Significance
Several bacteria that are pathogens of humans regulate the production of virulence factors in response to temperature changes, expressing them only at 37 °C. This thermoregulation is commonly due to the presence of RNA structures (RNA thermometers) in the 5′ regions of transcripts specifying regulatory proteins responsible for the expression of virulence-associated traits. At environmental conditions, RNA thermometers possess structures that block translation initiation of mRNAs, whereas at body temperature these structures are no longer stable, allowing the synthesis of their corresponding proteins. We report for the first time, to our knowledge, the molecular basis of thermoregulation of virulence-factor production in the opportunistic pathogen Pseudomonas aeruginosa, and have determined that this regulation is achieved by two RNA thermometers with previously unidentified characteristics not previously reported in bacteria.
Keywords: RNA thermometers, bacterial virulence, gene regulation, polarity, quorum sensing
Abstract
In a number of bacterial pathogens, the production of virulence factors is induced at 37 °C; this effect is often regulated by mRNA structures formed in the 5′ untranslated region (UTR) that block translation initiation of genes at environmental temperatures. At 37 °C, the RNA structures become unstable and ribosomes gain access to their binding sites in the mRNAs. Pseudomonas aeruginosa is an important opportunistic pathogen and the expression of many of its virulence-associated traits is regulated by the quorum-sensing (QS) response, but the effect of temperature on virulence-factor expression is not well-understood. The aim of this work is the characterization of the molecular mechanism involved in thermoregulation of QS-dependent virulence-factor production. We demonstrate that traits that are dependent on the QS transcriptional regulator RhlR have a higher expression at 37 °C, correlating with a higher RhlR concentration as measured by Western blot. We also determined, using gene fusions and point mutations, that RhlR thermoregulation is a posttranscriptional effect dependent on an RNA thermometer of the ROSE (Repression Of heat-Shock gene Expression) family. This RNA element regulates the expression of the rhlAB operon, involved in rhamnolipid production, and of the downstream rhlR gene. We also identified a second functional thermometer in the 5′ UTR of the lasI gene. We confirmed that these RNA thermometers are the main mechanism of thermoregulation of QS-dependent gene expression in P. aeruginosa using quantitative real-time PCR. This is the first description, to our knowledge, of a ROSE element regulating the expression of virulence traits and of an RNA thermometer controlling multiple genes in an operon through a polar effect.
A free-living γ-proteobacterium, Pseudomonas aeruginosa is one of the most clinically important opportunistic pathogens. It is responsible for acute infections in immune-compromised individuals and is the primary cause of death in patients with cystic fibrosis (1). The wide pathogenic capacity of this bacterium depends on its ability to produce and secrete multiple virulence factors that are regulated at the transcriptional level by the quorum-sensing (QS) response (2). Several mechanistic details of this intricate regulatory network remain to be elucidated at the molecular level.
The QS response is mediated by the bacterial production of acyl-homoserine lactones (autoinducers) that act as signal molecules interacting with transcriptional regulators of the LuxR family. In P. aeruginosa, Las and Rhl QS regulation is arranged in a hierarchical cascade. LasR interacts with 3-oxo-dodecanoyl-homoserine lactone (3O-C12-HSL) produced by the LasI enzyme, and activates the transcription of several genes encoding virulence factors and also of rhlR, lasI, and rhlI; rhlR encodes the second QS transcriptional regulator, whereas the product of the rhlI gene is the enzyme that produces butanoyl-homoserine lactone (C4-HSL), which is the autoinducer that interacts with RhlR (2, 3). RhlR/C4-HSL in turn promotes the expression of genes responsible for the production of several virulence factors. These include pyocyanin, whose biosynthetic enzymes are encoded by the two reiterated phz operons (4, 5), lectin PA-IL (product of lecA), and the rhlAB operon, encoding enzymes involved in the production of the biosurfactant rhamnolipids (2). Transcription of rhlR is driven by five promoters (SI Appendix, Fig. S1); four of the start sites are located immediately upstream of the gene (6), whereas the fifth promoter drives the synthesis of a large transcription unit that includes rhlA, rhlB, and rhlR (7). We will refer to this transcription unit as the rhlAB-R operon (SI Appendix, Fig. S1). We have demonstrated using genetic evidence and RT-PCR that rhlR can be part of an rhlABR polycistronic unit, implying that its transcription can be started from the rhlA promoter. Because RhlR activates this promoter, an RhlR-dependent positive autoregulatory loop is formed (7).
The P. aeruginosa QS response also involves the regulon that depends on the signal molecule 2-heptyl-3-hydroxy-4(1H)-quinolone (PQS) and the transcriptional regulator PqsR, which is not a member of the LuxR family of transcriptional regulators (2). The virulence traits that depend on PQS regulation are mainly produced by the enhancement of the RhlR-dependent QS response, giving rise to a complex network of positive and negative interactions that exist between the LasR- and RhlR-dependent QS regulons (2).
Several human pathogenic bacteria produce virulence-associated traits only under conditions of infection, and in various instances temperature has been shown to play a major role in the regulation of their expression (8–10). In these cases, the production of virulence determinants is completely abrogated at environmental temperatures (30 °C or lower), whereas the genes encoding these factors are fully expressed at 37 °C. In many organisms, the modulation of the expression of genes encoding virulence-associated traits is achieved by RNA structures present in the 5′ untranslated region (UTR) of genes encoding key transcriptional regulators (10, 11). Recently, it was reported that P. aeruginosa genes encoding virulence factors regulated by QS were induced at 37 °C, the human body temperature (12); however, the molecular mechanism for this thermoregulation has not yet been determined.
