ABSTRACT
The rodent arenavirus glycoprotein complex encodes a stable signal peptide (SSP) that is an essential structural component of mature virions. The SSP, GP1, and GP2 subunits of the trimeric glycoprotein complex noncovalently interact to stud the surface of virions and initiate arenavirus infectivity. Nascent glycoprotein production undergoes two proteolytic cleavage events: first within the endoplasmic reticulum (ER) to cleave SSP from the remaining precursor GP1/2 (glycoprotein complex [GPC]) glycoprotein and second within the Golgi stacks by the cellular SKI-1/S1P for GP1/2 processing to yield GP1 and GP2 subunits. Cleaved SSP is not degraded but retained as an essential glycoprotein subunit. Here, we defined functions of the 58-amino-acid lymphocytic choriomeningitis virus (LCMV) SSP in regard to glycoprotein complex processing and maturation. Using molecular biology techniques, confocal microscopy, and flow cytometry, we detected SSP at the plasma membrane of transfected cells. Further, we identified a sorting signal (FLLL) near the carboxyl terminus of SSP that is required for glycoprotein maturation and trafficking. In the absence of SSP, the glycoprotein accumulated within the ER and was unable to undergo processing by SKI-1/S1P. Mutation of this highly conserved FLLL motif showed impaired glycoprotein processing and secretory pathway trafficking, as well as defective surface expression and pH-dependent membrane fusion. Immunoprecipitation of SSP confirmed an interaction between the signal peptide and the GP2 subunit; however, mutations within this FLLL motif disrupted the association of the GP1 subunit with the remaining glycoprotein complex.
IMPORTANCE
Several members of the Arenaviridae family are neglected human pathogens capable of causing illness ranging from a nondescript flu-like syndrome to fulminant hemorrhagic fever. Infections by arenaviruses are mediated by attachment of the virus glycoprotein to receptors on host cells and virion internalization by fusion within an acidified endosome. SSP plays a critical role in the fusion of the virus with the host cell membrane. Within infected cells, the retained glycoprotein SSP plays a neglected yet essential role in glycoprotein biosynthesis. Without this 6-kDa polypeptide, the glycoprotein precursor is retained within the endoplasmic reticulum, and trafficking to the plasma membrane where SSP, GP1, and GP2 localize for glycoprotein assembly into infectious virions is inhibited. To investigate SSP contributions to glycoprotein maturation and function, we created an SSP-tagged glycoprotein to directly detect and manipulate this subunit. This resource will aid future studies to identify host factors that mediate glycoprotein maturation.
INTRODUCTION
Arenaviruses asymptomatically infect geographically specific rodent species and are capable of causing severe disease in humans (1). Human infections most frequently occur via close contact with rodent excreta and several arenaviruses, such as the Old World (OW) Lassa and Lujo viruses, as well as the New World (NW) arenaviruses Junín, Machupo, Guanarito, and Sabia, which are causative agents of hemorrhagic fever (2). Lymphocytic choriomeningitis virus (LCMV) is the prototype and most researched arenavirus. LCMV is capable of causing illness ranging from mild flu-like symptoms to aseptic meningitis. Additionally, LCMV has contributed to multiorgan failure and death in organ transplant recipients from undiagnosed, infected organs (3–6). The principal rodent host for LCMV is the ubiquitous Mus musculus, which allows for the near global distribution of LCMV (7, 8).
Arenaviruses are simple in their design, consisting of two segments of single-stranded RNA, the large (L) and small (s) segment. These segments each encode two nonoverlapping open reading frames. The large genome segment encodes the RNA-dependent RNA polymerase (L protein; ~220 kDa) and the matrix protein (Z protein; 11 kDa). The small genome segment encodes the glycoprotein complex (GPC; 75 kDa) and the nucleoprotein (NP; 63 kDa). The Z protein and the GP complex are expressed in a pseudopositive-sense orientation, while the L protein and the NP are expressed in a negative-sense orientation, giving the arenaviruses an ambisense gene expression strategy (9).
The arenavirus glycoprotein complex is expressed as a polyprotein precursor that undergoes two cleavage events. First, the stable signal peptide (SSP) is cleaved by cellular signal peptidase within the endoplasmic reticulum (ER). This 58-amino-acid protein is retained as a stable subunit and is critical for downstream, mature glycoprotein complex formation (10–12). Second, SSP traffics with immature GP1/2 precursor (amino acids 59 to 498), which undergoes cleavage by the SKI-1/S1P enzyme within the Golgi stacks to yield the GP1 and GP2 subunits (13, 14). The three independent subunits, consisting of the SSP, GP1, and GP2, traffic to the plasma membrane for viral assembly and egress (15–18). These three subunits interact noncovalently at the virion surface to bind with host cell receptors to initiate infectivity. The GP1 subunit interacts with the known cellular receptors: α-dystroglycan (α-DG) for the OW arenaviruses and transferrin receptor 1 (TfR1) for the NW viruses (19, 20). Recent studies using next-generation technologies provided evidence for the involvement of additional host factors for both Junín virus and Lassa virus entry (21, 22). The acidification of endocytic vesicles induces irreversible dissociation of the GP1 subunit from the remaining GP2 and SSP subunits, followed by a conformational change within the GP2 subunit to reveal the fusion domains which, in concert with SSP, induce fusion events between the virion and the host membrane to deliver the viral core to the newly infected cell cytoplasm (23, 24).
Generally, signal peptides are short, extended amino-terminal sequences found within secreted or transmembrane proteins whose function is to direct nascent proteins to the ER for maturation via the secretory pathway (25). Few viruses possess signal sequences that serve additional functions. The Ebola virus signal peptide was shown to modulate the extent of GP glycosylation, thereby moderating the extent of glycoprotein interaction with DC-SIGN/R (26). Additionally, the HIV-1 Env protein signal peptide is thought to regulate glycoprotein folding and maturation (27). In a nonviral model, the prion protein signal peptide interacts with the transmembrane domain for correct protein expression and membrane orientation (28).
Our current studies focus on the importance of SSP in terms of nascent glycoprotein complex expression, maturation, and function. Using Western blot analysis, confocal microscopy, and flow cytometry, we show that LCMV GP lacking the native SSP does not exit the endoplasmic reticulum or mature through the secretory pathway. We detect SSP at the plasma membrane, and mutations within SSP affect trafficking of the entire glycoprotein complex. Further, we define a conserved FLLL motif within the LCMV SSP that is the driving force for full-length and functional maturation and glycoprotein subunit association.
RESULTS
SSP mediates glycoprotein precursor processing.
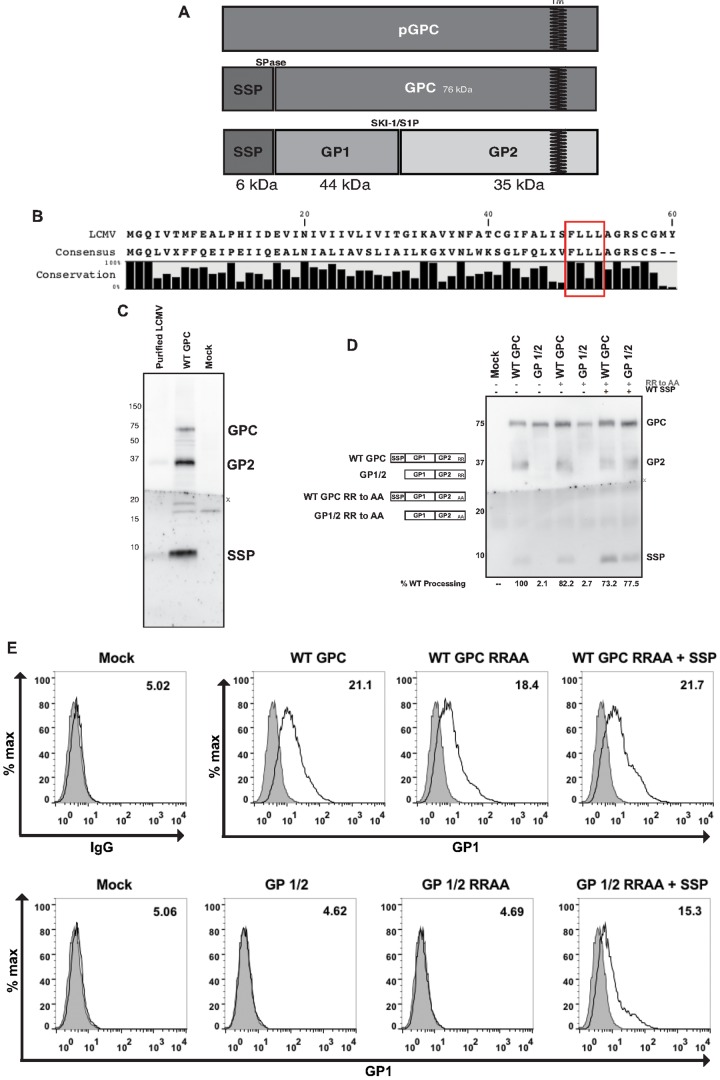
The glycoprotein complex is expressed as a polyprotein that undergoes two cleavage events (Fig. 1A). SSP retains a high degree of sequence conservation across Old World and New World signal peptides (Fig. 1B; see also Fig. S1 in the supplemental material) (29). This consensus alignment excludes the boa constrictor-derived arenaviruses, since this group of distantly related viruses expresses a filovirus-like glycoprotein (30). SSP is a key component of the glycoprotein complex that is not recycled within the endoplasmic reticulum but is retained as an essential subunit within purified virions (Fig. 1C). Previous studies have demonstrated a role of basic amino acids within transmembrane protein cytoplasmic domains for the intracellular transport and processing of proproteins into their mature forms (12, 31–33). We introduced alanine point mutations in the final two residues within the LCMV GP2 subunit and tested the ability of these constructs to produce a cleaved GP2 subunit, since GPC cleavage into GP1 and GP2 precedes membrane transport (29). Wild-type (WT) GPC with the RR-to-AA mutation, along with the in trans expression of wild-type SSP, resulted in the accumulation of the precursor GPC, which moderately affected levels of processed GP2, as quantified by densitometry analysis (Fig. 1D). The GP1/2 RR-to-AA mutant lacking the native SSP did not produce the cleaved GP2 subunit. We used flow cytometry to measure the surface expression of the glycoprotein using live cells stained with a conformation-specific GP1 antibody (Fig. 1E). Mutations within the WT GPC did not alter the amount of surface GP1 expression. The GP1/2 and the GP1/2 RRAA samples failed to produce a GP1 signal above background levels. Cotransfection of wild-type SSP with both the WT GPC RR-to-AA and with the GP1/2 RR-to-AA protein restored both GP2 processing and surface expression of GP1, suggesting the dibasic motif within the LCMV glycoprotein is not the driving force for secretory pathway trafficking, but rather this function resides within SSP.
