL-6 and the JAK/STAT and MAPK pathways are also linked to malignant glioblastoma through the expression of the US28 HCMV chemokine receptor (40). US28 activity might be responsible for the residual activation of STAT3 and ERK1/2 in TR stopUL7HA-infected cells compared to the mock-infected cell results (Fig. 6B).
In conclusion, we show for the first time that HCMV encodes a homologue structurally and functionally similar to CEACAM1. Ongoing and future work will be focused on the identification of the UL7 cellular receptor(s) as well as on the function of this viral gene in HCMV pathogenesis. Thus, alteration of UL7 binding or cell signaling may offer a novel strategy for the treatment of the angiogenic processes involved in HCMV-associated vascular diseases.
MATERIALS AND METHODS
Cells and virus preparation.
Human aortic endothelial cells (HAEC) (Lonza) were cultured in endothelial growth medium (EGM-2; Lonza), as previously described (12). Human fetal foreskin fibroblasts (HFFF) (purchased from Sigma) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Cellgro; Corning) supplemented with 10% fetal bovine serum (FBS) (HyClone; Thermo Scientific), 4.5 g/liter glucose, l-glutamine and sodium pyruvate, and antibiotics (penicillin [10 units/ml]-streptomycin [10 µg/ml]). The human embryonic kidney 293 cell line (HEK293) and HEK293/Cre4 cell line (Microbix Biosystems Inc.) were cultured in minimum essential medium (MEM) (Cellgro; Corning) supplemented with 10% FBS, penicillin, streptomycin, and l-glutamine. PD98058 was purchased from Cell Signaling Technology and STAT3 inhibitor XI (STX-0119) from Calbiochem. HCMV strain TR was derived originally from a patient with AIDS-related HCMV retinitis (41) and was cloned into a bacterial artificial chromosome (BAC) (NCBI accession no. AC146906) (42). Infectious virus was produced and titers were determined by plaque assays on HFFF as previously described (12).
Plasmids.
Full-length UL7 containing a hemagglutinin (HA) epitope tag in frame at the 3′ terminus of the ORF was obtained by PCR using DNA extracted from HCMV TR virions as the template and the primers listed in Table S1 in the supplemental material. UL7HA containing two stop codons, one at codon 6 and the second at codon 221, was generated by PCR using pGEM-UL7HA as the template and the primers listed in Table S1. pCDNA3-UL7HA was generated by PCR amplification of UL7HA from pGEM-T Easy using the primers listed in Table S1. For construction of UL7HA recombinant adenovirus, the respective ORF was amplified from TR DNA with the primers listed in Table S1, digested with EcoRI and BamHI, and then cloned into the pAdTet7 adenoviral vector. For construction of recombinant pUL7Fc, a PCR product was amplified from TR DNA using the primers listed in Table S1, digested with AscI and NotI, and then cloned into pCMV6-AC-Fc-S plasmid (Origene).
Construction of recombinant HCMV TR mutants.
Recombinant TRΔUL1–10 was constructed by E/T recombination using pBSKSII plus ΔNaeI/SspI/MCS vector as the template to generate the PCR product and the primers listed in Table S1 in the supplemental material as previously described (43). Restriction enzyme digestion combined with DNA sequence analysis confirmed deletion of UL1–10 from the HCMV TR-BAC genome.