There are several examples in bacteria of genes regulated by temperature-induced changes in an RNA structure at the 5′ UTR of their mRNAs (11, 13–16). Genes regulated by these so-called RNA thermometers code either for heat-shock proteins or for positive regulators of genes encoding virulence factors. There are several RNA structures that can function as thermometers (16). Some of the best-characterized microbial thermometers are the ROSE (Repression Of heat-Shock gene Expression) elements, which have only been reported in transcripts of genes encoding small heat-shock proteins present in several α- and γ-proteobacteria (17). The sequences of ROSE elements are not extensively conserved, but their structure consists of two, three, or four stem loops ranging from 60 to more than 100 nt in length. The 5′ hairpin(s) remains folded at higher temperatures, whereas the 3′ proximal stem loop that harbors the ribosome binding site or Shine–Dalgarno sequence (SD), and in some instances the AUG start codon, is stable only at low temperatures and melts as the temperature rises. Heat-induced structural changes to this 3′ proximal stem loop expose the SD sequence, allowing ribosome binding and mRNA translation (18).
In this report, we show that the production of P. aeruginosa RhlR-dependent virulence factors is thermoregulated, increasing at 37 °C, and that this increment is caused by a higher RhlR concentration at this temperature. The central element involved in thermoregulation is a ROSE element present in the 5′ UTR of rhlA. At environmental temperatures (30 °C or lower), this RNA thermometer is predicted to be highly structured, blocking rhlA translation, causing a polar effect on transcription of rhlB and rhlR initiated at the rhlA promoter. The presence of this RNA thermometer had been predicted by in silico analysis of the PAO1 genome (15), but no experimental evidence has been provided until now. In addition, we identified an RNA thermometer in the 5′ UTR of lasI, and we show that the translation of this gene is also increased at 37 °C, although this increase in translation results only in a marginal increment of 3O-C12-HSL levels. Finally, using quantitative real-time PCR, we verified that P. aeruginosa QS-dependent production of virulence factors is thermoregulated and that this response to temperature is dependent on the two RNA thermometers studied here, the ROSE element located in the 5′ UTR of rhlA and a ROSE-like element in the 5′ UTR of lasI.
Results and Discussion
Growth Temperature Modulates RhlR-Dependent Virulence-Factor Production in P. aeruginosa.
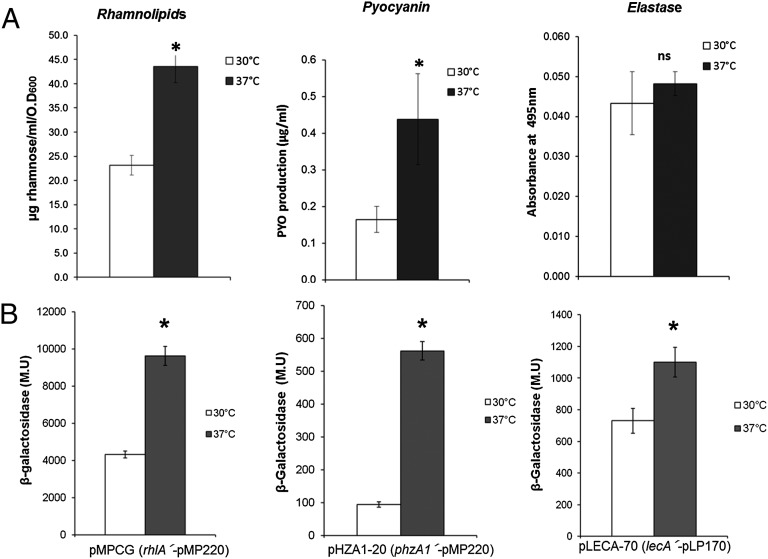
To determine whether virulence-factor production was affected by growth temperature in P. aeruginosa, we measured rhamnolipid production at 37 °C and 30 °C and found it to be considerably higher when this bacterium is cultivated at body temperature (Fig. 1A), and also that this difference is due to a higher level of transcription of the rhlAB-R operon at 37 °C (Fig. 1B).
Fig. 1.
Expression of RhlR-dependent virulence factors is thermoregulated. Production of several virulence-associated traits (A) and expression of several lacZ transcriptional fusions (B) when P. aeruginosa PAO1 was cultivated at 30 °C (white bars) and 37 °C (black bars) at OD600 1.5. Rhamnolipids and pyocyanin were determined from cultures grown in PPGAS medium, and elastase activity was determined from cultures grown in LB medium at an OD600 of 1.5. Data shown are the average of at least three independent determinations, and the error bars represent SDs. *P < 0.05 by Student t test; ns, not significant. M.U, Miller units.
Because transcription from the rhlAB-R promoter is strictly activated by RhlR/C4-HSL, we wondered whether the growth temperature also affected other traits regulated by this protein–autoinducer complex. We found that pyocyanin production was also higher at 37 °C (Fig. 1A) and that the transcription of the phzA1B1C1D1E1F1G1 operon, which is also dependent on RhlR/C4-HSL for its expression (4, 5), was strongly dependent on growth of the bacteria at 37 °C (Fig. 1B). Furthermore, transcription of lecA (the gene coding for PA-IL, a galactophilic lectin) is higher at 37 °C than at 30 °C (Fig. 1B). In contrast, the expression of virulence factors that are dependent on LasR/3O-C12-HSL was not significantly affected by growth temperature; for example, elastase expression was not significantly different when P. aeruginosa was grown at these temperatures (Fig. 1A).
Intracellular Concentration of RhlR Is Thermoregulated.