FIG 1 .
The conserved signal peptide mediates GP complex processing. (A) Schematic representation of the arenavirus glycoprotein complex, including the two cellular enzymes required for glycoprotein processing. Note the “TM” indicates the transmembrane domain. (B) Conservation of the glycoprotein signal peptide as compiled by alignment of all currently known rodent arenaviruses sequences. The consensus SSP sequence is compared to the lymphocytic choriomeningitis virus (LCMV) SSP sequence. Note the conserved FLLL motif near the carboxy terminus. (C) Western blot detection of the signal peptide from purified virus and from LCMV Armstrong strain GPC-transected cell lysates. Note the GPC band near 75 kDa is absent in purified virions. (D) Transfected BHK-21 cell lysates were probed with antibodies directed against the GP2 (83.6) and SSP (SP7) subunits. The extent of GP processing was quantified using ImageJ, and values were normalized to wild-type GPC. The “X” indicates the nitrocellulose membrane was sliced for antibody incubation, SSP (SP7) or GP2 (83.6), and then reassembled for chemiluminescence processing and image acquisition. (E) Detection of the GP1 subunit by flow cytometry using live, transfected, nonpermeabilized HEK 293T cells. Cells were incubated with the GP1-specific 2.11-10 antibody directly conjugated to Alexa Fluor 488. Gray shading indicates the mock IgG control population. The values located in the upper right corner of each histogram represent the mean fluorescence intensity (MFI) of GP1 expression. Shown are representative data from one of three independent experiments.
SSP C-terminal region is essential for GPC processing into GP1 and GP2 subunits.
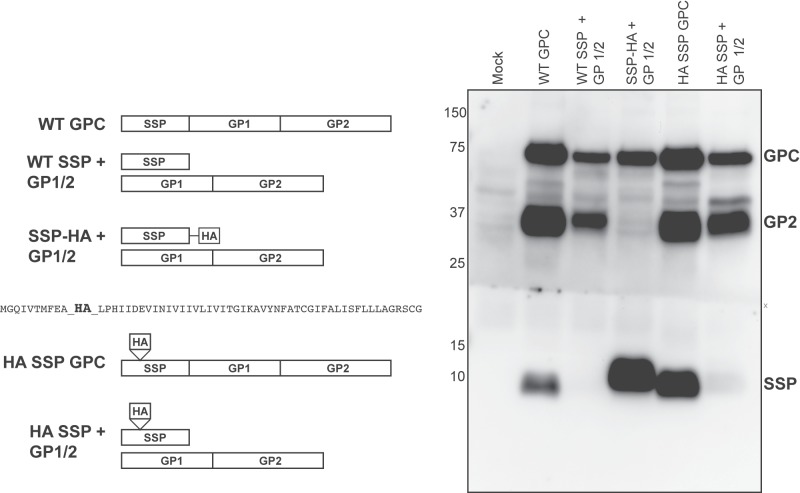
A native antibody directed against SSP that is applicable for immunofluorescence microscopy does not exist, as the SP7 antibody detects only SSP under denatured and reduced conditions (18, 34). We used a C-terminal hemagglutinin-tagged SSP (SSP-HA) plasmid construct to assess the significance of the SSP for GPC maturation. This construct was previously used to define SSP membrane topology, SSP’s role in pH-dependent fusion, and interactions between SSP and the Z matrix protein (35–38). Western blot analysis of HEK 293T cell lysates cotransfected with SSP-HA and the glycoprotein precursor lacking its cognate SSP (GP1/2) resulted in expression of SSP and the GPC but lacked the processed GP2 (Fig. 2). Digestion of transfected cell lysates with the peptide--N-glycosidase F (PNGase F) enzyme to rid N-linked glycans confirmed the GPC was glycosylated (see Fig. S1 in the supplemental material). Detection of the GP2 subunit was rescued upon cotransfection with wild-type SSP, suggesting the HA epitope interfered with SSP-GPC interactions needed for appropriate precursor processing. Since the presence of the epitope at the C terminus interfered with proper glycoprotein complex expression, we inserted the HA epitope in the N-terminal SSP region of the entire glycoprotein open reading frame, downstream of the myristoylation motif (HA SSP GPC). This internally tagged glycoprotein allowed for in cis detection of and experimentation with SSP relative to the remaining glycoprotein subunits. In trans expression of SSP-HA and GP1/2 allowed for precursor cleavage and the detection of the processed GP2 subunit, although this SSP, along with the wild-type SSP, was more rapidly degraded when provided on separate plasmids (Fig. 2).
FIG 2 .
Carboxy terminus of SSP is key for glycoprotein processing. Western blot analysis of transfected HEK 293T cell lysates to detect SSP (SP7) or the GP2 (83.6) subunits. The LCMV SSP amino acid sequence, including the location of the inserted HA epitope, is shown. The “X” indicates where the membrane was sliced for antibody incubation and then reassembled for chemiluminescence processing and image acquisition.
Blocking the SSP C-terminal region interferes with plasma membrane trafficking.
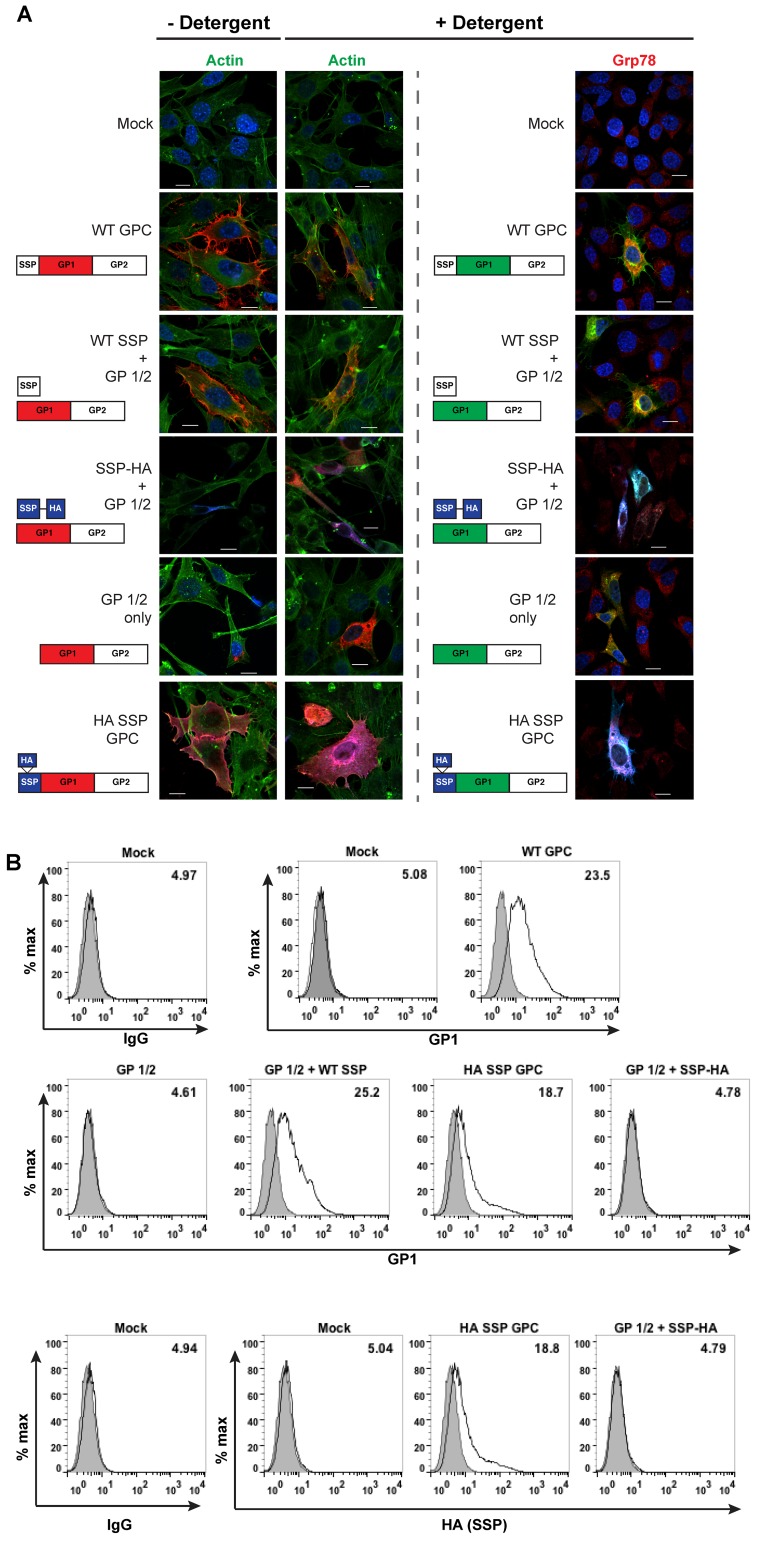
The signal peptide is required for GPC cleavage, a prerequisite for glycoprotein complex localization at the plasma membrane. We compared glycoprotein expression to phalloidin-defined cell membrane boundaries in the presence or absence of detergent permeabilization using confocal microscopy and a confirmation-specific GP1 antibody (39). In addition, we used the ER chaperone protein Grp78 to determine intracellular glycoprotein complex localization. Wild-type glycoprotein produced a plasma membrane signal with pseudopod-like membrane exvaginations with moderate intracellular Grp8 staining (Fig. 3A). A similar phenotype was produced with in trans coexpression of wild-type SSP and GP1/2 and with HA SSP GPC. Further, the HA SSP GPC allowed for SSP detection at the plasma membrane both with and without detergent treatment. In the absence of detergent permeabilization, the GP1/2 precursor lacking its SSP resulted in the lack of plasma membrane expression and a “leaky” ER staining, which was confirmed by Grp78 staining. Similarly, cotransfection of the SSP-HA and GP1/2 failed to yield plasma membrane detection of the GP complex. Rather, the entire complex was retained within the ER. Taking into account the defect in glycoprotein processing observed in transfected cell lysates (Fig. 2), blocking the SSP C-terminus interferes with the ability of the GP complex to traffic from the ER to the Golgi for processing by the cellular SKI-1/S1P enzyme to produce mature GP subunits (13, 14).
