Abstract
Most solid tumors, including pancreatic ductal adenocarcinoma (PDAC), exhibit structural and numerical chromosome instability (CIN). While often implicated as a driver of tumor progression and drug resistance, CIN also reduces cell fitness and poses a vulnerability which can be exploited therapeutically. The spindle assembly checkpoint (SAC) ensures correct chromosome-microtubule attachment, thereby minimizing chromosome segregation errors. Many tumors exhibit up-regulation of SAC components such as MPS1, which may help contain CIN within survivable limits. Prior studies showed that MPS1 inhibition with the small molecule, NMS-P715, limits tumor growth in xenograft models. In cancer cell lines, NMS-P715 causes cell death associated with impaired SAC function and increased chromosome mis-segregation. While normal cells appeared more resistant, effects on stem cells, which are the dose-limiting toxicity of most chemotherapeutics, were not examined. Elevated expression of 70 genes (CIN70), including MPS1, provides a surrogate measure of CIN and predicts poor patient survival in multiple tumor types. Our new findings show that the degree of CIN70 up-regulation varies considerably among PDAC tumors, with higher CIN70 gene expression predictive of poor outcome. We identified a 25 gene subset (PDAC CIN25) whose over-expression was most strongly correlated with poor survival and included MPS1. In vitro, growth of human and murine PDAC cells is inhibited by NMS-P715 treatment, whereas adipose-derived human mesenchymal stem cells are relatively resistant and maintain chromosome stability upon exposure to NMS-P715. These studies suggest that NMS-P715 could have a favorable therapeutic index and warrant further investigation of MPS1 inhibition as a new PDAC treatment strategy.
Keywords: CIN70, MPS1 kinase inhibitor, pancreatic ductal adenocarcinoma
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the fourth highest contributor to cancer-related death in the western world, with a lifetime risk of 1.47% and a 5 year survival rate of 6% (1). Detection typically occurs at an advanced stage of tumor progression when 80-85% of cases are inoperable. Even after tumor resection, 5 year survival is only 15-25% with existing forms of adjuvant therapy (2). The need for alternative therapeutic approaches to this deadly malignancy is thus imperative and recent insights into the causes and vulnerabilities of cancer (3-5), suggest that drugs targeting the high level of chromosome instability in PDAC could offer a new direction.
A hallmark of most solid tumors is chromosome instability (CIN) which is thought to accelerate tumor evolution, promote drug resistance and is linked to poor prognosis (6-10). CIN exists in two forms: 1) numerical CIN (nCIN) which is an increased frequency of chromosome segregation errors (resulting in an abnormal chromosome number termed aneuploidy) and 2) structural CIN reflecting sub-chromosomal rearrangements. Recent findings suggest that the benefits to the tumor of a CIN phenotype may be offset by negative consequences, which include altered protein stoichiometry and the risk of lethal chromosome imbalance (9, 11). In order to mitigate such deleterious effects, tumors may depend on the acquisition of compensatory mechanisms. An example of a CIN survival strategy is seen in changes to the mitotic spindle assembly checkpoint (SAC), which serves to limit chromosome segregation errors by ensuring that chromosomes have a bipolar attachment to the mitotic spindle prior to anaphase onset. Counterintuitively, loss of expression or mutational inactivation of SAC genes in tumors is rare. Instead, evidence suggests that tumors increase SAC activation and/or expression of SAC components, such as the kinase MPS1 (also known as TTK), which may contain segregation errors within survivable limits (12-15). Yeast mutants with faulty chromosome segregation are more sensitive to Mps1 inhibition than their wild-type counterparts (16) and targeted inhibition of MPS1 or other SAC associated proteins (namely, BUB1B/BUBR1, the aurora kinases or CENP-E), causes cancer cell death accompanied by massive chromosome mis-segregation. In keeping with the notion that tumors have increased SAC dependency, growth of non-tumorigenic cells was generally less affected by SAC inhibition (12, 17-26). MPS1 inhibitors have shown efficacy in xenograft models of multiple tumor types when administered alone (12, 20) or in combination with a microtubule targeting agent (27).
Carter et al., identified a set of 70 genes (CIN70) whose over-expression correlated with CIN and poor survival (7). Importantly, although some CIN70 genes are markers of proliferation, the prognostic utility of CIN70 was independent of tumor grade (8). CIN70 includes 35 genes that play a role in chromosome segregation, three encoding known SAC components. The CIN70 signature has proven to be a reliable surrogate for measuring CIN in numerous cancer types (7, 9) but has not been investigated in PDAC. In the present study, we examined CIN70 expression in three publically available PDAC microarray data sets (28-30) and found consistent up-regulation of CIN70 genes in PDAC compared to normal pancreas. This finding is in agreement with cytogenetic evidence for high levels of CIN in PDAC tissues and in genetically engineered mouse models of PDAC (8, 31, 32). Importantly, high CIN70 gene expression in resected PDAC was found to be a powerful predictor of poor patient survival. Included amongst the up-regulated CIN70 genes in PDAC, is the MPS1 kinase. Here, we show that treatment of PDAC cells with the MPS1 inhibitor, NMS-P715 (12), leads to significantly increased nCIN and cell death. At present, the mainstay PDAC chemotherapeutic is the DNA synthesis inhibitor, gemcitabine, whose dose-limiting toxicity is damage to stem cells of the hematopoietic system (33). Encouragingly, a previous study has shown untransformed cells to be more resistant to NMSP715 than many types of cancer cells (12). We extend these findings and demonstrate that adipose-derived mesenchymal stem cells (ASCs) (34) maintain chromosome stability in the presence of NMS-P715 and are markedly more resistant to its cytotoxic effects than PDAC cells. Conversely, gemcitabine exerted higher toxicity to the ASCs than the PDAC cells. Together, our results support the potential utility of a CIN gene expression signature in predicting PDAC patient survival and the promise of MPS1 inhibition as a new, cancer selective, targeted therapy for PDAC.
Materials and Methods
Gene expression and survival analysis
Four microarray datasets (GSE17891, GSE32676, GSE28735, GSE 45765), publically available at the NIH Gene Expression Omnibus (www.ncbi.nlm.gov/geo), were examined in this study. Dataset composition, procedures for dataset merging, survival analysis and statistical testing are detailed in supplementary information.