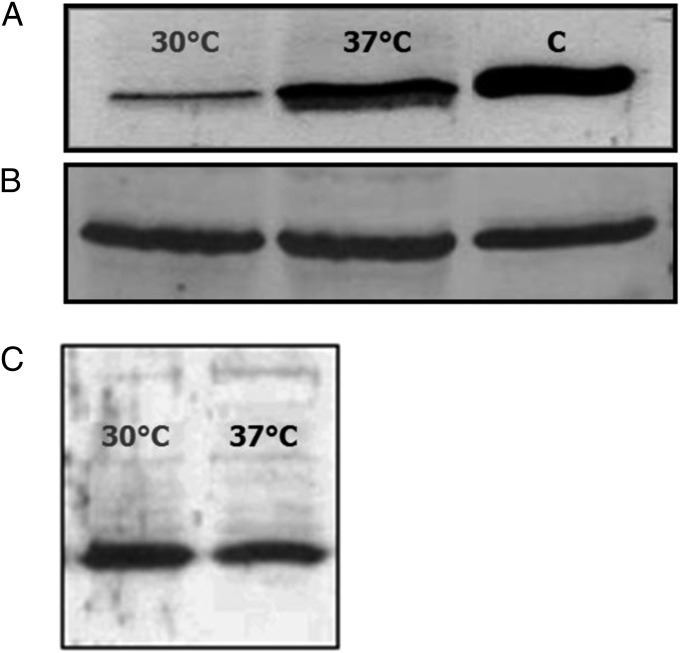
To determine whether the lower expression at 30 °C of the RhlR-dependent virulence-associated traits was due to a reduced RhlR concentration, we measured the level of this protein at 30 °C and 37 °C by means of western blot experiments. The results clearly show that RhlR concentration is considerably lower when P. aeruginosa is cultivated at 30 °C (Fig. 2A). This thermoregulation was not observed in the case of LasR (Fig. 2B), one of the main transcriptional regulators involved in rhlR expression as part of the QS response (SI Appendix, Fig. S1).
Fig. 2.
Effect of P. aeruginosa growth temperatures (30 °C and 37 °C) on RhlR and LasR intracellular concentration. Protein concentrations were determined by Western blot analysis. (A) RhlR concentration in extracts of PAO1 cells grown at OD600 1.5 on PPGAS medium at 30 °C and 37 °C. C, control. (B) LasR concentration in extracts of PAO1 cells grown to an OD600 of 1.5 on PPGAS medium at 30 °C and 37 °C. The same protein (RhlR or LasR) expressed in E. coli was used as the control. (C) RhlR concentration in extracts of PAO1-derived rhlA::Gm mutant grown at OD600 1.5 on PPGAS medium at 30 °C and 37 °C. Protein concentrations were determined in at least two independent experiments.
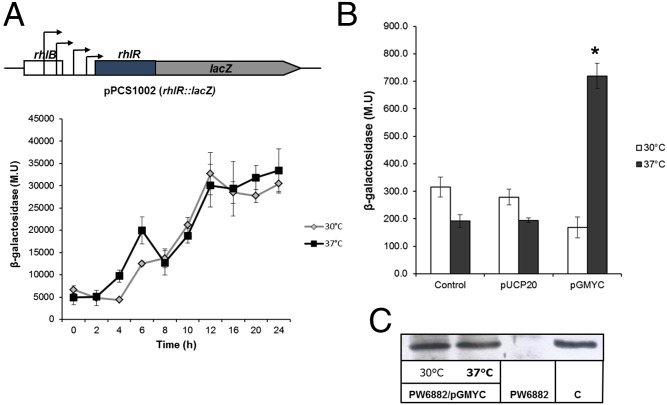
The reduced RhlR concentration at 30 °C is not due to a lower level of rhlR transcription from any of the four promoters located immediately upstream of this gene, because a strain that carries plasmid pSC1002 containing a lacZ fusion expressed from these four promoters produced similar levels of β-galactosidase along the P. aeruginosa growth curve at 30 °C and 37 °C (Fig. 3A).
Fig. 3.
Temperature-dependent expression of rhlR in P. aeruginosa depends on the transcription of the rhlAB-R operon at 37 °C. (A) Diagram of plasmid pSC1002, which encodes for an rhlR–lacZ transcriptional fusion (containing the four rhlR immediately upstream promoters), and the expression of this rhlR–lacZ fusion in P. aeruginosa PAO1 at 30 °C (gray line) and 37 °C (black line). (B) Expression of rhlR from the rhlA promoter in strain PW6882, which contains an in-frame Tn9-lacZ insertion inside rhlR (SI Appendix, Table S1) at 30 °C (white bars) and 37 °C (gray bars). The first set of bars at both temperatures represents the basal expression of chromosomal rhlR; the second set of bars corresponds to the expression of this gene in the presence of the promoter in strain PW6882, with cloning vector pUCP20 (used as control of rhlR LasR-dependent expression); and the third set of bars at each temperature represents the expression with plasmid pGMYC (SI Appendix, Table S1) expressing rhlR in trans. *P < 0.05 by Student t test. (C) RhlR concentration was determined by Western blot analysis, expressed from plasmid pGMYC. The samples were the same as those used for β-galactosidase determination in B.
As mentioned, the rhlAB-R operon is positively regulated by RhlR/C4-HSL, so a positive autoregulatory loop for rhlR expression is formed, resulting in full induction of the QS response (7). In the experiments of Fig. 3B, we show that this positive autoregulatory loop is only significant at 37 °C and not at 30 °C. In these experiments, we determined the expression of rhlR from the rhlA promoter in the presence of plasmid pGMYC, which codes for rhlR expressed under the lacZ promoter (SI Appendix, Table S1), by measuring the β-galactosidase activity in an rhlR::Tn′lacZ mutant (PW6882; SI Appendix, Table S1) that has an in-frame lacZ chromosomal insertion (Fig. 3B). The higher expression at 37 °C of chromosomal rhlR was not due to an increased expression of RhlR, because the concentration of this protein expressed from plasmid pGMYC was the same at both temperatures (Fig. 3C).