FIG 3 .
SSP is required for plasma membrane trafficking. (A) DBT cells were fixed 48 h posttransfection and either left untreated or treated with Triton X-100. Cells were stained using phalloidin-FITC to define the plasma membrane or with the Grp78 antibody to mark the endoplasmic reticulum. SSP was detected using the HA and the Alexa Fluor 405 antibodies. Cells were further incubated with the GP1 antibody (2.11-10) directly conjugated to Alexa Fluor 594. Scale bars indicate 20 µm. (B) Surface expression of live, unpermeabilized HEK 293T cells stained for GP1, SSP, or mock IgG and analyzed by flow cytometry. The gray shaded population indicates the mock IgG population, and the numbers in the upper right corner of each plot represent the MFI for GP1 or SSP expression. Representative data from one of three independent experiments.
We assessed plasma membrane trafficking of the glycoprotein complex on live, transfected cells using flow cytometry (Fig. 3B). The WT GPC, GP1/2 plus WT SSP, and HA SSP GPC samples supported the surface detection of the GP1 subunit. The GP1/2 and the GP1/2 plus SSP-HA samples resulted in mock levels of fluorescent intensity. Using the HA antibody to indirectly detect SSP, we detected wild-type GP1 levels of the HA signal with the HA SSP GPC sample, whereas the GP1/2 plus SSP-HA sample resulted in mock HA expression levels. Detection of the HA signal with the HA SSP GPC sample on live cells confirmed that our confocal microscopy results for SSP detection were not an artifact result from the fixation technique.
The conserved FLLL motif is key for glycoprotein expression.
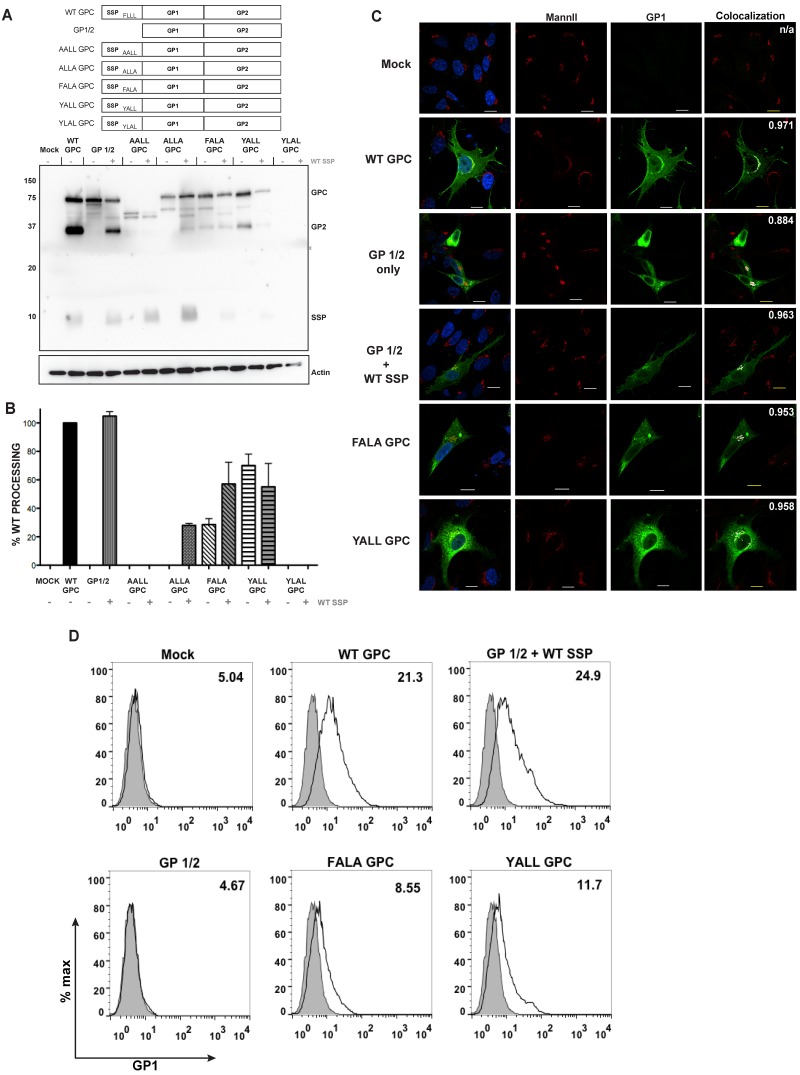
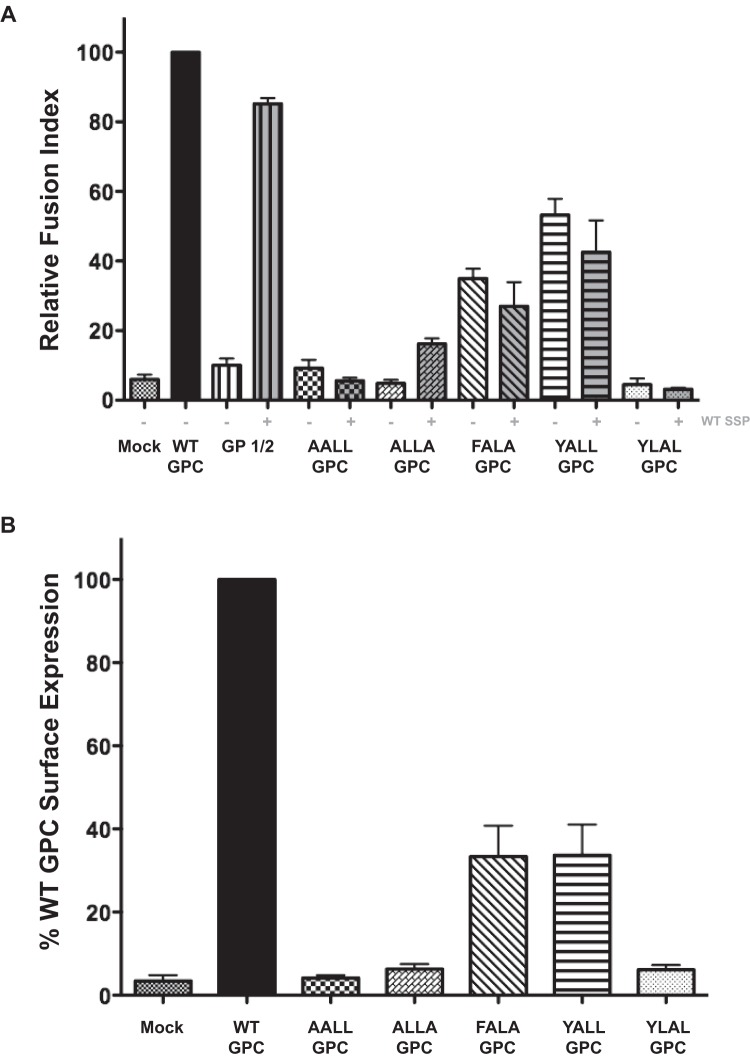
We introduced a panel of double point mutations within this FLLL motif in order to determine whether any one amino acid was key for a processed and mature GP complex. The AALL GPC was defective in its ability to allow posttranslational modifications and revealed a dominant negative maturation phenotype, since the addition of wild-type SSP did not rescue glycosylated glycoprotein expression or processing (Fig. 4A; see Fig. S2 in the supplemental material). The ALLA GPC supported precursor GP posttranslational modifications but was defective in producing a processed GP2 subunit. The ALLA GPC produced a cleavage GP2 subunit with the in trans addition of wild-type SSP (Fig. 4B). The FALA GPC retaining the conserved phenylalanine encoded by all known rodent arenaviruses and resulted in a reduced level of GP2 cleavage from the precursor GPC, while levels of GP2 were increased with the cotransfection of the wild-type SSP. Densitometry analysis of the FALA GPC and wild-type SSP cotransfection, from three independent replicates, resulted in an overall increase in the amount of GPC processed to yield the GP2 subunit, while the representative Western blot in Fig. 4A does not highlight this slight rescue in GP2 processing (Fig. 4B). The YALL GPC resulted in the highest percentage of processed glycoprotein of all SSP mutants we analyzed, though not at the level of the wild-type GP2 cleavage. Interestingly, the cotransfection of the wild-type SSP with YALL GPC resulted in the reduced expression of the total glycoprotein complex. The final variant we examined, YLAL GPC, was toxic to transfected cells and consistently yielded low levels of detectable glycoprotein. Overall, double point mutations within this conserved SSP motif indicate that this entire motif, and not one residue in particular, plays a dominant role in downstream processes that mediate processing of the precursor into mature glycoprotein subunits.