Cell lines
PANC-1 (ATCC® CRL-1469 TM) and BxPC-3 (ATCC® CRL-1687TM) human PDAC cell lines were grown in DMEM/10%FBS or RPMI/10%FBS, respectively, obtained from ATCC and verified by STS markers in 2013 by IDEXX RADIL (MO). hTERT-HPNE cells (ATCC® CRL-4023TM; purchased in 2012) were cultured according to the supplier's conditions. Murine 825-2 and 1170-4 KRC pancreatic cancer cells were newly derived from two distinct PDAC tumors arising in a genetically engineered mouse model of PDAC in which oncogenic Kras is combined with Rb deletion using Cre-recombinase (31), and cultured in RPMI/10%FBS. Human ASCs were collected from donors undergoing lipoaspiration using an approved protocol (IRB 0305-59) as described previously (35) and cultured in EGM2-MV medium (Lonza Cat#CC3202)/10%FBS. Population doubling times of BxPC-3, PANC-1, KRC, ASC and hTERT-HPNE cells were ~40-60h, ~50h, ~20h,~24-26h and ~40h respectively. Supplementary Table S1 provides cell line chromosome stability information.
Cell growth inhibition assays
The structurally defined inhibitor, NMS-P715, (12), was provided by Nerviano Medical Sciences or purchased from EMD Millipore (Cat#475949-5MG) and suspended in DMSO. Gemcitabine (Tocris Bioscience Cat#3259) was suspended in H2O. Drug dose-response assays were performed by plating 2,000 human or 1,000 KRC cells per well in triplicate in 96 well plates. Three replicate assays were performed per cell line. Compounds were added for 72h after which cells were methanol fixed and stained with 0.05% methylene blue (36). Optical density was measured at 620 nM after suspension in 0.5M HCl (36) on a Beckman-Coulter DTX880 MultiMode Detector. Proliferation was measured relative to vehicle control and IC50 determined using Compusyn software (37). Dose-response curves were generated using sigmoidal interpolation curve fitting in SigmaPlot 12.3. For clonogenic survival assays, cells were plated at the indicated densities in duplicate or triplicate in 12-well dishes and allowed to attach for 24h. For continuous treatment, NMS-P715 was replenished every three days. In the washout assays, cells received a 24h NMS-P715 treatment followed by culture in compound-free medium. Experiments were performed in duplicate. Cell growth quantification in the colony formation assay was by colorimetric methylene blue assay or manual counting. Growth inhibition was measured relative to vehicle control.
SAC override assays
Western analysis was performed using a phospho-Histone H3 (Ser10) (pS10H3) antibody (Millipore Cat#06-570). β-actin was used as a loading control (Sigma Cat#A5441). For immunofluorescence, BxPC-3, PANC-1 or KRC cells were plated at 10,000-20,000 cells/ well on chamber slides. Replicate cultures of BxPC-3 and PANC-1 cells were blocked in 75 nM nocodazole for 18 hr. 0.4 μmol/L NMS-P715 was added for the last 2h of the noc block, where indicated. For KRC cell treatment, see supplemental information. Cells were fixed in 4% formaldehyde/1XPBS and incubated with an Alexa-Fluor 488-labeled pS10H3 antibody (Cell Signaling Technologies Cat#9708S). ≥200 cells were scored per replicate. For FISH analysis, duplicate cell cultures were incubated with NMS-P715 or DMSO control then fixed in 3:1 (v/v) methanol: acetic acid. Chromosome numbers were counted in ≥ 50 cells per culture, with probes recognizing X chromosome or chromosome 17 centromeres (Abbott Molecular Cat#05-J08-033, 06-J37-027) in human cells or a chromosome 11qE1 probe (Kreatech Diagnostics Cat#30501) in mouse cells. Images of DAPI stained nuclei were captured using a Spot RTKE camera (Diagnostic Instruments) mounted on a Leica DM5000B fluorescence microscope (Leica Microsystems). Images were minimally processed using Adobe Photoshop to adjust brightness and/or contrast.
Flow cytometry
PANC-1 cells were prepared for flow cytometry following Annexin-V/ Propidium Iodide staining using a commercial kit (BD Biosciences Cat#556547). Cells were analyzed on a FACS-Calibur Flow Cytometer and data processed using FlowJo software.
Results
The CIN70 gene expression signature predicts survival in pancreatic ductal adenocarcinoma (PDAC)
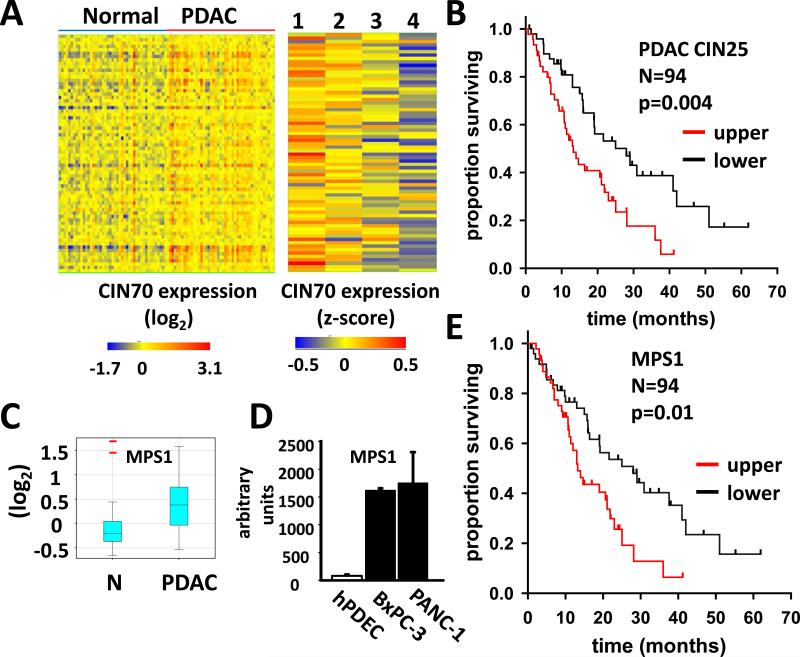
Elevated expression of the CIN70 genes has been associated with increased CIN in a range of human malignancies (7, 9). While cytogenetic studies have documented a high rate of CIN in PDAC (8), expression of the CIN70 gene signature in PDAC has not been reported. To assess CIN70 gene expression in PDAC, we examined a publically available gene expression microarray data set, comprising resected PDAC and adjacent normal tissue from 45 patients (28). As the matrix in Figure 1A (left panel) illustrates, the CIN70 signature shows a consistent pattern of up-regulation in PDAC relative to normal pancreas. Mean expression of 53/70 CIN70 genes was increased in PDAC relative to normal tissue, between 1.1 and 2.4 fold (p<0.05, paired t-test).