TR stopUL7HA and TR wtUL7HA BAC were constructed using the galactokinase (Galk) recombineering method (see Fig. S1 in the supplemental material) (44). In the first step, the galactokinase-kanamycin (Galk/Kan) ORF was amplified from pYD-C255, a GalK-Kan dual-expression plasmid (a gift from D. Yu, Washington University in St. Louis), using primers listed in Table S1. One of the single Gal-positive (Gal+) TR ΔUL7 BAC colonies was further characterized for UL7 replacement by PCR and used to initiate the counterselection step. To replace the Galk/Kan cassette in the second step, UL7 was amplified from pGEM-stopUL7HA plasmid using oligonucleotides listed in Table S1. Two independent TR stopUL7HA BAC clones were thus selected and produced to ensure that their phenotype did not result from an off-target mutation. To generate the revertant HCMV TR wtUL7HA-BAC, the insertion and substitution of the Galk/Kan cassette were repeated with a PCR product generated from the pGEM-UL7HA plasmid. Oligonucleotide primers used for the revertant TR wtUL7HA BAC are listed in Table S1. Recombinant viruses were screened by BAC digestion, PCR, and sequencing. Recombinant viruses were produced by electroporation of the corresponding BACs into HFFF. Virus stocks were propagated and stored and titers were determined as described above.
Adenovirus production.
Recombinant adenoviruses were produced by pAdTet7 UL7HA cotransfection of 293-Cre cells with Ad DNA (Ad5-ψ5) (45). Recombinant adenoviruses were expanded on 293-Cre cells, and the titers of the bulk stocks were determined on 293 cells by limiting dilution. Gene expression was driven by coinfection with Ad-Trans expressing the Tet-off transactivator as previously described (46). The recombinant adenoviral vector expressing green fluorescent protein (GFP) was a gift from D. Streblow at Oregon Health & Science University (OHSU) (47).
Preparation of HCMV and adenoviral secretomes.
HCMV TR secretomes were generated by infecting HAEC at a multiplicity of infection (MOI) of 3 PFU/cell for 96 h, as previously described (12). HCMV Towne secretomes were generated in fibroblasts infected at an MOI of 0.5 and harvested 96 hpi. For the adenovirus secretome, HAEC were starved for 12 h in serum-free endothelial basal medium-2 (SF-EBM2) and then coinfected with the appropriate adenovirus and transactivator (MOI of 250 PFU/cell). The supernatant from infections was harvested and clarified at 48 hpi.
Angiogenesis assay.
The ability of secretomes to induce angiogenic activity was measured using a modified version of the standard Matrigel in vitro tubule formation assay, as previously described (12). For experiments involving neutralization, the secretome containing anti-UL7 antibody or an anti-rabbit isotype control was incubated for 1 h at 37°C prior to stimulation of HAEC.
Fc fusion UL7 and antiserum production.
To express UL7 in a glycosylated form comparable to that of the native protein, stable clones were made by transfection of HEK293 with pCMV6-UL7Fc and selection with neomycin (G418; Gibco-BRL). Stable clones with G418 resistance were obtained by trypsinization of colonies, followed by plating at limiting dilutions onto a 96-well plate. Ten single clones were then expanded and analyzed for UL7 secretion by the use of mouse Fc and an IgG enzyme-linked immunosorbent assay (ELISA) kit (MednaBio). The clone producing the largest amounts of fusion protein was cultured in EX-Cell 293 serum-free medium (Sigma-Aldrich). The supernatant containing the fusion protein was purified using a HiTrap protein A HP column (GE Healthcare) according to the manufacturer’s specifications.
To generate an anti-UL7 serum, the recombinant protein was used for rabbit immunization. The serum obtained by bleeding at 10 days after the fourth immunization was purified on an HiTrap protein A HP column (GE Healthcare) according to the manufacturer’s specifications. An isotype control was generated by purifying rabbit serum prior immunization. Fc and CEACAM1 recombinant proteins were purchased at R&D Systems.
Immunoblotting.
Extracts were run on an 8% to 12% SDS-PAGE gel, transferred to Immobilon-P transfer membranes (Milipore Corp.), and visualized with antibodies specific for phospho-STAT3 (Tyr705 and 3E2; Cell Signaling), STAT3 (79D7; Cell Signaling), phospho-ERK1/2 (Thr202 and Tyr204; Cell Signaling Technology), ERK1 (C16; Santa Cruz), mouse monoclonal UL99 (pp28) and HA (HA-7; Sigma), rabbit UL7, and actin (C4; Millipore).