To further confirm that the higher RhlR concentration at 37 °C than at 30 °C in the PAO1 strain was determined by thermoregulation of rhlR expression when its transcription starts at the rhlA promoter, we determined the intracellular RhlR level in a PAO1 rhlA::Gm mutant (SI Appendix, Table S1). We used this mutant, because the inactivation of rhlA by the insertion of a Gm cassette causes a polar effect on the transcription of rhlB (19, 20) and its expression is abrogated. We reasoned that if rhlR induction at 37 °C is driven from the rhlA promoter, thermoregulation of rhlR would also be abrogated in the rhlA::Gm background. Indeed, in the rhlA::Gm mutant, the RhlR concentration was not dependent on the growth temperature, unlike in the wild-type strain (Fig. 2C). This result confirms that the increased rhlR expression at 37 °C is driven from the rhlA promoter. Furthermore, the concentration of RhlA itself is also thermoregulated (SI Appendix, Fig. S2).
A ROSE Element Is Responsible for Thermoregulation of the rhlAB-R Operon.
To determine whether an mRNA structure could determine thermoregulation, we analyzed the rhlA 5′ UTR using mfold version 3.2 (21) and the RNAfold program of the Vienna RNA package (22). These analyses revealed the presence of a putative ROSE RNA thermometer that contains all of the reported characteristics shown by these elements (14), including an unpaired G in the middle of the 3′ proximal stem (SI Appendix, Fig. S3A). As previously mentioned, this putative RNA thermometer has already been predicted by bioinformatic analysis of the P. aeruginosa genome (15) and is highly conserved in all of the available P. aeruginosa sequenced genomes (SI Appendix, Fig. S4) (23). In addition, this RNA structure in the rhlA 5′ UTR has been recently proposed to be the site of regulation of a small RNA that participates in nitrogen regulation (24).
The structure of one ROSE element has been elucidated using NMR. In this structure, the 3′ proximal stem loop is responsible for thermoregulation, and the deletion of a highly conserved unpaired G residue in this stem loop leads to the formation of a stable RNA helix without thermosensing ability (14, 18). Although there is no experimental evidence of participation of the 5′ proximal stem loop(s) in the functionality of ROSE elements, their conservation suggests that they might be important in the proper folding of the 3′ proximal temperature-responsive stem loop, which contributes to the regulation of expression of genes for small heat-shock proteins (11, 25, 26).
We therefore decided to determine whether the ROSE element present in the 5′ UTR of rhlA was indeed required for thermoregulation of the translation of rhlA by using a biosensor and by constructing point mutations that affect nucleotides that have been shown to be relevant for the function of other ROSE elements (17, 25).
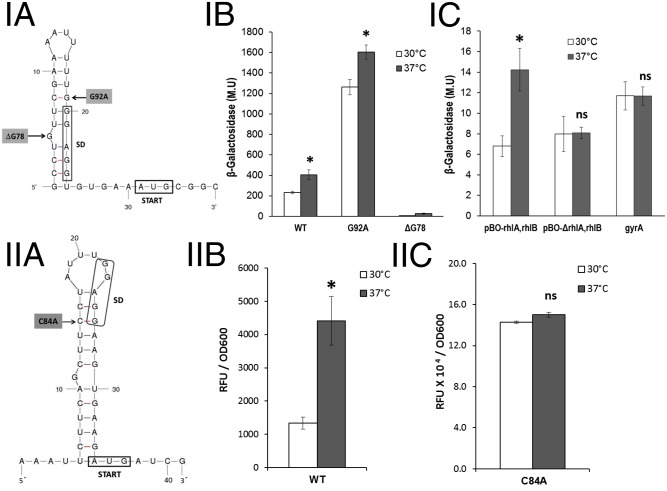
The pBAD18-lacZ481–based RNA-thermometer biosensor (17) that contains 36 nt of the putative rhlA ROSE element (we call this sequence “small ROSE”) is called plasmid pBO-rhlA; it includes the proximal 3′ stem loop and the AUG start codon (nucleotides 74–109 of the ROSE element; Fig. 4 I, A). We found that the lacZ reporter gene in pBO-rhlA was expressed at a reduced level at 30 °C and that expression was higher at 37 °C, as expected for a construct containing an RNA thermometer, because the translation of the lacZ reporter gene is thermoregulated (Fig. 4 I, B). On the other hand, we observed that a mutant in which the characteristic 3′ proximal stem loop of the ROSE element was disrupted by unpairing (G92A) expressed high levels of β-galactosidase at 30 °C and at 37 °C (Fig. 4 I, B). Furthermore, using this plasmid, we showed that a mutant lacking the unpaired G (ΔG78) presented an extremely reduced expression both at 30 °C and at 37 °C (Fig. 4 I, B). The much higher level of expression from the plasmid carrying the G92A mutation compared with plasmid pBO-rhlA at both temperatures shows that the ROSE element partially controls rhlA translation even at 37 °C (the G92A mutation does not modify the sequence of the ribosome binding site; SI Appendix, Fig. S3A).
Fig. 4.