Figure 1. CIN70 gene expression predicts PDAC patient survival.
A. Left panel Matrix of CIN70 gene expression in 45 matched pairs of normal pancreas and resected PDAC tissue. Rows and columns represent genes and samples, respectively. Right panel CIN70 expression in 59 PDAC patients segregated by survival quartile (detailed in text): 1, n=15: median survival 3.8 months; 2, n=17: median survival 10 months; 3, n= 12: median survival 16 months; 4, n=15: median survival 28 months. B. Survival analysis of 94 PDAC patients segregated by the PDAC CIN25 score. Upper and lower represent patients in the respective halves of the PDAC CIN25 score distribution. Upper: median survival 28 months. Lower: median survival 13.2 months. P value refers to log-rank test comparing survival curves. C. Box whisker plot of MPS1 expression in 45 matched pairs of normal (N) and PDAC tissue samples. Central band and lower and upper box limits denote median, first and third quartiles, respectively. Whiskers denote data within 1.5 interquartiles of upper and lower box limits. Red bars indicate outliers. D. MPS1 expression derived from affymetrix gene expression microarray analysis of primary human pancreatic ductal epithelial cells (hPDEC), BxPC-3 and PANC-1 cell lines. E. Survival analysis of 94 PDAC patient samples segregated by MPS1 expression. Upper and lower represent patients in the respective halves of the MPS1 expression distribution. Upper: median survival 28 months. Lower: median survival 13.2 months.
CIN has been linked to a poor prognosis (10). As a surrogate marker of a CIN phenotype, increased CIN70 gene expression has been similarly shown to predict poor patient outcome (7). To explore the relationship between CIN70 gene expression and patient survival in PDAC, the 45 patient data set was combined with two additional gene expression microarray studies (29, 30). Collectively, these studies comprise a total of 94 resected PDAC samples analyzed on the Affymetrix gene chip platform, for which subsequent patient survival data are provided. To overcome batch effects in the merged data, gene expression values within each dataset were first standardized by z score transformation, to have a mean of zero and unit variance, as previously described (38, 39). After excluding individuals who were lost to follow-up, or alive at study end, the remaining patients (n=59) were ranked by survival time and assigned to quartiles 1-4, in order of increasing survival. Figure 1A (right panel) is a matrix of CIN70 gene expression in each survival quartile. There is a striking association between increased CIN70 gene expression and a less favorable prognosis, with 62/70 genes significantly up-regulated in quartile 1 compared to quartile 4 (p<0.05, moderated t-test).
To further explore the prognostic value of the CIN70 gene signature in PDAC, all 94 patients were assigned a CIN70 score representing the mean expression of all CIN70 genes, as described (9). Kaplan Meier survival analysis of patients segregated by CIN70 score confirmed a significant relationship between increased CIN70 expression and poor prognosis (Supplementary Fig. S1; p=0.03, log-rank test). To enrich the CIN70 signature for genes with greatest prognostic significance in PDAC, genes were individually tested for their association with patient survival by univariate Cox regression and ranked by their Cox scores (Supplementary Table S2). As shown in Figure 1B, a mean gene expression score based on the subset of 25 CIN70 genes with the highest Cox scores (PDAC CIN25), was even more effective in predicting patient survival (p=0.004, log-rank test).
The mitotic kinase, MPS1, is up-regulated in PDAC and a potential therapeutic target
A number of CIN70 genes encode SAC components whose over-expression may support cancer cell survival by limiting chromosome segregation errors. Such “addiction” to overexpressed SAC genes, would make them potential therapeutic targets. The PDAC CIN25 subset includes the gene MPS1, encoding a mitotic kinase essential to SAC function (40). MPS1 is significantly over-expressed in PDAC relative to normal pancreas (1.4 fold, p=1.9X10−5, paired t-test) (Fig. 1C) and ranks 13th in its negative association with patient survival (p<0.001; Table S2). Consistent with the patient data, the PDAC cell lines, BxPC-3 and PANC-1, used in the present study, exhibit highly elevated MPS1 expression compared to primary pancreatic ductal epithelial cells (41) (Fig. 1D; p≤1.2X10−13, t-test). As shown in Fig. 1E, MPS1 expression effectively stratifies PDAC patients into prognostic categories (p=0.01, log-rank test), as previously reported in glioblastoma (27).
MPS1 inhibition reduces pS10H3 and induces apoptosis
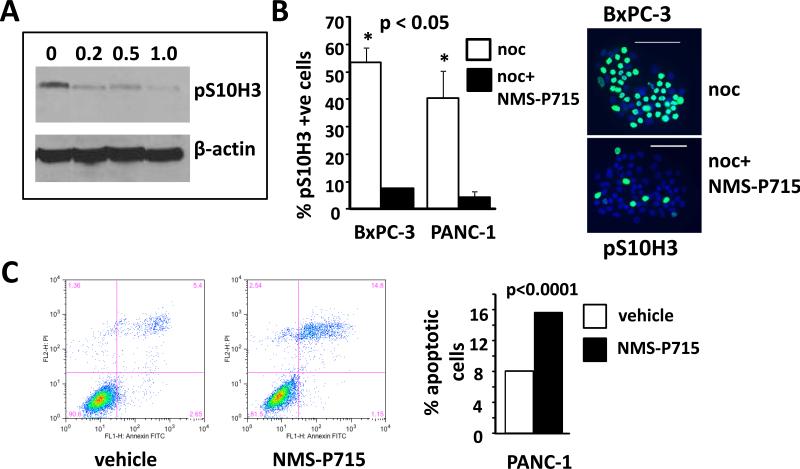
We tested the ability of the previously reported pharmacological MPS1 inhibitor, NMS-P715 (12), to attenuate SAC function in human PDAC cell lines. Western analysis showed a dose-dependent reduction of the mitotic marker pS10H3, in response to NMS-P715 treatment in PANC-1 cells (Fig. 2A), which is consistent with reduced SAC function and accelerated mitosis (12). NMS-P715-mediated SAC override was further confirmed in both PANC-1 and BxPC-3 cells arrested at pro-metaphase by nocodazole treatment. Here, the addition of NMS-P715 significantly reduced the proportion of nuclei positive for pS10H3 by immunofluorescence following nocodazole arrest (Fig. 2B). The effect of MPS1 inhibition on PANC-1 cell viability was examined by flow cytometry. A two-fold increase in the fraction of apoptotic cells was observed following exposure to 1 μmol/L of NMS-P715 for 40h (Fig.2C), which is indicative of reduced PANC-1 viability, concomitant with SAC override.