FIG 4 .
Mutations within the conserved FLLL motif affect GPC processing. (A) HEK 293T cell lysates were transfected with the indicated glycoprotein constructs and were probed with antibodies to detect the cleaved GP2 and SSP subunits. (B) Densitometry quantification of the processed GP2 subunit levels from the GPC precursor. All samples are normalized to the wild-type GPC, and error bars represent the standard errors of the means (SEM) from three independent experiments. (C) Confocal microscopy of DBT cell expression GP constructs and detection of the Golgi protein mannosidase II (MannII). Colocalization is shown, with the indicated areas depicted in white. Also included in the upper right corner of each image is the Manders’ overlay coefficient, quantified using ImageJ. (D) Surface expression of the GP1 subunit using flow cytometry. Gray shading indicates the mock IgG population. Values within each histogram plot represent the MFI of GP1 expression. Data represent results from one of three independent experiments.
Mutations within the FLLL motif inhibit glycoprotein trafficking.
The FLLL glycoprotein mutants that allowed GPC processing were examined for their ability to traffic to the plasma membrane using confocal microscopy. We used colocalization with the Golgi protein mannosidase II (MannII) as an indicator for glycoprotein exit from the ER. Wild-type GPC and the reconstituted wild-type GPC (SSP and GP1/2) allowed for GP1 localization with MannII as well as an extensive, defined plasma membrane (Fig. 4C). The GP1/2 protein, lacking the native SSP, produced less localization with the Golgi marker, showed extensive cytoplasmic accumulation of the precursor protein akin to retention within the ER, and failed to express surface GP1 above mock transfection levels via flow cytometry (Fig. 4D). The FALA GPC and the YALL GPC produced extensive cytoplasmic staining, indicating ER accumulation, as well as Golgi colocalization (Fig. 4C). While the FALA GPC and the YALL GPC generated cleaved GP2 and Golgi marker colocalization, these mutant glycoproteins produced a greater-than-2-fold reduction in plasma membrane localization compared to that of the WT GPC and the GP1/2 and WT SSP levels (Fig. 4D). The microscopy results strengthen the importance of an intact FLLL motif so SSP may orchestrate glycoprotein maturation processes downstream of targeting the nascent protein to the secretory pathway.
Mutations within the FLLL motif affect glycoprotein fusion activity.
In order to assess if the FLLL GPC mutants produced a functional glycoprotein complex, we performed a pH-dependent fusion assay (29, 40). Transfected DBT cells were subjected to acidic growth medium to induce syncytium formation via conformational changes within the glycoprotein (41). The mutants that allowed for GP2 detection, FALA GPC and YALL GPC, produced various levels of fusion activity. FALA GPC resulted in one-third the fusion activity of that of the wild-type glycoprotein. YALL GPC allowed for roughly half the level of fusion activity compared to that of the wild-type GPC (Fig. 5). Coexpression of the wild-type SSP resulted in a decrease in fusion activity for both FALA GPC and YALL GPC, whereas cotransfection slightly increased ALLA GPC fusion activity, although this amount was not appreciably higher than background levels.
FIG 5 .
FLLL motif acts in concert for acidic fusion activity. (A) Quantification of pH-dependent fusion activity using transfected DBT cells, as previously described (29, 40). (B) Flow cytometry quantification of GP1 expression using live HEK 293T cells. All values are normalized to WT GPC expression. Error bars represent the SEM of GP1 expression from three independent experiments.
We used flow cytometry to measure surface expression of each FLLL mutant glycoprotein (Fig. 5B). Only the FALA GPC and the YALL GPC mutants resulted in glycoprotein detection above mock transfection levels, although these two mutants had more than a 50% reduction in GP1 signal compared to that of WT GPC, indicating that fusion activity and plasma membrane trafficking are both affected by mutations within SSP.
SSP is vital for glycoprotein complex association.
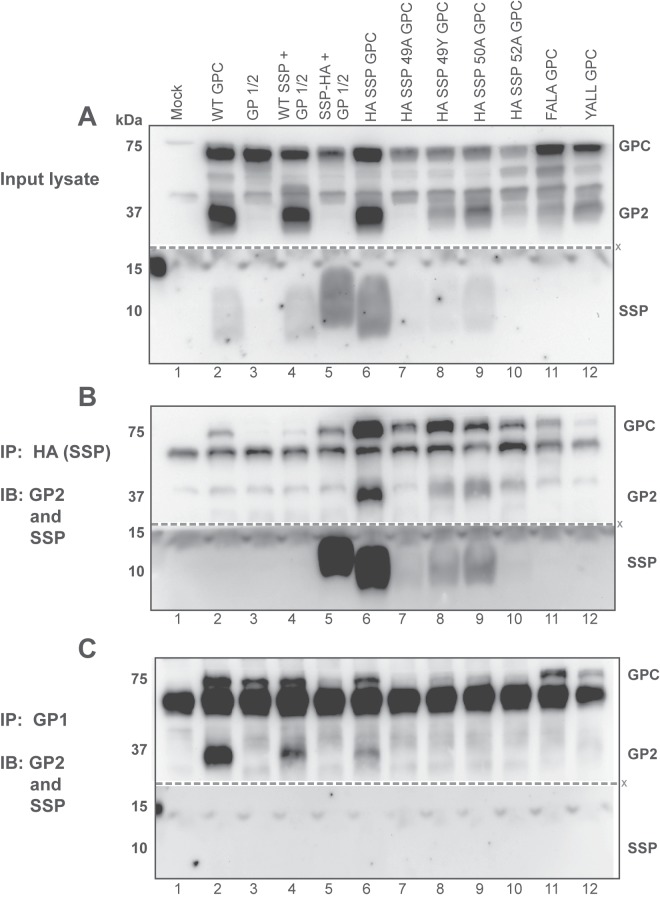
We performed immunoprecipitation (IP) assays to investigate the interaction between SSP and the rest of the GP complex, since mutations within SSP result in fusion activity deficiencies. Previous studies using GP2-targeted IP showed an interaction between the GP2 subunit and SSP (10, 34). We subjected a panel of point mutations within the SSP FLLL motif, as well as within the FALA GPC and YALL GPC double point mutants, to immunoprecipitation assays using both the HA and the confirmation-specific, neutralizing GP1 subunit 2.11-15 antibodies (Fig. 6). Cells transfected with the wild-type GPC, the cotransfected SSP and GP1/2, and the HA SSP GPC resulted in properly processed GP2 subunit detection (Fig. 6A, lanes 2, 4, and 6). The GP1/2 missing its native SSP, as well as the cotransfected SSP-HA plus GP1/2, did not result in processed GP2 (Fig. 6A, lanes 3 and 5). In order to directly assess SSP interactions by IP, we introduced FLLL motif point mutations within the context of the HA SSP GPC open reading frame. The HA SSP GPC 49A point mutant did not result in processed GP2 (Fig. 6A, lane 7). The HA SSP GPC 49Y, 50A, and 52A point mutants, as well as FALA GPC and YALL GPC double point mutants, all allowed GPC processing and GP2 detection (Fig. 6A, lanes 8 to 12).
FIG 6 .
Mutations within the FLLL motif affect inter-GP subunit association. (A) Western blot detection of HEK 293T cell lysate expression using the panel of glycoprotein variants. Nitrocellulose membranes were immunoblotted (IB) with antibodies directed against the GP2 (anti-83.6) and SSP (anti-SP7) subunits. Bound proteins from SSP immunoprecipitation (IP) using anti-HA agarose beads (B) and bound protein from GP1 immunoprecipitation (C) were separated using 12% SDS-PAGE. All membranes were probed for the GP2 (83.6) and SSP (SP7) subunits. For each blot, the “X” denotes where the nitrocellulose membranes were cut for antibody incubation prior to membrane reassembly for image acquisition.