IL-6 immunoassay.
A double-antibody sandwich enzyme-linked immunosorbent assay (ELISA) for the quantitative determination of human IL-6 levels in tissue culture media was performed on the stored supernatants according to the manufacturer’s recommended procedure (R&D Systems).
Sequence analysis.
Conserved domains in the UL7 sequence were identified using the NCBI Conserved Domain program (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Protein sequence alignment was performed using the NCBI Basic Local Alignment Search Tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Statistical analysis.
Data are expressed as means ± standard errors of the means (SEM). Statistical significance of differences was determined by Student’s t test and considered significant at P values of ≤0.05.
SUPPLEMENTAL MATERIAL
Schematic representation of TR stopUL7HA and TR wtUL7HA generation with the Galk/Kan recombineering method. Download
Multistep growth curves of TR stopUL7HA and TR wtUL7HA recombinant virus. Fibroblasts were infected at an MOI of 0.1, and the extent of viral replication was measured at the indicated days postinfection by determining the titers corresponding to the infectivity of cell suspension supernatants using plaque assays. The data shown are the averages of results of three independent biological replicates for each time point ± standard errors of the means (error bars). Download
UL7 expression in infected cells. Fibroblasts were mock infected or infected with TR wtUL7HA and TR stopUL7HA at an MOI of 0.5. pUL7 and IE2 were detected by indirect immunofluorescence at 72 hpi using a mouse anti-HA antibody and a rabbit IE2 antibody and visualized with fluorescent red Alexa Fluor 594 and fluorescent green Alexa Fluor 488, respectively. The nuclei were counterstained with fluorescent blue DAPI (4′,6-diamidino-2-phenylindole). Download
UL7 is a late gene. Analysis of RNA was performed by Northern blotting. HCMV-infected HAEC in the presence or absence of foscarnet (phosphonoformate [PFA]) were rinsed twice with ice-cold phosphate-buffered saline (PBS) at the indicated times, and total cellular RNA was extracted using a NucleoSpin RNAII kit (Macherey-Nagel). Fractionation of total RNA was performed on a 1% agarose, 2.2 M formaldehyde gel followed by blotting onto a nitrocellulose membrane (Hybond C-extra; Amersham). Hybridization was carried out with 106 cpm/ml of single-stranded RNA (ssRNA)-radiolabeled probe corresponding to the UL7 TR gene cloned in pGEM-T Easy vector (Promega). In vitro transcription with the Sp6 RNA polymerase (Ambion) was used to generate antisense RNA transcripts. Download
pUL7 is secreted in transfected cells. HEK293 cells were transfected with pCDNA3 (ctl) or pCDNA3UL7HA. After 48 h, cells (left panel) and supernatants (right panel) were harvested, treated with peptide N-glycosidase F (PNGase F), and then analyzed by Western blotting with antibodies specific for HA, UL7, and actin. In the right panel, the arrowhead indicates the high-molecular-weight pUL7 form. Download
pUL7 binds to the surface of HAEC. Data represent the results of flow cytometry analysis of pUL7Fc (blue line) and Fc (black line) interactions with HAEC (upper panel). Expression of hCD31 is shown in density plots, with rectangles indicating gates for cells analyzed for pUL7Fc and Fc binding (lower panel). HAEC (2 × 105) were incubated with 2 µg of pUL7Fc or Fc in blocking solution (5% mouse serum–2 mM EDTA–PBS) and stained with UL7 antibody followed by Alexa Fluor 594-labeled goat anti-rabbit. The HAEC population was confirmed using an antibody directed to CD31 (PB-conjugated anti-human CD31, clone WM59; BioLegend). All the steps were performed at 4°C. After surface staining with conjugated antibodies, polychromatic flow cytometric analysis was performed using an LSR II system (BD Biosciences), and data were analyzed by using FlowJo software (version 9.1; Tree Star, Inc.). Download
Oligonucleotides used for BAC mutagenesis and cloning.