Figure 2. NMS-P715 treated PDAC cell lines exhibit bypass of the spindle assembly checkpoint and apoptosis.
A. Western analysis using a pS10H3 antibody of human PANC-1 cells treated with either NMS-P715 (μmol/L) or vehicle (0) as indicated for 72h. B. Left panel: Cells were treated with nocodazole (noc) or noc plus NMS-P715 (noc+NMS-P715). Percentage pS10H3 positive cells in each treatment group is indicated in the graph. Right panel: representative image of BxPC-3 cells treated either with noc or noc + NMS-P715. pS10H3 positive cells are green. Nuclei (blue) are stained with DAPI. Scale bar =100 μm. C. Flow cytometric analysis of Annexin V (Annexin-FITC) and propidium iodide (PI) labeled PANC-1 cells treated with 1 μmol/L NMS-P715 for 40h. Histogram shows the percentage of apoptotic cells calculated by combining the fraction of Annexin V positive (bottom right) and double positive (top right) cells. Significance was measured using a t-test (B) or χ2 test (C).
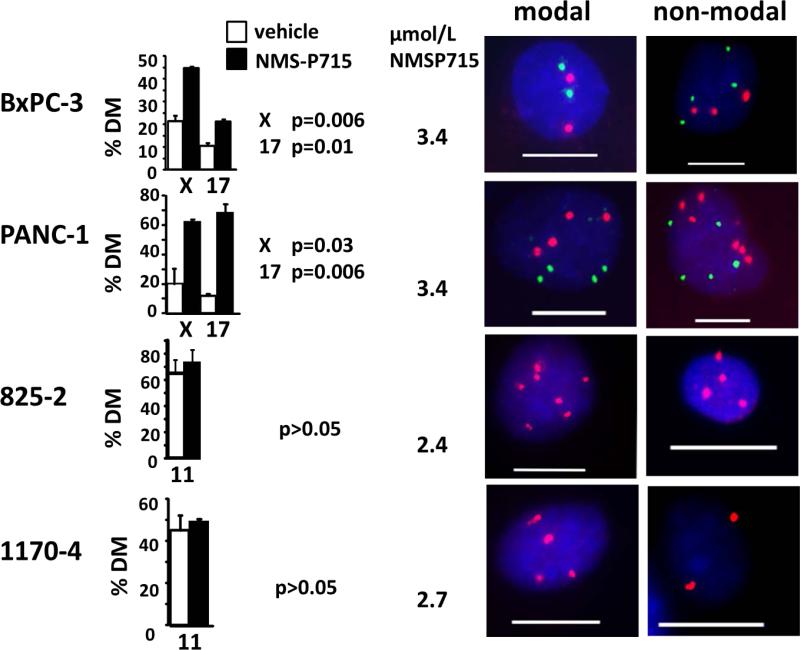
A FISH-based assay was used to measure the effects of NMS-P715 on nCIN, calculated as the percentage of cells exhibiting deviation from the modal number (% modal deviation) for chromosomes X and 17 (Fig. 3) (42). Both PANC-1 and BxPC-3 cells exhibited significantly increased nCIN after 48hr treatment with NMS-P715 at a concentration sufficient to cause significant growth inhibition (Fig. 3, Fig. 4A).
Figure 3. Elevated CIN in PDAC cells treated with the MPS1 inhibitor, NMS-P715.
FISH analysis with X chromosome (red) and chromosome 17 (green) centromeric probes in human BxPC3 or PANC-1 cells or a chromosome 11 probe (red) in mouse KRC cells (825-2 and 1170-4) after treatment with NMS-P715. Drug concentrations approximated preliminary IC50 values (data not shown). Cells were harvested after 48h (BxPC-3 and PANC-1) or 72h (825-2 and 1170-4). Percentage of cells exhibiting deviation from the modal chromosome number (%MD) provides an indirect measure of the CIN rate. Representative images of cells exhibiting either modal or non-modal signal numbers for each cell line are shown. Scale bar =10 μm. The modal chromosome numbers were: 2 for chromosomes X and 17 in BXPC-3 cells; 4 for chromosomes X and 17 in PANC-1 cells; 7 for chromosome 11 in 825-2 cells and 4 for chromosome 11 in 1170-4 cells. Significance was measured using a t-test.
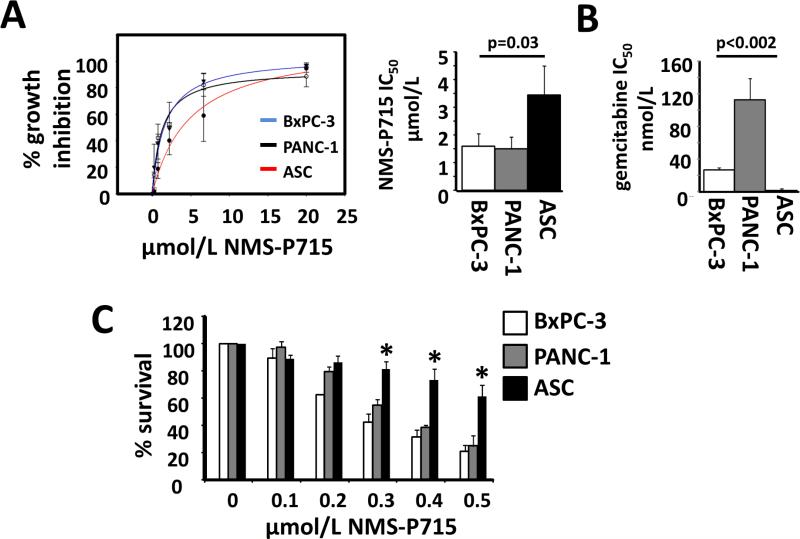
Figure 4. NMS-P715 selectively inhibits growth of PDAC cells. A. Left panel.