Immunoprecipitation of SSP by using HA agarose beads retained its association with the GP2 subunit. The wild-type HA SSP GPC immunoprecipitation results, containing only the inserted HA epitope with no point mutations, revealed the interaction with the GP2 subunit (Fig. 6B, lane 6). The HA SSP GPC 49A mutant did not allow for glycoprotein processing; thus, there was no detectable GP2 (Fig. 6B, lane 7). HA SSP GPC 49Y and HA SSP GPC 50A point mutants also resulted in varied GP2 subunit immunoprecipitation (Fig. 6B, lanes 8 and 9). The HA SSP GPC 52A sample, as well as FALA GPC and YALL GPC, did not reveal any GP2 pulldown product (Fig. 6B, lanes 10 to 12). FALA GPC and YALL GPC did not contain HA epitope expression for successful immunoprecipitation and served as additional agarose bead controls.
The GP1 subunit was immunoprecipitated using the 2.11-15 conformation-specific monoclonal antibody. The only samples to precipitate the cleaved GP2 subunit were the WT GPC, the in trans reconstituted GPC, and the HA SSP GPC (Fig. 6C, lanes 2, 4, and 6). None of the SSP single or double point mutant glycoprotein constructs allowed for glycoprotein subunit associations. In no sample did the GP1 immunoprecipitate SSP (Fig. 6C). FALA GPC and YALL GPC samples did not result in GP2 subunit detection, indicating that mutations within the SSP affect proper glycoprotein complex association. For both the HA and the GP1 IP assays, we did not interpret GPC pulldown results as positive IP interactions, since the antibodies are capable of interacting with the nascent, full-length, uncleaved precursor glycoprotein.
DISCUSSION
Given that the arenavirus signal peptide is an essential component of the glycoprotein complex, our goal was to dissect this subunit’s role in glycoprotein maturation. SSP was not degraded upon translocation into the secretory pathway but rather was retained and trafficked with the rest of the glycoprotein complex to the plasma membrane for viral packaging and egress. The requirement for SSP packaging within virions demonstrates two critical roles for this 58-amino-acid peptide. First, SSP acts as the leader protein for ER exit and GPC processing within the Golgi stacks, as deletion of either hydrophobic domain inhibits downstream GPC processing (29, 34). Second, SSP is an important structural component within virions alongside the spike GP1 and transmembrane GP2 subunits.
Using truncated glycoprotein constructs, previous authors reported that SSP is dispensable for GP ectodomain expression (42). Our current results contrast previous findings, as we used, or reconstituted, full-length glycoprotein expression vectors and not truncated plasmid constructs. The presence of the signal peptide was required for proper, full-length glycoprotein ectodomain processing (43). Our results further differed from previous research using both Old World and New World arenavirus glycoproteins in that the LCMV Armstrong strain (Arm-4) glycoprotein does not utilize basic residues within the GP2 cytoplasmic domain as the driving force for ER exit (12, 33). Our mutational analysis of the GP1/2 subunit containing a mutated basic motif at the ultimate portion of the GP2 cytoplasmic domain did not result in any cleaved GP2 subunit, indicating that this glycoprotein lacking its SSP was not capable of exiting the ER for proteolytic processing. When wild-type SSP was coexpressed with the mutated GP1/2, we detected the processed GP2 subunit. Additionally, our results contrasted Lassa glycoprotein studies which indicated the GP2 cytoplasmic domain played a role in GP maturation cleavage, although the authors did not use a full-length Lassa glycoprotein in their characterizations (44).
The SSP and the GP2 cytoplasmic domains have been shown to play critical roles in maturation. Recent studies using reverse genetics permitted infectious, chimeric Lassa/Junín-Candid1 virus production when the glycoprotein SSP and GP2 cytoplasmic domains were homologous and not of mismatched viral origin (45). This observation, together with our immunoprecipitation experiments, highlights the critical interactions between the SSP and GP2 subunits. Further, our confocal microscopy and flow cytometry analyses did not result in GP1/2 localization at the plasma membrane. Since we used only LCMV glycoprotein constructs for our experiments, it is possible the differing results we observed are due to slight differences between different arenavirus protein expression mechanisms.
Signal peptides, although diverse in their amino acid compositions, share a similar organization (25). Each signal peptide has a variable-length amino terminus, a hydrophobic center region, and a carboxyl terminus, including the signal peptidase cleavage site. In order to directly analyze the signal peptide, we took advantage of the variable amino-terminal region by inserting an HA epitope within this SSP region while in the context of the full-length glycoprotein open reading frame (HA SSP GPC). HA SSP GPC was an expansion of a tool previously used for Junín SSP membrane topology characterization, though in those studies SSP was expressed in trans with the remaining portion of the glycoprotein (37). Insertion of the HA epitope did not alter glycoprotein trafficking, as SKI-1/S1P-deficient cells were unable to cleave GPC into mature GP1 and GP2 unless functional SKI-1/S1P was supplemented in trans (data not shown). This HA SSP GPC behaved at wild-type levels in terms of its expression, GP2 cleavage, and plasma membrane localization. Additionally, this HA SSP GPC allowed us to target experiments, both biochemical and with microscopy, directly focusing on SSP while eliminating the technicalities of cotransfection.
We and others have shown the importance of the first signal peptide hydrophobic domain for precursor glycoprotein cleavage and pH-dependent membrane fusion (29, 34, 46). We were able to detect the HA epitope at the cell surface by flow cytometry using live, nonpermeabilized cells, indicating that the amino-terminal portion of SSP upstream of the first hydrophobic domain was presented on the exterior surface of the plasma membrane. This suggests SSP may exhibit more than one membrane orientation, and the lack of fusion activity resulting from the inserted epitope may be interfering with late-state SSP orientation to produce a fully assembled glycoprotein complex (34, 35, 47). This observation does not conflict with the proposed options for signal peptide hairpin-like orientation within the plasma membrane but merely provides for an alternative orientation during glycoprotein complex maturation and assembly (18, 35, 37).
In order to determine whether the carboxy-terminal region of SSP is genuinely mediating the intracellular trafficking of the glycoprotein complex and not a function of the inserted, foreign epitope, we focused on a highly conserved hydrophobic motif (FLLL) upstream of the signal peptidase recognition signal in the context of the full-length glycoprotein open reading frame. As stated previously, SSP is the most heavily conserved protein throughout the Arenaviridae family, although it is partnered with the GP2 subunit for phylogenetic analyses (7). Results from the expression of the FLLL GPC mutants indicate that each residue within this motif plays an equal role in glycoprotein maturation cleavage. Additionally, the lack of observed fusion activity observed by the HA SSP GPC indicates a defect in one of the final stages of glycoprotein complex assembly, an observation previously noted using internally tagged Junín SSP cotransfection assays (37). This lack of fusion activity serves as a limitation for the potential use of the HA SSP GPC in recombinant virus production.
Previous studies reported that single point mutations within this FLLL region, specifically the phenylalanine at position 49, affect both pH-dependent membrane fusion and glycoprotein infectivity (29, 37). Additionally, SSP residues at positions 5 and 50, the latter of which resides within this conserved FLLL motif, have been implicated to play roles in LCMV pathogenicity and virus propagation (48). Our microscopy results from the FALA GPC and YALL GPC mutants revealed defects in the progression through the secretory pathway. Levels of processed GP2 from the FALA GPC increased with the addition of supplemented wild-type SSP, although the presence of additional SSP failed to enhance the fusion activity of the glycoprotein. The FALA GPC and the YALL GPC, though producing the cleaved GP2 subunit, both resulted in significant ER and Golgi staining, as well as a greater than 50% reduction in both fusion activity and GP1 surface expression, indicating a defect in post-Golgi intracellular transport. The reduction in membrane localization exhibited by these mutants at 48 h posttransfection indicates an impaired maturation process, as the intracellular staining patterns observed were merely nascent glycoprotein at intermediate maturation stages, as transfection efficiencies were comparable under all conditions. Defects observed by mutating the FLLL motif indicate that this SSP functions as a sorting signal for secretory pathway trafficking.
In regard to the AALL GPC and the YLAL GPC, these two mutants lacked the ability to produce a glycosylated precursor GPC. Results from the GP1/2 expression, where the glycoprotein completely lacks its native SSP, demonstrated SSP was not required for the posttranslational N-linked glycan modification. AALL GPC and YLAL GPC produced a defective, nonfunctional protein whereby the presence of in trans wild-type SSP was unable to rescue downstream functions for cleavage and transport. These two GPC variants, especially the YLAL GPC, were cytotoxic, likely due to the induction of ER stress responses, as these two mutant GPCs lacked the ability to traffic to the Golgi stacks for precursor processing. In trans SSP expression was able to moderately rescue the ALLA GPC processing, since this mutant GP did produce a glycosylated precursor, thus allowing SSP to provide some chaperone function to an otherwise able (glycosylated) precursor. Based on our fusion assay results, the levels of processed GP2 from the cotransfection of wild-type SSP with the ALLA GPC were not enough to rescue a significant amount of functional glycoprotein. For the FALA GPC and the YALL GPC mutants, the decrease in fusion activity with the in trans wild-type SSP indicated a defect in SSP-mediated transport. This fusion activity defect was likely mediated by ER stress due to unequal ratios and overabundance of SSP relative to the rest of the glycoprotein complex.
Functional insertion of the HA epitope within the signal peptide region of the LCMV glycoprotein was further confirmed by the ability of the signal peptide to retain its interaction with the GP2 subunit, as determined by immunoprecipitation studies. We confirmed this SSP-GP2 interaction, as Schrempf et al. detected the signal peptide via GP2 pulldown experiments (34). Insertion of the FLLL double point mutations within the HA SSP GPC construct was detrimental to GPC expression, although single point mutations within this motif were tolerated. These mutations, while allowing SSP to retain its association with the transmembrane-spanning GP2 subunit, obstructed the interaction with the remaining extracellular spike GP1 subunit.