ACKNOWLEDGMENTS
This work was supported by grant AI21640 from the National Institute of Allergy and Infectious Diseases, NIH.
We thank Andrew Townsend for graphics assistance, Jamie Borton for the construction of the HCMV large-deletion panel, and Lindsey Crawford for the flow cytometry assistance and data analysis. We gratefully acknowledge Dong Yu and Daniel Streblow for providing reagents.
Footnotes
Citation MacManiman JD, Meuser A, Botto S, Smith PP, Liu F, Jarvis MA, Nelson JA, Caposio P. 2014. Human cytomegalovirus-encoded pUL7 is a novel CEACAM1-like molecule responsible for promotion of angiogenesis. mBio 5(6):e02035-14. doi:10.1128/mBio.02035-14.
REFERENCES
- 1. Almond PS, Matas A, Gillingham K, Dunn DL, Payne WD, Gores P, Gruessner R, Najarian JS. 1993. Risk factors for chronic rejection in renal allograft recipients. Transplantation 55:752–756. 10.1097/00007890-199304000-00013. [DOI] [PubMed] [Google Scholar]
- 2. Melnick JL, Adam E, DeBakey ME. 1998. The link between CMV and atherosclerosis. Infect. Med. 15:479–486. [Google Scholar]
- 3. Speir E, Modali R, Huang ES, Leon MB, Shawl F, Finkel T, Epstein SE. 1994. Potential role of human cytomegalovirus and p53 interaction in coronary restenosis. Science 265:391–394. 10.1126/science.8023160. [DOI] [PubMed] [Google Scholar]
- 4. Cobbs CS, Harkins L, Samanta M, Gillespie GY, Bharara S, King PH, Nabors LB, Cobbs CG, Britt WJ. 2002. Human cytomegalovirus infection and expression in human malignant glioma. Cancer Res. 62:3347–3350. [PubMed] [Google Scholar]
- 5. Orloff SL, Yin Q, Corless CL, Orloff MS, Rabkin JM, Wagner CR. 2000. Tolerance induced by bone marrow chimerism prevents transplant vascular sclerosis in a rat model of small bowel transplant chronic rejection. Transplantation 69:1295–1303. 10.1097/00007890-200004150-00015. [DOI] [PubMed] [Google Scholar]
- 6. Orloff SL, Streblow DN, Soderberg-Naucler C, Yin Q, Kreklywich C, Corless CL, Smith PA, Loomis CB, Mills LK, Cook JW, Bruggeman CA, Nelson JA, Wagner CR. 2002. Elimination of donor-specific alloreactivity prevents cytomegalovirus-accelerated chronic rejection in rat small bowel and heart transplants. Transplantation 73:679–688. 10.1097/00007890-200203150-00005. [DOI] [PubMed] [Google Scholar]
- 7. Streblow DN, Kreklywich C, Yin Q, De La Melena VT, Corless CL, Smith PA, Brakebill C, Cook JW, Vink C, Bruggeman CA, Nelson JA, Orloff SL. 2003. Cytomegalovirus-mediated upregulation of chemokine expression correlates with the acceleration of chronic rejection in rat heart transplants. J. Virol. 77:2182–2194. 10.1128/JVI.77.3.2182-2194.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Merigan TC, Renlund DG, Keay S, Bristow MR, Starnes V, O’Connell JB, Resta S, Dunn D, Gamberg P, Ratkovec RM, Richenbacher WE, Millar RC, DuMond C, DeAmond B, Sullivan V, Cheney T, Buhles W, Stinson EB. 1992. A controlled trial of ganciclovir to prevent cytomegalovirus disease after heart transplantation. N. Engl. J. Med. 326:1182–1186. 10.1056/NEJM199204303261803. [DOI] [PubMed] [Google Scholar]
- 9. Valantine HA, Gao SZ, Menon SG, Renlund DG, Hunt SA, Oyer P, Stinson EB, Brown BW, Jr, Merigan TC, Schroeder JS. 1999. Impact of prophylactic immediate post-transplant ganciclovir on development of transplant atherosclerosis: a post hoc analysis of a randomized, placebo-controlled study. Circulation 100:61–66. 10.1161/01.CIR.100.1.61. [DOI] [PubMed] [Google Scholar]