Characterization of RNA thermometers present in the 5′ UTR of rhlA and lasI. The proximal secondary structures of the 5′ UTRs of the rhlA (I, A) and lasI (II, A) mRNAs predicted by mfold program version 3.2 corresponding to the small-ROSE elements are shown; numbers shown for the mutated nucleotides correspond to positions in the complete versions of both structures that are shown in SI Appendix, Fig. S3 A and B. (I, B) β-Galactosidase activity levels of cells harboring plasmids with different versions of the small-ROSE element at the 5′ UTR region of rhlA fused to lacZ (pBO-rhlA). (I, C) Evaluation of the polar effect of rhlA on the temperature-dependent expression of rhlB and the negative control of the pBAD18-lacZ481–based RNA-thermometer biosensor carrying the E. coli gyrA 5′ UTR (pBOC-gyrA). (II, B) Characterization of the ROSE-like element at the 5′ UTR region of lasI. (II, C) Characterization of its derivative with the mutation C84A [pBOC-lasI (C84A)]. Because expression of the constructs containing the lasI ROSE-like element was low, it was necessary to use a β-galactosidase activity assay with a fluorescent substrate for its characterization (SI Appendix, Materials and Methods). The fluorescence detected is expressed as arbitrary fluorescence units (RFUs) and normalized with respect to OD600. Cells of E. coli DH5α transformed with the corresponding plasmids were grown at 30 °C in LB broth until exponential phase (optical density of 0.5 at 600 nm). After addition of 0.04% l-arabinose, half of the culture was maintained at 30 °C (white bars) and the other half was transferred to 37 °C (gray bars) for 60 min before measuring β-galactosidase activity. The results expressed in Miller units are the average of three independent experiments with the respective SDs. *P < 0.05 by Student t test.
Similar experiments were undertaken using plasmid pBOC-rhlA, which contains the complete ROSE element present in the rhlA 5′ UTR. These constructs gave results similar to those obtained with the small ROSE but with a lower level of expression (SI Appendix, Fig. S5).
The presence of the ROSE element explains the lower level of expression of rhlA at 30 °C due to a reduced level of translation at this temperature, which is reflected in a lower concentration of this protein (SI Appendix, Fig. S2). In addition, the thermoregulation observed in the expression of the downstream rhlR gene (Fig. 3B), and the lack of thermoregulation of RhlR concentration on an rhlA insertion mutant (Fig. 2C), was evidence of the presence of a mechanism that couples rhlA translation with the transcription of the rhlAB-R operon, presumably by a polar effect. To obtain additional evidence on the possible polar effect exerted by the rhlA ROSE element, we used a biosensor with a 5′ rhlA small-ROSE element followed by the entire rhlA coding region and an in-frame rhlB–lacZ fusion starting at the AUG codon of rhlB (pBO-rhlA,rhlB). This construct showed thermoregulation of lacZ expression (Fig. 4 I, C), making it evident that the polar effect on rhlB is due to the lack of translation of rhlA at 30 °C. It is important to stress that there is no apparent transcriptional terminator between rhlB and rhlR that might explain termination of transcription when rhlA translation is reduced.
A deletion derivative of this plasmid called pBO-ΔrhlA,rhlB that lacks 326 nt (+548 to +873), which corresponds to approximately one-third of the rhlA coding region, loses rhlB thermoregulation, because it is expressed at the same level at 30 °C and at 37 °C (Fig. 4 I, C). These results show that polarity exerted by lack of rhlA translation on rhlB expression is due to putative termination structures formed on the rhlA mRNA in the absence of attached ribosomes, which prematurely terminate transcription of the operon.
To show that the thermoregulation of the QS response was mainly due to different rhlR expression at different temperatures, and not to an indirect effect of LasR concentration, we used the same vector designed to characterize the rhlA RNA thermometers (pBAD18-lacZ481; SI Appendix, Table S1) with a cloned insert corresponding to 110 bp of the 5′ lasR UTR (pBOC-lasR; SI Appendix, Table S1) that is not predicted to contain an RNA thermometer. Furthermore, neither the LasR concentration, as measured by Western blot (Fig. 2B), nor lasR transcription, as determined by quantitative PCR (Fig. 5), was affected by growth temperature. As expected, the level of β-galactosidase activity encoded by plasmid pBOC-lasR was not affected by growth temperature (SI Appendix, Fig. S6).
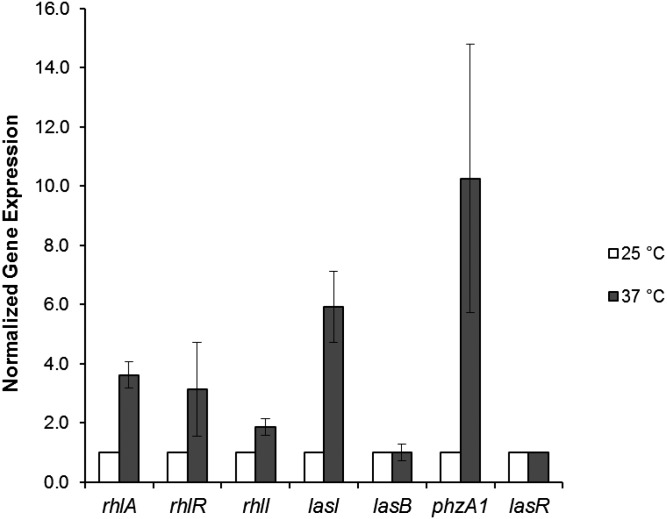
Fig. 5.
Gene expression at 25 °C and 37 °C determined by quantitative real-time PCR. Total RNAs were prepared from strain PAO1 cells grown at 25 °C (white bars) and 37 °C (gray bars) in PPGAS medium. Cultures were harvested at an OD600 of 1.5. The levels of transcripts were measured by quantitative real-time RT-PCR and normalized to lasR expression. The values represent the means and SDs of changes in comparison with the transcription level in PAO1 cells grown at 25 °C. All results are the average of at least three independent determinations, and the error bars represent SDs.
As a negative control for all experiments that measured thermoregulation based in plasmids derived from pBAD18-lacZ481, we used a construct that contains the 5′ UTR of the Escherichia coli gyrA gene that has already been used as a negative control (15). We found that β-galactosidase expression of this plasmid is not significantly different at 30 °C and 37 °C (Fig. 4 I, C). This result shows that β-galactosidase activity is not affected by growth temperature. However, different constructs based on plasmid pBAD18-lacZ481 show a different level of basal lacZ expression at 30 °C, presumably depending on specific features, including SD sequences, of each 5′ UTR tested in each biosensor.