Dose effect of NMS-P715 on viability of BxPC-3, PANC-1 and ASC cells after 72h. 0= vehicle control. Right panel: NMS-P715 IC50 values for BxPC-3, PANC-1 and ASCs after 72h. B. Gemcitabine IC50 values for BxPC-3, PANC-1 and ASCs after 72h. C. BxPC-3, PANC-1 and ASC cells were grown at low density (200 cells/ well) for 6 days in medium supplemented with NMS-P715 (μmol/L) or DMSO control (0) as indicated. Cell growth was measured by colorimetric assay. ASC data represent mean of 5 (A) or 3 (B, C) independent isolates. Data were collected from three (A, B) or two (C) independent experiments. Significance was measured by t-test.
In contrast, no nCIN increase was observed in two Rb deficient murine pancreatic cancer cell lines (KRC lines 825-2 and 1170-4) treated with a growth inhibitory dose of NMS-P715 (Fig. 3; see below). In agreement with earlier reports linking Rb loss to CIN via several mechanisms (43), KRC cells exhibit high basal nCIN (≥ 45% MD) shown by FISH (Fig. 3; Supplementary Table S1) and chromosome spread analysis (Supplementary Fig. S2; Supplementary Table S1). Elevated nCIN, together with their failure to arrest and reduced viability in nocodazole (Supplementary Fig. S2), raises the possibility that KRC cells have a weakened SAC (44). Further nCIN elevation in KRC cells after NMS-P715 treatment may be incompatible with their survival.
NMS-P715 selectively inhibits growth of PDAC cells
Therapeutic index (TI), defined as the ratio between the toxic and therapeutic doses of a drug, is an important determinant of clinical efficacy (45). The dose limiting toxicity of conventional chemotherapeutics in patients is typically damage to the stem cell compartments of the bone marrow or gastrointestinal tract. A more favorable TI may be achievable by targeting an enzyme which is more critical to the survival of a cancer cell than a stem cell (45, 46). A prior study has shown human fibroblasts and B-lymphocytes to be less sensitive to NMS-P715 in a standard proliferation assay than many cancer cell lines, including the PDAC cell line, BxPC3 (13). Here, to address effects on human stem cells, we compared the anti-proliferative action of NMS-P715 in pancreatic cancer cells to normal human adipose-derived mesenchymal stem cells (ASCs), obtained from donors undergoing lipoaspiration (35). In addition to being a uniquely accessible source of primary human cells with a stable diploid karyotype (35) and high proliferative capacity in vitro, ASCs are rich in multipotent stem cells (34). In comparison to 5 independent ASC isolates, PANC-1 and BxPC-3 were significantly more sensitive to NMS-P715 in a proliferation assay, with IC50 values of 1.5, 1.6 and 3.4 μM for PANC-1, BxPC-3 and ASCs respectively (Fig. 4A p=0.03). Notably, relative sensitivity to the standard PDAC chemotherapeutic, gemcitabine, was the reverse, with both PANC-1 and BxPC-3 showing markedly less growth inhibition than ASCs (Fig. 4B p<0.002). In clonogenic survival assays, a 24h treatment with 1 μmol/L NMSP715 was sufficient to greatly reduce survival of both BxPC-3 and PANC-1 cells and continuous treatment at 0.5 μmol/L for 9 days completely abolished their growth (Supplementary Fig. S3). Since ASCs are migratory and do not form colonies when plated at low density (Supplementary Fig. S4), a colorimetric methylene blue assay was used to compare growth of PDAC cells and three ASC isolates, seeded at 200 cells per well in a 12 well dish, over a 6 day period of treatment with NMS-P715. In this assay, the growth inhibition of BxPC-3 cells (78.83% ±4.49%) and PANC-1 cells (75.1% ±7.7%) was about twice that of the ASCs (38.2% ± 8.5%) at 0.5 μmol/L NMS-P715 (p<0.002) (Fig. 4C; Supplementary Fig. S4), further corroborating the increased sensitivity of PDAC cells to NMS-P715 relative to ASCs.
KRC cell growth was also sensitive to MPS1 inhibition, with anti-proliferative IC50 values of 1.3 and 2.2 μM for lines 825-2 and 1170-4, respectively (Supplementary Fig. S5). KRC cells showed >85% loss of viability after 24h treatment with 1 μmol/L NMS-P715 in a clonogenic survival assay, while continuous treatment for 5 days with 0.5-0.7 μmol/L NMS-P715 completely abolished cell growth (Supplementary Fig. S5).
Human adipose stem cells and normal pancreatic epithelial cells are resistant to SAC override following MPS1 inhibition
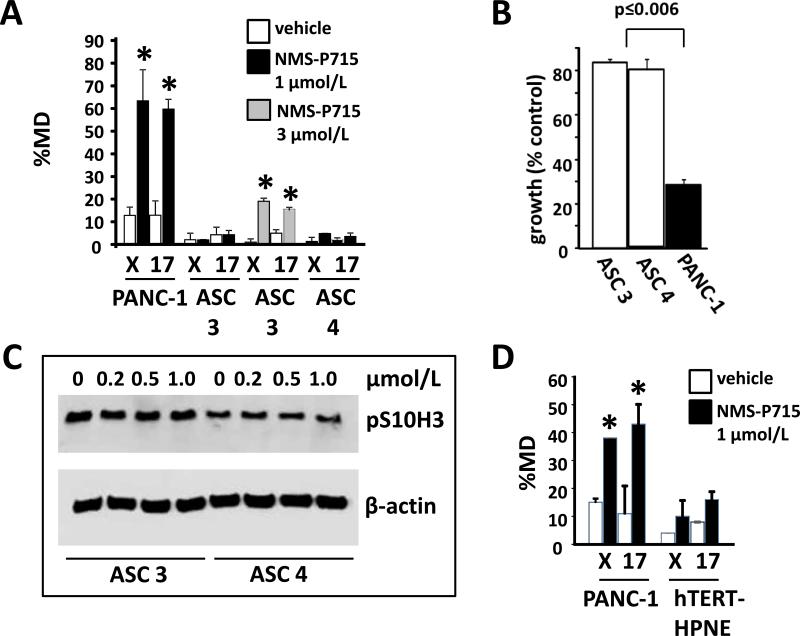
To explore the basis for the comparative resistance of ASCs to NMS-P715, two ASC isolates and PANC-1 cells were exposed for 72h to 1 μmol/L NMS-P715, a dose approximating the blood concentration of NMS-P715 achieved in preclinical mouse models (12). In contrast to PANC-1 cells, which exhibited significantly elevated nCIN levels after treatment, the ASCs maintained chromosomal stability and no significant nCIN increase was detected (Fig. 5A). A modest increase in nCIN in ASCs was observed upon exposure to 3 μmol/L NMS-P715 (Fig. 5A). Following treatment with 1 μmol/L NMS-P715 growth inhibition of PANC-1 cells in a clonogenic assay (72.2%) was markedly higher than ASC3 (16.1%) or ASC4 (23.2%) (p≤0.006; Fig. 5B, Supplementary Fig. S6). Consistent with the high chromosome stability observed after exposure to 1 μmol/L NMS-P715, ASCs did not exhibit a decrease in pS10H3 under these conditions (Fig. 5C), in contrast to PANC-1 (Fig. 2A). Consistent with the selectivity of NMSP715 towards PDAC cells, human telomerase immortalized pancreatic ductal epithelial cells (hTERT-HPNE) had an IC50 value of 3.4 μM in the proliferation assay (data not shown) and showed no significant nCIN increase after 72h treatment with 1 μmol/L NMS-P715 (Fig. 5D).