- 10. Streblow DN, Kreklywich CN, Andoh T, Moses AV, Dumortier J, Smith PP, DeFilippis V, Fruh K, Nelson JA, Orloff SL. 2008. The role of angiogenic and wound repair factors during CMV-accelerated transplant vascular sclerosis in rat cardiac transplants. Am. J. Transplant. 8:277–287. 10.1111/j.1600-6143.2007.02062.x. [DOI] [PubMed] [Google Scholar]
- 11. Dumortier J, Streblow DN, Moses AV, Jacobs JM, Kreklywich CN, Camp D, Smith RD, Orloff SL, Nelson JA. 2008. Human cytomegalovirus secretomes contains factors that induce angiogenesis and wound healing. J. Virol. 82:6524–6535. 10.1128/JVI.00502-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Botto S, Streblow DN, DeFilippis V, White L, Kreklywich CN, Smith PP, Caposio P. 2011. IL-6 in human cytomegalovirus secretome promotes angiogenesis and survival of endothelial cells through the stimulation of survivin. Blood 117:352–361. 10.1182/blood-2010-06-291245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Engel P, Pérez-Carmona N, Albà MM, Robertson K, Ghazal P, Angulo A. 2011. Human cytomegalovirus UL7, a homologue of the SLAM-family receptor CD229, impairs cytokine production. Immunol. Cell Biol. 89:753–766. 10.1038/icb.2011.55. [DOI] [PubMed] [Google Scholar]
- 14. Dunn W, Chou C, Li H, Hai R, Patterson D, Stolc V, Zhu H, Liu F. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. U. S. A. 100:14223–14228. 10.1073/pnas.2334032100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sekulin K, Görzer I, Heiss-Czedik D, Puchhammer-Stöckl E. 2007. Analysis of the variability of CMV strains in the RL11D domain of the RL11 multigene family. Virus Genes 35:577–583. 10.1007/s11262-007-0158-0. [DOI] [PubMed] [Google Scholar]
- 16. Davison AJ, Dolan A, Akter P, Addison C, Dargan DJ, Alcendor DJ, McGeoch DJ, Hayward GS. 2003. The human cytomegalovirus genome revisited: comparison with the chimpanzee cytomegalovirus genome. J. Gen. Virol. 84 (Pt 1):7–28. 10.1099/vir.0.18606-0. [DOI] [PubMed] [Google Scholar]
- 17. Gu A, Tsark W, Holmes KV, Shively JE. 2009. Role of Ceacam1 in VEGF induced vasculogenesis of murine embryonic stem cell-derived embryoid bodies in 3D culture. Exp. Cell Res. 315:1668–1682. 10.1016/j.yexcr.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ergün S, Kilik N, Ziegeler G, Hansen A, Nollau P, Götze J, Wurmbach JH, Horst A, Weil J, Fernando M, Wagener C. 2000. CEA-related cell adhesion molecule 1: a potent angiogenic factor and a major effector of vascular endothelial growth factor. Mol. Cell 5:311–320. 10.1016/S1097-2765(00)80426-8. [DOI] [PubMed] [Google Scholar]
- 19. Chee MS, Bankier AT, Beck S, Bohni R, Brown CM, Cerny R, Horsnell T, Hutchison CA, III, Kouzarides T, Martignetti JA, et al. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125–169. [DOI] [PubMed] [Google Scholar]
- 20. Dolan A, Cunningham C, Hector RD, Hassan-Walker AF, Lee L, Addison C, Dargan DJ, McGeoch DJ, Gatherer D, Emery VC, Griffiths PD, Sinzger C, McSharry BP, Wilkinson GW, Davison AJ. 2004. Genetic content of wild-type human cytomegalovirus. J. Gen. Virol. 85:1301–1312. 10.1099/vir.0.79888-0. [DOI] [PubMed] [Google Scholar]