Thermoregulation of Autoinducer Production.
The concentration of autoinducers is an important regulatory aspect of the P. aeruginosa QS response, so we were interested to determine whether it was affected by growth temperature. However, because the expression of autoinducer synthases LasI and RhlI is dependent on the transcriptional activation of the genes encoding these enzymes (lasI and rhlI) by LasR/3O-C12-HSL (2, 3) and because LasR concentration was not affected by growth temperature, we did not expect to see a significant increase in their concentration at 37 °C. Unexpectedly, we detected a slight increase in 3O-C12-HSL concentration and a substantial increase in C4-HSL concentration at 37 °C (SI Appendix, Fig. S7). To discard the effect of the increased concentration of RhlR at 37 °C on the production of autoinducers, we measured C4-HSL production in an rhlR mutant at both temperatures and observed a result similar to that obtained in the wild-type strain (SI Appendix, Fig. S7). These results suggest that some traits regulated by LasR/3O-C12-HSL, such as the production of autoinducers, and especially of C4-HSL, are also thermoregulated. However, because multiple metabolic steps are involved in the production of autoinducers, it could not be ruled out that the thermoregulation of their concentration might be due to higher metabolic rates at 37 °C.
A Second RNA Thermometer Is Present in the lasI 5′ UTR.
To determine whether the expression of the genes coding for the autoinducer synthases LasI and RhlI was affected by growth temperature, we measured the level of expression by real-time quantitative PCR of these genes and of those already studied using plasmid biosensors (Fig. 5). We corroborated the results obtained in previous experiments with plasmid biosensors, but unexpectedly we also detected considerable thermoregulation of mRNA concentration of lasI and to a lesser extent of rhlI (Fig. 5).
The higher lasI expression at 37 °C (Fig. 5) resulted only in a small increase in 3O-C12-HSL at this temperature (SI Appendix, Fig. S7), as previously mentioned, presumably due to metabolic conditions that modulate the level of LasI substrates. Furthermore, the small increase in 3O-C12-HSL did not have an effect on lasB expression (Fig. 5) or elastase production (Fig. 1), but may be the cause of the increase in rhlI expression (Fig. 5) and of the increased production of C4-HSL at 37 °C (SI Appendix, Fig. S7). The increase in C4-HSL produced by RhlI potentiates the effect of thermoregulation of RhlR-dependent gene expression.
The thermoregulation of lasI might be explained by the presence of an RNA secondary structure that could function as a thermometer in its 5′ UTR (Fig. 4 II, A and SI Appendix, Fig. S3B). This putative RNA thermometer contains the characteristic unpaired G in the middle of a predicted RNA stem. However, the unpaired G is not located in the sequence that is complementary to the SD sequence or the initiation codon (Fig. 4 II, A and SI Appendix, Fig. S3B), so we call it a “ROSE-like” element. This element at the 5′ UTR of the lasI transcript is highly conserved in all of the available P. aeruginosa sequenced genomes (SI Appendix, Fig. S4B) (23).
We followed a strategy similar to that described for rhlA ROSE-element analysis to determine whether this putative thermometer was functional (Fig. 4). In accordance with our prediction of the presence of an RNA thermometer, we found that the ROSE-like structure in the 5′ lasI UTR caused thermoregulation of its translation (Fig. 4 II, B), and that a mutation that disrupts the stem of the ROSE-like lasI thermometer (C84A) resulted in a considerable increase in expression of the lasI–lacZ fusion at both temperatures (Fig. 4 II, C). The increment of expression of the C84A mutant in the lasI RNA thermometer was significant at the two temperatures tested (Fig. 4 II, C), showing that this RNA thermometer inhibits lasI translation even at 37 °C. Because expression of the constructs containing the lasI ROSE-like element was low, it was necessary to use a β-galactosidase activity assay with a fluorescent substrate for its characterization (SI Appendix, Materials and Methods).
To obtain a global picture of the effect of growth temperature on P. aeruginosa gene expression, we carried out a transcriptome analysis of cells cultivated in proteose peptone-glucose-ammonium salts (PPGAS) media at 25 °C and 37 °C. We used commercially available genome-wide DNA microarrays (Affymetrix) (SI Appendix, Table S2), and the results obtained were deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) and are accessible through GEO Series accession no. GSE45695. We observed reduced levels of rhlA, rhlB, and rhlR mRNAs in bacteria grown at 25 °C compared with those cultured at 37 °C, but this lower expression was not statistically significant (SI Appendix, Table S3). These results were expected, considering the molecular mechanism involved in their regulation (RNA thermometer) and the low level of sensitivity of microarray analysis. In addition, the microarray data indicated a significant reduction at 25 °C of previously described RhlR-dependent genes (such as the operons encoding for the enzymes involved in pyocyanin synthesis), showing that the effect of thermoregulation of RhlR concentration is biologically significant. We also found in our microarray analysis that lasI was markedly thermoregulated (SI Appendix, Table S2).
We find in this work that, in contrast to the complete abrogation of expression of virulence genes by RNA thermometers at 30 °C in other bacterial pathogens (10, 11), P. aeruginosa still retains a considerable level of transcription of genes for virulence-associated traits at this temperature (Fig. 1) (12). This pattern of regulation is in agreement with the ability of P. aeruginosa to establish pathogenic interactions not only with mammals but also with different hosts including worms (27), flies (28), and even plants (29). Furthermore, our results show that P. aeruginosa displays a different strategy to establish pathogenic interactions with warm-blooded animals (with a full induction of QS response) than with other organisms. Therefore, research results of P. aeruginosa pathogenicity, or testing of possible therapeutic compounds to treat infections caused by this bacterium that uses insect, plant, or nematode models, might not be able to be extrapolated to clinical situations.