Figure 5. Human adipose stem cells and pancreatic ductal epithelial cells are comparatively resistant to nCIN elevation in the presence of NMS-P715. A, D.
Cells were hybridized with chromosome X or 17 centromeric probes following 72h exposure to NMS-P715 at the indicated concentrations or vehicle control. %MD was calculated as before (Fig. 3). *p≤0.037 (paired t-test). B. After treatment, cells were plated at 400 cells per well in triplicate in 12 well dishes and incubated for 6 days (PANC-1) or 7 days (ASCs) in compound-free medium to determine effects on cell growth. Percent growth inhibition of cells was determined relative to controls using a colorimetric assay (see Supplementary Fig. S6). C. Western analysis of ASC3 and ASC4 cells as outlined in Fig. 2A.
Discussion
We have shown that CIN70 genes are up-regulated in PDAC compared to normal pancreas, consistent with cytogenetic evidence for a high level of CIN in this malignancy (8). Expression of the CIN70 or a 25 gene subset (PDAC CIN25) which includes the SAC kinase MPS1, predicted clinical outcome in PDAC, with higher expression linked to shorter survival. Since tumor morphological criteria provide little clue as to prognosis in PDAC, it will be valuable to see whether the PDAC CIN25 signature could aid in PDAC patient stratification and the discrimination of candidates for resection.
Resistance to standard chemotherapeutic agents contributes to the dismal prognosis for PDAC patients, with only gemcitabine, the current standard of care, having useful efficacy (2). There is a clear need for new targeted therapeutic approaches. A major breakthrough in targeted cancer therapy has been the synthetic lethal approach to killing malignant cells, exemplified by the use of PARP inhibitors in BRCA deficient breast cancer (47). Synthetic lethality relies on delivering a second (pharmacological) hit that is lethal only in combination with a pre-existing mutation in the cancer cells and is thus well tolerated by normal cells (47). By extension, other vulnerabilities of the cancer cell, such as dependence on SAC gene over-expression to bolster a compromised SAC (13) or suppress other mitotic defects (23, 42, 43), may also provide an opportunity to selectively target cancer cells. In the present study, impaired PDAC cell growth following MPS1 inhibition was accompanied by override of SAC function and a marked increase in chromosome mis-segregation, consistent with death due to intolerable CIN. These findings are in agreement with prior studies on a range of other cancer cell types, using either pharmacological, or RNA interference based MPS1 inhibition (12, 17-20, 22).
Comparison with primary fibroblasts and non-tumorigenic cell lines has supported the possibility that cancer cells may be more sensitive to the effects of MPS1 inhibition than their normal counterparts (12, 17, 26). We further address this selectivity by testing the effects of MPS1 inhibition on normal human ASCs, which may better model the adult stem cells damaged by anti-mitotic drugs in vivo. In comparison to the PDAC cell lines, ASCs from multiple donors exhibited marked resistance to the anti-proliferative effects of NMS-P715. Moreover, unlike the PDAC cell lines, ASCs maintained SAC function in the presence of drug and exhibited no increase in the rate of chromosome mis-segregation. Combining previous (12) and present findings, 5 of 6 PDAC cell lines tested have shown greater sensitivity to the anti-proliferative effects of NMS-P715 compared to normal cells, consistent with heightened dependence on MPS1 function. Notably, we show the preferential activity of NMS-P715 against the PDAC cell lines to be the reverse of gemcitabine, that was more toxic to the ASCs.
Increasing awareness of the degree of genetic heterogeneity and plasticity, even within a single tumor (48), has highlighted the obstacles to targeted cancer therapy. It has been argued that the majority of targeted therapies may be destined to fail as tumors evolve new avoidance strategies and that an orthogonal approach combining multiple drugs with independent targets, will be necessary (3). In the face of such a challenge, attacking the underlying genomic instability that fuels genetic change in cancer cells, is an appealing alternative. Moreover, drugs which target CIN may prove useful in combination with standard therapies, to slow the emergence of resistance (49). Together, the current findings support the potential of MPS1 inhibition as a novel treatment strategy for PDAC. In xenograft studies of other human cancers, NMS-P715 has shown good efficacy and low toxicity (12). A critical next step will be to test the anti-tumorigenic effects of NMS-P715 in orthotopic transplants of human PDAC cells into pancreata of nude mice, or KRC cells into syngeneic mice with an intact immune system. Such studies will help determine whether NMS-P715 could offer an improved therapeutic index over current treatments, and/or be combined with such treatments to render them more effective. Importantly, the proposed link between CIN and increased dependence on MPS1 would predict that the most at-risk patients, with high CIN (and therefore high PDAC CIN25 gene expression), should respond well to MPS1 inhibition. The relationship between CIN and NMS-P715 sensitivity could be further explored in PDAC cell lines, since a recent microarray analysis of > 20 such lines reveals a range of PDAC CIN25 expression comparable to that seen in patients (Slee et al., unpublished; (50)).
Supplementary Material
Acknowledgements
We thank Drs. Catherine Steding, Changyu Shen and Gail Vance for helpful advice, the IUSCC Flow Cytometry Resource Facility for assistance and the IUSCC Center for Pancreatic Cancer Research for providing human PDAC cell lines.
Financial support: B. R. Grimes is supported by the Indiana Genomics Initiative (which is, in part, supported by the Lilly Endowment) and IUSM Diagnostic Genomics Division. J. Victorino is supported by a Bridges to the Doctorate Program (NIH grant R25-GM67592) and M. Korc, in part, by NIH grant R37-CA-075059.