- 21. Shikhagaie M, Mercé-Maldonado E, Isern E, Muntasell A, Albà MM, López-Botet M, Hengel H, Angulo A. 2012. The HCMV-specific UL1 gene encodes a late phase glycoprotein incorporated in the virion envelope. J. Virol. 86:4091–4101. 10.1128/JVI.06291-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gray-Owen SD, Blumberg RS. 2006. CEACAM1: contact-dependent control of immunity. Nat. Rev. Immunol. 6:433–446. 10.1038/nri1864. [DOI] [PubMed] [Google Scholar]
- 23. Horst AK, Ito WD, Dabelstein J, Schumacher U, Sander H, Turbide C, Brümmer J, Meinertz T, Beauchemin N, Wagener C. 2006. Carcinoembryonic antigen-related cell adhesion molecule 1 modulates vascular remodeling in vitro and in vivo. J. Clin. Invest. 116:1596–1605. 10.1172/JCI24340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kuespert K, Pils S, Hauck CR. 2006. CEACAMs: their role in physiology and pathophysiology. Curr. Opin. Cell Biol. 18:565–571. 10.1016/j.ceb.2006.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Turbide C, Kunath T, Daniels E, Beauchemin N. 1997. Optimal ratios of biliary glycoprotein isoforms required for inhibition of colonic tumor cell growth. Cancer Res. 57:2781–2788. [PubMed] [Google Scholar]
- 26. Singer BB, Scheffrahn I, Obrink B. 2000. The tumor growth-inhibiting cell adhesion molecule CEACAM1 (C-CAM) is differently expressed in proliferating and quiescent epithelial cells and regulates cell proliferation. Cancer Res. 60:1236–1244. [PubMed] [Google Scholar]
- 27. Duncan SR, Paradis IL, Yousem SA, Similo SL, Grgurich WF, Williams PA, Dauber JH, Griffith BP. 1992. Sequelae of cytomegalovirus pulmonary infections in lung allograft recipients. Am. Rev. Respir. Dis. 146:1419–1425. 10.1164/ajrccm/146.6.1419. [DOI] [PubMed] [Google Scholar]
- 28. Humbert M, Delattre RM, Fattal S, Rain B, Cerrina J, Dartevelle P, Simonneau G, Duroux P, Galanaud P, Emilie D. 1993. In situ production of interleukin-6 within human lung allografts displaying rejection or cytomegalovirus pneumonia. Transplantation 56:623–627. 10.1097/00007890-199309000-00024. [DOI] [PubMed] [Google Scholar]
- 29. Simmons P, Kaushansky K, Torok-Storb B. 1990. Mechanisms of cytomegalovirus mediated myelosuppression: perturbation of stromal cell function versus direct infection of myeloid cells. Proc. Natl. Acad. Sci. U. S. A. 87:1386–1390. 10.1073/pnas.87.4.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Müller MM, Klaile E, Vorontsova O, Singer BB, Obrink B. 2009. Homophilic adhesion and CEACAME1-S regulate dimerization of CEACAME1-L and recruitment of SHP-2 and c-Src. J. Cell Biol. 187:569–581. 10.1083/jcb.200904150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kamimura D, Ishihara K, Hirano T. 2003. IL-6 signal transduction and its physiological roles: the signal orchestration model. Rev. Physiol. Biochem. Pharmacol. 149:1–38. 10.1007/s10254-003-0012-2. [DOI] [PubMed] [Google Scholar]
- 32. Heinrich PC, Behrmann I, Müller-Newen G, Schaper F, Graeve L. 1998. Interleukin-6-type cytokine signaling through the gp130/Jak/STAT pathway. Biochem. J. 334:297–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Roca H, Varsos ZS, Sud S, Craig MJ, Ying C, Pienta KJ. 2009. CCL2 and interleukin-6 promote survival of human CD11b+ peripheral blood mononuclear cells and induce M2-type macrophage polarization. J. Biol. Chem. 284:34342–34354. 10.1074/jbc.M109.042671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moreno A, Villar ML, Cámara C, Luque R, Cespón C, González-Porqué P, Roy G, López-Jiménez J, Bootello A, Santiago ER. 2001. Interleukin-6 dimers produced by endothelial cells inhibit apoptosis of B-chronic lymphocytic leukemia cells. Blood 97:242–249. 10.1182/blood.V97.1.242. [DOI] [PubMed] [Google Scholar]