In summary, the results presented here show that the expression of P. aeruginosa PAO1 RhlR-dependent virulence-associated traits is considerably diminished at 30 °C due to the reduced expression of rhlR from the rhlA promoter. This thermoregulation is achieved by a ROSE RNA thermometer present in the rhlA 5′ UTR that interferes with its translation, and the coupling of this effect to rhlB and rhlR expression. This is the first report, to our knowledge, of a functional ROSE element that regulates the expression of virulence-associated genes, and it is also shown, to the authors’ knowledge, for the first time that a ROSE element thermoregulates the expression of an entire operon, presumably by means of a polar effect. In addition, we show that a second RNA thermometer in the 5′ UTR of lasI participates in the thermoregulation of virulence-factor production in P. aeruginosa, but the effect on the production of LasR-dependent virulence factors is only marginal. The only significant effect of the increased 3O-C12-HSL caused by the lasI RNA thermometer that we detected was the thermoregulation of rhlI and hence C4-HSL production.
The RNA thermometer that regulates RhlR-dependent virulence production is not exclusive to the PAO1 strain. We determined the presence of this conserved ROSE RNA thermometer in the available P. aeruginosa sequenced genomes (SI Appendix, Fig. S4) (23). The fact that this RNA thermometer is present in other P. aeruginosa strains suggests that it is important for the differential regulation of virulence trait production by P. aeruginosa growing in environmental conditions or when establishing infections in warm-blooded hosts.
Unraveling the way P. aeruginosa regulates the expression of its virulence-associated traits is of great importance for the development of strategies to control the morbidity and mortality caused by this bacterium. The results presented here contribute to the understanding of this complex phenomenon.
Materials and Methods
A brief description of the materials and methods used is as follows (see SI Appendix, Materials and Methods for a more detailed description).
Bacterial Strains, Plasmids, Media, and Growth Conditions.
The bacterial strains and plasmid vectors and their derivatives used in this study are summarized in SI Appendix, Table S1. A growth curve in PPGAS medium (30) of the PAO1 strain at 30 °C and 37 °C is shown in SI Appendix, Fig. S7.
β-Galactosidase Assays.
β-Galactosidase activity was measured either as described by Miller (31) or by using a fluorescent substrate as stated in the text.
DNA Manipulations and Genetic Techniques.
DNA manipulations were carried out as previously described (32) and standard genetic techniques were used.
Plasmid Construction.
Plasmids used to measure thermoregulation were constructed using a plasmid pBAD18-lacZ481–based biosensor (17) as vector using either annealed oligonucleotides or PCR products.
Other Techniques.
Standard techniques were used to perform RNA extraction and quantitative real-time PCR; microarray experiments and data analysis; SDS gel electrophoresis and Western blot analysis; determination of P. aeruginosa virulence-factor production; acyl-homoserine-lactone extraction; and analytical TLC (SI Appendix, Materials and Methods).
Computer Methods.
Sequences for computer analysis were retrieved from Pseudomonas Genome Database version 2 (www.pseudomonas.com). RNA secondary structures were predicted using mfold version 3.2 (21) and the RNAfold program of the Vienna RNA package (22).
Statistical Analysis.
Student t test was used to analyze statistical differences between two temperatures. A P value of <0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We acknowledge the technical assistance of Marisela Aguirre and Abigail Gonzalez-Valdez. We thank Prof. Franz Narberhaus and Birgit Klinkert for providing plasmid pBAD18-lacZ481. This work was partially financed by CONACYT (Grant 50201) and by DGAPA-Universidad Nacional Autónoma de México (UNAM) PAPIIT (Grant IN203613). The work published in this paper represents a substantial part of M.V.G.-B.'s research to obtain her PhD degree in the Programa de Maestria y Doctorado en Ciencias Bioquimicas, UNAM, and she received DGAPA-UNAM PAPIIT, CONACYT, and ICYT-DF scholarships during her studies.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. S.L. is a guest editor invited by the Editorial Board.
Data deposition: The microarray data presented in this paper are Minimum Information About a Microarray Experiment (MIAME)-compliant and have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE45695).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1402536111/-/DCSupplemental.
References
- 1.Driscoll JA, Brody SL, Kollef MH. The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs. 2007;67(3):351–368. doi: 10.2165/00003495-200767030-00003. [DOI] [PubMed] [Google Scholar]
- 2.Williams P, Cámara M. Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: A tale of regulatory networks and multifunctional signal molecules. Curr Opin Microbiol. 2009;12(2):182–191. doi: 10.1016/j.mib.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 3.Latifi A, Foglino M, Tanaka K, Williams P, Lazdunski A. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhIR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol Microbiol. 1996;21(6):1137–1146. doi: 10.1046/j.1365-2958.1996.00063.x. [DOI] [PubMed] [Google Scholar]
- 4.Mavrodi DV, et al. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J Bacteriol. 2001;183(21):6454–6465. doi: 10.1128/JB.183.21.6454-6465.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Recinos DA, et al. Redundant phenazine operons in Pseudomonas aeruginosa exhibit environment-dependent expression and differential roles in pathogenicity. Proc Natl Acad Sci USA. 2012;109(47):19420–19425. doi: 10.1073/pnas.1213901109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Medina G, Juárez K, Díaz R, Soberón-Chávez G. Transcriptional regulation of Pseudomonas aeruginosa rhlR, encoding a quorum-sensing regulatory protein. Microbiology. 2003;149(Pt 11):3073–3081. doi: 10.1099/mic.0.26282-0. [DOI] [PubMed] [Google Scholar]