Abbreviations
- PDAC
pancreatic ductal adenocarcinoma
- CIN
chromosome instability
- SAC
spindle assembly checkpoint
- ASCs
adipose derived mesenchymal stem cells
Footnotes
The authors have no conflict of interest. Riccardo Colombo is a full time employee of Nerviano Medical Sciences Srl.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Asuthkar S, Rao JS, Gondi CS. Drugs in preclinical and early-stage clinical development for pancreatic cancer. Expert Opin Investig Drugs. 2012;21:143–52. doi: 10.1517/13543784.2012.651124. [DOI] [PubMed] [Google Scholar]
- 3.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136:823–37. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colombo R, Moll J. Destabilizing aneuploidy by targeting cell cycle and mitotic checkpoint proteins in cancer cells. Curr Drug Targets. 2010;11:1325–35. doi: 10.2174/1389450111007011325. [DOI] [PubMed] [Google Scholar]
- 5.Preis M, Korc M. Signaling pathways in pancreatic cancer. Crit Rev Eukaryot Gene Expr. 2011;21:115–29. doi: 10.1615/critreveukargeneexpr.v21.i2.20. [DOI] [PubMed] [Google Scholar]
- 6.Lee AJ, Endesfelder D, Rowan AJ, Walther A, Birkbak NJ, Futreal PA, et al. Chromosomal instability confers intrinsic multidrug resistance. Cancer Res. 2011;71:1858–70. doi: 10.1158/0008-5472.CAN-10-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38:1043–8. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 8.Heim S, Mitelman F. Cancer Cytogenetics: Chromosomal and Molecular Genetic Abberations of Tumor Cells 3rd edition. Wiley-Blackwell.; New York: 2009. [Google Scholar]
- 9.Birkbak NJ, Eklund AC, Li Q, McClelland SE, Endesfelder D, Tan P, et al. Paradoxical relationship between chromosomal instability and survival outcome in cancer. Cancer Res. 2011;71:3447–52. doi: 10.1158/0008-5472.CAN-10-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McGranahan N, Burrell RA, Endesfelder D, Novelli MR, Swanton C. Cancer chromosomal instability: therapeutic and diagnostic challenges. EMBO Rep. 2012;13:528–38. doi: 10.1038/embor.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang YC, Williams BR, Siegel JJ, Amon A. Identification of aneuploidy-selective antiproliferation compounds. Cell. 2011;144:499–512. doi: 10.1016/j.cell.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colombo R, Caldarelli M, Mennecozzi M, Giorgini ML, Sola F, Cappella P, et al. Targeting the mitotic checkpoint for cancer therapy with NMS-P715, an inhibitor of MPS1 kinase. Cancer Res. 2010;70:10255–64. doi: 10.1158/0008-5472.CAN-10-2101. [DOI] [PubMed] [Google Scholar]
- 13.Yuan B, Xu Y, Woo JH, Wang Y, Bae YK, Yoon DS, et al. Increased expression of mitotic checkpoint genes in breast cancer cells with chromosomal instability. Clin Cancer Res. 2006;12:405–10. doi: 10.1158/1078-0432.CCR-05-0903. [DOI] [PubMed] [Google Scholar]
- 14.Schmidt M, Medema RH. Exploiting the compromised spindle assembly checkpoint function of tumor cells: dawn on the horizon? Cell Cycle. 2006;5:159–63. doi: 10.4161/cc.5.2.2309. [DOI] [PubMed] [Google Scholar]
- 15.Yang Z, Loncarek J, Khodjakov A, Rieder CL. Extra centrosomes and/or chromosomes prolong mitosis in human cells. Nat Cell Biol. 2008;10:748–51. doi: 10.1038/ncb1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dorer RK, Zhong S, Tallarico JA, Wong WH, Mitchison TJ, Murray AW. A small-molecule inhibitor of Mps1 blocks the spindle-checkpoint response to a lack of tension on mitotic chromosomes. Curr Biol. 2005;15:1070–6. doi: 10.1016/j.cub.2005.05.020. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt M, Budirahardja Y, Klompmaker R, Medema RH. Ablation of the spindle assembly checkpoint by a compound targeting Mps1. EMBO Rep. 2005;6:866–72. doi: 10.1038/sj.embor.7400483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janssen A, Kops GJ, Medema RH. Elevating the frequency of chromosome mis-segregation as a strategy to kill tumor cells. Proc Natl Acad Sci U S A. 2009;106:19108–13. doi: 10.1073/pnas.0904343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daniel J, Coulter J, Woo JH, Wilsbach K, Gabrielson E. High levels of the Mps1 checkpoint protein are protective of aneuploidy in breast cancer cells. Proc Natl Acad Sci U S A. 2011;108:5384–9. doi: 10.1073/pnas.1007645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tardif KD, Rogers A, Cassiano J, Roth BL, Cimbora DM, McKinnon R, et al. Characterization of the cellular and antitumor effects of MPI-0479605, a small-molecule inhibitor of the mitotic kinase Mps1. Mol Cancer Ther. 2011;10:2267–75. doi: 10.1158/1535-7163.MCT-11-0453. [DOI] [PubMed] [Google Scholar]
- 21.Kops GJ, Foltz DR, Cleveland DW. Lethality to human cancer cells through massive chromosome loss by inhibition of the mitotic checkpoint. Proc Natl Acad Sci U S A. 2004;101:8699–704. doi: 10.1073/pnas.0401142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwiatkowski N, Jelluma N, Filippakopoulos P, Soundararajan M, Manak MS, Kwon M, et al. Small-molecule kinase inhibitors provide insight into Mps1 cell cycle function. Nat Chem Biol. 2010;6:359–68. doi: 10.1038/nchembio.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding Y, Hubert CG, Herman J, Corrin P, Toledo CM, Skutt-Kakaria K, et al. Cancer-Specific requirement for BUB1B/BUBR1 in human brain tumor isolates and genetically transformed cells. Cancer Discov. 2013;3:198–211. doi: 10.1158/2159-8290.CD-12-0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wood KW, Lad L, Luo L, Qian X, Knight SD, Nevins N, et al. Antitumor activity of an allosteric inhibitor of centromere-associated protein-E. Proc Natl Acad Sci U S A. 2010;107:5839–44. doi: 10.1073/pnas.0915068107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stolz A, Vogel C, Schneider V, Ertych N, Kienitz A, Yu H, et al. Pharmacologic abrogation of the mitotic spindle checkpoint by an indolocarbazole discovered by cellular screening efficiently kills cancer cells. Cancer Res. 2009;69:3874–83. doi: 10.1158/0008-5472.CAN-08-3597. [DOI] [PubMed] [Google Scholar]