This study highlights the importance of this often-overlooked 58-amino-acid protein in orchestrating the transport and production of functional noncovalently interacting trimeric glycoprotein complexes. Components of all three glycoprotein subunits play a role in the proper glycosylation and proteolytic processing of the glycoprotein complex (43). Without SSP, the glycoprotein precursor is retained within the endoplasmic reticulum and is not capable of continuing its passage through the secretory pathway. Immunoprecipitation of both the SSP and the GP1 subunits revealed that SSP is the subunit that stabilizes the organization of the trimeric glycoprotein complex. Whole-genome small interfering RNA (siRNA) screens have implicated the involvement of the COP1 subunit for LCMV infection (49). Furthermore, proteomic analysis led to the identification of ERGIC-53 as a host factor involved in the production of infectious particles (50). Future experiments using the tools presented here may allow for the identification of host proteins that interact with SSP. A deeper understanding of the mechanism by which this family of viruses involves host pathways would define targets for antiviral therapeutics.
MATERIALS AND METHODS
Cell culture and virus stocks.
BHK-21, DBT, HEK 293T, and Vero E6 cell lines were maintained at 37°C, 5% CO2, with 1× Dulbecco’s modified Eagle’s medium (DMEM), 10% fetal bovine serum, penicillin-streptomycin, and l-glutamine. The LCMV Armstrong 4 (Arm-4) strain was used in this report (51, 52). Viral stocks were propagated in BHK-21 cells, and viral titers were determined via plaque assay using Vero E6 cells. Transient transfections were performed using Lipofectamine 2000 as per the manufacturer’s protocol (Life Technologies).
Plasmids.
Total RNA was isolated using Trizol from BHK-21 cells infected with LCMV Arm-4 at a multiplicity of infection (MOI) of 0.1 for 48 h at 37°C, 5% CO2. Viral glycoprotein cDNA was synthesized using a GP-specific primer (sequence provided upon request) flanked by XhoI recognition sequences using the GoScript reverse transcriptase as per the manufacturer’s protocol and cloned into the pTargeT vector (Promega). Point mutations were introduced using the site-directed QuikChange Lightning kit (Agilent Technologies), sequence verified using standard automated sequencing methods, and subcloned into the pCAGGS vector using the XhoI sites (53). The HA epitope tag (YPYDVPDYA), as well as the replacement of the native stable signal peptide with the signal sequence from the influenza A hemagglutinin protein (MEKIVLLFAIVSLVKS), was inserted into the GPC open reading frame using the overlap PCR technique (54).
For construction of the HA SSP GPC plasmid, the HA epitope was inserted between amino acids, A10 and L11, in the SSP region of the full-length glycoprotein open reading frame. The plasmid encoding only the tagged SSP was created by introduction of two consecutive stop codons at amino acids 59 and 60, the first two residues of the GP1 subunit, and immediately downstream of the SPase recognition site. Both these mutants were initially produced using the pTargeT plasmid and were shuttled into the pCAGGS backbone as stated above.
SSP alignment.
Amino acid sequences used to highlight the signal peptide conservation were aligned using CLC Sequence Viewer 6 and using the following arenaviruses: Allpahuayo virus (AY012687), Amapari virus (AF512834), Bear Canyon virus (AF512833), Big Brushy Tank virus (EF619036), Catarina virus (DQ865245), Chapare virus (EU260463), Cupixi virus (AF512832), Flexal virus (AF512831), Guanarito virus (NC_005077), Ippy virus (DQ328877), Junín virus (D10072), Lassa virus (AY628203), Latino virus (AF512830), Lujo virus (NC_012776), Luna virus (AB697691), LCMV (AY847350), Machupo virus (NC_005078), Mobala virus (AY342390), Mopeia virus (DQ328874), Morogoro virus (EU914103), Oliveros virus (U34248), Parana virus (AF512829), Pichinde virus (NC_006447), Pirital virus (AF277659), Sabia virus (NC_006317), Skinner Tank virus (EU123328), Tacaribe virus (NC_004293), Tamiami virus (AF512828), Tonto Creek virus (EF619033), and White water Arroyo virus (AF228063).
Western blotting.
Transfected cells were incubated for 48 h, washed with 1× phosphate-buffered saline (PBS) and 1 mM EDTA, collected (600 × g, 4°C, 15 min), and lysed on ice with 1% NP-40 lysis buffer and complete protease inhibitors (RPI Corp.). Clarified lysates (10,000 × g, 4°C, 15 min) were separated under reduced and denatured SDS-PAGE conditions and transferred to nitrocellulose membranes. For LCMV glycoprotein complex detection, the anti-GP2 (83.6) and anti-SSP (SP7) antibodies have been described previously (18, 55, 56). We also used anti-HA (Sigma), anti-actin (Millipore), goat anti-mouse horseradish peroxidase (HRP), and goat anti-rabbit HRP (Jackson ImmunoResearch) antibodies. Blots were processed using the Amersham ECL Prime detection reagents (GE Healthcare) and were developed using the Bio-Rad ChemiDoc XRS system. Densitometry quantification was performed using ImageJ (http://imagej.nih.gov/ij/).
Immunofluorescence.
DBT cells were plated onto flame-sterilized glass coverslips and transfected using Lipofectamine 2000 (Life Technologies). After 48 h of incubation, cells were fixed using 3.7% paraformaldehyde (PFA) at room temperature and, when relevant, permeabilized with PBS and 0.1% Triton X-100 for 5 min. Cells were blocked using 5% bovine serum albumin (BSA) in PBS-Tween 20 for 1 h prior to overnight primary antibody incubation at 4°C. The phalloidin-fluorescein isothiocyanate (FITC) and Alexa Fluor 405-, 488-, and 594-conjugated secondary antibodies (Life Technologies) were used to detect viral or cellular epitopes. For the triple labeling microscopy, cells were incubated with phalloidin-FITC overnight, followed by mouse anti-HA (Sigma) for an additional overnight staining prior to 1 h of incubation with Alexa Fluor 405. Ultimately, cells were stained with GP1 antibody (2.11-10) directly conjugated to Alexa Fluor 594 (Life Technologies). For the SSP-HA and GP1/2 and the HA SSP GPC samples, coverslips were mounted with a mounting medium lacking DAPI (4′,6-diamidino-2-phenylindole), while other samples used DAPI-containing mounting medium (Southern Biotechnologies). The mannosidase II antibody was kindly provided by K. Moreman (University of Georgia). Confocal microscopy images were acquired using the Nikon Eclipse Ti microscope. All images were processed linearly using ImageJ and cropped for assembly using Adobe Photoshop. Colocalization measurements were acquired using the ImageJ colocalization finder plugin (http://rsb.info.nih.gov/ij/plugins/colocalization-finder.html).
Flow cytometry.
Transfected HEK 293T cells were collected and disassociated using cold 5 mM EDTA in 1× PBS. Cells were resuspended in 5% fetal bovine serum (FBS), passed through a 26-gauge syringe, and blocked on ice for 30 min. Cells were incubated with either the anti-HA antibody (Sigma), the GP1 antibody directly conjugated to Alexa Fluor 488 (2/11/10-488), or the normal mouse IgG antibody (BioLegend) for 30 min on ice. Cells were pelleted (100 × g, 3 min, 10°C) and extensively washed with 5% FBS. For the HA antibody or normal IgG samples, cells were stained with the Alexa Fluor 488 secondary antibody for 30 min on ice. Cells were washed twice with 5% FBS prior to one wash with 1× PBS. Cells were transferred to prechilled fluorescence-activated cell sorting (FACS) tubes prior to 7-AAD viability staining (BioLegend). Flow cytometry was performed using the FACSCalibur and CellQuest Pro software (BD Biosciences). All data were analyzed using FlowJo software (Ashland, OR).
Fusion assay.
DBT cells were plated onto flame-sterilized glass coverslips and transfected using Lipofectamine 2000 (Life Technologies). After 48 h of incubation, cells were washed with 1× PBS and incubated with pH 5-adjusted DMEM growth medium for 1 h before returning to pH 7.4 DMEM maintenance medium for an additional 1 h. Cells were fixed with 3.7% paraformaldehyde and extensively washed with 1× PBS, and coverslips were mounted with DAPI-containing mounting medium. Images were acquired using the Nikon Eclipse Ti microscope. The fusion index was determined by counting the number of cells versus the total nuclei, as previously described (29).
Coimmunoprecipitation.
Transfected HEK 293T cells were lysed using 0.1% NP-40 lysis buffer (200 mM Tris [pH 6.8], 150 mM NaCl, 1 mM EDTA) with fresh protease inhibitors (RPI Corp.). Clarified lysates were incubated with protein G beads (GE Healthcare) for 30 min at 4°C and collected by mild centrifugation (8,200 × g, 4°C, 30 s) prior to overnight incubation with either anti-HA agarose beads (Sigma) or the GP1-specific 2.11-15 antibody at 4°C with continuous end-over-end rotation (39). With the GP1 immunoprecipitation samples, after overnight incubation protein G agarose beads were added to each sample for 1 h at 4°C. Beads were washed thrice with 0.1% NP-40 wash buffer and once without detergent.