- 35. Seino Y, Ikeda U, Ikeda M, Yamamoto K, Misawa Y, Hasegawa T, Kano S, Shimada K. 1994. Interleukin 6 gene transcripts are expressed in human atherosclerotic lesions. Cytokine 6:87–91. 10.1016/1043-4666(94)90013-2. [DOI] [PubMed] [Google Scholar]
- 36. Fischer P, Hilfiker-Kleiner D. 2007. Survival pathways in hypertrophy and heart failure: the gp130-STAT axis. Basic Res. Cardiol. 102:393–411. 10.1007/s00395-007-0674-z. [DOI] [PubMed] [Google Scholar]
- 37. Heinrich PC, Behrmann I, Haan S, Hermanns HM, Müller-Newen G, Schaper F. 2003. Principles of interleukin (IL)-6-type cytokine signaling and its regulation. Biochem. J. 374:1–20. 10.1042/BJ20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kurdi M, Booz GW. 2009. JAK redux: a second look at the regulation and role of JAKs in the heart. Am. J. Physiol. Heart Circ. Physiol. 297:H1545–H1556. 10.1152/ajpheart.00032.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lorenz K, Schmitt JP, Vidal M, Lohse MJ. 2009. Cardiac hypertrophy: targeting Raf/MEK/ERK1/2-signaling. Int. J. Biochem. Cell Biol. 41:2351–2355. 10.1016/j.biocel.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 40. Slinger E, Maussang D, Schreiber A, Siderius M, Rahbar A, Fraile-Ramos A, Lira SA, Söderberg-Nauclér C, Smit MJ. 2010. HCMV-encoded chemokine receptor US28 mediates proliferative signaling through the IL-6-STAT3 axis. Sci. Signal. 3:ra58. 10.1126/scisignal.2001180. [DOI] [PubMed] [Google Scholar]
- 41. Smith IL, Taskintuna I, Rahhal FM, Powell HC, Ai E, Mueller AJ, Spector SA, Freeman WR. 1998. Clinical failure of CMV retinitis with intravitreal cidofovir is associated with antiviral resistance. Arch. Ophthalmol. 116:178–185. [DOI] [PubMed] [Google Scholar]
- 42. Murphy E, Yu D, Grimwood J, Schmutz J, Dickson M, Jarvis MA, Hahn G, Nelson JA, Myers RM, Shenk TE. 2003. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc. Natl. Acad. Sci. U. S. A. 100:14976–14981. 10.1073/pnas.2136652100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Britt WJ, Jarvis M, Seo JY, Drummond D, Nelson J. 2004. Rapid genetic engineering of human cytomegalovirus using a lambda phage linear recombination system: demonstration that pp28 (UL99) is essential for production of infectious virus. J. Virol. 78:539–543. 10.1128/JVI.78.1.539-543.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33:e36. 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hsia DA, Mitra SK, Hauck CR, Streblow DN, Nelson JA, Ilic D, Huang S, Li E, Nemerow GR, Leng J, Spencer KS, Cheresh DA, Schlaepfer DD. 2003. Differential regulation of cell motility and invasion by FAK. J. Cell Biol. 160:753–767. 10.1083/jcb.200212114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Streblow DN, Soderberg-Naucler C, Vieira J, Smith P, Wakabayashi E, Ruchti F, Mattison K, Altschuler Y, Nelson JA. 1999. The human cytomegalovirus chemokine receptor US28 mediates vascular smooth muscle cell migration. Cell 99:511–520. 10.1016/S0092-8674(00)81539-1. [DOI] [PubMed] [Google Scholar]