- 26.Hsieh TC, Traganos F, Darzynkiewicz Z, Wu JM. The 2,6-disubstituted purine reversine induces growth arrest and polyploidy in human cancer cells. Int J Oncol. 2007;31:1293–300. [PMC free article] [PubMed] [Google Scholar]
- 27.Tannous BA, Kerami M, Van der Stoop PM, Kwiatkowski N, Wang J, Zhou W, et al. Effects of the Selective MPS1 Inhibitor MPS1-IN-3 on Glioblastoma Sensitivity to Antimitotic Drugs. J Natl Cancer Inst. 2013;105:1322–31. doi: 10.1093/jnci/djt168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang G, Schetter A, He P, Funamizu N, Gaedcke J, Ghadimi BM, et al. DPEP1 inhibits tumor cell invasiveness, enhances chemosensitivity and predicts clinical outcome in pancreatic ductal adenocarcinoma. PLoS One. 2012;7:e31507. doi: 10.1371/journal.pone.0031507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collisson EA, Sadanandam A, Olson P, Gibb WJ, Truitt M, Gu S, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17:500–3. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Donahue TR, Tran LM, Hill R, Li Y, Kovochich A, Calvopina JH, et al. Integrative survival-based molecular profiling of human pancreatic cancer. Clin Cancer Res. 2012;18:1352–63. doi: 10.1158/1078-0432.CCR-11-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carriere C, Gore AJ, Norris AM, Gunn JR, Young AL, Longnecker DS, et al. Deletion of Rb accelerates pancreatic carcinogenesis by oncogenic Kras and impairs senescence in premalignant lesions. Gastroenterology. 2011;141:1091–101. doi: 10.1053/j.gastro.2011.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–83. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 33.Abbruzzese JL, Grunewald R, Weeks EA, Gravel D, Adams T, Nowak B, et al. A phase I clinical, plasma, and cellular pharmacology study of gemcitabine. J Clin Oncol. 1991;9:491–8. doi: 10.1200/JCO.1991.9.3.491. [DOI] [PubMed] [Google Scholar]
- 34.Hong SJ, Traktuev DO, March KL. Therapeutic potential of adipose-derived stem cells in vascular growth and tissue repair. Curr Opin Organ Transplant. 2010;15:86–91. doi: 10.1097/MOT.0b013e328334f074. [DOI] [PubMed] [Google Scholar]
- 35.Grimes BR, Steiner CM, Merfeld-Clauss S, Traktuev DO, Smith D, Reese A, et al. Interphase FISH demonstrates that human adipose stromal cells maintain a high level of genomic stability in long-term culture. Stem Cells Dev. 2009;18:717–24. doi: 10.1089/scd.2008.0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oliver MH, Harrison NK, Bishop JE, Cole PJ, Laurent GJ. A rapid and convenient assay for counting cells cultured in microwell plates: application for assessment of growth factors. J Cell Sci. 1989;92:513–18. doi: 10.1242/jcs.92.3.513. [DOI] [PubMed] [Google Scholar]
- 37.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 38.Chen QR, Song YK, Wei JS, Bilke S, Asgharzadeh S, Seeger RC, et al. An integrated cross-platform prognosis study on neuroblastoma patients. Genomics. 2008;92:195–203. doi: 10.1016/j.ygeno.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo J, Schumacher M, Scherer A, Sanoudou D, Megherbi D, Davison T, et al. A comparison of batch effect removal methods for enhancement of prediction performance using MAQC-II microarray gene expression data. Pharmacogenomics J. 2010;10:278–91. doi: 10.1038/tpj.2010.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stucke VM, Sillje HH, Arnaud L, Nigg EA. Human Mps1 kinase is required for the spindle assembly checkpoint but not for centrosome duplication. EMBO J. 2002;21:1723–32. doi: 10.1093/emboj/21.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gysin S, Paquette J, McMahon M. Analysis of mRNA profiles after MEK1/2 inhibition in human pancreatic cancer cell lines reveals pathways involved in drug sensitivity. Mol Cancer Res. 2012;10:1607–19. doi: 10.1158/1541-7786.MCR-12-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Slee RB, Steiner CM, Herbert BS, Vance GH, Hickey RJ, Schwarz T, et al. Cancer-associated alteration of pericentromeric heterochromatin may contribute to chromosome instability. Oncogene. 2012;31:3244–53. doi: 10.1038/onc.2011.502. [DOI] [PubMed] [Google Scholar]
- 43.Manning AL, Dyson NJ. RB: mitotic implications of a tumour suppressor. Nat Rev Cancer. 2012;12:220–26. doi: 10.1038/nrc3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sihn CR, Suh EJ, Lee KH, Kim TY, Kim SH. p55CDC/hCDC20 mutant induces mitotic catastrophe by inhibiting the MAD2-dependent spindle checkpoint activity in tumor cells. Cancer Lett. 2003;201:203–10. doi: 10.1016/s0304-3835(03)00465-8. [DOI] [PubMed] [Google Scholar]
- 45.Kaelin WG., Jr The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer. 2005;5:689–98. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- 46.Rubin EH, Gilliland DG. Drug development and clinical trials--the path to an approved cancer drug. Nat Rev Clin Oncol. 2012;9:215–22. doi: 10.1038/nrclinonc.2012.22. [DOI] [PubMed] [Google Scholar]
- 47.Weidle UH, Maisel D, Eick D. Synthetic lethality-based targets for discovery of new cancer therapeutics. Cancer Genomics Proteomics. 2011;8:159–71. [PubMed] [Google Scholar]
- 48.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bakhoum SF, Compton DA. Chromosomal instability and cancer: a complex relationship with therapeutic potential. J Clin Invest. 2012;122:1138–43. doi: 10.1172/JCI59954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maupin KA, Sinha A, Eugster E, Miller J, Ross J, Paulino V, et al. Glycogene expression alterations associated with pancreatic cancer epithelial-mesenchymal transition in complementary model systems. PLoS One. 2010;5:e13002. doi: 10.1371/journal.pone.0013002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.