- 7.Croda-García G, Grosso-Becerra V, Gonzalez-Valdez A, Servín-González L, Soberón-Chávez G. Transcriptional regulation of Pseudomonas aeruginosa rhlR: Role of the CRP orthologue Vfr (virulence factor regulator) and quorum-sensing regulators LasR and RhlR. Microbiology. 2011;157(9):2545–2555. doi: 10.1099/mic.0.050161-0. [DOI] [PubMed] [Google Scholar]
- 8.Konkel ME, Tilly K. Temperature-regulated expression of bacterial virulence genes. Microbes Infect. 2000;2(2):157–166. doi: 10.1016/s1286-4579(00)00272-0. [DOI] [PubMed] [Google Scholar]
- 9.Johansson J. RNA thermosensors in bacterial pathogens. Contrib Microbiol. 2009;16:150–160. doi: 10.1159/000219378. [DOI] [PubMed] [Google Scholar]
- 10.Böhme K, et al. Concerted actions of a thermo-labile regulator and a unique intergenic RNA thermosensor control Yersinia virulence. PLoS Pathog. 2012;8(2):e1002518. doi: 10.1371/journal.ppat.1002518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johansson J, et al. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell. 2002;110(5):551–561. doi: 10.1016/s0092-8674(02)00905-4. [DOI] [PubMed] [Google Scholar]
- 12.Wurtzel O, et al. The single-nucleotide resolution transcriptome of Pseudomonas aeruginosa grown in body temperature. PLoS Pathog. 2012;8(9):e1002945. doi: 10.1371/journal.ppat.1002945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morita M, Kanemori M, Yanagi H, Yura T. Heat-induced synthesis of σ32 in Escherichia coli: Structural and functional dissection of rpoH mRNA secondary structure. J Bacteriol. 1999;181(2):401–410. doi: 10.1128/jb.181.2.401-410.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nocker A, et al. A mRNA-based thermosensor controls expression of rhizobial heat shock genes. Nucleic Acids Res. 2001;29(23):4800–4807. doi: 10.1093/nar/29.23.4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waldminghaus T, Gaubig LC, Narberhaus F. Genome-wide bioinformatic prediction and experimental evaluation of potential RNA thermometers. Mol Genet Genomics. 2007;278(5):555–564. doi: 10.1007/s00438-007-0272-7. [DOI] [PubMed] [Google Scholar]
- 16.Kortmann J, Narberhaus F. Bacterial RNA thermometers: Molecular zippers and switches. Nat Rev Microbiol. 2012;10(4):255–265. doi: 10.1038/nrmicro2730. [DOI] [PubMed] [Google Scholar]
- 17.Waldminghaus T, Fippinger A, Alfsmann J, Narberhaus F. RNA thermometers are common in α- and γ-proteobacteria. Biol Chem. 2005;386(12):1279–1286. doi: 10.1515/BC.2005.145. [DOI] [PubMed] [Google Scholar]
- 18.Chowdhury S, Maris C, Allain FH-T, Narberhaus F. Molecular basis for temperature sensing by an RNA thermometer. EMBO J. 2006;25(11):2487–2497. doi: 10.1038/sj.emboj.7601128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ochsner UA, Koch AK, Fiechter A, Reiser J. Isolation and characterization of a regulatory gene affecting rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. J Bacteriol. 1994;176(7):2044–2054. doi: 10.1128/jb.176.7.2044-2054.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rahim R, et al. Cloning and functional characterization of the Pseudomonas aeruginosa rhlC gene that encodes rhamnosyltransferase 2, an enzyme responsible for di-rhamnolipid biosynthesis. Mol Microbiol. 2001;40(3):708–718. doi: 10.1046/j.1365-2958.2001.02420.x. [DOI] [PubMed] [Google Scholar]
- 21.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31(13):3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grosso-Becerra MV, et al. Pseudomonas aeruginosa clinical and environmental isolates constitute a single population with high phenotypic diversity. BMC Genomics. 2014;15:318. doi: 10.1186/1471-2164-15-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wenner N, Maes A, Cotado-Sampayo M, Lapouge K. NrsZ: A novel, processed, nitrogen-dependent, small non-coding RNA that regulates Pseudomonas aeruginosa PAO1 virulence. Environ Microbiol. 2014;16(4):1053–1068. doi: 10.1111/1462-2920.12272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chowdhury S, Ragaz C, Kreuger E, Narberhaus F. Temperature-controlled structural alterations of an RNA thermometer. J Biol Chem. 2003;278(48):47915–47921. doi: 10.1074/jbc.M306874200. [DOI] [PubMed] [Google Scholar]
- 26.Waldminghaus T, Gaubig LC, Klinkert B, Narberhaus F. The Escherichia coli ibpA thermometer is comprised of stable and unstable structural elements. RNA Biol. 2009;6(4):455–463. doi: 10.4161/rna.6.4.9014. [DOI] [PubMed] [Google Scholar]
- 27.Tan M-W, Ausubel FM. Caenorhabditis elegans: A model genetic host to study Pseudomonas aeruginosa pathogenesis. Curr Opin Microbiol. 2000;3(1):29–34. doi: 10.1016/s1369-5274(99)00047-8. [DOI] [PubMed] [Google Scholar]
- 28.D’Argenio DA, Gallagher LA, Berg CA, Manoil C. Drosophila as a model host for Pseudomonas aeruginosa infection. J Bacteriol. 2001;183(4):1466–1471. doi: 10.1128/JB.183.4.1466-1471.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rahme LG, et al. Use of model plant hosts to identify Pseudomonas aeruginosa virulence factors. Proc Natl Acad Sci USA. 1997;94(24):13245–13250. doi: 10.1073/pnas.94.24.13245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Miller RM. Enhanced octadecane dispersion and biodegradation by a Pseudomonas rhamnolipid surfactant (biosurfactant) Appl Environ Microbiol. 1992;58(10):3276–3282. doi: 10.1128/aem.58.10.3276-3282.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller J. in Experiments in Molecular Genetics. Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 1972. pp. 352–355. [Google Scholar]
- 32.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.