SUPPLEMENTAL MATERIAL
SSP amino acid sequence conservation. Multiple-sequence alignment of each rodent arenavirus SSP clustered by serogroup and clade. Red stars indicate human pathogens. Download
Precursor GPC glycosylation. HEK 293T cell lysates were incubated with peptide-N-glycosidase F (PNGase F; Sigma) at 37°C for 5 min prior to SDS-PAGE. Nitrocellulose membrane was incubated with the 83.6 (GP2) antibody. Note the shift in molecular weight of both the precursor GPC (*) and the GP2 (**) subunit, indicating the PNGase F removal of N-linked glycans. Download
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (NIH) grant AI-065359 from the Pacific Southwest Regional Center for Excellence and by the National Science Foundation (NSF) grant DGE-0638751.
We are indebted to Bernhard Dobberstein (University of Heidelberg) for supplying the rabbit SP7 antibody. We thank Christine Sütterlin and Megan Angelini for helpful discussions and manuscript editing, respectively. We thank Melissa Lodoen, Norikiyo Ueno, and Andrea Tenner for flow cytometry assistance.
We declare no conflicts of interest.
Footnotes
Citation Bederka LH, Bonhomme CJ, Ling EL, Buchmeier MJ. 2014. Arenavirus stable signal peptide is the keystone subunit for glycoprotein complex organization. mBio 5(6):e02063-14. doi:10.1128/mBio.02063-14.
REFERENCES
- 1. Buchmeier M, de la Torre JC, Peters CF. 2013. Arenaviridae, p 1283–1302 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, vol 2, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2. Buchmeier MJ, de la Torre J, Peters CJ. 2007. Arenaviridae: the viruses and their replication, p 1792–1828 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 3. Jahrling PB, Peters CJ. 1992. Lymphocytic choriomeningitis virus. A neglected pathogen of man. Arch. Pathol. Lab. Med. 116:486–488. [PubMed] [Google Scholar]
- 4. Mets MB, Barton LL, Khan AS, Ksiazek TG. 2000. Lymphocytic choriomeningitis virus: an underdiagnosed cause of congenital chorioretinitis. Am. J. Ophthalmol. 130:209–215. 10.1016/S0002-9394(00)00570-5. [DOI] [PubMed] [Google Scholar]
- 5. Centers for Disease Control and Prevention (CDC) 2005. Lymphocytic choriomeningitis virus infection in organ transplant recipients—Massachusetts, Rhode Island. 2005. MMWR Morb. Mortal. Wkly. Rep. 54:537–539. [PubMed] [Google Scholar]
- 6. Fischer SA, Graham MB, Kuehnert MJ, Kotton CN, Srinivasan A, Marty FM, Comer JA, Guarner J, Paddock CD, DeMeo DL, Shieh WJ, Erickson BR, Bandy U, DeMaria A, Jr, Davis JP, Delmonico FL, Pavlin B, Likos A, Vincent MJ, Sealy TK, Goldsmith CS, Jernigan DB, Rollin PE, Packard MM, Patel M, Rowland C, Helfand RF, Nichol ST, Fishman JA, Ksiazek T, Zaki SR, LCMV in Transplant Recipients Investigation Team 2006. Transmission of lymphocytic choriomeningitis virus by organ transplantation. N. Engl. J. Med. 354:2235–2249. 10.1056/NEJMoa053240. [DOI] [PubMed] [Google Scholar]
- 7. Charrel RN, de Lamballerie X, Emonet S. 2008. Phylogeny of the genus arenavirus. Curr. Opin. Microbiol. 11:362–368. 10.1016/j.mib.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 8. Yama IN, Cazaux B, Britton-Davidian J, Moureau G, Thirion L, de Lamballerie X, Dobigny G, Charrel RN. 2012. Isolation and characterization of a new strain of lymphocytic choriomeningitis virus from rodents in southwestern France. Vector Borne Zoonotic Dis. 12:893–903. 10.1089/vbz.2011.0892. [DOI] [PubMed] [Google Scholar]
- 9. Auperin DD, Romanowski V, Galinski M, Bishop DH. 1984. Sequencing studies of Pichinde arenavirus S RNA indicate a novel coding strategy, an ambisense viral S RNA. J. Virol. 52:897–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Eichler R, Lenz O, Strecker T, Eickmann M, Klenk HD, Garten W. 2003. Identification of Lassa virus glycoprotein signal peptide as a trans-acting maturation factor. EMBO Rep. 4:1084–1088. 10.1038/sj.embor.7400002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. York J, Romanowski V, Lu M, Nunberg JH. 2004. The signal peptide of the Junín arenavirus envelope glycoprotein is myristoylated and forms an essential subunit of the mature G1-G2 complex. J. Virol. 78:10783–10792. 10.1128/JVI.78.19.10783-10792.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Agnihothram SS, York J, Nunberg JH. 2006. Role of the stable signal peptide and cytoplasmic domain of G2 in regulating intracellular transport of the Junín virus envelope glycoprotein complex. J. Virol. 80:5189–5198. 10.1128/JVI.00208-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lenz O, ter Meulen J, Klenk HD, Seidah NG, Garten W. 2001. The Lassa virus glycoprotein precursor GP-C is proteolytically processed by subtilase SKI-1/S1P. Proc. Natl. Acad. Sci. U. S. A. 98:12701–12705. 10.1073/pnas.221447598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Beyer WR, Pöpplau D, Garten W, von Laer D, Lenz O. 2003. Endoproteolytic processing of the lymphocytic choriomeningitis virus glycoprotein by the subtilase SKI-1/S1P. J. Virol. 77:2866–2872. 10.1128/JVI.77.5.2866-2872.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cordo SM, Cesio y Acuña M, Candurra NA. 2005. Polarized entry and release of Junin virus, a New World arenavirus. J. Gen. Virol. 86:1475–1479. 10.1099/vir.0.80473-0. [DOI] [PubMed] [Google Scholar]
- 16. Strecker T, Eichler R, Meulen J, Weissenhorn W, Dieter Klenk H, Garten W, Lenz O. 2003. Lassa virus Z protein is a matrix protein and sufficient for the release of virus-like particles [corrected]. J. Virol. 77:10700–10705. 10.1128/JVI.77.19.10700-10705.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Urata S, Noda T, Kawaoka Y, Yokosawa H, Yasuda J. 2006. Cellular factors required for Lassa virus budding. J. Virol. 80:4191–4195. 10.1128/JVI.80.8.4191-4195.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Froeschke M, Basler M, Groettrup M, Dobberstein B. 2003. Long-lived signal peptide of lymphocytic choriomeningitis virus glycoprotein pGP-C. J. Biol. Chem. 278:41914–41920. 10.1074/jbc.M302343200. [DOI] [PubMed] [Google Scholar]
- 19. Cao W, Henry MD, Borrow P, Yamada H, Elder JH, Ravkov EV, Nichol ST, Compans RW, Campbell KP, Oldstone MB. 1998. Identification of alpha-dystroglycan as a receptor for lymphocytic choriomeningitis virus and Lassa fever virus. Science 282:2079–2081. 10.1126/science.282.5396.2079. [DOI] [PubMed] [Google Scholar]
- 20. Radoshitzky SR, Abraham J, Spiropoulou CF, Kuhn JH, Nguyen D, Li W, Nagel J, Schmidt PJ, Nunberg JH, Andrews NC, Farzan M, Choe H. 2007. Transferrin receptor 1 is a cellular receptor for New World haemorrhagic fever arenaviruses. Nature 446:92–96. 10.1038/nature05539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lavanya M, Cuevas CD, Thomas M, Cherry S, Ross SR. 2013. siRNA screen for genes that affect Junínvirus entry uncovers voltage-gated calcium channels as a therapeutic target. Sci. Transl. Med. 5:204ra131. 10.1126/scitranslmed.3006827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jae LT, Raaben M, Herbert AS, Kuehne AI, Wirchnianski AS, Soh TK, Stubbs SH, Janssen H, Damme M, Saftig P, Whelan SP, Dye JM, Brummelkamp TR. 2014. Virus entry. Lassa virus entry requires a trigger-induced receptor switch. Science 344:1506–1510. 10.1126/science.1252480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Di Simone C, Buchmeier MJ. 1995. Kinetics and pH dependence of acid-induced structural changes in the lymphocytic choriomeningitis virus glycoprotein complex. Virology 209:3–9. 10.1006/viro.1995.1225. [DOI] [PubMed] [Google Scholar]
- 24. Klewitz C, Klenk HD, ter Meulen J. 2007. Amino acids from both N-terminal hydrophobic regions of the Lassa virus envelope glycoprotein GP-2 are critical for pH-dependent membrane fusion and infectivity. J. Gen. Virol. 88:2320–2328. 10.1099/vir.0.82950-0. [DOI] [PubMed] [Google Scholar]
- 25. Martoglio B, Dobberstein B. 1998. Signal sequences: more than just greasy peptides. Trends Cell Biol. 8:410–415. 10.1016/S0962-8924(98)01360-9. [DOI] [PubMed] [Google Scholar]
- 26. Marzi A, Akhavan A, Simmons G, Gramberg T, Hofmann H, Bates P, Lingappa VR, Pöhlmann S. 2006. The signal peptide of the ebolavirus glycoprotein influences interaction with the cellular lectins DC-SIGN and DC-SIGNR. J. Virol. 80:6305–6317. 10.1128/JVI.02545-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li Y, Luo L, Thomas DY, Kang CY. 2000. The HIV-1 Env protein signal sequence retards its cleavage and down-regulates the glycoprotein folding. Virology 272:417–428. 10.1006/viro.2000.0357. [DOI] [PubMed] [Google Scholar]