- 47. Murphy EA, Streblow DN, Nelson JA, Stinski MF. 2000. The human cytomegalovirus IE86 protein can block cell cycle progression after inducing transition into the S phase of permissive cells. J. Virol. 74:7108–7118. 10.1128/JVI.74.15.7108-7118.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic representation of TR stopUL7HA and TR wtUL7HA generation with the Galk/Kan recombineering method. Download
Multistep growth curves of TR stopUL7HA and TR wtUL7HA recombinant virus. Fibroblasts were infected at an MOI of 0.1, and the extent of viral replication was measured at the indicated days postinfection by determining the titers corresponding to the infectivity of cell suspension supernatants using plaque assays. The data shown are the averages of results of three independent biological replicates for each time point ± standard errors of the means (error bars). Download
UL7 expression in infected cells. Fibroblasts were mock infected or infected with TR wtUL7HA and TR stopUL7HA at an MOI of 0.5. pUL7 and IE2 were detected by indirect immunofluorescence at 72 hpi using a mouse anti-HA antibody and a rabbit IE2 antibody and visualized with fluorescent red Alexa Fluor 594 and fluorescent green Alexa Fluor 488, respectively. The nuclei were counterstained with fluorescent blue DAPI (4′,6-diamidino-2-phenylindole). Download
UL7 is a late gene. Analysis of RNA was performed by Northern blotting. HCMV-infected HAEC in the presence or absence of foscarnet (phosphonoformate [PFA]) were rinsed twice with ice-cold phosphate-buffered saline (PBS) at the indicated times, and total cellular RNA was extracted using a NucleoSpin RNAII kit (Macherey-Nagel). Fractionation of total RNA was performed on a 1% agarose, 2.2 M formaldehyde gel followed by blotting onto a nitrocellulose membrane (Hybond C-extra; Amersham). Hybridization was carried out with 106 cpm/ml of single-stranded RNA (ssRNA)-radiolabeled probe corresponding to the UL7 TR gene cloned in pGEM-T Easy vector (Promega). In vitro transcription with the Sp6 RNA polymerase (Ambion) was used to generate antisense RNA transcripts. Download
pUL7 is secreted in transfected cells. HEK293 cells were transfected with pCDNA3 (ctl) or pCDNA3UL7HA. After 48 h, cells (left panel) and supernatants (right panel) were harvested, treated with peptide N-glycosidase F (PNGase F), and then analyzed by Western blotting with antibodies specific for HA, UL7, and actin. In the right panel, the arrowhead indicates the high-molecular-weight pUL7 form. Download
pUL7 binds to the surface of HAEC. Data represent the results of flow cytometry analysis of pUL7Fc (blue line) and Fc (black line) interactions with HAEC (upper panel). Expression of hCD31 is shown in density plots, with rectangles indicating gates for cells analyzed for pUL7Fc and Fc binding (lower panel). HAEC (2 × 105) were incubated with 2 µg of pUL7Fc or Fc in blocking solution (5% mouse serum–2 mM EDTA–PBS) and stained with UL7 antibody followed by Alexa Fluor 594-labeled goat anti-rabbit. The HAEC population was confirmed using an antibody directed to CD31 (PB-conjugated anti-human CD31, clone WM59; BioLegend). All the steps were performed at 4°C. After surface staining with conjugated antibodies, polychromatic flow cytometric analysis was performed using an LSR II system (BD Biosciences), and data were analyzed by using FlowJo software (version 9.1; Tree Star, Inc.). Download
Oligonucleotides used for BAC mutagenesis and cloning.