- 28. Kim SJ, Rahbar R, Hegde RS. 2001. Combinatorial control of prion protein biogenesis by the signal sequence and transmembrane domain. J. Biol. Chem. 276:26132–26140. 10.1074/jbc.M101638200. [DOI] [PubMed] [Google Scholar]
- 29. Saunders AA, Ting JP, Meisner J, Neuman BW, Perez M, de la Torre JC, Buchmeier MJ. 2007. Mapping the landscape of the lymphocytic choriomeningitis virus stable signal peptide reveals novel functional domains. J. Virol. 81:5649–5657. 10.1128/JVI.02759-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stenglein MD, Sanders C, Kistler AL, Ruby JG, Franco JY, Reavill DR, Dunker F, Derisi JL. 2012. Identification, characterization, and in vitro culture of highly divergent arenaviruses from boa constrictors and annulated tree boas: candidate etiological agents for snake inclusion body disease. mBio 3(4):e00180-12. 10.1128/mBio.00180-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Feliciangeli S, Kitabgi P, Bidard JN. 2001. The role of dibasic residues in prohormone sorting to the regulated secretory pathway. A study with proneurotensin. J. Biol. Chem. 276:6140–6150. 10.1074/jbc.M009613200. [DOI] [PubMed] [Google Scholar]
- 32. McBride CE, Li J, Machamer CE. 2007. The cytoplasmic tail of the severe acute respiratory syndrome coronavirus spike protein contains a novel endoplasmic reticulum retrieval signal that binds COPI and promotes interaction with membrane protein. J. Virol. 81:2418–2428. 10.1128/JVI.02146-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kunz S, Edelmann KH, de la Torre JC, Gorney R, Oldstone MB. 2003. Mechanisms for lymphocytic choriomeningitis virus glycoprotein cleavage, transport, and incorporation into virions. Virology 314:168–178. 10.1016/S0042-6822(03)00421-5. [DOI] [PubMed] [Google Scholar]
- 34. Schrempf S, Froeschke M, Giroglou T, von Laer D, Dobberstein B. 2007. Signal peptide requirements for lymphocytic choriomeningitis virus glycoprotein C maturation and virus infectivity. J. Virol. 81:12515–12524. 10.1128/JVI.01481-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Eichler R, Lenz O, Strecker T, Eickmann M, Klenk HD, Garten W. 2004. Lassa virus glycoprotein signal peptide displays a novel topology with an extended endoplasmic reticulum luminal region. J. Biol. Chem. 279:12293–12299. 10.1074/jbc.M312975200. [DOI] [PubMed] [Google Scholar]
- 36. York J, Nunberg JH. 2007. Distinct requirements for signal peptidase processing and function in the stable signal peptide subunit of the Junín virus envelope glycoprotein. Virology 359:72–81. 10.1016/j.virol.2006.08.048. [DOI] [PubMed] [Google Scholar]
- 37. Agnihothram SS, York J, Trahey M, Nunberg JH. 2007. Bitopic membrane topology of the stable signal peptide in the tripartite Junín virus GP-C envelope glycoprotein complex. J. Virol. 81:4331–4337. 10.1128/JVI.02779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Capul AA, Perez M, Burke E, Kunz S, Buchmeier MJ, de la Torre JC. 2007. Arenavirus Z-glycoprotein association requires Z myristoylation but not functional RING or late domains. J. Virol. 81:9451–9460. 10.1128/JVI.00499-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Buchmeier MJ, Lewicki HA, Tomori O, Oldstone MB. 1981. Monoclonal antibodies to lymphocytic choriomeningitis and pichinde viruses: generation, characterization, and cross-reactivity with other arenaviruses. Virology 113:73–85. 10.1016/0042-6822(81)90137-9. [DOI] [PubMed] [Google Scholar]
- 40. Bonhomme CJ, Capul AA, Lauron EJ, Bederka LH, Knopp KA, Buchmeier MJ. 2011. Glycosylation modulates arenavirus glycoprotein expression and function. Virology 409:223–233. 10.1016/j.virol.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Di Simone C, Zandonatti MA, Buchmeier MJ. 1994. Acidic pH triggers LCMV membrane fusion activity and conformational change in the glycoprotein spike. Virology 198:455–465. 10.1006/viro.1994.1057. [DOI] [PubMed] [Google Scholar]
- 42. Burri DJ, Pasquato A, da Palma JR, Igonet S, Oldstone MB, Kunz S. 2013. The role of proteolytic processing and the stable signal peptide in expression of the Old World arenavirus envelope glycoprotein ectodomain. Virology 436:127–133. 10.1016/j.virol.2012.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Illick MM, Branco LM, Fair JN, Illick KA, Matschiner A, Schoepp R, Garry RF, Guttieri MC. 2008. Uncoupling GP1 and GP2 expression in the Lassa virus glycoprotein complex: implications for GP1 ectodomain shedding. Virol. J. 5:161. 10.1186/1743-422X-5-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schlie K, Strecker T, Garten W. 2010. Maturation cleavage within the ectodomain of Lassa virus glycoprotein relies on stabilization by the cytoplasmic tail. FEBS Lett. 584:4379–4382. 10.1016/j.febslet.2010.09.032. [DOI] [PubMed] [Google Scholar]
- 45. Albariño CG, Bird BH, Chakrabarti AK, Dodd KA, White DM, Bergeron E, Shrivastava-Ranjan P, Nichol ST. 2011. Reverse genetics generation of chimeric infectious Junin/Lassa virus is dependent on interaction of homologous glycoprotein stable signal peptide and G2 cytoplasmic domains. J. Virol. 85:112–122. 10.1128/JVI.01837-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Messina EL, York J, Nunberg JH. 2012. Dissection of the role of the stable signal peptide of the arenavirus envelope glycoprotein in membrane fusion. J. Virol. 86:6138–6145. 10.1128/JVI.07241-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. York J, Nunberg JH. 2009. Intersubunit interactions modulate pH-induced activation of membrane fusion by the Junin virus envelope glycoprotein GPC. J. Virol. 83:4121–4126. 10.1128/JVI.02410-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Takagi T, Ohsawa M, Morita C, Sato H, Ohsawa K. 2012. Genomic analysis and pathogenic characteristics of lymphocytic choriomeningitis virus strains isolated in Japan. Comp. Med. 62:185–192. [PMC free article] [PubMed] [Google Scholar]
- 49. Panda D, Das A, Dinh PX, Subramaniam S, Nayak D, Barrows NJ, Pearson JL, Thompson J, Kelly DL, Ladunga I, Pattnaik AK. 2011. RNAi screening reveals requirement for host cell secretory pathway in infection by diverse families of negative-strand RNA viruses. Proc. Natl. Acad. Sci. U. S. A. 108:19036–19041. 10.1073/pnas.1113643108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Klaus JP, Eisenhauer P, Russo J, Mason AB, Do D, King B, Taatjes D, Cornillez-Ty C, Boyson JE, Thali M, Zheng C, Liao L, Yates JR, III, Zhang B, Ballif BA, Botten JW. 2013. The intracellular cargo receptor ERGIC-53 is required for the production of infectious arenavirus, coronavirus, and filovirus particles. Cell Host Microbe 14:522–534. 10.1016/j.chom.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wright KE, Salvato MS, Buchmeier MJ. 1989. Neutralizing epitopes of lymphocytic choriomeningitis virus are conformational and require both glycosylation and disulfide bonds for expression. Virology 171:417–426. 10.1016/0042-6822(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 52. Beyer WR, Miletic H, Ostertag W, von Laer D. 2001. Recombinant expression of lymphocytic choriomeningitis virus strain WE glycoproteins: a single amino acid makes the difference. J. Virol. 75:1061–1064. 10.1128/JVI.75.2.1061-1064.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Niwa H, Yamamura K, Miyazaki J. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193–199. 10.1016/0378-1119(91)90434-D. [DOI] [PubMed] [Google Scholar]
- 54. Heckman KL, Pease LR. 2007. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protoc. 2:924–932. 10.1038/nprot.2007.132. [DOI] [PubMed] [Google Scholar]
- 55. Bruns M, Cihak J, Müller G, Lehmann-Grube F. 1983. Lymphocytic choriomeningitis virus. VI. Isolation of a glycoprotein mediating neutralization. Virology 130:247–251. 10.1016/0042-6822(83)90135-6. [DOI] [PubMed] [Google Scholar]
- 56. Weber EL, Buchmeier MJ. 1988. Fine mapping of a peptide sequence containing an antigenic site conserved among arenaviruses. Virology 164:30–38. 10.1016/0042-6822(88)90616-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SSP amino acid sequence conservation. Multiple-sequence alignment of each rodent arenavirus SSP clustered by serogroup and clade. Red stars indicate human pathogens. Download
Precursor GPC glycosylation. HEK 293T cell lysates were incubated with peptide-N-glycosidase F (PNGase F; Sigma) at 37°C for 5 min prior to SDS-PAGE. Nitrocellulose membrane was incubated with the 83.6 (GP2) antibody. Note the shift in molecular weight of both the precursor GPC (*) and the GP2 (**) subunit, indicating the PNGase F removal of N-linked glycans. Download